Abstract
Background
Alterations in brain-derived neurotrophic factor (BDNF) gene expression contribute to serious pathologies such as depression, epilepsy, cancer, Alzheimer's, Huntington and Parkinson's disease. Therefore, exploring the mechanisms of BDNF regulation represents a great clinical importance. Studying BDNF expression remains difficult due to its multiple neural activity-dependent and tissue-specific promoters. Thus, microarray data could provide insight into the regulation of this complex gene. Conventional microarray co-expression analysis is usually carried out by merging the datasets or by confirming the re-occurrence of significant correlations across datasets. However, co-expression patterns can be different under various conditions that are represented by subsets in a dataset. Therefore, assessing co-expression by measuring correlation coefficient across merged samples of a dataset or by merging datasets might not capture all correlation patterns.
Results
In our study, we performed meta-coexpression analysis of publicly available microarray data using BDNF as a "guide-gene" introducing a "subset" approach. The key steps of the analysis included: dividing datasets into subsets with biologically meaningful sample content (e.g. tissue, gender or disease state subsets); analyzing co-expression with the BDNF gene in each subset separately; and confirming co- expression links across subsets. Finally, we analyzed conservation in co-expression with BDNF between human, mouse and rat, and sought for conserved over-represented TFBSs in BDNF and BDNF-correlated genes. Correlated genes discovered in this study regulate nervous system development, and are associated with various types of cancer and neurological disorders. Also, several transcription factor identified here have been reported to regulate BDNF expression in vitro and in vivo.
Conclusion
The study demonstrates the potential of the "subset" approach in co-expression conservation analysis for studying the regulation of single genes and proposes novel regulators of BDNF gene expression.
Background
The accumulation of genome-wide gene expression data has enabled biologists to investigate gene regulatory mechanisms using system biology approaches. Recent developments in microarray technologies and bioinformatics have driven the progress of this field [1]. Moreover, publicly available microarray data provide information on human genome-wide gene expression under various experimental conditions, which for most researchers would be difficult to access otherwise.
BDNF (brain-derived neurotrophic factor) plays an important role in the development of the vertebrates' nervous system [2]. BDNF supports survival and differentiation of embryonic neurons and controls various neural processes in adulthood, including memory and learning [3], depression [4], and drug addiction [5]. Alterations in BDNF expression can contribute to serious pathologies such as epilepsy, Huntington, Alzheimer's, and Parkinson's disease [6]. Alteration in BDNF expression is associated with unfavorable prognosis in neuroblastoma [7], myeloma [8], hepatocellular carcinoma [9] and other tumors [10]. Apart from brain, expression of alternative BDNF transcripts has been detected in a variety of tissues (such as heart, muscle, testis, thymus, lung, etc.) [11,12]. Numerous studies have been conducted to unravel the regulation of BDNF expression in rodents and human. Data on the structure of human [11] and rodent [12]BDNF gene have been recently updated. Nevertheless, little is known about the regulation of human BDNF gene expression in vivo. Unraveling the regulation of BDNF expression remains difficult due to its multiple activity-dependent and tissue-specific promoters. Thus, analysis of the gene expression under various experimental conditions using microarray data could provide insight into the regulation of this complex gene.
Meta-coexpression analysis uses multiple experiments to identify more reliable sets of genes than would be found using a single data set. The rationale behind meta-coexpression analysis is that co-regulated genes should display similar expression patterns across various conditions. Moreover, such analysis may benefit from a vast representation of tissues and conditions [13]. A yeast study showed that the ability to correctly identify co-regulated genes in co-expression analysis is strongly dependent on the number of microarray experiments used [14]. Another study that examined 60 human microarray datasets for co-expressed gene pairs reports that gene ontology (GO) score for gene pairs increases steadily with the number of confirmed links compared to the pairs confirmed by only a single dataset [15]. Several studies have successfully applied meta-analysis approach to get important insights into various biological processes. For instance, microarray meta-analysis of aging and cellular senescence led to the observation that the expression pattern of cellular senescence was similar to that of aging in mice, but not in humans [16]. Data from a variety of laboratories was integrated to identify a common host transcriptional response to pathogens [17]. Also, meta-coexpression studies have displayed their efficiency to predict functional relationships between genes [18]. However, co-expression alone does not necessarily imply that genes are co-regulated. Thus, analysis of evolutionary conservation of co-expression coupled with the search for over-represented motifs in the promoters of co-expressed genes is a powerful criterion to identify genes that are co-regulated from a set of co-expressed genes [19,20].
In co-expression analysis, similarity of gene expression profiles is measured using correlation coefficients (CC) or other distance measures. If the correlation between two genes is above a given threshold, then the genes can be considered as «co-expressed» [1]. Co-expression analysis using a «guide-gene» approach involves measuring CC between pre-selected gene(s) and the rest of the genes in a dataset.
It is a common practice in meta-coexpression studies to assess co-expression by calculating the gene pair correlations after merging the datasets [20] or by confirming the re-occurrence of significant correlations across datasets [15]. However, it has been shown recently that genes can reveal differential co-expression patterns across subsets in the same dataset (e.g. gene pairs that are correlated in normal tissue might not be correlated in cancerous tissue or might be even anti-correlated) [21]. Therefore, assessing co-expression by measuring CC across merged samples of a dataset or by merging datasets may create correlation patterns that could not be captured using the CC measurement.
In this study, we performed co-expression analysis of publicly available microarray data using BDNF as a "guide-gene". We inferred BDNF gene co-expression links that were conserved between human and rodents using a novel "subset" approach. Then, we discovered new putative regulatory elements in human BDNF and in BDNF-correlated genes, and proposed potential regulators of BDNF gene expression.
Results
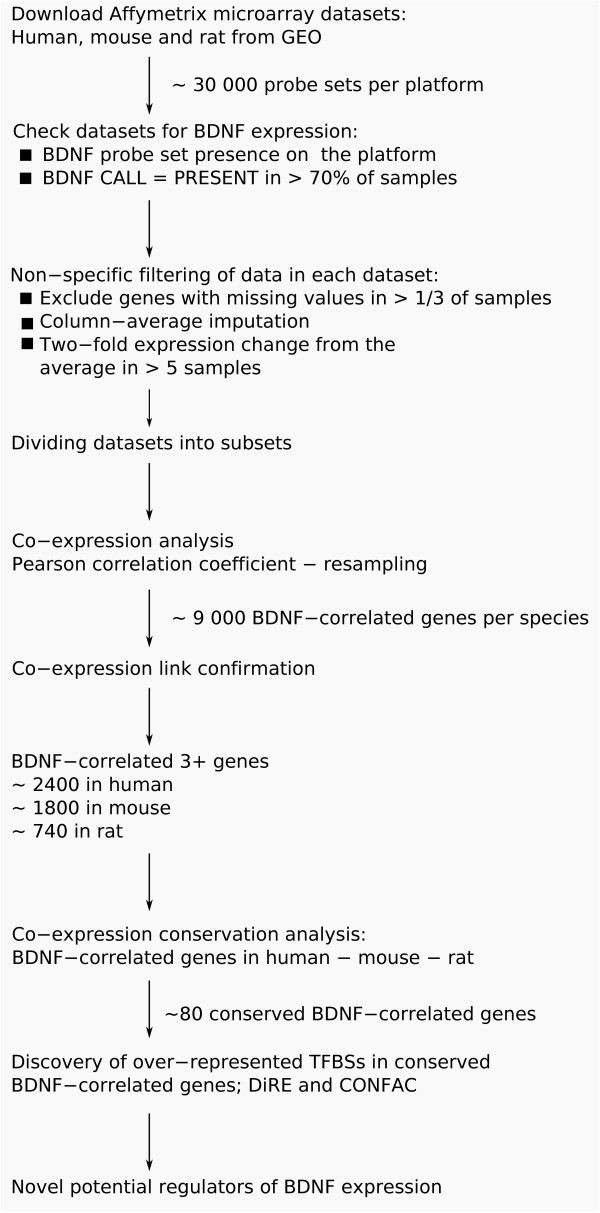
We analyzed 299 subsets derived from the total of 80 human, mouse and rat microarray datasets. In order to avoid spurious results that could arise from high-throughput microarray analysis methods, we applied successive filtering of genes. Then, we divided datasets into subsets with biologically meaningful sample content (e.g. tissue, gender or disease state subsets), analyzed co-expression with BDNF across samples separately in each subset and confirmed the links across subsets. Finally, we analyzed conservation in co-expression between human, mouse and rat, and sought for conserved TFBSs in BDNF and BDNF-correlated genes (Figure 1).
Figure 1.
Microarray data analysis flowchart. Altogether, 80 human, mouse and rat Affymetrix datasets were analyzed (dataset selection criteria: > 16 samples per dataset; BDNF detection call PRESENT in more than 70% of the samples). Data was subjected to non-specific filtering (missing values and 2-fold change filtering). Thereafter, datasets were divided into 299 corresponding subsets. Co-expression analysis in human, mouse and rat subsets allowed the detection of genes that co-expressed with BDNF in more than 3 subsets (~1000 genes for each species). As a result of co-expression conservation analysis, 84 genes were found to be correlated with BDNF in all three species. Discovery of over-represented motifs in the regulatory regions of these genes and in BDNF suggested novel regulators of BDNF gene expression.
Data filtering
Gene Expression Omnibus (GEO) from NCBI and ArrayExpress from EBI are the largest public peer reviewed microarray repositories, each containing about 8000 experiments. In order to avoid inaccuracies arising from measuring expression correlation across different microarray platforms [13] we used only Affymetrix GeneChips platforms for the analysis. Since ArrayExpress imports Affymetrix experiments from GEO http://www.ebi.ac.uk/microarray/doc/help/GEO_data.html, we used only GEO database to retrieve datasets.
A study examining the relationship between the number of analyzed microarray experiments and the reliability of the results reported that the accuracy of the analysis plateaus at between 50 and 100 experiments [14]. Another study demonstrated how the large amount of microarray data can be exploited to increase the reliability of inferences about gene functions. Links that were confirmed three or more times between different experiments had significantly higher GO term overlaps than those seen only once or twice (p < 10-15) [15]. Therefore, we performed meta-coexpression analysis using multiple experiments to increases the accuracy of the prediction of the co-expression links.
Since BDNF served as a guide-gene for our microarray study, qualitative and quantitative criteria were applied for selection of the experiments with respect to BDNF probe set presence on the platform [see Additional file 1: BDNF probe sets], BDNF signal quality and expression levels. In addition, non-specific filtering [19] was performed to eliminate the noise (see Methods/Microarray datasets). Consequently, 80 human, mouse and rat microarray experiments (datasets) from Gene Expression Omnibus (GEO) database met the selection criteria. Each dataset was split into subsets according to the annotation file included in the experiment [see Additional file 2: Microarray datasets and Additional file 3: Subsets]. In summary, 299 subsets were obtained from 38 human, 24 mouse and 18 rat datasets. From 38 human datasets, 8 were related to neurological diseases (epilepsy, Huntington's, Alzheimer's, aging, encephalitis, glioma and schizophrenia) and contained samples from human brain; another 9 datasets contained samples from human "normal" (non-diseased) tissues (non-neural, such as blood, skin, lung, and human brain tissues); 12 datasets had samples from cancerous tissues of various origins (lung, prostate, kidney, breast and ovarian cancer). The rest 9 datasets contained samples from diseased non-neural tissues (HIV infection, smoking, stress, UV radiation etc.). Out of 24 mouse datasets, 5 datasets were related to neurological diseases (brain trauma, spinal cord injury, amyotrophic lateral sclerosis, and aging); 15 datasets contained normal tissue samples (neural and peripheral tissues); 1 dataset contained lung cancer samples; 3 datasets were related to non-neural tissues' diseases (muscle dystrophy, cardiac hypertrophy and asthma). Among 18 rat datasets, 11 datasets were related to neurological diseases (spinal cord injury, addiction, epilepsy, aging, ischemia etc), 5 datasets were with "normal tissue samples" composition and 2 datasets examined heart diseases [see Additional file 2: Microarray datasets].
According to Elo and colleagues [22] the reproducibility of the analysis of eight samples approaches 55%. Selecting subsets with more than eight samples for the analysis could increase the reproducibility of the experiment however reducing the coverage, since subsets with lower number of samples would be excluded. Thus, we selected subsets with a minimum of eight samples for the analysis, in order to achieve satisfactory reproducibility and coverage. The expression information for human, mouse and rat genes obtained from GEO database, information about BDNF probe names used for each dataset, information about subsets derived from each experiment, and data on correlation of expression between BDNF and other genes for each microarray subset has been made available online and can be accessed using the following link: http://www.bio.lmu.de/~pavlidis/bmc/bdnf.
Differential expression of BDNF across subsets
Since the study was based on analyzing subsets defined by experimental conditions (gender, age, disease state etc) it was of biological interest to examine if BDNF is differentially expressed across subsets within a dataset. We used Kruskal-Wallis test [23] to measure differential expression. The results of this analysis are given in the Additional files 4, 5 and 6: Differential expression of the BDNF gene in human, mouse and rat datasets.
