Abstract
Three unusual tiglic acid-containing cyclodepsipeptides, possessing the revised regioisomeric structures for largamides A–C (1–3),have been isolated from the marine cyanobacterium Lyngbya confervoides collected from southeastern Florida. The two-dimensional structures were determined by NMR spectroscopy and the absolute configurations by chiral HPLC analysis of degradation products. Compounds 1–3 are moderate inhibitors of mammalian elastase activity in vitro with IC50 values ranging from 0.53 to 1.41 μM.
Keywords: Marine cyanobacteria, Lyngbya confervoides, cyclodepsipeptides, elastase
Introduction
Marine cyanobacteria produce secondary metabolites with unusual structural features most commonly incorporated in a peptidic framework or in peptide–polyketide hybrids [1]. The presence of unconventional or highly modified amino acid units is a characteristic biosynthetic signature of cyanobacterial metabolites [1,2]. We initiated screening of various collections of marine cyanobacteria from coastal Florida with the aim of discovering novel protease inhibitors. A preliminary bioassay-guided fractionation of the organic extract of Lyngbya confervoides led us to identify several active fractions. Our systematic chemical investigations of the active fractions of Lyngbya confervoides that previously yielded the potent elastase and chymotrypsin inhibitors lyngbyastatins 4–6 [3,4], trypsin inhibitor pompanopeptin A [5], and putative carboxypeptidase inhibitor pompanopeptin B [5] now also yielded largamides A–C (1–3), along with largamides D–H [6]. Here we describe the isolation, biological evaluation and revision of the proposed structures of largamides A–C (1–3) [6]. The natural products were tested for inhibitory activity against three proteolytic enzymes, namely elastase, chymotrypsin and trypsin. Compounds 1–3 demonstrated moderate inhibition of porcine pancreatic elastase (PPE), but no activity against the other two serine proteases up to 50 μM.
Elastase is associated with tissue destruction and inflammation characteristic of chronic obstructive pulmonary disease (COPD) such as emphysema. COPD is the fourth leading cause of death in the USA [7]. In a murine model of emphysema, PPE was capable of recruiting monocytes to the lung, increasing lung macrophage content to generate an inflammatory cell infiltrate [7]. PPE causes direct proteolytic injury to the lung and induces airspace enlargement; however, greater lesions develop related to subsequent endogenous inflammation and destruction [7]. Because of the critical role of elastase in inflammatory processes, elastase inhibitors might be of importance in the treatment of inflammation related syndromes.
Materials and Methods
General Experimental Procedures
Optical rotation was measured on a Perkin Elmer 341 polarimeter. UV spectra were recorded using a SpectraMax M5 (Molecular Devices). 1H and 2D NMR spectra were recorded in DMF-d7 on a Varian 500 MHz spectrometer operating at 500 MHz and 125 MHz using residual solvent signals as the internal standard (δH 8.02, δC 162.7). HSQC experiments were optimized for 1JCH = 145 Hz, and HMBC experiments were optimized for nJC,H = 7 Hz. HRMS data were obtained using an Agilent LC-TOF mass spectrometer equipped with an APCI/ESI multimode ion source detector.
Porcine pancreatic elastase (Elastase-high purity; EC134) and N-succinyl-Ala-Ala-Ala-p-nitroanilide (NS945, purity >95%) were purchased from EPC, Missouri, USA. Trypsin from porcine pancreas (T0303,Type IX-S, 13,000–20,000 BAEE units/mg protein), α-chymotrypsin from bovine pancreas (C4129, Type II, ≥40 units/mg protein), N-succinyl-Gly-Gly-Phe-p-nitroanilide (S1899, purity >98%), N-α-benzoyl-DL-arginine 4-nitroanilide hydrochloride (B4874, purity ≥98%), and PMSF (P7626, purity ≥98.5%) were procured from Sigma, USA. All other reagents and solvents used were of reagent grade or HPLC grades.
Biological material
Samples of Lyngbya confervoides were collected off the coast of Broward County (Fort Lauderdale, Florida, USA (26°01.1414′N, 80°05.9973′W; 26°15.134′N, 80°03.908′W) in July 2004 and August 2005. S. Golubic identified the cyanobacterium [8]. A voucher specimen is retained at the Smithsonian Marine Station.
