Abstract
Mutations in PARK8, encoding LRRK2, are the most common known cause of Parkinson's disease. The LRRK2 Roc-COR tandem domain exhibits GTPase activity controlling LRRK2 kinase activity via an intramolecular process. We report the interaction of LRRK2 with the dishevelled family of phosphoproteins (DVL1-3), key regulators of Wnt (Wingless/Int) signalling pathways important for axon guidance, synapse formation and neuronal maintenance. Interestingly, DVLs can interact with and mediate the activation of small GTPases with structural similarity to the LRRK2 Roc domain. The LRRK2 Roc-COR domain and the DVL1 DEP domain were necessary and sufficient for LRRK2–DVL1 interaction. Co-expression of DVL1 increased LRRK2 steady-state protein levels, an effect that was dependent on the DEP domain. Strikingly, LRRK2–DVL1-3 interactions were disrupted by the familial PARK8 mutation Y1699C, whereas pathogenic mutations at residues R1441 and R1728 strengthened LRRK2–DVL1 interactions. Co-expression of DVL1 with LRRK2 in mammalian cells resulted in the redistribution of LRRK2 to typical cytoplasmic DVL1 aggregates in HEK293 and SH-SY5Y cells and co-localization in neurites and growth cones of differentiated dopaminergic SH-SY5Y cells. This is the first report of the modulation of a key LRRK2-accessory protein interaction by PARK8 Roc-COR domain mutations segregating with Parkinson's disease. Since the DVL1 DEP domain is known to be involved in the regulation of small GTPases, we propose that: (i) DVLs may influence LRRK2 GTPase activity, and (ii) Roc-COR domain mutations modulating LRRK2–DVL interactions indirectly influence kinase activity. Our findings also link LRRK2 to Wnt signalling pathways, suggesting novel pathogenic mechanisms and new targets for genetic analysis in Parkinson's disease.
INTRODUCTION
The PARK8 locus encodes LRRK2, a 2527 amino acid cytosolic protein kinase. Mutations in PARK8 are the most common known cause of Parkinson's disease, with missense mutations found in patients with familial as well as apparently idiopathic Parkinson's disease (1–5). LRRK2 belongs to the ROCO family of proteins which are characterized by the unique combination of a Roc (Ras of complex proteins) domain with intrinsic GTPase activity and a COR (C-terminal of Roc) domain. The Roc-COR tandem domain controls LRRK2 kinase activity via an intramolecular process (6–12). The modification of LRRK2 GTPase and kinase activity by PARK8 mutations affecting residues in the Roc, COR and kinase domains is believed to lead to neuronal cell death, but the pathways involved remain elusive (1,2,7,8,11–16). The combination of GTPase activity mediated via the Roc-COR tandem domain and kinase activity of the mitogen-activated protein kinase kinase kinase domain suggests a complex role for LRRK2 in cell signalling. Additional protein–protein interaction domains, such as LRR (Leucine rich repeat) and WD40 propeller motifs provide additional complexity and could potentially localize LRRK2 to different subcellular compartments. The Roc domain shares sequence similarity with all five subfamilies of the Ras-related superfamily of small GTPases (Ras, Rho, Rab, Sar/Arf and Ran), has conserved amino acids involved in GTP binding/hydrolysis and exhibits intrinsic GTPase activity. Evidence suggests that the COR domain forms dimers, resulting in juxtaposition of the associated Roc domains (10). Since the isolated LRRK2 kinase domain was shown to be catalytically inactive (16,17), it is clear that the Roc and COR domains are vital for kinase activity and/or protein stabilization. One current theory suggests that mutations in the Roc and COR domains reduce GTPase activity, leading to higher kinase activity. This suggestion is based on reports that the R1441C/R1441G substitutions in the Roc domain reduce GTPase activity (9,10,14,15), whereas these mutations as well as the Y1699C mutation in the COR domain increase kinase activity (7,12).
Thus, the LRRK2 Roc domain is likely to serve as a molecular switch, regulating kinase activity by cycling between GDP-bound and GTP-bound states (6–17). However, it is currently unclear how this activity is controlled in vivo. Normally, guanine nucleotide exchange factors (GEFs) facilitate GTP binding and effector activation, whereas GTPase-activating proteins (GAPs) increase the intrinsic rate of GTP hydrolysis to terminate signalling (18). Interestingly, the GTPase activity of a Roc-COR tandem domain from Chlorobium tepidum, a prokaryotic homologue of LRRK2, shows a low affinity for nucleotide and fast GDP dissociation, suggesting that LRRK2 may not require a classical RhoGEF for GTPase activity (10). Rather, Roc GTPase activity was proposed to be stimulated solely by COR dimerization. However, this model seems simplistic, as it would not allow for up- or down-regulation of GTPase and kinase activity. Thus, the identification of proteins involved in regulating LRRK2 GTPase activity and mediating downstream signalling is of fundamental importance in understanding the pathogenesis of Parkinson's disease. Since the Roc-COR tandem domain controls LRRK2 kinase activity, we searched for interactors of the Roc-COR domain with likely relevance for GTPase and kinase activity. Here, we describe the discovery and characterization of the interaction of LRRK2 with all members of the dishevelled (DVL) family of phosphoproteins in yeast and mammalian cell systems. DVLs have a modular architecture consisting of DIX (Dishevelled/Axin), PDZ (PSD-95, DLG, ZO1) and DEP (Dishevelled, EGL-10, Pleckstrin) domains. Importantly, DVL proteins are key regulators of Wnt signalling pathways leading to multiple downstream effects (19), including the activation of small GTPases such as Rac1 that are structurally similar to the LRRK2 Roc domain (20). Recent studies have underlined the importance of DVLs in key processes in neuronal development, such as axon guidance, synapse formation and neuronal maintenance (19,21–27). The identification of DVL proteins as interactors of LRRK2 suggests a plausible physiological role for LRRK2 in these processes. DVL proteins altered the subcellular distribution of LRRK2 and co-localized in neurites and growth cones of differentiated dopaminergic SH-SY5Y cells. The LRRK2-DVL1 interaction stabilizes steady-state levels of LRRK2, but importantly LRRK2-DVL interactions are decreased or increased by selected PARK8 Roc-COR domain mutations segregating with Parkinson's disease.
Further functional characterization of LRRK2-DVL interactions is currently hindered by technical difficulties encountered in purifying intact DVL proteins, reproducibility and sensitivity of LRRK2 GTPase and kinase assays, and the lack of commercially available antibodies against LRRK2 and DVL1-3 proteins that function in immunoprecipitation experiments. Nonetheless, the dissemination of our results to a wider audience at this stage will highlight valuable leads for further research into the endogenous control of LRRK2 GTPase and kinase activity. We also suggest the relevance of Wnt signalling pathways to Parkinson's disease, underpin the importance of microtubule dynamics in neurodegeneration (28–34) and identify DVL proteins as therapeutic and genetic targets for future therapeutic and genetic research.
