Abstract
Common variants in the transcription factor 7-like 2 (TCF7L2) gene have been identified as the strongest genetic risk factors for type 2 diabetes (T2D). However, the mechanisms by which these non-coding variants increase risk for T2D are not well-established. We used 13 expression assays to survey mRNA expression of multiple TCF7L2 splicing forms in up to 380 samples from eight types of human tissue (pancreas, pancreatic islets, colon, liver, monocytes, skeletal muscle, subcutaneous adipose tissue and lymphoblastoid cell lines) and observed a tissue-specific pattern of alternative splicing. We tested whether the expression of TCF7L2 splicing forms was associated with single nucleotide polymorphisms (SNPs), rs7903146 and rs12255372, located within introns 3 and 4 of the gene and most strongly associated with T2D. Expression of two splicing forms was lower in pancreatic islets with increasing counts of T2D-associated alleles of the SNPs: a ubiquitous splicing form (P = 0.018 for rs7903146 and P = 0.020 for rs12255372) and a splicing form found in pancreatic islets, pancreas and colon but not in other tissues tested here (P = 0.009 for rs12255372 and P = 0.053 for rs7903146). Expression of this form in glucose-stimulated pancreatic islets correlated with expression of proinsulin (r2 = 0.84–0.90, P < 0.00063). In summary, we identified a tissue-specific pattern of alternative splicing of TCF7L2. After adjustment for multiple tests, no association between expression of TCF7L2 in eight types of human tissue samples and T2D-associated genetic variants remained significant. Alternative splicing of TCF7L2 in pancreatic islets warrants future studies. GenBank Accession Numbers: FJ010164–FJ010174.
INTRODUCTION
Common genetic variants within the transcription factor 7-like 2 gene (TCF7L2, previously known as T-cell factor 4, or TCF-4) have been strongly associated with increased risk of type 2 diabetes (T2D) (1). The association with T2D and impaired function of pancreatic islets was confirmed in several genome-wide association studies (GWAS) (2–4) and follow-up association studies (5–23). TCF7L2 is a ubiquitous protein that belongs to a family of TCF/lymphoid enhancer factor (LEF)) transcription factors (3). Prior to being associated with increased T2D risk, TCF7L2 was known as an important component of the WNT pathway (24,25) and as a regulator of the proliferative cellular compartment in the intestine (26). Several hypotheses have been proposed to explain how the genetic variants in TCF7L2 could increase T2D risk. TCF7L2 might affect the ability of pancreatic islets to produce insulin in response to stimulation by insulinotropic intestinal hormone Glucagon-Like Peptide 1 (GLP-1) or glucose. This hypothesis was based on the observations that TCF7L2 could directly regulate mRNA expression of proglucagon precursor in-vitro (27) and that the deletion of Tcf7l2 gene in a mouse knock-out model resulted in loss of gut epithelial secretory cells that produce GLP-1 and other hormones (26). In support of this hypothesis, it was demonstrated that carriers of risk variants of TCF7L2 have impaired incretin effect (28) and impaired GLP-1 stimulated insulin secretion (16,29). It is also possible that TCF7L2 has pleiotropic tissue-specific roles and alterations in one or more of these functions could lead to T2D.
The most significant genetic T2D association within TCF7L2 was detected for two intronic single nucleotide polymorphisms (SNPs), rs7903146 and rs12255372, located 50 kb from each other within a 92 kb linkage disequilibrium (LD) block spanning exon 4 and parts of introns 3 and 4 (1–15). All exons of TCF7L2 were sequenced in multiple patients and controls but no coding variation was identified (1) suggesting that T2D risk is associated with a non-coding variation. Genetic variations may affect levels of expression and splicing architecture of mRNA transcripts (30,31) thus several studies have tested the relationships between TCF7L2 mRNA expression and genotypes of rs7903146 and rs12255372 in human tissues (28,32–38). However, these studies have been limited by the use of small sets of samples and tissue types. Alternative exons reported for TCF7L2 (39–41) represent a potentially important source of functional molecular variation, yet the tissue specificity of the alternative splicing and its relationship with genetic variation has not been fully established.
Here, we evaluated the splicing diversity and expression of TCF7L2 in a broad range of human tissues. We provide evidence that expression of alternative exons of TCF7L2 is tissue-specific. One splicing form was unique to pancreatic islets, pancreas and colon and not found in other tissues studied. Expression of this form correlated with proinsulin expression in glucose-stimulated pancreatic islets. Our study does not provide strong evidence for association of T2D-associated SNPs rs7903146 and rs12255372 with expression of alternatively spliced forms of TCF7L2 in several tissues but points out a potentially functional biological effect in pancreatic islets. Future studies in larger sets of samples should confirm and explore this finding.
RESULTS
Expression of assay ‘ex7–8’ of TCF7l2 in human tissues and test for association with rs7903146 and rs12255372
First, we measured the mRNA expression of TCF7L2 using an assay that targeted exons 7 and 8 of the gene (assay ‘ex7–8’, Fig. 1), and detected signals in all human tissues tested (Supplementary Material, Table S1). When normalized to expression of two housekeeping genes, β2 microglobulin (B2M) and Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), the highest levels of expression were found in pancreas, colon, brain, small intestine and peripheral blood monocytes. The lowest levels of expression were found in resting and activated peripheral blood T and B cells and lymphoblastoid cell lines (Fig. 2 and Supplementary Material, Table S1). Expression of TCF7L2, measured with the assay ‘ex7–8’ in 311 samples of pancreas, pancreatic islets, colon, liver, monocytes and subcutaneous adipose tissue from individuals without T2D, was not associated with genotypes of the two most strongly T2D-associated SNPs, rs7903146 and rs12255372 (Table 1). On the basis of the number of samples and standard deviation in expression of assays in each tissue tested, we estimated the detectable fold difference between the groups of samples carrying the risk and non-risk alleles (Table 1). In this analysis, we had 80% power (with a type I error rate of 5%) to detect a 1.13-fold difference in expression in colon, a 1.33-fold difference in liver, a 1.37-fold difference in monocytes, 2.06-fold in skeletal muscle, a 2.34-fold difference in pancreatic islets and a 2.87-fold difference in adipose tissue (Table 1).
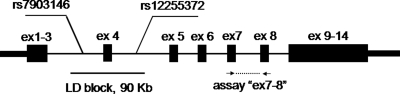
Figure 1.
Location of expression assay ‘ex7–8’ and associated LD block with SNPs rs7903146 and rs12255372 within TCF7L2 gene, exons are marked as black rectangles.
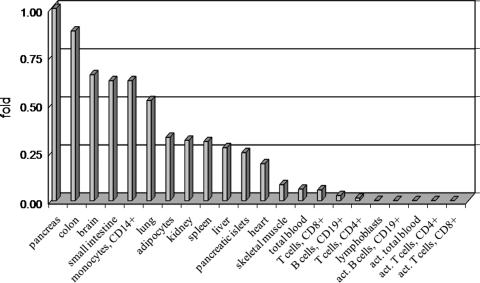
Figure 2.
Expression of assay ‘ex7–8’ of TCF7L2 in human tissues as fold difference compared with expression level in pancreas. TCF7L2 expression is normalized to the levels of endogenous controls B2M and GAPDH (Supplementary Material, Table S1). Each tissue is represented by 1 or 2–3 pooled samples.
Table 1.
