Abstract
In Saccharomyces cerevisiae, ARN1 encodes a transporter for the uptake of ferrichrome, an important nutritional source of iron. In the absence of ferrichrome, Arn1p is sorted directly from the trans-Golgi network (TGN) to the vacuolar lumen via the vacuolar protein-sorting pathway. Arn1p is mis-sorted to the plasma membrane in cells lacking Gga2p, a monomeric clathrin-adaptor protein involved in vesicular transport from the TGN. Although Ggas have been characterized as ubiquitin receptors, we show here that ubiquitin binding by Gga2 was not required for the TGN-to-endosome trafficking of Arn1, but it was required for subsequent sorting of Arn1 into the multivesicular body. In a ubiquitin-binding mutant of Gga2, Arn1p accumulated on the vacuolar membrane in a ubiquitinated form. The yeast epsins Ent3p and Ent4p were also involved in TGN-to-vacuole sorting of Arn1p. Amino-terminal sequences of Arn1p were required for vacuolar protein sorting, as mutation of ubiquitinatable lysine residues resulted in accumulation on the vacuolar membrane, and mutation of either a THN or YGL sequence resulted in mis-sorting to the plasma membrane. These studies suggest that Gga2 is involved in sorting at both the TGN and multivesicular body and that the first step can occur without ubiquitin binding.
The movement of small molecules across cellular membranes is a crucial process for cells and is entirely dependent on the activity of polytopic integral membrane proteins located in the plasma membrane or in the membranes of intracellular organelles. The activity of these transporters is subject to multiple levels of regulation. In both eukaryotes and prokaryotes, transporters are regulated at the transcriptional level. Eukaryotes also employ post-translational mechanisms to regulate the activity of transporters, frequently by altering their intracellular trafficking patterns. The mammalian glucose transporter GLUT4 (1) and the water channel AQP2 (2) undergo relocalization to the plasma membrane from intracellular vesicles in response to stimulation by the hormones insulin and vasopressin, respectively. The Menkes and Wilson copper transporters, ATP7A and ATP7B, relocalize in response to changes in intracellular copper (3). In yeast, transporters such as the general amino acid permease Gap1p (4), the manganese transporter Smf1p (5, 6), and the ferrichrome (FC)5 transporter Arn1p (7) undergo altered trafficking in response to changes in amino acid, manganese, and FC levels, respectively.
Arn1p is one of four homologous transporters in yeast that facilitate the uptake of extracellular siderophore-iron chelates, which are important nutritional sources of iron in unicellular organisms (8). Arn1p and the homologous Arn3p specifically recognize and take up FC-iron chelates, and both of these transporters demonstrate intracellular trafficking patterns that are controlled in part by the presence of their siderophore substrates (9, 10). ARN1 is transcriptionally activated under conditions of iron deficiency. If FC is not present in the medium, Arn1p is rapidly sorted from the trans-Golgi network (TGN) to the vacuole for degradation, via the vacuolar protein-sorting (VPS) pathway. If FC is present and gains access to the endosomal compartment via fluid phase endocytosis, FC binds to a receptor domain on the extracytosolic face of Arn1, which leads to the trafficking of a portion of Arn1p to the plasma membrane (11). At concentrations of FC high enough to stimulate transport, Arn1 cycles on and off the plasma membrane.
A common feature of the intracellular trafficking of Arn1p and other transporters is passage through the TGN, where cargo proteins are sorted into vesicles or tubular carriers destined for the plasma membrane, endosomes, or vacuoles (12). Trafficking from the TGN to endosomes and vacuoles involves the sorting of cargo proteins into clathrin-coated vesicles, and this sorting requires several types of adaptor proteins that localize to the TGN, recognize cargo destined for endosomes, and recruit the components of the clathrin coat (13). In yeast, these adaptors include the epsin-domain proteins Ent3 and Ent5 (14–16) and Gga1 and Gga2 (Golgi-localized, γ-adaptin ear-containing, Arf-binding proteins). These adaptor proteins are likely recruited to the TGN via interactions with phosphatidylinositol 4-phosphate (17) and the small GTPases of the ADP-ribosylation factor family.
The Gga proteins have a conserved, modular domain structure that mediates their interactions with protein and lipid-binding partners (Fig. 1A) (13, 18, 19). The amino-terminal VHS domain of mammalian Ggas binds to an acidic di-leucine sorting signal defined by the consensus motif DXXLL, which is located in the cytosolic domain of cargo proteins; however, in yeast, cargo proteins do not contain this type of motif, and the residues of the VHS domain of human Ggas that bind this motif are not conserved (20, 21). The GAT domain of human Gga3 also contains two binding sites for ubiquitin, and a number of conserved hydrophobic amino acid residues in the carboxyl-terminal half of the GAT domain have been shown to directly contribute to ubiquitin binding at the two sites (22–28).
FIGURE 1.
