Abstract
The packaging of the eukaryotic genome into chromatin represses gene expression by blocking access of the general transcription machinery to the underlying DNA sequences. Accordingly, eukaryotes have developed a variety of mechanisms to disrupt, alter, or disassemble nucleosomes from promoter regions and open reading frames to allow transcription to occur. Although we know that chromatin disassembly from the yeast PHO5 promoter is triggered by the Pho4 activator, the mechanism is far from clear. Here we show that the Pho4 activator can occupy its nucleosome-bound DNA binding site within the PHO5 promoter. In contrast to the role of Saccharomyces cerevisiae FACT (facilitates chromatin transcription) complex in assembling chromatin within open reading frames, we find that FACT is involved in the disassembly of histones H2A/H2B from the PHO5 promoter during transcriptional induction. We have also discovered that the proteasome is required for efficient chromatin disassembly and transcriptional induction from the PHO5 promoter. Mutants of the degradation function of the proteasome have a defect in recruitment of the Pho4 activator, whereas mutants of the ATPase cap of the proteasome do recruit Pho4 but are still delayed for chromatin assembly. Finally, we rule out the possibility that the proteasome or ATPase cap is driving chromatin disassembly via a potential ATP-dependent chromatin remodeling activity.
Eukaryotic chromatin is made up of a fundamental repeating unit, termed the nucleosome, which consists of 147 bp of DNA wrapped around the outside of an octamer of histone proteins (1). The histone octamer in turn comprises a heterotetramer of histone proteins H3/H4 and two heterodimers of histones H2A/H2B. In order to allow the transcription machinery to gain access to the DNA, the chromatin structure is altered by the concerted action of three processes (2): (i) post-translational modifications on the histones, (ii) the breakage of histone-DNA contacts by ATP-dependent chromatin remodeling machines, and (iii) the ultimate removal of histones from the DNA by histone chaperones. The sequence-specific transcriptional activators trigger these chromatin alterations occurring at promoters during transcriptional induction, but it is unclear whether transcriptional activators can first bind to their nucleosome-buried DNA recognition sequences to start this cascade of chromatin dynamics. Similarly, once transcription is complete, transcriptional activators leave the DNA, and promoters are repackaged with histones via the process of chromatin assembly.
The assembly and disassembly of nucleosomes appear to occur in a stepwise manner (3). This is due to the peripheral positions of the H2A/H2B dimers within the nucleosome (1), necessitating the removal of H2A/H2B prior to removal of the central H3/H4 tetramer. Conversely, H3/H4 must be deposited onto the DNA prior to H2A/H2B in order to assemble a nucleosome. Accordingly, histone chaperones exist that bind to either H2A/H2B or H3/H4 to mediate chromatin assembly and disassembly. We have previously shown that the histone chaperone Asf1 (anti-silencing function 1) promotes the disassembly of histones H3/H4 from multiple yeast promoter regions during transcriptional induction (4, 5), whereas the histone chaperone Spt6 promotes the deposition of H3/H4 onto promoters during transcriptional repression (6). However, we still do not know the identity of the histone chaperones that remove histones H2A/H2B from promoter regions during transcriptional induction or that replace H2A/H2B onto the promoter during transcriptional repression.
Dynamic chromatin assembly and disassembly processes also occur within the open reading frame during transcription. The histone chaperone Spt6 is required for the deposition of H3/H4 onto chromatin behind the RNA polymerase (7), whereas the histone chaperone FACT (facilitates chromatin transcription) assembles H2A/H2B onto the DNA behind the elongating RNA polymerase (7, 8). Yeast FACT is a heterodimeric protein complex of Spt16 and Pob3, although the HMG1-like protein Nhp6 also interacts and functionally cooperates with yFACT (9). Spt16 was originally identified to be a suppressor of Ty insertions into HIS4 and LYS2 (10). Although this implies that FACT has a role in the initiation of transcription, the molecular role of FACT at promoters has not yet been defined. The chromatin reassembly function of FACT within open reading frames requires ubiquitination of histone H2B on lysine 123 (11, 12). In turn, monoubiquitination of H2B Lys-123 is a result of the action of the ubiquitin ligase (E3)3 Bre1 and the ubiquitin-conjugating enzyme (E2) Rad6 (13) that travel with the elongating RNA polymerase (14). Notably, H2B Lys-123 ubiquitination is also required for the recruitment of the proteasome to the chromatin (15).
The proteasome is a multisubunit complex responsible for the selective degradation of almost all cytosolic proteins (16). Ubiquitinated proteins are recognized by the 26 S proteasome, which is a large proteolytic complex consisting of the 19 S cap complex and the 20 S catalytic core. The 19 S cap complex includes six ATPases, one of which is Sug1/Rpt6, and uses the energy provided by ATP hydrolysis to unwind proteins into unstructured chains. The 20 S core is composed of four stacked rings, which form a “tunnel” within which the unstructured protein chains undergo proteolytic cleavage into small peptides.
To better understand the molecular mechanisms of chromatin disassembly and reassembly at promoter regions, we have examined the role of FACT, the proteasome, and H2A Lys-123 ubiquitination on these processes at the well studied yeast PHO5 model promoter. Our studies reveal novel roles for FACT in promoter chromatin disassembly but not reassembly and a novel role for the 19 and 20 S proteasomes in transcriptional induction.
MATERIALS AND METHODS
Yeast Strains and Media
All media used were either with high (13.4 mm) phosphate or low (0.15 mm) phosphate. Media were prepared as follows. For 1 liter of medium, 0.7 g of yeast nitrogen base (without ammonium sulfate, phosphate, or amino acids), 2 g of glutamine, 100 ml of 20% glucose, and 3.9 g of MES were dissolved. Amino acids were added, and 1 m KH2PO4 and 1 m KCl were added to make the final ion concentration 13.4 mm. The pH of the medium was adjusted to 5.5, and the entire liter was filter-sterilized. The genotypes of all strains used are given in Table 1. The set of four strains used in Figs. 2E and 5E was generated by first making diploids that were heterozygous for the two temperature-sensitive (ts) mutations. Following meiosis and tetrad dissections, tetrads were identified that included one spore with both ts mutations, two spores with each individual ts mutation, and one spore with no ts mutation.
TABLE 1.