Estimated detectable differences in expression of assay ‘ex7–8’ of TCF7L2 in genotype groups of rs7903146 and rs12255372 in human tissue samples
| Tissue, number of samples | Detectable difference, 80% power, folda | Detectable difference, 95% power, folda | rs7903146, P-valueb | rs12255372, P-valueb |
|---|---|---|---|---|
| Pancreatic islets (n = 24) | 2.34 | 3.01 | 0.746 | 0.198 |
| Pancreas (n = 42) | 1.48 | 1.91 | 0.171 | 1.000 |
| Colon (n = 81) | 1.13 | 1.46 | 0.538 | 0.509 |
| Liver (n = 62) | 1.33 | 1.71 | 0.998 | 0.857 |
| Monocytes (n = 63) | 1.37 | 1.78 | 0.550 | 0.595 |
| Skeletal muscles (n = 25) | 1.60 | 2.06 | 0.749 | 0.049 |
| Subcutaneous adipose tissue (n = 14) | 2.87 | 3.70 | 0.523 | 0.539 |
aMinimal detectable difference in expression between groups of samples (with and without risk alleles), based on empirical standard deviation within each group and 5% of type 1 error.
bObserved P-values, univariate linear model.
Splicing diversity of TCF7l2
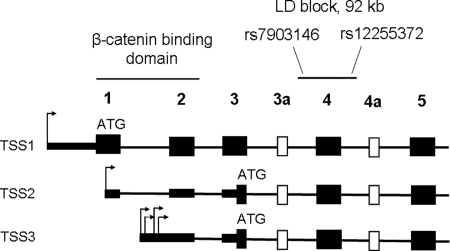
Since our analysis did not reveal any association between expression of TCF7L2 measured by assay ‘ex7–8’ and SNPs rs7903146 and rs12255372, we hypothesized that these genetic variants might be associated with alternative splicing. Members of the TCF/LEF family of transcription factors use alternative transcription start sites (TSSs) to produce functionally distinct proteins (42–44). To identify all TSS of TCF7L2, we performed 5′ rapid amplification of cDNA ends (5′RACE) in an RNA pool of eight tissues (pancreas, pancreatic islets, colon, liver, monocytes, skeletal muscles, subcutaneous adipose tissue and lymphoblastoid cell lines). We identified several TSS: (1) TSS1, located at −536 bp from the first translation start site and corresponding to RefSeq sequence (NM_030756); (2) TSS2, located at position +83 bp within exon 1; (3) a cluster collectively referred as TSS3 located in the first intron of the gene (+200, +239, +302, +338 bp, Fig. 3 and Supplementary Material, Table S2). Only transcripts initiated from the TSS1 can be translated from a start codon within exon 1 and produce protein isoforms with the ß-catenin binding domain encoded by exons 1 and 2. The next in-frame translation start site that can be used by transcripts expressed from the TSS2 and TSS3, is located within exon 3 (Fig. 3), and the resulting proteins would lack the ß-catenin binding domain. Both the main (exon 1) and the alternative translation start sites (exon 3) have weak Kozak consensus sequences, ‘aaaaaaATGC’ and ‘ttcatcATGA’, compared with a classical consensus site ‘gcca/gccATGG’ (45). We also performed 3′RACE in an attempt to detect alternative splicing forms of the 3′-UTRs. We did not detect any short PCR fragments corresponding to mRNA transcripts with significantly truncated 3′-UTR (data not shown). We PCR amplified, cloned and sequenced full-length cDNA fragments starting from the first translation start site within exon 1 through the end of the transcript in exon 14 (based on RefSeq NM_030756). We did not attempt to clone full-length cDNAs initiated from the TSS2 because this transcription start is embedded within exon 1 of the TSS1 form and the products would represent a mixture of transcripts. Because of low expression from the TSS3, further study of the transcripts initiated from this TSS was not pursued in detail. Sequencing of full-length cDNA clones reconfirmed several known alternative exons (3a, 4a, 12, 13, 13a and 13b) and minor in-frame inclusions of 3, 12 and 15 bp in exons 4a, 6 and 8, respectively (39–41) (Supplementary Material, Table S2). All sequences for full-length cDNA expression constructs were deposited in the GenBank (FJ010164–FJ010174, Supplementary Material, Table S3).
Figure 3.
Structure of the N-terminal part of TCF7L2: location of the β-catenin binding domain encoded by exons 1 and 2, and a T2D-associated LD block with SNPs rs7903146 and rs12255372. Shown: constitutive exons—black rectangles, alternative exons—white rectangles; alternative transcription starts sites TSS1, TSS2 and TSS3—arrows, potential translation starts—‘ATG’.
Tissue-specific expression of TCF7l2
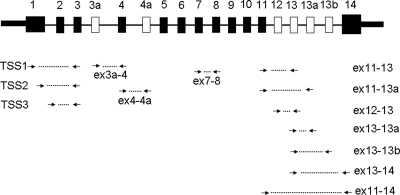
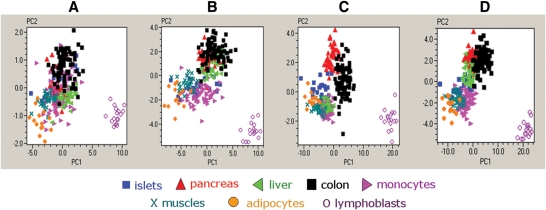
We designed 13 expression assays (Fig. 4) that targeted the majority of the TCF7L2 splicing forms, we observed in human tissues. Expression of these assays was quantified in samples from pancreas, pancreatic islets, colon, liver, monocytes, skeletal muscles, subcutaneous adipose tissue and lymphoblastoid cell lines. Expression of an alternative exon 13b (assay ‘ex13–13b’) was detected only in pancreatic islets, pancreas and colon and this assay was not used in the analysis that included all tissue samples. The expression of assay ‘ex4–4a’ was low in all tissues and was not studied further. Supplementary Material, Fig. S1 illustrates the diversity in expression of each assay in human tissues. We performed Principal Component Analysis (PCA) on samples from eight tissues using four sets of expression assays (Fig. 5A–D). We observed better discrimination between pancreatic islets and other tissues based on expression of assays for C-terminal alternative exons 12, 13, 13a and 14 of TCF7L2 (Fig. 5C) than those of assays detecting the N-terminal exons (Fig. 5A and B) or all expression assays together (Fig. 5D and Supplementary Material, Table S4 for relative expression of each splicing form in human tissues).
Figure 4.
Location of TaqMan expression assays within TCF7L2 gene. Shown: constitutive exons—black rectangles, alternative exons—white rectangles, expression assays—connected arrows.
Figure 5.
PCA in 289 samples from 8 human tissues. (A) PCA based on three N-terminal expression assays of TCF7L2: ‘TSS1’, TSS2’ and ‘ex7–8’. (B) PCA based on five N-terminal expression assays for TCF7L2: ‘TSS1’, ‘TSS2’, ‘TSS3’, ‘ex3a’ and ‘ex7–8’. (C) PCA based on six C-terminal expression assays for TCF7L2: ‘ex11–13’, ‘ex11–13a’, ‘ex11–14’, ‘ex12–13’, ‘ex13–13a’ and ‘ex13–14’. (D) PCA based on all eleven expression assays for TCF7L2.
