Abstract
There is a strong clinical imperative to identify discerning molecular biomarkers of disease to inform diagnosis, prognosis, and treatment. Ideally, such biomarkers would be drawn from peripheral sources non-invasively to reduce costs and lower potential for complication. Advances in high-throughput genomics and proteomics have vastly increased the space of prospective molecular biomarkers. Consequently, the search for molecular biomarkers of interest often entails genome- or proteome-wide discovery for candidate biomarkers. Here we present a novel framework for the identification of disease-specific protein biomarkers through the integration of biofluid proteomes and inter-disease genomic relationships using a network paradigm. We created a blood plasma biomarker network by linking genomic profiles from 136 diseases to 1,028 detectable blood plasma proteins. We created a urine biomarker network by linking genomic profiles from 127 diseases to 577 proteins detectable in urine. We find that, in both networks, the majority (> 80%) of putative protein biomarkers are linked to multiple disease conditions. Thus, putatively disease-specific protein biomarkers are found in only a small subset of the biofluids proteomes. These findings illustrate the importance of the context of inter-disease molecular networks in the focused discovery of molecular biomarkers for disease. The proposed framework is amenable to integration with complimentary network models of biology, which could further constrain the biomarker candidate space, and work to paint a larger picture for the role of inter-disease genomic relationships across varying physiological scales.
1. Introduction
Perhaps one of the most compelling prospects of translational genomics is the potential for the discovery of novel molecular biomarkers of disease that offer early detection of pathogenesis, inform prognosis, guide therapy, and monitor disease progression. Despite expectations, the elucidation of accurate and discriminating disease biomarkers has proved challenging1, and the widespread adoption of genomics-based biomarkers in the clinical management of disease remains to be realized2. There are many factors confounding the discovery and development of effective clinical biomarkers, including genetic variation between and among individuals and populations3, 4, deficiencies in biomolecule capture and quantification technologies5, transient shifts in proteome composition due to acute-phase reactants and environmental stress6-8, and logistical constraints related to associated costs and clinical acceptance9, 10. Such confounding factors can contribute to appreciable clinical heterogeneity for a particular disease with regards to diagnosis, treatment, and outcome.
Despite the relatively limited impact of genomics on the development of clinical biomarkers to date, there has been notable success in applying genomics techniques to better clarify and characterize the clinical heterogeneity observed for many complex diseases. In particular, high-throughput gene-expression profiling using microarrays has proven successful as a means by which genome-scale events can be linked to clinical metrics. Ramaswamy et al. demonstrated that gene expression signatures could accurately differentiate adenocarcinoma subtypes11. Chen et al. used microarray profiling of lung cancer tissues to derive a prognostic five-gene expression signature associated with relapse and survival. Potti et al. derived a set of gene expression signatures that were successful in predicting response to chemotherapeutic agents12. Although significant, the impact of such findings remains far removed from the clinic, as they often require undesirably invasive procurement of patient tissues, improved handling of unstable molecules (e.g. RNA), and improved consistency of measurements. Such factors have consequently impeded the customary use of microarrays in most clinical settings.
The desire for minimally invasive biomarker strategies has put a focus on established clinical biofluids, such as blood and urine, as sources of putative molecular biomarkers. Both blood and urine are easily and inexpensively obtained from patients as a conventional facet of clinical care, therefore biomarker strategies leveraging these fluids are particularly amenable to current clinical protocol13, 14. The advent of several blood plasma and urine proteome projects, with aims to identify the vast body of gene products comprising these biofluids, has generated new opportunities for genomics-based approaches to the elucidation of clinical molecular biomarkers15, 16. Microarray analyses of blood and urine have identified expression signatures symptomatic of diseases such as rheumatoid arthritis17, Alzheimer disease18, Chronic Fatigue Syndrome19, Huntington’s disease20, and glial brain tumors21.
