Abstract
Interaction specificity is a required feature of biological networks and a necessary characteristic of protein or small-molecule reagents and therapeutics. The ability to alter or inhibit protein interactions selectively would advance basic and applied molecular science. Assessing or modelling interaction specificity requires treating multiple competing complexes, which presents computational and experimental challenges. Here we present a computational framework for designing protein interaction specificity and use it to identify specific peptide partners for human bZIP transcription factors. Protein microarrays were used to characterize designed, synthetic ligands for all but one of 20 bZIP families. The bZIP proteins share strong sequence and structural similarities and thus are challenging targets to bind specifically. Yet many of the designs, including examples that bind the oncoproteins cJun, cFos and cMaf, were selective for their targets over all 19 other families. Collectively, the designs exhibit a wide range of novel interaction profiles, demonstrating that human bZIPs have only sparsely sampled the possible interaction space accessible to them. Our computational method provides a way to systematically analyze tradeoffs between stability and specificity and is suitable for use with many types of structure-scoring functions; thus it may prove broadly useful as a tool for protein design.
Designing peptides, proteins, or small molecules that bind to native protein targets is a promising route to new reagents and therapies. Yet dealing with the interaction specificity problem–i.e. achieving designs that are selective for their intended targets in preference to related alternatives–is difficult. Designing or assessing protein interaction specificity in a comprehensive manner is impeded by the challenges and costs inherent in modelling or measuring many competing complexes. Recent large-scale experiments that have characterized interaction specificity for a handful of protein families and/or domains represent significant progress in this area1–6. In particular, assays that provide a way to profile the interactions of a protein with many candidate partners offer an opportunity to explore how specificity can be introduced into proteins rationally, by design.
Computational design has led to remarkable advances in protein engineering over the past decade, including the design of protein-protein interactions7–15. Introducing considerations of specificity into protein-design calculations raises interesting theoretical challenges that have been addressed in a few prior studies7, 16, 17 and/or treated on a case-by-case basis in several applications7–10, 15. Most often, however, specificity is simply ignored in computational protein design. Several proteins or peptides that were optimized solely for binding to a native target were shown a posteriori to be specific for their intended interaction partner over a few related alternatives11–14. However, focusing only on the stability of the desired complex led to a lack of specificity, both in computational design and experimental selections, in other examples15, 16, 18. Strategies that can simultaneously consider affinity and multi-state specificity in the design process are therefore highly desirable7.
The basic-region leucine-zipper (bZIP) transcription factors provide an exciting but highly challenging opportunity to test strategies for interaction specificity design. The bZIPs homo- and/or heterodimerize by forming a parallel coiled coil (a “leucine zipper”) and bind DNA using a region rich in basic amino acids19. Approximately 53 human bZIP proteins that make up 20 families participate in a wide range of important biological processes and pose attractive targets for selective inhibition. Interest in inhibiting bZIPs dates to 1995, when Vinson and co-workers showed that heterodimers containing one bZIP subunit and one subunit with an acidic region replacing the basic region (A-ZIPs) are inactive. A-ZIPs have proven very useful for applications both in vitro and in vivo20, 21. However, these inhibitors mimic the interaction preferences of the proteins from which they are derived and typically associate with multiple bZIP families. Extensive sequence similarity among the leucine-zipper domains hampers efforts to make specific peptides that could provide more selective A-ZIPs or other inhibitors. For example, strong undesirable off-target interactions were observed when experimentally selecting synthetic partners for the cFos and cJun bZIP coiled coils out of peptide libraries18.
The bZIPs are also attractive design target s because experiments have probed sequence features that influence both structural and interaction specificity19, 22–24. Building upon these insights and taking advantage of large experimental data sets, computational models that provide useful predictions of bZIP interaction preferences have been developed4, 18, 25, 26. These prior studies afford a relatively mature understanding of bZIP partnering and provide the potential for specificity design.
We have developed a strategy for addressing specificity in protein-design calculations that rests on the trade-off between maximizing affinity and introducing specificity. The stability/specificity trade-off has been discussed previously7, 15–17, and has motivated the successful design of heterospecific coiled-coil pairs7. For our work, we note that a protein designed to bind optimally to a native target may also bind strongly to one or more undesired competitors, indicating that the difference in energy between forming undesired complexes and the design•target complex is not sufficiently large. New designs can be sought that increase this gap and are thus more selective for the target, but these will necessarily have reduced target affinities relative to the design that is optimal for target binding. The computational method presented here formalizes this trade-off by identifying sequences that minimize the stability sacrifice required to achieve increasing energy gaps from competing complexes. Such sequences posses the important property that they cannot simultaneously be improved both in predicted affinity and specificity.