Yeast strains used in this study
| Name | Genotype | Source/Reference |
|---|---|---|
| BY4741 | Mat a; his3D1; leu2D0; met15D0; ura3D0 | ResGen |
| BY4741asf1Δ | Mat a; his3D1; leu2D0; met15D0; ura3D0; asf1::KanMX4 | ResGen |
| BY4741bre1Δ | Mat a; his3D1; leu2D0; met15D0; ura3D0; bre1::KanMX4 | ResGen |
| BY4741rad6Δ | Mat a; his3D1; leu2D0; met15D0; ura3D0; rad6::KanMX4 | ResGen |
| FY56 | Mat α; his4-912d; lys2-128d; ura3-52 | Ref. 10 |
| L577 | Mat α; his4-912d; lys2-128d; ura3-52; spt16-197 | Ref. 10 |
| JLY096 | Mat α; his4-912d; lys2-128d; ura3-52; HTB1:6-HIS:HA:URA | Ref. 17 |
| JLY097 | Mat α; his4-912d; lys2-128d; ura3-52; spt16-197; HTB1:6-HIS:HA:URA | This study |
| Y865 | Mat α; ura3-52; trp1-289; his3Δ1; leu2-3,112; gal2; gal10 | Ref. 47 |
| Y869 | Mat α; ura3-52; trp1-289; his3Δ1; leu2-3,112; gal2; gal10; nhp6A::URA3; nhp6B::HIS3 | Ref. 47 |
| JR5-2A HTB1 | MATa ura-3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 [HTB1-CEN-TRP1] | Ref. 48 |
| JR5-2A htb1-KR | MATa ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 [htb1-K123R-CEN-TRP1] | Ref. 48 |
| MSY535 | Mat α; hht1-2 Δ(hht2hhf2) lys2-Δ201, leu2,3,112, ura3-52 | Ref. 49 |
| MYS536 | Mat α; hhf1-10 Δ(hht2hhf2) lys2-Δ201, leu2,3,112, ura3-52 | Ref. 49 |
| MSY559 | Mat α; HHF1 HHF2 Δ(hht2hhf2) lys2-Δ201, leu2,3,112, ura3-52 | Ref. 49 |
| SKW200 | Mat α; HHF1 HHF2 Δ(hht2hhf2) lys2-Δ201, leu2,3,112, ura3-52 asf1::KanMX | This study |
| SC733 | MAT a sug1-20 GAL4::HIS3 ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | Ref. 50 |
| SC727 | MAT a GAL4::HIS3 ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | Ref. 50 |
| JKT0018 | MAT a asf1::his+ ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | Ref. 4 |
| SC782 | MAT a GAL4::HIS3 ura3 leu2-3,112, his3-11,15 canR gal+ | Ref. 50 |
| SC779 | MAT a pre1-1 pre4-1 GAL4::HIS3 ura3 leu2-3,112, his3-11,15 canR gal+ | Ref. 50 |
FIGURE 2.
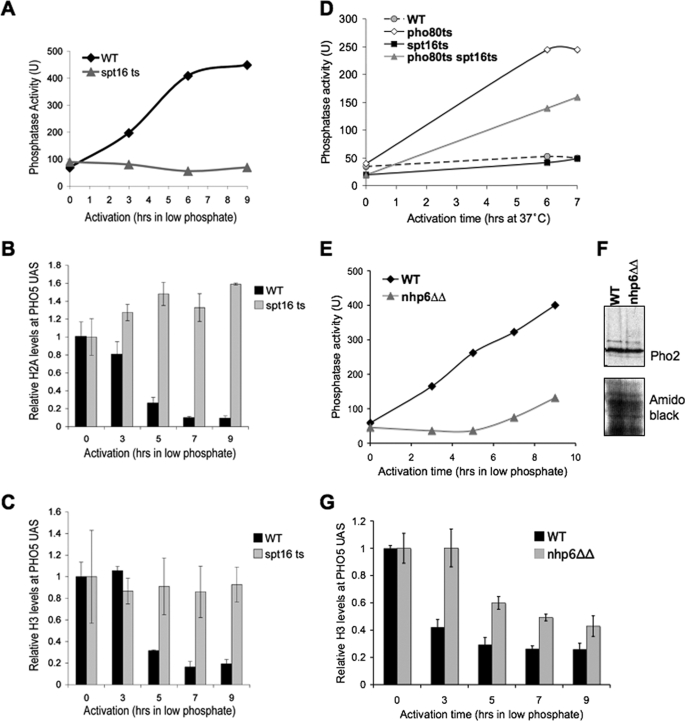
FACT promotes chromatin disassembly from the PHO5 promoter during transcriptional induction. A, strains JLY096 (WT) and JLY097 (spt16 ts) were switched to the non-permissive temperature of 39 °C, followed by phosphate depletion (inducing condition) and taking samples at the indicated times for acid phosphatase activity measurements as an indicator of PHO5 transcription. B, ChIP analysis of histone H2A at the UASp2 site of the PHO5 promoter in the same strains and time course as in A. The data were normalized to histone occupancy at the GAL1 promoter and the input samples. Average and S.D. of three independent experiments are plotted. C, ChIP analysis of histone H3 in the same strains and time course as in A and B. D, acid phosphatase activity of isogenic wild type, pho80, spt16, and pho80spt16 strains following shifting to the non-permissive temperature at time 0. E, acid phosphatase activity of stains Y865 (WT) and Y869 (nhp6ΔΔ) after the indicated times after switching to inducing conditions (low phosphate). F, Western blot analysis of the Pho2 activator in the same strains used in E. G, ChIP analysis of the same strains used in E for histone H3 occupancy at the UASp2 site of the PHO5 promoter at the indicated times after switching to inducing conditions (low phosphate). The data were normalized to histone occupancy at the GAL1 promoter and to the input samples. Average and S.D. of three independent experiments are plotted.
FIGURE 5.
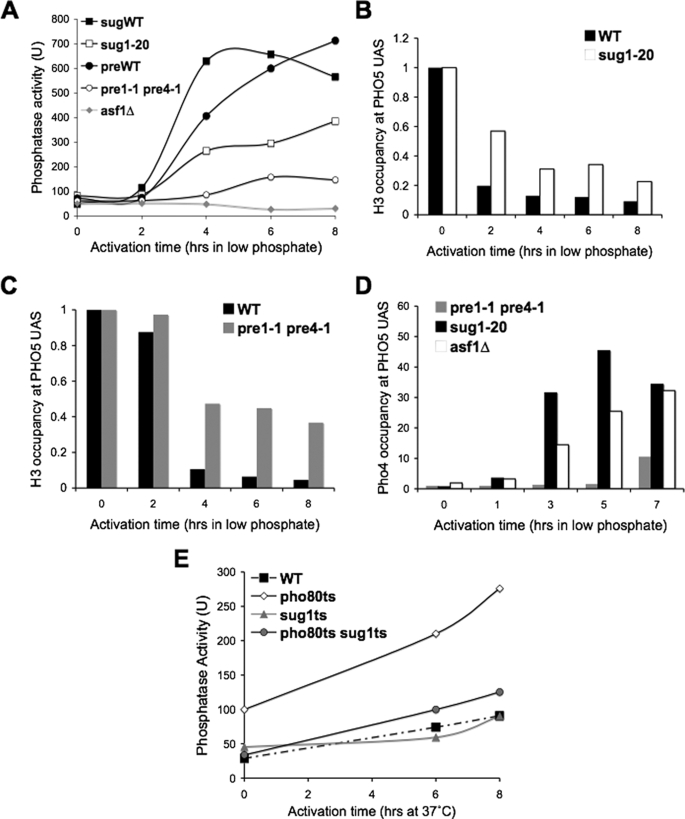
The proteasome promotes chromatin disassembly from the PHO5 promoter during transcriptional induction. A, acid phosphatase activity was measured as an indicator of PHO5 transcription in isogenic strains SC727 (sugWT), SC733 (sug1–20), and JKT0018 (asf1Δ) and also in isogenic strains SC782 (preWT) and SC779 (pre1-1 pre4-1) at the non-permissive temperature, followed by switching to inducing conditions (low phosphate) at the indicated times. B, ChIP analysis of histone H3 levels at the UASp2 site of the PHO5 promoter in strains SC727 (WT) and SC733 (sug1-20) from samples taken from the time course shown in A. The data were normalized to histone occupancy at the GAL1 promoter and the input samples. C, ChIP analysis of histone H3 from strains SC782 (preWT) and SC779 (pre1-1 pre4-1) taken from the same time course in A and analyzed as described for B. D, ChIP analysis of Pho4 in strains SC779 (pre1-1 pre4-1), JKT0018 (asf1Δ), and SC733 (sug1-20) at the non-permissive temperature at the indicated times after switching to inducing conditions (low phosphate). Samples were taken from the same time course shown in A, but the results were reproducible in independent time courses. E, acid phosphatase activity of isogenic wild type, pho80, sug1, and pho80sug1 strains following shifting to the non-permissive temperature at time 0.