Co-expression analysis
Since the expression of BDNF alternative transcripts is tissue-specific and responds to the variety of stimuli, seeking for correlated genes in each subset separately could help to reveal condition-specific co-expression. The term "subset" in this case must be understood as "a set of samples under the same condition".
We derived 119 human, 73 mouse and 107 rat subsets from the corresponding datasets. Pearson correlation coefficient (PCC) was chosen as a similarity measure since it is one of the most commonly used, with many publications describing analysis of Affymetrix platforms [13,24,25]. PCC between BDNF and other genes' probe sets was measured across samples for each subset separately. From each subset, probe sets with PCC r > 0.6 were selected. It was demonstrated by Elo and colleagues [22] that in the analysis of simulated datasets a cutoff value r = 0.6 showed both high reproducibility (~0.6 for profile length equal to 10) and low error. A "data-driven cutoff value" approach has been rejected because it is based on the connectivity of the whole network, whereas we focused only on the links between BDNF and other genes. A lower threshold of 0.4 generated a list of genes that showed no significant similarities when analyzed using g:Profiler tool that retrieves most significant GO terms, KEGG and REACTOME pathways, and TRANSFAC motifs for a user-specified group of genes [26]. The value r = 0.6 was chosen over more stringent PCC values because the lengths of the expression profiles were not too short (mean profile length ~17, standard deviation ~12). Moreover, the PCC threshold higher than 0.6 was not justified since we performed further filtering by selecting only conserved correlated genes, thus controlling the spurious results.
Each probe set correlation with BDNF that passed the threshold was defined as a "link". It has been previously shown that a link must be confirmed in at least 3 experiments (3+ link) in order to be called reliable [15]. Therefore, we selected (3+) genes for evolutionary conservation analysis, narrowing the list of correlated genes to eliminate the noise. g:Profiler analysis of these genes revealed that the results are statistically significant (low p-values) and the genes belong to GO categories that are relevant to biological functions of BDNF. For example, the list of human genes produced the following results when analyzed with g:Profiler (p-values for the GO categories are given in the parenthess): nervous system development (5.96·10-21), central nervous system development (3.29·10-07), synaptic transmission (4.40·10-11), generation of neurons (1.58·10-08), neuron differentiation (1.02·10-06), neurite development (4.11·10-07), heart development (1.67·10-09), blood vessel development (5.51·10-14), regulation of angiogenesis (7.16·10-09), response to wounding (1.32·10-11), muscle development (1.53·10-10), regulation of apoptosis (1.65·10-07), etc.
We have used r = 0.6 as a "hard" threshold value for the CC. A disadvantage of this approach is that there will be no connection between BDNF and other genes whose correlation with BDNF is 0.59 in a specific dataset [27]. Using multiple datasets was expected to remedy this effect. An alternative approach would be to use "soft" threshold approaches [27]. According to the soft threshold approach, a weight between 0 and 1 is assigned to the connection between each pair of genes (or nodes in a graph). Often, the weight between the nodes A and B is represented by some power of the CC between A and B. However, other similarity measures may be used given that they are restricted in [0, 1]. A drawback of the weighted CC approach is that it is not clear how to define nodes that are directly linked to a specific node [27] because the available information is related only to how strongly two nodes are connected. Thus, if neighbors to a node are requested, threshold should be applied to the connection strengths. Alternatively, Li and Horvath [28] have developed an approach to answer this question based on extending the topological overlap measure (TOM), which means that the nodes (e.g. genes) should be strongly connected and belong to the same group of nodes. However, this analysis requires the whole network of a set of genes. In the current analysis, we did not construct the co-expression network for all the genes of microarray experiments. Instead, we focused on a small part of it i.e. the BDNF gene and the genes linked to BDNF. Therefore, TOM analysis was not possible using our approach.
To see how the "weighted CC" method would affect the results of our study we used a simplified approach. Instead of applying "hard" threshold (0.6) for the CC we measured the strength of all the connections between BDNF and all the genes in a microarray experiment. The connection strength sj = [(1 + CCj)/2]b, where CCj denotes the CC between BDNF and the gene j, is between 0 and 1 and b is an integer. In order to define b, analysis of the scale-free properties of the network is required. However, we used the value 6. Great b values give lower weight to weak connections. Then we calculated the average sj(ave(sj)) among all the subsets. Finally, we sorted the genes based on their ave(sj) and calculated the overlap of the top of this list with our results for each species (human mouse and rat). When restricting the top of the weighted CC list to the same number of genes that we have obtained for the 3+ list for each species, we observed that the top-weighted CC genes overlap extensively with the 3+ list (overlapping > 80%) for each species. Therefore, even though the "soft" and "hard" thresholding approaches are considerably different we observe quite extensive overlap of the results. We would like to stress that we did not apply the full weighted CC and TOM methodology since it would require the construction of the whole network which was beyond the aims of our study. However, such investigation of the whole co-expression network could contribute to the understanding of BDNF regulation and function.
Correlation conservation and g:Profiler analysis
Co-expression that is conserved between phylogenetically distant species may reveal functional gene associations [29]. We searched for common genes in the lists of 2436 human, 1824 mouse and 740 rat genes (3+ genes, whose expression is correlated with BDNF). From these genes, 490 were found to be correlated with BDNF in human and mouse, 210 correlated with BDNF in human and rat, and 207 conserved between mouse and rat [see Additional file 7: Conserved BDNF-correlated genes]. We found a total of 84 genes whose co-expression with BDNF was conserved in all three organisms (Table 1) [see also Additional file 7: Conserved BDNF-correlated genes].
Table 1.
BDNF-correlated genes conserved between human, mouse and rat.
| GO category | Conserved correlated genes | |||||||||||
| protein tyrosine kinase PW * | ANGPT1 | BAIAP2 | DUSP1 | EPHA4 | EPHA5 | EPHA7 | FGFR1 | GAS6 | KALRN | IRS2 | NTRK2 | |
| PTPRF | FP106 | |||||||||||
| dendrite localization* | DBN1 | FREQ | GRIA3 | KCND2 | NTRK2 | |||||||
| signal transduction* | ANGPT1 | CREM | DUSP6 | EPHA5 | FGFR1 | IGFBP5 | KALRN | NR4A2 | PDE4B | PRKAG2 | PTPRF | TBX3 |
| BAIAP2 | CXCL5 | EGR1 | EPHA7 | GAS6 | IL6ST | KLF10 | NTRK2 | PENK | PRKCB | RGS4 | ZFP106 | |
| COL11A1 | DUSP1 | EPHA4 | FGF13 | GRIA3 | IRS2 | MYH9 | ODZ2 | PLAUR | PRKCE | SCG2 | ||
| hsa-miR-369-3p* | COL11A1 | DBC1 | DCN | DUSP1 | GAS6 | ITF-2 | KLF10 | NEUROD6 | PENK | TRPC4 | ||
| TF: CCCGCCCCCRCCCC (KROX) * | ATF3 | ATP1B1 | CCND2 | COL11A1 | DBN1 | DLGAP4 | EPHA7 | GAS6 | GRIA3 | IL6ST | IRS2 KCND2 | |
| KLF10 | NFIA | NPTXR | PCSK2 | SNCA | THRA | |||||||
| TF: GGGGAGGG (MAZ/SP1) * | ATF3 | CCND2 | DBC1 | DUSP6 | FREQ | ITF-2 | MBP | NPTXR | PCSK1 | PTGS2 | THRA | |
| BAIAP2 | COL4A5 | DBN1 | EGR1 | GRIA3 | KALRN | MDM2 | NR4A2 | PDE4B | PTPRF | TRPC4 | ||
| BASP1 | CREM | DLGAP4 | EPHA5 | HN1 | KLF10 | NFIA | NTRK2 | PRKCB1 | PURA | VCAN | ||
| CAMK2D | CXCL5 | DUSP1 | EPHA7 | IRS2 | LMO7 | NPTX1 | OLFM1 | PRSS23 | TBX3 | |||
| NS development* | BAIAP2 | EPHA4 | FGF13 | IRS2 | MBP | NEUROD6 | NR4A2 | OLFM1 | PTPRF | SMARCA4 | TBX3 | |
| DBN1 | EPHA7 | FGFR1 | KALRN | NEFL | NPTX1 | NTRK2 | PCSK2 | PURA | SNCA | |||
| angiogenesis | ANGPT1 | BAIAP2 | CYR61 | MYH9 | SCG2 | SERPINE1 | TBX3 | |||||
| apoptosis/anti-apoptosis | BIRC4 | KLF10 | NEFL | PLAGL1 | PRKCE | SCG2 | SNCA | TBX3 | ||||
| cell cycle | CAMK2D | CORO1A | DUSP1 | MDM2 | MYH9 | PPP3CA | ||||||
| synaptic transmission/plasticity | DBN1 | KCND2 | MBP | NPTX1 | NR4A2 | SNCA | ||||||
GO categories marked with a star (*) have been reported as statistically significant for this gene list by g:Profiler analysis tool. Human gene names are given representing mouse and rat orthologs whenever gene names for all three species are not the same. GO - gene ontology, PW - pathway, TF - transcription factor, NS - nervous system.
Due to a variety of reasons (e.g. sample size of a dataset/subset, probe set binding characteristics, sample preparation methods, etc.), when measured only in one dataset/subset, some of the co-expression links might occur by chance. Checking for multiple re-occurrence of a link is expected to reduce the number of false-positive links. More importantly, the conservation analysis should further reduce the number of artifacts. However, since our analysis comprised a multitude of subsets it was important to estimate the statistical significance of the results. To tackle this problem, we created randomized subsets similarly to what was described by Lee and colleagues [15] and calculated the distribution of correlated 3+ links for each species separately. The results showed that our co-expression link confirmation analysis resulted in a significantly higher number of links compared to the randomized data (p-value < 0.005 for each species). However, it should be mentioned that the number of 3+ links remained quite high in the randomized datasets: for human subsets it constituted about 58% of the observed 3+ links, for mouse about 43% and for rat 21%. These results justify the subsequent co-expression conservation analysis step. Indeed, in random human, mouse and rat subsets the number of correlated 3+ links was only about 9% of the discovered conserved BDNF-correlated links (that is ~7.5 genes out of 84).
Analysis of the list of 84 conserved BDNF-correlated genes using g:Profiler showed significantly low p-values for all the genes and revealed significant GO categories related to BDNF actions [see Additional file 8: g:Profiler analysis]. Statistically significant GO categories included: i) MYC-associated zinc finger protein (MAZ) targets (44 genes, p = 1.82·10-05); ii) signal transduction (36 genes, p = 3.51·10-06); iii) nervous system development (17 genes, p = 5.27·10-08); iv) Kruppel-box protein homolog (KROX) targets (18 genes, p = 1.21·10-04); v) transmembrane receptor protein tyrosine kinase pathway (7 genes, p = 3.56·10-06); vi) dendrite localization (5 genes, p = 1.82·10-05) (Table 1).
According to the Gene Ontology database, conserved BDNF-correlated gene products participate in axonogenesis (BAIAP2), dendrite development (DBN1), synaptic plasticity and synaptic transmission (DBN1, KCND2, MBP, NPTX1, NR4A2 and SNCA), regeneration (GAS6, PLAUR), regulation of apoptosis (XIAP (known as BIRC4), KLF10, NEFL, PLAGL1, PRKCE, SCG2, SNCA, and TBX3), skeletal muscle development (MYH9, PPP3CA, and TBX3) and angiogenesis (ANGPT1, BAIAP2, CYR61, MYH9, SCG2, SERPINE1 and TBX3) (Table 1). Out of 84, 24 BDNF-correlated genes are related to cancer and 14 are involved in neurological disorders (Table 2).
Table 2.