Extraction and Isolation
The freeze-dried sample (539.7 g)of Lyngbya confervoides was extracted with EtOAc–MeOH (1:1) and EtOH–H2O (1:1) to afford VPL–NP (78.8 g) and VPL–P (113.8 g), respectively. The crude extracts VPL–NP (26.6 g) and VPL–P (95.9 g) were then combined, suspended in H2O (500 mL) and defatted with hexanes (300 mL × 3; 0.67 g). The dried aqueous layer (~118 g) was further partitioned between n-BuOH (250 mL × 3) and H2O to afford n-BuOH extract (6.3 g) and an aqueous extract (~110 g) enriched with salt. The concentrated n-BuOH extract was applied on a column packed with diaion HP-20 (Supelco) polymeric resin (60 g), fractionated with H2O and increasing concentrations of MeOH, and then with MeCN to yield 7 fractions [Fr. 1: H2O (100%, 2 L, ~3 g); Fr. 2: H2O–MeOH (75:25 and 50:50), 2 L, 350 mg); Fr. 3: H2O–MeOH (25:75, 1 L, 175 mg); Fr. 4: H2O:MeOH (25:75, 1 L, 155 mg); Fr. 5: H2O:MeOH (25:75, 1 L, 368 mg); Fr. 6: MeOH (100%, 1 L, 496 mg); Fr. 7: MeCN (100%, 1 L, 162 mg)]. The fractions (Fr. 5 and Fr. 6) were subjected to reversed-phase preparative HPLC [Phenomenex Luna 10u (C18), 100 × 21.20 mm, 10.0 mL/min; UV detection at 220 and 240 nm, using a MeOH–H2O linear gradient (30–100% over 40 min and then 100% MeOH for 10 min)]. Fractions eluted with tR 16–23 min (58–70% MeOH) were then subjected to repeated semi-preparative reversed-phase HPLC (YMC-Pack ODS-AQ, 250 × 10 mm, 2.0 mL/min; UV detection at 220 and 240 nm) using a 0.05% TFA in MeOH (60–90% for 25 min, then 90–100% for 10 min and finally 100% MeOH for 10 min), to give largamide A (1), tR 21.0 min (1.2 mg), largamide B (2), tR 18.5 min (5.0 mg), and largamide C (3), tR 20.5 min (1.8 mg).
Largamide A (1): colorless, amorphous solid; [α]20D –55 (c 0.12, MeOH); UV (MeOH) λmax (log ε) 220 ( 4.34 ), 280 (3.32) nm; IR (film) νmax 3317 (br), 2959, 2924, 1725, 1659 (br), 1548, 1443, 1204 cm−1; 1H NMR, 13C NMR, and HMBC data, see Table 1; HR-ESI/APCI-MS m/z [M + H]+ 842.4303 (calcd for C41H60N7O12, 842.4300).
Table 1.
NMR data for largamide A (1) in DMF-d7 (500 MHz)
| Unit | C/H no. | δH (J in Hz) | δC, mult. | HMBCa |
|---|---|---|---|---|
| Leu | 1 | 172.0, qC | ||
| 2 | 4.67, ddd (10.0, 9.5, 4.5) | 49.8, CH | ||
| 3 | 1.70, m | 40.9, CH2 | 2, 4 | |
| 1.49, m | ||||
| 4 | 1.58, m | 24.8, CH | ||
| 5 | 0.86, d (6.5) | 23.6, CH3 | 3, 4, 6 | |
| 6 | 0.89, d (6.5) | 21.7, CH3 | 3, 4, 5 | |
| NH | 7.62, d (9.5) | 1 (Glu) | ||
| Glu | 1 | 171.6, qC | ||
| 2 | 4.51, ddd (9.5, 9.0, 4.5) | 53.0, CH | 1, 4 | |
| 3 | 2.53, m | 27.0, CH2 | 5 | |
| 2.14, m | ||||
| 4 | 2.54, m | 31.1, CH2 | 5 | |
| 2.42, m | ||||
| 5 | 175.2, qC | |||
| OH | not observed | |||
| NH | 7.54, d (9.0) | 1 (Abu) | ||
| Abu | 1 | 164.0, qC | ||
| 2 | 130.9, qC | |||
| 3 | 6.54, q (7.0) | 129.1, CH | 1 | |
| 4 | 1.76, d (7.0) | 12.7, CH3 | 1, 2, 3 | |