Test for association between expression of alternatively spliced forms of TCF7l2 and rs7903146 and rs12255372
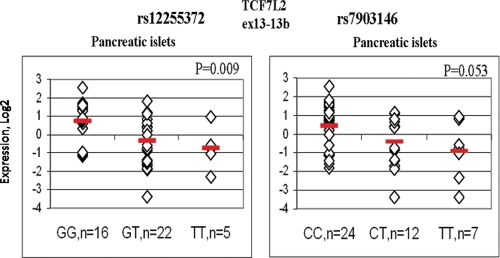
In each tissue, we used all available covariates (age, sex, BMI, etc.) to identify factors that could affect TCF7L2 expression (Supplementary Material, Tables S5A–G). Since the covariates were not available for all the samples and in the absence of strong and consistent effects of any of these factors, they were not used in the expression analysis. In pancreatic islets of non-T2D individuals, increasing counts of T2D-associated risk alleles of rs12255372 were associated with lower expression of several assays targeting C-terminal alternative exons of TCF7L2 in up to 26 samples of human pancreatic islets (with a significance for univariate linear model P = 0.002 for assay ‘ex13–14’, P = 0.003 for assay ‘13–13b’, P = 0.010 for assay ‘ex12–13’ and P = 0.022 for assay ‘ex13–13a’ (Supplementary Material, Table S6A). In a follow-up study, we performed expression analysis for seven assays detecting C-terminal exons of TCF7L2 in up to 24 additional samples of pancreatic islets (Supplementary Material, Table S6B). In the joint analysis of up to 50 samples, expression of assays ‘ex13–14’ and ‘ex13–13b’, adjusted for sample set, remained lower in samples with increasing counts of T2D-associated risk alleles: for assay ‘ex13–14’ P = 0.018 for rs12255372 and P = 0.020 for rs7903146 (Table 2) and for assay ‘ex13–13b’ P = 0.009 for rs12255372 and P = 0.053 for rs7903146 (Table 2 and Fig. 6).
Table 2.
Sample characteristics and T2F7L2 expression by genotypes of SNPs in pancreatic islets
| Traits, expression assays | rs7903146 |
rs12255372 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotypes | n | Mean | Fold | P-value* | Genotypes | n | Mean | Fold | P-value* | |
| BMI, kg/m2 | CC | 23 | 26.40 | ref | 0.812 | GG | 18 | 26.96 | ref | 0.562 |
| TT | 17 | 26.56 | 1.00 | TG | 24 | 26.13 | 0.97 | |||
| TT | 6 | 27.45 | 1.04 | TT | 4 | 27.80 | 1.03 | |||
| Age, years | CC | 23 | 48.37 | ref | 0.510 | GG | 18 | 51.77 | ref | 0.328 |
| TC | 17 | 47.68 | 0.99 | TG | 24 | 46.19 | 0.89 | |||
| TT | 6 | 55.50 | 1.15 | TT | 4 | 53.93 | 1.04 | |||
| Islets purity, % | CC | 22 | 76.95 | ref | 0.308 | GG | 17 | 74.02 | ref | 0.953 |
| TC | 16 | 67.94 | 0.88 | TG | 23 | 72.67 | 0.98 | |||
| TT | 6 | 72.19 | 0.94 | TT | 4 | 70.82 | 0.96 | |||
| ex11–13a | CC | 23 | −8.52 | ref | 0.171 | GG | 17 | −8.25 | ref | 0.502 |
| TC | 15 | −8.17 | 1.27 | TG | 23 | −8.59 | 0.79 | |||
| TT | 6 | −9.22 | 0.62 | TT | 4 | −8.96 | 0.61 | |||
| ex12–13a | CC | 21 | −7.45 | ref | 0.180 | GG | 15 | −7.17 | ref | 0.046 |
| TC | 14 | −7.21 | 1.18 | TG | 21 | −7.44 | 0.83 | |||
| TT | 5 | −8.46 | 0.50 | TT | 4 | −8.96 | 0.29 | |||
| ex13–13aa | CC | 24 | −9.53 | ref | 0.073 | GG | 18 | −9.29 | ref | 0.234 |
| TC | 15 | −9.23 | 1.23 | TG | 23 | −9.64 | 0.78 | |||
| TT | 6 | −10.79 | 0.42 | TT | 4 | −10.71 | 0.37 | |||
| ex11–13aa | CC | 20 | −6.81 | ref | 0.230 | GG | 14 | −6.57 | ref | 0.376 |
| TC | 14 | −6.72 | 1.06 | TG | 22 | −7.03 | 0.73 | |||
| TT | 6 | −7.56 | 0.59 | TT | 4 | −7.21 | 0.64 | |||
| ex11–14a | CC | 23 | −6.20 | ref | 0.156 | GG | 17 | −6.01 | ref | 0.559 |
| TC | 15 | −5.98 | 1.16 | TG | 23 | −6.35 | 0.79 | |||
| TT | 6 | −7.01 | 0.57 | TT | 4 | −6.57 | 0.68 | |||
| ex13–14a | CC | 23 | −6.52 | ref | 0.020 | GG | 17 | −6.15 | ref | 0.018 |
| TC | 15 | −6.65 | 0.91 | TG | 23 | −7.05 | 0.54 | |||
| TT | 6 | −8.13 | 0.33 | TT | 4 | −7.98 | 0.28 | |||
| ex13–13ba | CC | 24 | −10.90 | ref | 0.053 | GG | 16 | −10.48 | ref | 0.009 |
| TC | 12 | −11.56 | 0.63 | TG | 22 | −11.67 | 0.44 | |||
| TT | 7 | −12.24 | 0.40 | TT | 5 | −12.23 | 0.30 | |||
aExpression of assays, normalized to expression level of reference gene B2M, Log 2 scale, mean values are adjusted for sample set but not for multiple tests.
*P-values for univariate linear model under an additive genetic model; in bold P-values <0.05.
Figure 6.
Expression of assay ‘ex13–13b’ in human pancreatic islets by rs12255372 and rs7903146 genotypes. Normalized expression of assay ‘ex13–13b’ is shown in Log 2 scale relative to a mean value of the whole set. Association was tested with univariate linear regression under an additive genetic model for SNPs and with adjustment for two sample sets.
In pancreas (n = 46), colon (n = 128), monocytes (n = 65), liver (n = 62) and adipose tissue (n = 14), expression of TCF7L2 splicing forms was not associated with rs7903146 or rs12255372 (Supplementary Material, Table S6C–G). In skeletal muscle (n = 25), expression of several assays of TCF7L2 was significantly increased in carriers of risk alleles of rs12255372 (P = 0.049–0.005, Supplementary Material, Table S6H) but the effect was not proportional to the counts of risk alleles.
Test for association of expression of metabolic/endocrine factors with genotypes of rs7903146 and rs12255372
The counts of T2D-associated risk alleles of rs12255372 and rs7903146 were not associated with mRNA expression of several metabolic/endocrine factors important for T2D and expressed in pancreatic islets, pancreas or colon, such as proinsulin, proglucagon, ghrelin/obestatin, somatostatin, pancreatic and duodenum homeobox 1 (PDX1), proliferating cell nuclear antigen (PCNA) and vasoactive intestinal peptide (VIP) (Supplementary Material, Table S6A, C, D).
Test for correlation between expression of proinsulin and TCF7L2 assays in human pancreatic islets
We also correlated the expression of proinsulin and TCF7L2 assays in 23 samples of human pancreatic islets in unstimulated conditions and in 12 samples incubated for 24 h in the media with normal glucose concentration (5.5 mm) and with high glucose concentration (16.7 mm), corresponding to glucose blood level of T2D individuals. Expression of proinsulin most strongly correlated with expression of assay ‘ex13–13b’ both in 5.5 mm glucose condition, r2 = 0.90, P < 0.0001 and in 16.7 mm glucose condition, r2 = 0.84, P = 0.00063, but not in unstimulated samples (Table 3). Limited by small sample set of pancreatic islets (n = 12), we did not address affects of each of SNPs on glucose-stimulated expression.
Table 3.