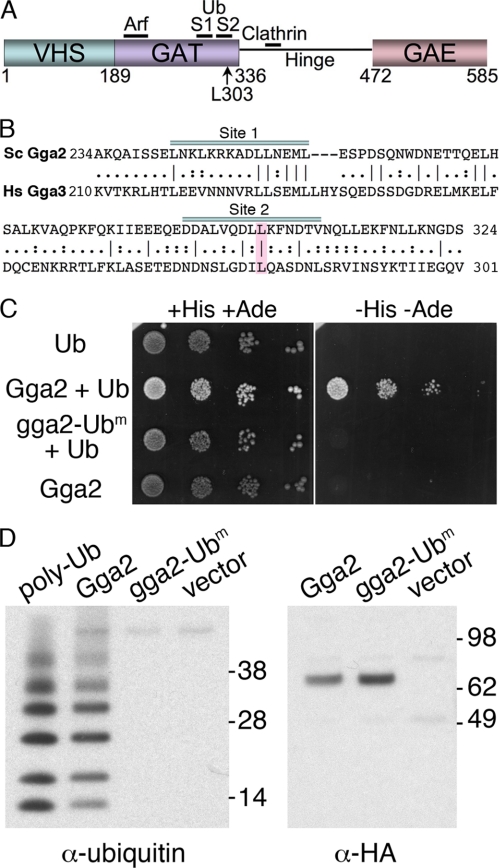
Loss of ubiquitin binding in Gga2 mutant L303A. A, domain structure of yeast Gga2. Binding sites for Arf, clathrin, and ubiquitin are indicated. B, sequence comparison of carboxyl-terminal GAT domains of yeast Gga2 and human Gga3. Ubiquitin-binding sites 1 and 2 are indicated. Mutated ubiquitin-binding residue Leu-303 is highlighted. C, loss of ubiquitin binding by yeast two-hybrid analysis. The AH109 strain was transformed with the empty vectors pGBKT7 and pGADT7-ubiquitin (Ub), pGBKT7-Gga2 VHS-GAT and pGADT7-ubiquitin (Gga2 + Ub), pGBKT7-Gga2 VHS-GAT L303A, and pGADT7-ubiquitin (gga2-Ubm + Ub), or pGBKT7-Gga2 VHS-GAT and the empty vector pGADT7 (Gga2). The resulting transformed strains were plated in serial 10-fold dilutions on SC medium lacking tryptophan and leucine but containing histidine and adenine (+HIS +ADE) or on SC medium lacking tryptophan, leucine, histidine, and adenine (−HIS −ADE). Plates were incubated for 3 days at 30 °C. ADE+ strains (e.g. Gga2 + Ub) exhibit a white colony morphology although ade2 strains are pink. D, loss of ubiquitin binding in the full-length Gga2 mutant L303A. The strain Y7096 was transformed with pGga2-HA, pGga2-Ubm-HA, or the empty vector and grown in selective medium at 30 °C. Cells were lysed; membranes were isolated, and solubilized membrane proteins were subjected to immunoprecipitation with anti-HA antibody. Immune complexes were incubated with 5 μm polyubiquitin chains (linked via Lys-63) and washed, and immune complexes with bound ubiquitin were eluted and subjected to SDS-PAGE and Western blotting with anti-ubiquitin (left panel) or anti-HA (right panel) antibodies. Polyubiquitin chains (15 pg) were included as a control.
Ubiquitination of certain cargo proteins, such as the yeast Gap1 and Fur4 (29) permeases or a mutant form of the Pma1 ATPase (30), is reportedly required for the efficient trafficking of the transporters from the TGN to the vacuole. Direct Gga binding to ubiquitinated lysine residues on the cargo proteins at the TGN is thought to permit the specific sorting of the proteins into clathrin-coated vesicles destined for endosomes and the vacuole. A recent report, however, indicated that ubiquitination of Gap1 was not required for Golgi to endosome sorting (31). Endosomal proteins destined for the vacuole are delivered via the multivesicular body (MVB) (32). Here, integral membrane proteins and lipids in the late endosome are sorted into invaginating membranes that form vesicles within the lumen of the MVB. The MVB then fuses with the vacuole and the intraluminal vesicles are delivered into the vacuolar lumen, where they are rapidly degraded. Proteins on the outer membrane of the MVB, which were not sorted into the luminal vesicles, then appear on the limiting membrane of the vacuole or are recycled back to the TGN or plasma membrane. Ubiquitin is the signal that allows cargo proteins to be sorted into the MVB (32, 33). Ubiquitinated integral membrane proteins in endosomes are recognized by the ubiquitin binding surfaces of the Vps27-Hse1 complex and the ESCRT-I complex, which then recruit the ESCRT-II and ESCRT-III complexes, directing the ubiquitinated cargo into invaginating vesicles of the MVB. Although the role of ubiquitin in the sorting of cargo protein into the MVB has been clearly demonstrated, recent work suggests that the mechanism of Gga-dependent sorting of cargo proteins into clathrin-coated vesicles at the TGN is not entirely explained on the basis of ubiquitin binding.
Arn1 also requires ubiquitination and the activity of Gga2 for efficient TGN-to-vacuole sorting; however, recent work raises the possibility that ubiquitination of Arn1 is not necessary for interaction with Gga2 (34). Cells carrying a temperature-sensitive form of Rsp5, the ubiquitin-protein isopeptide ligase that modifies integral membrane proteins in yeast, fail to sort Arn1 into the MVB and accumulate nonubiquitinated Arn1 on the vacuolar membrane. This finding indicates that in the absence of ubiquitination, Arn1 traffics from the TGN to the limiting membrane of the MVB, but it is not incorporated into the luminal vesicles of the MVB. In contrast, cells lacking Gga2 accumulate a large portion of Arn1 on the plasma membrane, and this Arn1 fails to enter the VPS pathway at all. Cells with mutations in both GGA2 and RSP5 also accumulate Arn1 on the plasma membrane. These findings indicate that Gga2 is required for Arn1 to efficiently enter the VPS pathway and suggest that Gga2 might be able to interact with Arn1 at the TGN without binding ubiquitin.
In these studies we address the question of how Gga2 recognizes Arn1 at the TGN. We have examined the trafficking of Arn1 in cells expressing a mutant form of Gga2 that does not bind ubiquitin and the trafficking of mutant forms of Arn1 that exhibit a loss of ubiquitin modification. We find that Gga2 acts at two sequential steps in the TGN-to-vacuole trafficking of Arn1 as follows: ubiquitin-independent transport from the TGN to the vacuoles and ubiquitin-dependent targeting to the MVB pathway.
MATERIALS AND METHODS
Strains, Plasmids, Media, and Antibodies
Strains and plasmids used in this study are listed in Table 1. The gga2Δ strains were described previously (34). Strains deleted for ENT3, ENT4, and ENT5 expressing Arn1-GFP were isolated by crossing the deletion mutant collection with the query strain YYG001 and generating haploid segregants using the synthetic genetic array approach (35).
TABLE 1.