Conserved correlated genes are associated with various types of cancer and neurological disorders.
| Disease | Associated genes | References |
| Schizophrenia | BDNF RGS4 NR4A2 | Schmidt-Kastner et al. (2006) |
| Parkinson's disease | BDNF PTGS2 SNCA NR4A2 | Murer et al. (2001) Chae et al. (2008) Pardo and van Duijn (2005) |
| Alzheimer's | BDNF KALRN |
Murer et al. (2001) Youn et al. (2007) |
| Polyglutamine neurodegeneration | NEFL BAIAP2 |
Mosaheb et al. (2005) Thomas et al. (2001) |
| alpha-mannosidosis | MAN1A1 | D'Hooge et al. (2005) |
| Ophthalmopathy | CYR61 DUSP1 EGR1 PTGS2 | Lantz et al. (2005) |
| Epilepsy | BDNF DUSP6 EGR1 | Binder and Scharfman (2004) Rakhade et al. (2007) |
| Depression | BDNF DUSP1 | Russo-Neustadt and Chen (2005) Rakhade et al. (2007) |
| Ischemia | BDNF CD44 PTGS2 | Binder and Scharfman (2004) Murphy et al. (2005) |
| Ovarian carcinoma | BDNF ITF2 DUSP1 RGS4 | Yu et al. (2008) Kolligs et al. (2002) Puiffe et al. (2007) |
| Breast cancer | BDNF FGFR1 CCND2 PLAU SERPINE1 PLAUR MAZ DUSP6 EGR1 KFL10 PTRF |
Tozlu et al. (2006) Koziczak et al. (2004) Grebenchtchikov et al. (2005) Cui et al. (2006) Liu et al. (2007) Reinholz et al. (2004) Levea et al. (2000) |
| Lung cancer | BDNF ODZ2 CCND2 GFI1 | Ricci et al. (2005) Kan et al. (2006) |
| Prostate cancer | BDNF IGFBP5 PLAUR p75NTR | Bronzetti et al. (2008) Nalbandian et al. (2005) |
| Pheochromocytoma | PCSK1 PCSK2 SCG2 | Guillemot et al. (2006) |
| Endometrial cancer | CXCL5 OLFM1 | Wong et al. (2007) |
| Leukemia | PKCB1 CCND2 | Hans et al. (2005) |
Interactions among correlated genes
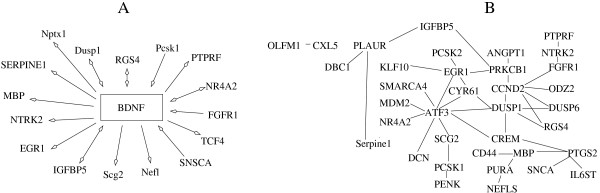
We searched if any of the correlated genes had known interactions with BDNF using Information Hyperlinked over Proteins gene network (iHOP). iHOP allows navigating the literature cited in PubMed and gives as an output all sentences that connect gene A and gene B with a verb http://www.ihop-net.org/[30]. We constructed a "gene network" using the iHOP Gene Model tool to verify BDNF-co-expression links with the experimental evidences reported in the literature (Figure 2). For the URL links to the cited literature see Additional file 9: iHOP references.
Figure 2.
Reported interactions between conserved correlated genes in human. Connections between the genes were created by accessing the literature using iHOP tool. (A) Interactions between correlated genes and BDNF. Arrows: "↔" co-expression or co-regulation; "BDNF← "regulation of BDNF; "BDNF→" regulation by BDNF. (B) Connections among correlated genes.
According to the literature, 17 out of 84 conserved correlated genes have been reported to have functional interaction or co-regulation with BDNF (Figure 2A). IGFBP5 [31], NR4A2, RGS4 [32] and DUSP1 [33] have been previously reported to be co-expressed with human or rodent BDNF. Other gene products, such as FGFR1 [34] and SNCA [35] are known to regulate BDNF expression. Proprotein convertase PCSK1 is implied in processing of pro-BDNF [36]. PTPRF tyrosine phosphatase receptor associates with NTRK2 and modulates neurotrophic signaling pathways [37]. Thyroid hormone receptor alpha (THRA) induces expression of BDNF receptor NTRK2 [38]. Finally, expression of such genes like EGR1 [39], MBP [40], NEFL [41], NPTX1 [42], NTRK2, SERPINE1 [43], SCG2 [44], SNCA [45] and TCF4 (also known as ITF2) [46] is known to be regulated by BDNF signaling. CCND2, DUSP1, DUSP6, EGR1 and RGS4 gene expression is altered in cortical GABA neurons in the absence of BDNF [47].
iHOP reports the total of 250 interactions with human BDNF. In order to assess the probability of observing 17/84 or more functional interactions between BDNF and other genes, we had to make an assumption regarding the total number of human genes that iHOP uses. A lower number of total genes would result in higher p-values whereas a higher number of total genes would produce lower p-values. We assumed that the total number of human genes is N = 5000, 10000, 20000 or 30000. Furthermore, the total number of genes linked to BDNF is m = 250 based on iHOP data. Thus, the p-values were obtained using the right-tail of the hypergeometric probability distribution. For N = 5000, 10000, 20000 or 30000, the p-values are 1.0 × 10-07, 1.7 × 10-12, 1.3 × 10-17, 1.18 × 10-20 respectively.
By analyzing the iHOP network indirect connections with BDNF could be established for the genes that did not have known direct interactions with BDNF (Figure 2B). For example, SCG2 protein is found in neuroendocrine vesicles and is cleaved by PCSK1 [48] - protease that cleaves pro-BDNF. BDNF and NTRK2 signaling affect SNCA gene expression and alpha-synuclein deposition in substantia nigra [49]. ATF3 gene is regulated by EGR1 [50], which expression is activated by BDNF [39]. For more interactions see Figure 2.
Motif discovery
Assuming that genes with similar tissue-specific expression patterns are likely to share common regulatory elements, we clustered co-expressed genes according to their tissue-specific expression using information provided by TiProd database [51]. Each tissue was assigned a category and the genes expressed in corresponding tissues were clustered into the following categories: i) CNS, ii) peripheral NS (PNS), ii) endocrine, iii) gastrointestinal, and iv) genitourinary. We applied DiRE [52] and CONFAC [53] motif-discovery tools to search for statistically over-represented TFBSs in the clusters and among all conserved BDNF-correlated genes. DiRE can detect regulatory elements outside of proximal promoter regions, as it takes advantage of the full gene locus to conduct the search. The software predicts function-specific regulatory elements (REs) consisting of clusters of specifically associated and conserved TFBSs, and it also scores the association of individual TFs with the biological function shared by the group of input genes [52]. DiRE selects a set of candidate REs from the gene loci based on the inter-species conservation pattern which is available in the form of precomputed alignments of genomic sequence from fish, rodent, human and other vertebrate lineages [54]. This type of the alignment enables the tool to detect regulatory elements that are phylogenetically conserved at the same genomic positions in different species. CONFAC software [53] enables the identification of conserved enriched TFBSs in the regulatory regions of sets of genes. To perform the search, human and mouse genomic sequences from orthologous gene pairs are compared by pairwise BLAST, and only significantly conserved (e-value < 0.001) regions are analyzed for TFBSs.
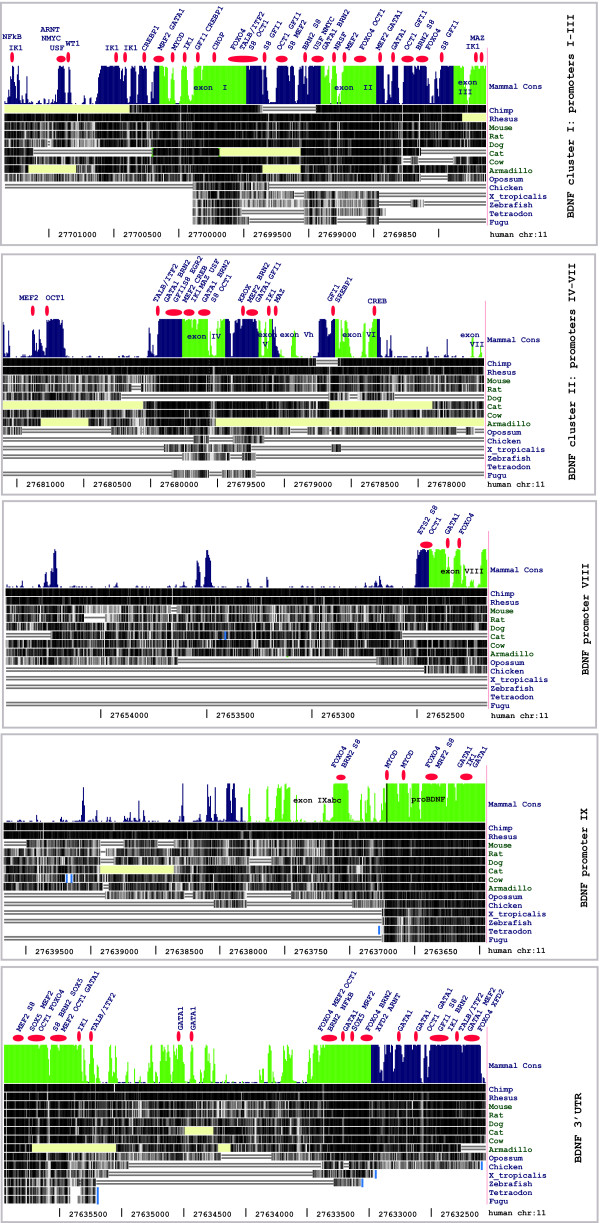
Using DiRE we discovered two regulatory regions at the human BDNF locus that were enriched in TFBSs (Figure 3) [see also Additional file 10: DiRE motif discovery results for BDNF and 84 conserved correlated genes]. The first regulatory region spans 218 bp and is located 622 bp upstream of human BDNF exon I transcription start site (TSS). The second putative regulatory region is 1625 bp long and located 2915 bp downstream of the BDNF stop-codon. Analysis of mouse and rat gene lists produced similar results. Significant over-representation of binding sites for WT1, KROX, ZNF219, NFkB, SOX, CREB, OCT, MYOD and MEF2 transcription factors was reported by DiRE in BDNF and BDNF-correlated genes when all the genes were analyzed as one cluster [see Additional file 10: DiRE motif discovery results for BDNF and 84 conserved correlated genes]. Also, the following cluster-specific over-representation of TFBSs was detected: i) CNS - KROX; ii) endocrine - TAL1beta/TCF4, ETS2, SOX5, and ARID5B (known as MRF2); iii) gastrointestinal - MMEF2, and SREBF1; iv) genitourinary - ATF4/CREB, and GTF3 (TFIII) (Table 3) [see also Additional file 11: DiRE motif discovery results for conserved BDNF-correlated genes clustered by tissue-specific expression].
Figure 3.
Novel regulatory elements in the BDNF gene. Highly conserved TFBSs in the BDNF locus as predicted by DiRE and CONFAC tools. Given TFBSs were also found to be over-represented in the BDNF-correlated genes. Histograms represent evolutionary conservation across 9 mammal species (adapted from UCSC Genome Browser at http://genome.ucsc.edu/) (39). The height of the histogram reflects the size of the conservation score. Conservation for each species is shown in grayscale using darker values to indicate higher levels of overall conservation. Missing sequences are highlighted by regions of yellow. Single line: no bases in the aligned species; double line: aligning species has one or more unalignable bases in the gap region. Transcribed regions (BDNF exons and 3'UTR) are highlighted in green; non-transcribed regions (BDNF promoters and introns) are highlighted in blue. Red ovals represent TFBSs mapped to the BDNF gene sequences. Mapped TFBSs have Matrix Similarity score >0.85 and Core Similarity score >0.99. Core elements of presented TFBSs have 100% of conservation across mammals. For the structure of human BDNF see Pruunsild et al., 2007 [11].
Table 3.
Over-represented conserved TFBSs in human BDNF and in the BDNF-correlated genes as predicted by DiRE and CONFAC.