RESULTS
LRRK2 associates with DVL family proteins via a Roc-COR–DEP domain interaction
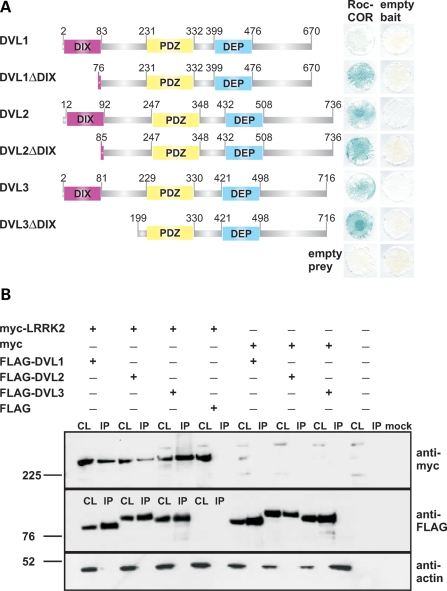
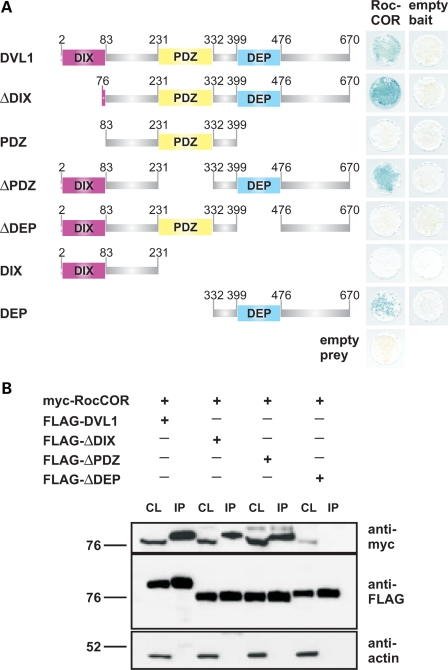
In order to identify LRRK2 accessory proteins potentially regulating kinase activity, we screened an embryonic human brain cDNA library (Clontech) using the LexA yeast two-hybrid (YTH) system and the LRRK2 Roc-COR tandem domain (Roc-COR) as ‘bait’. This resulted in the identification of several overlapping partial cDNAs encoding DVL2 and DVL3, members of the dishevelled family of phosphoproteins (Fig. 1A), which contain single DIX, PDZ and DEP domains. None of the encoded proteins harboured an intact DIX domain, suggesting that this motif was not necessary for LRRK2 binding. Q-PCRs confirmed that DVL1–DVL3 transcripts are detectable in the adult human brain, including the substantia nigra (Supplementary Material, Fig. S1). DVLs were considered promising candidates for further analysis since they are known to interact with and mediate the activation of small GTPases, such as Rac1 and RhoA. Interestingly, the DVL1/DVL2 DEP domain alone is sufficient for Rac1 activation, whereas both PDZ and DEP domains are required for RhoA activation (20,35). Further analysis demonstrated that LRRK2 interacts with full-length DVL1-3 proteins in yeast (Fig. 1A) and HEK293 cells, as demonstrated by co-immunoprecipitation of myc-tagged LRRK2 and FLAG-tagged DVL constructs (Fig. 1B). It is also noteworthy that the DVLs appear to differ in the nature of their interaction with LRRK2. Although the Roc-COR bait demonstrated an interaction with all three full-length DVL proteins (DVL1-3; Fig. 1A), the interaction between the LRRK2 Roc-COR domain and DVL1 was consistently weaker compared with DVL2 or DVL3 (Fig. 1A). However, deletion constructs lacking the N-terminal DVL1 DIX domain (DVL1ΔDIX) showed a robust interaction with the Roc-COR bait, equivalent to DVL2 or DVL3 (Fig. 1A). Expressing selected subdomains or deletions of DVL1 in yeast (Fig. 2A) or HEK293 cells (Fig. 2B) demonstrated that removal of the DIX and/or PDZ domains did not abolish DVL interactions with the LRRK2 Roc-COR tandem domain, whereas constructs lacking the DEP domain were no longer associated with LRRK2 (Fig. 2A and B). Hence, DVL1 is capable of interacting with the LRRK2 Roc-COR region in yeast and mammalian cells via the DEP domain, which can interact with and mediate the activation of small GTPases such as Rac1 (20).
Figure 1.
The LRRK2 Roc-COR tandem domain interacts with dishevelled proteins DVL1, DVL2 and DVL3. (A) Full-length or partial DVL1, DVL2 and DVL3 preys were tested for interactions with the LRRK2 Roc-COR tandem domain bait in the YTH system using LacZ freeze-fracture assays. All negative controls show yeast growth but no blue colouration in the LacZ assay, demonstrating that the co-expression of bait and prey plasmids with empty prey or bait vectors does not result in transcription of reporter genes, i.e. no autoactivation was observed. Note that deletion of the DVL1 DIX domain (DVL1ΔDIX) increases the strength of the Roc-COR-DVL1 interaction. (B) Co-immunoprecipitation of LRRK2 and DVL1, DVL2 and DVL3 in HEK293 cells co-transfected with full-length myc-tagged LRRK2 and full-length FLAG-tagged DVL1, DVL2 or DVL3 constructs. Note that myc-LRRK2 is present in the cell lysates (CL) and FLAG-DVL1, FLAG-DVL2 and FLAG-DVL3 immunoprecipitation (IP) samples purified using FLAG beads, but not in IP samples from cells co-transfected with the empty FLAG vector.
Figure 2.
The LRRK2 Roc-COR tandem domain interacts with the DVL1 DEP domain. (A) DVL1 deletion constructs lacking key domains (ΔDIX, ΔPDZ or ΔDEP), or encoding individual DIX, PDZ or DEP domains were tested for interactions with the LRRK2 Roc-COR tandem domain bait using the YTH system. LacZ freeze-fracture assays demonstrate that only constructs expressing the DVL1 DEP domain are able to interact with the Roc-COR bait, whereas the DIX and PDZ domains were dispensable. (B) Confirmation of the LRRK2-binding site on DVL1 by co-immunoprecipitation in HEK293 cells co-transfected with constructs encoding the myc-tagged LRRK2 Roc-COR tandem domain and FLAG-tagged DVL1 variants. Note that the myc-tagged LRRK2 Roc-COR tandem domain is present in the cell lysates (CL) and FLAG-DVL1, FLAG-DVL1ΔDIX, FLAG-DVL1ΔPDZ immunoprecipitation (IP) samples purified using FLAG beads, but not in IP samples from cells co-transfected with FLAG-DVL1ΔDEP.
DVL co-expression alters the subcellular distribution of LRRK2
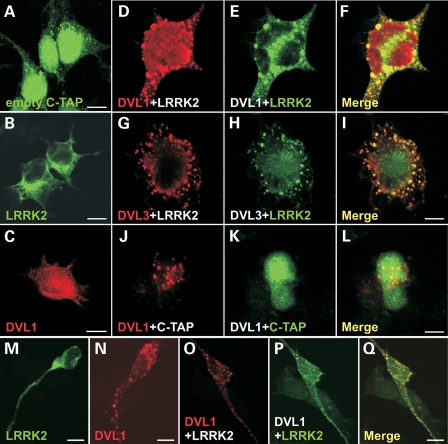
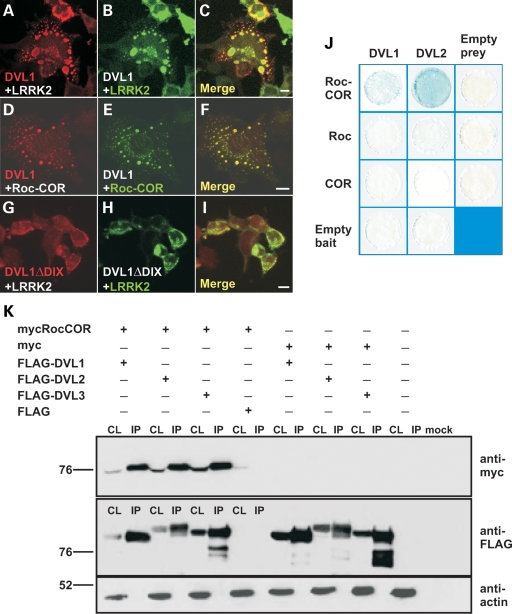
Transfection of C-terminally TAP-tagged LRRK2 in HEK293 cells results in cytoplasmic distribution of the protein (Fig. 3B) and perinuclear aggregates in 5–10% of the transfected cells similar to that previously reported for myc- or EGFP-tagged LRRK2 (13). In contrast, expression of FLAG-tagged DVL1 results in cytoplasmic round or doughnut-shaped protein aggregates (Fig. 3C). Interestingly, the co-expression of TAP-LRRK2 and FLAG-DVL1 or FLAG-DVL3 results in a redistribution of LRRK2 to DVL aggregates and changes in cell morphology (Fig. 3D–I), a result that was not observed using the TAP epitope tag alone (Fig. 3J–L). A similar redistribution of myc-LRRK2 to FLAG-DVL1 aggregates was also observed in non-differentiated dopaminergic SH-SY5Y cells (Fig. 3O–Q). As expected, this redistribution relies on LRRK2 Roc-COR-dishevelled interactions, since the full-length myc-tagged LRRK2 (Fig. 4A–C) and the myc-tagged Roc-COR tandem domain (Fig. 4D–F) both redistributed to FLAG-DVL1 aggregates. The unusual distribution of DVL proteins relies on the N-terminal DIX domain, since deletion of this region resulted in the loss of cytoplasmic DVL1 aggregates and the redistribution of FLAG-DVL1ΔDIX and myc-LRRK2 throughout the cytoplasm with some enrichment at the cell membrane (Fig. 4G–I). However, experiments in yeast suggested that neither the Roc nor COR domains alone interacted with DVL1 and DVL2 (Fig. 4J), suggesting that an intact tandem domain is required for LRRK2-DVL interactions. We confirmed this by co-immunoprecipitation experiments showing that FLAG-tagged DVL1-3 were able to co-precipitate the myc-tagged LRRK2 Roc-COR tandem domain from co-transfected HEK293 cells (Fig. 4K).