| NH | 10.16, s | 1 (Ala) | ||
| Ala | 1 | 175.8, qC | ||
| 2 | 4.33, qd (6.5, 2.5) | 50.7, CH | 1, 1 (Thr) | |
| 3 | 1.38, d (6.5) | 16.8, CH3 | 1, 2 | |
| NH | 8.83, d (2.5) | 1 (Thr) | ||
| Thr | 1 | 170.3, qC | ||
| 2 | 4.74, dd (8.5, 3.0) | 55.8, CH | 1, 1 (Tyr) | |
| 3 | 5.37, qd (6.3, 3.0) | 72.9, CH | 4 | |
| 4 | 1.18, d (6.3) | 16.1, CH3 | 2, 3 | |
| NH | 7.90, d (8.5) | 1 (Tyr) | ||
| Tyr | 1 | 172.7, qC | ||
| 2 | 4.78, ddd (10.0, 9.5, 4.5) | 55.9, CH | 1, 4 | |
| 3 | 3.07, dd (−14.0, 4.5) | 38.1, CH2 | 2, 4, 5/9 | |
| 2.82, dd (−14.0, 10.0) | ||||
| 4 | 128.6, qC | |||
| 5/9 | 7.12, d (8.5) | 131.1, CH | 7 | |
| 6/8 | 6.70, d (8.5) | 115.6, CH | 7 | |
| 7 | 157.1, qC | |||
| OH | 9.34, s | 7 | ||
| NH | 8.10, br | 1 (Val) | ||
| Val | 1 | 171.9, qC | ||
| 2 | 4.29, dd (9.0, 7.0) | 59.3, CH | 1, 3, 4, 5 | |
| 3 | 2.01, m | 31.6, CH | 2, 4, 5 | |
| 4 | 0.75, d (6.5) | 19.7, CH3 | 2, 3 | |
| 5 | 0.72, d (6.5) | 18.4, CH3 | 2, 3 | |
| NH | 7.26, brd (7.0) | 1 (Tig) | ||
| Tig | 1 | 169.3, qC | ||
| 2 | 132.7, qC | |||
| 3 | 6.42, qq (7.0, 1.0) | 130.4, CH | 1 | |
| 4 | 1.71, br d (7.0) | 13.8, CH3 | 2, 3 | |
| 5 | 1.80, br s | 12.5, CH3 | 1, 2, 3 |
Protons showing long-range correlation with indicated carbon.
Largamide B (2): colorless, amorphous solid; [α]20D –43 (c 0.12, MeOH); UV (MeOH) λmax (log ε) 220 ( 4.09 ), 280 (3.09) nm; IR (film) νmax 3331 (br), 2958, 2923, 1721, 1656, 1640, 1597 (br), 1461 cm−1; 1H NMR, 13C NMR, and HMBC data, see Table 2; HR-ESI/APCI-MS m/z [M + H]+ 920.4404 (calcd for C46H62N7O13, 920.4406).
Table 2.
NMR data for largamides B (2) and C (3) in DMF-d7 (500 MHz)
| Largamide B (2) | Largamide C (3) | ||||||
|---|---|---|---|---|---|---|---|
| Unit | C/H no. | δH (J in Hz) | δC, mult. | HMBCa | δH (J in Hz) | δC, mult. | HMBCa |
| Ahppab/Ahphac | 1 | 171.7, qC | 171.7, qC | ||||
| 2 | 4.65, ddd (9.5, 9.5, 5.5) | 51.1, CH | 1, 3, 4 | 4.61, ddd (9.5, 9.0, 5.0) | 51.2, CH | 1, 3, 4, 4a | |
| 3 | 1.88, m 1.56, m |
31.2, CH2 | 2, 4, 5 | 1.91, m 1.57, m |
31.4, CH2 | 5 | |
| 4 | 1.60, m 1.58, m |
27.9, CH2 | 2, 3, 5, 6 | 1.36, m | 25.6, CH2 | ||
| 4ac | – | – | 1.47, m 1.53, m |
32.0, CH2 | 2, 3, 5 | ||
| 5 | 2.56, m 2.48, m |
34.7, CH2 | 3, 4, 6, 7 | 2.43, m | 35.1, CH2 | 4, 4a, 6, 7/11 | |
| 6 | 133.1, qC | 133.4, qC | |||||
| 7/11 | 7.01, d (8.5) | 129.9, CH | 5, 6, 8/10, 9 | 7.01, d (8.5) | 129.7, CH | 5, 8/10, 9 | |
| 8/10 | 6.73, d (8.5) | 115.7, CH | 6, 7/11, 9 | 6.74, d (8.5) | 115.6, CH | 6, 9 | |
| 9 | 156.5, qC | 156.4, qC | |||||
| OH | 9.29, sd | 9.28, s | 8/10, 9 | ||||
| NH | 7.65, d (9.5) | 1, 1 (Glu) | 7.64, d (9.5) | ||||
| Glu | 1 | 171.6, qC | 171.6, qC | ||||
| 2 | 4.50, ddd (10.0, 9.0, 5.0) | 53.2, CH | 1, 3, 4, 1 (Abu) | 4.52, ddd (10.0, 8.5, 4.5) | 53.1, CH | 1, 3, 4, 1 (Abu) | |
| 3 | 2.51, m 2.18, m |