Disease conditions are most often signified by the dysregulation of complex biological pathways involving multiple, interacting gene products. Thus integrative approaches linking gene expression activity with proteomics and physiopathology are needed to identify highly discerning subsets of molecular biomarkers from the vast combinatorial space of candidates. One such approach is to frame the space of biomarker candidates within the context of inter-disease relationships. The current approach to biomarker discovery is based on the implicit assumption that the heterogeneity of clinical disease classifications is subsumed by the underlying molecular pathophysiology of the disease condition. However, recent studies have shed light on widespread genomic and genetic correspondence between diseases previously thought to be dissimilar based on anatomy and manifest symptoms22-24. In fact, the similarity of responses across diseases and tissues raises concerns about the specificity of putative biomarkers derived under the consideration of a single disease condition.
Here we propose an integrative, network-based model for biomarker prioritization that identifies putatively high-specificity biomarkers in blood and urine proteomes using inter-disease relationships derived from gene expression profiles across hundreds of diseases and nearly ten thousand microarrays. We find that a majority of detectable protein biomarkers (>80%) exhibit non-discerning disease connectivity in the biomarker network, potentially impacting their clinical utility for a single disease. Our findings highlight the importance of integrating the context of broad inter-disease relationship profiling into future molecular biomarker discovery and prioritization efforts.
2. Methods
2.1. Discovery and annotation of disease experiments
Microarray experiments characterizing human disease conditions were automatically identified using a previously developed method25. In brief, microarrays were obtained from the NCBI Gene Expression Omnibus (GEO)26. We have previously shown that the experimental context for GEO Series (GSE), or collections of microarrays, can be obtained using MeSH terms from PUBMED records associated with GEO experiments. MeSH terms derived in this manner were evaluated for disease concepts using the Unified Medical Language System (UMLS)27. Each GSE determined to be relevant to a human disease was subject to automated annotation of the disease condition, the tissue or biological substance from which the samples were derived, and whether or not the experiment measured a normal control state complimentary to the annotated disease state by means of a previously described method28. We only used microarray experiments in which disease and normal tissues were measured in the same experiment. The disease and tissue annotations were manually reviewed in a post-processing step to ensure accuracy. This process yielded 383 disease-related experiments (238 unique diseases) totaling 8,435 microarrays.
2.2. Microarray data preparation and analysis
For each microarray platform associated with the annotated disease experiments we updated the mappings between the platform-specific probe identifiers and the Entrez GeneID identifiers in an automated manner using the AILUN system29. Microarrays were only compared to other microarrays within their original experiments (or GSE). For each disease experiment we derived a set of significantly differentially expressed genes using SAMR30. SAMR was configured to estimate the False Discovery Rate (FDR) using 1,000 rounds of measurement permutations. Genes were considered to be significantly differentially expressed if the estimated fold-change was > 1.5 and the estimated FDR was < 5%.
2.3. Construction and analysis of the proteome biomarker networks
A database of human blood plasma proteomes was constructed using data from the HUPO Plasma Proteome Project15 and a non-redundant list from the Plasma Proteome Institute31. Only the 3020 proteins from the high-confidence set of identified peptides in the HUPO PPP dataset were included in the analysis. Urine proteome data was obtained from the MAPU Proteome database32 and the Urinary Exosome database33. The original data sets were parsed into a MySQL database and the International Protein Index (IPI) identifiers were mapped to Entrez GeneID identifiers using AILUN29. Disease-associated genes from microarray studies were associated with protein biomarkers using Entrez GeneID as the associative identifier. Networks were constructed such that diseases and genes (proteins) were nodes, and edges between gene and disease nodes were formed when a gene was found to be significantly differentially expressed in the disease state. The networks rendering and analysis was performed using the yED graph editor (http://www.yworks.com).
2.4. Functional annotation enrichment analysis
Functional annotation enrichment for disease-associated protein biomarkers was conducted using the DAVID system34. For each biomarker network, genes linked to at least one disease were considered to be the “gene list” and the entire list of gene identifiers associated with the respective proteomes were used as the background population. P-values were adjusted using Benjamini-Hochberg correction35.
3. Results
In linking proteome biomarkers with disease, we find that 1,028 (38.5%) plasma and 577 (39.9%) urine proteins were found to be significantly differentially expressed in one or more of the 238 distinct disease states represented in the microarray data. Of those, 846 (82.2%) plasma and 490 (84.9%) urine proteins are significantly differentially expressed in more than one disease state. Thus, less than 20% of putative proteome disease markers exhibit specificity for a single disease.