Acid Phosphatase Activity Assays
Approximately 5 ml of cells were collected by centrifugation and washed with cold 0.1 m sodium acetate, pH 3.6, and then resuspended in 500 μl of the same buffer. To determine the number of cells used for each reaction, 100 μl of the cells were diluted 1:10 in double-distilled H20 and read at A600 nm. For each sample reaction, another 100 μl of washed cells were diluted 1:5 for a total volume of 500 μl in the same sodium acetate buffer and prewarmed for 10 min at 30 °C. A 500-μl sample of buffer alone was also included as a control as well as an appropriate volume (500 μl/reaction) of freshly made substrate, NPP (nitrophenyl phosphate (0.0742 g/10 ml), 0.1 m sodium acetate, pH 3.6). After warming, 500 μl of substrate was added to each reaction sample and incubated at 30 °C for 10 min, at which time 250 μl of stop solution, 1 m Na2CO3, was added. Samples were centrifuged for 1 min and then read at A410 nm. Phosphatase activity was equated as (A420 × 1000)/(A600 × volume of cell lysate used (μl) × incubation time (min). Single time courses are shown in each case, but comparable results were obtained for all of the phosphatase assays in independent time courses.
Chromatin Immunoprecipitation (ChIP) Analysis
ChIP analyses were performed as described previously (17), using either 2.5 μl of the C-terminal anti-histone H3 (catalog number ab1791; Abcam), 2 μl of anti-HA (catalog number mms-101r; Covance), or 2 μl of anti-Pho4 (courtesy of E. O'Shea) overnight at 4 °C. The sequential ChIP analyses utilized micrococcal nuclease-digested mononucleosomes as the template, whereas all other analyses utilized sonicated chromatin fragments. All ChIP quantitation was performed by real time PCR using a Roche Applied Sciences Light Cycler 480. The linear range of PCR templates was determined by performing a 10-fold serial dilution standard curve, which usually proved that a 1:10 dilution was sufficient. Each sample was analyzed in triplicate using 10-μl reactions in a 384-well plate format. The thermal profile was as follows: 1) denaturation at 95 °C for 10 min; 2) run cycle of 95 °C for 15 s and then 60 °C for 1 min for 50–60 cycles; 3) cooling at 40 °C for 30 s. Each immunoprecipitation sample was normalized to its respective input samples (to account for the number of cells taken) as well as a control region called GAL1/10, whose histone occupancy is regulated by glucose, not phosphate, levels.
Primers and Taqman probes used were as follows: PHO5 UASp2 A, GAATAGGCAATCTCTAAATGAATCGA; PHO5 UASp2 B, GAAAACAGGGACCAGAATCATAAATT; PHO5 UASp2 probe, FAM-ACCTTGGCACTCACACGTGGGACTAGC-MGB; GAL1/10 A, GACGCACGGAGGAGAGTCTT; GAL1/10 B, CGCTTAACTGCTCATTGCTATATTG; GAL1/10 probe, FAM-CGCTCGGCGGCTTCTAATCCG-MGB.
Western Blotting
Samples were prepared by washing ∼50 μl of actively dividing cells twice with double-distilled H2O, resuspending in 100 μl of yeast sample buffer, and then boiling for 5 min. Samples were separated on a 10% acrylamide gel and transferred to a polyvinylidene fluoride membrane. The membrane was probed with 1:1000 anti-Pho2 antibody (courtesy of E. O'Shea) and then 1:50,000 IgG peroxidase conjugate antibody (catalog number A-1949; Sigma). Processing of the membrane was performed with the ECL Western blotting detection reagents (catalog number RPN2209; GE Healthcare).
Nucleosome Remodeling Assays
Homogenous nucleosomes were reconstituted by salt dilution using Xenopus laevis recombinant histone octamers and 601 nucleosome-positioning sequences. The DNA used for nucleosome reconstitution was synthesized by PCR, which comprises 601 positioning DNA sequence at the center flanked by 69 and 59 bp of extranucleosomal DNA. About one-tenth of the DNA was end-labeled with p32.
For the nucleosome remodeling assay by gel shift, the indicated amounts of RSC (remodels the structure of chromatin), PA700, and 26 S were incubated with mononucleosomes (12 nm) for 30 min at 30 °C in the presence of 2 mm ATP. The purified yeast RSC remodeling complex was used as a control for remodeling reactions. The bound proteins were competed off, and reactions were terminated by adding excessive competitor DNA and 2 mm γ-thio-ATP. The samples were analyzed on 5% PAGE with 0.2× TBE with buffer recirculation. Buffer conditions used for PA700 were as follows: 20 mm Na-HEPES, pH 7.8, 60 mm NaCl, 3 mm MgCl2, 0.4 mm EDTA, 4 mm β-mercaptoethanol, 6% glycerol, 100 μg/ml bovine serum albumin, 0.2 mm phenylmethylsulfonyl fluoride, 0.08% Nonidet P-40, 2 mm ATP. Buffer conditions for 26 S were as follows: 20 mm Na-HEPES, pH 7.8, 60 mm NaCl, 5 mm MgCl2, 2.8 mm β-mercaptoethanol, 6% glycerol, 100 μg/ml bovine serum albumin, 0.2 mm phenylmethylsulfonyl fluoride, 0.08% Nonidet P-40, 2 mm ATP. Buffer conditions for RSC were as follows: 20 mm Na-HEPES, pH 7.8, 60 mm NaCl, 3 mm MgCl2, 2 mm β-mercaptoethanol, 7% glycerol, 100 μg/ml bovine serum albumin, 0.2 mm phenylmethylsulfonyl fluoride, 0.08% Nonidet P-40, 2 mm ATP.
For the nucleosome remodeling assay by restriction accessibility assay, 6 nm concentrations of labeled mononucleosomes were incubated with RSC (3 nm), PA700 (3–81 nm), or 3–54 nm 26 S for 30 min at 30 °C with 3.75 units of RsaI. The reactions were stopped and deproteinized by adding an equal volume of stop solution (20 mm Tris-HCl, pH 8, 1.2% SDS, 80 mm EDTA, 5% glycerol, 0.2 mg/ml proteinase K) and incubating at 50 °C for 20 min.
RESULTS
Pho4 Binds to a Nucleosome Prior to Nucleosome Disassembly
Our studies of chromatin disassembly during transcriptional induction use ChIP analysis of histone and factor occupancy at the well characterized budding yeast PHO5 gene promoter (4–6, 17). PHO5 encodes the major acid phosphatase in Saccharomyces cerevisiae. PHO5 transcription is induced by phosphate depletion, which causes Pho81 to inhibit the Pho80-Pho85 cyclin-dependent kinase complex, leading to relocalization of the Pho4 activator to the nucleus (18). The Pho4 binding site within the PHO5 upstream activating sequence, UASp2, is occluded by a nucleosome (nucleosome −2) in repressing conditions (Fig. 1A). The localization of Pho4 to the nucleus is required for the subsequent disassembly of four nucleosomes (nucleosomes −1 to −4) from the PHO5 promoter (19, 20), which is required in turn to allow the subsequent recruitment of the general transcription machinery (5). Whether Pho4 gains access to its nucleosome-occluded UASp2 DNA binding site to trigger this cascade of events is not known.
FIGURE 1.