Correlation between mRNA expression of proinsulin and TCF7L2 in pancreatic islets
| TCF7L2 assay | Unstimulated, n = 23 |
Stimulated 5.5 mm glucose, n = 12 |
Stimulated 16.7 mm glucose, n = 12 |
|||
|---|---|---|---|---|---|---|
| r2 | P-value | r2 | P-value | r2 | P-value | |
| TSS3 | −0.296 | NS | 0.14 | NS | 0.17 | NS |
| Ex3a–4 | −0.407 | NS | 0.35 | NS | 0.29 | NS |
| Ex11–13 | −0.333 | NS | 0.61 | 0.035 | 0.57 | NS |
| Ex11–13a | −0.481 | 0.020 | 0.46 | NS | 0.29 | NS |
| Ex11–14 | −0.436 | 0.038 | 0.63 | 0.028 | 0.46 | NS |
| Ex12–13 | −0.021 | NS | 0.73 | 0.007 | 0.54 | NS |
| Ex13–13a | −0.158 | NS | 0.69 | 0.013 | 0.46 | NS |
| Ex13–14 | −0.292 | NS | 0.53 | NS | 0.44 | NS |
| Ex13–13b | 0.189 | NS | 0.90 | <0.0001 | 0.84 | 0.00063 |
r2—Pearson correlation coefficient, NS—not significant, in bold—significant correlations, P < 0.05; Glucose-stimulated islets were incubated in 5.5 or 16.7 mm glucose media at 37°C for 24 h.
DISCUSSION
Family-based linkage studies identified a region on chromosome 10q25.3 as a locus associated with T2D and fine-mapping studies within this region identified TCF7L2 as a candidate gene for the association (1,46,47). Recent GWAS and follow-up studies confirmed several genetic variants within the TCF7L2 gene as the strongest risk factors for development of T2D (2–15) and impaired function of pancreatic islets (16–22). As the first step towards understanding the biology of this genetic association, we performed studies on splicing and mRNA expression of TCF7L2 in human tissues. We show here: (1) the expression of TCF7L2 alternative exons represents a tissue-specific signature; (2) a unique splicing form of TCF7L2 is expressed in pancreatic islets, pancreas and colon but not in other tissues examined here; (3) expression of this splicing form correlates with expression of proinsulin in glucose-stimulated pancreatic islets; (4) expression of alternatively spliced forms of TCF7L2 in eight human tissues examined here is not associated with T2D-associated risk variants rs12255372 and rs7903146.
Non-coding genetic variations have been shown to increase disease risk by altering levels of mRNA expression and splicing architecture of mRNA transcripts (30,31). In this study, we identified several types of potentially functional splicing variations within TCF7L2: alternative TSSs, alternative exons 3a and 4a encompassing the associated LD block; and alternative C-terminal exons 12, 13, 13a and 13b. Alternative TSSs can generate mRNA transcripts encoding proteins with opposing biological functions such as proliferation versus differentiation as described for LEF/TCF, c-Myc and p73 proteins (48–50). In the case of TCF7L2, the alternative promoters produce protein isoforms that can activate or repress the WNT pathway dependent on the presence of β-catenin binding domain encoded by exons 1 and 2 (51). Alternative exons 3a and 4a are located within a protein region responsible for interaction with a Groucho repressor protein (52). The combinatorial tissue-specific inclusion of alternative C-terminal exons 12, 13, 13a and 13b can activate alternative stop codons within exons 13a, 13b or 14 and create protein isoforms with short, medium and long reading frames (39,41). The function of these protein isoforms is unclear; however, only the long isoforms have binding sites for the C-terminal binding protein (CTBP), a negative regulator of the WNT pathway (53–55). Therefore, all alternative forms of TCF7L2 could be functionally important. However, the splicing variations tested here were not significantly associated with T2D-associated risk variants rs12255372 and rs7903146 in the human tissues tested.
A potentially biologically important observation in this study, though not strongly supported by statistics after adjustment for multiple tests, was a decrease in expression of two expression assays of TCF7L2, ‘ex13–14’ and ‘ex13–13b’, in pancreatic islets of carriers of T2D-associated risk variants rs12255372 and rs7903146. This observation is in line with a notion that depletion of TCF7L2 in pancreatic islets leads to increased apoptosis, decrease in proliferation and glucose-induced insulin secretion (56) found in carriers of risk variants of TCF7L2 (19–21). The assay ‘ex13–14’ detects a splicing form that encodes a protein with long reading frame and a CRARF protein domain formed upon splicing of exons 13 and 14. The CRARF domain is a potent activator of the WNT pathway and is the most conserved part of the TCF7L2 protein in all species (54,57). The assay ‘ex13–13b’ detects the most tissue-specific splicing form of TCF7L2 expressed in pancreatic islets, colon and at a lower level in pancreas but not in multiple samples representing liver, skeletal muscles, adipocytes, monocytes or lymphoblastoid cell lines tested here. Expression of this form in pancreatic islets was ∼30-fold higher than in pancreas (Supplementary Material, Fig. S3). In pancreatic islets cultured at normal (5.5 mm) or high glucose (16.7 mm) conditions, mRNA expression of proinsulin most strongly correlated with expression of assay ‘ex13–13b’ (r2 = 0.84–0.90, P < 0.00063), but not with other assays of TCF7L2. Perhaps, the splicing form detected by this assay marks a specific glucose-sensing or secretory cell type within pancreatic islets. Limited by a small sample set of glucose-induced islets (n = 12), we did not address effects of these SNPs on correlation with insulin expression. We did not observe association between counts of risk alleles of rs7903146 and rs12255372 and mRNA levels of proinsulin, proglucagon (the precursor of the insulinotropic hormone GLP-1) and several other metabolic/endocrine factors in pancreatic islets, pancreas and colon but this does not exclude the possibility that protein expression and secretion of these factors may be affected in carriers of the TCF7L2 risk variants.
The reports on association of TCF7L2 expression in human pancreatic islets and genotypes of T2D-associated variants within TCF7L2 are few. Lyssenko et al. (28) used the assay ‘ex7–8’ of TCF7L2 in 15 samples of human pancreatic islets and reported a significantly higher expression in carriers of rs7903146 risk genotypes. Osmark et al. (38) addressed the expression of several splicing forms in a set of 17 pancreatic islets but did not observe any association between expression of several TCF7L2 assays and genotypes of rs7903146. For several assays that targeted the same splicing forms (‘ex3a–4’, ‘ex11–14’, ‘ex11–13’, ‘ex11–13a’) we see the same results as reported by Osmark et al. The assays ‘ex13–14’ and ‘ex13–13b’ whose expression showed some evidence for association with T2D-associated variants of TCF7L2 in our study have not been previously used in any tissue. Additionally, we used larger sample sets for each tissue and for the first time studied expression of TCF7L2 in tissues such as pancreas, peripheral blood monocytes and colon.