Among the putative biomarker proteins associated with disease we identified a number of enriched gene annotation terms (Table 2). Disease-associated plasma biomarker proteins were enriched for plasma membrane proteins, and proteins involved in sugar and carbohydrate metabolism. Disease-associated urine biomarker proteins were enriched for extracellular proteins, and proteins involved in amine metabolism and biotic stimulus response.
Table 2.
Annotation enrichment for disease-associated biomarkers
| GO Term | P-value |
|---|---|
| Plasma | |
| (GO:0005975) carbohydrate metabolic process | 3.1E-5 |
| (GO:0019318) hexose metabolic process | 1.1E-4 |
| (GO:0006066) alcohol metabolic process | 4.6E-4 |
| (GO:0044459) plasma membrane part | 5.3E-4 |
| Urine | |
| (GO:0009308) amine metabolic process | 7.7E-3 |
| (GO:0044421) extracellular region part | 1.4E-2 |
| (GO:0050896) response to stimulus | 1.8E-2 |
We found that a majority of diseases could not be linked to a disease-specific protein biomarker in either the blood plasma or urine proteomes. Among the distinct disease conditions represented in the microarray data, 136 (57.1%) were linked to plasma proteins, while 127 (53.4%) were mapped to urine proteins. Of these, 65.4% and 72.4% link exclusively to biomarkers shared by other diseases in plasma and urine respectively. A selection of disease conditions associated with multiple disease-specific biomarker proteins are listed in Table 3.
Table 3.
A subset of diseases associated with multiple disease-specific protein biomarkers
| Disease | Disease-specific protein biomarkers |
|---|---|
| Plasma | |
| Idiopathic cardiomyopathy | MACF1, SF3B2, RFX5, TLN1, FSHR, PCCA, PGK2, NEK1, RGS3, RGN, CYP3A43 |
| Thrombocytopenia | CYLC2, PIGK, AASS, PANX2, DSPP, XPC, TBL1X, TCERG1 |
| Malignant melanoma | PDE3A, CALR, PDCD6IP, CHAC, KIAA0586 |
| AIDS | PAPPA, TRADD, KIAA0649, APRIN, MAP3K5 |
| Huntington’s disease | MAML1, PLGL, RNF10, KIAA0913, OAS1 |
| Urine | |
| Idiopathic cardiomyopathy | DEFA3, ALDH1L1, CD177, TLN1, SLURP1, BPI, APOH, C8B |
| Glioblastoma | WISP2, PRDX3, TIMP2, ACO1 |
| Breast cancer | ENPP4, PFKP, THBD, IGFALS |
| Acute promyelocytic leukemia | CSPG3, LGALS7, HSPA5 |
| Adenovirus infection | VGF, AGA, UMOD |
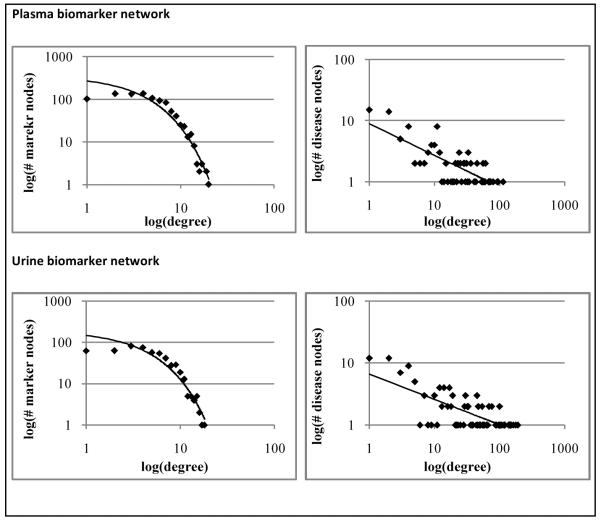
The mean disease linkage degree for a protein biomarker node was 5.09 in the plasma network and 5.06 in the urine network. The mean biomarker linkage degree for a disease node was 36.19 in the plasma network and 22.57 in the urine network. The distribution of disease connectivity across biomarker nodes was found to follow an exponential model in both the blood (R2 = 0.94) and urine (R2 = 0.93) networks, suggesting a scale-variance in attachment (Figure 1). The distribution of biomarker connectivity across disease nodes was found to follow a weak power-law model in both the blood plasma (R2 = 0.59) and urine (R2 = 0.53) biomarker networks, suggesting a scale-free property. This suggests that diseases with many biomarkers preferentially gain more biomarkers. The equivalent graph of the connectivity of the biomarkers matches an exponential curve. The shape of the curve actually splits into two parts. At low connectivity, biomarkers gain connections to diseases randomly as more diseases are added. At higher connectivity, biomarkers then gain connections to diseases preferentially if they are already connected. The two segments to the log-log plots of biomarker node degree distributions in Figure 1 thus suggest there are two separate populations of biomarkers.