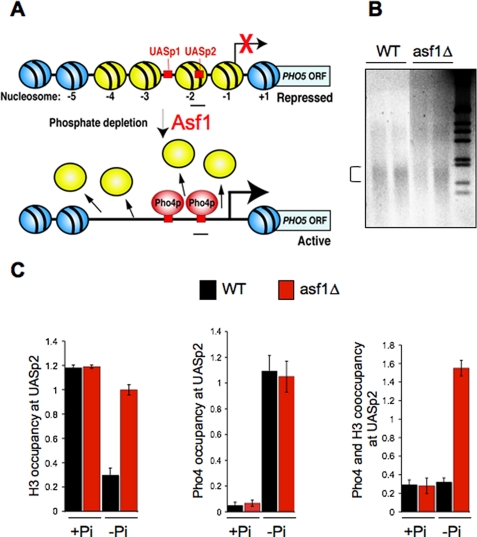
Pho4 and a histone octamer can co-occupy nucleosome −2 of the PHO5 promoter in vivo. A, schematic of the PHO5 promoter. The four yellow nucleosomes are disassembled during transcriptional induction, a process that is facilitated by Asf1. Phosphate depletion is the signal for nuclear localization of the Pho4 activator. B, micrococcal nuclease digestion of chromatin to mononucleosomes, used for the ChIP analyses shown in C, from strain BY4741 (WT) and BY4741asf1Δ (asf1Δ) for the −Pi samples. The bracket indicates the mononucleosome. The right lane contains the 1-kb ladder. C, ChIP analysis of H3 and Pho4 occupancy over the UASp2 binding site. Samples were taken from the strains in B, grown in PHO5-repressing conditions (+Pi), or 8 h after switching to inducing conditions, low phosphate medium (−Pi). The left panel shows histone occupancy over UASp2, normalized to histone occupancy at the GAL1 promoter and input samples. The average and S.D. of three independent analyses are shown. The middle panel shows Pho4 occupancy at UASp2 in the same samples. The right panel shows co-occupancy of Pho4 and H3 as determined by sequential ChIP analysis from the same samples.
Our earlier analyses had provided circumstantial evidence that Pho4 activator can bind to its UASp2 site that is very close to the dyad axis of symmetry of nucleosome −2 of the PHO5 promoter (Fig. 1A) while histones H3 and H2A are still present (4, 21). This potential intermediate state was achieved by slowing down the chromatin disassembly process by deleting the gene encoding the Asf1 histone chaperone in PHO5-inducing (low phosphate) conditions (4, 21). Given that the helix-loop-helix DNA binding domain of Pho4 makes intimate contacts with opposite faces of the DNA duplex (22), it was unexpected to find the histone octamer and Pho4 coexisting on the same piece of DNA in vivo. Because our previous ChIP analyses had used sonication to generate ∼500-bp chromatin fractions, we revisited these experiments using micrococcal nuclease- generated mononucleosomes (Fig. 1B). Disassembly of PHO5 nucleosome −2 is apparent 8 h after switching to low phosphate medium in wild type yeast, whereas nucleosome −2 is largely intact in the asf1 mutant strain (Fig. 1C, left). By contrast, the amount of Pho4 recruited to UASp2 8 h after switching to low phosphate medium is equivalent in both the wild type and asf1 mutant strain (Fig. 1C, middle). It is important to note that all analyses of factor occupancy at the PHO5 promoter are internally normalized to another region of the yeast genome (the GAL1 promoter), where there are no changes in histone occupancy or Pho4 occupancy in response to changes in phosphate concentrations (4, 5). In our previous ChIP analyses with the same antibodies used here, we have also shown that these factors are not detectably present at nonspecific DNA, such as the mitochondrial COX3 gene (4, 5). It is extremely unlikely that the Pho4 bound to nucleosome −2 in the asf1 mutant reflects binding to the adjacent UASp1 site, because the UASp2 within the nucleosome −2 site is 13 times stronger than the UASp1 site (23). As such, the Pho4 occupancy in the asf1 mutant in the inducing condition (−Pi) would have been 8% of that seen in the wild type strain if it were due to binding to the UASp1 site alone, not the equivalent binding that we see with the asf1 mutant and wild type strain in Fig. 1C (middle). In addition, the UASp1 site is in a nucleosome-free region and therefore is likely to have been destroyed by digestion to mononucleosomes. To provide additional proof of co-occupancy of histones H3 and Pho4 at nucleosome −2, we performed sequential ChIP analysis of the mononucleosomes. First we immunoprecipitated for Pho4, followed by elution of the immunoprecipitates and their subsequent immunoprecipitation for H3. As shown in the right panel of Fig. 1C, we found that H3 and Pho4 co-occupy nucleosome −2 in the asf1 mutant in inducing conditions (−Pi). These results establish that the Pho4 activator does co-occupy the same stretch of DNA as a histone octamer in vivo, clearly demonstrating that histone removal occurs after activator binding.
FACT Promotes Promoter Chromatin Disassembly and Subsequent Transcriptional Induction
We sought to investigate the mechanism whereby activator binding triggers the subsequent removal of histones H2A and H2B from the PHO5 promoter during transcriptional induction. To ask whether the H2A/H2B chaperone FACT is implicated in chromatin disassembly from promoter regions, we examined transcriptional induction in a temperature-sensitive mutant of the Spt16 subunit of FACT following phosphate removal after FACT had been first inactivated by shifting to the restrictive temperature. Activation of the PHO5 gene was greatly delayed upon inactivation of Spt16 (Fig. 2A). In agreement, disassembly of histones H2A/H2B from the PHO5 promoter is greatly delayed when FACT is inactivated prior to the addition of the signal for PHO5 induction (−Pi) (Fig. 2B). As expected, the removal of H3/H4 from the PHO5 promoter was also delayed when FACT was inactivated, given that H3/H4 cannot be removed from the DNA until after H2A/H2B are removed (Fig. 2C). These data indicate that the histone chaperone FACT is important for the disassembly of H2A/H2B from the PHO5 promoter and its subsequent transcriptional induction.
Induction of the PHO5 gene in response to phosphate removal requires that the cell first use up its endogenous phosphate and polyphosphate stores, which itself requires growth. Given that Spt16 is an essential protein, it was possible that the failure of spt16 mutant yeast to induce PHO5 transcription might be due to its growth defect at the non-permissive temperature. To rule out this possibility, we inactivated the Pho80 protein with a conditional allele, which results in Pho4 being constitutively nuclear under all phosphate conditions at the non-permissive temperature (24). As such, phosphate depletion and growth are not required for induction of PHO5 transcription in the absence of Pho80. Following a shift to the non-permissive temperature in high phosphate medium, we found that the pho80 mutant was able to induce PHO5 transcription as expected, whereas the wild type strain could not (Fig. 2D). By contrast, the pho80 spt16 double mutant gave an intermediate result, being able to partially induce PHO5 (Fig. 2D). These data confirm that FACT promotes chromatin disassembly and the subsequent transcriptional induction from the PHO5 promoter.
To provide additional confirmation of a role for FACT in promoter chromatin disassembly, we examined the HMG-1-like protein Nhp6, which functionally potentiates the ability of FACT to alter nucleosome structure (25, 26). Accordingly, we found that induction of PHO5 transcription was greatly delayed in the absence of Nhp6, which was achieved by the deletion of the functionally redundant NHP6A and NHP6B genes (Fig. 2E). This delay in PHO5 transcriptional induction in the absence of Nhp6 was not a consequence of altered levels of the Pho2 activator that is required for PHO5 induction, since its levels were indistinguishable between wild type and Nhp6 mutants (Fig. 2F). Rather, the delay in transcriptional induction in the absence of Nhp6 was most likely due to the delay in chromatin disassembly that is apparent in the absence of Nhp6 (Fig. 2G). Taken together, these results demonstrate that FACT has a novel function in promoting histone H2A/H2B removal from promoter regions during transcriptional induction.