| TFBS |
p-value CONFAC |
Target genes |
| ARNT | 0.012 | BDNF pI-II, BDNF 3'UTR; PRKCE, USP2, CAMK2D, CCND2, NEUROD6, THRA, DUSP1, CBX6, ATP1B1, FREQ, ITF-2 |
| POU3F2 (BRN2) |
< 0.001 | BDNF pII-V, BDNF exon II, IV, IX, BDNF3'UTR; USP2, CAMK2D, THRA, NFIA, PRSS23, CBX6, CUGBP2, EPHA5, EPHA7, BAIAP2, RKCE, CPD, EPHA4, IL6ST, CCND2, DUSP6, KCND2, MAN1A1, SCG2, GRIA3, COL11A1, TRPC4, FGF13, HN1, ANGPT1, TCF4, MYH9, PCSK1 |
| CHOP | NA | BDNF I, COL11A1, CD44, BAIAP2, PPP3CA, IL6ST, NEUROD6, SCG2, CYR61, IGFBP5, THRA, NFIA, FGF13, ATP1A2, ANGPT1, DBC1, CUGBP2, EGR1 |
| CREB | 0.013 | BDNF pI, IV, VI, BDNF exon I; BAIAP2, PRKCE, USP2, EPHA4, CAMK2D, CCND2, FGFR1, CYR61, GRIA3, THRA, DUSP1, PENK, PCSK1, PCSK2, HN1, ATP1B1, EGR1, COL4A5, KLF10, EPHA4, FGF13, CBX6, CUGBP2, EPHA5 |
| ETS2 | NA | BDNF pII, VIII; THRA, EPHA7, FGF13, BAIAP2 and NFIA promoters, and in COL11A1, PLAGL1, and XIAP intergenic regions |
| FOXO4 | < 0.001 | BDNF exon I, II, VIII, IX, BDNF pIII, IV, BDNF 3'UTR; CD44, TBX3, BAIAP2, PPP3CA, CPD, USP2, PRKCB, EPHA4, CORO1A, CAMK2D, NEUROD6, FGFR1, SCG2, CYR61, GRIA3, THRA, NFIA, COL11A1, DUSP1, TRPC4, PRSS23, PCSK2, ANGPT1, FREQ, PRKAG2, TCF4, MYH9, PCSK1, DBC1, CUGBP2, EGR1, EPHA5 |
| GATA1 | < 0.001 | BDNF pI, III-V, BDNF exon I, II, VIII, IX, BDNF 3'UTR; CD44, TBX3, SNCA, PPP3CA, PRKCE, COL4A5, USP2, EPHA4, IL6ST, SLC4A7, CAMK2D, ATF3, CCND2, NEUROD6, DUSP6, KCND2, SCG2, CYR61, IGFBP5, THRA, NFIA, COL11A1, PENK, FGF13, PRSS23, ATP1B1, ATP1A2, ANGPT1, DBC1, CUGBP2, EGR1 |
| GFI1 | < 0.001 | BDNF exon I, BDNF pII-VI, BDNF 3'UTR; SNCA, ATP1A2, MYH9, DBC1, CD44, BAIAP2, PPP3CA, PRKCE, COL4A5, CPD, USP2, EPHA4, IL6ST, SLC4A7, CAMK2D, CCND2, NEUROD6, KCND2, SCG2, CYR61, IGFBP5, THRA, NFIA, COL11A1, DUSP1, TRPC4, PENK, FGF13, PRSS23, PCSK2, ATP1B1, PTPRF, ANGPT1, TCF4, CUGBP2, EGR1, EPHA5, EPHA7 |
| IK1 (ikaros) | < 0.001 | BDNF pI, BDNF exon I-V, IX, BDNF 3'UTR; PRKCB, KLF10, KCND2, THRA, NFIA, COL11A1, FGF13, ATP1A2, MYH9, PCSK1, CUGBP2, EPHA7 |
| KROX family | NA | BDNF pV, BDNF exon IV; PPP3CA, NFIA, DBN1, KCND2, IRS2, MAN1A2, CCND2, PVRL3, XIAP, DLGAP4, CYR61, ATP1B1, PURA, SMARCA4, MYH9, GRIA3, EPHA4, DUSP6, EGR1, COL4A5, TRPC4, PRKCB, NPTX1, PTGS2, EPHA5, FGFR1, CBX6, PRKCE, KLF10, THRA, ATP1A2, BAIAP2, CPD, CORO1A, CAMK2D, IGFBP5, DUSP1, PTPRF, FREQ, PRKAG2 |
| MAZ | NA | BDNF pVh, BDNF exon III, IV; CD44, PPP3CA, PRKCE, COL4A5, USP2, PRKCB, KLF10, EPHA4, CAMK2D, CCND2, DUSP6, GRIA3, THRA, COL11A1, PENK, FGF13, CBX6, ATP1B1, PTPRF, ATP1A2, FREQ, DBN1, CUGBP2, EGR1, EPHA7 |
| MEF2 | NA | BDNF pII-V, BDNF exon II, IX, BDNF 3'UTR; CD44, TBX3, BAIAP2, PPP3CA, PRKCE, COL4A5, EPHA4, IL6ST, CAMK2D, CCND2, NEUROD6, DUSP6, MAN1A1, IGFBP5, COL11A1, TRPC4, PRSS23, ANGPT1, FREQ, PURA, MYH9, PCSK1, CUGBP2, EPHA7, SNCA, FGF13 |
| MYC/MAX | NA | BDNF pI, II, IV; CD44, TBX3, PRKCE, USP2, CAMK2D, CCND2, NEUROD6, THRA, NFIA, DUSP1, CBX6, ATP1B1, FREQ, ITF-2, EGR1 |
| MYCN | NA | BDNF pI, II; PRKCE, USP2, CAMK2D, CCND2, NEUROD6, THRA, DUSP1, CBX6, ATP1B1, FREQ, ITF-2 |
| MYOD | < 0.001 | BDNF exon I, IX; CD44, PRKCE, USP2, PRKCB, EPHA4, DUSP6, SCG2, SMARCA4, THRA, PRSS23, ATP1B1, CUGBP2 |
| NFkB | < 0.001 | BDNFI, BDNF 3'UTR; PPP3CA, KLF10, PCSK2, ATP1B1, ANGPT1, MYH9, USP2, DUSP6, FGF13, PURA, BAIAP2, CAMK2D, CCND2, FGFR1, CYR61, PCSK2, MYH9, CUGBP2, EGR1, EPHA7 |
| NRSF | NA | BDNFII, EPHA4, IRS2, EPHA5, NPTX1, PRKCB, TRPC4, COL4A5 |
| S8 | < 0.001 | BDNF pII-IV, BDNF exon II, IV, VIII, IX, BDNF 3'UTR; CD44, BAIAP2, PRKCE, NPTX1, EPHA4, CAMK2D, CCND2, NEUROD6, DUSP6, FGFR1, KCND2, MAN1A1, SCG2, THRA, NFIA, COL11A1, PENK, PCSK2, ANGPT1, PURA, ITF-2, MYH9, DBC1, CUGBP2, EGR1, EPHA5 |
| SOX5 | 0.001 | BDNF exon I, BDNF 3'UTR; EPHA4, THRA and PLAGL1 3'UTR; NFIA and OLFM1 promoters; SCG2 intergenic region; KCND2 intron |
| TAL1/TCF4 | NA | BDNF pIV, BDNF exon I, BDNF 3'UTR; ATP1B1 3'UTR, MYH9 3'UTR and XIAP 3'UTR; SCG2, CD44, SERPINE1, SLC4A7, CCND2, NEUROD6, FGFR1, THRA, COL11A1, PCSK2, ANGPT1, DBC1, CUGBP2 |
| WT1 | NA | BDNF pI, BASP1, PPP3CA, NFIA, DBN1, EPHA7, BAIAP2, XIAP, DLGAP4, PURA, IRS2, ATP1B1, KCND2, GRIA3, HN1, EPHA4, EGR1, COL4A5, TRPC4, ATP1A2, PRKCB, NPTX1, DBC1, EPHA5 |
In BDNF, TFBSs were found in promoters (p), exons or 3'UTR of the gene. In the correlated genes, TFBSs were searched for and discovered mostly in promoters (unless indicated otherwise). P-values are given for the TFBSs discovered using CONFAC. NA - not applicable for the TFBSs discovered using DiRE [see Additional files 10 and 11 for TFBS importance score].
To cross-check the results obtained with DiRE, we repeated the analysis using the CONFAC tool. CONFAC results overlapped with DiRE results and suggested novel regulatory elements in human BDNF promoters/exons I-IX and in BDNF 3'UTR, which were highly conserved among mammals and over-represented in the BDNF-correlated genes. Then, evolutionary conservation across mammals was checked for the core element of each TFBS discovered in the BDNF gene using UCSC Genome Browser. Based on MW test results [see Additional file 12: The results of Mann-Whitney tests (CONFAC)], on the Importance score [see Additional file 10: DiRE motif discovery results for BDNF and 84 conserved correlated genes] and on the conservation data (UCSC), we propose potential regulators of BDNF (Figure 3 and Table 3) [see also Additional file 13: Highly conserved TFBSs in the BDNF gene (according to DiRE and CONFAC)]. It is remarkable, that the TFBSs discovered in the BDNF gene are highly conserved: most of the TFBSs are 100% conserved across mammals from human to armadillo, some of them being conserved even in fish (Figure 3).
Discussion
Microarray meta-analysis has proved to be useful for constructing large gene-interaction networks and inferring evolutionarily conserved pathways. However, it is rarely used to explore the regulatory mechanisms of a single gene. We have exploited microarray data from 80 experiments for the purpose of the detailed analysis of the conservation of BDNF gene expression and regulation. Analysis of co-expression conservation combined with motif discovery allowed us to predict potential regulators of BDNF gene expression as well as to propose novel gene interactions. Several transcription factors that were identified here as potential regulators of human BDNF gene have been previously shown to regulate rodent BDNF transcription in vitro and in vivo. These transcription factors include REST (also known as NRSF) for BDNF promoter II [55], CREB for BDNF promoter I and IV [56,57], USF [58], NFkB [59], and MEF2 for BDNF promoter IV [60]. The support of the bioinformatics findings by experimental evidence strongly suggests that the potential regulatory elements discovered in this study in the BDNF locus may be involved in the regulation of BDNF expression.
According to g:Profiler, 44 out of 84 conserved correlated genes identified in this study (including BDNF) carry MYC-associated zinc finger protein (MAZ) transcription factor binding sites. Our study revealed putative binding sites for MAZ in BDNF promoter Vh and in exons III and IV, suggesting that MAZ could be involved in BDNF gene regulation from promoters III, and possibly from promoters IV, V, Vh and VI that lie in close proximity in the genome. It has been shown that MAZ is a transcriptional regulator of muscle-specific genes in skeletal and cardiac myocytes [61]. Histone deacetylation and DNA methylation might be involved in the regulation of expression of target genes by MAZ [62]. BDNF mRNA expression in the heart is driven by promoters IV, Vh and VI [11]. Epigenetic regulation of the BDNF gene expression is achieved in a cell-type and promoter-specific manner [12,63]. This could be a possible regulation mechanism of the BDNF gene by MAZ. Also, MAZ drives tumor-specific expression of PPARG in breast cancer cells, a nuclear receptor that plays a pivotal role in breast cancer [64]. Expression levels of BDNF and BDNF-correlated genes CCND2, DUSP6, EGR1, KLF10 and PTPRF are altered in breast cancer (see Table 2). These genes were identified as putative targets of MAZ in the present study suggesting potential role for MAZ in their regulation in breast cancer cells.
Our analysis revealed that Wilms' tumor suppressor 1 (WT1) transcription factor binding sites are overrepresented in the BDNF-correlated genes. WT1 binding sites were detected in BDNF promoter I, in IRS2 (insulin receptor substrate 2), EGR1, BAIAP2 (insulin receptor substrate p53) and PURA promoters and in 19 other genes. WT1 acts as an oncogene in Wilms' tumor (or nephroblastoma), gliomas [65] and various other human cancers [66]. WT1 activates the PDGFA gene in desmoplastic small round-cell tumor, which contributes to the fibrosis associated with this tumor [67]. Puralpha (PURA), a putative WT1 target gene identified in this study, has also been reported to enhance transcription of the PDGFA gene [68]. WT1 regulates the expression of several factors from the insulin-like growth factor signaling pathway [69]. WT1 was also shown to bind the promoter of EGR1 gene [70]. Neurotrophins and their receptors also may be involved in the pathogenesis of some Wilms' tumors [71]. Transcriptional activation of BDNF receptor NTRK2 by WT1 has been shown to be important for normal vascularization of the developing heart [72]. Moreover, WT1 might have a role in neurodegeneration, observed in Alzheimer's disease brain [73]. We hypothesize that BDNF and other WT1 targets identified in this study, can play a role in normal development and tumorigenesis associated with WT1.
KROX family transcription factors' binding sites were found to be abundant in the promoters of BDNF and BDNF-correlated genes. KROX binding motif was detected in BDNF promoter V and EGR2 binding site was found in BDNF promoter IV. Also, EGR1 gene expression was correlated with BDNF in human, mouse and rat. KROX family of zinc finger-containing transcriptional regulators, also known as Early Growth Response (EGR) gene family, consists of EGR1-EGR4 brain-specific transcription factors [74] that are able to bind to the same consensus DNA sequence (KROX motif) [75]. EGR1 is involved in the maintenance of long-term potentiation (LTP) and is required for the consolidation of long-term memory [76]. EGR3 is essential for short-term memory formation [77] and EGR2 is necessary for Schwann cell differentiation and myelination [78,79]. Since BDNF plays a significant role in the above mentioned processes, it would be intriguing to study the regulation of BDNF by EGR factors.
Binding sites for GFI1 and MEF2 were found in BDNF promoters, exons and 3'UTR, and in the promoter of the SNCA gene. GFI1 binding sites were detected in BDNF promoters II-VI and in exon I. MEF2 sites were found in BDNF promoters II-V and in exons II and IX. SNCA overexpression and gene mutations that lead to SNCA protein aggregation cause Parkinson's disease (PD) [80]. BDNF and SNCA expression levels change conversely in the nigro-striatal dopamine region of the PD brain [80,81]. The myocyte enhancer factor-2 (MEF2) is known to be necessary for neurogenesis and activity-dependent neuronal survival [82,83]. Inactivation of MEF2 is responsible for dopaminergic loss in vivo in an MPTP mouse model of PD [84]. MEF2 recruits transcriptional co-repressor Cabin1 and class II HDACs to specific DNA sites in a calcium-dependent manner [85]. MEF2 is one of the TFs that contribute to the activity-dependent BDNF transcription from promoter IV [60]. The growth factor independence-1 (GFI1) transcription factor is essential for the development of neuroendocrine cells, sensory neurons, and blood. Also, GFI1 acts as an oncogene in human small cell lung cancer (SCLC), the deadliest neuroendocrine tumor [86]. GFI1 mediates reversible transcriptional repression by recruiting the eight 21 corepressor (ETO), histone deacetylase (HDAC) enzymes and the G9a histone lysine methyltransferase [87]. It has also been shown that GFI1 Drosophila homolog Senseless interacts with proneural proteins and functions as a transcriptional co-activator suggesting that GFI1 also cooperates with bHLH proteins in several contexts [88]. Our findings are impelling to explore inverse regulation of BDNF and SNCA genes by GFI1 and MEF2 in neurons generally and in Parkinson's disease models in particular.