27.0, CH2 | 2, 4, 5 | 2.50, m 2.17, m |
27.0, CH2 | 1, 2, 4, 5 | |
| 4 | 2.59, m 2.47, m |
31.0, CH2 | 2, 3, 5 | 2.57, m 2.46, m |
31.0, CH2 | 2, 3, 5 | |
| 5 | 175.1, qC | 175.1, qC | |||||
| OH | 12.51, br s | not observed | |||||
| NH | 7.50, d (9.0) | 1 (Abu) | 7.51, d (8.5) | 2, 1 (Abu) | |||
| Abu | 1 | 164.1, qC | 164.0, qC | ||||
| 2 | 131.1, qC | 130.9, qC | |||||
| 3 | 6.55, qd (7.0, 1.0) | 129.2, CH | 1, 4 | 6.56, qd (7.0, 1.5) | 129.2, CH | 1, 4 | |
| 4 | 1.77, d (7.0) | 12.7, CH3 | 1, 2, 3 | 1.76, d (7.0) | 12.5, CH3 | 1, 2, 3 | |
| NH | 10.16, s | 1, 1 (Ala) | 10.16, s | 1, 1 (Ala) | |||
| Ala | 1 | 175.8, qC | 175.7, qC | ||||
| 2 | 4.30, qd (7.0, 2.5) | 50.7, CH | 1, 3 | 4.32, qd (7.0, 2.5) | 50.6, CH | 1, 3 | |
| 3 | 1.37, d (7.0) | 16.8, CH3 | 1, 2 | 1.38, d (7.0) | 16.7, CH3 | 1, 2 | |
| NH | 8.83, d (2.5) | 1, 2, 3 | 8.85, d (2.5) | 1, 2, 3 | |||
| Thr | 1 | 170.4, qC | 170.3, qC | ||||
| 2 | 4.73, dd (8.5, 2.5) | 56.0, CH | 1, 1 (Tyr) | 4.73, dd (8.5, 3.0) | 55.8, CH | 1, 1 (Tyr) | |
| 3 | 5.37, qd (6.5, 2.5) | 73.4, CH | 1, 4, 1 (Ahppa) | 5.38, qd (6.5, 3.0) | 73.3, CH | 1, 1 (Ahpha) | |
| 4 | 1.18, d (6.5) | 16.2, CH3 | 2, 3 | 1.18, d (6.5) | 16.1, CH3 | 2, 3 | |
| NH | 7.96, d (8.5) | 2, 1(Tyr) | 7.91, d (8.5) | 2, 1 (Tyr) | |||
| Tyr | 1 | 172.8, qC | 172.7, qC | ||||
| 2 | 4.79, ddd (10.0, 8.5, 4.5) | 55.5, CH | 1, 3, 4, 1 (Val) | 4.78, ddd (10.0, 8.0, 4.5) | 55.5, CH | 1, 4, 1 (Val) | |
| 3 | 3.07, dd (–14.0, 4.5) 2.82, dd (–14.0, 10.0) |
38.2, CH2 | 1, 2, 4, 5/9 | 3.07, dd (–14.0, 4.5) 2.82, dd (–14.0, 10.0) |
38.0, CH2 | 1, 2, 4, 5/9 | |
| 4 | 128.6, qC | 128.5, qC | |||||
| 5/9 | 7.11, d (8.5) | 131.0, CH | 6/8, 7 | 7.11, d (8.5) | 131.0, CH | 3, 6/8, 7 | |
| 6/8 | 6.70, d (8.5) | 115.6, CH | 5/9, 7 | 6.70, d (8.5) | 115.6, CH | 5/9, 7 | |
| 7 | 157.1, qC | 157.0, qC | |||||
| OH | 9.34, sd | 9.35, s | 6/8, 7 | ||||
| NH | 8.09, d (8.5) | 1, 2, 1 (Val) | 8.08, d (8.0) | 1, 1 (Val) | |||
| Val | 1 | 172.0, qC | 172.0, qC | ||||
| 2 | 4.28, dd (8.5, 7.0) | 59.2, CH | 1, 3, 4, 5, 1 | 4.28, dd (8.5, 7.0) | 59.2, CH | 1, 3, 4, 5, 1(Tig) | |
| (Tig) | |||||||
| 3 | 2.01, m | 31.6, CH | 1, 2, 4, 5 | 2.01, m | 31.5, CH | 1, 2, 4, 5 | |
| 4 | 0.74, d (7.0) | 19.8, CH3 | 2, 3, 5 | 0.74, d (7.0) | 19.6, CH3 | 2, 3, 5 | |
| 5 | 0.71, d (7.0) | 18.3, CH3 | 2, 3, 4 | 0.72, d (7.0) | 18.2, CH3 | 2, 3, 4 | |
| NH | 7.25, d (8.5) | 1, 2, 1 (Tig) | 7.26, d (8.5) | 1, 2, 1 (Tig) | |||
| Tig | 1 | 169.3, qC | 169.3, qC | ||||
| 2 | 132.7, qC | 132.6, qC | |||||
| 3 | 6.42, qq (6.7, 1.5) | 130.4, CH | 1, 4, 5 | 6.42, qd (7.0, 1.0) | 130.4, CH | 1, 4, 5 | |
| 4 | 1.71, dq (6.7, 1.5) | 13.8, CH3 | 2, 3 | 1.71, dq (7.0, 1.0) | 13.7, CH3 | 2, 3 | |
| 5 | 1.79, br s | 12.6, CH3 | 1, 2, 3 | 1.80, br s | 12.4, CH3 | 1, 2, 3 | |
Protons showing long-range correlation with indicated carbon.