FACT Is Not Required for Promoter Chromatin Reassembly or Transcriptional Repression
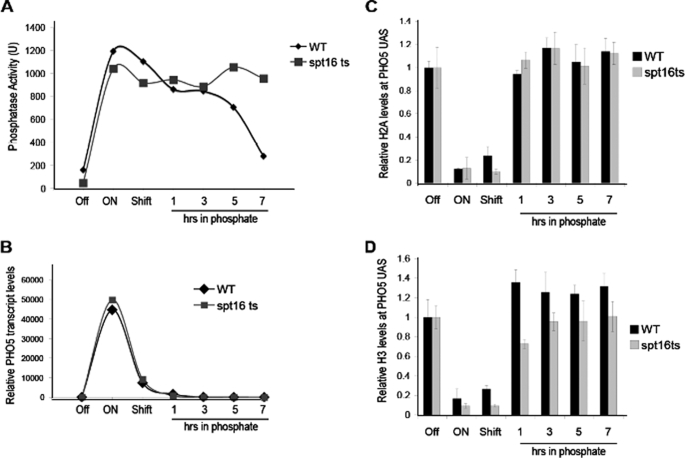
Given that FACT mediates chromatin reassembly within open reading frames, we asked whether FACT might also be involved in reassembling chromatin onto promoters during transcriptional repression. After fully activating the PHO5 gene at the permissive temperature to enable promoter chromatin disassembly (which requires functional FACT; Fig. 2), we shifted the spt16 temperature-sensitive mutant to the non-permissive temperature and then added phosphate as a signal for repression. When we assayed the activity of the Pho5 phosphatase as an indicator of transcriptional repression, we found that in contrast to the wild type strain treated in the same manner, the phosphatase activity was not reduced in the spt16 mutant (Fig. 3A). To investigate whether the maintenance of Pho5 protein activity in the absence of FACT under repressing conditions actually reflected a lack of transcriptional repression, we analyzed the PHO5 RNA levels by reverse transcription-PCR. Quite unexpectedly, we found that the reduction in the Pho5 RNA transcript levels upon adding the repression signal (phosphate) was indistinguishable between wild type and spt16 mutant strains (Fig. 3B). In agreement, the spt16 mutant was fully able to reassemble chromatin onto the PHO5 promoter during transcriptional repression (Fig. 3, C and D), a process that we have previously shown to be essential for repression of PHO5 transcription (6). Taken together, these data indicate that FACT is not required for the reassembly of chromatin onto the PHO5 promoter in order to achieve its transcriptional repression. However, in the absence of FACT, more Pho5 protein is present, which could potentially be due to either protein stabilization or increased translation of the PHO5 transcript.
FIGURE 3.
FACT does not mediate chromatin reassembly onto the PHO5 promoter during transcriptional repression. A, acid phosphatase activity in strain JLY096 (WT) and JLY097 (spt16 ts). The PHO5 gene was fully induced at the permissive temperature, followed by a “shift” to the non-permissive temperature and the subsequent addition of phosphate as the signal for transcriptional repression. B, quantitative reverse transcription analysis of PHO5 transcript levels during the same time course as in A. C, ChIP analysis of histone H2A levels at the UASp2 site of the PHO5 promoter in the same strains and time course as in A. The data were normalized to histone occupancy at the GAL1 promoter and the input samples. Average and S.D. value of three independent experiments are plotted. D, as for C but with analysis for histone H3 levels.
Contribution of Histone Modifications to Promoter Chromatin Disassembly
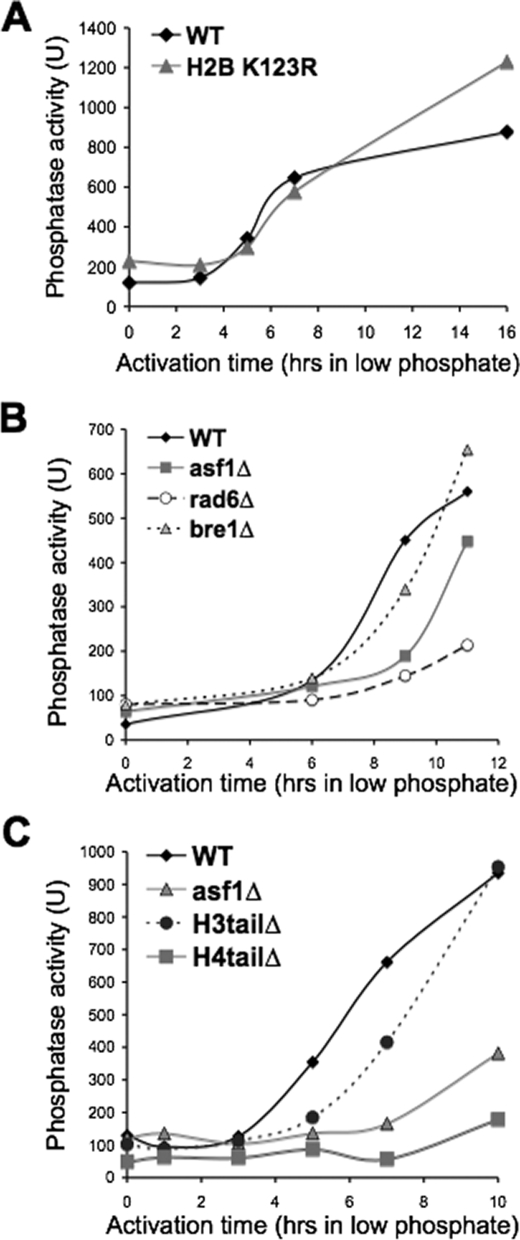
Because monoubiquitination of histone H2B on Lys-123 is required for FACT-mediated chromatin reassembly within open reading frames (11), we asked whether H2B Lys-123 ubiquitination is also important for FACT-mediated chromatin disassembly at promoters. To ask this question, we used a strain that has Lys-123 of H2B replaced with an arginine to prevent ubiquitination. We found that the H2B K123R strain was fully capable of inducing PHO5 transcription (Fig. 4A), indicating that H2B Lys-123 ubiquitination is not required for FACT-mediated promoter chromatin disassembly. In agreement, deletion of BRE1 encoding the E3 ubiquitin ligase for H2B Lys-123, also had no effect on induction of PHO5 transcription (Fig. 4B). Interestingly, deletion of the gene encoding the E2 Rad6 that works with Bre1 to ubiquitinate H2B Lys-123 resulted in delayed PHO5 induction (Fig. 4B). These data indicate that although H2B Lys-123 ubiquitination is irrelevant for promoter chromatin disassembly and subsequent PHO5 activation, Rad6-mediated ubiquitination of another protein is likely to be required for PHO5 induction.
FIGURE 4.
Rad6 and the N terminus of H4 are required for transcriptional induction of PHO5. A, acid phosphatase activity in strain JR5–2A HTB1 (WT) and JR5–2A htb1-KR (H2B K123R) following phosphate depletion as the signal for transcriptional induction. B, acid phosphatase activity in strain BY4741 (WT), BY4741asf1Δ (asf1Δ), BY4741bre1Δ (bre1Δ), and BY4741rad6Δ (rad6Δ) following phosphate depletion as the signal for transcriptional induction. C, acid phosphatase activity in strain MSY536 (WT), SK200 (asf1Δ), MSY559 (H3 tail Δ), and MSY535 (H4 tail Δ) following phosphate depletion as the signal for transcriptional induction.
