SUMMARY
We propose a similarity-based regression method to detect associations between traits and multimarker genotypes. The model regresses similarity in traits for pairs of ”unrelated” individuals on their haplotype similarities, and detects the significance by a score test for which the limiting distribution is derived. The proposed method allows for covariates, uses phase-independent similarity measures to bypass the needs to impute phase information, and is applicable to traits of general types (e.g., quantitative and qualitative traits). We also show that the gene-trait similarity regression is closely connected with random effects haplotype analysis, although commonly they are considered as separate modeling tools. This connection unites the classic haplotype sharing methods with the variance component approaches, which enables direct derivation of analytical properties of the sharing statistics even when the similarity regression model becomes analytically challenging.
Keywords: Haplotype-based association test, Haplotype sharing, Haplotype similarity
1. Introduction
A haplotype is an ordered allele sequence of multiple markers on the same chromosome. Compared to a single marker, a haplotype may better capture genetic variations because it incorporates information from multiple markers simultaneously and conserves the joint linkage disequilibrium (LD) structure among them (International HapMap Consortium 2003; de Bakker et al. 2005). However, modeling haplotype variations tends to require a large number of parameters or run into sparsity problems, as the majority of the polymorphism is concentrated on a relatively small number of haplotypes while the rest is sparsely spread over a fair number of categories. The existence of the many categories and sparse categories often leads to either power loss or unstable statistical inference, and consequently, limits the efficiency of haplotype-based association analysis in practice.
Modeling haplotype similarity instead of haplotype variants have been frequently considered as a tool to bypass the problems caused by many haplotypes and rare haplotypes. To detect the potential regions that may contain functional variants, approaches based on haplotype similarity look for unusual sharing of chromosomal segments within homogeneous trait groups (Houwen et al. 1994; McPeek and Strahs 1999). The underlying rationale is that affected individuals tend to share similar genetic materials in close vicinity to the disease mutation; even with complex diseases that exhibit greater etiologic heterogeneity, one can still expect to find disproportionately large clusters of cases sharing common haplotypes in the region flanking a disease mutation (Feder et al. 1996; Puffenberger et al. 1994).
The excess similarity among cases can be identified by either comparing to the similarity level expected from genealogical process (Durham and Feingold 1997; Sevice et al. 1999), or by contrasting with control haplotypes (Van der Meulen and te Meerman 1997; Bourugain et al. 2000, 2001, 2002; Tzeng et al. 2003a,b; Yu et al. 2004). The former approaches were used in the successful positional cloning of Mendelian disorders since the 1980’s. However, extension of these methods to complex diseases makes comparisons to the genealogical expectations less practical. The latter approaches bypass the need to model the evolutionary process by which the observed haplotypes were produced. However, these methods tend to be feasible only with binary traits and do not incorporate covariate information. Moreover, these methods limit similarity calculations to the concordant samples only (i.e., case-case similarity and control-control similarity) and do not use information obtained from case-control similarity. Recently, Sha et al. (2007) addressed the latter concern by contrasting similarity of concordant pairs (case-case and control-control) with similarity of discordant pairs (case-control), and showed that such contrast can improve power.
Current developments have shifted the focus from two-sample tests to regression models that correlate trait similarity with genetic similarity (Beckmann et al. 2005; Wessel and Schork 2007). This new direction incorporates similarity comparison between discordant pairs, and establishes a model-based framework that is ready for accommodating covariates and various trait types. The idea of gene-trait similarity was pioneered by Qian and Thomas (2001) with pedigree data. Qian and Thomas (2001) quantified the similarities of phenotypes and of haplotypes within each family, and correlated these family statistics using the Mantel statistics. Beckmann et al. (2005) extended the framework for population-based samples. Although not accounting for covariate information yet, their methods can work on qualitative and quantitative traits. Wessel and Schork (2007) took one step further and developed a general regression framework for dissimilarity analysis between phenotypes and genotypes. Their model treats genetic similarity as the response variable, and treats trait similarity and environmental covariates as explanatory variables. However, because covariates tend to affect the disease risk rather than the genetic variants, it would be more desirable to switch the roles of genetic similarity and trait similarity. Finally, one major challenge of the gene-trait similarity approaches is the complex correlation structure introduced by the pairwise samples, as the observation unit in the regression is now pairs of individuals instead of single individuals. Consequently, the distributions of the test statistics are hard to derive analytically, and permutation is needed to find p-values.
Our proposed similarity method also follows this direction. For trait similarity, we measure the trait covariance of all distinct individual pairs conditional on covariates. For haplotype similarity, we measure the sharing level of haplotype pairs of two individuals. We then propose to regress trait similarity on haplotype similarity, and study gene-trait association by testing for zero regression coefficient of haplotype similarity. In section 2, we formulate the gene-trait similarity regression model, construct a score test for association, and derive its limiting distribution to facilitate hypothesis testing in large scale. To tackle the issue of missing phases, we use the similarity metrics that can measure haplotype similarity directly from genotypes. In section 3, we show that the similarity regression is closely connected to an alternative haplotype analysis approach, the variance component method. In Section 4 we investigate the performance of the proposed method using simulations. In Section 5, we apply it to the case-control data obtained from the amyotrophic lateral sclerosis study of Schymick et al. (2007). Finally we conclude with discussions and remarks in Section 6.
2. Methods
2.1 The Gene-Trait Similarity Model
Let Yi denote the trait value, Xi denote the K × 1 covariate vector including the intercept term, and Hi denote the L × 1 haplotype vector of the ith individual in a sample of n subjects. The h th element of Hi, denoted by Hi,h, records the number of copies of haplotype h that subject i carries.
For genetic similarity, define Sij to be the haplotype similarity between subjects i and j (i ≠ j). From the definition of H, we have
| (1) |
where s (h, k) is a certain similarity metric that is used to measure the similarity level between haplotypes h and k. Let under the condition of no haplotype effects, in which γ is the covariate effects including the intercept. We assume that the conditional mean can be modeled by some specific function of such as that specified by the generalized linear model. Then for trait similarity, which is denoted by Zij, we define
| (2) |
which is the weighted cross product of the trait residuals with some weight ωi. The cross product of residuals has been used to describe the level of trait similarity between a pair of subjects in linkage studies (Elston et al. 2000; Thomas et al. 1999). Here the residual is defined with respect to the covariate-adjusted mean for each subject. The weight ωi may be used to account for the fact that Yi is not necessarily homogeneous. In principle, ωi can be any pre-specified positive values such as 1, or some function of the trait variance. As we will illustrate later, optimal ωi can be identified if a model is imposed on trait values and haplotype effects.