We recognize the limitations of the approaches used in this study. Human tissue samples consist of multiple cell types that can exhibit diverse pattern of TCF7L2 expression. Tissue biopsies were taken during surgeries preceded by fasting and stress, thus these samples may not represent optimal physiological conditions for studies on T2D-related phenotypes. We attempted to identify and quantify the majority of splicing variation within TCF7L2. However, the analysis did not cover several minor splicing variations and yet unknown splicing forms of TCF7L2 that can exist and contribute to disease. The P-values for association between the counts of T2D-associated alleles and TCF7L2 expression were not adjusted for multiple testing. A Bonferroni correction would be overly stringent given the correlation between the 13 assays within each tissue. Our goal was to identify potential biologically important differences that should be tested in other sets of samples and with additional methods. We understand that with relatively small sample sets representing each tissue type, some of our results might be false-positives or false-negatives. Larger numbers of samples from each tissue type and, particularly, pancreatic islets, would increase the power to detect association between TCF7L2 expression and TCF7L2 SNP variants. Risk variants of TCF7L2 SNPs might be associated with several types of molecular changes relevant for T2D and our study addressed only one of these types, mRNA expression of alternative splicing forms. It is possible that there might be other important molecular phenotypes of TCF7L2 yet to be identified and examined. The C-terminal exons of TCF7L2 are located >100 kb away from the associated LD block. How the associated DNA variants can affect the mRNA expression and splicing at this distance, is not yet understood. A potential mechanism could be related to long-distance co-transcriptional regulation of mRNA expression and splicing (58,59).
In conclusion, our findings provide new evidence of tissue-specific patterns of alternative splicing of TCF7L2. Moreover, we provide the first suggestive evidence that decreased expression of one ubiquitous and one rare tissue-specific alternative splicing forms of the gene in pancreatic islets may be associated with T2D-associated genetic variants of TCF7L2. Future studies should confirm and explore the observed association between genetic variation within TCF7L2, tissue-specific alternative splicing and risk of T2D.
MATERIALS AND METHODS
Tissue samples
Purified human pancreatic islets in the US set were obtained from the Islet Cell Resource Centers (IRB exemption number 3072) and the National Disease Research Interchange (IRB exemption number 3269) under IRB exemption by the National Institutes of Health Office of Human Subjects Research. Pancreatic islets were provided by Washington University, St. Louis, MO, USA; City of Hope National Medical Center, Duarte, CA, USA; University of Miami, Miami, FL, USA and the Joslin Diabetes Center, Boston, MA, USA. Information on age, sex, BMI, islets purity and viability, duration of cold ischemic time (time between pancreas removal and islet preparation), cause of death, T2D status and CMV infection was available for these samples. Pancreatic islets in the Swedish set were provided by the Nordic Network for Clinical Islet Transplantation by the courtesy of Dr Olle Korsgren at Uppsala University, Sweden and Dr Jalal Taneera at the Human Tissue laboratory at Lund University Diabetes Centre, Sweden.
Human pancreas, colon and liver samples were biopsies obtained from the University of Minnesota Tissue Procurement Center, Minneapolis, MN, USA, and Children's Hospital, Boston, MA, USA. For the majority of these samples clinical reports indicating primary diagnosis, T2D status, age, sex and BMI were available. All tissue biopsies were non-malignant tissues as confirmed by pathology reports. Skeletal muscles biopsies were collected from healthy individuals and patients with Duchenne or Becker Muscular dystrophy at the Research Center for Genetic Medicine, Children's National Medical Center, Washington, DC, USA. Information about age, sex, BMI and disease status was available for these samples. Samples of fresh blood were from anonymous human blood donors and information about age and sex of the subjects were provided by the NIH Clinical Center Division of Transfusion Medicine. Monocytes were separated from fresh blood with CD14+-coated magnetic MicroBeads with AutoMacs (Miltenyi Biotec). Frozen samples of human subcutaneous adipose tissue were purchased from Zen-Bio, Inc. and information on age, sex, BMI and T2D status was available for these samples. IRB approval for de-identified tissues was obtained from all Institutions involved. All samples with known T2D were excluded (five samples in the whole set of all tissues). 293T (human embryonic kidney), HeLa (human cervical carcinoma), HepG2 (human hepatocellular carcinoma) were purchased from ATCC (the American Type Culture Collection) and lymphoblastoid cell lines from CEPH (Centre d' Etude du Polymorphisme Humain) individuals were purchased from Coriell cell repositories.
Glucose-stimulated pancreatic islets
Twelve samples from the US set were split in six aliquots each. Three aliquots were incubated in CMRL-1066 cell culture media with 5.5 mm glucose while other three aliquots of the same samples were incubated with CMRL-1066 cell culture media with 16.7 mm glucose for 24 h.
Preparation of DNA and RNA
DNA from tissue samples was prepared with DNeasy Kit (Qiagen) except for adipose tissue where DNA was recovered after RNA preparation according to Trizol protocol (Invitrogen). RNA was prepared with Trizol (Invitrogen) followed by RNeasy kit (Qiagen). RNA from the second set of pancreatic islets RNA was isolated using the AllPrep DNA/RNA Mini Kit (Qiagen). No sign of RNA degradation was observed using agarose gel electrophoresis and Experion DNA 1 K gel chips (Bio-Rad). RNA samples from all additional human tissues and cDNA from purified blood fractions (Multiple Tissue cDNA panels) were purchased from Clontech.
Genotyping
Genotyping was performed with an allelic-discrimination TaqMan assays for rs12255372 and rs7903146 and the Universal Genotyping Master Mix (Applied Biosystems). Alternatively or as an additional control, genotyping was performed by primer extension method using a Sequenom MALDI-TOF platform (Broad Institute).
cDNA preparation and qPCR
cDNA was prepared in a 20 µl volume with 500 ng of DNase-treated total RNA with use of random hexamers and Superscript III reverse transcriptase (Invitrogen) (set 1) or with the Qiagen Omniscript RT kit with 1 µm dT18 oligomer and 3 µm random hexamer primers (set 2 (38)). DNA was diluted with water to 200 µl and an equivalent of 5–10 ng of total RNA was used for each reaction (except for lymphoblastoid cell lines where 2 µg of total RNA was used as a starting amount). All reactions were run in three or four technical replicates. Standard deviations of below 0.2 Ct between the replicates were acceptable for high-expressing assays and 0.5 Ct for low-expressing assays. Endogenous controls B2M (ß-2 microglobulin) or GAPDH (Glyceraldehyde-3-phosphate dehydrogenase) were measured from the same cDNA preparations with expression assays Hs00187842_m1 for B2M and 4333764F for GAPDH (Applied Biosystems). All primers for custom-designed assays were created to amplify only from cDNA and TaqMan probes were placed on the junctions between specific exons (Supplementary Material, Table S7 for primer sequences and assay information). All quantitative PCR reactions were performed in a 10 µl volume using the 7900 Real-Time PCR System (ABI) in 384 well plates. Universal Expression Master Mix was used for all expression studies with TaqMan probes according to standard conditions (Applied Biosystems). The reactions with 2×SYBR Green PCR mix (Qiagen) and specific primers were carried out for 15 min at 95°C, followed by 40 cycles of 15 s at 95°C, 15 sec at 59°C and 45 s at 72°C. Post-PCR melting curve analysis was used after each run. Initially, gel-purified PCR fragments were sequenced to ensure the specificity of amplification and correct splicing.
Data analysis
Values from three to four technical replicates for each expression assay were averaged and normalized to the expression levels of BM2 and GAPDH run in separate reactions from the same cDNA preparation. Relative differences in expression were calculated according to formula: dCt = Ctreference − Cttarget, where ‘reference’ represents levels of expression (Ct values) of B2M, GAPDH or both genes and ‘target’ represents level of expression of TCF7L2 assays; negative Ct means that a target is expressed lower than a reference gene; fold difference in expression between a reference and a target can be calculated as 2(dCt). Within each tissue and assay, the log-transformed expression values were tested for normality. The associations between specified variables were tested using univariate linear model with inclusion of covariates where indicated (SPSS 16.0 software). The SNP-TCF7L2 expression or SNP-T2D related gene expression analyses were used according to an additive genetic model. The PCA analysis on mean-centered expression values for selected assays was performed with GenEx software (MultiD). We estimated the fold-difference in gene expression between samples with and without the T2D associated risk genotype that could be detected with 80 or 95% power with a type 1 error rate of 5%. For the power calculations for assay ‘ex7–8’ have been performed with observed standard deviation for this assay in each tissue and the number of samples with and without T2D associated risk genotypes (StatMate software, GraphPad). The power analysis differs from our regression analysis of gene expression and allele count, but is a good approximation when the less frequent genotype homozygote samples are rare.
