Abstract
Retrotransposition of processed mRNAs is a frequent source of novel sequence acquired during the evolution of genomes. The vast majority of retroposed gene copies are inactive pseudogenes that rapidly acquire mutations that disrupt the reading frame, while precious few are conserved to become new genes. Utilizing a multi-breed association analysis in the domestic dog, we demonstrate that a recently acquired fgf4 retrogene causes chondrodysplasia, a short-legged phenotype that defines several common dog breeds including the dachshund, corgi and basset hound. The discovery that a single evolutionary event underlies a breed-defining phenotype for 19 diverse dog breeds demonstrates the importance of unique mutational events in constraining and directing phenotypic diversity in the domestic dog.
The domestic dog is arguably the most morphologically diverse species of mammal and theories abound regarding the source of its extreme variation (1). Two such theories rely on the structure and instability of the canine genome, either in an excess of rapidly mutating microsatellites (2) or an abundance of overactive SINEs (3), to create increased variability from which to select for new traits. Another theory suggests that domestication has allowed for the buildup of mildly deleterious mutations that, when combined, create the variation observed in the domestic dog (4). The notion of gene duplication as a major cause of morphologic diversity has received little attention.
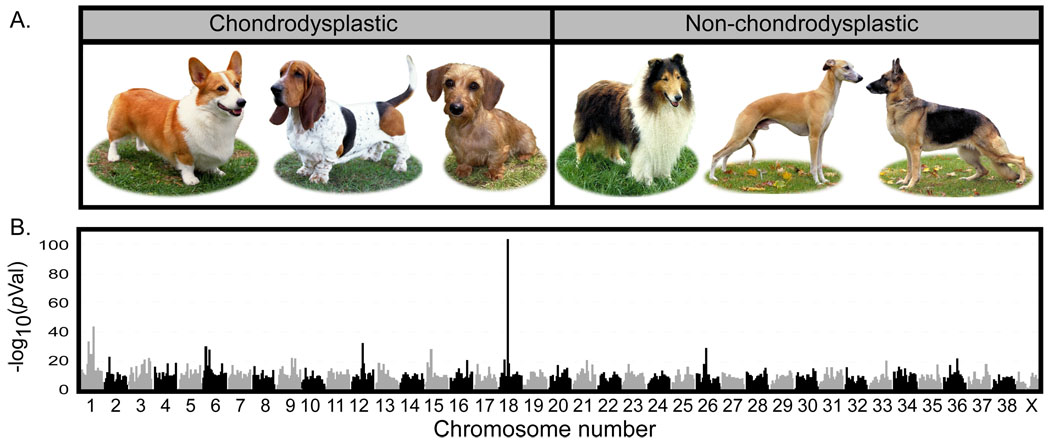
The majority of phenotypic variation in domestic dogs is found among, rather than within, the over 350 recognized domestic dog breeds. One aspect of interbreed variation is leg length, with some of the most striking short-legged breeds displaying limb morphology characteristic of chondrodysplasia, also known as short-limbed or disproportional dwarfism (Table S1). The trait is a primary requirement in the American Kennel Club (AKC) “breed standard” for over a dozen domestic breeds including the dachshund, Pekingese, and basset hound, where it was found to be dominant and allelic based on arranged crosses (5). The phenotype primarily affects the length of the long bones, with growth plates calcifying early in development, thus producing shortened bones with a curved appearance (Figure 1A) (6, 7).
Figure 1.
Results of a whole genome association analysis for chondrodysplasia across 72 breeds of dog. A). Examples of breeds used as cases (Pembroke Welsh corgi, basset hound, and dachshund pictured) and controls (collie, whippet, and German shepherd dog) in this analysis. B) Alternating shades of gray and black designate the chromosomal boundaries. The two highest peaks are found on chromosome 18 at bases 23,298,242 and 23,729,786 in CanFam2 assembly (www.genome.ucsc.edu). The peaks are less than .5 Mb apart and appear merged in the graph. Dog photos courtesy of Mary Bloom ©AKC.