BDNF promoters II-V and BDNF exons II, IV and IX contain BRN2 (brain-specific homeobox/POU domain POU3F2) binding sequences. BRN2 is driving expression of the EGR2 gene - an important factor controlling myelination in Schwann cells [78,79]. BRN2 also activates the promoter of the Notch ligand Delta1, regulating neurogenesis. It also regulates the division of neural progenitors, as well as differentiation and migration of neurons [89]. Considering a prominent role of BDNF in myelination and neurogenesis, it is reasonable to hypothesize that BRN2 fulfills its tasks in part by regulating BDNF gene expression.
Evidence is emerging that not only proximal promoters, but also distant elements upstream and downstream from TSS can regulate transcription [90,91]. We found that BDNF 3'UTR contains potential binding sites for TCF4 (also known as ITF2), GFI1, BRN2, NFkB and MEF2.
Finally, we have discovered multiple binding sites in human BDNF promoters for the transcription factors that have been shown to participate in neuronal activity-dependent transcription of rodent BDNF gene. BDNF promoters I and IV are the most highly induced following neuronal activation. BDNF promoter I was shown to be regulated by cAMP-responsive element (CRE) and the binding sequence for upstream stimulatory factor 1/2 (USF) in response to neuronal activity and elevated calcium levels [92]. Several TFs (USF [58], CREB [57], MEF2 [60], CaRF [93] and MeCP2 [63]) regulate BDNF promoter IV upon calcium influx into neurons. Rat BDNF promoter II has also shown induction by neuronal activity, though to a lesser extent compared to promoters I and IV [12,94]. However, calcium responsive elements have not been yet studied in BDNF promoter II and it was believed that its induction is regulated by the elements located in the promoter I. Our analysis of human BDNF gene detected CREBP1 and USF binding sites in BDNF promoter I, USF and MEF2 binding sites in promoter II and USF, MEF2 and CREB binding sites in promoter IV. We suggest that MEF2 and USF elements might contribute to BDFN promoter II induction by neuronal activity. In addition, we have detected conserved TCF4 (ITF2) binding sequences in BDNF promoter IV, and in exon I. It has been shown that calcium-sensor protein calmodulin can interact with the DNA binding basic helix-loop-helix (bHLH) domain of TCF4 inhibiting its transcriptional activity [95]. Preliminary experimental evidence (Sepp and Timmusk, unpublished data) suggests that TCF4 transcription factor is involved in the regulation of BDNF transcription. TCF4 might play in concert with CREB, MEF2 and other transcription factors to modulate BDNF levels following neuronal activity.
In our study we performed the analysis of a well-known gene and it served as a good reference to evaluate the results of the "subset" approach. However, the "subset" method coupled with the analysis of evolutionary conservation of co-expression is suitable for studying poorly annotated genes as well. This approach examines co-expression across a variety of conditions, which helps to discover novel biological processes and pathways that the guide-gene and its co-expressed genes are related to. Also, searching for conserved TFBS modules in co-expressed genes helps to discover functionally important genomic regions and this does not require detailed prior knowledge of the guide-gene's structure. However, when attempting to study less known genes, additional in silico analysis of genomic sequences using bioinformatics tools for prediction of promoters, TSSs and exon-intron junctions would be useful. Also, sequence alignment with co-expressed genes' promoters would be informative.
Conclusion
A major impediment of meta-coexpression analysis is the differences among experiments. So far, analyzing gene expression across different microarray platforms remains a challenge. Discrepancies in the expression measurements among different platforms originate from different probe sequences used, different number of genes on the platform, etc. Therefore, in order to obtain reliable results, we used only one microarray platform type for the analysis. In addition, we introduced a new approach to increase the accuracy of the analysis: we divided datasets into subsets and sought for correlated genes for each subset, implying that each subset represents an independent experimental condition. We have also performed correlation link confirmation among subsets and correlation conservation analysis to discover functionally related genes.
One of the limitations of the co-expression conservation analysis is the fact that it detects only phylogenetically conserved co-expression events. Human-specific phenomena cannot be captured by this kind of analysis. In relation to BDNF this means, for example, that regulation of human BDNF gene by antisense BDNF RNA (BDNFOS gene) [11,96] could not be studied by co-expression conservation analysis, since BDNFOS gene is not expressed in rodents [12,97]. Also, co-expression analysis using microarray experiments is limited by the number of genes included in the microarray platforms. For example, since BDNFOS probe sets were absent from microarray platforms, we could not study co-expression, anti-coexpression or differential expression of BDNF and BDNFOS. In addition, our list of correlated genes did not include all possible correlation links with BDNF due to the fact that our analysis was deliberately limited to Affymetrix microarray platforms. Moreover, in our analysis we included only those experiments that met certain requirements regarding the BDNF gene expression. However, biologically meaningful results justify our rigorous filtering approach: correlated genes identified in this study are known to regulate nervous system development, and are associated with various types of cancer and neurological disorders. Also, experimental evidence supports the hypothesis, that transcription factor identified here can act as potential BDNF regulators.
In summary, we have discovered a set of genes whose co-expression with BDNF was conserved between human and rodents. Also, we detected new potential regulatory elements in BDNF-correlated genes and in the BDNF locus using bioinformatics analysis, in which BDNF was playing a role of a guide-gene. The presented concept of co-expression conservation analysis can be used to study the regulation of any other gene of interest. The study provides an example of using high-throughput advancements in studying single genes and proposes hypotheses that could be tested using molecular biology techniques.
Methods
Microarray datasets and data filtering
Homo sapiens, Mus Musculus and Rattus Norvegicus microarray datasets were downloaded from (GEO) [98]. We selected Affymetrix GeneChips experiments that comprised a minimum of 16 samples. Datasets which contained BDNF Detection call = Absent [99] in more than 30% of the samples were not selected [see Additional file 2: Microarray datasets] for the list of datasets used in the analysis. Since the arrays contained normalized data, no additional transformation was performed. To reduce the noise, we carried out non-specific filtering of data in each dataset. Genes that had missing values in more than 1/3 of the samples of a given dataset were excluded from the analysis in order to avoid data over-imputation [100]. For the remaining genes, we followed a column-average imputation method. Totally, only 0.098% of the gene expression values were imputed with this approach. Further, we selected the genes whose expression changes were greater than two-fold from the average (across all samples) in at least five samples in a dataset [19,49]. Additionally, datasets were eliminated from the study if BDNF probe sets' expression failed to meet the above mentioned criteria [see Additional file 1: BDNF probe sets]. Out of 72 human datasets, only 38 passed non-specific filtering, whereas 24 out of 82 mouse and 18 out of 35 rat datasets passed the filtering and were used for the analysis.
Each dataset was split into subsets (i.e. normal tissue, disease tissue, control, treatment, disease progression, age, etc.) so that subsets of the same dataset would not have any overlapping samples [see Additional file 3: Subsets]. The division into subsets was performed manually, according to the information included in the experiment. In some cases subsets could be further subdivided into biologically appropriate sub-subsets [see Additional file 2: Microarray datasets and Additional file 3: Subsets]. Subsets that contained less than eight samples were excluded from analysis to avoid inaccuracy in the estimation of genic correlations. Biological and technical replicates were handled as equal. From all human datasets, one (GDS564 dataset) contained one technical replicate per male sample and one technical replicate for all female samples except one. For the mouse datasets no technical replicates' data accompanied the dataset information. Finally, in rat GDS1629 dataset one technical replicate has been used for each biological replicate.
Differential expression
We used Kruskal-Wallis test [23] to measure differential expression of BDNF across subsets in each dataset. Kruskal-Wallis test is a non-parametric method for testing equality of population medians within different groups. It is similar to one-way analysis of variance (ANOVA). However, it does not require the normality assumption. Alternatively, it represents an extension of Mann-Whitney U test [101,102] for more than 2 samples. Since we used multiple datasets we applied the false discovery rate approach (FDR) at the 0.05 level as it is described by Benjamini and Hochberg (1995) [103].
Co-expression analysis
For each gene standard Pearson correlation coefficient (PCC) was calculated across samples. We followed a resampling strategy, which allows the calculation of the standard deviation of the PCC between a pair of probe sets. PCC was calculated for each subset separately. The PCC was calculated following a resampling bootstrap approach. For example, in order to calculate the CCj between BDNF and gene j when data consisted of m points, we resampled the m points with replacement creating 2000 re-samples [104]. Then the CCj was calculated as the average CC for the 2000 re-samples and the 95% bootstrap confidence interval was estimated. The average CC is very close to the sample CC. However, when m is a small number and outliers are contained in the sample then the bootstrap confidence interval may be large. The motivation behind the bootstrap approach is to avoid genes with large bootstrap confidence intervals. Thus, when we request the links between BDNF and the genes in the microarray experiment we ask for the genes j, whose CCj is greater than 0.6 and the 95% bootstrap confidence interval contains only positive numbers. If instead of the bootstrapping approach we would use just the sample CC, which is more efficient computationally, then a larger set of links would be obtained which would contain some genes with very large bootstrap confidence intervals.
A threshold value of r = 0.6 was used to retrieve a list of probe sets that were co-expressed with the BDNF probe set [22,49]. Each probe set correlation with BDNF that passed the threshold was termed as a "link". It should be noted that the PCC was calculated between probe set pairs and not between gene-name pairs. Thus, when more than one probe set-pair was associated with the same gene-pair we excluded all the links except the one with the highest PCC value.
Co-expression link confirmation
We defined a "co-expression link confirmation" as a re-occurrence of links in multiple subsets. In order to avoid artifacts and biologically irrelevant links, we performed link confirmation to select the genes that were correlated with BDNF in three or more subsets [15]. It should be noticed that systematic differential expression within a subset could result in high PCC values. However, high PCC values in this case do not reveal any relationship between genes and represent a by-product of the differential expression of genes within a heterogeneous subset. We used a minimum between 1000 and 10% of all the probe sets within the subset as a threshold. Subsets that yielded more co-expression links between BDNF and other genes than an arbitrary threshold were excluded from further analysis. Thus, 5% of all the subsets were excluded.
Probe set re-annotation and ortholog search
Prior to the identification of the links that are conserved between human, mouse and rat, we transformed the probe set-pair links to gene-pair links. We used g:Profiler [26] to transform the probe set names to Ensemble gene names (ENSG). However, since many probe sets are currently related to the expressed sequence tags (ESTs), not all the probe sets could be mapped to the known genes using g:Profiler. For each dataset, we used its annotation file (see: ftp://ftp.ncbi.nih.gov/pub/geo/DATA/annotation/platforms/). To assign Ensemble gene names to the "unmapped" probe sets, we obtained the probe set sequence identifier (GI number) using the annotation file. Then, we retrieved RefSeq accession for each GI number from NCBI database. Finally, we continued with a best-hit blast approach for all three species.
Co-expression conservation and g:Profiler analysis
By performing a co-expression conservation analysis we identified the links that have passed prior filters (PCC threshold and link confirmation) and are conserved among human, mouse and rat.
Genes which co-expression with BDNF was found to be conserved between human, mouse, and rat constituted the input list for the g:Profiler. g:Profiler http://biit.cs.ut.ee/gprofiler/[26] is a public web server used for characterizing and manipulating gene lists resulting from mining high-throughput genomic data. It detects gene-ontology categories that are overrepresented by the input list of genes or by sorted sublists of the input. g:Profiler is using the "Set Count and Sizes" (SCS) method to calculate p-values [26].
Correlated genes' interactions
We used iHOP resource (Information Hyperlinked over Proteins, http://www.ihop-net.org/) [30] to find reports in the literature about known interaction between BDNF-correlated genes. iHOP generates a network of genes and proteins by mining the abstracts from PubMed. A link in such a network does not mean a specific regulatory relationship, but any possible interaction between two genes (such as protein activation, regulation of transcription, co-expression, etc). Each reference was verified manually to ensure the citation of valid interactions.
Motif discovery
We clustered BDNF-correlated genes according to their tissue-specific expression using gene expression information available in the TiProD database [51] (BDNF gene was included in every cluster). The TiProD database contains information about promoter tissue-specific expression for human genes. For each gene the list of tissues where the gene expression has been detected can be obtained from TiProD together with the tissue specificity score. For each gene we extracted information on tissue expression, selecting tissues with specificity score higher than 0.2. Each tissue was assigned a category according to its anatomy and function and the genes expressed in corresponding tissues were clustered into CNS, peripheral NS, endocrine, gastrointestinal or genitourinary cluster. Then, we searched for combinations of over-represented TFBS among the list of correlated genes, as well as the tissue clusters discovered by TiProD.