Our framework, CLASSY (Cluster expansion and Linear programming-based Analysis of Specificity and Stability), makes use of two computational techniques to implement the above idea. The first is integer linear programming (ILP), an optimization method that has been applied to the energy-minimization problem in protein design27. The second is cluster expansion (CE), which we use to convert a structure-based interaction model into a sequence-based scoring function that is very fast to evaluate28,29. Importantly, CE allows us to apply ILP at the sequence level, rather than at the structure level. This makes it possible to impose constraints on the energies of design undesired partner interactions during optimization of the design•target energy, which is the keystone of the CLASSY approach. The power of CE and ILP mean that arbitrary numbers of desired and undesired states and relationships between them can be included in CLASSY designs. Thus, CLASSY can deal with problems beyond the scope of traditional design methods, making it an appropriate approach for designing specific anti-bZIP peptides.
As one example of how CLASSY can be used, we implemented a procedure called a specificity sweep to identify sequences of optimal stability that satisfy increasing requirements on specificity. For this purpose, the quantity Δ was defined as the energy gap between the lowest-energy undesired state and the desired target state (Fig. 1A). A specificity sweep begins by using ILP to find the sequence with the highest binding affinity for the target, ignoring specificity. An initial value for the quantity Δ is then computed by predicting the energies of all possible complexes involving this design. The ILP optimization is repeated, this time designing a protein that optimizes binding with the target subject to the constraint that all undesired states have energy gaps to the designed state that are larger than Δ plus a small increment. This is repeated, gradually increasing the value of Δ, until it is no longer possible to find design sequences that satisfy the constraints. Although CLASSY can be run with any value assigned to Δ, one advantage of the specificity sweep exploring a broad range of Δ values is that no assumption of how much stability or specificity is “enough” need be made prior to the calculation.
Figure 1.
Designing specific peptides using CLASSY. A) Specificity sweep scheme. A sequence (black) is sought that binds a target (red) but not several undesired partners (gray) or itself. Panels from left to right illustrate iterations of the CLASSY procedure, during which the specificity gap Δ is increased. B) and C) A specificity sweep with MafG as the target and all other human bZIP coiled coils (except MafK, in the same family as MafG) and the design homodimer as undesired complexes. The plot in B corresponds to the cartoon in A. Red dots, black bars and gray bars represent energies of the design•target, design•design, and design•other bZIP complexes, respectively. C plots design•target complex stability vs. specificity (Δ). Portions of several designed complexes are shown using helical wheels (orange highlights amino-acid changes from the previously shown sequence). The rightmost solution is anti-SMAF.
Candidate designs from a specificity sweep list may be selected for testing by a user, after considering predicted stability:specificity tradeoffs and the sequence changes that bring these about. Other considerations may be included, as CLASSY provides the ability to restrict arbitrary linear functions of sequence. In our application, a bias for the bZIP coiled-coil fold was imposed by constraining designs to be leucine-zipper like according to a position-specific scoring matrix (PSSM). Similar constraints could also be used, for example, to place requirements on predicted solubility. Such considerations, which are often included in designs in an ad hoc manner or by employing manual post-evaluation and filtering, can be naturally incorporated into the CLASSY procedure.
We applied CLASSY to design partners for nearly all human bZIPs and used our computational results to assess the difficulty of the bZIP interaction specificity design problem. We sought anti-bZIP designs predicted to bind their targets and yet interact minimally with themselves and with members of the 19 non-target bZIP families. Because of the extremely high sequence similarity within families, we did not require that the designs discriminate between siblings in the target family. The desired design•target heteromeric complex, as well as undesired design•design and design•off-target complexes, were modelled as coiled-coil dimers on a fixed-backbone template and evaluated using energy functions similar to that of reference 26, which was shown previously to give good performance predicting native bZIP interaction preferences26 (also see Supplementary Information).
Specificity sweeps were computed for the 46 bZIPs in reference 4. These calculations predicted that specificity will arise only rarely among bZIP partners optimized for stability alone. Such designs are almost all predicted to form strong homodimers, regardless of the family they are targeted against (Fig. S2). Negative design is also required to disfavour complexes with undesired bZIP competitors. Approximately 65% of 46 designs optimized for affinity alone were judged to face significant competition from non-target families; this can be addressed in CLASSY by sacrificing stability, as shown in Supplementary Fig. 2. We carried out additional computational analyses to estimate how candidate bZIP partners are distributed in stability-specificity space (Supplementary Fig. 12). Even when the design design homodimer is the only undesired state, the vast majority of sequence space is predicted to be non-specific. Thus, addressing specificity is critical, but the drastic reduction this imposes on acceptable sequences makes the design problem challenging.
We next tested 48 peptides designed to bind representative targets from all 20 bZIP families, using a protein microarray assay that has been validated for measuring interaction preferences for bZIPs4. Sequences to be tested were selected from the specificity sweeps by hand, considering the magnitude of Δ, the amount of stability lost relative to the most stable design, and sequence features such as excessive loss of hydrophobic interactions in the core (see Fig. 1C for the example of anti-SMAF; Supplementary Table 1 provides detailed descriptions of the origin of each design). In a few of the cases where we designed more than one peptide against a given target, experimental results for initial designs were incorporated to guide the CLASSY design procedure. For example, anti-ZF was designed using a modified specificity sweep that up-weighted the influence of XBP-1 in determining Δ, after this protein was experimentally determined to be a problematic competitor. The ability to easily incorporate information about known competitors is one advantage of CLASSY.