Figure 3.
Redistribution of LRRK2 to cytoplasmic aggregates containing DVL proteins and changes in cell morphology. Confocal microscopy showing the distributions of (A) TAP-epitope, (B) TAP-tagged full-length LRRK2 and (C) FLAG-tagged full-length DVL1. Note that the TAP epitope has a predominantly nuclear and cytoplasmic distribution, whereas TAP-LRRK2 is distributed evenly in the cytoplasm including cell processes. In contrast, FLAG-DVL1 is expressed in the cytoplasm forming round structures that represent DVL protein polymers (36). TAP-LRRK2 re-distributes to FLAG-DVL1 (D–F) and FLAG-DVL3 (G–I) aggregates upon co-transfection, and cells show unique changes in morphology, spreading and flattening out to cover a larger surface area. In contrast, the TAP epitope tag does not co-distribute with FLAG-DVL1 aggregates (J–L), and changes in cell morphology are not observed. Similar results were obtained with myc-LRRK2, LRRK2-EGFP and LRRK2-V5-His (data not shown), suggesting that the redistribution of LRRK2 to DVL aggregates is independent on the nature of the epitope tag. A similar distribution of myc-LRRK2 (M) and FLAG-DVL1 (N) and the redistribution of myc-LRRK2 to FLAG-DVL1 aggregates (O–Q) are seen in non-differentiated dopaminergic SH-SY5Y cells, demonstrating that this effect also occurs in different cell lines. Scale bars: 10 µm.
Figure 4.
The cytoplasmic distribution of DVL–LRRK2 complexes is dependent on the DVL1 DIX domain and the intact LRRK2 Roc-COR tandem domain. Confocal microscopy showing the co-distribution of FLAG-tagged full-length DVL1 with myc-tagged full-length LRRK2 in cytoplasmic DVL protein polymers (A–C). Since a myc-tagged Roc-COR construct shows similar targeting to cytoplasmic FLAG-DVL1 aggregates, the Roc-COR tandem domain is sufficient for this interaction (D–F). Deleting the DIX domain by site-directed mutagenesis (DVL1ΔDIX) resulted in the loss of cytoplasmic DVL1 aggregates and the redistribution of FLAG-DVL1ΔDIX and myc-LRRK2 throughout the cytoplasm, but with apparent enrichment at the cell membrane (G–I). Scale bars: 10 µm. (J) LacZ freeze-fracture assays demonstrate that the intact Roc-COR tandem domain, but not individual Roc or COR domains, binds DVL1 and DVL2 effectively. (K) Co-immunoprecipitation of the myc-tagged LRRK2 Roc-COR tandem domain by FLAG-tagged DVLs confirms that the LRRK2 Roc-COR tandem domain is sufficient for DVL binding. Note that the myc-tagged LRRK2 Roc-COR tandem domain is present in the cell lysates (CL) and enriched in FLAG-bead purified immunoprecipitation (IP) samples from co-transfections with FLAG-DVL1, FLAG-DVL2 and FLAG-DVL3, but not in control experiments using empty FLAG vector.
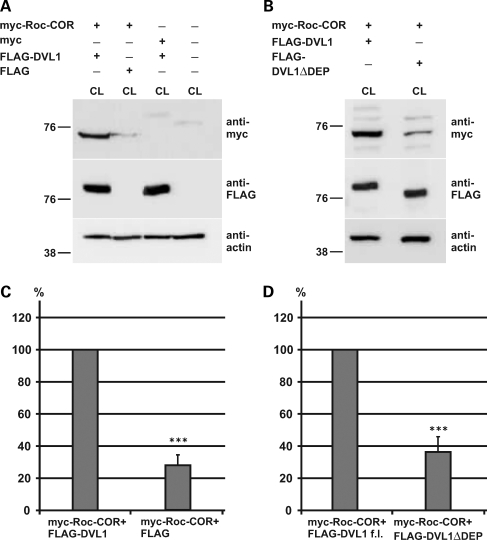
DVL1 stabilizes LRRK2 protein expression
In order to assess the functional effects of DVL1 on LRRK2 activity, we attempted both LRRK2 kinase assays (13) and GTPase (14) assays, although these have been unsuccessful to date. As might have been predicted, given the involvement of DVLs in diverse Wnt signalling pathways (19–27), immunoprecipitation of FLAG-DVLs alone from transfected HEK293 cells resulted in GTPase and kinase activity that is likely to be unrelated to LRRK2 (data not shown). Experiments with recombinant LRRK2 and DVL proteins will be required to resolve this issue. However, we were able to show clear effects of FLAG-DVL1 on the stability of the myc-tagged Roc-COR domain in cell lysates from co-transfected cells (Fig. 5A), with quantification showing that co-expression of FLAG-DVL1 increased levels of myc-Roc-COR by approximately 4-fold (Fig. 5C). This effect was lost upon co-expression of the myc-Roc-COR protein with FLAG-DVL1ΔDEP, implying that the DEP domain is crucial for mediating this effect (Fig. 5B and D).
Figure 5.
Co-expression of DVL1 stabilizes the expression of the LRRK2 Roc-COR tandem domain. (A) Co-expression of the myc-tagged Roc-COR tandem domain with FLAG-DVL1 results in a stabilization of the Roc-COR domain in cell lysates. (B) The stabilization of LRRK2 is dependent on the DVL1 DEP domain interaction with the Roc-COR tandem domain, since DVL1 lacking the interacting DEP domain (FLAG-DVL1ΔDEP) was not capable of stabilizing myc-Roc-COR. (C and D) Quantification of these effects after normalization to levels of cellular actin shows that FLAG-DVL1 increases myc-Roc-COR ∼4-fold compared with controls with an empty FLAG vector or FLAG-DVL1ΔDEP. Statistical significance was determined using a Student's t-test (two-tailed). Error bars represent the standard deviation of the mean. ***P < 0.001.