Figure 1.
Independent log-log plots of node degree distributions for biomarker and disease nodes.
4. Discussion
In this study we propose an integrative network model for biomarker prioritization using inter-disease relationships derived from microarray studies, and putative protein biomarkers from large-scale biofluids proteome studies. Unlike traditional biomarker prioritization approaches, our approach first considers all possible (i.e. measureable) protein biomarkers in a biofluid proteome and places them within the context of inter-disease relationships across the broad spectrum of human disease to identify putative protein biomarkers that are likely to be highly discerning for a disease of interest.
Our approach finds validation in the finding that a majority proportion (> 80%) of measurable proteins in both the blood plasma and urine proteomes are non-specific for any single disease condition. Interestingly, disease-specific biomarkers appear to be associated preferentially with a subset of diseases. Given the vast resources required for both identifying and biologically validating putative molecular biomarkers, it might be prudent to focus biomarker discovery efforts on the diseases enriched for disease-specific biomarker associations. Such enriched associations could imply that a novel and discriminating pathway is involved in the pathogenesis of the disease, and could lead to the identification of highly discriminating upstream biomarker candidates.
Although many of the discriminating disease-biomarker associations predicted by our approach remain to be biologically and clinically validated, there is, in several cases, a compelling degree of biological continuity between the predicted disease-specific biomarker and the understood molecular phenomena underlying the disease. One such example is our prediction that Collectin Sub-family Member 10 (COLEC10) as a putative disease-specific biomarker for Crohn’s disease. Crohn’s disease is a chronic, debilitating inflammatory bowel disorder that can affect any portion of the digestive tract36. Recent genome-wide association studies and other investigations into the pathogenesis of Crohn’s disease have revealed a number of susceptibility genes37, 38 and the major role of the body’s innate immune response against enteric microbiota39, 40. Collectins have been implicated as significant regulators of the innate immune system, particularly with regards to host defense response to microorganisms41. Collectins are known to induce pro-inflammatory cytokines and participate in activation of the compliment system via the lectin pathway during the microorganism defense reponse42. Therefore COLEC10 could serve as a novel biomarker that is sensitive to the episodic manifestations of Crohn’s disease to inform ongoing disease management, whereas current biomarkers for the disease are primarily diagnostic43.
Another interesting finding is the identification of GDP dissociation inhibitor 1 (GDI1) as a disease-specific biomarker for Hypercholesterolemia. GDI1 is a known regulator of the GDP/GTP exchange reactions of Rab proteins and a participant in the vesicle mediated cellular transport44. GDI and Rab are also known to participate in the cellular transport of lipids, and GDI/Rab dysregulation has been observed in the presence of cholesterol accumulation45.