In total, 48 peptides designed against 20 targets were tested for interaction with 33 representative human bZIP coiled coils and for self-association. Fluorescence intensities measured on bZIP arrays have previously been shown to reflect relative interaction strengths measured in solution4. Each peptide in turn (both designed and native) was labelled with the fluorescent dye Cy-3 and used to probe aldehyde-derivatized slides printed with potential partners. Of the 48 designs tested, 40 bound to their intended target, as assessed by fluorescence signal above background (Supplementary Fig. 1). The probability of this occurring by chance, given the distribution of design-human interaction signals from the arrays, was ~10−11. Self-association of the designs was also evaluated. Only 40% of the designs showed detectable self-interactions using the same criterion, and all but 6 interacted with a human bZIP more strongly than they interacted with themselves (Fig. 2A and Supplementary Fig. 1).
Figure 2.
Experimental testing of anti-bZIP designs. A) Peptide array results for the most specific design identified for each human bZIP family. Columns show experiments using the indicated protein to probe an array. For the Specificity panel (left), designs in solution were used to probe human bZIPs and designs on the surface. In the Relative Stability panel (right), human bZIP targets were used to probe an array containing the cognate design of each target and 33 human bZIPs. Data are plotted as -log(F/Fmax), with F the fluorescence signal on the array, such that the strongest interaction has a value of zero. Values of -log(F/Fmax) above 1.0 were set to 1.0. Thick red circles – design•target; thin red circles – design interactions with siblings in the target family; grey squares – interactions with other human bZIPs; black squares – design•design. Designs are named using the family of their target. B) Solution testing of anti-SMAF complexes assayed using circular dichroism. In each panel, anti-SMAF alone is shown with dashed lines, the partner being tested with a solid line, the numerical average of these two signals with open circles (◦) and the mixture of the two peptides with closed circles (•). (B, C) Anti-SMAF interacts with target MafG (Tm ~ 38 °C). (D) Anti-SMAF interacts, at most, very weakly with cJun, the closest competitor according to microarray data. (E) There is no evidence for anti-SMAF interacting with MafB, a sequence closely related to the target. CD spectra in (B) were collected at 25 °C. Anti-SMAF unfolds with Tm ~12 °C. Similar data for other complexes are included in Supplemenatary Figures 3–8.
To determine the interaction specificity of the designed molecules, we used Cy-3 labelled designed peptides and compared the array signal for interaction with the target to that for interaction with non-target competitors. Results for the most specific design identified for each of the 20 families are shown in Fig. 2A. These designs are named using the target family name. For 10 designs, the strongest interaction observed was with the intended target. Strikingly, 8 of these designs bound their targets with array signals distinctly greater than for any other non-target-family partner (targets: ZF, cFos, MafG, ATF-2, cJun, cMaf, XBP-1, ATF-4, leftmost in the Specificity panel of Fig. 2A). This indicates measurable interaction specificity on the arrays. For 2 more designs, fluorescence signal for interaction with the target was only marginally greater than that for interaction with 1–2 other proteins (targets: ATF-3, C/EBPγ). Nine other designs bound their targets, but less strongly than they bound to members of other families. For one target family (PAR), the designed peptide did not show detectable binding above background.
To assess the stability of each design•target interaction, we labelled each native bZIP target with Cy-3 and probed an array containing 33 representative human bZIP peptides as well as the anti-target design. This experiment assayed design•target stability relative to interactions of the target with its native partner(s). The strongest signal was often from the design•target complex, indicating that many designs can be expected to out-compete native partners of the targets, using modest concentrations (summarized in Fig. 2A, complete data in Supplementary Information). Less stable designs can likely be improved through generic strategies such as the addition of acidic extensions, as for the A-ZIPs20.
To validate the array assay, 28 mixtures involving the 7 best designs were characterized in solution using thermal denaturation monitored by circular dichroism. Each designed peptide was tested for interaction with (1) its target, (2) its next-best interaction partner, as reported by the array, (3) a protein closely related by sequence to the target, and (4) itself. We monitored whether the mixtures showed an increase in the temperature of denaturation (Tm) compared to that expected from the average of the signals of the individual components (Figs 2B–E and Supplementary Figs 3–8). In all cases, the Tm studies supported binding of each design to its intended target. For the 21 undesired complexes tested, 18 either showed no evidence for interaction or a Tm that was clearly lower than that of the design•target complex. For the remaining 3 undesired complexes, formation of mixtures complicated the analyses, although these are probably also weaker than the corresponding design•target complexes (Supplementary Figs 4, 5, 6). Solution data were also examined for consistency with the array measurements and supported the same relative ordering of stabilities for 35 of 41 comparable cases (see Supplementary Information).