PARK8 mutations segregating with Parkinson's disease alter LRRK2–DVL interactions
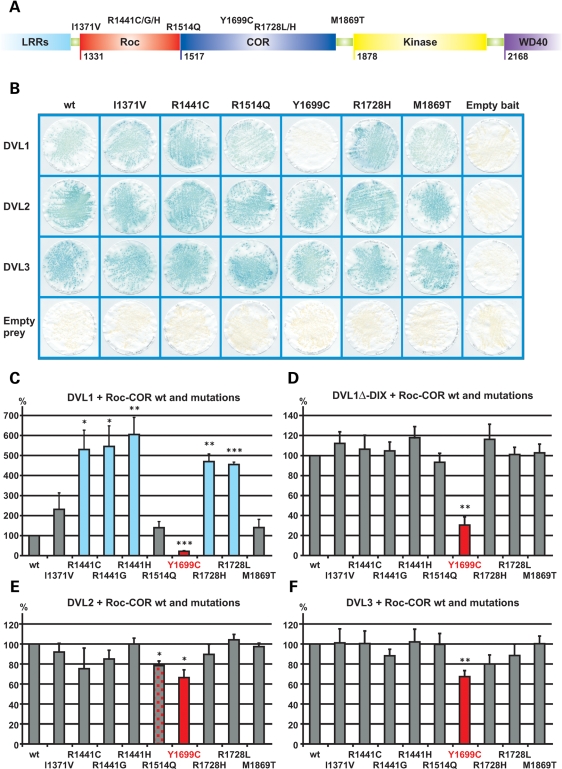
Numerous PARK8 mutations affect residues in the Roc-COR tandem domain (Fig. 6), but the pathogenic mechanism underlying these changes is as yet unclear. Most mutations are assumed to alter the folding or intrinsic GTPase activity of the Roc-COR tandem domain, so in turn influencing LRRK2 kinase activity (9–16). For this reason, we decided to assess whether selected familial Parkinson's disease mutations (I1371V, R1441C, R1441G, R1441H, R1514Q, Y1699C, R1728H, R1728L or M1869T) in the Roc-COR domain (Fig. 6A) have any influence on LRRK2 interactions with DVL1, DVL2 or DVL3. Surprisingly, the COR domain mutation Y1699C weakened the interaction of LRRK2 with DVL1, 2 and 3 as well as DVL1ΔDIX (Fig. 6B–F). From freeze-fracture filters (Fig. 6B), this effect is most evident for DVL1, although quantitative YTH assays revealed that Y1699C also influences DVL2 and DVL3 interactions (Fig. 6E and F). In contrast, all known substitutions at residues R1441 and R1728 appear to strengthen the LRRK2–DVL1 interaction (Fig. 6B and C). Curiously, these mutations do not influence Roc-COR bait interactions with DVL1ΔDIX, DVL2 or DVL3 preys (Fig. 6B and D–F), i.e. the observed effect is specific for full-length DVL1. Other substitutions including I1371V, R1514Q and M1869Q did not alter the interaction between the LRRK2 Roc-COR domain and any of the DVL1 or DVL3 baits. However, the R1514Q substitution caused a small but significant reduction of the Roc-COR interaction with DVL2.
Figure 6.
Modulation of the interaction between the LRRK2 Roc-COR tandem domain and dishevelled proteins (DVL1-3) by familial Parkinson's disease mutations. (A) Locations of amino acids affected by familial Parkinson's disease mutations in the Roc-COR tandem domain. (B) LacZ freeze-fracture assays of LRRK2-DVL interactions. Using this semi-quantitative assay, the most striking effect observed is that the Y1699C mutation in the COR domain clearly disrupts the LRRK2–DVL1 interaction. All negative controls show yeast growth but no blue colour in the LacZ assay, demonstrating that the co-expression of bait and prey plasmids with empty prey or empty bait vectors does not result in transcription of reporter genes, i.e. no autoactivation was observed. (C–F) Quantitative liquid YTH assays using CPRG as substrate for β-galactosidase expression reveal that substitutions at R1441 and R1728 show a strengthened interaction between the Roc-COR domain bait and DVL1 prey, whereas Y1699C disrupted interactions between the Roc-COR domain bait and DVL1, DVL2 and DVL3 preys. Statistical significance was determined using a Student's t-test (two-tailed). Error bars represent the standard deviation of the mean. ***P < 0.001, **P < 0.01, *P < 0.05.
DVL1 and LRRK2 co-localize in neurites and growth cones of differentiated SH-SY5Y cells
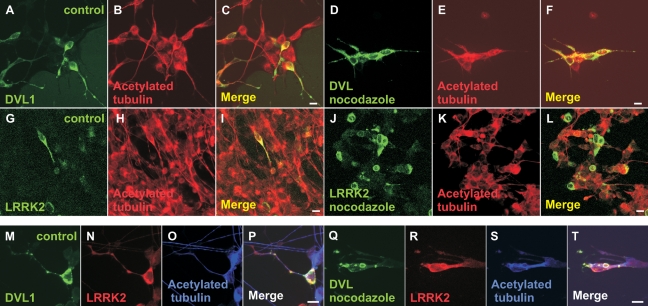
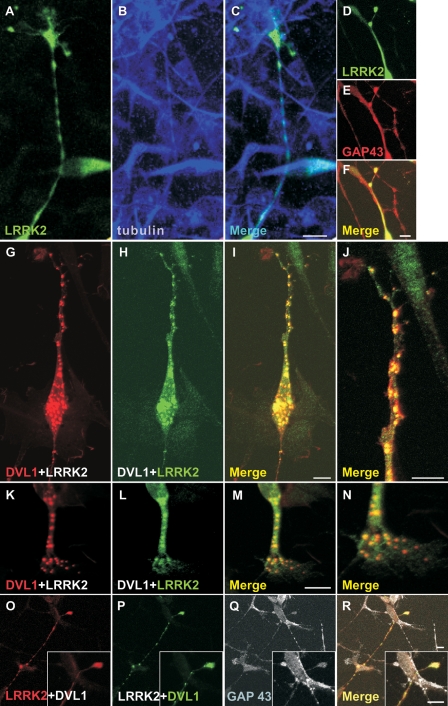
Since DVLs were previously shown to stabilize microtubules in nocodazole-treated differentiated neurones (22), we examined whether (i) a similar effect could be observed upon transfection of DVL1 into differentiated SH-SY5Y cells and (ii) whether co-expression of LRRK2 enhances or opposes DVL-induced microtubule stabilization. The formation of a stable microtubule network was assessed by staining for acetylated tubulin (22). As predicted, differentiating SH-SY5Y cells expressing FLAG-DVL1 showed an abundance of stable microtubules and neurite projections (Fig. 7A–C) that were resistant to nocodazole treatment (Fig. 7D–F). In contrast, SH-SY5Y cells expressing myc-tagged full-length LRRK2 contained acetylated tubulin (Fig. 7G–I), but this was not resistant to nocodazole treatment (Fig. 7J–L). Nocodazole-resistant acetylated tubulin was also detected in cells co-expressing FLAG-DVL1 and myc-LRRK2 (Fig. 7M–T). Because of the low co-transfection efficiency in these experiments, we could not assess whether LRRK2-DVL1 co-expression affected the length or number of neuronal processes. However, DVL1 still appeared to be able to stabilize microtubules against nocodazole treatment in the presence of LRRK2. After differentiation with retinoic acid treatment, myc-LRRK2 is observed in the cytoplasm and neuronal processes (visualized by labelling for tubulin) and appears to be enriched in growth cones as shown by double-labelling with antibodies directed against GAP43 (Fig. 8A–F), a neuronal growth cone protein thought to be involved in pathfinding. Co-transfection of myc-LRRK2 and FLAG-DVL1 resulted in co-localization of both proteins in puncta within neurites (Fig. 8G–J) and growth cones (Fig. 8K–R).
Figure 7.
DVL1, but not LRRK2, stabilizes microtubules treated with nocodazole. (A–L) Confocal microscopy showing stable microtubules stained for acetylated tubulin in SH-SY5Y cells 5 days after treatment with retinoic acid transfected with FLAG-DVL1 or myc-LRRK2. Note that in cells transfected with FLAG-DVL1, stable microtubules were observed both before (A–C) and after (D–F) nocodazole treatment. However, while acetylated tubulin was observed in cells expressing myc-LRRK2 before nocodazole treatment (G–I), LRRK2 alone was unable to protect microtubules from nocodazole treatment (J–L). Interestingly, cells co-transfected with FLAG-DVL1 and myc-LRRK2 showed stable microtubules both before (M–P) and after (Q–T) nocodazole treatment. Scale bars: 10 µm.
Figure 8.