We recognize several caveats in our approach. Foremost, our approach makes the naïve assumption that if a gene is significantly differentially expressed in a disease condition that this differential will be reflected in either blood plasma or urine regardless of the anatomical locus of the disease site. While quantifications of mRNA expression can be far removed from the modulation of protein fragments in biofluids, there is reason to believe that such an assumption can hold true in a sufficient number of cases. Interestingly, notable proportions of the proteins identified by blood plasma and urine proteome projects are annotated with Gene Ontology terms signifying intracellular localization, including: intracellular part (55.4%), intracellular organelle part (20.3%), cytoskeleton (9.6%), and nuclear part (6.4%). Such phenomenon may be accounted for by sufficient secretion of intracellular proteins inside small-membrane vesicles known as exosomes by various tissue types46-50. Furthermore, cells undergoing destruction as a consequence of pathogenesis are likely to emanate intracellular matter into biofluids. We also recognize that the specificity of a protein biomarker in our networks is subject to the available disease-condition data available. The addition of novel disease conditions into future versions of the biomarker network could even further reduce the proportions of disease-specific protein biomarkers. Our study is also limited by the available number and quality of microarray datasets across diseases
The framework proposed in this study is not intended to serve as an unequivocal means for biomarker elucidation. Rather we suggest that the integration of our approach with other forms of biomarker network biology is likely to lead to even more sophisticated approaches to informatics-based biomarker discovery. Alterovitz et al. proposed an information theoretic framework for biomarker discovery that identified high-quality peripheral biomarker candidates by identifying significant tissue-biofluid channels across a wide range of tissues and biofluids proteomes51. Our approach could be used in combination with their biofluids channel approach to find optimal intersections between disease-specificity space and biofluid-tissue interaction space to even further refine the scope of putative biomarker proteins for a particular disease condition.
5. Conclusion
The discovery of discerning molecular biomarkers for a disease state of interest is encumbered by the vast combinatorial space of prospective candidate markers. Our work provides a novel framework for reducing the space of candidate markers using a network of genome-scale disease conditions and biofluids proteomes. While a more traditional biomarker discovery endeavor might start with the disease condition of interest to identify biomarker candidates in a “bottom-up” approach, we offer a “top-down” approach that begins with the broad space of human disease and full compliments of biofluid proteomes to quickly discern candidate protein biomarkers discriminately associated with the a disease condition. This work establishes the importance of genome-wide, inter-disease relationships in biomarkers discovery and paves the way for novel integrative methods that incorporate inter-disease network models to further refine biomarker discovery.
Figure 2.

A rendering of the plasma biomarker network is shown. Disease-specific biomarkers (green) are found extending from diseases (blue) at the periphery of the network.
Table 1.
A subset of indiscriminate, highly-connected biomarker nodes and their disease targets
| Biomarker | Diseases |
|---|---|
| Plasma | |
| AZGP1 | Cardiac hypertrophy, Spinal cord injury, Idiopathic cardiomyopathy, Idiopathic thrombocytopenic purpura, E. coli infection of the CNS, Hypercholesterolemia, Clear cell carcinoma of kidney, Hypertrophy, Glioblastoma, Adenoma of small intestine, Thrombocytopenia, Carcinoma in situ of small intestine, AML, Huntington’s disease, Porcine nephropathy, Allergic asthma, Cirrhosis of liver, Adenovirus infection, Squamous cell carcinoma, Duchenne muscular dystrophy |