Three of our best designs target cJun, cFos, and ATF-2. These proteins are constituents of the AP-1 transcription factor complexes involved in cell proliferation and oncogenesis. The cJun•cJun, cJun•cFos, and cJun•ATF-2 dimers are involved in these important processes in ways that have not been fully elucidated. Complexes involving cJun have previously been targeted for disruption using a dominant-negative A-ZIP version of cFos20. But because cFos also binds ATF-2 and its family members4, the A-ZIP strategy is not as specific as might be desired. The same is true for cJun and ATF-2: native partners of these targets also bind to additional families. Attempts to identify new partners for cFos and cJun using experimental selection strategies gave peptides that strongly self-associated and also bound bZIPs non-specifically (i.e. the intended anti-cFos and anti-Jun peptides bound both FOS and JUN family members tightly)18, 30. Our designed peptides provide a way to introduce specificity, e.g. to disrupt cJun•cFos but not cJun•cJun or cJun•ATF-2, using anti-FOS.
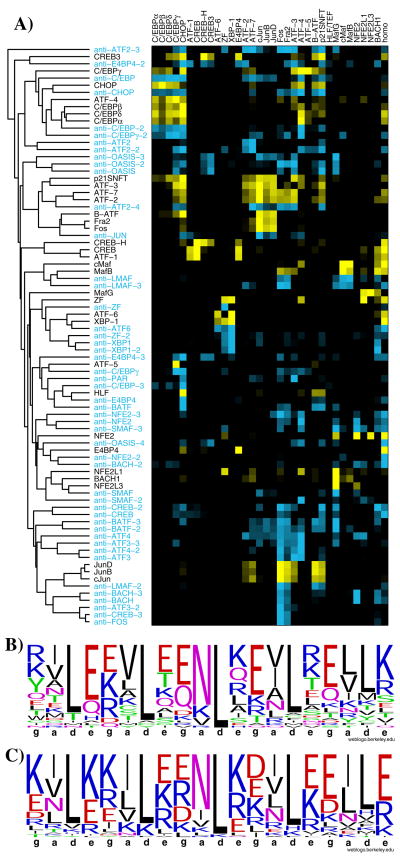
Fig. 3A shows the interaction profiles of native bZIP leucine zippers and the designed anti-bZIP peptides. The native proteins exhibit diverse interaction properties, despite their limited sequence variability (Fig. 3B)4. The designed peptides are even more limited in sequence diversity, yet they encode many additional, novel specificity profiles, suggesting that bZIP-like coiled-coil interaction space is only sparsely sampled by the human proteins (Fig. 3C). Based on the frequency of success of our interaction prediction model, and results from CLASSY analysis, we conservatively estimate that >1,900 very distinct interaction profiles can be encoded using the restricted sequence space employed in our designs. This may prove useful for applications in synthetic biology (see Supplementary Information).
Figure 3.
Properties of designed peptides compared to human bZIP leucine-zippers. A) Hierarchical clustering of interaction profiles for 33 human peptides and 48 designs; an interaction profile consists of the array signals for interactions with 33 surface-bound human peptides. Proteins on the surface are in columns and those in solution are in rows, with designed proteins and their interaction profiles in blue and human bZIP interaction profiles in yellow. B), C) Sequence logos for a, d, e, and g positions from the first 5 heptads of the 33 human bZIP leucine zippers in B) and the 48 designed peptides in C) (http://weblogo.berkeley.edu).
CLASSY designs exhibited canonical bZIP specificity determinants, such as a preference for Asn residues at a positions to pair across helices, and charge complementarity at g-to-e′ pairs (see Fig. 1C for coiled-coil heptad positions; a prime indicates a residue on the opposite helix, see Supplementary Fig. 15)19, 24. Interestingly, g-to-a′ pairs were predicted to make a comparable, if not larger, contribution to specificity than g-to-e′ pairs. Other unanticipated specificity patterns also emerged, involving steric interactions between a and d′ sites (see Supplementary Information for a fuller discussion). The significance of such interactions has not been broadly recognized in parallel coiled coils, although recent studies suggest their importance in anti-parallel dimers31.
CLASSY provides a way to analyze and optimize stability/specificity tradeoffs in protein design. The CE/ILP procedure imposes few formal requirements on the type of scoring function that can be used or the type of specificity problem that can be addressed. However, measuring and predicting interaction specificity for proteins generally remains challenging. Here, the bZIPs provided several advantages. The bZIP microarray assay benefits from reversible folding of short coiled coils, and data from prior array measurements of many bZIP transcription factor pairs were critical for developing predictive models4, 25, 26. Experimental helix propensities contributed to the quality of these models, and knowledge of particular specificity determinants (e.g. the special role of Asn pairs) improved predictions and also disfavoured the formation of higher-order oligomers19. Finally, symmetric fixed-backbone models proved adequate for this application26. This facilitated both structural modelling and cluster-expansion training, although CE can also be used for asymmetric structures and with flexible backbones32. Further details about features specific to bZIP modelling are in Methods and Supplementary Discussion.