In order to identify the genetic foundations of breed-defining phenotypes such as canine chondrodysplasia, we developed a multi-breed approach for mapping fixed canine traits. A total of 835 dogs from 76 distinct breeds that provided maximal coverage of phenotypic variation were genotyped using the Affymetrix version 2.0 SNP chip (8, 9). Chondrodysplastic breeds, or “cases”, were defined based on specific morphologic criteria set forth in each breed standard (8, 10) and comprised 95 dogs from eight breeds. The “control” or non-chondrodysplastic group included 702 dogs from 64 breeds lacking the above features (Figure 1A, Table S1).
Single marker analysis revealed a strong association (odds ratio (OR) =33.54) between a SNP on chromosome 18 (CFA18) at base position 23,298,242 (CanFam2) and the chondrodysplasia phenotype (χ2 = 437; p-value = 9×10−104 uncorrected; Figure 1B). The second best peak of association was found at position 23,729,786; 431 kb telomeric to the first, with a p-value of 2×10−57. Because the p-values are inflated due to population structure (4% of p-values < 10−7), we also performed independent Mann-Whitney U-tests on the distribution of allele frequencies within the chondrodysplastic and control breeds. The two SNPs on CFA18 retained the strongest association with p-values of 1.15×10−5 and 2.74×10−5, respectively. The best haplotype across the chromosome spanned the five SNPs beginning at position 23,298,242 and ending at position 23,729,786 (uncorrected p-value =1.9×10−111) (Table S1).
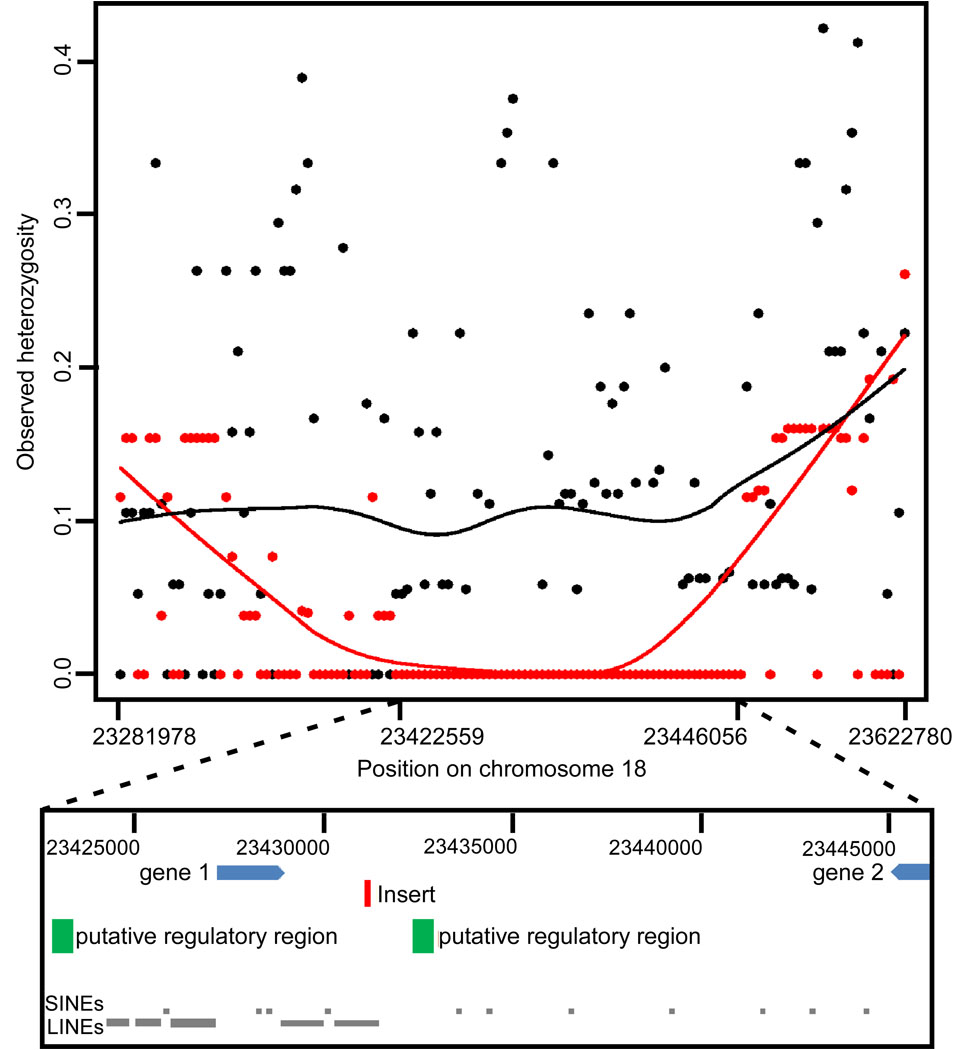
Because registered members of a breed are expected to meet specific morphologic criteria, we hypothesized that breed-defining traits such as chondrodysplasia would be under strong selective pressure. We compared heterozygosity in 139 cases and 173 controls genotyped at an additional 64 SNPs that spanned the associated region (Table S2) and observed 125 kb (23,320,831–23,445,875) in which the cases displayed considerably lower levels of heterozygosity than the controls, indicative of a selective sweep (case average = 1.9%, control = 19.6%, p=6×10−6, paired t-test), (11–14).