To extend our analyses of the histone modifications and requirements for promoter chromatin disassembly, we examined yeast deleted for either the N terminus of histone H3 or H4. We found that although yeast lacking the N-terminal tail of H3 are fully competent for PHO5 induction, the N-terminal tail of H4 is essential for PHO5 induction (Fig. 4C). These data demonstrate that histone modifications within the N-terminal tail of H3, but not the H4 tail, are dispensable for promoter chromatin disassembly and subsequent transcriptional induction. The requirement for the H4 N-terminal tail for PHO5 induction is consistent with the previously published role of H4 Lys-16 acetylation in recruitment of the Pho2 activator, which subsequently recruits Pho4 to the UASp2 binding site within the PHO5 promoter (27).
Distinct Functions for the 19 and 20 S Proteasome for Promoter Chromatin Disassembly and Transcriptional Activation
Given that we had found a requirement for the ubiquitin-conjugating (E2) enzyme Rad6 in PHO5 transcriptional induction (Fig. 4B), we asked whether the proteasome is also required for PHO5 gene induction. To do this, we used temperature-sensitive mutants of the Sug1 component of the 19 S proteasomal cap and a double temperature sensitive mutant of the Pre1 and Pre4 components of the 20 S proteasomal core. We found that both the sug1-20 mutant and the pre4-1 pre1-1 double mutant had delayed activation of PHO5 at the non-permissive temperature after phosphate depletion (Fig. 5A), although the defect was greater with the pre4-1 pre1-1 mutant. These data indicate that both the 19 and 20 S components of the proteasome are required for efficient PHO5 induction. Furthermore, we find that H3 removal from the PHO5 promoter was delayed in both the sug1-20 mutant and the pre4-1 pre1-1 double mutant, although the defect was more pronounced in the pre4-1 pre1-1 (Fig. 5, B and C). These data indicate that both the 19 and 20 S components of the proteasome are required for efficient PHO5 promoter chromatin disassembly and the subsequent transcriptional induction of PHO5. Given that promoter binding by the Pho4 activator is a prerequisite for promoter chromatin disassembly, we examined Pho4 recruitment to the PHO5 promoter. We have previously shown that Pho4 recruitment is not altered in asf1 mutants (Fig. 1B) (4, 5). By comparison with the asf1Δ strain, we find that there is no defect in Pho4 recruitment to the PHO5 promoter in the sug1-20 mutant but that there is a severe defect in Pho4 recruitment in the pre4-1 pre1-1 double mutant (Fig. 5D). These data demonstrate that the 20 S proteasome but not the 19 S proteasome is required for Pho4 recruitment. This result suggests that either degradation of an unstructured protein or ubiquitin-independent protein degradation by the 20 S proteasome is required for Pho4 recruitment to the PHO5 promoter and the subsequent chromatin disassembly and transcriptional induction.
Given that the proteasome is essential, it was possible that the delay of the proteasome mutants to induce PHO5 transcription might be due to its growth defect at the non-permissive temperature. To rule out this possibility, we inactivated Pho80, which allows PHO5 induction in the absence of growth, as described above. We found that the pho80 mutant was able to induce PHO5 transcription as expected, whereas the wild type strain could not (Fig. 5E). By contrast, the pho80 sug1-1 double mutant still showed a clear delay in induction of PHO5 (Fig. 5E). These data confirm that the proteasome promotes chromatin disassembly and the subsequent transcriptional activation from the PHO5 promoter.
The Proteasome Is Not an ATP-dependent Chromatin Remodeler
We were intrigued by the fact that Pho4 is recruited to the PHO5 promoter in the absence of function of the 19 S proteasome, yet chromatin disassembly is defective. This raised the possibility that perhaps the ATPase activity of the 19 S proteasome may be serving an important function to break histone-DNA contacts during chromatin disassembly from promoter regions. This idea is consistent with the observations that 1) the proteasome is recruited to promoter regions (15) and stimulates transcription, 2) the proteasome has a non-proteolytic role in transcription (28–30), 3) the Sug1 component of the 19 S cap genetically and physically interacts with FACT (31, 32), and 4) the bacterial homolog of the Sug1 ATP-dependent helicase can destabilize protein-DNA interactions in a non-proteolytic manner (33). As such, we decided to test whether the 19 S proteasome functions as a novel ATP-dependent chromatin remodeler to break the interactions between histones and DNA, which would in turn enable subsequent modifications to the chromatin structure, such as histone removal.
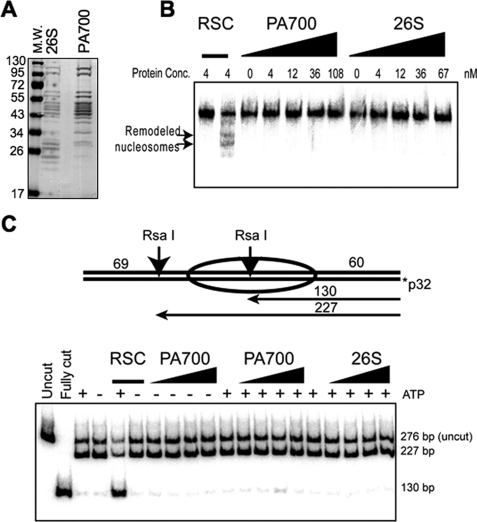
Bovine 19 S (PA700) and 26 S proteasome were purified to near homogeneity, as described previously (34) (Fig. 6A), and were confirmed to be active for ATPase activity (data not shown) (35). Mononucleosomes were assembled in vitro onto the 601 nucleosome-positioning sequence by salt gradient dialysis of X. laevis recombinant histones. The chromatin remodeling activity of the ATP-dependent chromatin remodeler yeast RSC was apparent by its ability to move the histone octamer to alternative positions on the DNA, as assayed by an electrophoretic mobility shift assay (Fig. 6B). By contrast, neither PA700 nor 26 S proteasome was able to move the histone octamer in vitro (Fig. 6B). As a second assay for ATP-dependent chromatin remodeling activity, we measured accessibility of a Rsa1 restriction site located within the positioned nucleosome (Fig. 6C). Although the RSC complex enabled the Rsa1 enzyme to gain access to its site in an ATP-dependent manner, this was not the case with either PA700 or 26 S proteasome. These results demonstrate that neither the 19 S proteasome cap nor the 26 S proteasome can function as an ATP-dependent chromatin remodeler.
FIGURE 6.
The proteasome is not an ATP-dependent chromatin remodeler. A, Coomassie-stained SDS-PAGE analysis of the purified bovine 26 S proteasome and the 19 S cap (PA700) used for the remodeling assays. B, gel shift analysis of mononucleosome remodeling by the indicated amounts of RSC, PA700, or 26 S proteasome. The arrows indicate the remodeled nucleosomes. C, restriction accessibility analysis of mononucleosome remodeling by RSC, PA700, or 26 S proteasome. The 130-bp fragment is only produced if the RsaI restriction enzyme can gain access to its site that is otherwise buried by the nucleosome.