Strains and plasmids used in this study
| Strain | Genotype | Ref. |
|---|---|---|
| AH109 | MATatrp1-901 leu2-3 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1UAS-GAL1TATA-HIS3 GAL2UAS-GAL2TATA-ADE2 URA3::MEL1UAS-MEL1TATA-lacZ MEL1 | Clontech |
| BY4742 | MATahis3Δ1 leu2Δ0; lys2Δ0; ura3Δ0 | ATCC |
| YPH499 | MATaura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 | Stratagene |
| YKY102 | MATaura3 leu2 trp1 ade2 bar1 | 34 |
| YDY001 | MATaura3 leu2 trp1 ade2 bar1 gga2Δ::KanMX4 | 34 |
| YKY103 | M MATaura3 leu2 trp1 ade2 bar1 rsp5-1 | 34 |
| YDY002 | MATaura3 leu2 trp1 ade2 bar1 rsp5-1gga2Δ::KanMX4 | 34 |
| ARN1-GFP | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ARN1::eGFP-HIS5 | 52 |
| ARN4-GFP | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ARN4::eGFP-HIS5 | 52 |
| YDY003 | ARN1-GFP gga2Δ::KanMX4 | 34 |
| CWY101 (arn1-4D) | YPH499 arn1::HISG arn2::HISG arn3::HISG arn4::HISG | 47 |
| YDY004 | arn1-4Δ gga2Δ::KanMX4 | 34 |
| CMY119 | MATα gga1Δ::TRP1 gga2Δ::HIS3 ura3-52 leu2-3,112 his3-200 trp1-901 lys2-801 suc2-9 | 53 |
| Y7096 | MATα leu2-Δ0 his3-Δ1 ura3-Δ0 met15-Δ0 lyp1Δ cyh2 can1Δ::STE2pr-SpHIS5 | 35 |
| YYG001 | MATα leu2-Δ0 his3-Δ1 ura3-Δ0 met15-Δ0 lyp1Δ cyh2 can1Δ::STE2pr-SpHIS5 ARN1::eGFP-NatMX | This study |
| YYG011 | YYG001 ent3Δ::KanMX4 | This study |
| YYG012 | YYG001 ent4Δ::KanMX4 | This study |
| YYG013 | YYG001 ent5Δ::KanMX4 | This study |
| Plasmid | Vector and insert information | Ref. |
|---|---|---|
| pGBKT7 | Clontech | |
| pGBKT7-Gga2 VHS-GAT | This study | |
| pGBKT7-Gga2 VHS-GAT L303A | This study | |
| pGADT7 | Clontech | |
| pUBI4 | pGADT7-ubiquitin | 26 |
| pCM53-3 | pYX112-GGA2-HA | 54 |
| pHW40 | pYX112-GGA2-UbM-HA | This study |
| pYD001 | pRS316-ARN1-GFP | This study |
| pYD002 | pRS316-ARN111–13A-GFP | This study |
| pYD003 | pRS316-ARN114–16AGFP | This study |
| pYD004 | pRS316-ARN129–31A-GFP | This study |
| pYD005 | pRS316-ARN138–40AGFP | This study |
| pYD006 | pRS316-ARN1-N,M/ARN4-C-GFP | This study |
| pYD007 | pRS316-ARN4-N/ARN1-M,C-GFP | This study |
The plasmids pGBKT7-Gga2 VHS-GAT L303A and pHW40 were constructed by site-directed mutagenesis of their respective wild type parent plasmids using the QuikChange kit (Stratagene, La Jolla, CA), and mutations were confirmed by sequencing. The plasmid pYD001 expressing ARN1-GFP was constructed in pRS316. Briefly, the plasmid pRS316-ARN1-HA was linearized by SpeI, and PCR was performed using pFa6a-GFP(S65T)-KanMX6 (36) as template and an appropriate primer pair designed for homologous recombination at ARN1 locus on pRS316 resulting in eliminating the HA tag. The products of PCR and the linearized pRS316-ARN1-HA were co-transformed into BY4742 using the lithium acetate method. The plasmid was rescued and confirmed by DNA sequencing. Alanine-scanning mutagenesis of the Arn1 amino terminus was performed in pRS316-ARN1-GFP. Groups of three amino acid residues from the Arn1 amino terminus (amino acids 2–73) were mutated to alanine by two-step PCR. Briefly, the first step PCR was performed using pRS316-ARN1-HA as a template and a primer pair designed for replacing the target amino acid residues with Ala, and the second step PCR was performed using the products of the first step PCR as template and a primer pair designed for homologous recombination at ARN1 locus on pRS316. The plasmid pRS316-ARN1-GFP was linearized with NruI. The linearized pRS316-ARN1-GFP and the products of the second step PCR were co-transformed into BY4742 using the lithium acetate method. Plasmids were rescued and confirmed by DNA sequencing.
Plasmids expressing Arn1-N,M(1–582)-Arn4C(572–606)-GFP were constructed in pRS316-Arn1-GFP. Briefly, the plasmid pRS316-Arn1-GFP was linearized by BamHI, and Arn4C(572–606) was amplified by PCR using pRS426-Arn4 as template and an appropriate primer pair designed for homologous recombination. The products of the PCR and the linearized pRS316-ARN1-GFP were co-transformed into BY4742 using the lithium acetate method. The plasmid was rescued and confirmed by DNA sequencing.
Rich medium and synthetic complete medium were prepared as described (37). Iron-poor medium containing 10 μm ferrous ammonium citrate and 1 mm ferrozine was prepared as described previously (38). FC was added at the indicated concentrations as the ferric chelate.
The following mouse monoclonal antibodies were used: HA.11 (Covance), anti-GFP (Roche Applied Science), and anti-ubiquitin (P4D1) (Covance). Anti-Pma1 and anti-Vps10 antibodies were purchased from Invitrogen. Horseradish peroxidase-conjugated (Amersham Biosciences) or Cy3-conjugated (Jackson ImmunoResearch) anti-mouse IgG was used as secondary antibodies.
Yeast Two-hybrid Assays
Strain AH109 was transformed with the indicated plasmids. Transformants were spotted in serial 10-fold dilutions on synthetic complete media lacking the indicated amino acids and incubated for 3 days at 30 °C.
Immunoprecipitation Assays in Yeast
Yeast were grown at 30 °C to an optical density of 0.6 in 100 ml of iron-poor medium with no FC added. Yeast cells were harvested and resuspended in 500 ml of ice-cold lysis buffer (50 mm Tris/HCl, pH 7.5, 300 mm NaCl, 5 mm EDTA, 1 mm dithiothreitol, 5 mm N-ethylmaleimide) containing 2.5× complete protease inhibitor mixture (Roche Applied Science) and 1 mm phenylmethylsulfonyl fluoride, and then 1 volume of acid-washed glass beads was added. The cells were disrupted by vigorous vortexing 10 times at 1 min each and with 1 min of chilling on ice between each vortexing. The cell lysate was centrifuged at 500 × g, and the supernatant then subjected to an 18,000 × g centrifugation at 4 °C for 1 h. The membrane pellet was solubilized in lysis buffer containing 2.5× complete protease inhibitor mixture, 1 mm phenylmethylsulfonyl fluoride, and 5% dodecyl maltoside at 4 °C for 30 min and then subjected to an 18,000 × g centrifugation at 4 °C for 10 min. Solubilized membrane proteins were quantitated, and equal amounts were incubated with anti-GFP or anti-HA antibody at 4 °C for 1 h, followed by incubation with 20 μl of pre-equilibrated Dynabeads Protein G (Invitrogen) at 4 °C for 1 h. The resin was then washed six times with lysis buffer and resuspended in 40 μl of NuPAGE LDS sample buffer (Invitrogen). Bound proteins were resolved by SDS-PAGE (NuPAGE Norvex 4–12% BisTris gradient gel, Invitrogen) and revealed by immunoblotting followed by chemiluminescent detection (Amersham Biosciences). For ubiquitin binding experiments, Lys-63 linked polyubiquitin chains (Enzo Life Sciences) at 5 μm (as monomer) were incubated with immune complexes at 4 °C for 1 h. Following washes the bound proteins were eluted in SDS samples buffer and subjected to Western blotting.