We propose a similarity regression model of the following form to study and test the gene-trait association:
| (3) |
where eij’s are some mean-zero error terms. By the definition of Zij (which has been adjusted for the effects of baseline and other covariates), the proposed regression has a zero intercept. Intuitively, in a chromosomal region that contains disease genes, one would expect that b > 0 as higher genetic similarity would lead to higher trait similarity. In ”null” regions, b ≈ 0 as genetic similarity and trait similarity would have little correlation.
Similar to Elston et al. (2000), one may estimate b in (3) using the least-square method and conduct inference accordingly. However, this approach is complicated by the need to invert a large variance-covariance matrix of eij’s. To bypass this issue, in this work we focus on the score test of b = 0, where the inverse of the covariance matrix takes place only under the null hypothesis and the covariance matrix becomes diagonal. In general, the estimation of b can be accomplished by using the variance-component estimation in a generalized linear mixed model if such model is posited on trait values.
2.2 The Score Test of H0 : b = 0
To construct the score test for testing H0 : b = 0, we further assume that under the condition of no haplotype effects, where mi is a known prior weight, such as the binomial denominator, ϕ is the dispersion parameter, and υ (μi) is the variance function. With the two moment restrictions on and in our model, we use the following estimating equations for (b, γ, ϕ) to construct the score test for testing H0 : b = 0:
Assume that Yi’s are independent under the null hypothesis of no haplotype effect (i.e., b = 0). An intuitive explanation of this independence assumption is that, under H0, the trait dependence should reflect the baseline relationship between the pair of the subjects. Here such relationship is zero because the study subjects are unrelated. This assumption will be more rigorously justified after we postulate a model on haplotype effects (Section 3).
Following the above null independence assumption of Yi’s and by the definition of variance, we have that under H0, cov(Zij, Yl | X, H) = 0 ∀i ≠ j and ∀l, and Thus Ub, the score statistic for b, is
where γ̂ is the maximum likelihood estimate of γ under H0, ϕ̂ is the restricted maximum likelihood type of estimate of ϕ under H0 and To derive its distribution, we rewrite Ub in a quadratic form as
| (4) |
In the above equation, S0 is a matrix with diagonal elements equal to 0 and off-diagonal elements equal to Sij, and Ω = diag {ωi}. We show in Web Appendix A that (4) has approximately the same distribution as the weighted chi-square random variable where ’s are independent chi-squared variables, and λ1 ≥ λ2 ≥ … ≥ λn are the ordered eigenvalues of the matrix C defined as
where Q is the projection matrix V−1 – V−1 DX (XT DV−1 DX)−1 XT DV−1 under H0 with matrix D = diag {∂δ (ηi)/∂ηi} and With this result, we approximate the distribution of Ub by the three-moment approximation method of Imhof (1961) as did in Allen and Satten (2007). The level-α significance threshold is estimated by
where and χα is the αth quantile of (i.e., chi-squared distribution with h′ degrees of freedom). Alternatively, one can report the p-value of the observed statistic Ub by where
2.3 Analysis with Unphased Genotype Data
To bypass the issues involved in phase missing and phase imputation, we use measures that do not require phase information to quantify haplotype similarity. One possible choice of such s (h, k) is the ”counting measure”, scount (h, k), of Tzeng et al. (2003a), which calculates the proportion of alleles in common between haplotype h and haplotype h. Define to be a 2 × 1 vector whose elements record the number of major alleles and the number of minor alleles for subject i at marker m, m = 1, 2, …, M. It can be shown that
That is, haplotype similarity score between subjects i and j is equal to the average allelic sharing across markers. With this transformation, it is possible to bypass the need of imputing haplotype phase, because all we need to measure haplotype similarity is the allele counts at each SNP.
Concerns may arise in terms of choosing between phase-dependent and phase-independent metrics for performing haplotype-similarity analysis. In theory, metrics using the phase information should be more powerful as they may capture the identical-by-descent (IBD) sharing more precisely. However, these metrics are not robust to practical complications such as genotyping errors and recent marker mutations that often limit their performance in reality. Indeed, previous and recent works have found that phase-dependent and phase-independent metrics have very similar performance (Tzeng et al. 2003a; Sha et al. 2007).
3. Connection with the Variance-Component Score Test
Similar to the linkage analysis where the regression-based approaches have been shown equivalent to the variance-component methods (Sham and Purcell 2001), the proposed gene-trait similarity regression is also connected with the variance-component approaches of haplotype analysis. The connection can be obtained by the fact that E(Zij | X, H) ≈ ωiωj × cov(Yi, Yj | X, H). To see this, consider a generalized linear mixed model (GLMM):
| (5) |
where μi = E(Yi | β, Xi, Hi), g (·) is the link function, γ is the fixed effect of environmental covariates, and β is the random effect of the haplotypes. Conditional on Xi, Hi and β, Yi’s are assumed to be independent with conditional mean μi and conditional variance Under this model, and and under H0, and Matrix Rβ = {rhk} describes the correlation between effects of haplotypes h and k. One common correlation structure imposed on β is to allow evolutionarily close haplotypes to be more correlated, such as to set rhk = s(h, k) with rhk being the (h, k) element of matrix Rβ, and s (h, k) (0 ≤ s(h, k) ≤ 1) being the similarity metric that measures the similarity level between haplotypes h and k (Schaid 2004; Tzeng and Zhang 2007).