Co-localization of LRRK2 and DVL1 in the cytoplasm, neurites and growth cones in differentiated dopaminergic cells. (A–F) Confocal microscopy showing expression of myc-tagged LRRK2 in SH-SY5Y cells 5 days after treatment with retinoic acid. Note that LRRK2 co-localizes with tubulin staining in neurites (A–C) and GAP43 immunostaining in growth cones (D–F). Co-expression of FLAG-tagged DVL1 with myc-tagged LRRK2 in differentiated SH-SY5Y cells shows co-localization of both proteins in multiple cytoplasmic aggregates (G–J). (J) Magnification of the process shown in (G–I). Co-localization of LRRK2 and DVL1 in an extended neuronal terminal, suggestive of a growth cone (K–N) and in growth cones co-stained for GAP43 (O–R). Scale bars: 10 µm.
DISCUSSION
The aim of our study was to characterize potential modulators of LRRK2 GTPase and kinase activity by identifying interactors of the LRRK2 Roc-COR tandem domain. Using the YTH system, we found that the LRRK2 Roc-COR tandem domain interacts with dishevelled family proteins (DVL1, DVL2, DVL3). We found that DVL1 interacts with LRRK2 via the DEP domain, which is found in various signalling proteins such as GEFs, GAPs and Roco family proteins (6). DEP domains have also been shown to interact with and mediate the activation of small GTPases such as Rac1 and RhoA, which share structural similarity with the LRRK2 Roc domain (20,35). Quantitative PCRs confirmed that transcripts for DVL1-3 are expressed in brain and detectable in substantia nigra. Co-immunoprecipitation of epitope-tagged constructs confirmed that LRRK2 associates with DVL1, DVL2 and DVL3 in mammalian cells and that DVL proteins increase the steady-state level of the LRRK2 Roc-COR domain. Although the exact mechanism responsible for this observation remains to be determined, it is tempting to speculate that DVLs stabilize the Roc-COR dimer, which according to current models (10) would enhance LRRK2 GTPase activity. DVLs may also hinder the binding of the E3 ubiquitin ligase CHIP (36,37) which interacts with the Roc domain and decreases LRRK2 levels via ubiquitination and proteasome-dependent degradation.
Interestingly, DVL proteins show a distinct subcellular localization when expressed in mammalian cells, forming doughnut-shaped cytoplasmic aggregates that are thought to represent DVL polymers that are able to mediate Wnt signalling (38). This unique expression pattern appears to be mediated by the DVL DIX domain, which is necessary for DVL polymerization. Co-expression of LRRK2 and DVLs resulted in a re-distribution of LRRK2 to DVL aggregates and changes in cell morphology, supporting a direct interaction of LRRK2 with DVL polymers. Co-localization of LRRK2 and DVL1 aggregates was also observed in neurites and growth cones of differentiated dopaminergic SH-SY5Y cells. These findings support the idea of an important interplay between LRRK2 and DVLs in the development and maintenance of neurites via Wnt signalling pathways. In humans, 19 spatially and temporally expressed Wnt ligands signal through their Frizzled (FZ) receptors and their co-receptors in the cell membrane, leading to hyperphosphorylation of DVLs and the subsequent downstream activation of one of three Wnt signalling pathways (19). These include the canonical Wnt pathway, the non-canonical or planar cell polarity cascade and the Wnt/Ca2+ pathway. These different pathways control cell fate and tissue polarity and affect several neuronal processes such as CNS development, axon guidance, neurite outgrowth/branching and synapse formation (19–27). DVL proteins are also involved in the differentiation of dopaminergic cells in the ventral midbrain, and in the release and recycling of dopaminergic vesicles at presynaptic sites (24–27) and have been linked to neurodegeneration (39–41). A potential link between LRRK2 and Wnt signalling was also suggested by a recent LRRK2 RNAi study (42), which revealed that LRRK2 knockdown resulted in the up-regulation of several genes involved in the canonical Wnt/β-catenin signalling pathway, including DVL2.
A key finding of this study is the modulation of a LRRK2–accessory protein interaction of significance for LRRK2 GTPase and kinase activity by PARK8 Roc-COR domain mutations segregating with Parkinson's disease. For example, the Y1699C substitution in the COR domain, resulting from a key familial PARK8 mutation (5), disrupts LRRK2 interactions with DVL1, DVL2 and DVL3. This could result from direct disruption of the DVL1-binding site, or alternatively Y1699C might alter the conformation of the Roc-COR dimer, making the structure less accessible to DVL1-3. Little is known about the influence of the Y1699C mutation on LRRK2 GTPase or kinase activity (7,10,12,13,16), but individuals with the Y1699C mutation show unique clinical aspects including a ‘prominent behavioural disorder’ with unexpectedly high prevalence of anxiety and depression that can pre-date the onset of Parkinson's disease symptoms (5). The Y1699C mutation also seems fully penetrant, which is not the case for other pathogenic mutations in LRRK2 such as G2019S, suggesting a more severe genetic defect for Y1699C (1–5). In contrast, the LRRK2 Roc-COR interaction with DVL1 is strengthened by pathogenic mutations causing R1441C/G/H changes and the R1728H/L substitutions. Although the R1728H/L substitutions have not been proved to be pathogenic, the likelihood that two different substitutions occur at exactly the same amino acid position is low, unless these are disease-causing. Our study supports a potential pathogenic mechanism for R1728H/L (4). Interestingly, the effects of the R1441C/G/H and R1728H/L substitutions were not observed for DVL1ΔDIX, DVL2 and DVL3. These results imply that DVL1 multimerization has a subtle effect on DVL1–LRRK2 interactions, and that DVL1 appears to interact differently with LRRK2 compared with DVL2 and DVL3. Intriguingly, substitutions that do not show clear segregation with Parkinson's disease (e.g. R1514Q and M1869T (3,4)) did not show clear-cut effects on interactions with DVL1 or DVL3, although a slight reduction in DVL2–LRRK2 interactions was observed for the R1514Q mutant. In summary, all investigated pathogenic mutations (Y1699C, R1441C/G/H and R1728H/L) in the Roc-COR tandem domain have a specific influence on the interaction of LRRK2 with DVL1, but only the Y1699C substitution alters interactions with DVL1, DVL2 and DVL3. If DVL proteins enhance LRRK2 GTPase activity, these results imply that Y1699C would reduce intrinsic GTPase activity mediated by DVL1–DVL3, whereas R1441C/G/H and R1728H/L would stabilize LRRK2–DVL1 interactions. The effect of such a stable complex on GTPase and kinase activity remains uncertain, as this could either reduce or enhance LRRK2 activity. Since dishevelled proteins are known to co-precipitate with small GTPases such as Rac1 and RhoA (20,35), studies with recombinant LRRK2 and DVL proteins will be required to provide clear evidence of an influence of DVLs on LRRK2 GTPase and kinase activity.
Lastly, this study also suggests that the dishevelled genes (DVL1, DVL2 and DVL3) represent excellent candidates for genetic screening in individuals with familial and idiopathic Parkinson's disease. Interestingly, DVL1 knockout mice are viable and fertile and demonstrate no gross physical abnormalities. However, they show an impaired social behaviour phenotype (43), with lower huddling behaviour, nest building, social dominance and whisker trimming. These behaviours could be interpreted as reflecting increased anxiety in social contexts, and whether this phenotype mirrors the ‘prominent behavioural disorder’ seen in patients with the LRRK2 Y1699C mutation remains to be seen. In contrast, both DVL2 and DVL3 knockout mice exhibit cardiovascular outflow tract defects, although DVL2 knockouts also exhibit vertebral and rib malformations and neural tube closure defects (44,45). Although DVL1 has been assessed as a candidate gene in Alzheimer's disease (40), as yet, no human disease has been associated with human DVL1, DVL2 or DVL3. Given our results, we suggest that it would be timely to re-examine this gene family in unresolved cases of familial or idiopathic Parkinson's disease. In particular, we note that the DVL1 gene maps to chromosome 1p36 (http://genome.ucsc.edu/), a region of the human genome enriched in Parkinson's disease genes, including PARK6 (PINK1), PARK7 (DJ-1) and PARK9 (ATP13A2). In addition, several studies have implicated DVL proteins in dopaminergic synapse formation and transmitter release (21,24–27). Over-expression of Parkinson's disease-associated LRRK2 mutations in neuronal cultures induces a progressive ‘reduction’ in neurite length and branching, especially for axons, the longest neuronal processes (28,29). Similar effects are observed when DVL1 is down-regulated via RNAi, which results in abrogation of axon differentiation (23). Lastly, DVL1 is one of a total of 24 genes that have been found to display a significantly higher rate of protein evolution in primates than in rodents, implying a specialized function in the human brain (46).