Determinants of protein interaction specificity are not yet as well understood for other complexes, but significant progress in this area is evident. Zinc-finger/DNA, SH2/peptide and PDZ/peptide complexes have been extensively studied, and both assays and interaction models have been developed that make these good candidates for design using CLASSY (see Supplementary Information for further discussion)2, 3, 12, 33,34. Large-scale interaction experiments are becoming more common, and general-purpose models to describe protein structures and energies are under development33, 35–37. Advances in these areas will expand the problems that can be addressed using CLASSY. In the long term, we hope this approach will help address how interaction crosstalk can be controlled in both evolved and designed protein systems.
Methods Summary
Structure-based modelling of coiled-coil interactions was done as previously described, with modifications detailed in the Methods and Supplementary Information 26. Using the technique of cluster expansion, structure-based models were converted to functions of sequence that included constant, single-residue and residue-pair terms. Training of the cluster expansion used 61,780 random bZIP-like sequences that were modelled structurally28, 29. A limited amino-acid alphabet was considered, which included the 10 residues most frequently found at each coiled-coil heptad position in native bZIPs. Constrained optimization employing integer linear programming (ILP) was used to design a, d, e and g sites. ILP optimization minimized the energy of design•target complexes, subject to constraints on the energy gap with respect to undesired complexes and the match of the design sequence to a position-specific scoring matrix derived from 432 native bZIP leucine zippers. Other positions in the coiled-coil repeat (b, c and f positions) were chosen to be consistent with the designed interface a, d, e and g residues, using a probabilistic framework. For each design target, the ILP optimization was repeated with increasing values of the specificity gap parameter Δ, in a procedure termed a specificity sweep. Sequences for experimental testing were selected manually from candidates generated using the specificity sweeps.
For experimental testing, His6-tagged peptides were expressed in RP3098 cells and purified by Ni-NTA followed by reverse-phase HPLC. Coiled-coil microarrays were printed, processed and probed as described previously4. Fluorescence signals from the arrays were processed to remove background and normalized. Circular dichroism measurements were performed using standard techniques to measure spectra between 195 and 280 nm at 25 °C or thermal stability by monitoring ellipticityat 222 nm. Data were fit to appropriate thermodynamic equations to obtain apparent Tms. Detailed descriptions of all procedures are included in the Methods and the Supplementary Information.
Methods
Modelling bZIP leucine-zipper interactions
Two variants of the previously described energy function HP/S/C were used to evaluate the relative stability of coiled-coil dimer structures26. Models were constructed using a single backbone, with rotameric sampling and continuous relaxation used to position side chains. HP/S/Ca is the model as published26, with scale factor s = 0 such that intra-chain interactions in the folded structure do not directly contribute to stability (though there are indirect contributions). HP/S/Ca replaces core a-a′ and d-d′terms derived from structure-based calculations with weights from a machine-learning algorithm26. In the variant model HP/S/Cv, structure-based a-a′ interactions were replaced with a-a′ experimental coupling energies for 55 amino-acid combinations22 and the d-d′ interaction for Leu-Leu was replaced with the empirical value − 2 kcal/mol. Following cluster expansion (see below), a-position point contributions were adjusted such that 100 folding free energies measured by Acharya et al.22 were predicted optimally (in the least squares sense, see Supplementary Fig. 10). The following 10 amino acids were allowed: V, L, N, I, K, A, R, T, S, and E for a positions; L, V, I, M, H, Y, T, A, K, and F for d positions; E, K, R, Q, L, S, T, A, V, and I for e positions; E, K, Q, R, L, Y, T, D, A, and I for g positions. These are the 10 amino acids most frequently encountered in the respective positions in bZIPs. Additionally, for the a position, these are also the 10 amino acids for which Vinson and co-workers have measured coupling energies 22.
Cluster expansion
Cluster expansion (CE) provides a way to express the energy of a sequence adopting a particular backbone structure as an algebraic function of the sequence itself28. The formal basis of the technique is described in the Supplementary Methods. In this study, the desired and undesired structures had the same backbone, and thus one cluster expansion (for parallel, coiled-coil dimers) was sufficient. CE calculations for both HP/S/Ca and HP/S/Cv included single-residue and residue-pair terms. A training set was built by randomly generating 61,780 coiled-coil sequences with heptad position-specific amino-acid probabilities taken from a multi-species alignment of 432 bZIPs (personal communication with Mona Singh, Princeton University). Gly and Pro were not included. Pair contributions were included only for amino-acid pairs ≤ 7 residues apart, resulting in 9,929 possible effective cluster interactions (ECI): 1 constant, 68 point and 9,860 pair terms. After the fitting procedure, 2,544 and 2,470 ECI survived the statistical significance test (e.g. lowered the cross-validated error28) for HP/S/Ca and HP/S/Cv, respectively. The performance of the resulting cluster expansions on a similarly generated test set of 10,000 sequences not used in training is shown in Supplementary Fig. 11.