Let g′ (μ) = ∂g (μ) /∂μ. Then by Taylor expansion on the mean function with respect to β around E(β) = 0, we have
Therefore
Consequently, the expected trait similarity is
| (6) |
| (7) |
The result indicates that trait similarity is (approximately) in a linear relationship with haplotype similarity if we choose (In that case, E(Zij | X, H) ≈ τSij.) For the canonical link g(·), this choice is equivalent to Comparing (7) to (3), we see that testing b = 0 in the similarity regression is the same as testing for τ = 0 in a variance component model. This implies that under the null hypothesis of b = 0, τ is zero and hence Yi’s are independent. The connection also suggests that the test of H0 : b = 0 should be one-sided, and that the gene-trait regression model (3) can use a zero intercept.
Recently Tzeng and Zhang (2007) constructed the variance component score test for testing H0 : τ = 0 based on the GLMM (5) as
where matrix S = {Sij}, W = diag {wi} with and Δ = diag {g′ (μi)}. Notice that under GLMM (5), Ub of equation (4) becomes
| (8) |
as V−1 = ΔW Δ. Comparing Tτ to Ub in (8), we notice that both statistics incorporate genetic information solely through the form of haplotype similarity (i.e., S or S0). We also note that both statistics share analogous quadratic forms; the forms are almost identical if ωi = g′ (μi) is used, in which case Thus while haplotype sharing (i.e., to detect unusual sharing of haplotypes among homogeneous trait groups) and haplotype smoothing (i.e., to smooth the haplotype effects by introducing correlation structure on similar haplotypes) are commonly considered as separate modeling strategies in haplotype analysis, they are unified through the framework of similarity regression and random effects haplotype analysis. With this comparison, we also learn the subtle difference between the two is that Ub of similarity regression uses information from between-individual comparison (i.e., i ≠ j), while Tτ of variance component includes the comparison of between and within individuals. Relatively speaking, the amount of information contributed by the comparison of the two haplotypes within a person is small (relative to the between-individual comparison) because two out of the four comparisons are self-comparison and hence not informative. As a result, although more data information is utilized in the variance-component test, we expect a similar performance of the two approach in detecting haplotype-phenotype association.
4. Simulation Studies
We performed simulation studies to investigate the behaviors of the proposed gene-trait similarity regression. We follow the same simulation scheme as Tzeng and Zhang (2007), where a coalescent process is first used to generate the SNP sequences, and then a causal SNP (rather than causal haplotypes or haplotype-similarity levels) is used to determine the trait values. Specifically, we implemented the coalescent program of Wall and Prichard (2003) to generate SNP sequences using the following parameters: an effective population size of 104, a scaled mutation rate of 5.6 × 10−4 (per bp), and a scaled recombination rate around 6 × 10−3 (per bp) for the cold spots and 45 times greater for the hot spots. These parameters are chosen to produce a similar number of common SNPs to the European American sample in the SeattleSNP database and to mimic the linkage disequilibrium pattern of the SELP gene observed in it. A total number of 100 sequences were generated from this model. We selected certain SNPs as the disease loci and form a haplotype region by including the two SNPs to its left and the three SNPs to its right. The disease SNP is selected based on the disease allele frequency (0.1 and 0.3) and the pairwise LD pattern between the disease SNP and its flanking SNPs (R2). We focus mostly on regions with max R2 > 0.7, i.e., at least one of the neighboring SNPs is highly correlated with the unobserved disease SNP. Such scenario reflects the common study designs where the disease SNP variation is captured by at least one tagSNPs. In the simulation studies, we further classify the haplotype regions by the level of average R2, after seeing exploratory results that suggest the relevancy of the average LD level across markers. The average R2 is obtained by averaging the 5 R2 ’s of the disease SNP and one of the nearby SNPs.
We next determined the trait value of an individual using the regression model g (μi) = γ0 + γ1Xi + θGi, where μi ≡ E(Yi | Xi, Gi) and Xi is a standard normal variable. The genetic value Gi is determined by the genotype of the disease locus (AA, Aa, or aa) and the disease effect (additive, dominant, and recessive). For genotypes AA, Aa, and aa, Gi = (2, 1, 0) for additive effect, (1, 1, 0) for dominant effect, and (1, 0, 0) for recessive effect. We considered two types of traits: binary traits and normal traits. For binary traits, we use the logit link and set γ0 = −4.5, γ1 = 0; and θ = log 2, resulting a disease rate of 1%. For normal traits, we use the identity link and set γ0 = 0, γ1 = 1, and θ = 1. We set the trait variance Var (Yi | Xi, Gi) to be 2 and 4 for allele frequency q = 0.1 and q = 0.3, respectively. The resulting heritability is about 0.1 under the additive model, 0.06 ~ 0.08 under the dominant model, and 0.01 ~ 0.02 under the recessive model. For both traits, we set θ = 0 when evaluating the sizes of the proposed test.
Under each simulation scenario, we used balanced case-control sampling to obtain 100 cases and 100 controls, and used random sampling to obtain 200 individuals with normal traits. We then removed the disease SNP information and converted the remaining haplotype data to unphased genotypes. We performed haplotype association analysis using three approaches: the standard haplotype regression of Schaid et al. (2002) (referred to as HAP method), the gene-trait similarity regression method with ωi = 1/υ (μi) (referred to as HS method), and the single SNP analysis using 1 degree of freedom (referred to as SNP-1 method) and 2 degrees of freedom (referred to as SNP-2 method). The results of the SNP methods were obtained using the minimum p-value among the 5 SNPs, whose significant thresholds were determined using the multiple-testing correction method of Moskvina and Schmidt (2008). This method estimates the effect number of independent tests for correlated SNPs at a given overall type I error rate, and estimates the significance level for each individual test accordingly. In the HAP method, haplotypes with frequencies less than the program default threshold (i.e., 5/(2×No. of individuals)) will be pooled into the category of the reference haplotype.
Type I Error Rates
Table 1 shows the type I error rates for the binary traits and normal traits at the nominal levels of 0.05 and 0.01. The results are obtained via 10,000 replications. For both trait types, the type I error rates are somehow conservative, especially at the 0.05 nominal level. There could be two possible causes for the differences: (a) the use of the quadratic form ZT ΛZ (see Web Appendix A) to approximate the distribution of Ub, and (b) the use of the 3-moment approximation to approximate the distribution of the quadratic form. We ran simulation to investigate and found that (a) was the more plausible cause. To reach this conclusion, we obtained the empirical p-values, for each replication, by resampling 104 Ub’s using the fact that The resulting type I error rates do not differ substantially from the 3-moment approximation, which indicates that the 3-moment method approximates the distribution of the quadratic form reasonably well. We also examined the type I error rates with an increased sample size of 1000. The resulting type I error rates based on 2,000 replications are shown in Table 1. When the sample size increases, the type I error rates approach to the nominal levels. This suggests that the numerical differences we observed are likely due to the loss of accuracy in approximating Ub with ZT ΛZ when using moderate sample sizes. This is possibly caused by the fact that matrix C (and hence Λ) is not positive definite, and the use of large samples can improve the accuracy and alleviate the over-conservative situation.