In summary, we propose that dishevelled proteins are key LRRK2 interactors, since they have the potential to influence LRRK2 GTPase and kinase activity. Since mutations in the Roc-COR tandem domain segregating with Parkinson's disease either weaken or strengthen LRRK2-DVL1 interactions, this implies that the correct level of LRRK2 activity is key to the health of dopaminergic neurones. We also suggest that DVL proteins and other components of Wnt signalling pathways may represent novel therapeutic targets in the treatment of Parkinson's disease, whereas the DVL1 gene on chromosome 1p36 should be considered a high-priority candidate for genetic screening. Our data also link LRRK2 to Wnt signalling pathways that control the development and maintenance of neuronal processes, suggesting novel potential mechanisms in the pathogenesis of Parkinson's disease.
MATERIALS AND METHODS
Cloning of LRRK2/DVL expression constructs and site-directed mutagenesis
LRRK2 and DVL cDNAs were amplified from human whole-brain first-strand cDNA (Clontech) and cloned into the YTH bait vector pDS-BAIT (pDS; Dualsystems Biotech), the prey vector pACT2 (Clontech) or the mammalian expression vectors pRK5myc or pRK5FLAG. This results in an in-frame fusion of the LexA-BD, GAL4-AD, myc or FLAG epitope tags to the N-termini of LRRK2 or DVL proteins, respectively. Mutations were made in different constructs using the QuikChange site-directed mutagenesis kit (Stratagene). Amplifications were performed using Pfx DNA polymerase, and all plasmids were sequenced to confirm the introduction of the desired mutations.
YTH assays
The yeast strain L40 (Invitrogen) was co-transformed with various pDS-LRRK2 and pACT2-DVL constructs. Transformations were plated on selective dropout media lacking either leucine, tryptophan and histidine supplemented with 0.5 mm 3-AT (for suppression of ‘leaky’ histidine expression) for nutritional selection, or leucine and tryptophan for transformation controls. After incubation at 30°C for 3–6 days to allow prototropic colonies to emerge, LacZ reporter gene assays were performed as described previously (47). Quantitative YTH assays were performed by resuspending cell pellets in Z-buffer containing 40 mm β-mercaptoethanol, followed by lysis in 0.1% (w/v) SDS and 0.1% (v/v) chloroform (48). All protein interactions were assayed in three to four independent experiments in triplicate. After the addition of chloropheno-red-β-d-galactopyranoside (CPRG), the colour change was recorded at 540 nm and readings adjusted for turbidity of the yeast suspension at 620 nm. The background signal (bait plus empty pACT2 vector) was subtracted from each reading and values were normalized to the wild-type Roc-COR response, which was set at 100%. Statistical significance was determined using a Student's t-test (two-tailed). Error bars represent the standard deviation of the mean.
Cell culture and immunocytochemistry
HEK293 cells (ATCC CRL1573) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal bovine serum, 2 mm glutamine, 100 U/ml penicillin G and 100 µg/ml streptomycin at 37°C in 95% air−5% CO2 (49). Exponentially growing cells were transfected with constructs encoding epitope-tagged DVL and LRRK2 using Lipofectamine LTX reagent (Invitrogen). After 24 h, cells were fixed in 4% (w/v) PFA and stained with antibodies recognizing myc (Sigma) and FLAG (Sigma) and secondary anti-mouse and anti-rabbit antibodies conjugated to Alexa 488, Alexa 546 or Cy5 fluorochromes (Molecular Probes).
Co-immunoprecipitation and Western blotting
HEK293 cells were transfected as described earlier and harvested 48 h post-transfection, lysed in a solution containing 50 mm NaCl/complete protease inhibitor cocktail (Roche) and homogenized using a tissue grinder. Following centrifugation (4°C, 40 min, 100 000g), cell lysates were recovered and the concentration of NaCl was increased to 150 mm. One millilitre of cell lysate containing ∼700 µg of protein was added to 40 µl of anti-FLAG M2 affinity gel (Sigma) and incubated overnight at 4°C on a turning disk in order to purify the FLAG-tagged proteins. The affinity gel was subjected to centrifugation (4°C, 100g, 3 min), followed by two washes in 50 mm NaCl, 50 mm Tris, 0.1% Triton X-100, two washes in 150 mm NaCl, PBS, 0.1% Triton X-100 and two washes in PBS, 0.1% Triton X-100. The FLAG fusion proteins were eluted with 150 ng of 3× FLAG peptide (Sigma) for 30 min at RT. The eluates were analysed by SDS–PAGE and immunoblotting. Approximately 10 µg of protein was loaded into 4–12% (w/v) BisTris pre-cast gels (Invitrogen). Proteins were transferred to polyvinylidine fluoride membranes (Millipore) and non-specific bands blocked with 5% (w/v) skimmed milk in PBS plus 0.1% (v/v) Tween 20 or with 20% (v/v) horse serum in PBS. Anti-myc antibody (Sigma) was used at 1:2000, and anti-FLAG antibody (Sigma) was used at 1:3000 at 4°C overnight. For detection, an HRP-conjugated anti-rabbit secondary antibody (Santa Cruz) was used at a final dilution of 1:2000, together with the SuperSignal West Pico Chemiluminescent Substrate (Pierce).
Culture and immunocytochemistry of SH-SY5Y cells
SH-SY5Y cells were cultured in DMEM:F12 (1:1 Gibco) supplemented with 10% (v/v) FBS and 1% (v/v) penicillin/streptomycin. SH-SY5Y cells were differentiated by treatment with 10 µm retinoic acid and subsequent incubation for 4–7 days in neurobasal medium supplemented with 2 mm of l-glutamine, 1% (v/v) penicillin/streptomycin and B27 supplement (Gibco). SH-SY5Y cells were transiently transfected using Neuromag (Oz Biosciences) or FuGene 6 (Roche). After 48 h, cells were fixed in methanol and stained with antibodies recognizing GAP-43 (mAb347; Chemicon), NeuN (mAb377; Chemicon), beta-tubulin (NB600-1470; Novus Biologicals) or acetylated tubulin (6-11B-1; Sigma). In order to destabilize microtubules, cells were treated with 5 µm nocodazole (Sigma) for 30 min prior to fixation and immunocytochemistry.
Confocal microscopy and image analysis
Confocal microscopy was performed using a Zeiss LSM 510 META. All images were taken with a ×63 objective. Fluorescence excited by the 488, 543 and 633 nm laser lines of argon and helium/neon lasers was detected separately using only one laser at the time (multitrack function) and a combination of band pass filters (BP 505-530, BP 560-615), long pass (LP 560) filters and meta function (649–798) dependent on the combination of fluorochromes used.
Q-PCR experiments
A relative quantification study of LRRK2/DVL gene expression in substantia nigra was performed using the comparative method (ΔΔCT), commercially available TaqMan primer sets and an ABI 7500 real-time PCR system (Applied Biosystems, Warrington, UK). Total RNA was converted into first-strand cDNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems). Cycling conditions were 50°C for 2 min, 95°C for 10 min and 40 cycles of 95°C for 15 s, followed by 60°C for 1 min. Data were analysed using the SDS 7500 system software (v1.3.1, Applied Biosystems) before being exported into Microsoft Excel. Experiments were repeated four times in triplicate using GAPDH and PPIA as endogenous controls for normalizing gene expression.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by grants from the Royal Society, the British Medical Association (Dawkins and Lawson award) and the Wellcome Trust (WT088145) to K.H. and two School of Pharmacy PhD studentships. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. Funding to pay the Open Access publication charges for this article was provided by The School of Pharmacy.