Surface Binding Assays in Yeast
Yeast were grown in 100 ml of iron-poor medium at 30 °C to an optical density of 0.3 and grown for an additional 2 h with either no FC or with 0.02 or 5 μm FC added. Surface binding assays were performed as described (39). Briefly, yeast cells were harvested, treated with 20 mm NaN3/KF on ice for 15 min, and then washed with 5 ml of ice-cold binding buffer (50 mm sodium citrate, pH 6.5, 5% glucose, 1 mm bathophenanthroline disulfonate, 20 mm NaN3/KF). 500 μl of 10 A600 cells were mixed with 100 nm [55Fe-FC] in binding buffer on ice with gently shaking for 15 min. Cells were again washed with binding buffer, and retained [55Fe-FC] was measured.
Microscopy of Living Cells
To visualize cells expressing GFP-tagged proteins, yeast cells were grown overnight in iron-poor medium at 30 °C to early log phase (A600 = 0.3) and grown for an additional 2 h with either no FC or 0.02 or 5 μm FC added. Images were obtained on Zeiss fluorescence microscope with Hamamatsu charge-coupled digital camera controlled by IPLabs software. The GFP signal was visualized by excitation at 488 nm and collection with a 505-nm filter. Fluorescence images were processed using Adobe Photoshop.
Subcellular Fractionation and Sucrose Density Gradient Analysis
Subcellular fractionation and sucrose density gradient analysis were performed as described previously (40). Briefly, 250 ml of yeast cultures were grown to mid-logarithmic phase (A600 = 0.5) in iron-poor medium at 30 °C. Cells were harvested and resuspended in 200 ml of ice-cold STE10 (25 mm Tris/HCl, pH 7.4, 5 mm EDTA, 10% (w/v) sucrose) containing 2.5× complete protease inhibitor mixture (Roche Applied Science) and 1 mm dithiothreitol, and then 1 volume of acid-washed glass beads was added. The cells were disrupted by vigorous vortexing 8 times for 1 min each and 1 min chilling on ice between each vortexing. After a clearing spin at 500 × g, the lysates were loaded onto a 12-ml sucrose gradient (20–60%) and spun at 100,000 × g for 17 h at 4 °C. Fourteen fractions were collected from the top of the gradient. Aliquots were loaded onto SDS-polyacrylamide gels and subjected to immunoblot analysis.
RESULTS
Loss of Ubiquitin Binding with Mutation of a Site 2 Leucine Residue
The GAT domains of human Gga1 and Gga3 have been shown, using various approaches, to bind ubiquitin with differing affinities, whereas human Gga2 has been shown to bind ubiquitin only very weakly (22, 26–28). Crystallographic studies of the human Gga3 GAT domain in complex with ubiquitin indicate that a hydrophobic patch on helices α1 and α2 forms a binding site for ubiquitin referred to as site 1 (23, 25). Although not present in crystal structures, NMR chemical shift perturbations, surface plasmon resonance analyses, pulldown experiments, and in vivo mutational analyses indicate that a second site for ubiquitin -binding, site 2, is formed by a hydrophobic patch on helices α2 and α3. Mutation of leucine 280 in site 2 of the human Gga3 GAT domain results in a complete loss of interaction with mono-ubiquitin-agarose (28) or GST-ubiquitin (22), although a GST-Gga3-GAT fusion protein with this mutation demonstrated reduced but detectable binding to mono- and polyubiquitin by pulldown and surface plasmon resonance (23). Yeast Gga1 and Gga2 also have been shown to bind ubiquitin (22, 27), but the ubiquitin-binding sites have not been mapped in the yeast Gga proteins. We introduced a mutation in the GAT domain of yeast Gga2 (L303A) that corresponds to the Leu-280 site 2 mutation of human Gga3 (Fig. 1B) and analyzed the ubiquitin binding activity of the mutant by yeast two-hybrid assay (Fig. 1C) and by in vitro binding of polyubiquitin (Fig. 1D). The VHS-GAT domains of yeast Gga2 were fused to the Gal4 DNA binding domain and ubiquitin to the Gal4 activation domain. The wild type Gga2 VHS-GAT fusion interacted strongly with ubiquitin, as indicated by vigorous growth on plates lacking histidine and adenine (Fig. 1C, −His −Ade). Mutation of Gga2 L303A (gga2-Ubm) resulted in a complete loss of detectable interaction between the VHS-GAT fusion and ubiquitin.
To confirm that the L303A mutation conferred significant loss of ubiquitin binding in the full-length Gga2 protein, we examined the capacity of immunoprecipitated Gga2 and Gga2-L303A to bind polyubiquitin chains linked at lysine 63 of ubiquitin (Fig. 1D). As human Gga3 has been shown to contain two binding sites for ubiquitin and yeast Gga2 exhibited very low affinity for mono-ubiquitin-agarose (data not shown), we examined the binding of yeast Gga2 to polyubiquitin. Because we had previously shown that Arn1 was modified with Lys-63-linked polyubiquitin chains (34) and because a similar modification was required for the MVB sorting of Gap1 (31), we selected Lys-63-linked polyubiquitin as a substrate. Yeast cells were transformed with plasmids bearing wild type Gga2-HA, Gga2-L303A-HA (gga2-Ubm), or the empty vector. Membrane protein extracts were subjected to immunoprecipitation; the immune complexes were incubated with polyubiquitin, and the bound proteins were eluted and analyzed by Western blotting. Although wild type Gga2 bound polyubiquitin well, ubiquitin binding by Gga2-L303A was undetectable (Fig. 1D, left panel). We confirmed that both wild type and mutant Gga2 were expressed at similar levels in cells and present at similar levels in the immune complexes (Fig. 1D, right panel). Thus, substitution of leucine 303 with alanine effectively eliminates ubiquitin binding by yeast Gga2.