We used DiRE http://dire.dcode.org/[52] and CONFAC http://morenolab.whitehead.emory.edu/[53] tools for the discovery of TFBSs in the conserved co-expressed genes. DiRE uses position weight matrices (PWM) available from version 10.2 of the TRANSFAC Professional database [105]. In DiRE, up to 5000 background genes can be used. Only those TFBSs are extracted that occur less frequently in 95% of permutation tests than in the original distribution (corresponding to a p-value < 0.05 to observe the original distribution by chance) and that corresponds to at least a twofold increase in their density in the original distribution as compared with an average pair density in permutation tests. To correct for multiple hypothesis testing, the hypergeometric distribution with Bonferroni correction is used in the DiRE tool [106]. For each discovered TFBS DiRE defines the 'importance score' as the product of the transcription factor (TF) occurrence (percentage of tissue-specific TF with the particular TFBS) and its weight (tissue-specificity importance) in a tissue-specific set of candidate TF. Thus, the importance score is based on the abundance of the TFBS in tissue-specific TF and on the specificity of the TF that contain the particular TFBS.
Conserved transcription factor binding site (CONFAC) software [53] enables the high-throughput identification of conserved enriched TFBSs in the regulatory regions of sets of genes using TRANSFAC matrices. CONFAC uses the Mann-Whitney U-test to compare the query and the background set. It uses a heuristic method for reducing the number of false positives while retaining likely important TFBSs by applying the mean-difference cutoff which is similar to the use of fold change cutoffs in SAM analyses [107] of DNA microarray data [53]. According to the data provided by CONFAC, 50 random gene sets were compared to random sets of 250 control genes. Only one TFBS exceeded 5% false positive rate for the set of 250 random control genes that we used in our analysis with the parameters advised by the authors [53]. We used promoter sequences of BDNF-correlated genes and the sequences of BDNF promoters, exons, introns and the 3'UTR for the analysis. Matrix Similarity cut-off 0.85 and Core Similarity cut-off 0.95 were used for motif discovery; and the parameters recommended by authors - for Mann-Whitney tests (p-value cutoff 0.05 and mean-difference cutoff 0.5) [53].
Evolutionary conservation across mammals was confirmed manually for the 5-nucleotide core element of each TFBS discovered in the BDNF gene using UCSC Genome Browser [108].
Authors' contributions
TA and PP made equal contribution to conception and design of the study. PP performed computational analysis of data; TA and TT performed interpretation of the results. TA and PP were involved in drafting the manuscript; TT revised the manuscript for important intellectual content. TA, PP and TT have given final approval of the version to be published.
Supplementary Material
BDNF probe sets. Affymetrix microarray probe sets for BDNF gene. BDNF probe set target sequences are given for each platform type that was used in the co-expression conservation analysis.
Microarray datasets. Datasets that passed non-specific filtering and were used in the analysis (38 human microarray datasets, 24 mouse datasets and 18 rat datasets). Each dataset was divided into subsets (disease state, age, agent, etc) according to experimental annotations. When possible, subsets were subdivided further (marked by *). Experiments were classified based on their description and the tissue origin. GDS refers to GEO Datasets.
Subsets. Dataset: GDS1018/1368678_at/Bdnf/Rattus norvegicus. Expression profiling of brain hippocampal CA1 and CA3 neurons of Sprague Dawleys subjected to brief preconditioning seizures. According to the dataset annotation, dataset could be divided into three subsets by cell type (A) or into two subsets by protocol (B). In addition, subsets could be subdivided further into cell type.protocol sub-subsets: CA1 pyramidal neuron.control, CA1 pyramidal neuron.preconditioning seizure, CA3 pyramidal neuron.control, etc. After filtering, subset containing less than eight samples (CA3 pyramidal neuron.preconditioning seizure) was excluded from the analysis.
Differential expression of the BDNF gene in human datasets. Differential expression of BDNF was measured across subsets in each dataset using Kruskal-Wallis test. Only statistically significant results are presented.
Differential expression of the BDNF gene in mouse datasets. Differential expression of BDNF was measured across subsets in each dataset using Kruskal-Wallis test. Only statistically significant results are presented.
Differential expression of the BDNF gene in rat datasets. Differential expression of BDNF was measured across subsets in each dataset using Kruskal-Wallis test. Only statistically significant results are presented.
Conserved BDNF-correlated genes. Genes, whose correlation with BDNF was confirmed in at least 3 subsets (3+ genes) and was conserved between i) human, mouse and rat; ii) human and rat; iii) human and mouse; iv) mouse and rat.
g:Profiler analysis. Functional profiling of the list of BDNF-correlated genes conserved between human, mouse and rat using g:G:Profiler. For details see also http://biit.cs.ut.ee/gprofiler/.
iHOP references. Interactions between conserved correlated genes in human and mouse (URL links to the literature cited in iHOP).
DiRE motif discovery results for BDNF and 84 conserved correlated genes. Over-represented TFBSs are given together with the Importance Score (cut-off 0.1 recommended by DiRE). Numbers 1 and 2 (in All 1 and 2) refer to the different ways that DiRE tool analyzes evolutionary conserved regions (ECR): 1) top 3 ECRs + promoter ECRs; 2) UTR ECRs + promoter ECRs.
DiRE motif discovery results for conserved BDNF-correlated genes clustered by tissue-specific expression. TFBSs over-represented in each tissue cluster are given together with the Importance Score (cut-off 0.1 recommended by DiRE). CNS - central nervous system, PNS - peripheral nervous system.
The results of Mann-Whitney tests (CONFAC). Overrepresented TFs in the conserved BDNF-correlated gene list. Bar graphs show the average conserved TFBS frequencies for the sample gene set (conserved BDNF-correlated genes, blue bars) and control gene set (random 250 genes, red bars). A minimum threshold for the differences in the average TFBS frequencies between the two groups was set by p-value cutoff 0.05 and a mean-difference cutoff 0.5.
Highly conserved TFBSs in the BDNF gene (according to DiRE and CONFAC). Represented TFBSs have Matrix Similarity score >0.85 and Core Similarity score >0.99. TFBS sequences are highlighted in blue; "+" or "-" mark the DNA strand orientation; BDNF exons and 3'UTR are highlighted in green; the regulatory region in BDNF downstream from polyadenylation sites identified by DiRE is highlighted yellow.
Acknowledgments
Acknowledgements
This work was supported by the following grants: the Wellcome Trust International Senior Research Fellowship [grant number 067952]; Estonian Ministry of Education and Research [grant number 0140143]; Estonian Enterprise [grant number EU27553]; and Estonian Science Foundation [grant number 7257] to TT; the Volkswagen-Foundation [grant number 824234-1] to PP.
We thank Jüri Reimand and Marko Piirsoo for critical comments on the manuscript. Mari Sepp, Indrek Koppel, Priit Pruunsild and other members of our lab are acknowledged for useful suggestions and discussions.
Contributor Information
Tamara Aid-Pavlidis, Email: tamara.aid@gmail.com.
Pavlos Pavlidis, Email: pavlidis@zi.biologie.uni-muenchen.de.
Tõnis Timmusk, Email: tonis.timmusk@ttu.ee.
References
- Aoki K, Ogata Y, Shibata D. Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol. 2007;48:381–390. doi: 10.1093/pcp/pcm013. [DOI] [PubMed] [Google Scholar]
- Bibel M, Barde YA. Neurotrophins: key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev. 2000;14:2919–2937. doi: 10.1101/gad.841400. [DOI] [PubMed] [Google Scholar]
- Bekinschtein P, Cammarota M, Izquierdo I, Medina JH. BDNF and memory formation and storage. Neuroscientist. 2008;14:147–156. doi: 10.1177/1073858407305850. [DOI] [PubMed] [Google Scholar]
- Martinowich K, Manji H, Lu B. New insights into BDNF function in depression and anxiety. Nat Neurosci. 2007;10:1089–1093. doi: 10.1038/nn1971. [DOI] [PubMed] [Google Scholar]
- Bolanos CA, Nestler EJ. Neurotrophic mechanisms in drug addiction. Neuromolecular Med. 2004;5:69–83. doi: 10.1385/NMM:5:1:069. [DOI] [PubMed] [Google Scholar]
- Hu Y, Russek SJ. BDNF and the diseased nervous system: a delicate balance between adaptive and pathological processes of gene regulation. J Neurochem. 2008;105:1–17. doi: 10.1111/j.1471-4159.2008.05237.x. [DOI] [PubMed] [Google Scholar]
- Li Z, Tan F, Thiele CJ. Inactivation of glycogen synthase kinase-3beta contributes to brain-derived neutrophic factor/TrkB-induced resistance to chemotherapy in neuroblastoma cells. Mol Cancer Ther. 2007;6:3113–3121. doi: 10.1158/1535-7163.MCT-07-0133. [DOI] [PubMed] [Google Scholar]
- Hu Y, Wang YD, Guo T, Wei WN, Sun CY, Zhang L, Huang J. Identification of brain-derived neurotrophic factor as a novel angiogenic protein in multiple myeloma. Cancer Genet Cytogenet. 2007;178:1–10. doi: 10.1016/j.cancergencyto.2007.05.028. [DOI] [PubMed] [Google Scholar]
- Yang ZF, Ho DW, Lam CT, Luk JM, Lum CT, Yu WC, Poon RT, Fan ST. Identification of brain-derived neurotrophic factor as a novel functional protein in hepatocellular carcinoma. Cancer Res. 2005;65:219–225. [PubMed] [Google Scholar]
- Ricci A, Graziano P, Mariotta S, Cardillo G, Sposato B, Terzano C, Bronzetti E. Neurotrophin system expression in human pulmonary carcinoid tumors. Growth Factors. 2005;23:303–312. doi: 10.1080/08977190500233813. [DOI] [PubMed] [Google Scholar]
- Pruunsild P, Kazantseva A, Aid T, Palm K, Timmusk T. Dissecting the human BDNF locus: bidirectional transcription, complex splicing, and multiple promoters. Genomics. 2007;90:397–406. doi: 10.1016/j.ygeno.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res. 2007;85:525–535. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith OL, Pleasance ED, Fulton DL, Oveisi M, Ester M, Siddiqui AS, Jones SJ. Assessment and integration of publicly available SAGE, cDNA microarray, and oligonucleotide microarray expression data for global coexpression analyses. Genomics. 2005;86:476–488. doi: 10.1016/j.ygeno.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Yeung KY, Medvedovic M, Bumgarner RE. From co-expression to co-regulation: how many microarray experiments do we need? Genome Biol. 2004;5:R48. doi: 10.1186/gb-2004-5-7-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Hsu AK, Sajdak J, Qin J, Pavlidis P. Coexpression analysis of human genes across many microarray data sets. Genome Res. 2004;14:1085–1094. doi: 10.1101/gr.1910904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennmalm K, Wahlestedt C, Larsson O. The expression signature of in vitro senescence resembles mouse but not human aging. Genome Biol. 2005;6:R109. doi: 10.1186/gb-2005-6-13-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner RG, Young RA. Insights into host responses against pathogens from transcriptional profiling. Nat Rev Microbiol. 2005;3:281–294. doi: 10.1038/nrmicro1126. [DOI] [PubMed] [Google Scholar]
- Wolfe CJ, Kohane IS, Butte AJ. Systematic survey reveals general applicability of "guilt-by-association" within gene coexpression networks. BMC Bioinformatics. 2005;6:227. doi: 10.1186/1471-2105-6-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causton HC, Quackenbush J, Brazma A. Microarray Gene Expression Data Analysis: A Beginner's Guide. Blackwell Publishing, Chichester, West Sussex; 2003. [Google Scholar]
- Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- Choi JK, Yu U, Yoo OJ, Kim S. Differential coexpression analysis using microarray data and its application to human cancer. Bioinformatics. 2005;21:4348–4355. doi: 10.1093/bioinformatics/bti722. [DOI] [PubMed] [Google Scholar]
