Abstract
Gaucher disease (GD), the most prevalent lysosomal storage disease, is caused by a deficiency of glucocerebrosidase (GCase). The identification of small molecules acting as agents for enzyme enhancement therapy is an attractive approach for treating different forms of GD. A thermal denaturation assay utilizing wild type GCase was developed to screen a library of 1,040 Food and Drug Administration-approved drugs. Ambroxol (ABX), a drug used to treat airway mucus hypersecretion and hyaline membrane disease in newborns, was identified and found to be a pH-dependent, mixed-type inhibitor of GCase. Its inhibitory activity was maximal at neutral pH, found in the endoplasmic reticulum, and undetectable at the acidic pH of lysosomes. The pH dependence of ABX to bind and stabilize the enzyme was confirmed by monitoring the rate of hydrogen/deuterium exchange at increasing guanidine hydrochloride concentrations. ABX treatment significantly increased N370S and F213I mutant GCase activity and protein levels in GD fibroblasts. These increases were primarily confined to the lysosome-enriched fraction of treated cells, a finding confirmed by confocal immunofluorescence microscopy. Additionally, enhancement of GCase activity and a reduction in glucosylceramide storage was verified in ABX-treated GD lymphoblasts (N370S/N370S). Hydrogen/deuterium exchange mass spectrometry revealed that upon binding of ABX, amino acid segments 243–249, 310–312, and 386–400 near the active site of GCase are stabilized. Consistent with its mixed-type inhibition of GCase, modeling studies indicated that ABX interacts with both active and non-active site residues. Thus, ABX has the biochemical characteristics of a safe and effective enzyme enhancement therapy agent for the treatment of patients with the most common GD genotypes.
Gaucher disease (GD),3 caused by deficiency of a lysosomal enzyme glucocerebrosidase (GCase), an acidic β-glucosidase (EC 3.2.1.45), encompasses a continuum of clinical spectrum from a perinatal lethal disorder to an asymptomatic form. Three major clinical subtypes (1–3) have been broadly defined, primarily based on the degree of neurological involvement. These are useful in determining prognosis and management (1, 2). Type 1 (GD-1), known as the non-neuronopathic form, is characterized by the presence of bone disease, hepatosplenomegaly, anemia, and thrombocytopenia. Type 2 and 3 (GD-2 and GD-3), also known as neuronopathic forms, are characterized by the presence of primary neurological disease. In the past, GD-2 and -3 were distinguished by age of onset and rate of disease progression, but these distinctions are not absolute (3). Generally, patients with GD-2 have an age of onset from days to months with a rapidly progressive neurological course. Individuals with GD-3 may have onset before reaching 2 years of age, often have a more slowly progressive neurological course, and may live into the 3rd or 4th decade. Interestingly, carriers of GD are also considered to have an increased risk of developing Parkinson disease (4–6).
Skin fibroblasts from GD-1 patients contain 10–25% of residual GCase activity. The level of 25–30% is believed to represent the “critical threshold” of GCase activity, under which symptoms associated with GD are observed (7). Over 250 mutations have been reported in the GBA gene (encoding GCase) to cause GD. Of these, 203 are missense mutations (8). The most prevalent mutations found are N370S in GD-1 patients and L444P in GD-3 patients (9). However, L444P can also be found in GD-2 patients (10). Most of these missense mutations are believed to affect the proper folding of GCase in the endoplasmic reticulum (ER), re-directing the misfolded protein to the ER-associated degradation pathway. The mutations that have been demonstrated to affect the early folding of GCase are G202R, N370S, and F213I (11–14). In GD patient fibroblasts, particularly those with an N370S or F213I allele, growth in media containing a sub-inhibitory concentration of N-nonyl-deoxynojirimycin, N-octyl-β-valienamine, or the iminosugar isofagomine (IFG), specific competitive inhibitors of GCase, has been demonstrated to increase levels of lysosomal GCase activity 2–4-fold (7, 11, 13, 15, 16). These compounds function as pharmacological chaperones (PCs). PCs act by stabilizing the native conformation of the mutant GCase in the ER, allowing more functional molecules to form and evade the ER-associated degradation pathway by instead being passed onto the ER transport machinery (17), ultimately resulting in increased amounts in the lysosome. It is believed that once the complex enters the lysosomes of cells containing stored substrate, the inhibitors will be displaced allowing GCase to hydrolyze the substrate (18). However, the ideal PC for any lysosomal storage disease (LSD) should bind maximally at the neutral pH of the ER and minimally at the acidic pH of lysosomes.
The current enzyme replacement therapy (ERT) for GD is limited to the treatment of non-neurological symptoms, because of the inability of the enzyme to cross the blood-brain barrier (19). This therapeutic approach is also very expensive (∼$200,000/year/patient) and must be administered intravenously. Small molecules are more likely to cross the blood-brain barrier, are less expensive to manufacture, and can be taken orally. Most of the small molecules, currently identified as PCs for GCase, are chemical compounds that have never been tested in humans and therefore are not approved by the drug regulation authorities. The screen of chemical libraries of Food and Drug Administration (FDA)-approved compounds has already proven to be a practical and successful strategy to identify small molecules that can behave as EET agents for specific lysosomal enzymes deficient in LSDs (20). To date, all reported approaches to library screening have utilized an assay aimed at identifying novel inhibitors of the target enzyme (20–23). In this context, we describe here the identification of ambroxol (ABX) as an EET agent for GCase using a more general thermal denaturation assay to screen the NINDS, National Institutes of Health, library of 1,040 drugs that have been previously used to treat humans. We then characterize the biochemical effects of ABX treatment on three GD patient cell lines, L444P/L444P, N370S/N370S, and F213I/L444P. Only the latter two cell lines responded with enhanced lysosomal GCase activity and protein levels. Additionally, substrate storage was reduced as demonstrated by liquid chromatography-electrospray ionization-tandem mass spectrometry after 15 days of treatment of an N370S/N370S lymphoblast line with ABX.
EXPERIMENTAL PROCEDURES
Chemical Reagents and Antibodies
The United States drug collection of 1040 compounds (NINDS Library at National Institutes of Health) was purchased from Microsource Discovery Systems, Inc., and stored at −20 °C in a 96-well format. The two synthetic fluorogenic substrates, 4-methylumbelliferyl-β-d-glucopyranoside (MUbGlc) and 4-methylumbelliferyl-β-N-acetylglucosaminide (MUG), purchased from Sigma, were used to assay the lysosomal enzymes GCase and total β-hexosaminidase (Hex), respectively. Cerezyme, modified human recombinant GCase (rGCase), was purchased from Genzyme Corp. Taurodeoxycholic acid sodium salt (TDC) was from Calbiochem EMD Biosciences; conduritol B epoxide (CBE) and IFG chloride were from Toronto Research Chemicals Inc. (Canada). Ambroxol hydrochloride (ABX), ambroxol derivatives, fluphenazine dihydrochloride (FZN), and Triton X-100 were purchased from Sigma. The bicinchoninic acid (BCA)-containing kit, purchased from Pierce, was used for protein determination. Primary antibodies used were rabbit polyclonal IgG anti-human GCase (raised by ourselves against rGCase), mouse monoclonal IgG1 anti-human Lamp-1 (lysosomal associated membrane protein-1) and Lamp-2 from the Developmental Studies Hybridoma Bank, and mouse monoclonal IgG1 anti-rat protein-disulfide isomerase (PDI) from StressGen Bioreagents. Secondary antibodies donkey anti-rabbit and donkey anti-mouse were purchased from the Jackson ImmunoResearch; Alexa Fluor 488 chicken anti-rabbit IgG and Alexa Fluor 594 chicken anti-mouse IgG were from Molecular Probes, Inc. MACS® columns (Miltenyi Biotec GmbH, Germany) were used to isolate lysosomes by magnetic chromatography as described previously (22, 24, 25). Other chemicals used were analytical grade reagents from general laboratory suppliers.
Cell Lines and Tissue Culture Conditions
Primary skin fibroblasts derived from a patient with GD-1 homozygous for the N370S mutation (c.1226A→/c.1226A→G) was provided by the Hospital for Sick Children Tissue Culture Facility. The GD-2/3 fibroblast line homozygous for the L444P/L444P mutation (c.1448T→/c.1448T→C) was purchased from Coriell Institute Biorepository as GM07968. The GD-2/3 fibroblast line with the genotype F213I/L444P (c.754T→A/c.1448T→C) was a gift from F. Choy (University of Victoria, British Columbia, Canada). Epstein-Barr virus immortalized human lymphoblast cell lines homozygous for the N370S GD-1 mutation or a wild type control, used for the measurement of GC storage, were both provided by the Hospital for Sick Children Tissue Culture Facility.
Fibroblast cells were grown in complete α-minimal essential medium from Wisent Inc. (Canada) in the presence of 5% antibiotics (penicillin and streptomycin, Invitrogen), supplemented with 10% fetal bovine serum (FBS, Wisent Inc,. Canada), and incubated at 37 °C in a humidified atmosphere with 5% CO2. Lymphoblast cell lines were grown as suspensions in RPMI 1640 medium (Wisent Inc., Canada) supplemented with antibiotics and 20% FBS.
Primary Screening Conditions
The NINDS library comprised of 1,040 FDA-approved compounds, that were dissolved in DMSO at 10 mm, was screened manually for compounds that stabilized rGCase activity against thermal denaturation. Each of the library compounds was diluted to 200 μm by addition of McIlvaine citrate-phosphate buffer (CP; 20 mm citrate, pH 5.5) containing rGCase (0.3 μm) in a set of master plates and kept on ice. From each master plate, two identical aliquots (25 μl) were prepared. One was kept on ice, and the second aliquot was incubated at 51 °C for exactly 12 min and then returned to ice for 5 min. Following equilibration of both sets of plates to room temperature (5 min), the GCase enzyme assay was carried out at room temperature for 20 min after the addition of 25 μl of 1.6 mm MUbGlc (in CP 20 mm, plus TDC 0.4% (w/v), pH 5.5). The reaction was stopped by addition of 200 μl of 2-amino-2-methyl-1-propanol (MAP; 0.1 m, pH 10.5). The fluorescence of the 4-methylumbelliferone (MU) reporter group liberated following hydrolysis of the MU-based substrate was measured with an M2 Microplate spectrofluorimeter (Molecular Devices Inc.) with excitation and emission wavelengths set at 365 and 450 nm, respectively. Residual GCase activity, remaining after thermal denaturation in the presence of each of the compounds, was calculated by dividing the fluorescence units measured in each well incubated at 51 °C by the fluorescence units of the corresponding well kept at 0 °C. Compounds were classified as “hits” if they raised the residual GCase enzyme activity 3 S.D. above the residual enzyme activity measured in the presence of DMSO only. A previous screen of the same NINDS library (final concentration 20 μm) for GCase inhibitors using previously described conditions (20) identified FZN as a potent inhibitor. Although FZN was capable of stabilizing GCase at 51 °C, it did not significantly enhance GCase enzyme activity in vivo using GD-1 patient fibroblasts described above (data not shown). FZN was thus used as a convenient positive control at the same concentration as the other library compounds (200 μm) in the primary screen.