Refers to compound 2.
Refers to compound 3.
No HMBC observed. Assigned by comparison of data for 2 with data for 1 and 3.
Largamide C (3): colorless, amorphous solid; [α]20D –46 (c 0.11, MeOH); UV (MeOH) λmax (log ε) 220 ( 4.27 ), 280 (2.97) nm; IR (film) νmax 3318 (br), 2967, 2924, 1725, 1662, 1641, 1597 (br), 1465 cm−1; 1H NMR, 13C NMR, and HMBC data, see Table 2; HR-ESI/APCI-MS m/z [M + H]+ 934.4559 (calcd for C47H64N7O13, 934.4562).
Absolute Configuration
Compounds 1, 2, and 3 (0.1 mg each) were treated with 6 N HCl (0.3 mL) and heated at 110 ºC for 24 h. The hydrolyzates were evaporated to dryness and the residue resuspended in H2O (100 μL) and then subjected to chiral HPLC analysis (Phenomenex, Chirex 3126 N,S-dioctyl-(D)-penicillamine, 250 mm × 4.60 mm, 5 μm; solvent, 2 mM CuSO4–MeCN, 95:5); UV detection 254 nm; flow rate 1.0 mL/min). The retention times of amino acids in the hydrolyzates were compared with those of authentic standards. The retention times for the standards were as follows (tR, min): L-Ala (6.8), D-Ala (8.5), L-Thr (7.2), L-allo-Thr (9.4), D-Thr (8.0), D-allo-Thr (9.4), L-Val (17.2), D-Val (22.2), L-Tyr (53.0), D-Tyr (60.5), L-Leu (47.0), D-Leu (58.0), L-Glu (53.0), and D-Glu (57.0). The hydrolyzates of 1–3 yielded peaks corresponding to L-Ala (6.8) and L-Thr (7.2), L-Val (17.2), D-Glu (57.0) and D-Tyr (60.5). The hydrolyzate of compound 1 also generated an additional peak (tR 47.0 min), thus verifying the presence of L-Leu.
Compounds 2 and 3 (0.1 mg each) were each dissolved in 3 mL of MeOH and ozone was bubbled through the solution at 25 °C for 30 min. The solvent was evaporated, and the products were suspended in H2O2–HCOOH (1:2) and heated for 20 min at 70 oC. Then the mixtures were concentrated to dryness and subjected to acid hydrolysis and chiral HPLC analysis as described above, confirming the presence of L-Ala, L-Val, L-Thr, and D-Glu in both compounds. An additional peak for D-Asp due to ozonolysis (tR 35.5 min; solvent, 2 mM CuSO4–MeCN, 95:5) must have originated from Tyr, indicating D configuration of Tyr in the parent molecules (tR of L-Asp standard, 28.0 min). Furthermore, L-2-aminoadipic acid (tR 58.5 min; solvent, 2 mM CuSO4–MeCN, 95:5) and L-2-aminopimelic acid (tR 41.0 min; solvent, 2 mM CuSO4–MeCN, 90:10) were detected in the degradation product mixtures of compounds 2 and 3, respectively. The corresponding D-isomers were not present. Retention times of standards (tR, min) using the matching solvent systems: D-2-aminoadipic acid (84.0), D-2-aminopimelic acid (53.0).
Serine Protease Inhibition Assays
Compounds 1–3 dissolved in DMSO were tested for serine protease-inhibitory activity against porcine pancreatic elastase, α-chymotrypsin from bovine pancreas, or trypsin from porcine pancreas, in the presence of chromogenic substrates (N-succinyl-Ala-Ala-Ala-p-nitroanilide for elastase, N-succinyl-Gly-Gly-Phe-p-nitroanilide for chymotrypsin, N-α-benzoyl-DL-arginine 4-nitroanilide hydrochloride for trypsin). Activity was determined [3–5] by measuring the increase in absorbance at 405 nm over 30 min in a microplate reader using phenylmethylsulfonyl fluoride (PMSF) and lyngbyastatin 4 (purity ≥ 95%)[3] as positive controls. All assays were performed in quadruplicate and the data were expressed as mean ± S.D. and IC50 values calculated.