ACKNOWLEDGEMENTS
K.H. thanks Dr Mark R. Cookson and Professor Robert J. Harvey for constructive comments and suggestions on the study.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Paisán-Ruíz C., Jain S., Evans E.W., Gilks W.P., Simón J., van der Brug M., López de Munain A., Aparicio S., Gil A.M., Khan N., et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 2.Zimprich A., Biskup S., Leitner P., Lichtner P., Farrer M., Lincoln S., Kachergus J., Hulihan M., Uitti R.J., Calne D.B., et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Healy D.G., Falchi M., O'Sullivan S.S., Bonifati V., Durr A., Bressman S., Brice A., Aasly J., Zabetian C.P., Goldwurm S., et al. Phenotype, genotype, and worldwide genetic penetrance of LRRK2-associated Parkinson's disease: a case–control study. Lancet Neurol. 2008;7:583–590. doi: 10.1016/S1474-4422(08)70117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paisán-Ruíz C., Nath P., Washecka N., Gibbs J.R., Singleton A.B. Comprehensive analysis of LRRK2 in publicly available Parkinson's disease cases and neurologically normal controls. Hum. Mutat. 2008;29:485–490. doi: 10.1002/humu.20668. [DOI] [PubMed] [Google Scholar]
- 5.Khan N.L., Jain S., Lynch J.M., Pavese N., Abou-Sleiman P., Holton J.L., Healy D.G., Gilks W.P., Sweeney M.G., Ganguly M., et al. Mutations in the gene LRRK2 encoding dardarin (PARK8) cause familial Parkinson's disease: clinical, pathological, olfactory and functional imaging and genetic data. Brain. 2005;128:2786–2796. doi: 10.1093/brain/awh667. [DOI] [PubMed] [Google Scholar]
- 6.Bosgraaf L., Van Haastert P.J. Roc, a Ras/GTPase domain in complex proteins. Biochim. Biophys. Acta. 2003;1643:5–10. doi: 10.1016/j.bbamcr.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 7.Smith W.W., Pei Z., Jiang H., Dawson V.L., Dawson T.M., Ross C.A. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat. Neurosci. 2006;9:1231–1233. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- 8.Mata I.F., Wedemeyer W.J., Farrer M.J., Taylor J.P., Gallo K.A. LRRK2 in Parkinson's disease: protein domains and functional insights. Trends Neurosci. 2006;29:286–293. doi: 10.1016/j.tins.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 9.Guo L., Gandhi P.N., Wang W., Petersen R.B., Wilson-Delfosse A.L., Chen S.G. The Parkinson's disease-associated protein, leucine-rich repeat kinase 2 (LRRK2), is an authentic GTPase that stimulates kinase activity. Exp. Cell Res. 2007;313:3658–3670. doi: 10.1016/j.yexcr.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gotthardt K., Weyand M., Kortholt A., Van Haastert P.J., Wittinghofer A. Structure of the Roc-COR domain tandem of C. tepidum, a prokaryotic homologue of the human LRRK2 Parkinson kinase. EMBO J. 2008;27:2239–2249. doi: 10.1038/emboj.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito G., Okai T., Fujino G., Takeda K., Ichijo H., Katada T., Iwatsubo T. GTP binding is essential to the protein kinase activity of LRRK2, a causative gene product for familial Parkinson's disease. Biochemistry. 2007;46:1380–1388. doi: 10.1021/bi061960m. [DOI] [PubMed] [Google Scholar]
- 12.West A.B., Moore D.J., Choi C., Andrabi S.A., Li X., Dikeman D., Biskup S., Zhang Z., Lim K.L., Dawson V.L., Dawson T.M. Parkinson's disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum. Mol. Genet. 2007;16:223–232. doi: 10.1093/hmg/ddl471. [DOI] [PubMed] [Google Scholar]
- 13.Greggio E., Jain S., Kingsbury A., Bandopadhyay R., Lewis P., Kaganovich A., van der Brug M.P., Beilina A., Blackinton J., Thomas K.J., et al. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol. Dis. 2006;23:329–341. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 14.Lewis P.A., Greggio E., Beilina A., Jain S., Baker A., Cookson M.R. The R1441C mutation of LRRK2 disrupts GTP hydrolysis. Biochem. Biophys. Res. Commun. 2007;357:668–671. doi: 10.1016/j.bbrc.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X., Tan Y.C., Poulose S., Olanow C.W., Huang X.Y., Yue Z. Leucine-rich repeat kinase 2 (LRRK2)/PARK8 possesses GTPase activity that is altered in familial Parkinson's disease R1441C/G mutants. J. Neurochem. 2007;103:238–247. doi: 10.1111/j.1471-4159.2007.04743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaleel M., Nichols R.J., Deak M., Campbell D.G., Gillardon F., Knebel A., Alessi D.R. LRRK2 phosphorylates moesin at threonine-558: characterization of how Parkinson's disease mutants affect kinase activity. Biochem. J. 2007;405:307–317. doi: 10.1042/BJ20070209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greggio E., Zambrano I., Kaganovich A., Beilina A., Taymans J.M., Daniëls V., Lewis P., Jain S., Ding J., Syed A., et al. The Parkinson disease-associated leucine-rich repeat kinase 2 (LRRK2) is a dimer that undergoes intramolecular autophosphorylation. J. Biol. Chem. 2008;13:16906–16914. doi: 10.1074/jbc.M708718200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bos J.L., Rehmann H., Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 19.Wallingford J.B., Habas R. The developmental biology of Dishevelled: an enigmatic protein governing cell fate and cell polarity. Development. 2005;132:4421–4436. doi: 10.1242/dev.02068. [DOI] [PubMed] [Google Scholar]
- 20.Habas R., Dawid I.B., He X. Coactivation of Rac and Rho by Wnt/Frizzled signaling is required for vertebrate gastrulation. Genes Dev. 2003;17:295–309. doi: 10.1101/gad.1022203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmad-Annuar A., Ciani L., Simeonidis I., Herreros J., Fredj N.B., Rosso S.B., Hall A., Brickley S., Salinas P.C. Signaling across the synapse: a role for Wnt and Dishevelled in presynaptic assembly and neurotransmitter release. J. Cell Biol. 2006;174:127–139. doi: 10.1083/jcb.200511054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ciani L., Salinas P.C. c-Jun N-terminal kinase (JNK) cooperates with Gsk3beta to regulate dishevelled-mediated microtubule stability. BMC Cell. Biol. 2007;8:27. doi: 10.1186/1471-2121-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X., Zhu J., Yang G.Y., Wang Q.J., Qian L., Chen Y.M., Chen F., Tao Y., Hu H.S., Wang T., Luo Z.G. Dishevelled promotes axon differentiation by regulating atypical protein kinase C. Nat. Cell Biol. 2007;9:743–754. doi: 10.1038/ncb1603. [DOI] [PubMed] [Google Scholar]
- 24.Schulte G., Bryja V., Rawal N., Castelo-Branco G., Sousa K.M., Arenas E. Purified Wnt-5a increases differentiation of midbrain dopaminergic cells and dishevelled phosphorylation. J. Neurochem. 2005;92:1550–1553. doi: 10.1111/j.1471-4159.2004.03022.x. [DOI] [PubMed] [Google Scholar]
- 25.Castelo-Branco G., Sousa K.M., Bryja V., Pinto L., Wagner J., Arenas E. Ventral midbrain glia express region-specific transcription factors and regulate dopaminergic neurogenesis through Wnt-5a secretion. Mol. Cell. Neurosci. 2006;31:251–262. doi: 10.1016/j.mcn.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 26.Bryja V., Schulte G., Rawal N., Grahn A., Arenas E. Wnt-5a induces Dishevelled phosphorylation and dopaminergic differentiation via a CK1-dependent mechanism. J. Cell Sci. 2007;120:586–595. doi: 10.1242/jcs.03368. [DOI] [PubMed] [Google Scholar]