Secondary Screening Conditions
For secondary screening, the rate of thermal denaturation of rGCase, incubated at 51 °C over a 60-min period in the presence of increasing amounts of the test drug, was determined. Assays were done in duplicate, and all the steps were carried out on ice until the final enzyme assays. The rGCase working mixtures (0.3 μm in CP, pH 5.5, plus human serum albumin (0.5% w/v) and TDC (0.2% w/v)) were prepared containing increasing concentrations of the test compounds and distributed in duplicate. A thermal denaturation assay was carried out for each drug (0–1.2 mm, final) as described above (primary screening conditions). Similarly, the remaining GCase enzyme activity was calculated as described above.
GCase Enzyme Kinetics in the Presence of ABX or One of Its Derivatives
The inhibitory activity of ABX or any of its derivatives was determined at different pH values using compound stocks dissolved and diluted in DMSO. Dose-response curves were generated using increasing concentrations of inhibitor in the presence of GCase in CP (100 mm), and were carried out at four different pH values (4.3, 4.7, 5.6, or 6.7). To optimize the levels of hydrolysis to within the most accurate range of the plate reader (10,000–100,000 fluorescence units), different amounts of GCase were used at each of the pH values; i.e. 87, 43, 9, or 13 nm, respectively. Reactions were initiated by the addition of MUbGlc (0.8 or 2.5 mm final concentration), incubated in a 37 °C water bath for 15 or 30 min, and terminated by the addition of MAP, and the fluorescence was measured (as above). Assays were performed in 96-well plates in triplicate. IC50 values were extracted from dose-response curves via nonlinear regression analysis of the log of the inhibitor concentration versus the measured residual enzyme activity using Prism 5.0 (Graph Pad Software, Inc.).
To determine the mode of inhibition (competitive, mixed, or noncompetitive), Michaelis-Menten curves were generated at acidic (pH 5.6) and neutral pH (pH 7). Stocks of rGCase (5.2 or 8.8 nm in 100 mm CP, pH 5.6 or 7.0), MubGlc substrate dilutions (0.625–20 mm in water), and ABX dilutions (∼0.5–4 times its Ki in DMSO) or DMSO alone were prepared and aliquoted in 96-well plates. Enzyme reactions (in duplicate) were initiated by addition of the diluted substrate, incubated for 30 min in a 37 °C water bath, and terminated by the addition of MAP, and fluorescence was determined (as above). Using Prism 5.0, Km and Vmax values were extracted from the Michaelis-Menten curves (hydrolysis rate in nmol/h/mg rGCase versus substrate concentration) following nonlinear regression. Similarly, Ki and K′i values were obtained by nonlinear regression analysis of these data fitted to either a competitive, mixed, or noncompetitive model of inhibition. The data were found to fit the mixed model of inhibition best, following comparisons of the fit of the data using the F-statistic analysis program within Prism 5.0.
In Cellulo Enzyme Enhancement Activity Assays
To evaluate the enzyme-enhancing effect of ABX or IFG, GD-1–3 fibroblasts were grown in complete media (α-minimal essential medium with 10% FBS and antibiotics) containing ABX (10–60 μm), IFG (10–60 μm), ABX derivatives, or DMSO alone (0.5% v/v) for 5 days. Subsequently, media were removed, and cells were washed twice with phosphate-buffered saline (Ca2+- and Mg2+-free) and harvested by scraping. Following centrifugation, cells were lysed by the addition of NaH2PO4 (10 mm, pH 6.0) plus TDC (0.2% w/v) and Triton X-100 (0.1% v/v) and left on ice for 30 min. After a second centrifugation, lysate was diluted in CP (20 mm, pH 5.5) plus TDC (0.4% w/v), Triton X-100 (0.2% v/v), and human serum albumin (0.5% w/v). GCase activity assays (performed in triplicate) were initiated by the addition of MUbGlc (20 mm) after the dilution, incubated at 37 °C for 1 h, and stopped by addition of MAP, and the fluorescence was measured (as above) (26). The contribution of nonlysosomal GCase activity was determined by re-assaying in the presence of irreversible inhibitor of acid GCase, CBE (27). Toxicity of the test drug was monitored by measuring total Hex activity in cell lysates using the substrate MUG (1.6 mm) (22). Enzyme activities are expressed as nanomoles of 4-methylumbelliferone generated per h and milligrams of total protein in the cell lysate. Increases in GCase relative activity were expressed as ratios of enzyme activity measured in fibroblasts treated with ABX or IFG versus the activity of mock-treated (DMSO/buffer) cells. GD-1 lymphoblasts were similarly treated with either ABX or IFG, but only at a single concentration (30 μm).
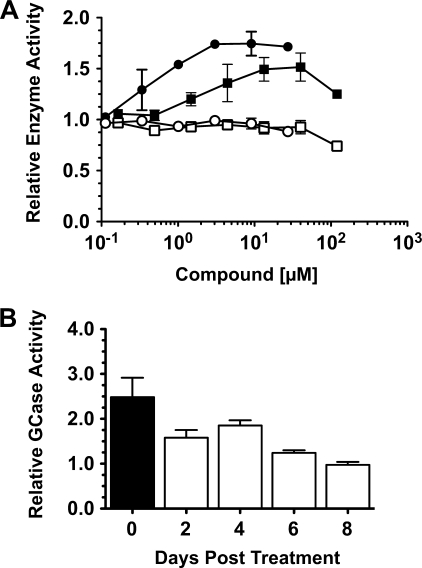
To examine the “chaperoning” half-life of ABX, GD-1 fibroblasts were grown in complete α-minimal essential medium containing 60 μm ABX or DMSO (mock-treated) for 5 days. Subsequently, the cells were “chased” for 8 days by replacing the ABX/DMSO-containing media with fresh media lacking the compounds and thereafter every other day. The GCase activity was measured in cell lysates at 0, 2, 4, 6, and 8 days after growth in drug-containing media and expressed relative to similarly processed mock-treated cells.
Western Blot Analysis
Aliquots of lysates (20 μg of total protein) or the enriched lysosomal fractions (5 μg of total protein) from treated and untreated fibroblasts (as indicated in respective figure legends) were subjected to Western blot analysis as reported previously (22).
Purification of Iron-Dextran-labeled Lysosomes by Magnetic Chromatography
An enriched lysosomal fraction was prepared from GD-1 fibroblasts treated either with ABX (60 μm), IFG (30 μm), or DMSO alone for 5 days, as reported previously (22, 25).
Indirect Immunofluorescence Staining and Confocal Microscopy Imaging
Indirect immunofluorescence and confocal microscopy imaging were performed as described previously (28). Primary and secondary antibodies were as described above in chemical reagents, whereas nuclear staining was as reported previously (22).
Glucosylceramide Analysis
A modification of a combined high performance liquid chromatography (HPLC) and electrospray ionization-tandem mass spectrometry method, as described previously (29, 30), was used to measure the total lipid content, including six individual forms (based on variations in the carbon chain length of their fatty acid moiety) of GC in wild type (untreated, 12 days of growth) or GD-1 lymphoblasts (N370S/N370S) (10 or 15 days of growth) in the absence or presence of ABX (20 μm) or IFG (20 μm). Each lipid species in the mock-, ABX-, and IFG-treated groups was measured three times independently from two different extracts of treated cells, i.e. either 10 or 15 days in culture. Media (RPMI with 20% FBS ± ABX or IFG) were replaced every 5 days. At the end of the treatment period, cells were chased for 24 h in fresh media alone. Subsequently, the cells were pelleted by centrifugation (1000 × g) and freeze-dried. Extraction of GC from the dried lymphoblast cell pellets (∼200 μg of total cell protein) was performed according to the method of Folch et al. (31), incorporating 400 pmol of N-palmitoyl-d3-glucosylceramide (GC 16:0(d3)) (Matreya Inc.) as an internal standard.
Lipid species in the dried extracts (resuspended in 100 μl of methanol containing 5 mm ammonium formate, pH 4.0) were resolved by HPLC (Agilent) using a C18 (3 μm, 50 mm × 2.1 mm) column (Alltech) maintained at ambient temperature (21 °C). The HPLC mobile phases consisted of mobile phase A, aqueous 5 mm ammonium formate, pH 4.0, methanol, tetrahydrofuran (5:2:3, v/v), and mobile phase B, aqueous 5 mm ammonium formate, pH 4.0, methanol, tetrahydrofuran (1:2:7, v/v). A gradient program at a flow rate of 200 μl/min was used with the initial composition consisting of 70% phase A and then linearly changed to 100% phase B over 7 min and maintained there for 3 min. Re-equilibration at 70% phase A was established (3 min) prior to the next injection. Following chromatography, individual species of GC in the collected fractions were quantified using multiple reaction monitoring on a SCIEX API 3000 triple-quadrupole mass spectrometer as described previously (29, 30).
Statistical Method Used
All GC measurements (three independent measurements for each of six species of GC from two independent cell extracts for the three groups of mutant cells, and one cell extract from the wild type control) were included in the analysis. A linear regression model using maximum likelihood algorithm for parameter estimation was used to determine the significance of the differences between all four groups (normal control, ABX, IFG, and mock) using ABX as the reference category (32). All regression models were adjusted for timing of samples (10 or 15 days in culture) and for triplicate measurements of each form of GC. An exponential transformation was applied to all GC values. Parameter estimates were back-transformed by solving regression equations to obtain an effect estimate and 95% confidence interval. Finally, a separate model was created using the mock group as a reference to determine whether there were any differences between it and the IFG group.
Hydrogen/Deuterium Exchange (H/D-Ex) Mass Spectrometry Studies
The H/D-Ex method was performed as described in detail previously (22, 33). In this study, ligand stocks of ABX, FZN, and IFG at 47 mm in DMSO were used at 213-fold molar excess relative to GCase (22.2 μm). The data acquisition and exchange corrections and calculations were performed as reported earlier (22, 34).