Results
The organic extract of Lyngbya confervoides collected near Ft. Lauderdale (Florida, USA) was subjected to HP-20 chromatography, crude reversed-phase fractionation and two HPLC purification steps to yield compounds 1–3 as colorless, amorphous solids. The planar structures of 1–3 (Figure 1) were determined by a combination of NMR (1H, 13C, COSY, 1D TOCSY, ROESY, HSQC, and HMBC) spectroscopic analysis and mass spectrometry as described below.
Fig. 1.
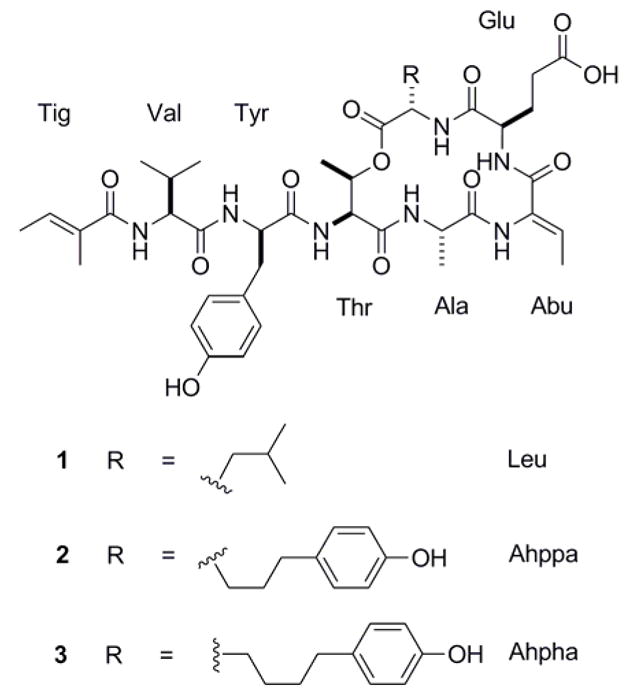
Structures of largamides A–C (1–3)
Compound 1 gave a [M + H]+ peak at m/z 842.4303 in the HR-ESI/APCI-MS, which, combined with 13C NMR data, suggested a molecular formula of C41H59N7O12. A detailed 1H NMR, COSY, HSQC and HMBC analysis in DMF-d7 (Table 1) indicated that compound 1 was a heptapeptide, consisting of six regular amino acid units (alanine, valine, glutamic acid, tyrosine, threonine, leucine) and one modified amino acid moiety, 2-amino-2-butenoic acid (Abu). Remaining 1H NMR signals for two deshielded methyl groups (δH 1.80 and 1.71) and one olefinic methine proton (δH 6.42) could be rationalized by an additional carboxylic acid residue, 2-methyl-2-butenoic acid, supported by COSY and HMBC data. In particular, signal multiplicity (qq) and coupling constants (7.0 and 1.0 Hz) for the olefinic methine proton H-3 (δH 6.42) were indicative of vicinal and allylic coupling to two methyl groups. Lack of a ROESY correlation between both the 2-methyl group resonating at δH 1.80 and the methine resonating at δH 6.42 within this carboxylic acid unit in combination with a weak ROESY correlations between both methyl groups (δH 1.71, 1.80) indicated E configuration and thus the trivial name tiglic acid (Tig) for this unit. The geometry of the Abu unit was deduced as Z based on an NOE between the Abu NH (δH 10.16) and Abu methyl group (δH 1.76). HMBC supported by ROESY correlations unambiguously established the linear sequence of the amino acid units and tiglic acid moiety. The cyclic depsipeptide core was proposed due to the low-field shift of the Thr β proton (δH 5.37) indicative of an ester functionality which involved the C-terminal leucine moiety, consistent with weak ROESY correlations between both units (Thr δH-2 4.74 and Leu δH-2 4.67). Furthermore, an absorption at νmax 1725 cm−1 characteristic of an ester carbonyl stretching vibration was observed in the IR spectrum of 1 in addition to the strong amide stretching bands for peptides (νmax 1659 cm−1), supporting the proposed planar structure.