- Elo LL, Jarvenpaa H, Oresic M, Lahesmaa R, Aittokallio T. Systematic construction of gene coexpression networks with applications to human T helper cell differentiation process. Bioinformatics. 2007;23:2096–2103. doi: 10.1093/bioinformatics/btm309. [DOI] [PubMed] [Google Scholar]
- Kruskal WH, Wallis WA. Use of ranks in one-criterion variance analysis. Journal of the American Statistical Association. 1953;47:583–621. doi: 10.2307/2280779. [DOI] [Google Scholar]
- Williams EJ, Bowles DJ. Coexpression of neighboring genes in the genome of Arabidopsis thaliana. Genome Res. 2004;14:1060–1067. doi: 10.1101/gr.2131104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mecham BH, Klus GT, Strovel J, Augustus M, Byrne D, Bozso P, Wetmore DZ, Mariani TJ, Kohane IS, Szallasi Z. Sequence-matched probes produce increased cross-platform consistency and more reproducible biological results in microarray-based gene expression measurements. Nucleic Acids Res. 2004;32:e74. doi: 10.1093/nar/gnh071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler--a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007:W193–200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- Li A, Horvath S. Network neighborhood analysis with the multi-node topological overlap measure. Bioinformatics. 2007;23:222–231. doi: 10.1093/bioinformatics/btl581. [DOI] [PubMed] [Google Scholar]
- Oti M, van Reeuwijk J, Huynen MA, Brunner HG. Conserved co-expression for candidate disease gene prioritization. BMC Bioinformatics. 2008;9:208. doi: 10.1186/1471-2105-9-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann R, Valencia A. A gene network for navigating the literature. Nat Genet. 2004;36:664. doi: 10.1038/ng0704-664. [DOI] [PubMed] [Google Scholar]
- Hausman GJ, Poulos SP, Richardson RL, Barb CR, Andacht T, Kirk HC, Mynatt RL. Secreted proteins and genes in fetal and neonatal pig adipose tissue and stromal-vascular cells. J Anim Sci. 2006;84:1666–1681. doi: 10.2527/jas.2005-539. [DOI] [PubMed] [Google Scholar]
- Schmidt-Kastner R, van Os J, H WMS, Schmitz C. Gene regulation by hypoxia and the neurodevelopmental origin of schizophrenia. Schizophr Res. 2006;84:253–271. doi: 10.1016/j.schres.2006.02.022. [DOI] [PubMed] [Google Scholar]
- Kwon J, Wang YL, Setsuie R, Sekiguchi S, Sato Y, Sakurai M, Noda M, Aoki S, Yoshikawa Y, Wada K. Two closely related ubiquitin C-terminal hydrolase isozymes function as reciprocal modulators of germ cell apoptosis in cryptorchid testis. Am J Pathol. 2004;165:1367–1374. doi: 10.1016/S0002-9440(10)63394-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto I, Rosenthal JJ, Blagburn JM, Blanco RE. Fibroblast growth factor 2 applied to the optic nerve after axotomy up-regulates BDNF and TrkB in ganglion cells by activating the ERK and PKA signaling pathways. J Neurochem. 2006;96:82–96. doi: 10.1111/j.1471-4159.2005.03510.x. [DOI] [PubMed] [Google Scholar]
- Kohno R, Sawada H, Kawamoto Y, Uemura K, Shibasaki H, Shimohama S. BDNF is induced by wild-type alpha-synuclein but not by the two mutants, A30P or A53T, in glioma cell line. Biochem Biophys Res Commun. 2004;318:113–118. doi: 10.1016/j.bbrc.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Marcinkiewicz M, Savaria D, Marcinkiewicz J. The pro-protein convertase PC1 is induced in the transected sciatic nerve and is present in cultured Schwann cells: comparison with PC5, furin and PC7, implication in pro-BDNF processing. Brain Res Mol Brain Res. 1998;59:229–246. doi: 10.1016/S0169-328X(98)00141-7. [DOI] [PubMed] [Google Scholar]
- Yang T, Massa SM, Longo FM. LAR protein tyrosine phosphatase receptor associates with TrkB and modulates neurotrophic signaling pathways. J Neurobiol. 2006;66:1420–1436. doi: 10.1002/neu.20291. [DOI] [PubMed] [Google Scholar]
- Pastor R, Bernal J, Rodriguez-Pena A. Unliganded c-erbA/thyroid hormone receptor induces trkB expression in neuroblastoma cells. Oncogene. 1994;9:1081–1089. [PubMed] [Google Scholar]
- Pollak DD, Herkner K, Hoeger H, Lubec G. Behavioral testing upregulates pCaMKII, BDNF, PSD-95 and egr-1 in hippocampus of FVB/N mice. Behav Brain Res. 2005;163:128–135. doi: 10.1016/j.bbr.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Djalali S, Holtje M, Grosse G, Rothe T, Stroh T, Grosse J, Deng DR, Hellweg R, Grantyn R, Hortnagl H, et al. Effects of brain-derived neurotrophic factor (BDNF) on glial cells and serotonergic neurones during development. J Neurochem. 2005;92:616–627. doi: 10.1111/j.1471-4159.2004.02911.x. [DOI] [PubMed] [Google Scholar]
- Kitagawa A, Nakayama T, Takenaga M, Matsumoto K, Tokura Y, Ohta Y, Ichinohe M, Yamaguchi Y, Suzuki N, Okano H, et al. Lecithinized brain-derived neurotrophic factor promotes the differentiation of embryonic stem cells in vitro and in vivo. Biochem Biophys Res Commun. 2005;328:1051–1057. doi: 10.1016/j.bbrc.2005.01.063. [DOI] [PubMed] [Google Scholar]
- Ring RH, Alder J, Fennell M, Kouranova E, Black IB, Thakker-Varia S. Transcriptional profiling of brain-derived-neurotrophic factor-induced neuronal plasticity: a novel role for nociceptin in hippocampal neurite outgrowth. J Neurobiol. 2006;66:361–377. doi: 10.1002/neu.20223. [DOI] [PubMed] [Google Scholar]
- Sun CY, Hu Y, Wang HF, He WJ, Wang YD, Wu T. Brain-derived neurotrophic factor inducing angiogenesis through modulation of matrix-degrading proteases. Chin Med J (Engl) 2006;119:589–595. [PubMed] [Google Scholar]
- Fujita Y, Katagi J, Tabuchi A, Tsuchiya T, Tsuda M. Coactivation of secretogranin-II and BDNF genes mediated by calcium signals in mouse cerebellar granule cells. Brain Res Mol Brain Res. 1999;63:316–324. doi: 10.1016/S0169-328X(98)00299-X. [DOI] [PubMed] [Google Scholar]
- von Bohlen und Halbach O, Minichiello L, Unsicker K. Haploinsufficiency for trkB and trkC receptors induces cell loss and accumulation of alpha-synuclein in the substantia nigra. Faseb J. 2005;19:1740–1742. doi: 10.1096/fj.05-3845fje. [DOI] [PubMed] [Google Scholar]
- Carter CJ. Multiple genes and factors associated with bipolar disorder converge on growth factor and stress activated kinase pathways controlling translation initiation: implications for oligodendrocyte viability. Neurochem Int. 2007;50:461–490. doi: 10.1016/j.neuint.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Glorioso C, Sabatini M, Unger T, Hashimoto T, Monteggia LM, Lewis DA, Mirnics K. Specificity and timing of neocortical transcriptome changes in response to BDNF gene ablation during embryogenesis or adulthood. Mol Psychiatry. 2006;11:633–648. doi: 10.1038/sj.mp.4001835. [DOI] [PubMed] [Google Scholar]
- Laslop A, Weiss C, Savaria D, Eiter C, Tooze SA, Seidah NG, Winkler H. Proteolytic processing of chromogranin B and secretogranin II by prohormone convertases. J Neurochem. 1998;70:374–383. doi: 10.1046/j.1471-4159.1998.70010374.x. [DOI] [PubMed] [Google Scholar]
- Hovatta I, Kimppa K, Laine MM, Lehmussola A, Pesanen T, Saarela J, Saarikko I, Saharinen J, Tiikkainen P, Toivanen T, et al. DNA microarray data analysis. Helsinki: CSC; 2005. [Google Scholar]
- Bottone FG, Jr, Moon Y, Alston-Mills B, Eling TE. Transcriptional regulation of activating transcription factor 3 involves the early growth response-1 gene. J Pharmacol Exp Ther. 2005;315:668–677. doi: 10.1124/jpet.105.089607. [DOI] [PubMed] [Google Scholar]
- Chen X, Wu JM, Hornischer K, Kel A, Wingender E. TiProD: the Tissue-specific Promoter Database. Nucleic Acids Res. 2006:D104–107. doi: 10.1093/nar/gkj113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotea V, Ovcharenko I. DiRE: identifying distant regulatory elements of co-expressed genes. Nucleic Acids Res. 2008:W133–139. doi: 10.1093/nar/gkn300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanam S, Moreno CS. CONFAC: automated application of comparative genomic promoter analysis to DNA microarray datasets. Nucleic Acids Res. 2004:W475–484. doi: 10.1093/nar/gkh353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004:W280–286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmusk T, Palm K, Lendahl U, Metsis M. Brain-derived neurotrophic factor expression in vivo is under the control of neuron-restrictive silencer element. J Biol Chem. 1999;274:1078–1084. [PubMed] [Google Scholar]
- Shieh PB, Hu SC, Bobb K, Timmusk T, Ghosh A. Identification of a signaling pathway involved in calcium regulation of BDNF expression. Neuron. 1998;20:727–740. doi: 10.1016/S0896-6273(00)81011-9. [DOI] [PubMed] [Google Scholar]
- Tao X, Finkbeiner S, Arnold DB, Shaywitz AJ, Greenberg ME. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron. 1998;20:709–726. doi: 10.1016/S0896-6273(00)81010-7. [DOI] [PubMed] [Google Scholar]
- Chen WG, West AE, Tao X, Corfas G, Szentirmay MN, Sawadogo M, Vinson C, Greenberg ME. Upstream stimulatory factors are mediators of Ca2+-responsive transcription in neurons. J Neurosci. 2003;23:2572–2581. doi: 10.1523/JNEUROSCI.23-07-02572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsky RH, Xu K, Zhu D, Kelly C, Terhakopian A, Novelli A, Marini AM. Nuclear factor kappaB is a critical determinant in N-methyl-D-aspartate receptor-mediated neuroprotection. J Neurochem. 2001;78:254–264. doi: 10.1046/j.1471-4159.2001.00386.x. [DOI] [PubMed] [Google Scholar]
- Greer PL, Greenberg ME. From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron. 2008;59:846–860. doi: 10.1016/j.neuron.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Himeda CL, Ranish JA, Hauschka SD. Quantitative proteomic identification of MAZ as a transcriptional regulator of muscle-specific genes in skeletal and cardiac myocytes. Mol Cell Biol. 2008;28:6521–6535. doi: 10.1128/MCB.00306-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Ugai H, Nakata-Tsutsui H, Kishikawa S, Suzuki E, Murata T, Yokoyama KK. Transcriptional regulation by zinc-finger proteins Sp1 and MAZ involves interactions with the same cis-elements. Int J Mol Med. 2003;11:547–553. [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- Wang X, Southard RC, Allred CD, Talbert DR, Wilson ME, Kilgore MW. MAZ drives tumor-specific expression of PPAR gamma 1 in breast cancer cells. Breast Cancer Res Treat. 2008;111:103–111. doi: 10.1007/s10549-007-9765-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiba T, Izumoto S, Kagawa N, Suzuki T, Hashimoto N, Maruno M, Yoshimine T. Expression of WT1 protein and correlation with cellular proliferation in glial tumors. Neurol Med Chir (Tokyo) 2007;47:165–170. doi: 10.2176/nmc.47.165. discussion 170. [DOI] [PubMed] [Google Scholar]
- Yang L, Han Y, Suarez Saiz F, Minden MD. A tumor suppressor and oncogene: the WT1 story. Leukemia. 2007;21:868–876. doi: 10.1038/sj.leu.2404624. [DOI] [PubMed] [Google Scholar]
- Lee SB, Kolquist KA, Nichols K, Englert C, Maheswaran S, Ladanyi M, Gerald WL, Haber DA. The EWS-WT1 translocation product induces PDGFA in desmoplastic small round-cell tumour. Nat Genet. 1997;17:309–313. doi: 10.1038/ng1197-309. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Pedigo N, Shenoy S, Khalili K, Kaetzel DM. Puralpha activates PDGF-A gene transcription via interactions with a G-rich, single-stranded region of the promoter. Gene. 2005;348:25–32. doi: 10.1016/j.gene.2004.12.050. [DOI] [PubMed] [Google Scholar]
- Werner H, Re GG, Drummond IA, Sukhatme VP, Rauscher FJ, 3rd, Sens DA, Garvin AJ, LeRoith D, Roberts CT., Jr Increased expression of the insulin-like growth factor I receptor gene, IGF1R, in Wilms tumor is correlated with modulation of IGF1R promoter activity by the WT1 Wilms tumor gene product. Proc Natl Acad Sci USA. 1993;90:5828–5832. doi: 10.1073/pnas.90.12.5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma PM, Yang X, Bowman M, Roberts V, Sukumar S. Molecular cloning of rat Wilms' tumor complementary DNA and a study of messenger RNA expression in the urogenital system and the brain. Cancer Res. 1992;52:6407–6412. [PubMed] [Google Scholar]
- Eggert A, Grotzer MA, Ikegaki N, Zhao H, Cnaan A, Brodeur GM, Evans AE. Expression of the neurotrophin receptor TrkB is associated with unfavorable outcome in Wilms' tumor. J Clin Oncol. 2001;19:689–696. doi: 10.1200/JCO.2001.19.3.689. [DOI] [PubMed] [Google Scholar]