Multi-state design optimization
Design sequences were optimized for interaction with the target using integer linear programming (ILP), imposing constraints on the design interaction energy with competitors and a degree of match to a bZIP position-specific scoring matrix (PSSM). The ILP and PSSM are detailed in the Supplementary Methods. We performed two types of CLASSY calculations. The first, a specificity sweep, starts by using ILP to identify the design sequence that produces the provably lowest predicted binding energy to a target. Given this sequence, energy gaps between the design•target dimer and al design•competitor dimers, including design•design, are calculated as gap=Edesign:competitor − Edesign:t arg et. The minimal gap (which may be negative) is defined as Δ. In the next iteration of the specificity sweep, the design•target energy is re-optimized, this time imposing constraints to require that all gaps be greater than Δ + 1 kcal/mol. In each round, Δ is updated and this procedure is repeated until no more solutions exist (Fig. 1A). Designs to be tested are chosen from this list of optimized sequences, as discussed in the main text.
Anti-bZIP designs were tested in three rounds of microarray experiments. When we sought to improve upon a previously tested design, we sometimes used experimental results to formulate biased specificity sweeps. In these calculations, custom offsets were applied to enhance or diminish the significance of some gaps relative to others; the remainder of the procedure was identical to that for a standard specificity sweep. For example, a biased specificity sweep was used to design anti-ZF after the first design tested (named as anti-ZF-2) interacted with XBP-1 more strongly than with ZF, contrary to predictions of the model. This is illustrated and explained further in Supplementary Fig. 9. Supplementary Table 1 contains a list of all designs and the procedures by which they were obtained, including the details of any biased specificity sweeps employed.
In all CLASSY procedures, except where noted in Supplementary Table 1, 46 human bZIPs were considered (sequences take from ref 25), and the modelled states were as follows: the design•target complex was the only desired state; design off-target bZIP complexes for all bZIPs not in the family of the target bZIP were treated as undesired; the design design homodimer was also an undesired state.
Further details on the theory behind CLASSY, as well as other computational analyses performed in this study, are in Supplementary Methods and Supplementary Discussion.
Choosing 33 representative human bZIPs
To avoid redundancy and conserve resources and time we used a representative set of 33 human bZIPs that covered all 20 families (see Supplementary Fig. 13). Representatives were chosen based on sequence similarity and reagent availability and described well the distinct interaction profiles reported by Newman and Keating4. Computational design was nevertheless conducted with 46 human bZIPs taken from Newman and Keating4.
Plasmid construction and peptide expression, purification and labelling
Synthetic genes encoding all designs were constructed using DNAWorks38 to design primers that contained flanking BamHI and XhoI restriction sites. A two-step PCR method was used to assemble the primers and the PCR products were digested with BamHI and XhoI and cloned into a modified pDEST17 vector4. All synthetic genes were confirmed to be correct by sequencing. Plasmids encoding human leucine-zipper peptides have been previously published in reference 4 with the exceptions of modified Jun family constructs that are described in the Supplementary Methods.
Plasmids were transformed into RP3098 cells and 1 L cultures in LB were grown to 0.4–0.6 OD and induced at 37 °C for 3–4 hours with addition of 1mM IPTG. Peptides were purified under denaturing conditions (guanidine hydrochloride, GuHCl) by binding to Ni-NTA resin and eluted with 60% acetonitrile/1% TFA. Following reduction with 10 mM TCEP in 5% acetic acid for 3 minutes at 65 °C, peptides were further purified using reverse-phase HPLC. The molecular weights of all designed peptides were confirmed as correct to within 0.15% by mass spectrometry. To generate dye labelled-peptides, 10 molar excess of Cy3 NHS ester in 6 M GuHCl/100 mM phosphate (pH 7.5) was added to lyophilized aliquots of protein and incubated for 2 hours at room temperature. Free dye was removed using size-exclusion spin columns. Labelled peptides were stored at −80 °C.
Preparation and probing of arrays
Lyophilized aliquots of protein were resuspended to a concentration of 40 μM in 6 M GuHCl/100 mM phosphate (pH 7.5)/0.04% Tween-20/10 μM Alexa Fluor 633 hydrazide. Proteins were printed on aldehyde-presenting glass slides (Thermo Fisher Scientific) using a Microgrid TAS Arrayer. Twelve identical subarrays were printed on each slide. Each protein was spotted twice, in two different print orders, for a total of four spots for each protein per subarray. After printing, slides were divided into subarrays by drawing a hydrophobic boundary (PAP pen, Electron Microscopy Sciences). Slides were stored at −80 °C for up to 1 month.