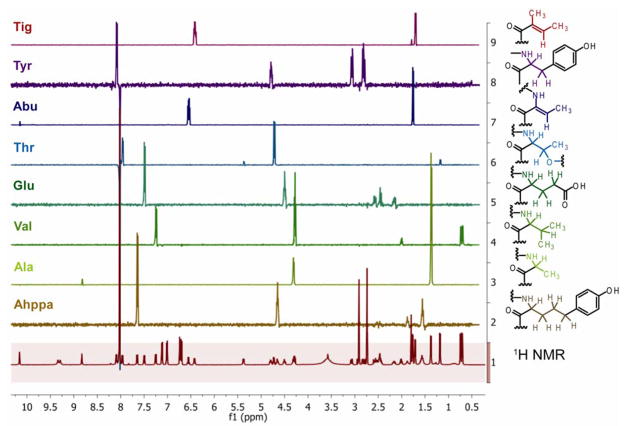
An [M + H]+ peak at m/z 920.4404 in the HR-ESI/APCI-MS of 2 in conjunction with 13C NMR data suggested a molecular formula of C46H61N7O13 for compound 2. The 1H NMR spectrum indicated that compound 2 is closely related to 1. Further NMR analysis (1H, COSY, HSQC, HMBC, ROESY) as carried out for 1 and additional 1D TOCSY experiments in DMF-d7 (Figure 2) revealed the presence of eight spin systems as for 1, except that the leucine moiety was replaced by a 2-amino-5-(4′-hydroxy-phenyl)pentanoic acid (Ahppa) residue (Table 2).
Fig. 2.
1H NMR (front) and 1D TOCSY spectra of largamide B (2) in DMF-d7.
The 1H NMR spectrum of 3 was virtually identical to the one of 2. HR-ESI/APCI-MS analysis for compound 3 provided an [M + H]+ peak at m/z 934.4559, suggesting a molecular formula of C47H63N7O13 and thus that compound 3 differs from 2 only by the presence of an additional methylene group. Closer inspection of NMR data revealed that the Ahppa unit was homologated to a 2-amino-5-(4′-hydroxy-phenyl)hexanoic acid (Ahpha) unit in 3 (Table 2), in agreement with the molecular formula requirements. HMBC analysis verified the units’ connectivity and the proposed planar structure.
The absolute configuration of all amino acid units in compounds 1–3 was deduced by chiral HPLC analysis of the acid hydrolysis products, revealing D configuration for glutamic acid and tyrosine and L configuration for all other usual amino acids. For compounds 2 and 3, additionally, acid hydrolysis was preceded by ozonolysis with oxidative work-up, thereby allowing us to detect 2-aminoadipic acid and 2-aminopimelic acid instead of Ahppa and Ahpha, respectively, to take advantage of commercially available standards. This analysis confirmed that L-Leu in 1 was replaced by L-Ahppa in 2 and L-Ahpha in 3.
Compounds 1–3 were tested for serine protease-inhibitory activity. They showed moderate activity against porcine pancreatic elastase activity in vitro in a dose-dependent manner with IC50 values of 1.41 ± 0.28, 0.53 ± 0.19, and 1.15± 0.46 μM, respectively. To largely eliminate the possibility that the elastase activity is due to trace amounts of the potent inhibitors lyngbyastatins 4, 5, or 6, [3,4] compounds 1–3 were repeatedly purified by HPLC without loss of activity. Compounds 1–3 were inactive against trypsin and chymotrypsin (tested up to 50 μM). They also did not affect the viability of any of the cancer cell lines tested (HT29, U2OS, IMR-32) for growth inhibitory activity.
Discussion
A tiglic acid moiety present as a key feature in 1–3 had not been previously described in a cyanobacterial metabolite; however, it has been found in the bacterial metabolite cytosaminomycin D from Streptomyces sp. KO-8119 [9]. It is commonly encountered in higher plants [10] where it is presumably biosynthesized from isoleucine [11]. However, compounds 1–3 differ from largamides A–C, recently isolated from an Oscillatoria sp. from Key Largo (Florida), only by the presence of the tiglic acid instead of the senecioic acid residue [6]. The physical description of the largamide-producing cyanobacterium closely matches the morphology and appearance of the organism we have been investigating. Close inspection and comparison of NMR data in DMF-d7, DMSO-d6 and CD3OD (Supporting Information) with those reported for largamides A–C in CD3OD and DMSO-d6 [6] suggested that the senecioic acid residue in these compounds was assigned incorrectly, that compounds 1–3 and largamides A–C are identical [12] and that DMF-d7 was a superior solvent since there was no signal overlap in the low-field region of the 1H NMR spectra. Furthermore, our collections yielded a common set of largamides (D–H) [6], while lyngbyastatins 4–6 and pompanopeptins A and B may be unique to our samples from the Ft. Lauderdale area [3–5]. This high secondary metabolite content of a single organism demonstrates the enormous biosynthetic potential of cyanobacteria.