Table 1.
Type I error rates obtained by the 3-moment approximation (3-M) with sample size n = 200, obtained by the resampling methods (Resamp) with n = 200, and obtained by the 3-moment approximation with n = 1000.
| α = 0.05 |
α = 0.01 |
||||||
|---|---|---|---|---|---|---|---|
| n = 200 | n = 200 | n = 1000 | n = 200 | n = 200 | n = 1000 | ||
| q | avg R2 | 3-M | Resamp | 3-M | 3-M | Resamp | 3-M |
| Binary Traits | |||||||
| 0.1 | 0.2 | 0.042 | 0.046 | 0.046 | 0.012 | 0.010 | 0.009 |
| 0.3 | 0.035 | 0.039 | 0.041 | 0.010 | 0.009 | 0.011 | |
| 0.4 | 0.022 | 0.024 | 0.050 | 0.006 | 0.006 | 0.014 | |
| 0.6 | 0.040 | 0.046 | 0.043 | 0.011 | 0.010 | 0.010 | |
| 0.3 | 0.2 | 0.028 | 0.032 | 0.052 | 0.008 | 0.008 | 0.011 |
| 0.3 | 0.032 | 0.038 | 0.043 | 0.009 | 0.008 | 0.013 | |
| 0.5 | 0.043 | 0.047 | 0.051 | 0.008 | 0.008 | 0.007 | |
| 0.7 | 0.047 | 0.050 | 0.049 | 0.010 | 0.010 | 0.008 | |
| Normal Traits | |||||||
| 0.1 | 0.2 | 0.042 | 0.046 | 0.053 | 0.011 | 0.009 | 0.009 |
| 0.3 | 0.033 | 0.037 | 0.044 | 0.011 | 0.010 | 0.009 | |
| 0.4 | 0.021 | 0.024 | 0.049 | 0.007 | 0.007 | 0.013 | |
| 0.6 | 0.037 | 0.041 | 0.049 | 0.010 | 0.009 | 0.009 | |
| 0.3 | 0.2 | 0.027 | 0.029 | 0.044 | 0.008 | 0.008 | 0.008 |
| 0.3 | 0.036 | 0.042 | 0.043 | 0.009 | 0.008 | 0.008 | |
| 0.5 | 0.042 | 0.047 | 0.051 | 0.012 | 0.012 | 0.009 | |
| 0.7 | 0.045 | 0.049 | 0.045 | 0.009 | 0.008 | 0.010 | |
Power
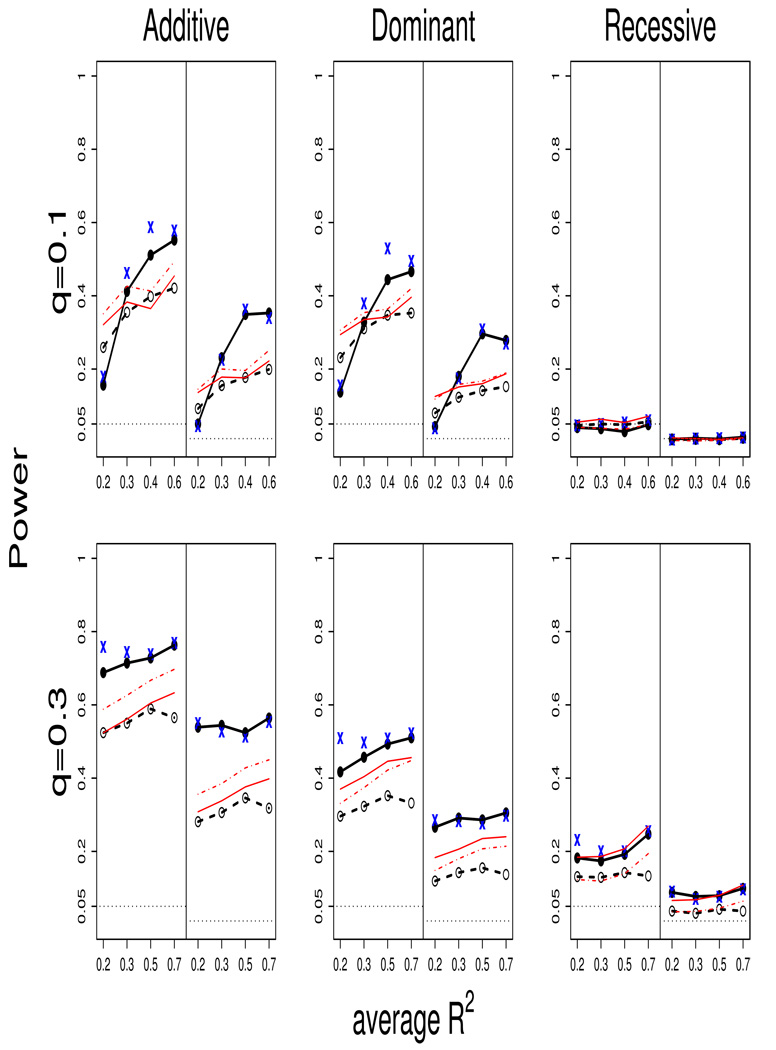
Results of the power comparisons are shown in Figure 1 (for binary traits) and Figure 2 (for normal trait) at the nominal levels 0.05 and 0.01. We considered the scenarios that the true genetic effects are either additive, dominant or recessive. The data were analyzed with an additive model, with the exception to the SNP-2 method. The power calculations are based on 5,000 replications. The regions studied here have max R2 > 0.9.
Figure 1.