Normal and Abnormal Sorting of Arn1 in a Gga2 Ubiquitin-binding Mutant
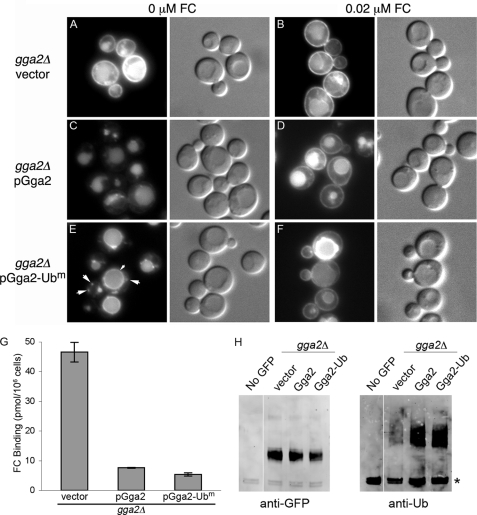
Although the Gga1 and Gga2 proteins of yeast have been described as being functionally interchangeable, they are probably not redundant, as deletion of GGA2 alone results in the mis-sorting of Arn1 and Arn3 (9, 34). We examined whether pGga2-Ubm could complement the Arn1 trafficking defect of the gga2Δ strain (Fig. 2). When no FC is present, Arn1 normally traffics directly from the TGN to the vacuole, via the MVB, for degradation (Fig. 2C). In strains expressing Arn1 with a carboxyl-terminal GFP, fluorescent signal accumulates in the lumen of the vacuole because of the protease resistance of the GFP domain, which remains after the rapid degradation of Arn1. Although Arn1 transiently accumulates in the endosomal compartment, this is under-represented in the Arn1-GFP constructs because of the relatively slow folding of the GFP domain (>30 min) (41) and the short half-life of endosomal Arn1 (<10 min) (34). Addition of low concentrations of FC to the growth medium results in a redistribution of a portion of Arn1 to the cell surface. This pattern of trafficking was seen in the gga2Δ strain transformed with the wild type plasmid, pGga2 (Fig. 2D). In the gga2Δ strain transformed with the empty vector, Arn1 exhibited a strong mislocalization to the plasma membrane, even in the absence of FC, with some signal still present in the vacuolar lumen (Fig. 2, A and B). However, when the gga2Δ strain was transformed with the ubiquitin-binding mutant, pGga2-Ubm, Arn1 was no longer present on the cell surface in the absence of FC (Fig. 2E). In contrast to both wild type Gga2 and no Gga2, however, cells expressing pGga2-Ubm accumulated Arn1 on the limiting membrane of the vacuole and in bright punctate intracellular vesicles. These images indicated that a mutant Gga2 with defects in ubiquitin binding complemented the TGN to MVB trafficking of Arn1, but they failed to direct a significant fraction of Arn1 into the luminal vesicles of the MVB.
FIGURE 2.
Ubiquitin-independent sorting of Arn1p by Gga2p. A–F, localization of Arn1-GFP to the vacuolar membrane in a gga2Δ strain carrying the Gga2 ubiquitin-binding mutant. The strain YDY003 (gga2Δ ARN1-GFP) was transformed with either vector (pYX112), pGga2-HA, or pGga2-Ubm and grown overnight in iron-poor medium. Either no FC or 0.02 μm FC was added during the final 2 h of growth. Epifluorescent images and DIC images were shown in parallel. E, small arrowheads indicate the vacuolar membrane, and large arrowheads indicate endosomes. G, complementation of Arn1 surface mislocalization in a gga2Δ strain carrying the Gga2 ubiquitin-binding mutant. Strain YDY002 (rsp5-1 gga2Δ) carrying either vector (pYX112), pGga2, or pGga2-Ubm was grown in iron-poor medium overnight at 25 °C to induce expression of Arn proteins and shifted to 37 °C for the final 2 h of growth; no FC was added. Cells were treated with 20 mm NaN3/KF, harvested, washed, and mixed with 100 nm [55Fe-FC] on ice for 15 min. Cells were again washed, and retained [55Fe-FC] was measured. H, ubiquitination of Arn1-GFP in a gga2Δ strain carrying the Gga2 ubiquitin-binding mutant. Congenic gga2Δ strains carrying either vector (pYX112), pGga2-HA, or Gga2-Ubm were grown in iron-poor medium overnight to induce expression of Arn1-GFP, and no FC was added. An equivalent number of cells were lysed, and membranes were isolated and solubilized with detergent. Equivalent amounts of membrane protein were subjected to immunoprecipitation with anti-GFP antibody. Immunoprecipitates were then subjected to SDS-PAGE and Western blotting using an anti-GFP antibody to detect Arn1-GFP (left) or an anti-ubiquitin antibody to detect ubiquitin (right). The asterisk indicates immunoglobulin heavy chain.
To confirm that Arn1 was not mis-sorted to the plasma membrane in cells expressing pGga2-Ubm, we assessed the amount of Arn1 localized to the plasma membrane by measuring the amount of radiolabeled FC that bound to Arn1 on the cell surface (Fig. 2G). To ensure that no ubiquitin binding occurred, we used the rsp5-1 strain at the restrictive temperature, which lacks ubiquitination of integral membrane proteins (42). In a gga2Δ rsp5-1 strain transformed with an empty vector, large amounts of FC binding were detected on the cell surface. Transformation of this strain with either pGga2 or pGga2-Ubm resulted in a 6.1- and an 8.7-fold reduction in FC binding, respectively, indicating that the ubiquitin-binding mutant of Gga2 was equivalent to the wild type in its capacity to complement the surface mislocalization phenotype of Arn1p.