Slides were prepared for probing by: (1) washing face up in −80 °C ethanol for 30 seconds; (2) transferring to 80% ethanol/10 mM NaOH and incubating with shaking for 15 minutes; (3) washing in H2O for 15 seconds; (4) incubating in PBS/0.1% Tween- 20 for 15 minutes with shaking; (5) drying by centrifugation. Slides were then immediately probed by diluting labelled peptide in 6 M GuHCl/100 mM phosphate (pH 7.5)/6 mM TCEP 6-fold into 1.2X Buffer (1.2% BSA, 1.2X PBS, 0.12% Tween-20). The resulting solution was mixed and 35 μl was immediately pipetted onto each subarray. Each sample was probed in duplicate on adjacent subarrays, for a total of 8 spots used to detect each interaction. Slides were covered with a box and incubated for 1 hour. Slides were washed in PBS/0.1% Tween-20 for 15 seconds and then H2O for 15 seconds and were then dried by centrifugation. Slides were scanned using a DNA Microarray Scanner (Agilent) at several photo-multiplier tube voltage levels. The concentration of probe was 160 nM unless otherwise indicated.
Additional details on experimental techniques and data analysis are provided in Supplementary Methods.
Supplementary Material
Acknowledgments
This work was supported by NIH award GM67681 and used computer equipment purchased under NSF award 0216437. We thank the MIT BioMicro center for arraying instrumentation and R.T. Sauer, M. Singh, B. Tidor, M. Laub, T.A. Baker, J.H. Davis, M.S. Kay, J.R.S. Newman, W. F. DeGrado and members of the Keating lab, especially O. Ashenberg and T.C.S. Chen, for thoughtful comments on the manuscript.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions GG, AWR and AEK conceived the project. GG developed, implemented and applied the CLASSY formalism and carried out all computational analyses. AWR designed and performed all experiments. All authors analyzed data and guided the research plan. GG and AEK wrote the paper, in consultation with AWR.
Author Information Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. Correspondence and requests for materials should be addressed to AEK (keating@mit.edu).
References
- 1.Stiffler MA, et al. PDZ domain binding selectivity is optimized across the mouse proteome. Science. 2007;317:364–9. doi: 10.1126/science.1144592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones RB, Gordus A, Krall JA, MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439:168–74. doi: 10.1038/nature04177. [DOI] [PubMed] [Google Scholar]
- 3.Wiedemann U, et al. Quantification of PDZ domain specificity, prediction of ligand affinity and rational design of super-binding peptides. J Mol Biol. 2004;343:703–18. doi: 10.1016/j.jmb.2004.08.064. [DOI] [PubMed] [Google Scholar]
- 4.Newman JR, Keating AE. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science. 2003;300:2097–101. doi: 10.1126/science.1084648. [DOI] [PubMed] [Google Scholar]
- 5.Landgraf C, et al. Protein interaction networks by proteome peptide scanning. PLoS Biol. 2004;2:E14. doi: 10.1371/journal.pbio.0020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 2005;3:e334. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Havranek JJ, Harbury PB. Automated design of specificity in molecular recognition. Nat Struct Biol. 2003;10:45–52. doi: 10.1038/nsb877. [DOI] [PubMed] [Google Scholar]
- 8.Kortemme T, et al. Computational redesign of protein-protein interaction specificity. Nat Struct Mol Biol. 2004;11:371–9. doi: 10.1038/nsmb749. [DOI] [PubMed] [Google Scholar]
- 9.Ali MH, et al. Design of a heterospecific, tetrameric, 21-residue miniprotein with mixed alpha/beta structure. Structure. 2005;13:225–34. doi: 10.1016/j.str.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 10.van der Sloot AM, et al. Designed tumor necrosis factor-related apoptosis-inducing ligand variants initiating apoptosis exclusively via the DR5 receptor. Proc Natl Acad Sci U S A. 2006;103:8634–9. doi: 10.1073/pnas.0510187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin H, et al. Computational design of peptides that target transmembrane helices. Science. 2007;315:1817–22. doi: 10.1126/science.1136782. [DOI] [PubMed] [Google Scholar]
- 12.Reina J, et al. Computer-aided design of a PDZ domain to recognize new target sequences. Nat Struct Biol. 2002;9:621–7. doi: 10.1038/nsb815. [DOI] [PubMed] [Google Scholar]
- 13.Shifman JM, Mayo SL. Exploring the origins of binding specificity through the computational redesign of calmodulin. Proc Natl Acad Sci U S A. 2003;100:13274–9. doi: 10.1073/pnas.2234277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu X, Apgar JR, Keating AE. Modeling backbone flexibility to achieve sequence diversity: the design of novel alpha-helical ligands for Bcl-xL. J Mol Biol. 2007;371:1099–117. doi: 10.1016/j.jmb.2007.04.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolon DN, Grant RA, Baker TA, Sauer RT. Specificity versus stability in computational protein design. Proc Natl Acad Sci U S A. 2005;102:12724–9. doi: 10.1073/pnas.0506124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kangas E, Tidor B. Electrostatic specificity in molecular ligand design. JComp Phys. 2000;112:9120–9131. [Google Scholar]