For each sample of glucose-stimulated pancreatic islets, expression values of technical triplicates were averaged and then normalized to expression of endogenous control, B2M. The normalized values were averaged for three aliquots of the same samples. Therefore, for each sample and experimental condition expression was measured nine times. The Pearson correlation coefficients between expression of TCF7L2 assays and proinsulin were calculated with SPSS 16.0.
5′RACE and analysis of splicing forms
5′RACE cDNA from multiple tissues was prepared from 200 to 500 ng of total RNA with use of the BD SMART RACE cDNA amplification kit (Clontech). PCR fragments were amplified with universal primers included in the kit and several gene-specific specific primers. PCR fragments were gel-purified (Qiaquick, Qiagen) and sequenced with BD Terminator chemistry (Applied Biosystems). The full-length cDNA fragments from the first ATG translation start site in exon 1 and to the most distant stop codon within exon 14 were amplified by a proof-reading KOD polymerase (Novagen), cloned into pFC8A vector (Promega) and multiple individual clones were sequenced. Expression constructs for TCF7L2 splicing forms are available upon request.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
The project was supported by NIH grant 1R21DK078029-01 (JH), Intramural Research Programs of NHGRI and NCI of NIH. Work at LUDC was funded by grants from the Swedish Research Council, the Wallenberg Foundation and the Novo Nordisk Foundation. Funding to pay the Open Access publication charges for this article was provided by the intramural research program of NCI/NIH.
ACKNOWLEDGEMENTS
A special thank you to Marjorie Carlson and Mona Svärdh for technical assistance, Sara Bowell and Drs Ralph Powell, Amy Skubitz, Stephen Schmechel, and Charlie Moldow for facilitation of the access to tissue samples through the University of Minnesota Tissue Procurement Facility, to Islet Cell Resource Centers and the National Disease Research Interchange Network for human islets. Human islets (Swedish cohort) were provided by the Nordic Network for Clinical Islet Transplantation by the courtesy of Dr Olle Korsgren at Uppsala University, Sweden and Dr Jalal Taneera at the Human Tissue laboratory at Lund University Diabetes Centre, Sweden. LPO was a Wenner-Gren Fellow.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Grant S.F., Thorleifsson G., Reynisdottir I., Benediktsson R., Manolescu A., Sainz J., Helgason A., Stefansson H., Emilsson V., Helgadottir A., et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat. Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 2.Scott L.J., Mohlke K.L., Bonnycastle L.L., Willer C.J., Li Y., Duren W.L., Erdos M.R., Stringham H.M., Chines P.S., Jackson A.U., et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saxena R., Voight B.F., Lyssenko V., Burtt N.P., de Bakker P.I., Chen H., Roix J.J., Kathiresan S., Hirschhorn J.N., Daly M.J., et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 4.Zeggini E., Weedon M.N., Lindgren C.M., Frayling T.M., Elliott K.S., Lango H., Timpson N.J., Perry J.R., Rayner N.W., Freathy R.M., et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang C., Qi L., Hunter D.J., Meigs J.B., Manson J.E., van Dam R.M., Hu F.B. Variant of transcription factor 7-like 2 (TCF7L2) gene and the risk of type 2 diabetes in large cohorts of U.S. women and men. Diabetes. 2006;55:2645–2648. doi: 10.2337/db06-0643. [DOI] [PubMed] [Google Scholar]
- 6.van Vliet-Ostaptchouk J.V., Shiri-Sverdlov R., Zhernakova A., Strengman E., van Haeften T.W., Hofker M.H., Wijmenga C. Association of variants of transcription factor 7-like 2 (TCF7L2) with susceptibility to type 2 diabetes in the Dutch Breda cohort. Diabetologia. 2007;50:59–62. doi: 10.1007/s00125-006-0477-z. [DOI] [PubMed] [Google Scholar]
- 7.Scott L.J., Bonnycastle L.L., Willer C.J., Sprau A.G., Jackson A.U., Narisu N., Duren W.L., Chines P.S., Stringham H.M., Erdos M.R., et al. Association of transcription factor 7-like 2 (TCF7L2) variants with type 2 diabetes in a Finnish sample. Diabetes. 2006;55:2649–2653. doi: 10.2337/db06-0341. [DOI] [PubMed] [Google Scholar]
- 8.Saxena R., Gianniny L., Burtt N.P., Lyssenko V., Giuducci C., Sjogren M., Florez J.C., Almgren P., Isomaa B., Orho-Melander M., et al. Common single nucleotide polymorphisms in TCF7L2 are reproducibly associated with type 2 diabetes and reduce the insulin response to glucose in nondiabetic individuals. Diabetes. 2006;55:2890–2895. doi: 10.2337/db06-0381. [DOI] [PubMed] [Google Scholar]
- 9.Mayans S., Lackovic K., Lindgren P., Ruikka K., Agren A., Eliasson M., Holmberg D. TCF7L2 polymorphisms are associated with type 2 diabetes in northern Sweden. Eur. J. Hum. Genet. 2007;15:342–346. doi: 10.1038/sj.ejhg.5201773. [DOI] [PubMed] [Google Scholar]
- 10.Marzi C., Huth C., Kolz M., Grallert H., Meisinger C., Wichmann H.E., Rathmann W., Herder C., Illig T. Variants of the transcription factor 7-like 2 gene (TCF7L2) are strongly associated with type 2 diabetes but not with the metabolic syndrome in the MONICA/KORA surveys. Horm. Metab. Res. 2007;39:46–52. doi: 10.1055/s-2007-957345. [DOI] [PubMed] [Google Scholar]
- 11.Humphries S.E., Gable D., Cooper J.A., Ireland H., Stephens J.W., Hurel S.J., Li K.W., Palmen J., Miller M.A., Cappuccio F.P., et al. Common variants in the TCF7L2 gene and predisposition to type 2 diabetes in UK European Whites, Indian Asians and Afro-Caribbean men and women. J. Mol. Med. 2006;84:1–10. doi: 10.1007/s00109-006-0108-7. [DOI] [PubMed] [Google Scholar]
- 12.Helgason A., Palsson S., Thorleifsson G., Grant S.F., Emilsson V., Gunnarsdottir S., Adeyemo A., Chen Y., Chen G., Reynisdottir I., et al. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat. Genet. 2007;39:218–225. doi: 10.1038/ng1960. [DOI] [PubMed] [Google Scholar]
- 13.Groves C.J., Zeggini E., Minton J., Frayling T.M., Weedon M.N., Rayner N.W., Hitman G.A., Walker M., Wiltshire S., Hattersley A.T., et al. Association analysis of 6,736 U.K. subjects provides replication and confirms TCF7L2 as a type 2 diabetes susceptibility gene with a substantial effect on individual risk. Diabetes. 2006;55:2640–2644. doi: 10.2337/db06-0355. [DOI] [PubMed] [Google Scholar]
- 14.Damcott C.M., Pollin T.I., Reinhart L.J., Ott S.H., Shen H., Silver K.D., Mitchell B.D., Shuldiner A.R. Polymorphisms in the transcription factor 7-like 2 (TCF7L2) gene are associated with type 2 diabetes in the Amish: replication and evidence for a role in both insulin secretion and insulin resistance. Diabetes. 2006;55:2654–2659. doi: 10.2337/db06-0338. [DOI] [PubMed] [Google Scholar]