Fifty-four amplicons were sequenced in 44 dogs from 20 breeds (nine case and 11 control) with a goal of 1) identifying additional SNPs; 2) searching for causative mutations; and 3) finding the smallest haplotype shared among chondrodysplastic breeds (Table S3). Fifty of the 123 SNPs identified formed a single continuous homozygous haplotype in all 26 chondrodysplastic dogs tested, covering approximately 24 kb (23,422,559 to 23,446,056) (Figure 2A). A portion of the 3’UTR of semaphorin 3c (sema3c), a putative thioredoxin-domain containing 1 (txndc1) pseudogene, and two evolutionarily conserved sequences are contained within the shared haplotype (Figure 2B).
Figure 2.
Observed heterozygosity in chondrodysplastic (red) and non-chondrodysplastic (black) breeds within the associated region on chromosome 18. A) Graph of observed heterozygosity (Ho) across a 34 kb region on CFA18. Each point is the average Ho at one marker across all individuals within the group. The X axis shows the position on chromosome 18. The lines, red for chondrodysplastic and black for non-chondrodysplastic, show the trend in heterozygosity across the region by LOWESS (locally weighted least squares) best fit to the data. The average Ho for controls across the 24 kb homozygous region is 0.10. B) Schematic of the region that is homozygous and identical in chondrodysplastic breeds. Gene 1 is a pseudogene similar to thiorodoxin domain containing 1 (txndc1). Gene 2 is the 3’ end of semaphorin 3c (sema3c). The green boxes labeled putative regulatory regions are conserved in both sequence and context in all mammals for which genome data are available. A five kb insertion (red rectangle) was found within the fourth LINE between the two putative regulatory elements. The insertion contains an fgf4 retrogene. Arrangement of genes and conserved regions are per the CanFam2 assembly (www.genome.ucsc.edu).
An insert of approximately five kb starting at position 23,431,136 (Figure S1) was found by tiling PCR amplicons across the homozygous region. This insert was present in all dogs from the original eight breeds and 11 of 12 additional breeds that fit at least two of the three chondrodysplastic criteria (175 dogs from 19 breeds) (8). Seven of the 175 short-legged dogs were heterozygous for the insert (Table S4). The insert was not found in all 204 medium to long-legged dogs from 41 breeds that do not display the trait (Table S4).
Although the insertion was unambiguously associated with chondrodysplasia, the initial analysis did not address whether the position of the insert or its specific content was causative. We therefore sequenced the insert using an Illumina Genome Analyzer. A library was first created from a gel-extracted long-range PCR product that spanned the entire insert from two unrelated chondrodysplastic dogs (dachshund and Scottish terrier). The sequence data were assembled using Velvet algorithms (15). BLAT analysis (genome.ucsc.edu) revealed a single contig with complete alignment at 100% identity to fibroblast growth factor 4 (FGF4), which is located on CFA18 at position 51,439,516; approximately 30 Mb from the insert.