Power comparisons for binary traits. The power are calculated based on 5,000 replications. In each plot, the left (right) panels are for nominal level 0.05 (0.01), and the horizontal dotted lines in the bottom indicates the corresponding nominal level. The upper (lower) panel is the results for allele frequency q = 0.1 (0.3). Lines with circles are results for haplotype-based analysis, and lines without circles are results for SNP-based analysis. Specifically, the solid lines with filled circles indicate the HS method, the dashed lines with open circles indicate the HAP method, the dotted-dashed lines indicate the SNP-1 method, and the solid lines indicate the SNP-2 method. The “x” signs indicate the VC method.
Figure 2.
Power comparisons for normal traits. The power are calculated based on 5,000 replications. In each plot, the left (right) panels are for nominal level 0.05 (0.01), and the horizontal dotted lines in the bottom indicates the corresponding nominal level. The upper (lower) panel is the results for allele frequency q = 0.1 (0.3). Lines with circles are results for haplotype-based analysis, and lines without circles are results for SNP-based analysis. Specifically, the solid lines with filled circles indicate the HS method, the dashed lines with open circles indicate the HAP method, the dotted-dashed lines indicate the SNP-1 method, and the solid lines indicate the SNP-2 method. The “x” signs indicate the VC method.
We first compare the HS method vs. the other methods. The general power patterns are similar for both trait types so we can concentrate the comparison on either type. With allele frequency of 0.1 (the upper panel) and under either additive or dominant effect, the HS method has higher power when the average R2 is high, but exhibits a power drop when average R2 is low (i.e., the case where only 1 SNP is correlated with the disease SNP). Under recessive effect, most of the methods have no power to detect the effect and hence all power is around the nominal level.
When the allele frequency is 0.3 (the lower panel), the HS method exhibits a clear power gain over the other methods (even in cases where the average R2 is low) for the additive and dominant effect scenarios. Under the recessive effect scenario, the SNP-2 methods have comparable or better power than the HS methods.
In the cases presented here, the SNP method tends to have higher power than the HAP method. This is likely due to the fact that these selected regions contain disease SNPs that are in extremely high correlation with at least one neighboring SNPs (i.e., max R2 > 0.9). The SNP method is expected to perform well when one SNP is a perfect surrogate to the disease SNP. However, the standard haplotype analysis, which regresses the trait values on multiple haplotype polymorphisms, must spend extra degrees of freedom on the less or not significant variants and hence becomes less efficient than the SNP method in this case.
We also compared the HS method with the variance-component (VC) method of Tzeng and Zhang (2007), and observed similar power performance for these two approaches. This reconfirms the findings reported in section 3.
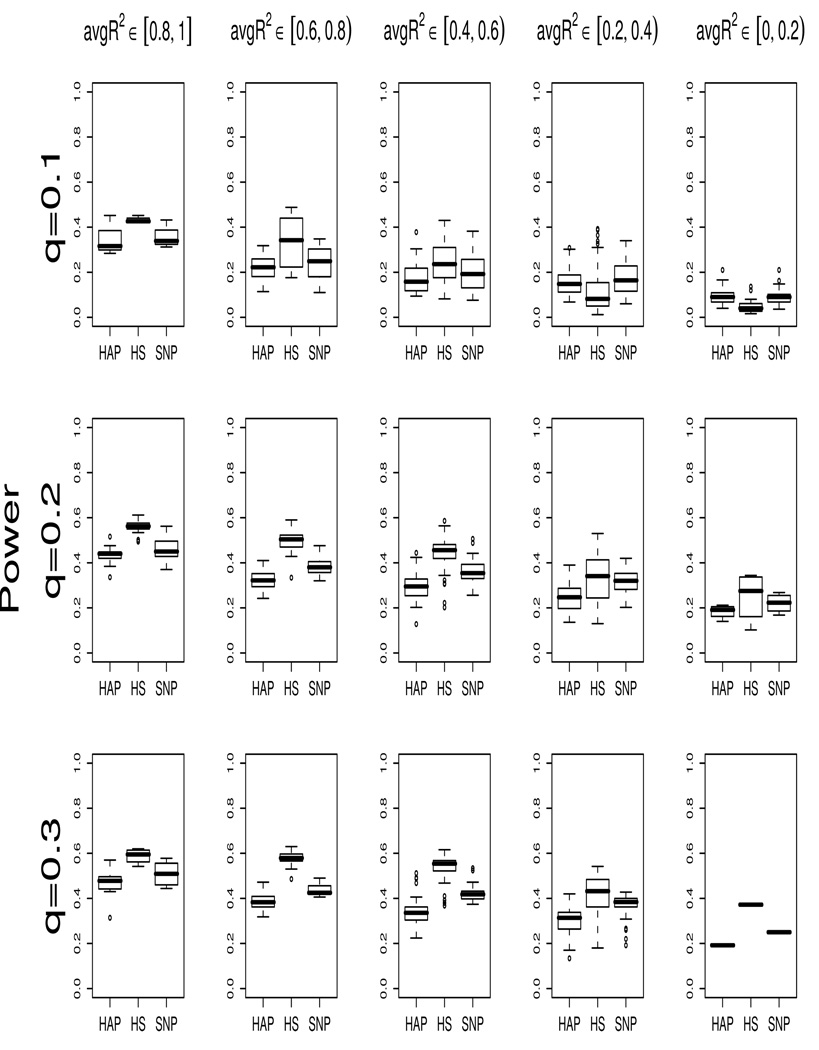
To make sure that the power pattern observed above was not subject to the regions chosen, we ran additional simulations to systematically examine the power performance of each method. In this set of simulations, only the binary trait under the additive effect was considered, due to the similarity in results between different trait types and effects. Regions with max R2 > 0.7 were examined and the disease allele frequency was set to be less than 0.4. There are a total of 207 regions with q ∈ 0.1 ± 0.045, 208 regions with q ∈ 0.2 ± 0.045, and 121 regions with q ∈ 0.3 ± 0.045. Figure 3 shows boxplots of power across all regions for each method. The power was obtained based on 1,000 replications at the nominal level of 0.01.
Figure 3.