SUPREX Analysis
SUPREX was performed using rGCase (26.6 μm) at pH 7.0 or pH 5.5 in the presence or absence of ligands, ABX, and IFG. The molar ratio of GCase and ligands (dissolved in DMSO, 4% v/v final) in the stock solutions was 1:72 and 1:386 for IFG and ABX, respectively. Stock solutions (with or without a ligand) were diluted 10-fold into a series of deuterated exchange buffers containing either HEPES (50 mm, pH 7.0) or sodium acetate (50 mm, pH 5.5), plus varying concentrations of guanidine hydrochloride (GdnHCl). For each sample, four specific exchange time points (5–120 min) were selected. Aliquots of exchanged samples (5 μl) were quenched with 7 or 7.5 μl of a chilled urea (2 m) and formic acid (0.8%, v/v). Samples were digested (1 min, 0 °C) by adding ice-chilled immobilized pepsin (4 mg/ml) slurry (5 μl), and centrifuged (10 s). The aliquoted supernatant was stored on dry ice. Prior to matrix-assisted laser desorption/ionization (MALDI)-mass spectra acquisition, room temperature-thawed aliquots (1.5 μl) of digests were mixed with 9 μl of α-cyano-hydroxycinnamic acid solution (2.5 mg/ml), containing 50% acetonitrile and 0.1% trifluoroacetic acid. Samples (0.7 μl) were spotted on a MALDI plate and dried in vacuo. Spectra were acquired on a MALDI-TOF-TOF ABI 4700 analyzer mass spectrometer (Applied Biosystems).
Initially, up to 10 GCase peptides were used to follow mass increases of deuterium incorporation at each of the four time intervals and at different GdnHCl concentrations. Each of the 10 peptides yielded SUPREX curves (mass increase plotted as a function of GdnHCl concentration at each of the four exchange times) with similar transition midpoint values. Thus, in subsequent experiments only two representative GCase peptides (positions 67–91 and 106–127) were selected to generate the SUPREX curves to derive the m values (35, 36). The averaged m values for the two peptides were then used to calculate the folding free energy, and the dissociation constant (Kd value) of the protein-ligand complex was determined at pH 5.5 and 7, as described previously (33, 34).
Docking Studies
Ligands FZN and ABX were docked into the GCAse-IFG complex (Protein Data Bank code 2NSX) using the flexible docking protocol (37) within Discovery Studio 2.1 (Accelrys). Prior to docking, water and IFG were removed from chain B of 2NSX, and residues within 4 Å of the bound IFG were selected as flexible. Binding of IFG, FZN, and ABX was studied using two docking experiments, taking IFG as a control ligand. All ligand conformations were generated using CatConf component of Discovery Studio (Accelrys). CHARMm force field was used for both protein and ligands. For ABX or FZN, two poses were selected for further analysis that were consistent with H/D-Ex data and in the list of top 10 poses based on CDOCKER energy. For studying π-π interactions AutoDockTools was used (38).
RESULTS
Screening of NINDS Library and Identification of ABX
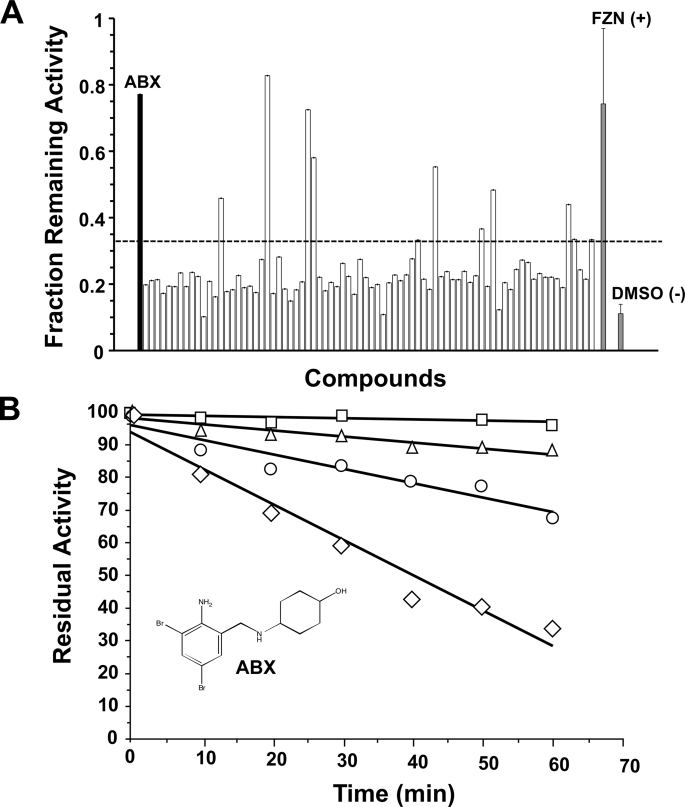
A fraction of the results from a screening of the 1,040 small molecules from the NINDS library, based on the ability of each compound to attenuate thermal denaturation of GCase, is shown in Fig. 1A. In this screening assay GCase residual activity was standardized by the GCase activity obtained from the corresponding well of the twin plate that was kept on ice during the thermal denaturation stress period. FZN was previously identified as the primary “hit” in a classical inhibitory-based screening of the library, with an IC50 of 15 μm using 0.8 mm of substrate (MUbGlc), pH 5.5. FZN was also found to be a potent stabilizer of GCase against thermal denaturation in the secondary screen and to have the characteristics of a mixed-type GCase inhibitor (data not shown). However, in the final GD cell-based assay, FZN failed to enhance mutant GCase activity in cellulo (data not shown). Despite its failure as a PC, FZN was useful as a positive control when we re-screened the library using the more general approach of attenuation of thermal denaturation as the primary assay. In this screen ABX was identified by its ability to confer a protective effect on GCase activity comparable with that of FZN (Fig. 1A). A secondary screen confirmed that ABX stabilizes GCase against thermal denaturation in a dose-dependent manner (Fig. 1B). The other potential hits identified in the primary screen were found to be either fluorogenic compounds or, like FZN, they failed to behave as EET agents for GCase in the final GD cell-based assay (data not shown).
FIGURE 1.
Identification of ABX as a potential PC for GD using an attenuation of GCase thermal denaturation assay. A, primary screening data are shown for 80 of the 1,040 chemical compounds (one 96-well plate), including ABX (black bar). Each plate contained 80 compounds from the library tested at 200 μm (open bars). One column (8 wells) contained only solvent (DMSO (−), negative control, gray bar), and another column FZN (FZN (+), positive control, gray bar). The y axis indicates the relative enzymatic activity after thermal denaturation compared with the corresponding well of an identical plate that was kept on ice. Those wells that retained a residual GCase activity of >3 S.D. (bars above the dotted line) of the mean of values from wells containing only DMSO were considered candidate PCs for GCase and were further investigated. B, secondary screening of ABX, with its molecular structure depicted, confirming that it stabilizes GCase toward heat inactivation in a dose-dependent manner (0 mm ABX, diamonds; 0.15 mm ABX, circles; 0.3 mm ABX, triangles; 1.2 mm ABX, squares). Data are expressed as the mean of duplicate assays.
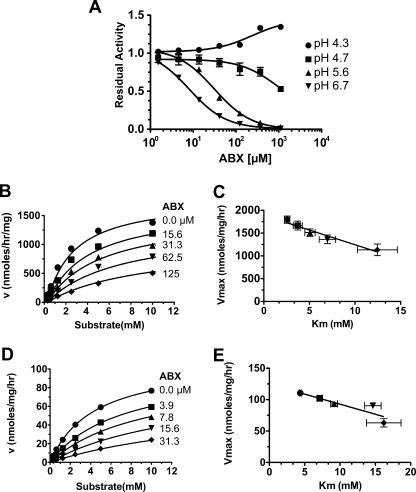
Further characterization of ABX (C13H18Br2N2O), trans-4-(2-amino-3,5-dibromobenzylamino)-cyclohexanol hydrochloride, found that it also acted as an inhibitor of rGCase (Fig. 2), but with an IC50 value of about twice that of FZN at pH 5.6. Interestingly ABX demonstrated an unusual degree of pH dependence in regards to its IC50 values (Fig. 2A), which represents a highly desirable property for a potential EET agent. At pH 4.3, ABX was noninhibitory, and even became an apparent activator of GCase as it neared millimolar concentrations. At pH 4.7 ABX had IC50 values of 810 or 950 μm against 0.8 or 2.5 mm substrate (MUbGlc), respectively. At pH 5.6, the IC50 values dropped to 31 ± 3 μm at either substrate concentration, and at pH 6.7 they dropped further to 3.7 ± 0.5 (0.8 mm substrate) and 8.1 ± 0.7 μm (2.5 mm substrate). Detailed kinetic analyses at pH 5.6 and 7.0 (Fig. 2, B and D) determined that ABX functions as a mixed inhibitor of GCase (Fig. 2, C and E) with an α value of ∼10 (Table 1). There was also an ∼4-fold increase in its Ki value between pH 7.0 and pH 5.6 (Table 1).
FIGURE 2.
Characterization of ABX as a pH-dependent, mixed inhibitor of GCase. A, inhibitory curves of increasing concentrations of ABX versus residual GCase activity using 0.8 mm MUbGlc at the indicated pH values. B and D, nonlinear regression analysis of GCase velocities at pH 5.6 (B) or pH 7.0 (D) versus increasing MUbGlc concentrations in the presence of the indicated concentrations of ABX (μm). This multivariate dataset (v, [I], [S]) was then fitted to the equation describing mixed type inhibition using GraphPad Prism version 5.0b (Table 1). C and E, plot of apparent Vmax versus apparent Km values at pH 5.6 (C) or pH 7.0 (E) demonstrates the validity of the mixed inhibitor model. The resulting kinetic constants are given in Table 1.
TABLE 1.
Summary of the kinetic parameters of the effects of ABX on GCase
| pH | 5.6 | 7.0 |
|---|---|---|
| Vmaxa | 1800 ± 51b | 110 ± 10 |
| Kmc (μm) | 2.6 ± 0.2 | 4.3 ± 0.1 |
| Kid (μm) | 23 ± 3 | 5.2 ± 0.2 |
| αe | 7 ± 3 | 10 ± 2 |
a Values are given in nanomoles of MU/h/mg (GCase).
b Values ± 1 S.D. from triplicate assays are shown.
c Value for MUbGlc in the absence of ABX is given.
d The affinity−1 of ABX for free enzyme is shown.
e K′i/Ki is shown, where K′i = the affinity−1 of ABX for the enzyme-substrate complex.