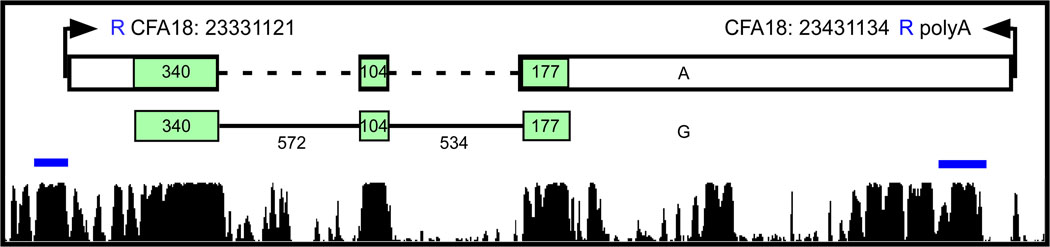
Using Sanger sequencing with primers designed from the annotated FGF4 gene sequence, together with the sequence surrounding the insertion site (Table S5), we were able to demonstrate that the insert contained a conserved fgf4 retrogene. Neither the introns nor the upstream promoter sequences of the gene were present in the insert, however all exons were present, with no alterations in the coding sequence, as well as the 3’ UTR and poly-A tail characteristic of retrotransposition of processed mRNA (Figure 3).
Figure 3.
Comparison of insert to source FGF4 gene. The first row on the figure displays the alignment of the insert sequence to the source FGF4 sequence. FGF4 has three coding exons represented by the green boxes on the graph and begins at CFA18 position 51439420 and ends at position 51441146. All three exons are present in the insert which aligns between positions 51439178 and 51442902. Tthe insert includes 242 bases upstream of the start site and 1756 bases downstream of the stop codon followed by a polyA repeat. A 13 base sequence (AAGTCAGACAGAG) derived from the insert site, indicated by a blue R on the figure, is repeated at both ends of the insert. The second line shows the coding sequence of FGF4 with the size of the exons and introns labeled. Alignment of the mouse promoter and enhancer sequences are indicated by the blue lines directly above the dog/human/mouse/rat conservation track shown at the bottom of the figure (www.genome.ucsc.edu; TRED: http://rulai.cshl.edu/cgi-bin/TRED/tred.cgi?process=home)(34). Coding sequence is predicted based on sequence similarity of translated proteins (accession # XM_540801).
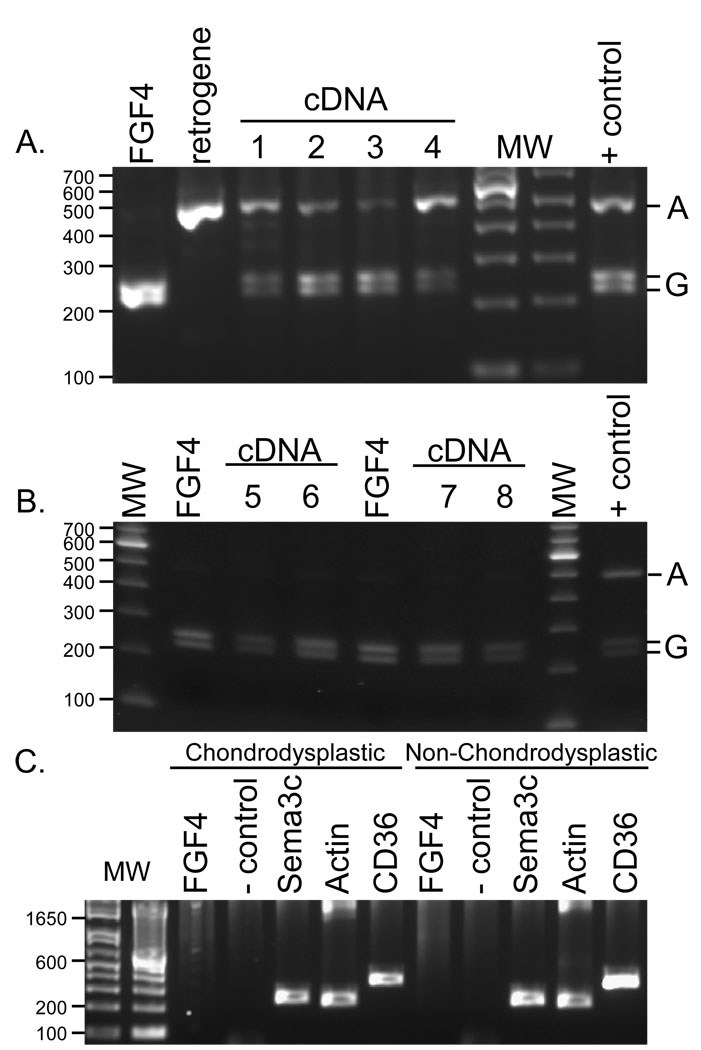
To determine if the retrogene was expressed we searched for retrogene specific sequences in complete cDNA of chondrodysplastic dogs. A single base at a position syntenic to chr18:51441601, 455 bp distal to the coding sequence of FGF4, differed between the retrogene and the source gene, with the former displaying an A nucleotide and the latter a G, in all samples tested. Both A and G alleles were observed in cDNA created from articular cartilage of the long bones of chondrodysplastic dogs (Figure 4A), while cDNA and genomic DNA samples were homozygous for the G allele in non-chondrodysplastic dogs as demonstrated in the restriction enzyme assay in Figure 4B.