Boxplots of power of the HAP/HS/SNP-1 methods for all simulated genome regions with max R2 > 0.7. The data consist of 100 cases and 100 control which were generated under an additive model. The power are evaluated based on 1,000 replications at nominal level 0.01. The upper/middle/lower panels are the results for allele frequency q ≈ 0.1/0.2/0.3, respectively.
We present the power by different ranges of the average R2: (a) > 0.8, (b) (0.6, 0.8], (c) (0.4, 0.6]; (d) (0.2, 0.4], and (e) ≤ 0.2. The scenarios (a) and (e) are considered to be cases at the end of the spectrum while the rest are intermediate cases. In scenario (a), all neighboring SNPs are in very high LD with the disease SNP (all R2 > 0.8), and in (e), the disease SNP has high correlation with only 1 nearby SNP and has 0 correlation with the rest 4 SNPs. Intermediate levels of multilocus LD are represented in cases (b) to (d). Given a certain disease allele frequency, we see in Figure 3 that the median power of the HAP method (left boxplots) and the HS method (middle boxplots) increase with average R2. This trend is expected: when more SNPs start to capture a greater amount of information about the disease locus, the power of those methods that utilize multimarker information should improve as well. We also note that the HS power increases more sharply than the HAP method, suggesting that the HS method could be more sensitive to the changes in the joint LD structure of multiloci. In contrast to the increase pattern of the HS and HAP methods, the median power of the SNP method (right boxplots) stays relatively constant.
The boxplots support the general pattern identified in Figure 1 and Figure 2: the HS method mostly has higher power than the HAP method and the SNP-1 method, and the exception is the cases with low average R2 and low allele frequency. This low power scenario of the HS method can be seen in the top right two panels of Figure 3. We suspect that this is related to the HS method being more sensitive to multilocus LD structure, and are actively investigating the underlying mechanisms.
5. Analysis of the ALS study of Schymick et al.
We applied our method to a case-control study of amyotrophic lateral sclerosis (ALS) obtained from the National Institute of Neurological Disorders and Stroke (NINDS) neurogenetics Repository at the Coriell Institute. The ALS study was conducted by Schymick et al. (2007) and one main objective of this study is to identify genetic factors that could contribute in the pathogenesis of sporadic ALS. The study recruited 276 patients with sporadic ALS and 271 neurologically normal controls, and genotyped 555,352 SNPs across the genome. Schymick et al. (2007) performed a genome-wide association analysis and reported the 34 most significant SNPs with p-values less than 0.0001 based on the single SNP tests. Although none of the 34 SNPs was significant after the Bonferroni correction for multiple testing, the mot significant SNP (rs4363506) lives in the close proximity to the dedicator of cytokinesis 1 gene (DOCK1), which is recognized to play an important role in motorneuron disease.
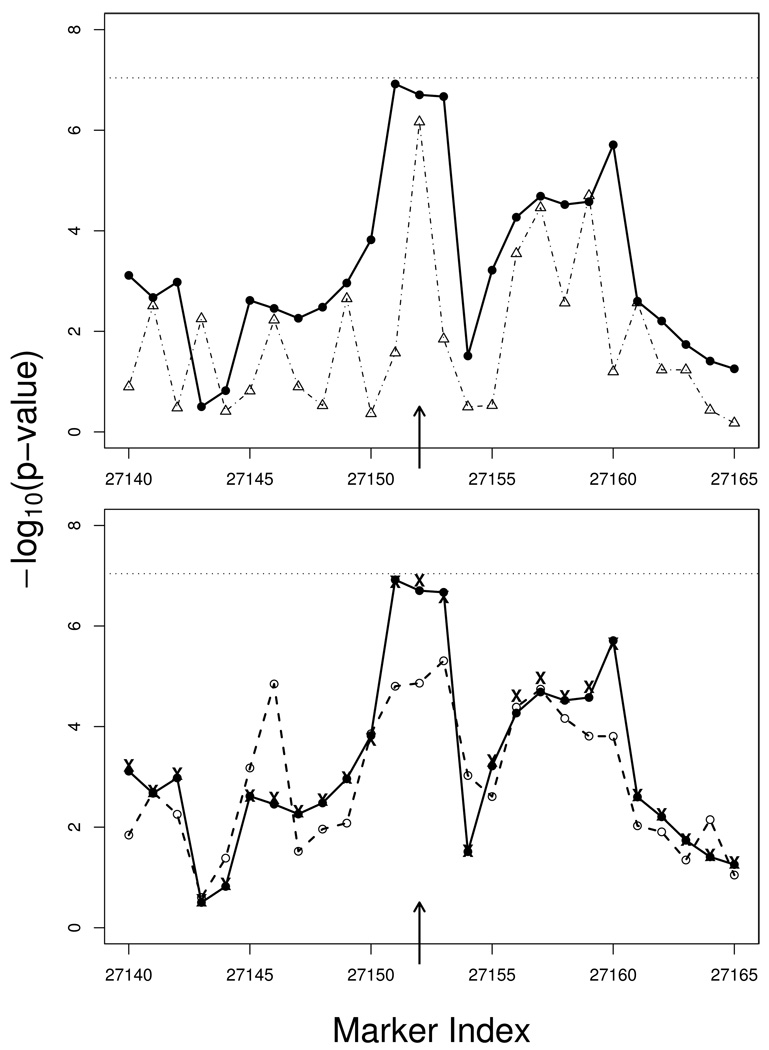
To illustrate the proposed HS method, we analyzed a portion of the ALS data. We concentrate on Chromosome 10 where the most significant SNP is located. Due to ambiguous marker information, we exclude the 26,258th SNP of Chromosome 10 and worked with the remaining 28,817 SNPs. We replicated the single SNP test (SNP-2 method) of Schymick et al’s, and followed their haplotype definition to perform 3-SNP sliding window haplotype association tests using the HS method. The HS method identified the most significant association SNP right around rs4363506 (p-value=1.2 × 10−7), and the p-values for the region around rs4363506 are shown in Figure 4 on the scale of negative logarithm of base 10 (upper panel). We see that haplotype analyses exhibited a smoother association signals across SNPs than the single SNP tests. Although the less-noisy haplotypic signals came at the cost of modeling multimarker variations, we see that the HS method achieved a level of significance that is comparable to single SNP analyses. To compare, we also implemented the HAP method and the VC method. The results are presented in the lower panel of Figure 4. We observed that the p-values of the HAP method are slightly less significant around rs4363506, and as expected, the VC method yields very similar results as the HS method.