The elastase-inhibitory activity of largamides A–C (1–3) is inferior to the activity of lyngbyastatins 4–6, which bear the 3-amino-6-hydroxy-2-piperidone (Ahp) residue critical for their serine protease-inhibitory activity [3–5]. Yet the largamide A–C core structure represents a new structural scaffold for elastase inhibition. Since numerous fatty acids have been shown to inhibit elastase [13], it is possible that the carboxylic acid moiety of the glutamic acid residue in largamides A–C (1–3) may play a role.
Supplementary Material
Acknowledgments
This work was funded by the Florida Sea Grant College Program with support from NOAA, Office of Sea Grant, U.S Department of Commerce, Grant No. NA06OAR4170014; and NIGMS grant P41GM086210. We thank L. Fisher and K. Banks (Broward County Environmental Protection Department) and C. Ross, A. Capper, and R. Ritson-Williams (Smithsonian Marine Station) for assistance with collections and W. Yoshida (University of Hawaii) for providing NMR data. We also thank C. Bewley for helpful discussions and comparison with authentic largamides A–C from Oscillatoria sp. from Key Largo. Mass spectral analyses were performed at the UCR Mass Spectrometry Facility, Department of Chemistry, University of California at Riverside. This is contribution # 759 from the Smithsonian Marine Station at Fort Pierce.
Footnotes
1H NMR, 13C NMR, and 1D TOCSY spectra of compounds 1–3 are available at http://www.thieme-connect.de/ejournals/toc/plantamedica.
References
- 1.Gerwick WH, Tan LT, Sitachitta N. Nitrogen-containing metabolites from marine cyanobacteria. Alkaloids Chem Biol. 2001;57:75–184. doi: 10.1016/s0099-9598(01)57003-0. [DOI] [PubMed] [Google Scholar]
- 2.Welker M, von Döhren H. Cyanobacterial peptides – nature’s own combinatorial biosynthesis. FEMS Microbiol Rev. 2006;30:530–563. doi: 10.1111/j.1574-6976.2006.00022.x. [DOI] [PubMed] [Google Scholar]
- 3.Matthew S, Ross C, Rocca JR, Paul VJ, Luesch H. Lyngbyastatin 4, a dolastatin 13 analogue with elastase and chymotrypsin inhibitory activity from the marine cyanobacterium Lyngbya confervoides. J Nat Prod. 2007;70:124–127. doi: 10.1021/np060471k. [DOI] [PubMed] [Google Scholar]
- 4.Taori K, Matthew S, Rocca JR, Paul VJ, Luesch H. Lyngbyastatins 5–7, potent elastase inhibitors from Floridian marine cyanobacteria, Lyngbya spp. J Nat Prod. 2007;70:1593–1600. doi: 10.1021/np0702436. [DOI] [PubMed] [Google Scholar]
- 5.Matthew S, Ross C, Paul VJ, Luesch H. Pompanopeptins A and B, new cyclic peptides from the marine cyanobacterium Lyngbya confervoides. Tetrahedron. 2008;64:4081–4089. [Google Scholar]
- 6.Plaza A, Bewley CA. Largamides A–H, Unusual cyclic peptides from the marine cyanobacterium Oscillatoria sp. J Org Chem. 2006;71:6898–6907. doi: 10.1021/jo061044e. [DOI] [PubMed] [Google Scholar]
- 7.Houghton AM, Quintero PA, Perkins DL, Kobayashi DK, Kelley DG, Marconcini LA, et al. Elastin fragments drive disease progression in a murine model of emphysema. J Clin Invest. 2006;116:753–759. doi: 10.1172/JCI25617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paul VJ, Thacker RW, Banks K, Golubic S. Benthic cyanobacterial bloom impacts the reefs of South Florida (Broward County, USA) Coral Reefs. 2005;24:693–697. [Google Scholar]
- 9.Shiomi K, Haneda K, Tomoda H, Iwai Y, Omura S. Cytosaminomycins, new anticoccidial agents produced by Streptomyces sp. KO-8119. II. Structure elucidation of cytosaminomycins A, B, C and D. J Antibiot (Tokyo) 1994;47:782–786. doi: 10.7164/antibiotics.47.782. [DOI] [PubMed] [Google Scholar]
- 10.Mann J. Secondary Metabolism. 2. Oxford: Clarendon Press; 1987. pp. 95–172. [Google Scholar]
- 11.Rhee SW, Leete E, McGaw BA. Stereospecificity of enzymic dehydrogenation during tiglate biosynthesis. J Am Chem Soc. 1980;102:7344–7348. [Google Scholar]
- 12.Bewley C. Personal communication [Google Scholar]
- 13.Rennert B, Melzig MF. Free fatty acids inhibit the activity of Clostridium histolyticum collagenase and human neutrophil elastase. Planta Med. 2002;68:767–769. doi: 10.1055/s-2002-34411. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.