- 15.Chandak G.R., Janipalli C.S., Bhaskar S., Kulkarni S.R., Mohankrishna P., Hattersley A.T., Frayling T.M., Yajnik C.S. Common variants in the TCF7L2 gene are strongly associated with type 2 diabetes mellitus in the Indian population. Diabetologia. 2007;50:63–67. doi: 10.1007/s00125-006-0502-2. [DOI] [PubMed] [Google Scholar]
- 16.Schafer S.A., Tschritter O., Machicao F., Thamer C., Stefan N., Gallwitz B., Holst J.J., Dekker J.M., t'Hart L.M., Nijpels G., et al. Impaired glucagon-like peptide-1-induced insulin secretion in carriers of transcription factor 7-like 2 (TCF7L2) gene polymorphisms. Diabetologia. 2007;50:2443–2450. doi: 10.1007/s00125-007-0753-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raitakari O.T., Ronnemaa T., Huupponen R., Viikari L., Fan M., Marniemi J., Hutri-Kahonen N., Viikari J.S., Lehtimaki T. Variation of the transcription factor 7-like 2 (TCF7L2) gene predicts impaired fasting glucose in healthy young adults: the Cardiovascular Risk in Young Finns Study. Diabetes Care. 2007;30:2299–2301. doi: 10.2337/dc07-0539. [DOI] [PubMed] [Google Scholar]
- 18.Palmer N.D., Lehtinen A.B., Langefeld C.D., Campbell J.K., Haffner S.M., Norris J.M., Bergman R.N., Goodarzi M.O., Rotter J.I., Bowden D.W. Association of TCF7L2 gene polymorphisms with reduced acute insulin response in Hispanic Americans. J. Clin. Endocrinol. Metab. 2008;93:304–309. doi: 10.1210/jc.2007-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munoz J., Lok K.H., Gower B.A., Fernandez J.R., Hunter G.R., Lara-Castro C., De Luca M., Garvey W.T. Polymorphism in the transcription factor 7-like 2 (TCF7L2) gene is associated with reduced insulin secretion in nondiabetic women. Diabetes. 2006;55:3630–3634. doi: 10.2337/db06-0574. [DOI] [PubMed] [Google Scholar]
- 20.Watanabe R.M., Allayee H., Xiang A.H., Trigo E., Hartiala J., Lawrence J.M., Buchanan T.A. Transcription factor 7-like 2 (TCF7L2) is associated with gestational diabetes mellitus and interacts with adiposity to alter insulin secretion in Mexican Americans. Diabetes. 2007;56:1481–1485. doi: 10.2337/db06-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J., Kuusisto J., Vanttinen M., Kuulasmaa T., Lindstrom J., Tuomilehto J., Uusitupa M., Laakso M. Variants of transcription factor 7-like 2 (TCF7L2) gene predict conversion to type 2 diabetes in the Finnish Diabetes Prevention Study and are associated with impaired glucose regulation and impaired insulin secretion. Diabetologia. 2007;50:1192–1200. doi: 10.1007/s00125-007-0656-6. [DOI] [PubMed] [Google Scholar]
- 22.Stolerman E.S., Manning A.K., McAteer J.B., Fox C.S., Dupuis J., Meigs J.B., Florez J.C. TCF7L2 variants are associated with increased proinsulin/insulin ratios but not obesity traits in the Framingham Heart Study. Diabetologia. 2009;52:614–620. doi: 10.1007/s00125-009-1266-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stancakova A., Kuulasmaa T., Paananen J., Jackson A.U., Bonnycastle L.L., Collins F.S., Boehnke M., Kuusisto J., Laakso M. Association of 18 confirmed susceptibility loci for type 2 diabetes with indices of insulin release, proinsulin conversion, and insulin sensitivity in 5 327 non-diabetic Finnish men. Diabetes. 2009 doi: 10.2337/db09-0117. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clevers H., van de Wetering M. TCF/LEF factor earn their wings. Trends Genet. 1997;13:485–489. doi: 10.1016/s0168-9525(97)01305-x. [DOI] [PubMed] [Google Scholar]
- 25.Brantjes H., Barker N., van Es J., Clevers H. TCF: Lady Justice casting the final verdict on the outcome of Wnt signalling. Biol. Chem. 2002;383:255–261. doi: 10.1515/BC.2002.027. [DOI] [PubMed] [Google Scholar]
- 26.Korinek V., Barker N., Moerer P., van Donselaar E., Huls G., Peters P.J., Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat. Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 27.Yi F., Brubaker P.L., Jin T. TCF-4 mediates cell type-specific regulation of proglucagon gene expression by beta-catenin and glycogen synthase kinase-3beta. J. Biol. Chem. 2005;280:1457–1464. doi: 10.1074/jbc.M411487200. [DOI] [PubMed] [Google Scholar]
- 28.Lyssenko V., Lupi R., Marchetti P., Del Guerra S., Orho-Melander M., Almgren P., Sjogren M., Ling C., Eriksson K.F., Lethagen A.L., et al. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J. Clin. Invest. 2007;117:2155–2163. doi: 10.1172/JCI30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pilgaard K., Jensen C.B., Schou J.H., Lyssenko V., Wegner L., Brons C., Vilsboll T., Hansen T., Madsbad S., Holst J.J., et al. The T allele of rs7903146 TCF7L2 is associated with impaired insulinotropic action of incretin hormones, reduced 24 h profiles of plasma insulin and glucagon, and increased hepatic glucose production in young healthy men. Diabetologia. 2009;52:1298–1307. doi: 10.1007/s00125-009-1307-x. [DOI] [PubMed] [Google Scholar]
- 30.Zhang W., Duan S., Bleibel W.K., Wisel S.A., Huang R.S., Wu X., He L., Clark T.A., Chen T.X., Schweitzer A.C., et al. Identification of common genetic variants that account for transcript isoform variation between human populations. Hum. Genet. 2008;125:81–93. doi: 10.1007/s00439-008-0601-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graham R.R., Kozyrev S.V., Baechler E.C., Reddy M.V., Plenge R.M., Bauer J.W., Ortmann W.A., Koeuth T., Gonzalez Escribano M.F., Pons-Estel B., et al. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat. Genet. 2006;38:550–555. doi: 10.1038/ng1782. [DOI] [PubMed] [Google Scholar]
- 32.Ahlzen M., Johansson L.E., Cervin C., Tornqvist H., Groop L., Ridderstrale M. Expression of the transcription factor 7-like 2 gene (TCF7L2) in human adipocytes is down regulated by insulin. Biochem. Biophys. Res. Commun. 2008;370:49–52. doi: 10.1016/j.bbrc.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 33.Wegner L., Hussain M.S., Pilgaard K., Hansen T., Pedersen O., Vaag A., Poulsen P. Impact of TCF7L2 rs7903146 on Insulin Secretion and Action in Young and Elderly Danish Twins. J. Clin. Endocrinol. Metab. 2008;93:4013–4019. doi: 10.1210/jc.2008-0855. [DOI] [PubMed] [Google Scholar]
- 34.Elbein S.C., Chu W.S., Das S.K., Yao-Borengasser A., Hasstedt S.J., Wang H., Rasouli N., Kern P.A. Transcription factor 7-like 2 polymorphisms and type 2 diabetes, glucose homeostasis traits and gene expression in US participants of European and African descent. Diabetologia. 2007;50:1621–1630. doi: 10.1007/s00125-007-0717-x. [DOI] [PubMed] [Google Scholar]