- 27.Andersson E.R., Prakash N., Cajanek L., Minina E., Bryja V., Bryjova L., Yamaguchi T.P., Hall A.C., Wurst W., Arenas E. Wnt5a regulates ventral midbrain morphogenesis and the development of A9-A10 dopaminergic cells in vivo. PLoS ONE. 2008;3:e3517. doi: 10.1371/journal.pone.0003517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacLeod D., Dowman J., Hammond R., Leete T., Inoue K., Abeliovich A. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron. 2006;52:587–593. doi: 10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 29.Plowey E.D., Cherra S.J., III, Liu Y.J., Chu C.T. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J. Neurochem. 2008;105:1048–1056. doi: 10.1111/j.1471-4159.2008.05217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sämann J., Hegermann J., Gromoff E.V., Eimer S., Baumeister R., Schmidt E. Caenorhabditis elegans LRK-1 and PINK-1 act antagonistically in stress response and neurite outgrowth. J. Biol. Chem. 2009;284:16482–16491. doi: 10.1074/jbc.M808255200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lesnick T.G., Papapetropoulos S., Mash D.C., Ffrench-Mullen J., Shehadeh L., de Andrade M., Henley J.R., Rocca W.A., Ahlskog J.E., Maraganore D.M. A genomic pathway approach to a complex disease: axon guidance and Parkinson disease. PLoS Genet. 2007;3:e98. doi: 10.1371/journal.pgen.0030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LeBoeuf A.C., Levy S.F., Gaylord M., Bhattacharya A., Singh A.K., Jordan M.A., Wilson L., Feinstein S.C. FTDP-17 mutations in Tau alter the regulation of microtubule dynamics: an ‘alternative core’ model for normal and pathological Tau action. J. Biol. Chem. 2008;283:36406–36415. doi: 10.1074/jbc.M803519200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feinstein S.C., Wilson L. Inability of tau to properly regulate neuronal microtubule dynamics: a loss-of-function mechanism by which tau might mediate neuronal cell death. Biochim. Biophys. Acta. 2005;1739:268–279. doi: 10.1016/j.bbadis.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 34.Braak H., Ghebremedhin E., Rüb U., Bratzke H., Del Tredici K. Stages in the development of Parkinson's disease-related pathology. Cell Tissue Res. 2004;318:121–134. doi: 10.1007/s00441-004-0956-9. [DOI] [PubMed] [Google Scholar]
- 35.Habas R., Kato Y., He X. Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel Formin homology protein Daam1. Cell. 2001;107:843–854. doi: 10.1016/s0092-8674(01)00614-6. [DOI] [PubMed] [Google Scholar]
- 36.Ko H.S., Bailey R., Smith W.W., Liu Z., Shin J.H., Lee Y.I., Zhang Y.J., Jiang H., Ross C.A., Moore D.J., et al. CHIP regulates leucine-rich repeat kinase-2 ubiquitination, degradation, and toxicity. Proc. Natl Acad. Sci. USA. 2009;106:2897–2902. doi: 10.1073/pnas.0810123106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding X., Goldberg M.S. Regulation of LRRK2 stability by the E3 ubiquitin ligase CHIP. PLoS One. 2009;4:e5949. doi: 10.1371/journal.pone.0005949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwarz-Romond T., Fiedler M., Shibata N., Butler P.J., Kikuchi A., Higuchi Y., Bienz M. The DIX domain of Dishevelled confers Wnt signaling by dynamic polymerization. Nat. Struct. Mol. Biol. 2007;14:484–492. doi: 10.1038/nsmb1247. [DOI] [PubMed] [Google Scholar]
- 39.De Ferrari G.V., Chacón M.A., Barría M.I., Garrido J.L., Godoy J.A., Olivares G., Reyes A.E., Alvarez A., Bronfman M., Inestrosa N.C. Activation of Wnt signaling rescues neurodegeneration and behavioral impairments induced by beta-amyloid fibrils. Mol. Psychiatry. 2003;8:195–208. doi: 10.1038/sj.mp.4001208. [DOI] [PubMed] [Google Scholar]
- 40.Russ C., Lovestone S., Powell J.F. Identification of genomic organisation, sequence variants and analysis of the role of the human dishevelled 1 gene in late onset Alzheimer's disease. Mol. Psychiatry. 2002;7:104–109. doi: 10.1038/sj.mp.4000941. [DOI] [PubMed] [Google Scholar]
- 41.Toledo E.M., Colombres M., Inestrosa N.C. Wnt signalling in neuroprotection and stem cell differentiation. Prog. Neurobiol. 2008;86:281–296. doi: 10.1016/j.pneurobio.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 42.Häbig K., Walter M., Poths S., Riess O., Bonin M. RNA interference of LRRK2-microarray expression analysis of a Parkinson's disease key player. Neurogenetics. 2008;9:83–94. doi: 10.1007/s10048-007-0114-0. [DOI] [PubMed] [Google Scholar]
- 43.Long J.M., LaPorte P., Paylor R., Wynshaw-Boris A. Expanded characterization of the social interaction abnormalities in mice lacking Dvl1. Genes Brain Behav. 2004;3:51–62. doi: 10.1046/j.1601-183x.2003.00045.x. [DOI] [PubMed] [Google Scholar]
- 44.Hamblet N.S., Lijam N., Ruiz-Lozano P., Wang J., Yang Y., Luo Z., Mei L., Chien K.R., Sussman D.J., Wynshaw-Boris A. Dishevelled 2 is essential for cardiac outflow tract development, somite segmentation and neural tube closure. Development. 2002;129:5827–5838. doi: 10.1242/dev.00164. [DOI] [PubMed] [Google Scholar]
- 45.Etheridge S.L., Ray S., Li S., Hamblet N.S., Lijam N., Tsang M., Greer J., Kardos N., Wang J., Sussman D.J., Chen P., Wynshaw-Boris A. Murine dishevelled 3 functions in redundant pathways with dishevelled 1 and 2 in normal cardiac outflow tract, cochlea, and neural tube development. PLoS Genet. 2008;4:e1000259. doi: 10.1371/journal.pgen.1000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dorus S., Vallender E.J., Evans P.D., Anderson J.R., Gilbert S.L., Mahowald M., Wyckoff G.J., Malcom C.M., Lahn B.T. Accelerated evolution of nervous system genes in the origin of Homo sapiens. Cell. 2004;119:1027–1040. doi: 10.1016/j.cell.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 47.Fuller K.J., Morse M.A., White J.H., Dowell S.J., Sims M.J. Development of a yeast trihybrid screen using stable yeast strains and regulated protein expression. Biotechniques. 1998;25:85–92. doi: 10.2144/98251st04. [DOI] [PubMed] [Google Scholar]
- 48.Ramamoorthy K., Wang F., Chen I.C., Norris J.D., McDonnell D.P., Leonard L.S., Gaido K.W., Bocchinfuso W.P., Korach K.S., Safe S. Estrogenic activity of a dieldrin/toxaphene mixture in the mouse uterus, MCF-7 human breast cancer cells, and yeast-based estrogen receptor assays: no apparent synergism. Endocrinology. 1997;138:1520–1527. doi: 10.1210/endo.138.4.5056. [DOI] [PubMed] [Google Scholar]
- 49.Harvey K., Duguid I.C., Alldred M.J., Beatty S.E., Ward H., Keep N.H., Lingenfelter S.E., Pearce B.R., Lundgren J., Owen M.J., et al. The GDP-GTP exchange factor collybistin: an essential determinant of neuronal gephyrin clustering. J. Neurosci. 2004;24:5816–5826. doi: 10.1523/JNEUROSCI.1184-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.