ABX as an Enzyme Enhancement Agent for GCase
To identify the range of optimum concentrations of ABX that produces enhanced residual GCase activity in treated GD cells, a dose-response curve was generated using a GD-1 (N370S/N370S) fibroblast line. IFG was used to compare the effects on residual GCase activity generated by ABX treatment of these cells (Fig. 3A) (7). The optimal range of ABX concentrations that produced significant enhancement of mutant GCase activity without noticeable toxic effects on the cells (as judged by a lack of change in the level of total Hex activity) was 5–60 μm. IFG had the best response at the previously reported concentration of ∼30 μm (7). Treatment of the cells with DMSO alone had no effect on mutant GCase activity. Similarly, when GD-1 (N370S/N370S) lymphoblasts were treated for 5 days with 30 μm of either ABX or IFG, their GCase activities were increased by ∼2- or ∼3-fold, respectively (data not shown).
FIGURE 3.
Determination of the levels of enhanced GCase activity and the length of its retention after treatment of N370S/N370S GD-1 fibroblast cell line with ABX. A, relative GCase (filled circles, IFG-treated; or squares, ABX-treated) and Hex (open circles, IFG-treated; or squares, ABX-treated) activities (y axis, 1 = no change from untreated cells) of cells treated with various concentrations of each drug (x axis). B, GD-1 cells were grown in media containing 60 μm ABX for 5 days (black rectangle) and then in normal media for the 2–8-day chase (open rectangles), and the relative GCase activity was determined. Bars represent S.E.
To evaluate the persistence of GCase enhancement produced by ABX after drug removal in N370S/N370S fibroblasts, several plates of cells were first treated (pulsed) for 5 days with ABX at 60 μm, and then the media were replaced with normal media and the enhanced GCase activity chased over a period of 8 days. This “pulse-chase” experiment demonstrated that the enhanced N370S GCase activity gradually decreases back to base-line levels within ∼6 days (Fig. 3B). This suggests a reasonably long half-life of ABX in cells, as wild type GCase in skin fibroblasts has been reported to have a half-life of ∼60 h (39, 40).
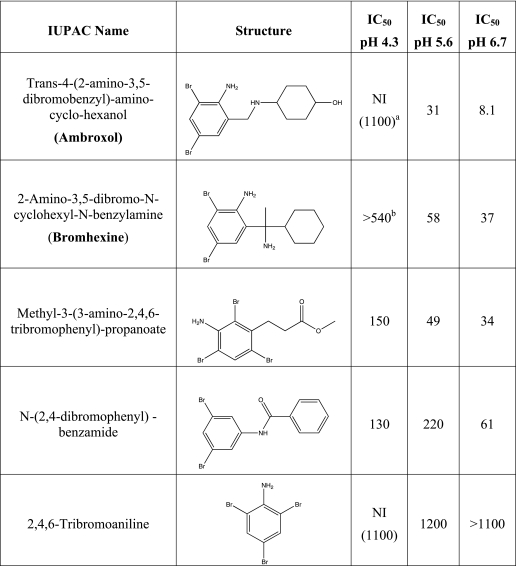
Analyses of ABX Derivatives as pH-dependent Inhibitors of GCase
The IC50 values of four compounds that have molecular structures similar to that of ABX were determined at concentrations ranging from 9 to 600 μm and at pH 4.3, 5.6, and 6.7, against 2.5 mm MUbGlc. None of these derivatives produced IC50 values lower than ABX, nor did any of them exhibit a pH dependence more appropriate for use as a PC than ABX (Table 2).
TABLE 2.
Names, structures, and IC50 values (μm) of ABX derivatives at various pH values toward GCase assayed with 2.5 mm MUbGlc
NI indicates not inhibitory.
a NI, not inhibitory (highest concentration evaluated, μm).
b Estimated IC50.
As ABX is a known metabolite of bromhexine, N370S/N370S GD-1 fibroblasts were grown in the presence of this derivative at concentrations ranging from 0.018 to 120 μm for 5 days. However, none of these treatments produced significant enhancement of mutant GCase activity (data not shown), possibly because the levels of the ABX produced following metabolism of bromhexine in cells may have been insufficient to enable enhancement of endogenous mutant GCase (41).
Responses of GD Cell Lines with Different GCase Mutations to Treatment with ABX
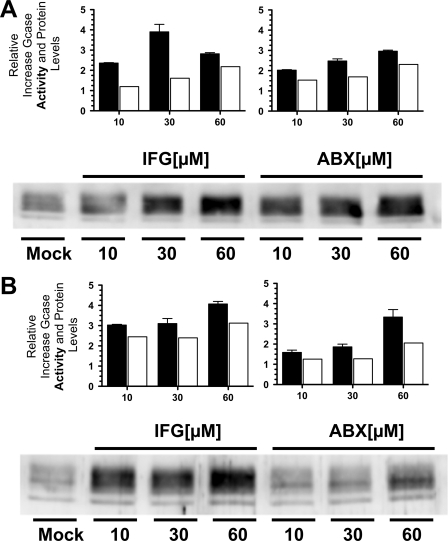
Three GD patient fibroblast cell lines containing different missense mutations were tested at 10, 30, and 60 μm ABX or IFG (Fig. 4). Consistent with the dose-response data (Fig. 3), the N370S/N370S GD-1 line, with an initial residual activity of ∼20% of normal (230 nmol of MUGlc/h/mg of total protein), showed significant increases in both residual GCase activity and protein levels (Fig. 4A). A GD-2/3 cell line, F213I/L444P (∼5% residual GCase activity), also showed significant enhancements of both relative GCase activity and protein levels (Fig. 4B). However another GD-2/3 fibroblast cell line homozygous for the L444P mutation (∼1% residual activity), treated with either ABX or IFG, produced no significant increases in relative GCase activity or protein levels (data not shown). The contribution of nonlysosomal GCase activity was measured in N370S/N370S fibroblasts indirectly by specifically inactivating acid GCase with CBE (42). After CBE treatment the remaining GCase activity in these cells was ∼1% of normal, as described earlier (27, 43) and was not affected by either ABX or IFG treatment (data not shown).
FIGURE 4.
Comparison of the levels of enhanced GCase activity and protein in GD-1 and GD-2/3 fibroblasts treated with either ABX or IFG. A, N370S/N370S GD-1 fibroblasts treated with IFG (left panels) or ABX (right panels). B, F213I/L444P GD-2/3 fibroblasts treated with IFG (left panels) or ABX (right panels). In both N370S and F213I mutant cell lines, ABX generated comparable levels of enhancements of both GCase enzyme activity (black rectangles) and protein levels (open rectangles), derived from densitometry scans of the Western blot below the graphs, to those obtained at equivalent concentrations of IFG. In the Western blot panels, Mock represents the untreated cell lines. Bars represent S.E.
GCase Activity and Protein Levels Are Increased in Lysosomal Enriched Fractions of GD Cells Treated with ABX or IFG
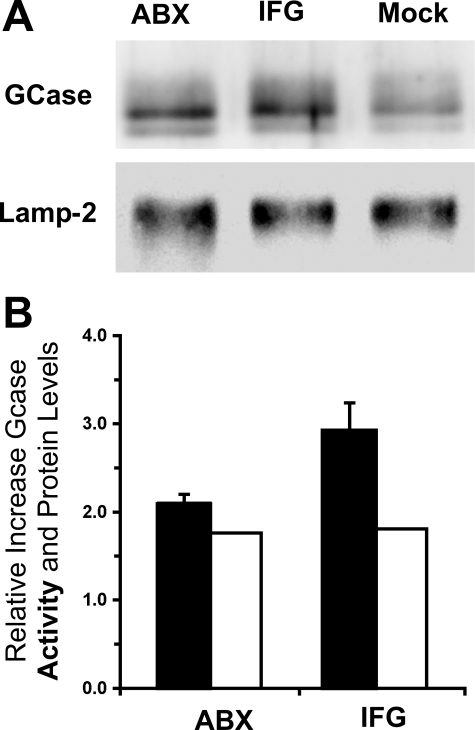
To confirm the intracellular location of the mutant GCase before and after ABX treatment, cells were grown in medium containing ferrous-dextran colloid, which is incorporated into lysosomes through bulk endocytosis. A magnetic separation was performed to produce an enriched lysosomal fraction from a post-nuclear supernatant as judged by an ∼10-fold increase in the specific activity of lysosomal Hex activity (24, 25). The ABX-treated (60 μm) or IFG-treated (30 μm) N370S/N370S cell line showed significantly higher levels of GCase protein and activity, as compared with untreated controls, in the enriched lysosomal fractions (Fig. 5).
FIGURE 5.
Enhancement of both GCase activity and protein levels in an enriched lysosomal fraction isolated from GD-1 fibroblasts (N370S/N370S) after treatment with either ABX or IFG. A, Western blot of GCase protein in the lysosomes from cells treated with either 60 μm ABX (left lane) or 30 μm IFG (middle lane) or untreated cells (Mock, right lane). A Western blot for lysosomal Lamp-2 is shown as a loading control. B, relative increases of GCase activity (black rectangles) and protein (densitometry scans of A, open rectangles) are represented in the histogram. Bars represent S.E.
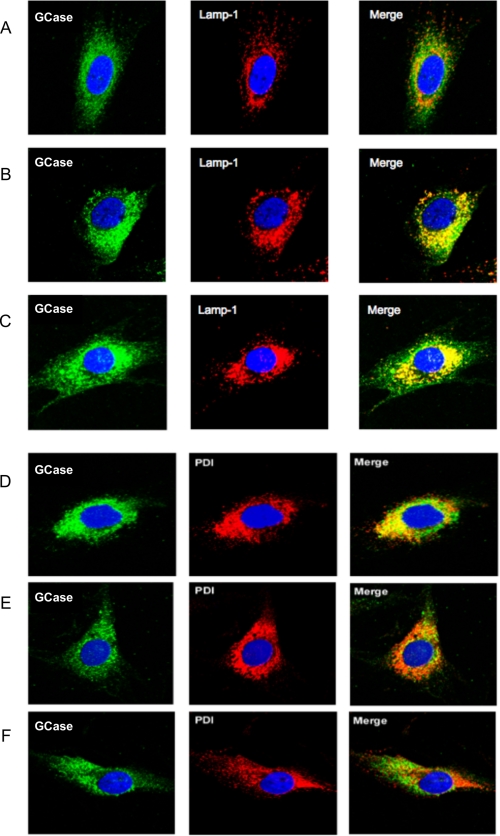
To further confirm that the intracellular location of the enhanced GCase activity was indeed lysosomal, we analyzed the cellular co-localization of GCase with Lamp-1 or PDI, both accepted markers of either the lysosomal or ER compartment, respectively. Indirect immunofluorescence staining and confocal microscopy imaging were performed on N370S/N370S fibroblasts treated for 5 days with either DMSO (1% v/v) (Fig. 6, A and D), ABX (40 μm) (Fig. 6, B and E), or IFG (30 μm) (Fig. 6, C and F). Localization of GCase signals (green) was first compared with those corresponding to Lamp-1 (red) (Fig, 6, A–C). During the recording of the immunofluorescence signals for GCase and Lamp-1 by confocal imaging, the microscope settings remained the same to give a semi-quantitative sense of protein levels through the intensity of the respective signals. The intensity of the GCase signals, as well as the levels of co-localization of GCase with Lamp-1 (yellow in Merge), was much more pronounced in treated cells (Fig. 6, B and C) than in the mock control cells (Fig. 6A).