DISCUSSION
Activator Binding to the Nucleosome Triggers Subsequent Chromatin Disassembly
Although it is clear that activator binding is the trigger for the removal of histones from the underlying DNA sequence, the mechanism whereby the activators start this process was far from clear. With a few exceptions, all of the studies of transcription factors binding to their sites within nucleosomes have been performed on in vitro-assembled chromatin templates, and virtually all of these studies have used binding sites near the edges of the nucleosome that are transiently exposed or multiple binding sites that allow invasion from the edge of the nucleosome (reviewed in Ref. 36). Only a few examples exist where the binding of a transcription factor to a site near the nucleosomal dyad was demonstrated either in vivo (37) or in vitro (38) (39, 40). In these cases, the binding was greatly reduced by the histones and/or required the action of ATP-dependent chromatin remodeling factors or poly(dA·dT) DNA elements. Histone remodeling and/or displacement presumably occurs so rapidly following activator binding in vivo that the mechanism whereby activators initially gain access to and destabilize the nucleosome prior to nucleosome remodeling/disassembly remains unclear. However, we fortuitously trapped a novel intermediate of an activator occupying a nucleosome-bound DNA sequence in vivo when we inactivated the chromatin disassembly factor Asf1 (Fig. 1A). Even more striking than our discovery that the Pho4 activator and the histone octamer co-occupy the same piece of DNA in vivo is the fact that the Pho4 binding site is very close to, if not at, the dyad axis of symmetry of the nucleosome, which is the least accessible part of the nucleosomal DNA. Notably, the DNA sequence occupied by the histone octamer at PHO5 nucleosome −2 does not notably change upon Pho4 binding in the asf1 mutant (4). Many of the activators whose nucleosome binding has been studied to date, such as the glucocorticoid receptor, only recognize one face of the DNA double helix (41, 42), making it understandable how these solvent-accessible sites could be bound within a nucleosome. By contrast, Pho4 is a basic helix-loop-helix DNA binding domain protein that embraces the DNA from both sides (22). It is hard to imagine that the nucleosome structure is not altered by the binding of Pho4 to the nucleosomal dyad. However, in the absence of the Asf1 histone chaperone that facilitates chromatin disassembly, this altered nucleosome structure is clearly stable enough to allow retention of all the histones on the DNA after Pho4 binding. Although future studies will be required to map the exact nature of the changes to the nucleosome structure that occur upon Pho4 binding, we propose that these activator-induced nucleosome changes are the trigger that drives histone removal from promoters.
FACT Promotes Transcriptional Initiation via Its Role in Promoter Chromatin Disassembly
Exactly how nucleosomes are removed from promoter regions to allow the general transcription machinery to access the DNA sequence is not entirely clear. We had shown previously that the histone H3/H4 chaperone Asf1 (4) and weakening of the intrinsic nucleosome structure via acetylation of histone H3 on lysine 56 (17) was involved in this process. In the current study, we have discovered a novel role for the H2A/H2B chaperone FACT in chromatin disassembly from the yeast PHO5 promoter. This activity of FACT is in contrast to the well established role of FACT in chromatin reassembly behind the elongating RNA polymerase II within open reading frames (12, 43). However, recent studies provide precedent for a promoter-specific role for FACT to drive transcriptional activation, because FACT was required for TBP recruitment to the GAL1 and HO promoters (8, 44). Given that we have previously shown that promoter chromatin disassembly is required for TBP recruitment to the PHO5 promoter (5), the role of FACT in chromatin disassembly from the PHO5 promoter (Fig. 2) can explain the requirement for FACT for TBP recruitment (8, 44). Our data also provide a molecular basis for the observations made over 10 years ago demonstrating that Nhp6A/B potentiates promoter-specific transcriptional activation in vivo (45).
It was interesting to see a significant increase in H2A/H2B occupancy at the PHO5 promoter upon inactivation of FACT (Fig. 2B). We think that this indicates that there is a high degree of H2A/H2B exchange at the PHO5 promoter when FACT is around, with FACT mediating the H2A/H2B removal from the DNA and another factor mediating the H2A/H2B replacement. The dynamic nature of H2A/H2B would lead to a suboptimal occupancy of H2A/H2B relative to H3/H4 on the PHO5 promoter. This idea of H2A/H2B being highly dynamic is consistent with our observation that when we delay H3/H4 removal from the PHO5 promoter by inactivation of the histone chaperone Asf1, this additionally delays H2A/H2B removal from the PHO5 promoter (17). As such, we proposed that H3/H4 removal is the rate-limiting and regulated step of the chromatin disassembly process and that H2A/H2B can keep returning to the PHO5 promoter as long as H3/H4 tetramers are present on the DNA. Accordingly, if H2A/H2B occupancy is normally suboptimal due to its highly dynamic nature, then inactivation of the H2A/H2B removal factor FACT will result in higher H2A/H2B occupancy, as seen in Fig. 2B.
FACT function within the open reading frame is enhanced by monoubiquitination of lysine 123 of histone H2B (11). This does not appear to be the case for FACT-mediated chromatin disassembly from the PHO5 promoter, because mutation of lysine 123 of H2B or inactivation of the E3 ligase that mediates this ubiquitination, Bre1, has no effect on chromatin disassembly from the PHO5 promoter (Fig. 4B). Interestingly, however, we did find a role for the E2 Rad6 in chromatin disassembly, suggesting that ubiquitination of proteins other than H2B may be important for chromatin disassembly.
Dual Roles for the Proteasome in Stimulating Activator Binding and Promoter Chromatin Disassembly
Our studies have uncovered two novel functions for the proteasome within promoters. First, we have found that the 20 S but not the 19 S is important for recruitment of the Pho4 activator to the PHO5 promoter. It is not clear whether this is a direct or indirect consequence of inactivation of the 20 S proteasome. Notably, inactivation of the 20 S but not the 19 S proteasome leads to the defect in activator recruitment. The 20 S proteasome does have proteolytic functions in the absence of the 19 S cap (35) (e.g. in the degradation of unstructured protein regions). Alternatively, given that the 19 S cap mediates ubiquitin recognition, the function of the 20 S at promoter regions is likely to be mediated via proteolysis of non-ubiquitinated substrates.
In addition, we have found a role for the 19 S proteasome in chromatin disassembly from the PHO5 promoter. This activity is likely to be quite distinct from the previously reported role for the 19 S proteasome in recruitment of SAGA to promoters (46), because we have previously shown that SAGA is required for Pho4 activator binding (5). However, because we see no defect in Pho4 activator binding in our 19 S proteasome mutant (Fig. 5D), this indicates that SAGA recruitment to the PHO5 promoter is not defective in the 19 S proteasome mutant. It is also possible that promoter chromatin disassembly is potentiated by the intact 26 S proteasome, not just the 19 S cap, but this is difficult to discern given the role of the 20 S proteasome in the upstream event of Pho4 recruitment. Exactly how the 19 or 26 S proteasome contributes to chromatin disassembly from the PHO5 promoter is unclear. Clearly, however, the 19 or 26 S proteasome is not facilitating chromatin disassembly via a potential role in breaking histone-DNA contacts, because neither the 19 S subunit nor the entire 26 S proteasome has ATP-dependent chromatin remodeling activity. Future studies will reveal whether the proteasome-mediated chromatin disassembly from promoters is related to the Bre1-independent involvement of the E2 Rad6 in chromatin disassembly.
Acknowledgments
We are very grateful for the generous gifts of strains from Mary-Anne Osley, Fred Winston, Michael Snyder, Mitch Smith, and Stephan Johnston. We thank Erin O'Shea for the Pho2 and Pho4 antibody.
This work was supported, in whole or in part, by National Institutes of Health Grant GM64475 (to J. K. T.) and Training Grant T32 CA082086 (to M. R.).