| CD46 | Malignant neoplasm of prostate, Complex dental cavity, Fracture of bone, MODY, Dermatomyositis, Bacterial infection, Clear cell carcinoma of kidney, Spinal cord injury, Status epilepticus, Senescence, Fracture of femur, Barrett’s ulcer of esophagus, Rheumatoid arthritis, Urothelial carcinoma, Astrocytoma, Glioblastoma, Congestive cardiomyopathy, Obesity, Lung transplant rejection |
| LAMA2 | Breast cancer, Dermatomyositis, Malignant neoplasm of stomach, Acute lung injury, Malignant melanoma, Glioblastoma, Adenovirus infection, Duchenne muscular dystrophy, Acute promyelocytic leukemia, Senescence, Barrett’s ulcer of esophagus, AML, Hypercholesterolemia, Hepatic lipidosis, Acute pancreatitis, Idiopathic thrombocytopenic purpura, Porcine nephropathy, Urothelial carcinoma, AIDS |
| Urine | |
| AKR1C1 | Acute lung injury, Acute arthritis, Essential thrombocythemia, Ulcerative colitis, Lung transplant rejection, Malignant melanoma, Carcinoma in situ of small intestine, Dehydration, Adenoma of small intestine, Bacterial infection, Glioblastoma, Oligodendroglioma, Urothelial carcinoma, Progeria syndrome, Atrial fibrillation, Huntington’s disease, SARS, Adenocarcinoma of lung |
| PRG4 | Multiple benign melanocytic nevi, Urothelial carcinoma, Type 2 diabetes mellitus, Actinic keratosis, Adenocarcinoma of lung, Thrombocytopenia, Acute myeloid leukemia, Huntington’s disease, Cardiomyopathy, Ventilator-associated lung injury, Macular degeneration, Congestive cardiomyopathy, Polycystic ovary syndrome, Dermatomyositis, Adenovirus infection, Acute pancreatitis |
| AQP2G4 | Clear cell carcinoma of kidney, Dermatomyositis, Breast cancer, Duchenne muscular dystrophy, Hepatocellular carcinoma, Bacterial infection, Barrett’s ulcer of esophagus, Helicobacter pylori GI infection, Macular degeneration, MODY, Urothelial carcinoma, AML, Crohn’s disease, Ulcerative colitis, Epithelial proliferation |
Acknowledgements
The work was supported by grants from the Lucile Packard Foundation for Children’s Health, National Institute of General Medical Sciences (R01 GM079719), Howard Hughes Medical Institute, and the Pharmaceutical Research and Manufacturers of America Foundation. The authors would like to thank Alex Skrenchuk for computing support.
References
- 1.Baker M. Nature biotechnology. 2005;23:297–304. doi: 10.1038/nbt0305-297. [DOI] [PubMed] [Google Scholar]
- 2.Bell J. Nature. 2004;429:453–6. doi: 10.1038/nature02624. [DOI] [PubMed] [Google Scholar]
- 3.Pharoah PD, et al. The New England journal of medicine. 2008;358:2796–803. doi: 10.1056/NEJMsa0708739. [DOI] [PubMed] [Google Scholar]
- 4.Radich JP, et al. Genomics. 2004 [Google Scholar]
- 5.Johnson CJ, et al. Proteomics. 2008;8:715–30. doi: 10.1002/pmic.200700665. [DOI] [PubMed] [Google Scholar]
- 6.Nesatyy VJ, Suter MJ. Mass spectrometry reviews. 2008 doi: 10.1002/mas.20177. [DOI] [PubMed] [Google Scholar]
- 7.Mendrick DL. Toxicology. 2008;245:175–81. doi: 10.1016/j.tox.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 8.Nascimento MM, et al. Nephrol Dial Transplant. 2004;19:2803–9. doi: 10.1093/ndt/gfh493. [DOI] [PubMed] [Google Scholar]
- 9.Williams SA, et al. Nature reviews. 2006;5:897–902. doi: 10.1038/nrd2174. [DOI] [PubMed] [Google Scholar]
- 10.Etzioni R, et al. Nat Rev Cancer. 2003;3:243–52. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- 11.Ramaswamy S, et al. Proc Natl Acad Sci U S A. 2001;98:15149–54. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Potti A, et al. Nature medicine. 2006;12:1294–300. doi: 10.1038/nm1491. [DOI] [PubMed] [Google Scholar]
- 13.Liotta LA, et al. Nature. 2003;425:905. doi: 10.1038/425905a. [DOI] [PubMed] [Google Scholar]
- 14.Thongboonkerd V, et al. Contributions to nephrology. 2004;141:292–307. doi: 10.1159/000074606. [DOI] [PubMed] [Google Scholar]
- 15.Omenn GS, et al. Proteomics. 2005;5:3226–45. doi: 10.1002/pmic.200500358. [DOI] [PubMed] [Google Scholar]