FIGURE 6.
Changes in the intracellular localization of N370S/N370S GCase after treatment of GD-1 fibroblasts with either ABX or IFG. A and D, mock-treated cells were compared with cells treated with 40 μm ABX (B and E) or 30 μm IFG (C and F). Co-localization of signals (yellow in the Merge panels) for GCase (green in the GCase panels) and the lysosomal marker Lamp-1 (red in the Lamp-1 panels) was increased over mock-treated cells (A) in both ABX-treated (B) and IFG-treated cells (C). At the same time co-localization of signals from GCase and the ER-resident chaperone protein, PDI (red in the “PDI” panels), decreased after treatment with either ABX treatment (E) or IFG treatment (F), as compared with mock-treated cells (D). Cell nuclei were stained with 4′,6-diamidino-2-phenylindoledihydrochloride (blue).
The movement of mutant GCase from the ER to lysosomes after treatment with ABX or IFG was confirmed by comparing the intracellular localization of the resident ER protein PDI (red) with that of GCase (green), before (Fig. 6D) and after (Fig. 6, E and F) drug treatment. Untreated GD cells (N370S/N370S) are shown with the GCase signal artificially enhanced through confocal microscope settings (Fig. 6D) over treated cells (Fig. 6, E and F) to better evaluate and compare its cellular localization. Untreated cells produced a substantial level of yellow in the “Merged” image indicating co-localization of mutant GCase with PDI in the ER (Fig. 6D). In contrast, treatment with ABX (Fig. 6E) induced a remarkable reduction in the co-localization of GCase with PDI (little yellow visible in Merge), as did cells treated with IFG (Fig. 6F).
ABX Treatment of a GD-1 Lymphoblast Cell Line Results in a Reduction in Glucosylceramide Levels
A lymphoblast cell line homozygous for the N370S mutation was continually treated with ABX (20 μm) or IFG (20 μm) and compared with mock-treated and normal control cells, to evaluate the effects of the PCs on the levels of the major GCase substrate GC. A general lipid analysis was carried out on these four groups of cells (supplemental Figs. S1A and S2). This included quantifying six individual forms of GC, defined by the length of their fatty acid chain. The level of each form of GC was measured by liquid chromatography-electrospray ionization-tandem mass spectrometry (29) in triplicate for mutant cells grown for 10 (supplemental Fig. S1B and S2) or 15 days. The levels of individual GC species and summed totals in picomoles of lipid/mg of total protein are shown in Table 3 along with results of the statistical analysis comparing ABX with the mock-treated group. Additional statistical analyses comparing ABX with IFG and the wild type control, as well as IFG compared with mock, are included as supplemental Table 1S. GC measurements demonstrated that five of its six forms and the combined total GC level were significantly (p < 0.001) higher in ABX-treated than in normal control cells. All six forms were significantly lower in ABX-treated compared with IFG-treated cells. Most importantly, five of the six species and total GC levels were significantly lower in ABX-treated than mock-treated mutant cells. Interestingly, total and five GC forms were also significantly higher in IFG than mock-treated cells (data not shown). Therefore, for any form of GC (except the minor GC 24:1) control cells have the lowest values, followed by ABX-treated, then mock-treated, and finally IFG-treated with the highest value (supplemental Table 1S).
TABLE 3.
GC levels in normal (control) lymphoblasts and N370S/N370S lymphoblasts treated for 10 or 15 days with ABX, IFG, or DMSO (mock) expressed in picomoles of lipid/mg of total protein (average ± S.E., n = 3 for wild type and n = 6 for all others)
| GC species | Control (wild type) | IFG | ABX | Mock (DMSO) | ABX versus Mocka | p |
|---|---|---|---|---|---|---|
| GC 16b | 359 ± 17 | 1092 ± 77 | 685 ± 55 | 939 ± 116 | −404 (−362, −447) | <0.001 |
| GC 18 | 45 ± 2 | 159 ± 14 | 111 ± 9 | 137 ± 13 | −26 (−13, −41) | <0.001 |
| GC 20 | 59 ± 3 | 133 ± 8 | 90 ± 5 | 114 ± 10 | −24 (−12, −37) | <0.001 |
| GC 22 | 131 ± 5 | 242 ± 11 | 191 ± 8 | 221 ± 17 | −30 (−13, −48) | <0.001 |
| GC 24 | 93 ± 4 | 214 ± 10 | 174 ± 7 | 187 ± 15 | −14 (−30, +2) | 0.08 |
| GC 24/1c | 185 ± 6 | 201 ± 10 | 162 ± 12 | 190 ± 11 | −28 (−12, −45) | <0.001 |
| GC total | 873 ± 32 | 2131 ± 157 | 1482 ± 117 | 1876 ± 196 | −374 (−324, −508) | <0.001 |
a A linear regression model using maximum likelihood algorithm for parameter estimation was used to determine the significance of the differences between ABX and mock-treated cells. The first value represents the average difference between the ABX and mock groups, followed by the 95% confidence interval. If all three values are positive or all three values are negative, the difference is statistically significant. For example, on average the major GC 16 form is 404 pmol/mg protein lower in the ABX group than in mock, with a confidence interval of 362–447, which is statistically significant (highlighted in boldface type). Only the ABX versus mock difference for the GC 24 form is not significant. For a complete statistical comparison of ABX with the IFG and wild type groups see supplemental Table 1S.
b Data refer to the carbon chain length of the fatty acid moiety, which defines the individual forms of GC.
c GC contains a 24-carbon fatty acid chain with 1 double bond.
Identification of Segments of GCase That Are Stabilized through Binding of the Enzyme to ABX, FZN, or IFG by H/D-Ex MS/MS
The H/D-Ex MS/MS analysis resulted in 82% (408/497 residues) coverage of the protein sequence, as was reported previously (16). The four sequence gaps corresponded to known N-glycosylation sites (Asn-19, Asn-59, Asn-146, and Asn-270). A nonglycosylated peptide corresponding to the predicted glycosylation site Asn-462 was identified, indicating a population of GCase is not glycosylated at Asn-462.
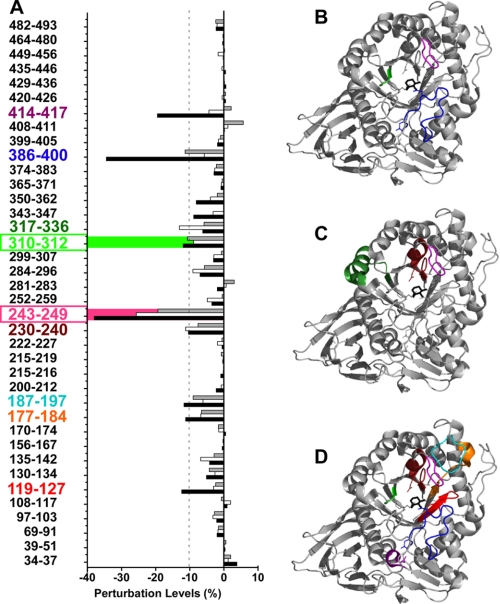
Reductions in the rate of deuteration of GCase upon incubation with ABX, FZN, or IFG (Fig. 7A) were determined and mapped onto the crystallographic structure of GCase complexed with IFG (Protein Data Bank code 2NSX) (44) (Fig. 7, B–D). ABX significantly (95% confidence level) decreased the degree of deuteration, as represented by the negative perturbations ≥10% in three sequence segments, 243–249 (−19%), 310–312 (−11%), and 386–400 (−11%) from domain III, which are located at the mouth of the 8-stranded β/α barrel (TIM barrel) and active site (Fig. 7B and supplemental Table 1S). Whereas these same segments were also stabilized (−38, −12, and −34%, respectively) by IFG binding (16), FZN significantly stabilized only segment 243–249 (−26%). However, FZN did affect segment 310–312 (β8) to almost the same extent as ABX and IFG, but its −9% change was slightly below the cut off for significance (Fig. 7A and supplemental Table 2S). In the presence of ABX, two other segments 187–197 (−9%) and 230–240 (−8%) were also stabilized, but they demonstrated weaker perturbations. The former segment was significantly stabilized by IFG (−12%), but not FZN, and the latter was stabilized by both IFG (−10%) and FZN (−11%). IFG also significantly stabilized segments 119–127 (−12%) and 414–417 (−19%). These segments were not stabilized by either ABX or FZN (Fig. 7A and supplemental Table 2S).
FIGURE 7.
Identification and mapping of the segments of GCase that are stabilized upon formation of the ABX, FZN, or IFG enzyme-ligand complex by H/D-Ex MS. A, slower rates of H/D-Ex represented by negative perturbations (x axis) indicate rigidification (stabilization) of the associated segments of the protein (y axis) (supplemental Table 1S). Bars (ABX, gray; FZN, open; and IFG, black) that exceed the dotted line are considered as indicative of a statistically significant change in the mobility of the associated segment of the protein. B–D, ribbon diagrams based on the crystal structure of IFG bound to GCase (Protein Data Bank code 2NSX) are color-coded to indicate areas of GCase identified in A that are stabilized by binding to ABX (B), FZN (C), or IFG (D); segments 119–127 (IFG; red); 177–184 (IFG; orange); 187–197 (IFG; cyan); 230–240 (IFG and FZN; burgundy); 243–249 (IFG, FZN, and ABX; pink); 310–312 (IFG, FZN, and ABX; green); 317–336 (FZN; dark green); 386–400 (IFG and ABX; blue); and 414–417 (IFG; purple).