Figure 4.
P-values from the ALS data analysis around the most promising SNP reported in Schymick et al. (i.e., SNP rs4363506 with location indicated by the arrow). The p-values are presented on the scale of negative logarithm of base 10, and the horizontal lines indicate the Bonferroni genome-wide threshold. Upper panel: p-values of the HS method based on 3-SNP haplotypes (solid line with filled circle) vs. p-values of the SNP-2 method (dotted-dashed line with open triangle). Lower panel: p-values of the HS method (solid line with filled circle) vs. p-values of the HAP method (dashed line with open circle). The “x” signs indicate the p-values of the VC method.
6. Discussion
In this article we introduced a regression model of trait similarity and haplotype similarity to study haplotype association. We set trait similarity to be the weighted mean-corrected trait cross products, and haplotype similarity to be the sharing degrees of the haplotypes between two subjects. We constructed a score statistic based on the similarity regression to test the null hypothesis of zero association between genotypes and traits. The score statistic is shown to follow a weighted chi-squared distribution under the null hypothesis, which can be approximated by the three-moment approximation method of Imhof (1961). The proposed method uses a simple statistic, eliminates the needs to perform permutation, and can be easily scaled up to the whole genome scale. In addition, the proposed method applies to both qualitative and quantitative traits, and can incorporate covariate effects such as population structure and other environmental confounders. The test could be conservative with moderate sample size, but the conservativeness is alleviated with an increased sample size. Through simulation studies, we explored its performance and observed that the proposed HS method result in power increase in many cases.
The HS method assumes that the adjacent SNPs have a high probability to be passed down together from some common ancestors in a region containing the disease SNP, and hence models the sharing level of the genetic materials instead of the variants themselves. On one hand, this rationale indicates that the HS method is more effective/sensitive in taking advantage of the joint correlation of the SNPs. On the other hand, this also means that the performance of the HS method can be more vulnerable to a low multilocus LD. For example, Figure 1 to Figure 3 show that a combination of low average LD and low allele frequency of the disease SNP could diminish the performance of the HS method. While Figure 1 to Figure 3 are based on regions with max R2 > 0.7 (i.e., the disease SNP is ”tagged” by at least one of the neighboring SNPs), we found that a similar high/low power patterns by the allele frequency and the average LD are observed for regions with max R2 ∈ [0.5, 0.7) (i.e., moderately tagged) and for regions with max R2 < 0.5 (i.e., not tagged) (results not shown). We are examining whether this feature of low power implies any of the following potential causes: (i) there is a joint acting effect between the allele frequency and the multilocus LD, (ii) the conditions required in the HS method is jeopardized with low average LD level and low allele frequency, or (iii) the performance of the HS method can be better predicted by other high-order LD structure than average R2, such as the multiple order Markov chains of Kim et al. (2008). Nevertheless, the findings suggest that the HS methods would be most ideal if coupled with a careful determination of haplotype regions based on the LD structure of multimarkers so to obtain the power gain.
The idea of our haplotype-trait similarity regression can be traced back to the Haseman-Elston regression model for linkage analysis (Haseman and Elston 1972; Elston et al. 2000), where the trait similarity of sib pairs is regressed on their IBD sharing probability. In our case, we treat the entire population as a family, and regress trait similarity of pair-wise samples on their identical-by-state status. Just like the Haseman-Elston regression, which is shown to be equivalent to the VC methods in linkage analysis (Sham and Purcell 2001), our gene-trait similarity regression is also analytically united with the VC approaches. Specifically, we showed that testing the regression coefficient of the haplotype similarity is the same as testing the genetic variance component in a mixed model. In addition, from the score statistics of the two approaches, we see both methods utilize haplotype similarity, with a difference that the within-individual haplotype comparison is used in the variance-component model but not in the gene-trait similarity regression. These connections suggest that the two approaches should have similar performance; this conjecture is supported by the simulations and data analysis. This equivalence offers a new way of looking at haplotype-sharing methods: it enables direct derivation of analytical solutions of the sharing statistics under the framework of VC methods. In the cases studied here, both the similarity regression and the VC model yield closed-form solutions. But for more complicated models (i.e., incorporating interaction effects), the similarity regression model becomes much more analytically challenging yet the VC method offers an efficient and reliable technique to draw inference.
Supplementary Material
Web Appendices A and B, referenced in Sections 2, 3, and 4, are available under the Paper Information link at the Biometrics website http://www.biometrics.tibs.org.
Acknowledgments
The authors are grateful to the NINDS Human Genetics Resource Center DNA and Cell Line Repository at Coriell and the laboratory for supplying the data of the ALS study used in this work. They also thank the associate editor and two reviewers for their helpful and constructive comments, and Drs. Kathryn Roeder, Silviu-Alin Bacanu, Andrew Allen, and Dahlia Nielsen for their careful discussions and suggestions on this work. J.Y.T. was supported by NSF grant DMS-0504726 and NIH grant R01 MH074027. D.Z. and M.D. were supported by NIH grant R01 CA085848. S.M.C was supported by NSC grant 96-2119-M-006-010. D.C.T. was supported by NIH grants R01 CA52862 and U01 ES015090.