- Wagner N, Wagner KD, Theres H, Englert C, Schedl A, Scholz H. Coronary vessel development requires activation of the TrkB neurotrophin receptor by the Wilms' tumor transcription factor Wt1. Genes Dev. 2005;19:2631–2642. doi: 10.1101/gad.346405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell MA, Xie C, Xiong S, Markesbery WR. Wilms' tumor suppressor (WT1) is a mediator of neuronal degeneration associated with the pathogenesis of Alzheimer's disease. Brain Res. 2003;983:84–96. doi: 10.1016/S0006-8993(03)03032-4. [DOI] [PubMed] [Google Scholar]
- Beckmann AM, Wilce PA. Egr transcription factors in the nervous system. Neurochem Int. 1997;31:477–510. doi: 10.1016/S0197-0186(96)00136-2. discussion 517-476. [DOI] [PubMed] [Google Scholar]
- Swirnoff AH, Milbrandt J. DNA-binding specificity of NGFI-A and related zinc finger transcription factors. Mol Cell Biol. 1995;15:2275–2287. doi: 10.1128/mcb.15.4.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MW, Errington ML, French PJ, Fine A, Bliss TV, Garel S, Charnay P, Bozon B, Laroche S, Davis S. A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nat Neurosci. 2001;4:289–296. doi: 10.1038/85138. [DOI] [PubMed] [Google Scholar]
- Li L, Yun SH, Keblesh J, Trommer BL, Xiong H, Radulovic J, Tourtellotte WG. Egr3, a synaptic activity regulated transcription factor that is essential for learning and memory. Mol Cell Neurosci. 2007;35:76–88. doi: 10.1016/j.mcn.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan R, Svaren J, Le N, Araki T, Watson M, Milbrandt J. EGR2 mutations in inherited neuropathies dominant-negatively inhibit myelin gene expression. Neuron. 2001;30:355–368. doi: 10.1016/S0896-6273(01)00282-3. [DOI] [PubMed] [Google Scholar]
- Ghislain J, Charnay P. Control of myelination in Schwann cells: a Krox20 cis-regulatory element integrates Oct6, Brn2 and Sox10 activities. EMBO Rep. 2006;7:52–58. doi: 10.1038/sj.embor.7400573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin AC, Westerlund M. Parkinson's disease: a genetic perspective. Febs J. 2008;275:1377–1383. doi: 10.1111/j.1742-4658.2008.06301.x. [DOI] [PubMed] [Google Scholar]
- Howells DW, Porritt MJ, Wong JY, Batchelor PE, Kalnins R, Hughes AJ, Donnan GA. Reduced BDNF mRNA expression in the Parkinson's disease substantia nigra. Exp Neurol. 2000;166:127–135. doi: 10.1006/exnr.2000.7483. [DOI] [PubMed] [Google Scholar]
- Skerjanc IS, Wilton S. Myocyte enhancer factor 2C upregulates MASH-1 expression and induces neurogenesis in P19 cells. FEBS Lett. 2000;472:53–56. doi: 10.1016/S0014-5793(00)01438-1. [DOI] [PubMed] [Google Scholar]
- Li H, Radford JC, Ragusa MJ, Shea KL, McKercher SR, Zaremba JD, Soussou W, Nie Z, Kang YJ, Nakanishi N, et al. Transcription factor MEF2C influences neural stem/progenitor cell differentiation and maturation in vivo. Proc Natl Acad Sci USA. 2008;105:9397–9402. doi: 10.1073/pnas.0802876105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PD, Mount MP, Shree R, Callaghan S, Slack RS, Anisman H, Vincent I, Wang X, Mao Z, Park DS. Calpain-regulated p35/cdk5 plays a central role in dopaminergic neuron death through modulation of the transcription factor myocyte enhancer factor 2. J Neurosci. 2006;26:440–447. doi: 10.1523/JNEUROSCI.2875-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han A, Pan F, Stroud JC, Youn HD, Liu JO, Chen L. Sequence-specific recruitment of transcriptional co-repressor Cabin1 by myocyte enhancer factor-2. Nature. 2003;422:730–734. doi: 10.1038/nature01555. [DOI] [PubMed] [Google Scholar]
- Kazanjian A, Gross EA, Grimes HL. The growth factor independence-1 transcription factor: new functions and new insights. Crit Rev Oncol Hematol. 2006;59:85–97. doi: 10.1016/j.critrevonc.2006.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Zarebski A, Montoya-Durango D, Grimes HL, Horwitz M. Gfi1 coordinates epigenetic repression of p21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol Cell Biol. 2005;25:10338–10351. doi: 10.1128/MCB.25.23.10338-10351.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acar M, Jafar-Nejad H, Giagtzoglou N, Yallampalli S, David G, He Y, Delidakis C, Bellen HJ. Senseless physically interacts with proneural proteins and functions as a transcriptional co-activator. Development. 2006;133:1979–1989. doi: 10.1242/dev.02372. [DOI] [PubMed] [Google Scholar]
- Castro DS, Skowronska-Krawczyk D, Armant O, Donaldson IJ, Parras C, Hunt C, Critchley JA, Nguyen L, Gossler A, Gottgens B, et al. Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev Cell. 2006;11:831–844. doi: 10.1016/j.devcel.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Dresser DW, Guerrier D. Candidate Sertoli cell specific promoter element for a TGFbeta family member (Amh) and a 3' UTR enhancer/repressor for the same gene. Gene. 2005;363:159–165. doi: 10.1016/j.gene.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Spinelli G, Birnstiel ML. The modulator is a constitutive enhancer of a developmentally regulated sea urchin histone H2A gene. Bioessays. 2002;24:850–857. doi: 10.1002/bies.10143. [DOI] [PubMed] [Google Scholar]
- Tabuchi A, Sakaya H, Kisukeda T, Fushiki H, Tsuda M. Involvement of an upstream stimulatory factor as well as cAMP-responsive element-binding protein in the activation of brain-derived neurotrophic factor gene promoter I. J Biol Chem. 2002;277:35920–35931. doi: 10.1074/jbc.M204784200. [DOI] [PubMed] [Google Scholar]
- Tao X, West AE, Chen WG, Corfas G, Greenberg ME. A calcium-responsive transcription factor, CaRF, that regulates neuronal activity-dependent expression of BDNF. Neuron. 2002;33:383–395. doi: 10.1016/S0896-6273(01)00561-X. [DOI] [PubMed] [Google Scholar]
- Metsis M, Timmusk T, Arenas E, Persson H. Differential usage of multiple brain-derived neurotrophic factor promoters in the rat brain following neuronal activation. Proc Natl Acad Sci USA. 1993;90:8802–8806. doi: 10.1073/pnas.90.19.8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarikettu J, Sveshnikova N, Grundstrom T. Calcium/calmodulin inhibition of transcriptional activity of E-proteins by prevention of their binding to DNA. J Biol Chem. 2004;279:41004–41011. doi: 10.1074/jbc.M408120200. [DOI] [PubMed] [Google Scholar]
- Liu QR, Walther D, Drgon T, Polesskaya O, Lesnick TG, Strain KJ, de Andrade M, Bower JH, Maraganore DM, Uhl GR. Human brain derived neurotrophic factor (BDNF) genes, splicing patterns, and assessments of associations with substance abuse and Parkinson's Disease. Am J Med Genet B Neuropsychiatr Genet. 2005;134B:93–103. doi: 10.1002/ajmg.b.30109. [DOI] [PubMed] [Google Scholar]
- Liu QR, Lu L, Zhu XG, Gong JP, Shaham Y, Uhl GR. Rodent BDNF genes, novel promoters, novel splice variants, and regulation by cocaine. Brain Res. 2006;1067:1–12. doi: 10.1016/j.brainres.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affymetrix . Statistical Algorithms Reference Guide. Affymetrix Santa Clara, CA; 2002. [Google Scholar]
- Troyanskaya OG, Botstein D, Altman RB. Missing value estimation. In: Berrar DP, Dubitzky W, Granzow M, editor. A practical approach to microarray data analysis. Dortrecht: Kluwer Academic Publishers; 2003. pp. 65–76. [Google Scholar]
- Wilcoxon F. Individual comparisons by ranking methods. Biometrics. 1945;1:80–83. doi: 10.2307/3001968. [DOI] [Google Scholar]
- Mann HB, Whitney DR. On a test of whether one of two random variables is stochastically larger than the other. Annals of Mathematical Statistics. 1947;18:50–60. doi: 10.1214/aoms/1177730491. [DOI] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B. 1995;57:289–300. [Google Scholar]
- Wood M. Statistical inference using bootstrap confidence intervals. Significance. 2004;1:180–184. doi: 10.1111/j.1740-9713.2004.00067.x. [DOI] [Google Scholar]
- Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V, Meinhardt T, Pruss M, Reuter I, Schacherer F. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Loots GG, Nobrega MA, Ovcharenko I. Predicting tissue-specific enhancers in the human genome. Genome Res. 2007;17:201–211. doi: 10.1101/gr.5972507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
BDNF probe sets. Affymetrix microarray probe sets for BDNF gene. BDNF probe set target sequences are given for each platform type that was used in the co-expression conservation analysis.
Microarray datasets. Datasets that passed non-specific filtering and were used in the analysis (38 human microarray datasets, 24 mouse datasets and 18 rat datasets). Each dataset was divided into subsets (disease state, age, agent, etc) according to experimental annotations. When possible, subsets were subdivided further (marked by *). Experiments were classified based on their description and the tissue origin. GDS refers to GEO Datasets.
Subsets. Dataset: GDS1018/1368678_at/Bdnf/Rattus norvegicus. Expression profiling of brain hippocampal CA1 and CA3 neurons of Sprague Dawleys subjected to brief preconditioning seizures. According to the dataset annotation, dataset could be divided into three subsets by cell type (A) or into two subsets by protocol (B). In addition, subsets could be subdivided further into cell type.protocol sub-subsets: CA1 pyramidal neuron.control, CA1 pyramidal neuron.preconditioning seizure, CA3 pyramidal neuron.control, etc. After filtering, subset containing less than eight samples (CA3 pyramidal neuron.preconditioning seizure) was excluded from the analysis.
Differential expression of the BDNF gene in human datasets. Differential expression of BDNF was measured across subsets in each dataset using Kruskal-Wallis test. Only statistically significant results are presented.
Differential expression of the BDNF gene in mouse datasets. Differential expression of BDNF was measured across subsets in each dataset using Kruskal-Wallis test. Only statistically significant results are presented.
Differential expression of the BDNF gene in rat datasets. Differential expression of BDNF was measured across subsets in each dataset using Kruskal-Wallis test. Only statistically significant results are presented.
Conserved BDNF-correlated genes. Genes, whose correlation with BDNF was confirmed in at least 3 subsets (3+ genes) and was conserved between i) human, mouse and rat; ii) human and rat; iii) human and mouse; iv) mouse and rat.
g:Profiler analysis. Functional profiling of the list of BDNF-correlated genes conserved between human, mouse and rat using g:G:Profiler. For details see also http://biit.cs.ut.ee/gprofiler/.
iHOP references. Interactions between conserved correlated genes in human and mouse (URL links to the literature cited in iHOP).
DiRE motif discovery results for BDNF and 84 conserved correlated genes. Over-represented TFBSs are given together with the Importance Score (cut-off 0.1 recommended by DiRE). Numbers 1 and 2 (in All 1 and 2) refer to the different ways that DiRE tool analyzes evolutionary conserved regions (ECR): 1) top 3 ECRs + promoter ECRs; 2) UTR ECRs + promoter ECRs.
DiRE motif discovery results for conserved BDNF-correlated genes clustered by tissue-specific expression. TFBSs over-represented in each tissue cluster are given together with the Importance Score (cut-off 0.1 recommended by DiRE). CNS - central nervous system, PNS - peripheral nervous system.
The results of Mann-Whitney tests (CONFAC). Overrepresented TFs in the conserved BDNF-correlated gene list. Bar graphs show the average conserved TFBS frequencies for the sample gene set (conserved BDNF-correlated genes, blue bars) and control gene set (random 250 genes, red bars). A minimum threshold for the differences in the average TFBS frequencies between the two groups was set by p-value cutoff 0.05 and a mean-difference cutoff 0.5.
Highly conserved TFBSs in the BDNF gene (according to DiRE and CONFAC). Represented TFBSs have Matrix Similarity score >0.85 and Core Similarity score >0.99. TFBS sequences are highlighted in blue; "+" or "-" mark the DNA strand orientation; BDNF exons and 3'UTR are highlighted in green; the regulatory region in BDNF downstream from polyadenylation sites identified by DiRE is highlighted yellow.