Figure 4.
Restriction fragment length polymorphism genotyping of FGF4, the fgf4 retrogene, and the fgf4 transcript from chondrodysplastic dogs. A) A 505 bp fragment was amplified from gel extracted PCR products containing the fgf4 retrogene, the source FGF4 3’UTR, and from messenger cDNA created from articular cartilage of the distal humerus (lanes 1–2) and the proximal humerus (lanes 3–4) of a 4 week old chondrodysplastic dog. Each fragment was cut to completion with restriction enzyme BsrB1 and run on a 2% agarose gel. The cDNA shows alleles specific to both the source gene and the retrogene verifying expression of the latter. B) The same experiment was done on non-chondrodysplastic fetal dogs (a spaniel mix in lanes 5–6, hound mix in lanes 7–8). Lanes 5 and 7 are amplified from the cDNA from proximal tibia. Lanes 6 and 8 are from cDNA from distal femur. Genotypes from the source gene and cDNA are identical as no other copy of FGF4 is present. C) Genes were amplified in cDNA from articular cartilage from the proximal humerus in an adult chondrodysplastic (Shih Tzu) and non-chondrodysplastic (Siberian Husky) dog. Though RNA levels were low in these tissues, expression of CD36 and Sema3C were strong, but neither the source FGF4 nor the fgf4 retrogene could be detected.
Gene duplication through retrotransposition differs from a tandem duplication that may simply double the gene dosage (16) as the retrogene must acquire a new promoter, likely with a different expression profile, in order to be active. To accomplish this, retrogenes often borrow contextual regulatory elements (17). We therefore assessed the expression of thrombospondin receptor (CD36) and Sema3c, which are upstream and downstream of the insert. A PCR-based assay on cDNA from the articular cartilage of fetal and neonatal dogs revealed expression of both genes in the growing limb (Figure S2). Further examination of expression in cartilage tissues from adult dogs shows that though the surrounding genes were expressed, neither the source FGF4 gene nor the fgf4-retrogene were still expressed (Figure 3C), suggesting that the gene does not follow the expression pattern of its surroundings nor is it ubiquitously expressed and implying it has a specific time-sensitive role. The retrogene is inserted in the middle of a LINE with both LINEs and SINEs upstream (Figure 2B). These transposable elements likely provide the regulatory machinery necessary to promote expression of the fgf4 retrogene (18) with localization and temporal control coming from the intact 3’UTR (19).
We hypothesize that atypical expression of the FGF4 transcript in the chondrocytes may be causing inappropriate activation of one or more of the fibroblast growth factor receptors such as FGFR3. An activating mutation in FGFR3 is responsible for > 95% of achondroplasia cases, the most common form of dwarfism in humans, and 60–65% of hypochondroplasia cases, a human syndrome that is more similar in appearance to breed defining chondrodysplasia (reviewed in (20)). FGF4 has been shown to induce the expression of sprouty genes, which interfere with the ubiquitin mediated degradation of the FGF receptors including FGFR3, and over-expression of the sprouty genes can cause chondrodysplastic phenotypes in both mice and humans (21, 22).