- E3
- ubiquitin-protein isopeptide ligase
- E2
- ubiquitin carrier protein
- MES
- 4-morpholineethanesulfonic acid
- ts
- temperature-sensitive
- ChIP
- chromatin immunoprecipitation.
REFERENCES
- 1.Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Nature 389,251–260 [DOI] [PubMed] [Google Scholar]
- 2.Li B., Carey M., Workman J. L. (2007) Cell 128,707–719 [DOI] [PubMed] [Google Scholar]
- 3.Smith S., Stillman B. (1991) EMBO. J. 10,971–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adkins M. W., Howar S. R., Tyler J. K. (2004) Mol. Cell. 14,657–666 [DOI] [PubMed] [Google Scholar]
- 5.Adkins M. W., Williams S. K., Linger J., Tyler J. K. (2007) Mol. Cell. Biol. 27,6372–6382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adkins M. W., Tyler J. K. (2006) Mol. Cell. 21,405–416 [DOI] [PubMed] [Google Scholar]
- 7.Kaplan C. D., Laprade L., Winston F. (2003) Science 301,1096–1099 [DOI] [PubMed] [Google Scholar]
- 8.Mason P. B., Struhl K. (2003) Mol. Cell. Biol. 23,8323–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Formosa T., Eriksson P., Wittmeyer J., Ginn J., Yu Y., Stillman D. J. (2001) EMBO J. 20,3506–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malone E. A., Clark C. D., Chiang A., Winston F. (1991) Mol. Cell. Biol. 11,5710–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pavri R., Zhu B., Li G., Trojer P., Mandal S., Shilatifard A., Reinberg D. (2006) Cell 125,703–717 [DOI] [PubMed] [Google Scholar]
- 12.Fleming A. B., Kao C. F., Hillyer C., Pikaart M., Osley M. A. (2008) Mol. Cell. 31,57–66 [DOI] [PubMed] [Google Scholar]
- 13.Kao C. F., Hillyer C., Tsukuda T., Henry K., Berger S., Osley M. A. (2004) Genes Dev. 18,184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao T., Kao C. F., Krogan N. J., Sun Z. W., Greenblatt J. F., Osley M. A., Strahl B. D. (2005) Mol. Cell. Biol. 25,637–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ezhkova E., Tansey W. P. (2004) Mol. Cell 13,435–442 [DOI] [PubMed] [Google Scholar]
- 16.Wolf D. H., Hilt W. (2004) Biochim. Biophys. Acta. 1695,19–31 [DOI] [PubMed] [Google Scholar]
- 17.Williams S. K., Truong D., Tyler J. K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105,9000–9005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komeili A., O'Shea E. K. (2000) Curr. Opin. Cell Biol. 12,355–360 [DOI] [PubMed] [Google Scholar]
- 19.Boeger H., Griesenbeck J., Strattan J. S., Kornberg R. D. (2003) Mol. Cell. 11,1587–1598 [DOI] [PubMed] [Google Scholar]
- 20.Reinke H., Hörz W. (2003) Mol. Cell. 11,1599–1607 [DOI] [PubMed] [Google Scholar]
- 21.Williams S. K., Tyler J. K. (2007) Curr. Opin. Genet. Dev. 17,88–93 [DOI] [PubMed] [Google Scholar]
- 22.Shimizu T., Toumoto A., Ihara K., Shimizu M., Kyogoku Y., Ogawa N., Oshima Y., Hakoshima T. (1997) EMBO J. 16,4689–4697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maerkl S. J., Quake S. R. (2007) Science 315,233–237 [DOI] [PubMed] [Google Scholar]
- 24.O'Neill E. M., Kaffman A., Jolly E. R., O'Shea E. K. (1996) Science 271,209–212 [DOI] [PubMed] [Google Scholar]
- 25.Ruone S., Rhoades A. R., Formosa T. (2003) J. Biol. Chem. 278,45288–45295 [DOI] [PubMed] [Google Scholar]
- 26.Rhoades A. R., Ruone S., Formosa T. (2004) Mol. Cell. Biol. 24,3907–3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nourani A., Utley R. T., Allard S., Côté J. (2004) EMBO J. 23,2597–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferdous A., Kodadek T., Johnston S. A. (2002) Biochemistry 41,12798–12805 [DOI] [PubMed] [Google Scholar]
- 29.Ferdous A., Gonzalez F., Sun L., Kodadek T., Johnston S. A. (2001) Mol. Cell. 7,981–991 [DOI] [PubMed] [Google Scholar]
- 30.Gonzalez F., Delahodde A., Kodadek T., Johnston S. A. (2002) Science 296,548–550 [DOI] [PubMed] [Google Scholar]
- 31.Sun L., Johnston S. A., Kodadek T. (2002) Biochem. Biophys. Res. Commun. 296,991–999 [DOI] [PubMed] [Google Scholar]
- 32.Xu Q., Singer R. A., Johnston G. C. (1995) Mol. Cell. Biol. 15,6025–6035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burton B. M., Baker T. A. (2005) Protein. Sci. 14,1945–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeMartino G. N., Proske R. J., Moomaw C. R., Strong A. A., Song X., Hisamatsu H., Tanaka K., Slaughter C. A. (1996) J. Biol. Chem. 271,3112–3118 [DOI] [PubMed] [Google Scholar]
- 35.Liu C. W., Li X., Thompson D., Wooding K., Chang T. L., Tang Z., Yu H., Thomas P. J., DeMartino G. N. (2006) Mol. Cell. 24,39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morse R. H. (2003) Biochem. Cell. Biol. 81,101–112 [DOI] [PubMed] [Google Scholar]
- 37.Zhu Z., Thiele D. J. (1996) Cell 87,459–470 [DOI] [PubMed] [Google Scholar]
- 38.Wechsler D. S., Papoulas O., Dang C. V., Kingston R. E. (1994) Mol. Cell. Biol. 14,4097–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imbalzano A. N. (1998) Methods 15,303–314 [DOI] [PubMed] [Google Scholar]
- 40.White C. L., Luger K. (2004) J. Mol. Biol. 342,1391–1402 [DOI] [PubMed] [Google Scholar]
- 41.Perlmann T., Wrange O. (1988) EMBO J. 7,3073–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Q., Wrange O. (1995) Mol. Cell. Biol. 15,4375–4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Belotserkovskaya R., Oh S., Bondarenko V. A., Orphanides G., Studitsky V. M., Reinberg D. (2003) Science 301,1090–1093 [DOI] [PubMed] [Google Scholar]
- 44.Biswas D., Dutta-Biswas R., Mitra D., Shibata Y., Strahl B. D., Formosa T., Stillman D. J. (2006) EMBO J. 25,4479–4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paull T. T., Carey M., Johnson R. C. (1996) Genes Dev. 10,2769–2781 [DOI] [PubMed] [Google Scholar]
- 46.Lee D., Ezhkova E., Li B., Pattenden S. G., Tansey W. P., Workman J. L. (2005) Cell 123,423–436 [DOI] [PubMed] [Google Scholar]
- 47.Costigan C., Kolodrubetz D., Snyder M. (1994) Mol. Cell. Biol. 14,2391–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robzyk K., Recht J., Osley M. A. (2000) Science 287,501–504 [DOI] [PubMed] [Google Scholar]
- 49.Glowczewski L., Yang P., Kalashnikova T., Santisteban M. S., Smith M. M. (2000) Mol. Cell. Biol. 20,5700–5711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Russell S. J., Johnston S. A. (2001) J. Biol. Chem. 276,9825–9831 [DOI] [PubMed] [Google Scholar]