SUPREX Analysis of ABX and IFG Binding to GCase at Acidic or Neutral pH
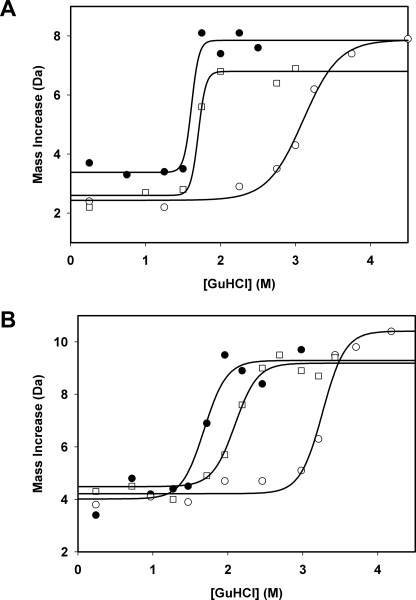
SUPREX protocol was used as an orthogonal method for measuring the change in the stability of GCase (Cerezyme) upon binding of ABX or IFG. With this procedure increasing concentrations of GdnHCl were used as the denaturant, and increasing average rates of deuteration, measured by H/D-Ex MS, were used to monitor unfolding (Fig. 8). Initial SUPREX experiments analyzed up to 10 GCase peptides that provided a SUPREX-like (sigmoidal) curve. All these curves produced similar transition midpoint (m values) at the same exchange time (data not shown). To facilitate throughput, data from two representative peptides over four exchange time periods were used to complete the SUPREX analyses (Fig. 8). The transition midpoint value of each SUPREX curve was then extracted. As shown in Fig. 8, IFG shifted the transition midpoint of peptide 67–91 up to 1.4 m GdnHCl at both pH 7.0 and 5.5. In contrast, ABX induced a marginal transition midpoint shift at pH 7.0 and only a slight shift at pH 5.5 for this peptide. After extraction of transition midpoint values, a linear extrapolation method was followed to determine the m values for these two peptides. From these values the binding energy and Kd constants of GCase for each ligand at an acidic or neutral pH were calculated (35, 36). IFG showed a significantly higher binding energy than ABX at both pH conditions (Table 4). Although the binding energy for ABX dramatically decreases at the more acidic pH, there is almost no change in the binding energy for IFG between pH 7.0 and 5.5 (Table 4). These data were also reflected in the calculated Kd values for ABX and IFG (Table 4).
FIGURE 8.
Determination of the binding energies and dissociation constants at acidic or neutral pH of the GCase-ABX or -IFG complex by SUPREX. Representative SUPREX curves (one of two GCase peptides, 67–91, examined at 1 of 4 H/D-Ex points, 30 min) of the change in mass, due to deuteration (y axis), in the SUPREX experiment in the presence of increasing concentrations of GdnHCl (x axis) are shown. The denaturation curve in the absence of ligand (filled circles), in the presence of ABX (open squares), or IFG (open circles) at pH 5.5 (A) or pH 7.0 (B) are shown. The solid lines represent the best fit of the data from each SUPREX curve to a four-parameter sigmoidal equation using SigmaPlot.
TABLE 4.
SUPREX-derived binding energies and dissociation constants for ABX and IFG to human rGCase
| Biophysical parameters | pH 5.5 |
pH 7.0 |
||
|---|---|---|---|---|
| ABX | IFG | ABX | IFG | |
| Binding energy (kcal/mol) | −0.2 ± 0.5a | −2.6 ± 0.4 | −1.0 ± 0.3 | −2.5 ± 0.3 |
| Kd (μm) | NDb | 1.9 | 182 | 2.2 |
a Data show ± 1 S.D.
b ND means not detected.
Characterization of the Binding Interactions of ABX and FZN with GCase by Molecular Modeling
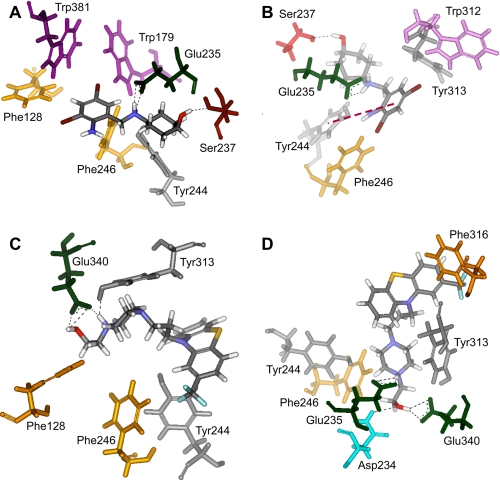
Two of the top 10 poses identified from each modeling study of GCase with ABX or FZN were selected based on their consistency with H/D-Ex data. This approach successfully identified the hydrogen bond interactions between IFG and residues Asp-127, Trp-179, Glu-235, Glu-340, Trp-381, and Asn-396 in the active site of GCase (data not shown), previously determined by co-crystallization of GCase with IFG (44).
Two hydrogen bonding interactions, i.e. with GCase residues Glu-235 and Ser-237, were predicted in each of the two top poses for ABX (Fig. 9, A and B). Only residue Glu-235 is located at the active site (44). In one of the poses a π-π interaction with residue Tyr-244 is also present, as well as hydrophobic interactions with residues Phe-246, Trp-312 and Tyr-313 (Fig. 9B). In the other pose the cyclohexane ring (Fig. 9A) can interact hydrophobically with Tyr-244 and Phe-246. In the FZN-binding poses there are either two (Fig. 9C) or three (Fig. 9D) hydrogen bonding interactions. In one pose hydrogen bonding occurs between FZN and GCase at residues Glu-340 and Tyr-313 (Fig. 9C). In the alternative pose hydrogen bonding interactions occur at residues Glu-235, Asp-234, and Glu-340 (Fig. 9D). Two of these residues (Glu-235 and Glu-340) are located at the active site. Residues Phe-246, Tyr-244, Phe-316, and even Tyr-313 may also generate hydrophobic interactions with FZN (Fig. 9, C and D). It is interesting to observe from the modeling studies that both ABX and FZN are surrounded by aromatic and hydrophobic residues. The nature of interactions in the case of ABX seems to be a combination of hydrogen bonding and π-π and hydrophobic interactions. FZN seems to be interacting mainly with the help of hydrogen bonding and hydrophobic interactions.
FIGURE 9.
Two most likely structures, based on their consistency with H/D-Ex and previous co-crystallization (GCase and IFG) data, of the GCase-ABX or -FZN complex, determined by the flexible docking methodology. A and B, two poses detailing the predicted interactions of ABX with GCase. Both poses predict hydrogen bonding with Glu-235 and Ser-237, indicated by dashed lines. A, there are hydrophobic interactions between the cyclohexane ring and Phe-246 and Tyr-244. B, orientation selected for clarity of the π-π interaction (red dashed line) predicted with the ABX molecule and Tyr-244 of GCase, as well as other hydrophobic interactions with residues Phe-246, Trp-312, and Tyr-313. C and D, two poses are shown of the interaction of FZN with GCase. C, FZN molecule has two hydrogen bonding interactions with GCase at residues Glu-340 and Tyr-313 (dashed lines). D, in another orientation, FZN is shown to interact with GCase via three hydrogen bonding interactions with Asp-234, Glu-235, and Glu-340 (dashed lines). C and D, residues Phe-246, Tyr-244, Phe-316, and even Tyr-313 may also generate hydrophobic interactions with FZN.
DISCUSSION
ERT has been successfully used to treat GD-1 patients (45, 46). However, as a surprisingly low critical threshold of lysosomal enzyme activity is needed in most LSDs to prevent storage of substrate (18), therapies based on small molecules that target the endogenous synthesis of the stored substrate, SRT, and/or the folding efficiency of the mutant enzyme, EET, are attractive and less expensive alternatives with the potential to also treat the neurological forms of LSDs. Both of these approaches have yielded encouraging results in some clinical and preclinical trials (47–49). EET has also been shown to produce a synergistic effect when combined with ERT in fibroblasts and a mouse model of Pompe disease (50) and to increase the half-life of exogenously added GCase in macrophages (51). Additionally by preventing the aggregation and/or storage of mutant misfolded enzyme in the ER, EET could lower the levels of the unfolded protein response, which can lead to ER stress. ER stress has been associated with the pathogenesis of some LSDs (52–54).
The screening of a medium size FDA-approved drug library has been shown to yield potential small molecules that function as EET agents for different LSDs, e.g. late-onset GM2 gangliosidosis (20), as well as potential pharmacological agents for neurodegenerative diseases (55–57). An initial screening assay based on the ability of each compound to inhibit GCase identified FZN as the major hit. Although FZN was demonstrated in a secondary screen to function as a strong stabilizer of GCase toward thermal denaturation, it failed in the final cell-based assay to enhance mutant GCase activity. The ability of FZN to stabilize GCase in vitro was confirmed via H/D-Ex and molecular modeling experiments. The in cellulo failure of FZN to act as a PC may have been the result of poor bio-availability or the rapid metabolism of the drug by cells. These may be the same reasons for the need to use similar concentrations of IFG to accomplish the same levels of enhancement of mutant GCase activity in cells as ABX, despite IFG being an ∼1000-fold stronger inhibitor of GCase in vitro (7). These data clearly demonstrate that the ability of a compound to act as a PC cannot be predicted solely on the basis of its in vitro binding and/or inhibitory activity, but it must be shown to be effective in enhancing mutant enzyme activity in patient cells.
We next decided to use thermal denaturation as the primary screening assay for the library to determine whether we could identify compounds that may interact allosterically with the enzyme, i.e. that bound to the active site and/or other regions of GCase. ABX was identified in this screen (Fig. 1A). Although weaker than FZN in terms of inhibitory activity, it was also found to be a mixed type inhibitor of GCase (Fig. 2 and Table 1) (58). Interestingly, the IC50 values of ABX were highly pH-dependent (Fig. 2A), with maximal inhibition occurring at the neutral pH. This is an ideal characteristic for an EET agent, which must work best at the pH of the ER. At pH 4.3, near that of lysosomes, ABX did not inhibit the enzyme and actually became an apparent activator of its activity at high, nonphysiological concentrations, i.e. greater than the maximal concentration evaluated in humans (Fig. 2A). This same type of pH dependence for a GCase inhibitor was recently demonstrated for diltiazem, an L-type Ca2+ channel blocker, which was also found to act as a PC in GD cells. However, as compared with ABX, diltiazem requires ∼10-fold higher concentrations to achieve the same levels of inhibition at neutral pH (59). The pH dependence of ABX binding to GCase was further confirmed in SUPREX analyses (Fig. 8). Because of its low binding energy, it was not possible to obtain a dissociation constant for ABX at pH 5.5, using SUPREX (Table 4). On the other hand, IFG, a classical carbohydrate-based competitive inhibitor, retained its ability to bind GCase over a wide pH range, which could result in residual inhibition of the enzyme once it reached the lysosomal compartment (Fig. 8 and Table 4). Another major advantage of ABX as an EET agent is that for many years ABX has been used to treat humans for airway mucus hyper-secretion diseases (chronic bronchitis, asthma, and cystic fibrosis). Furthermore, it has been used as an alternative preventative treatment for hyaline membrane disease in newborns, because it increases surfactant secretion in the lungs (60–62). It also has been shown to have anti-inflammatory and antioxidant properties (61, 63, 64), both of which may also be useful in tackling other aspects of the pathogenesis of LSDs (54). Paradoxically, the specific mechanism of action of ABX has yet to be elucidated (64). Given the extensive pharmacokinetic and safety studies of ABX in humans (65, 66), testing ABX in phase I/II clinical trials to evaluate it as an EET agent for GD might easily be expedited.