References
- Allen A, Satten G. Statistical models for haplotype sharing in case-parent trio data. Human Heredity. 2007;64:35–44. doi: 10.1159/000101421. [DOI] [PubMed] [Google Scholar]
- Beckmann L, Thomas DC, Fischer C, Chang-Claude J. Haplotype sharing analysis using Mantel statistics. Human Heredity. 2005;59:67–78. doi: 10.1159/000085221. [DOI] [PubMed] [Google Scholar]
- Durham LK, Feingold E. Genome scanning for segments shared identical by descent among distant relatives in isolated populations. The American Journal of Human Genetics. 1997;61:830–842. doi: 10.1086/514891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. ”Efficiency and power in genetic association studies”. Nature Genetics. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, Dormishian F, Domingo R, Ellis MC, Fullan A, Hinton LM, Jones NL, Kimmel BE, Kronmal GS, Lauer P, Lee VK, Loeb DB, Mapa FA, McClelland E, Meyer NC, Mintier GA, Moeller N, Moore T, Morikang E, Prass CE, Quintana L, Starnes SM, Schatzman RC, Brunke KJ, Drayna DT, Risch NJ, Bacon BR, Wolff RK. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nature Genetics. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- Haseman JK, Elston RC. “The investigation of linkage between a quantitative trait and a marker locus”. Behavior Genetics. 1972;2:3–19. doi: 10.1007/BF01066731. [DOI] [PubMed] [Google Scholar]
- Houwen RHJ, Baharloo S, Blankenship K, Raeymaekers P, Juyn J, Sandkuijl LA, Freimer NB. Genome screening by searching for shared segments: Mapping a gene for benign recurrent intrahepatic cholestasis. Nature Genetics. 1994;8:380–386. doi: 10.1038/ng1294-380. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Imhof J. Computing the distribution of quadratic forms in normal variables. Biometrika. 1961;48:419–426. [Google Scholar]
- Kim Y, Feng S, Zeng ZB. Measuring and partitioning the high-order linkage disequilibrium by multiple order Markov chains. Genetic Epidemiology. 2008;32:301–312. doi: 10.1002/gepi.20305. [DOI] [PubMed] [Google Scholar]
- Molitor J, Marjoram P, Thomas D. Application of Bayesian spatial statistical methods to analysis of haplotypes effects and gene mapping. Genetic Epidemiology. 2003;25:95–105. doi: 10.1002/gepi.10251. [DOI] [PubMed] [Google Scholar]
- Moskvina V, Schmidt KM. On multiple-testing correction in genome-wide association studies. Genetic Epidemiology. 2008;32:567–573. doi: 10.1002/gepi.20331. [DOI] [PubMed] [Google Scholar]
- Pe’er I, de Bakker PI, Maller J, Yelensky R, Altshuler D, Daly MJ. Evaluating and improving power in whole-genome association studies using fixed marker sets. Nature Genetics. 2006;38:663–667. doi: 10.1038/ng1816. [DOI] [PubMed] [Google Scholar]
- Puffenberger EG, Kauffman ER, Bolk S, Matise TC, Washington SS, Angrist M, Weissenbach J, Garver KL, Mascari M, Ladda R, SIaugenhaupt SA, Chakravarti A. Identity-by-descent and association mapping of a recessive gene for Hirschsprung disease on human chromosome 13q22. Human Molecular Genetics. 1994;3:1217–1225. doi: 10.1093/hmg/3.8.1217. [DOI] [PubMed] [Google Scholar]
- Qian D, Thomas DC. Genome scan of complex traits by haplotype sharing correlation. Genetic Epidemiology. 2001;21 Suppl 1:S582–S587. doi: 10.1002/gepi.2001.21.s1.s582. [DOI] [PubMed] [Google Scholar]
- Schaid DJ. Evaluating associations of haplotypes with traits. Genetic Epidemiology. 2004;27:348–364. doi: 10.1002/gepi.20037. [DOI] [PubMed] [Google Scholar]
- Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. he American Journal of Human Genetics. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymick JC, Scholz SW, Fung HC, Britton A, Arepalli S, Gibbs JR, Lombardo F, Matarin M, Kasperaviciute D, Hernandez DG. Genome-wide genotyping in amyotrophic lateral sclerosis and neurologically normal controls: first stage analysis and public release of data. Lancet Neurology. 2007;6:322–328. doi: 10.1016/S1474-4422(07)70037-6. [DOI] [PubMed] [Google Scholar]
- Service SK, Lang DW, Freimer NB, Sandkuijl LA. Linkage-disequilibrium mapping of disease genes by reconstruction of ancestral haplotypes in founder populations. The American Journal of Human Genetics. 1999;64:1728–1738. doi: 10.1086/302398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha Q, Chen HS, Zhang S. A new association test using haplotype similarity. Genetic Epidemiology. 2007;31:577–593. doi: 10.1002/gepi.20230. [DOI] [PubMed] [Google Scholar]
- Thomas DC, Qian D, Gauderman WJ, Siegmund K, Morrison JL. A generalized estimating equations approach to linkage analysis in sibships in relation to multiple markers and exposure factors. Genetic Epidemiology. 1999;17 Suppl 1:S737–S742. doi: 10.1002/gepi.13701707121. [DOI] [PubMed] [Google Scholar]
- Thomas DC, Morrison JL, Clayton DG. Bayes estimates of haplotype effects. Genetic Epidemiology. 2001;21 Suppl 1:S712–S717. doi: 10.1002/gepi.2001.21.s1.s712. [DOI] [PubMed] [Google Scholar]
- Tzeng JY, Devlin B, Wasserman L, Roeder K. On the identification of disease mutations by the analysis of haplotype similarity and goodness of fit. The American Journal of Human Genetics. 2003a;72:891–902. doi: 10.1086/373881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng JY, Byerley W, Devlin B, Roeder K, Wasserman L. Outlier detection and false discovery rates for whole-genome DNA matching. Journal of the American Statistical Association. 2003b;98:236–246. [Google Scholar]
- Tzeng JY, Zhang D. Haplotype-based association analysis via variance component score test. The American Journal of Human Genetics. 2007;81:927–938. doi: 10.1086/521558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Meulen MA, Te Meerman GJ. Haplotype sharing analysis in affected individuals from nuclear families with at least one affected offspring. Genetic Epidemiology. 1997;14:915–920. doi: 10.1002/(SICI)1098-2272(1997)14:6<915::AID-GEPI59>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Wall JD, Pritchard JK. Assessing the performance of the haplotype block model of linkage disequilibrium. The American Journal of Human Genetics. 2003;73:502–515. doi: 10.1086/378099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel J, Schork NJ. Generalized genomic distance-based regression methodology for multilocus association analysis. The American Journal of Human Genetics. 2006;79:792–806. doi: 10.1086/508346. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Web Appendices A and B, referenced in Sections 2, 3, and 4, are available under the Paper Information link at the Biometrics website http://www.biometrics.tibs.org.