- 16.Adachi J, et al. Genome Biol. 2006;7:R80. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olsen N, et al. British Medical Journal. 2004 [Google Scholar]
- 18.Maes OC, et al. Neurobiol Aging. 2007;28:1795–809. doi: 10.1016/j.neurobiolaging.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 19.Kaushik N, et al. British Medical Journal. 2005 [Google Scholar]
- 20.Borovecki F, et al. Proceedings of the National Academy of Sciences. 2005 [Google Scholar]
- 21.Smith ER, et al. Clin Cancer Res. 2008;14:2378–86. doi: 10.1158/1078-0432.CCR-07-1253. [DOI] [PubMed] [Google Scholar]
- 22.Lamb J. Nat Rev Cancer. 2007;7:54–60. doi: 10.1038/nrc2044. [DOI] [PubMed] [Google Scholar]
- 23.Goh KI, et al. Proc Natl Acad Sci U S A. 2007;104:8685–90. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Segal E, et al. Nature genetics. 2004;36:1090–8. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- 25.Butte AJ, Chen R. AMIA ... Annual Symposium proceedings / AMIA Symposium; 2006; pp. 106–10. [PMC free article] [PubMed] [Google Scholar]
- 26.Barrett T, et al. Nucleic acids research. 2005;33:D562–6. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bodenreider O. Nucleic acids research. 2004;32:D267–70. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dudley J, Butte AJ. Pacific Symposium on Biocomputing; 2008.pp. 580–91. [Google Scholar]
- 29.Chen R, et al. Nature methods. 2007;4:879. doi: 10.1038/nmeth1107-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tusher VG, et al. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson NL, et al. Mol Cell Proteomics. 2004;3:311–26. doi: 10.1074/mcp.M300127-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, et al. Nucleic acids research. 2007;35:D771–9. doi: 10.1093/nar/gkl784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pisitkun T, et al. Proc Natl Acad Sci USA. 2004;101:13368–73. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dennis G, Jr., et al. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 35.Benjamini Y, et al. Behavioural brain research. 2001;125:279–84. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 36.Baumgart DC, Carding SR. Lancet. 2007;369:1627–40. doi: 10.1016/S0140-6736(07)60750-8. [DOI] [PubMed] [Google Scholar]
- 37.Rioux JD, et al. Nature genetics. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barrett JC, et al. Nature genetics. 2008 [Google Scholar]
- 39.Muller CA, et al. Cell Mol Life Sci. 2005;62:1297–307. doi: 10.1007/s00018-005-5034-2. [DOI] [PubMed] [Google Scholar]
- 40.Abreu MT, Sparrow MP. The Mount Sinai journal of medicine, New York. 2006;73:1067–73. [PubMed] [Google Scholar]
- 41.van de Wetering JK, et al. European journal of biochemistry / FEBS. 2004;271:1229–49. doi: 10.1111/j.1432-1033.2004.04040.x. [DOI] [PubMed] [Google Scholar]
- 42.Haagsman HP, et al. Neonatology. 2008;93:288–94. doi: 10.1159/000121454. [DOI] [PubMed] [Google Scholar]
- 43.Papadakis KA, et al. Inflammatory bowel diseases. 2007;13:524–30. doi: 10.1002/ibd.20106. [DOI] [PubMed] [Google Scholar]
- 44.Bachner D, et al. Human molecular genetics. 1995;4:701–8. doi: 10.1093/hmg/4.4.701. [DOI] [PubMed] [Google Scholar]
- 45.Lebrand C, et al. The EMBO journal. 2002;21:1289–300. doi: 10.1093/emboj/21.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caby MP, et al. Int Immunol. 2005;17:879–87. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 47.Lamparski HG, et al. J Immunol Methods. 2002;270:211–26. doi: 10.1016/s0022-1759(02)00330-7. [DOI] [PubMed] [Google Scholar]
- 48.Yu X, et al. Cancer Res. 2006;66:4795–801. doi: 10.1158/0008-5472.CAN-05-4579. [DOI] [PubMed] [Google Scholar]
- 49.van Niel G, et al. J Biochem. 2006;140:13–21. doi: 10.1093/jb/mvj128. [DOI] [PubMed] [Google Scholar]
- 50.van Niel G, et al. Gastroenterology. 2001;121:337–49. doi: 10.1053/gast.2001.26263. [DOI] [PubMed] [Google Scholar]
- 51.Alterovitz G, et al. Pacific Symposium on Biocomputing. 2008:231–42. [PubMed] [Google Scholar]