The ABX concentrations that produced the highest GCase enhancements in cell culture, 30 and 60 μm (12.4 and 24.8 μg/ml), are ∼15–150 times the human plasma concentrations (0.15–0.8 μg/ml) derived from previous clinical pharmacokinetics studies (67, 68). However, ABX produced enhancement of GCase activity in cells between 5 and 60 μm (Fig. 3A), which suggests a wide therapeutic window when testing the drug in phase I/II trials with GD patients. There is also very limited information on tissue distribution of ABX available in humans. However, a 25-fold accumulation in lungs compared with plasma was shown to occur in patients and the “volume of distribution,” i.e. total ABX in the body/ABX blood concentration, is high (560 liters) (69). Pharmacokinetics studies in rats have shown that ABX is widely distributed in lung, liver, kidney, and brain (62), i.e. after a 7.5 mg/kg intravenous infusion, ABX was found at ∼70 mg/g of lung tissue and ∼15 mg/g of brain tissue. Therefore, in humans the ABX concentration is most likely to be much higher in tissues than in plasma. This property may also be an advantage in the treatment of the neuronopathic forms of GD, where crossing blood-brain barrier is critical. However, the use of ABX as a central nervous system drug remains controversial (70).
During the short 10 or 15 days of growth in ABX-containing media, GC levels were significantly reduced in N370S/N370S GD-1 lymphoblasts but were not normalized (Table 3). It is not surprising that complete normalization did not occur given the short treatment time and the fact that the enhanced GCase activity only approached levels found in the GD carrier. Additionally, whereas a decrease in plasma GC levels is the most widely used biomarker for validating the efficacy of various therapies for GD, normalization of GC levels often was not achieved even after 2 years of ERT or SRT (71, 72).
The observation that 10 or 15 days of continuous growth in ABX-containing media reduced the GC level in GD-1 lymphoblasts but that IFG treatment slightly increased their GC levels may reflect the balance that must be maintained in IFG-treated cells, between enhancement of GCase folding in the ER and inhibition of the enzyme in the lysosome. Continuous treatment of cells with IFG may not allow such a balance to be achieved. In contrast, because ABX ceases to be an inhibitor of GCase below pH 4.5 (Figs. 2 and 8 and Tables 1 and 4), its enhancement of GCase folding in the ER would not be counterbalanced by its presence in the lysosome.
Comparison of the changes in the H/D-Ex rates of various segments from the wild type GCase that occur upon binding of candidate PCs demonstrated that all of the ligands herein (ABX, FZN, and IFG (16)), as well as of two other small molecules, identified by high throughput screening of a 50,000 compound library (22), stabilize segments 243–249 (major) and 310–312 (minor) of the enzyme. Whereas all of these ligands except FZN also stabilized segment 386–400, FZN was the only ligand to stabilize segment 317–336 (Fig. 7A and supplemental Table 1S).
The three-dimensional docking models, taking into account the data from the present H/D-Ex studies (Fig. 9), predict that both ABX and FZN interact with GCase not only by hydrogen bonding, as is the case with IFG (44), but also hydrophobic and π-π interactions. These additional interactions with non-active site residues could explain the mixed-type inhibition observed for ABX and FZN.
In conclusion, this study has introduced a thermal denaturation assay as an alternative method for screening small molecule libraries for candidate PCs. ABX, a drug with a long history of use in humans, was identified as a potent stabilizer of GCase and characterized as a pH-dependent, mixed inhibitor of the enzyme. This small molecule was shown to function as an EET agent for patient cell lines with two common GCase mutations associated with GD-1 and GD-2/3. The treatment of these lines resulted in increased levels of GCase activity and protein in lysosomes and the reduction of GC storage. These data indicate that ABX could be rapidly developed as an effective EET agent for GD, either as an alternative or an adjunct to ERT or SRT.
Supplementary Material
Acknowledgments
We acknowledge the technical assistance of Sayuri Yonekawa, Amy Leung, Daphne Benedict, and Maria F. Da Rosa with Dr. Clifford Lingwood (Research Institute, Hospital for Sick Children) and Jan Blanchard with Eric D. Brown (McMaster University). We thank Cedric Manlhiot (Clinical Research Program Manager, Cardiovascular Clinical Research Unit, Labatt Family Heart Centre, Hospital for Sick Children) for help with statistical analysis of the data presented in Table 3. We thank Dr. John Callahan (Research Institute, Hospital for Sick Children) for helpful and critical suggestions. We thank Drs. Tracy Stockley and Peter Ray (Molecular Diagnostic Laboratory, Hospital for Sick Children) who made available one of the cell lines studied. We also thank Jimmy Ortins and Hongwei Huang of Accelrys Inc. for kindly providing software for evaluation. The LAMP-1 and -2 antibodies were obtained from the Developmental Studies Hybridoma Bank maintained by the Department of Biological Sciences, University of Iowa, Iowa City. This line of research was made possible by an initial bequest from the Uger Estate (to D. M.).
This work was supported by the David M. C. Ju Foundation (to B. R.), the Life for Luke Foundation (to G. M.), and Canadian Institutes of Health Research Team Grant CTP-82944 (to D. M.).
 The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1S and 2S and Figs. 1S and 2S.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1S and 2S and Figs. 1S and 2S.
- GD
- Gaucher disease
- ABX
- ambroxol
- EET
- enzyme enhancement therapy
- ER
- endoplasmic reticulum
- ERT
- enzyme enhancement therapy
- FDA
- Food and Drug Administration
- FZN
- fluphenazine
- GC
- glucosylceramide
- GCase
- glucocerebrosidase
- GD-1
- type 1 Gaucher disease
- GD-2
- type 2 Gaucher disease
- GD-3
- type 3 Gaucher disease
- GdnHCl
- guanidine hydrochloride
- Hex
- β-hexosaminidase
- H/D-Ex
- hydrogen/deuterium exchange
- IFG
- isofagomine
- LSD
- lysosomal storage disease
- Lamp-1 or -2
- lysosomal associated membrane protein-1 or -2
- MUbGlc
- 4-methylumbelliferyl-β-d-glucopyranoside
- MUG
- 4-methylumbelliferyl-β-N-acetylglucosaminide
- PDI
- protein-disulfide isomerase
- PC
- pharmacological chaperone
- SRT
- substrate reduction therapy
- SUPREX
- Stability of Unpurified Proteins from Rates of hydrogen/deuterium Exchange
- TDC
- sodium taurodeoxycholic acid
- HPLC
- high pressure liquid chromatography
- IFG
- isofagomine
- FBS
- fetal bovine serum
- CBE
- conduritol B epoxide
- MAP
- 2-amino-2-methyl-1-propanol
- rGCase
- human recombinant GCase
- MALDI
- matrix-assisted laser desorption ionization.
REFERENCES
- 1.Sidransky E., Ginns E. I. (1997) J. Med. Genet. 34,876–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sidransky E. (1997) Adv. Pediatr. 44,73–107 [PubMed] [Google Scholar]
- 3.Zhao H., Grabowski G. A. (2002) Cell. Mol. Life Sci. 59,694–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goker-Alpan O., Lopez G., Vithayathil J., Davis J., Hallett M., Sidransky E. (2008) Arch. Neurol. 65,1353–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goker-Alpan O., Schiffmann R., LaMarca M. E., Nussbaum R. L., McInerney-Leo A., Sidransky E. (2004) J. Med. Genet. 41,937–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gan-Or Z., Giladi N., Rozovski U., Shifrin C., Rosner S., Gurevich T., Bar-Shira A., Orr-Urtreger A. (2008) Neurology 70,2277–2283 [DOI] [PubMed] [Google Scholar]
- 7.Steet R. A., Chung S., Wustman B., Powe A., Do H., Kornfeld S. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103,13813–13818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hruska K. S., LaMarca M. E., Scott C. R., Sidransky E. (2008) Hum. Mutat. 29,567–583 [DOI] [PubMed] [Google Scholar]
- 9.Koprivica V., Stone D. L., Park J. K., Callahan M., Frisch A., Cohen I. J., Tayebi N., Sidransky E. (2000) Am. J. Hum. Genet. 66,1777–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goker-Alpan O., Hruska K. S., Orvisky E., Kishnani P. S., Stubblefield B. K., Schiffmann R., Sidransky E. (2005) J. Med. Genet. 42,e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin H., Sugimoto Y., Ohsaki Y., Ninomiya H., Oka A., Taniguchi M., Ida H., Eto Y., Ogawa S., Matsuzaki Y., Sawa M., Inoue T., Higaki K., Nanba E., Ohno K., Suzuki Y. (2004) Biochim. Biophys. Acta 1689,219–228 [DOI] [PubMed] [Google Scholar]
- 12.Ron D., Walter P. (2007) Nat. Rev. Mol. Cell Biol. 8,519–529 [DOI] [PubMed] [Google Scholar]
- 13.Sawkar A. R., Cheng W. C., Beutler E., Wong C. H., Balch W. E., Kelly J. W. (2002) Proc. Natl. Acad. Sci. U.S.A. 99,15428–15433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zimmer K. P., le Coutre P., Aerts H. M., Harzer K., Fukuda M., O'Brien J. S., Naim H. Y. (1999) J. Pathol. 188,407–414 [DOI] [PubMed] [Google Scholar]