The chondrodysplastic breeds were developed in many different countries for a variety of occupations (10). Based on genomic analysis of population structure, they do not share a recent common ancestry (23, 24). However, since we find a common haplotype of 24Kb surrounding the fgf4 retrogene in 19 short legged breeds it is likely the chondrodysplastic phenotype arose only once, before the division of early dogs into modern breeds. Thereafter, the retrogene and its associated phenotype were both maintained and propagated by breeders for purposes specific to each breed.
To further understand the origin of the fgf4 retrogene, we compared haplotypes from the source gene, the retrogene, and the insertion site in both dogs and their wild progenitor, the gray wolf. The ancestor of all chondrodysplastic breeds would have needed to carry both a source gene with the rare haplotype found in the retrogene, and the 24 Kb haplotype that defines the insertion site (Figure S3, Table S6). This combination was not found in any of the dogs that we tested but was identified in wolves from Europe and the Middle East, supporting fossil evidence that these populations contributed to the early development of the dog (25, 26).
Though retrogenes are recognized as an important source of novel functional elements found between recently diverged species (27–29), little is known about the relationship between retrotransposition and phenotypic variation within species (29, 30). We have found a single retrotransposition event producing a conserved, expressed retrogene that has strongly focused the evolutionary direction of morphological change in the dog, as at least 12% of American breeds share a common phenotype and the retrogene. This retrogene is actively segregating within the species, has a coding sequence that is identical to that of the source gene, and is the only example of a functional retrogene found in morphologically distinct populations of a single species that is actively maintained by selection. If such rare mutational events or “sports”, as Charles Darwin referred to them in The Origin of Species (31), happen only in the evolution of domestic animals, then these systems may be less informative for understanding the origin of evolutionary novelty in wild species. However, if the type of molecular phenomenon we have observed represents a class of genomic change associated with dramatic phenotypic evolution, such as that characteristic of adaptive radiation (17, 32, 33), then such genetic changes might be keystone molecular innovations.
Supplementary Material
Supporting Online Material
Materials and Methods
Figure S1, S2, S3
Tables S1, S2, S3, S4, S5
References
- 1.Wayne RK, Ostrander EA. Trends Genet. 2007 Nov;23:557. doi: 10.1016/j.tig.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 2.Fondon JW, 3rd, Garner HR. Proc Natl Acad Sci U S A. 2004 Dec 28;101:18058. doi: 10.1073/pnas.0408118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirkness EF, et al. Science. 2003;301:1898. doi: 10.1126/science.1086432. [DOI] [PubMed] [Google Scholar]
- 4.Cruz F, Vila C, Webster MT. Mol Biol Evol. 2008 Nov;25:2331. doi: 10.1093/molbev/msn177. [DOI] [PubMed] [Google Scholar]
- 5.Stockard CR. The Genetic and Endocrinic Basis for Differences in Form and Behavior. Philadelphia: The Wistar Institute of Anatomy and Biology; 1941. [Google Scholar]
- 6.Heuertz S, et al. Eur J Hum Genet. 2006;14:1240. doi: 10.1038/sj.ejhg.5201700. [DOI] [PubMed] [Google Scholar]
- 7.Palmer N. In: Pathology of Domestic Animals. Jubb KVF, Kennedy PC, Palmer N, editors. vol. 1. San Diego: Acedemic Press, Inc; 1993. p. 779. in. [Google Scholar]
- 8.Information on materials and methods is available online on Science Online.