Ubiquitin Modification of Mislocalized Arn1
Sorting of integral membrane proteins into the luminal vesicles of the MVB is dependent on ubiquitin modification of these proteins, and this modification is blocked in cells carrying a temperature-sensitive mutation in RSP5 (rsp5-1) (33, 42). In the rsp5-1 strain, Arn1 fails to sort into the MVB, accumulating instead on the limiting membrane of the vacuole (34). We questioned whether the Arn1 that accumulated on the vacuolar membrane of the gga2Δ pGga2-Ubm strain was ubiquitinated. A gga2Δ strain carrying Arn1-GFP was transformed with pGga2, pGga2-Ubm, or empty vector and grown in iron-poor medium without FC to induce Arn1-GFP expression. Arn1-GFP was immunoprecipitated from cellular membranes using antibodies directed against GFP, and the precipitated Arn1-GFP was probed with either anti-GFP or anti-Ub antibodies (Fig. 2H). In the gga2Δ strain transformed with empty vector (in which Arn1 is located on the plasma membrane), ubiquitinated Arn1 was barely detectable. In contrast, similar amounts of ubiquitinated Arn1 were detected in the strains transformed with pGga2 or pGga2-Ubm, although the overall amount of modified Arn1 was low in both cases (higher molecular weight species were not apparent in the anti-GFP blot). Elevated expression levels of Gga2 from the TPI1 promoter of pGga2 and pGga2-Ubm were associated with increased ubiquitination of Arn1. These results suggest that the failure to sort Arn1 into the MVB in the gga2Δ pGga2-Ubm strain was not because of a lack of ubiquitination or a failure to recruit Rsp5 to Arn1-containing membranes.
Involvement of Yeast Epsins Ent3 and Ent4 in Trafficking of Arn1
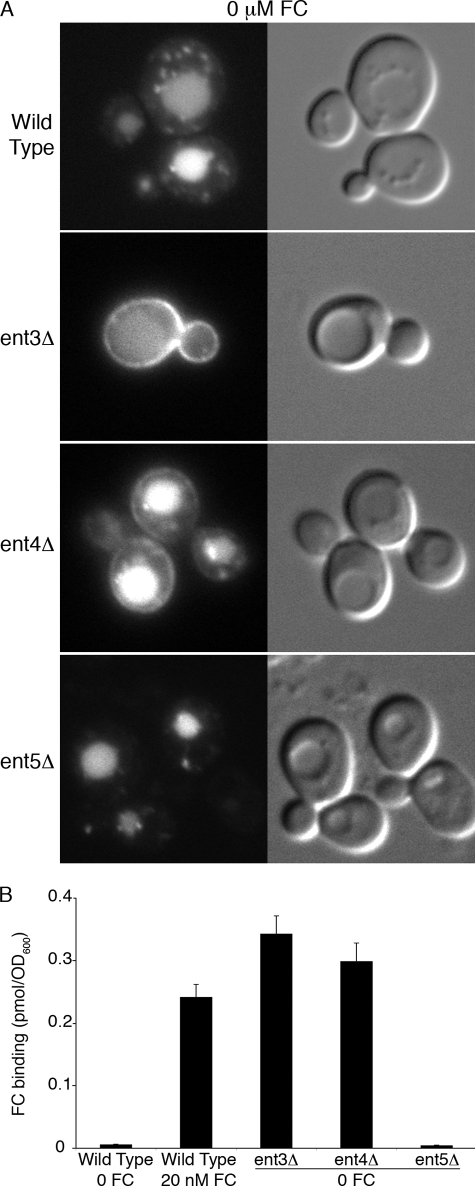
The yeast genome contains five proteins with homology to mammalian epsins, and four of these are known to be involved in the formation of clathrin-coated vesicles during endocytosis (Ent1 and Ent2) (43) and TGN-to-endosome trafficking (Ent3 and Ent5). Both Ent3 and Ent5 bind Gga2 in vivo, and Ent3 is dependent on Gga2 for localization to the TGN (15, 44). Cells lacking both Ent3 and Ent5 display defects in the trafficking of cargo proteins from the TGN to endosomes (14) and into the MVB (16). Protein-protein and genetic interaction data indicate that Ent3 and Gga2 may cooperate in the binding and recruitment of cargo proteins to clathrin-coated vesicles at the TGN. We screened the deletion mutant collection for strains that exhibited defects in the trafficking of Arn1 and found that strains lacking Ent3 or the previously uncharacterized Ent4 mislocalized a portion of Arn1 to the cell surface in the absence of FC (Fig. 3A), although a strain lacking Ent5 did not. We confirmed the mislocalization of Arn1 to the plasma membrane in the ent3Δ and ent4Δ strains by measuring the binding of radiolabeled Fe-FC to the cell surface (Fig. 3B). In a wild type strain, Arn1 was detected on the cell surface only in the presence of FC and not when FC was absent. In cells lacking Ent3 and Ent5, substantial amounts of Arn1 were detected on the surface in the absence of FC, whereas cells lacking Ent5 did not mislocalize Arn1. The similarity of the Arn1 trafficking defect in the gga2Δ strain and the ent3Δ and ent4Δ strains supports a model in which Gga2 and Ent3 and Ent4 exhibit cargo-specific binding and recruitment of Arn1 at the TGN. Furthermore, this observation indicated that Ent4, similar to the paralogous Ent3 and Ent5, was involved in trafficking at the TGN.
FIGURE 3.
Plasma membrane mislocalization of Arn1-GFP in strains lacking Ent3 or Ent4. Strains YYG001 (Arn1-GFP), YYG011 (ent3Δ Arn1-GFP), YYG012 (ent4Δ Arn1-GFP), and YYG013 (ent5Δ Arn1-GFP) were grown in iron-poor medium and subjected to fluorescent imaging (A) or the FC binding assay (B). In the FC binding assay, strain YYG001 (wild type) treated with 20 nm FC for 2 h was included as a positive control.