- 15.Chang H. H., Asano N., Ishii S., Ichikawa Y., Fan J. Q. (2006) FEBS J. 273,4082–4092 [DOI] [PubMed] [Google Scholar]
- 16.Kornhaber G. J., Tropak M. B., Maegawa G. H., Tuske S. J., Coales S. J., Mahuran D. J., Hamuro Y. (2008) ChemBioChem. 9,2643–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiseman R. L., Powers E. T., Buxbaum J. N., Kelly J. W., Balch W. E. (2007) Cell 131,809–821 [DOI] [PubMed] [Google Scholar]
- 18.Tropak M. B., Mahuran D. (2007) FEBS J. 274,4951–4961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grabowski G. A. (2008) Expert Opin. Emerg. Drugs 13,197–211 [DOI] [PubMed] [Google Scholar]
- 20.Maegawa G. H., Tropak M., Buttner J., Stockley T., Kok F., Clarke J. T., Mahuran D. J. (2007) J. Biol. Chem. 282,9150–9161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tropak M. B., Blanchard J. E., Withers S. G., Brown E. D., Mahuran D. (2007) Chem. Biol. 14,153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tropak M. B., Kornhaber G. J., Rigat B. A., Maegawa G. H., Buttner J. D., Blanchard J. E., Murphy C., Tuske S. J., Coales S. J., Hamuro Y., Brown E. D., Mahuran D. J. (2008) ChemBioChem. 9,2650–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng W., Padia J., Urban D. J., Jadhav A., Goker-Alpan O., Simeonov A., Goldin E., Auld D., LaMarca M. E., Inglese J., Austin C. P., Sidransky E. (2007) Proc. Natl. Acad. Sci. U.S.A. 104,13192–13197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diettrich O., Mills K., Johnson A. W., Hasilik A., Winchester B. G. (1998) FEBS Lett. 441,369–372 [DOI] [PubMed] [Google Scholar]
- 25.Tropak M. B., Reid S. P., Guiral M., Withers S. G., Mahuran D. (2004) J. Biol. Chem. 279,13478–13487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown C. A., Mahuran D. J. (1993) Am. J. Hum. Genet. 53,497–508 [PMC free article] [PubMed] [Google Scholar]
- 27.Overkleeft H. S., Renkema G. H., Neele J., Vianello P., Hung I. O., Strijland A., van der Burg A. M., Koomen G. J., Pandit U. K., Aerts J. M. (1998) J. Biol. Chem. 273,26522–26527 [DOI] [PubMed] [Google Scholar]
- 28.Martin D. R., Rigat B. A., Foureman P., Varadarajan G. S., Hwang M., Krum B. K., Smith B. F., Callahan J. W., Mahuran D. J., Baker H. J. (2008) Mol. Genet. Metab. 94,212–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hein L. K., Meikle P. J., Hopwood J. J., Fuller M. (2007) Mol. Genet. Metab. 92,336–345 [DOI] [PubMed] [Google Scholar]
- 30.Fuller M., Rozaklis T., Lovejoy M., Zarrinkalam K., Hopwood J. J., Meikle P. J. (2008) Mol. Genet. Metab. 93,437–443 [DOI] [PubMed] [Google Scholar]
- 31.Folch J., Lees M., Sloane Stanley G. H. (1957) J. Biol. Chem. 226,497–509 [PubMed] [Google Scholar]
- 32.Liu C. (2003) in Advanced Medical Statistics ( Lu Y., Fang J.-O., eds) pp. 1051–1072, World Scientific Publishing Co., Hackensack, NJ [Google Scholar]
- 33.Hamuro Y., Coales S. J., Southern M. R., Nemeth-Cawley J. F., Stranz D. D., Griffin P. R. (2003) J. Biomol. Tech. 14,171–182 [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z., Smith D. L. (1993) Protein Sci. 2,522–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Powell K. D., Fitzgerald M. C. (2003) Biochemistry 42,4962–4970 [DOI] [PubMed] [Google Scholar]
- 36.Tang L., Roulhac P. L., Fitzgerald M. C. (2007) Anal. Chem. 79,8728–8739 [DOI] [PubMed] [Google Scholar]
- 37.Koska J., Spassov V. Z., Maynard A. J., Yan L., Austin N., Flook P. K., Venkatachalam C. M. (2008) J. Chem. Inf. Model 48,1965–1973 [DOI] [PubMed] [Google Scholar]
- 38.Sanner M. F. (1999) J. Mol. Graph. Model 17,57–61 [PubMed] [Google Scholar]
- 39.Jonsson L. M., Murray G. J., Sorrell S. H., Strijland A., Aerts J. F., Ginns E. I., Barranger J. A., Tager J. M., Schram A. W. (1987) Eur. J. Biochem. 164,171–179 [DOI] [PubMed] [Google Scholar]
- 40.Leonova T., Grabowski G. A. (2000) Mol. Genet. Metab. 70,281–294 [DOI] [PubMed] [Google Scholar]
- 41.Meijer L. A., Verstegen J. C., Bull S., Fink-Gremmels J. (2004) J. Vet. Pharmacol. Ther. 27,219–225 [DOI] [PubMed] [Google Scholar]
- 42.Premkumar L., Sawkar A. R., Boldin-Adamsky S., Toker L., Silman I., Kelly J. W., Futerman A. H., Sussman J. L. (2005) J. Biol. Chem. 280,23815–23819 [DOI] [PubMed] [Google Scholar]
- 43.Yu Z., Sawkar A. R., Whalen L. J., Wong C. H., Kelly J. W. (2007) J. Med. Chem. 50,94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lieberman R. L., Wustman B. A., Huertas P., Powe A. C., Jr., Pine C. W., Khanna R., Schlossmacher M. G., Ringe D., Petsko G. A. (2007) Nat. Chem. Biol. 3,101–107 [DOI] [PubMed] [Google Scholar]
- 45.Beck M. (2007) Hum. Genet. 121,1–22 [DOI] [PubMed] [Google Scholar]
- 46.Bembi B., Deegan P. (2008) Acta Paediatr. Suppl. 97,81–82 [DOI] [PubMed] [Google Scholar]
- 47.Cox T. M., Aerts J. M., Andria G., Beck M., Belmatoug N., Bembi B., Chertkoff R., Vom Dahl S., Elstein D., Erikson A., Giralt M., Heitner R., Hollak C., Hrebicek M., Lewis S., Mehta A., Pastores G. M., Rolfs A., Miranda M. C., Zimran A. (2003) J. Inherited Metab. Dis. 26,513–526 [DOI] [PubMed] [Google Scholar]
- 48.Cox T. M., Aerts J. M., Belmatoug N., Cappellini M. D., vom Dahl S., Goldblatt J., Grabowski G. A., Hollak C. E., Hwu P., Maas M., Martins A. M., Mistry P. K., Pastores G. M., Tylki-Szymanska A., Yee J., Weinreb N. (2008) J. Inherited Metab. Dis. 31,319–336 [DOI] [PubMed] [Google Scholar]
- 49.Yu Z., Sawkar A. R., Kelly J. W. (2007) FEBS J. 274,4944–4950 [DOI] [PubMed] [Google Scholar]
- 50.Porto C., Cardone M., Fontana F., Rossi B., Tuzzi M. R., Tarallo A., Barone M. V., Andria G., Parenti G. (2009) Mol. Ther. 17,964–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen J. S., Edwards N. J., Hong Y. B., Murray G. J. (2008) Biochem. Biophys. Res. Commun. 369,1071–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ron I., Horowitz M. (2005) Hum. Mol. Genet. 14,2387–2398 [DOI] [PubMed] [Google Scholar]
- 53.Schmitz M., Alfalah M., Aerts J. M., Naim H. Y., Zimmer K. P. (2005) Int. J. Biochem. Cell Biol. 37,2310–2320 [DOI] [PubMed] [Google Scholar]
- 54.Wei H., Kim S. J., Zhang Z., Tsai P. C., Wisniewski K. E., Mukherjee A. B. (2008) Hum. Mol. Genet. 17,469–477 [DOI] [PubMed] [Google Scholar]
- 55.Boston-Howes W., Williams E. O., Bogush A., Scolere M., Pasinelli P., Trotti D. (2008) Exp. Neurol. 213,229–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Masuda N., Peng Q., Li Q., Jiang M., Liang Y., Wang X., Zhao M., Wang W., Ross C. A., Duan W. (2008) Neurobiol. Dis. 30,293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang H., Guan Y., Wang X., Smith K., Cormier K., Zhu S., Stavrovskaya I. G., Huo C., Ferrante R. J., Kristal B. S., Friedlander R. M. (2007) Eur. J. Neurosci. 26,633–641 [DOI] [PubMed] [Google Scholar]
- 58.Copeland R. (2000) Enzymes: A Practical Introduction to Structure, Mechanism, and Data Analysis, 2nd Ed., pp. 266–304, Wiley-VCH, Inc., New York [Google Scholar]
- 59.Rigat B., Mahuran D. (2009) Mol. Genet. Metab. 96,225–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Molina G., Holguin E., Teran E. (2004) Am. J. Obstet. Gynecol. 191,2177; author reply 2178 [DOI] [PubMed] [Google Scholar]
- 61.Yang B., Yao D. F., Ohuchi M., Ide M., Yano M., Okumura Y., Kido H. (2002) Eur. Respir. J. 19,952–958 [DOI] [PubMed] [Google Scholar]
- 62.Wauer R. R., Schmalisch G., Hammer H., Buttenberg S., Weigel H., Huth M. (1989) Eur. Respir. J. 3,57S–65S [PubMed] [Google Scholar]
- 63.Bianchi M., Mantovani A., Erroi A., Dinarello C. A., Ghezzi P. (1990) Agents Actions 31,275–279 [DOI] [PubMed] [Google Scholar]
- 64.Malerba M., Ragnoli B. (2008) Expert Opin. Drug Metab. Toxicol. 4,1119–1129 [DOI] [PubMed] [Google Scholar]
- 65.Couet W., Girault J., Reigner B. G., Ingrand I., Bizouard J., Acerbi D., Chiesi P., Fourtillan J. B. (1989) Int. J. Clin. Pharmacol. Ther. Toxicol. 27,467–472 [PubMed] [Google Scholar]
- 66.Lee H. J., Joung S. K., Kim Y. G., Yoo J. Y., Han S. B. (2004) Pharmacol. Res. 49,93–98 [DOI] [PubMed] [Google Scholar]
- 67.Oosterhuis B., Storm G., Cornelissen P. J., Su C. A., Sollie F. A., Jonkman J. H. (1993) Eur. J. Clin. Pharmacol. 44,237–241 [DOI] [PubMed] [Google Scholar]
- 68.Rojpibulstit M., Kasiwong S., Juthong S., Phadoongsombat N., Faroongsarng D. (2003) Clin. Drug Invest. 23,273–280 [DOI] [PubMed] [Google Scholar]
- 69.Colombo L., Marcucci F., Marini G. M., Pierfederici P., Mussini E. (1990) J. Chromatogr. 513,141–147 [DOI] [PubMed] [Google Scholar]
- 70.Weiser T. (2008) CNS Neurosci. Ther. 14,17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsai P., Lipton J. M., Sahdev I., Najfeld V., Rankin L. R., Slyper A. H., Ludman M., Grabowski G. A. (1992) Pediatr. Res. 31,503–507 [DOI] [PubMed] [Google Scholar]
- 72.Groener J. E., Poorthuis B. J., Kuiper S., Helmond M. T., Hollak C. E., Aerts J. M. (2007) Clin. Chem. 53,742–747 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.