- 17.Deutsch JM, Kurosky T. New algorithm for protein design. Phys Rev Lett. 1996;76:323–326. doi: 10.1103/PhysRevLett.76.323. [DOI] [PubMed] [Google Scholar]
- 18.Mason JM, Schmitz MA, Muller KM, Arndt KM. Semirational design of Jun-Fos coiled coils with increased affinity: Universal implications for leucine zipper prediction and design. Proc Natl Acad Sci U S A. 2006;103:8989–94. doi: 10.1073/pnas.0509880103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vinson C, Acharya A, Taparowsky EJ. Deciphering B-ZIP transcription factor interactions in vitro and in vivo. Biochim Biophys Acta. 2006;1759:4–12. doi: 10.1016/j.bbaexp.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 20.Gerdes MJ, et al. Activator protein-1 activity regulates epithelial tumor cell identity. Cancer Res. 2006;66:7578–88. doi: 10.1158/0008-5472.CAN-06-1247. [DOI] [PubMed] [Google Scholar]
- 21.Krylov D, Olive M, Vinson C. Extending dimerization interfaces: the bZIP basic region can form a coiled coil. Embo J. 1995;14:5329–37. doi: 10.1002/j.1460-2075.1995.tb00217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Acharya A, Rishi V, Vinson C. Stability of 100 homo and heterotypic coiled-coil a-a′ pairs for ten amino acids (A, L, I, V, N, K, S, T, E, and R) Biochemistry. 2006;45:11324–32. doi: 10.1021/bi060822u. [DOI] [PubMed] [Google Scholar]
- 23.Krylov D, Barchi J, Vinson C. Inter-helical interactions in the leucine zipper coiled coil dimer: pH and salt dependence of coupling energy between charged amino acids. J Mol Biol. 1998;279:959–72. doi: 10.1006/jmbi.1998.1762. [DOI] [PubMed] [Google Scholar]
- 24.Lupas AN, Gruber M. The structure of alpha-helical coiled coils. Adv Protein Chem. 2005;70:37–78. doi: 10.1016/S0065-3233(05)70003-6. [DOI] [PubMed] [Google Scholar]
- 25.Fong JH, Keating AE, Singh M. Predicting specificity in bZIP coiled-coil protein interactions. Genome Biol. 2004;5:R11. doi: 10.1186/gb-2004-5-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grigoryan G, Keating AE. Structure-based prediction of bZIP partnering specificity. J Mol Biol. 2006;355:1125–42. doi: 10.1016/j.jmb.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 27.Kingsford CL, Chazelle B, Singh M. Solving and analyzing side-chain positioning problems using linear and integer programming. Bioinformatics. 2005;21:1028–36. doi: 10.1093/bioinformatics/bti144. [DOI] [PubMed] [Google Scholar]
- 28.Grigoryan G, et al. Ultra-fast evaluation of protein energies directly from sequence. PLoS Comput Biol. 2006;2:e63. doi: 10.1371/journal.pcbi.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou F, et al. Coarse-Graining Protein Energetics in Sequence Variables. Phys Rev Lett. 2005;95:148103. doi: 10.1103/PhysRevLett.95.148103. [DOI] [PubMed] [Google Scholar]
- 30.Mason JM, Muller KM, Arndt KM. Positive aspects of negative design: simultaneous selection of specificity and interaction stability. Biochemistry. 2007;46:4804–14. doi: 10.1021/bi602506p. [DOI] [PubMed] [Google Scholar]
- 31.Hadley EB, Testa OD, Woolfson DN, Gellman SH. Preferred side-chain constellations at antiparallel coiled-coil interfaces. Proc Natl Acad Sci U S A. 2008;105:530–5. doi: 10.1073/pnas.0709068105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Apgar JR, Hahn S, Grigoryan G, Keating AE. Cluster-expansion models flexible-backbone protein energetics. J Comp Chem. doi: 10.1002/jcc.21249. (in press) [DOI] [PubMed] [Google Scholar]
- 33.Sanchez IE, et al. Genome-wide prediction of SH2 domain targets using structural information and the FoldX algorithm. PLoS Comput Biol. 2008;4:e1000052. doi: 10.1371/journal.pcbi.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaplan T, Friedman N, Margalit H. Ab initio prediction of transcription factor targets using structural knowledge. PLoS Comput Biol. 2005;1:e1. doi: 10.1371/journal.pcbi.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boas FE, Harbury PB. Potential energy functions for protein design. Curr Opin Struct Biol. 2007;17:199–204. doi: 10.1016/j.sbi.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 36.Das R, Baker D. Macromolecular modeling with ROSETTA. Annu Rev Biochem. 2008;77:363–82. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 37.Zhou H, Zhou Y. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 2002;11:2714–26. doi: 10.1110/ps.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoover DM, Lubkowski J. DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res. 2002;30:e43. doi: 10.1093/nar/30.10.e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.