Loss of Ubiquitin Modification Associated with MVB Sorting Defect
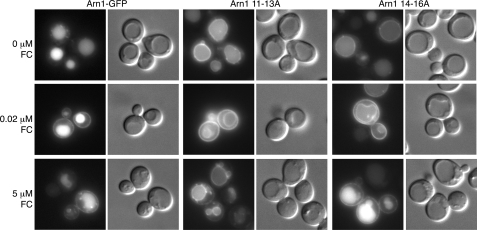
We hypothesized that if ubiquitin modification of Arn1 was required for Gga2 binding at the TGN, then loss of ubiquitin modification would result in an Arn1 trafficking pattern similar to that of a gga2Δ strain. We have already reported that the trafficking of Arn1 in cells carrying the rsp5-1 allele does not resemble that of the gga2Δ strain, but the argument could be made that this difference is because of indirect effects or to a more widespread defect in the rsp5-1 strain. We therefore examined the trafficking of mutant alleles of Arn1 that have reduced ubiquitination. Lysine residues 11, 14, and 15 in the cytosolic amino terminus of Arn1 are preferentially ubiquitinated by Rsp5 (34). Alanine-scanning mutagenesis of the amino terminus of Arn1-GFP produced two mutant alleles with alanine substitutions for these lysine residues. Arn111–13A and Arn114–16A contain alanine substitutions for lysine 11 and lysines 14 and 15, respectively. Fluorescence imaging of cells expressing wild type and mutant alleles of Arn1-GFP demonstrated that, in the absence of FC, both Arn111–13A and Arn114–16A were detected on the limiting membrane of the vacuole, as well as in the vacuolar lumen, although wild type Arn1-GFP was detected exclusively in the lumen of the vacuole (Fig. 4). In neither of these mutants was Arn1-GFP detected on the cell surface, as is the case in the gga2Δ strain. Addition of 20 nm FC to the growth medium of these strains resulted in the characteristic detection of both wild type and mutant alleles of Arn1-GFP on the plasma membrane, but the Arn111–13A and Arn114–16A strains persisted in localizing a portion of Arn1 to the vacuolar membrane. Addition of higher (>1 μm) FC to the growth medium typically resulted in a lesser amount of Arn1 on the cell surface, as the protein cycles on and off the surface through recycling endosomes. Here the addition of 5 μm FC to the growth medium resulted in persistence of the Arn1 signal on the cell surface and on the vacuolar membrane in the Arn111–13A strain but a loss of Arn1 signal on the vacuolar membrane in the Arn114–16A strain, suggesting that ubiquitination of these lysine residues was not functionally equivalent. Again, mutations that prevented ubiquitination were associated with a defect in sorting Arn1 into the MVB without a defect in TGN to late endosome trafficking.
FIGURE 4.
Mislocalization to the vacuolar membrane of Arn1 mutants that lack ubiquitin modification. YPH499 was transformed with either wild type Arn1-GFP or mutant forms of Arn1-GFP that are poorly ubiquitinated (Arn111–13A and Arn114–16A) and grown overnight in iron-poor medium with either no FC, 0.02 μm FC, or 5 μm FC added during the final 2 h of growth. Epifluorescent images and DIC images are shown in parallel.
Requirement of Residues in the Arn1 Amino Terminus for TGN-to-Endosome Trafficking
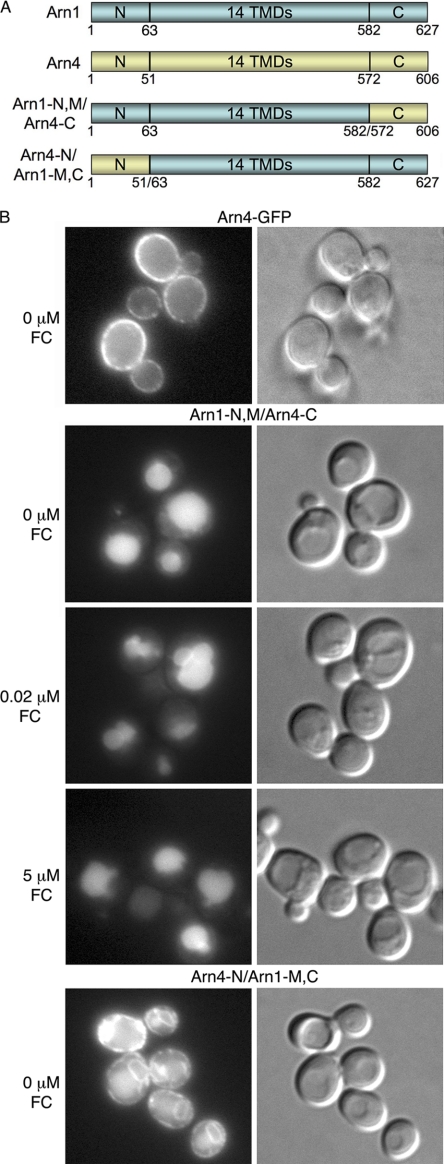
Our data indicated that Arn1 was not directed from the TGN to endosomes because of ubiquitin modification. Yeast Gga proteins do not recognize DXXLL-type sorting motifs (which are not present in Arn1 or Arn3). Similarly, the FSDSPEF peptide involved in the Gga-dependent sorting of Pep12 into the late endosome is not present in Arn1 or Arn3 (45). We attempted to map the domains of Arn1 that were required for interaction with adaptor proteins at the TGN by constructing Arn1/Arn4-GFP chimeric proteins and examining their intracellular trafficking patterns (Fig. 5A). Arn4 is a paralog of Arn1 and facilitates the uptake of the bacterial siderophore enterobactin in yeast (46, 47). Unlike Arn1 and Arn3, Arn4 is expressed exclusively on the plasma membrane, whether enterobactin is present in the growth medium or not (Fig. 5B) (48). We constructed a transporter that contained the Arn1 amino-terminal and transmembrane domains fused to the carboxyl-terminal domain of Arn4 (Arn1-N,M/Arn4-C). In the absence of FC, this transporter trafficked to the vacuolar lumen and was not detected on the plasma membrane (Fig. 5B). Addition of FC to the medium did not produce any detectable redistribution of the transporter to the plasma membrane. This observation was consistent with previous findings that a cluster of phenylalanine residues in the carboxyl terminus of Arn1 was required for the FC-induced redistribution to the plasma membrane (11). The data also suggested that sequences that conferred vacuolar protein sorting to Arn1 could be located in the amino terminus and that the carboxyl terminus of Arn4 was not sufficient to confer plasma membrane localization. We also constructed a transporter in which the amino terminus of Arn4 was fused to the transmembrane and carboxyl-terminal domains of Arn1-GFP (Arn4-N/Arn1-M,C). This chimeric transporter exhibited GFP signal in a peripheral and perinuclear pattern, indicating the endoplasmic reticulum, as well as a diffuse cytosolic signal (Fig. 5B). This pattern of GFP was most likely indicative of an unfolded or poorly folded protein, and this chimera was not analyzed further.
FIGURE 5.
Mis-sorting of Arn1/Arn4 chimeric transporters. A, domain organization of Arn1, Arn4, and Arn1/4 chimeras. Cytosolic amino-terminal (N), carboxyl-terminal (C), and membrane-spanning domains containing 14 predicted transmembrane domains (14 TMDs) are indicated. B, YPH499 was transformed with a plasmid expressing amino acids 1–582 of Arn1 fused to amino acids 572–606 of Arn4 followed by GFP (Arn1-N,M/Arn4-C) or a plasmid expressing amino acids 1–51 of Arn4 fused to residues 63–627 of Arn1 followed by GFP (Arn4-N/Arn1-M,C). Strain Arn4-GFP contained ARN4 with the GFP coding sequence integrated at the carboxyl terminus. Cells were grown as in Fig. 4. Epifluorescent images and DIC images were shown in parallel.