- 35.Cauchi S., Meyre D., Dina C., Choquet H., Samson C., Gallina S., Balkau B., Charpentier G., Pattou F., Stetsyuk V., et al. Transcription factor TCF7L2 genetic study in the French population: expression in human beta-cells and adipose tissue and strong association with type 2 diabetes. Diabetes. 2006;55:2903–2908. doi: 10.2337/db06-0474. [DOI] [PubMed] [Google Scholar]
- 36.Huertas-Vazquez A., Plaisier C., Weissglas-Volkov D., Sinsheimer J., Canizales-Quinteros S., Cruz-Bautista I., Nikkola E., Herrera-Hernandez M., Davila-Cervantes A., Tusie-Luna T., et al. TCF7L2 is associated with high serum triacylglycerol and differentially expressed in adipose tissue in families with familial combined hyperlipidaemia. Diabetologia. 2008;51:62–69. doi: 10.1007/s00125-007-0850-6. [DOI] [PubMed] [Google Scholar]
- 37.Hindle A.K., Brody F., Tevar R., Kluk B., Hill S., McCaffrey T., Fu S. TCF7L2 expression in diabetic patients undergoing bariatric surgery. Surg. Endosc. 2008;23:1292–1297. doi: 10.1007/s00464-008-0001-2. [DOI] [PubMed] [Google Scholar]
- 38.Osmark P., Hansson O., Jonsson A., Ronn T., Groop L., Renstrom E. Unique splicing pattern of the TCF7L2 gene in human pancreatic islets. Diabetologia. 2009;52:850–854. doi: 10.1007/s00125-009-1293-z. [DOI] [PubMed] [Google Scholar]
- 39.Duval A., Busson-Leconiat M., Berger R., Hamelin R. Assignment of the TCF-4 gene (TCF7L2) to human chromosome band 10q25.3. Cytogenet. Cell Genet. 2000;88:264–265. doi: 10.1159/000015534. [DOI] [PubMed] [Google Scholar]
- 40.Howng S.L., Huang F.H., Hwang S.L., Lieu A.S., Sy W.D., Wang C., Hong Y.R. Differential expression and splicing isoform analysis of human Tcf-4 transcription factor in brain tumors. Int. J. Oncol. 2004;25:1685–1692. [PubMed] [Google Scholar]
- 41.Shiina H., Igawa M., Breault J., Ribeiro-Filho L., Pookot D., Urakami S., Terashima M., Deguchi M., Yamanaka M., Shirai M., et al. The human T-cell factor-4 gene splicing isoforms, Wnt signal pathway, and apoptosis in renal cell carcinoma. Clin. Cancer Res. 2003;9:2121–2132. [PubMed] [Google Scholar]
- 42.Van de Wetering M., Castrop J., Korinek V., Clevers H. Extensive alternative splicing and dual promoter usage generate Tcf-1 protein isoforms with differential transcription control properties. Mol. Cell Biol. 1996;16:745–752. doi: 10.1128/mcb.16.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hovanes K., Li T.W., Waterman M.L. The human LEF-1 gene contains a promoter preferentially active in lymphocytes and encodes multiple isoforms derived from alternative splicing. Nucleic Acids Res. 2000;28:1994–2003. doi: 10.1093/nar/28.9.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hovanes K., Li T.W., Munguia J.E., Truong T., Milovanovic T., Lawrence Marsh J., Holcombe R.F., Waterman M.L. Beta-catenin-sensitive isoforms of lymphoid enhancer factor-1 are selectively expressed in colon cancer. Nat. Genet. 2001;28:53–57. doi: 10.1038/ng0501-53. [DOI] [PubMed] [Google Scholar]
- 45.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reynisdottir I., Thorleifsson G., Benediktsson R., Sigurdsson G., Emilsson V., Einarsdottir A.S., Hjorleifsdottir E.E., Orlygsdottir G.T., Bjornsdottir G.T., Saemundsdottir J., et al. Localization of a susceptibility gene for type 2 diabetes to chromosome 5q34-q35.2. Am. J. Hum. Genet. 2003;73:323–335. doi: 10.1086/377139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duggirala R., Blangero J., Almasy L., Dyer T.D., Williams K.L., Leach R.J., O'Connell P., Stern M.P. Linkage of type 2 diabetes mellitus and of age at onset to a genetic location on chromosome 10q in Mexican Americans. Am. J. Hum. Genet. 1999;64:1127–1140. doi: 10.1086/302316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li T.W., Ting J.H., Yokoyama N.N., Bernstein A., van de Wetering M., Waterman M.L. Wnt activation and alternative promoter repression of LEF1 in colon cancer. Mol. Cell Biol. 2006;26:5284–5299. doi: 10.1128/MCB.00105-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benassayag C., Montero L., Colombie N., Gallant P., Cribbs D., Morello D. Human c-Myc isoforms differentially regulate cell growth and apoptosis in Drosophila melanogaster. Mol. Cell Biol. 2005;25:9897–9909. doi: 10.1128/MCB.25.22.9897-9909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zaika A.I., Slade N., Erster S.H., Sansome C., Joseph T.W., Pearl M., Chalas E., Moll U.M. DeltaNp73, a dominant-negative inhibitor of wild-type p53 and TAp73, is up-regulated in human tumors. J. Exp. Med. 2002;196:765–780. doi: 10.1084/jem.20020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Korinek V., Barker N., Morin P.J., van Wichen D., de Weger R., Kinzler K.W., Vogelstein B., Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 52.Brantjes H., Roose J., van De Wetering M., Clevers H. All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Res. 2001;29:1410–1419. doi: 10.1093/nar/29.7.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fang M., Li J., Blauwkamp T., Bhambhani C., Campbell N., Cadigan K.M. C-terminal-binding protein directly activates and represses Wnt transcriptional targets in Drosophila. EMBO J. 2006;25:2735–2745. doi: 10.1038/sj.emboj.7601153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Atcha F.A., Syed A., Wu B., Hoverter N.P., Yokoyama N.N., Ting J.H., Munguia J.E., Mangalam H.J., Marsh J.L., Waterman M.L. A unique DNA binding domain converts T-cell factors into strong Wnt effectors. Mol. Cell Biol. 2007;27:8352–8363. doi: 10.1128/MCB.02132-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang W., Dodge M., Gundapaneni D., Michnoff C., Roth M., Lum L. A genome-wide RNAi screen for Wnt/beta-catenin pathway components identifies unexpected roles for TCF transcription factors in cancer. Proc. Natl. Acad Sci. USA. 2008;105:9697–9702. doi: 10.1073/pnas.0804709105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shu L., Sauter N.S., Schulthess F.T., Matveyenko A.V., Oberholzer J., Maedler K. Transcription factor 7-like 2 regulates beta-cell survival and function in human pancreatic islets. Diabetes. 2008;57:645–653. doi: 10.2337/db07-0847. [DOI] [PubMed] [Google Scholar]
- 57.Atcha F.A., Munguia J.E., Li T.W., Hovanes K., Waterman M.L. A new beta-catenin-dependent activation domain in T cell factor. J. Biol. Chem. 2003;278:16169–16175. doi: 10.1074/jbc.M213218200. [DOI] [PubMed] [Google Scholar]
- 58.Listerman I., Sapra A.K., Neugebauer K.M. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat. Struct. Mol. Biol. 2006;13:815–822. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- 59.Kornblihtt A.R., de la Mata M., Fededa J.P., Munoz M.J., Nogues G. Multiple links between transcription and splicing. RNA. 2004;10:1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.