- 9.Drogemuller C, et al. Science. 2008;321:1462. doi: 10.1126/science.1162525. [DOI] [PubMed] [Google Scholar]
- 10.American Kennel Club. The Complete Dog Book. ed. 19th Edition Revised. New York, NY: Howell Book House; 1998. Official Publication of the American Kennel Club; p. 790. [Google Scholar]
- 11.Simonsen KL, Churchill GA, Aquadro CF. Genetics. 1995;141:413. doi: 10.1093/genetics/141.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colby C, Williams SM. Genetics. 1995;140:1129. doi: 10.1093/genetics/140.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sutter NB, et al. Science. 2007 April 6;316:112. doi: 10.1126/science.1137045. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pollinger JP, Bustamante CD, Fledel-Alon A, Schmutz S, Gray MM, Wayne RK. Genome Res. 2005;15:1809. doi: 10.1101/gr.4374505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zerbino DR, Birney E. Genome Res. 2008;18:821. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salmon Hillbertz NHC, et al. Nat Genet. 2007;39:1318. doi: 10.1038/ng.2007.4. [DOI] [PubMed] [Google Scholar]
- 17.Vinckenbosch N, Dupanloup I, Kaessmann H. Proc Natl Acad Sci U S A. 2006 Feb 28;103:3220. doi: 10.1073/pnas.0511307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feschotte C. Nat Rev Genet. 2008 May;9:397. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fraidenraich D, Lang R, Basilico C. Dev Biol. 1998 Dec 1;204:197. doi: 10.1006/dbio.1998.9053. [DOI] [PubMed] [Google Scholar]
- 20.Horton WA, Lunstrum GP. Rev Endocr Metab Disord. 2002;3:381. doi: 10.1023/a:1020914026829. [DOI] [PubMed] [Google Scholar]
- 21.Minowada G, et al. Development. 1999 Oct;126:4465. doi: 10.1242/dev.126.20.4465. [DOI] [PubMed] [Google Scholar]
- 22.Guo C, et al. Cell Signal. 2008 Aug;20:1471. doi: 10.1016/j.cellsig.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parker HG, et al. Genome Research. 2007;17:1562. doi: 10.1101/gr.6772807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker HG, et al. Science. 2004 May 21;304:1160. [Google Scholar]
- 25.Germonpre M, et al. Journal of Archaeological Science. 2009 Feb;36:473. [Google Scholar]
- 26.Sablin MV, Khlopachev GA. Current Anthropology. 2002;43:795. [Google Scholar]
- 27.Brosius J. Genetica. 1999;107:209. [PubMed] [Google Scholar]
- 28.Marques AC, Dupanloup I, Vinckenbosch N, Reymond A, Kaessmann H. PLoS Biol. 2005 Nov;3:e357. doi: 10.1371/journal.pbio.0030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaessmann H, Vinckenbosch N, Long M. Nat Rev Genet. 2009 Jan;10:19. doi: 10.1038/nrg2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohno S. Semin Cell Dev Biol. 1999;10:517. doi: 10.1006/scdb.1999.0332. [DOI] [PubMed] [Google Scholar]
- 31.Darwin CR. The Origin of Species by Means of Natural Selection. London: John Murray; 1872. [Google Scholar]
- 32.Betran E, Bai Y, Motiwale M. Molecular Biology and Evolution. 2006;23:2191. doi: 10.1093/molbev/msl090. [DOI] [PubMed] [Google Scholar]
- 33.Rohozinski J, Bishop CE. Proc Natl. Acad Sci U.S.A. 2004;101:695. doi: 10.1073/pnas.0401130101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nowling T, Bernadt C, Johnson L, Desler M, Rizzino A. J Biol Chem. 2003 Apr 18;278:13696. doi: 10.1074/jbc.M207567200. [DOI] [PubMed] [Google Scholar]
- 35.We acknowledge NSF grants 0733033 (RKW) and 516310 (CDB), NIH grant 1R01GM83606 (CDB), and the Foundation for Fighting Blindness grant EY06855 (GMA). We thank Darcie Babcock and Catherine Degnin for technical assistance, Dr. James L. Cook for assistance with tissue identification and acquisition, Dr. Lee Niswander for sharing her insights regarding the role of FGF4 in limb development, and Dr. Edward Giniger for careful reading and suggestions regarding the manuscript. We gratefully acknowledge the dog owners who generously provided samples as well as the American Kennel Club-Canine Health Foundation, and the Affymetrix Corporation. Finally, we thank the Intramural Program of the National Human Genome Research Institute of NIH for their continued support.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Online Material
Materials and Methods
Figure S1, S2, S3
Tables S1, S2, S3, S4, S5