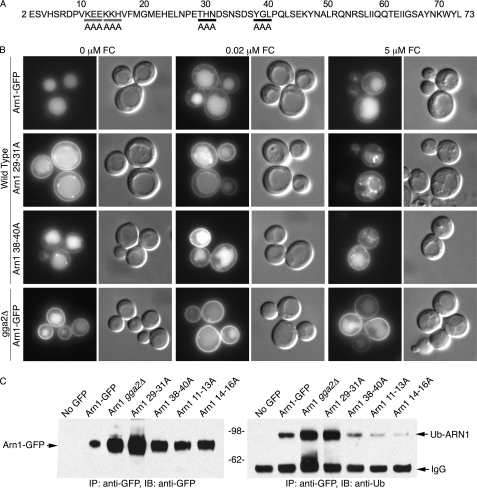
On the basis of the trafficking of the Arn1/Arn4 chimera, we performed alanine-scanning mutagenesis of the amino terminus of Arn1, substituting alanine for groups of three sequential amino acids, and we examined each of the resulting 24 plasmids (Fig. 6A). Of these, the two plasmids containing mutations of ubiquitinated lysine residues (Fig. 4) and two plasmids containing other mutations exhibited altered trafficking of the expressed mutant Arn1. In the Arn129–31A mutant, a threonine-histidine-asparagine (THN) sequence was substituted with alanines, and the resulting Arn1-GFP allele was mislocalized to both the plasma membrane and the vacuolar membrane in cells grown without FC (Fig. 6B). Addition of 20 nm FC to the medium did not result in an appreciable change in localization, although addition of 5 μm FC was associated with an increase in the punctate intracellular and vacuolar membrane staining. Mutation of the tyrosine-glycine-leucine (YGL) sequence in Arn138–40A resulted in a mutant Arn1-GFP allele that was expressed at lower levels, but with a similar mislocalization to the plasma membrane. Again the addition of 20 nm FC did not result in an appreciable change in Arn1 distribution, but addition of 5 μm FC was associated with increased intracellular punctate staining. For comparison, in cells grown without FC, no Arn1 was detected on the surface in the wild type Arn1-GFP, although expression of wild type Arn1 in the gga2Δ strain led to the mislocalization of large amounts of Arn1 to the plasma membrane. As Gga2 appeared to be involved in Arn1 sorting at both the TGN and late endosomal membrane, the trafficking of the Arn129–31A mutant and the Arn138–40A mutant was consistent with these mutations leading to a defect in Gga2 and/or Ent3 binding. We examined the expression levels of the amino-terminal mutants of Arn1 and the capacity of these mutants to undergo ubiquitin modification (Fig. 6C). As expected, mutations that interfered with the delivery of Arn1 to the vacuole for degradation led to the accumulation of more Arn1 protein in the cell (Fig. 6C, left panel). As shown previously, the Arn111–13A and Arn114–16A strains lacking ubiquitinated lysine residues exhibited very low levels of ubiquitination. The Arn138–40A mutant exhibited a lower level of expression than the Arn129–31A mutant and a reduced level of ubiquitination, although the trafficking patterns of the Arn138–40A mutant differed from those of the lysine mutants. The Arn129–31A mutant exhibited no ubiquitination defect and was expressed at high levels within the cell, indicating that the folding of the amino terminus of this mutant was sufficient to support recognition by the Rsp5 ubiquitin ligase.
FIGURE 6.
Mislocalization of amino-terminal mutant forms of Arn1-GFP. A, sequence of amino terminus of Arn1. Alanine-scanning mutants of ubiquitinated lysine residues, Arn111–13A, and Arn114–16A are indicated with gray bars. Arn129–31A and Arn138–40A mutants are indicated with black bars. B, YPH499 was transformed with either wild type Arn1-GFP or a mutant form of Arn1-GFP (Arn129–31A or Arn138–40A), and a congenic gga2Δ strain was transformed with wild type Arn1-GFP. Cells were grown as in Fig. 4. Epifluorescent images and DIC images are shown in parallel. Longer exposure times were required to image Arn138–40A. C, ubiquitination of amino-terminal mutant forms of Arn1-GFP. Transformants from B as well as YPH499 transformed with poorly ubiquitinated mutants Arn111–13A and Arn114–16A were quantitatively subjected to immunoprecipitation (IP) and Western blotting as in Fig. 2C. N-Ethylmaleimide was included in buffers to inhibit de-ubiquitination.
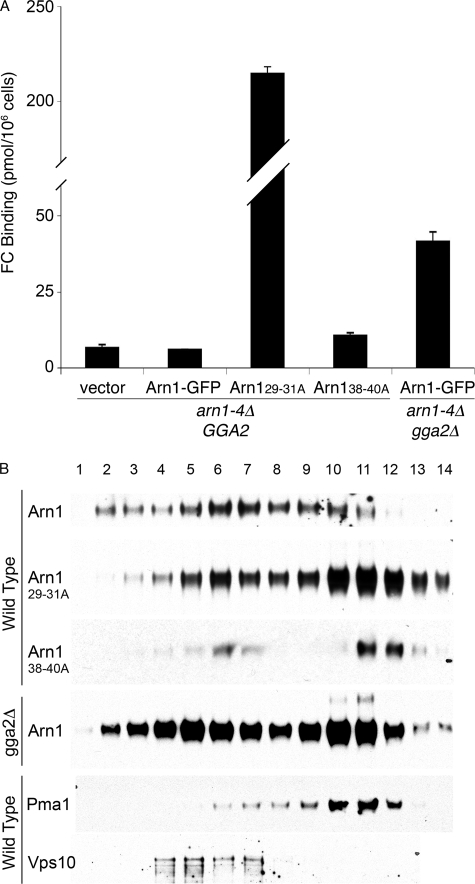
To quantitatively determine the degree of mislocalization exhibited by the Arn129–31A and Arn138–40A mutants, we measured the amount of Arn1-GFP on the cell surface using the FC binding assay. A strain from which ARN1, -2, -3, and -4 have been deleted was also deleted for GGA2, and the resulting GGA2+ and gga2Δ strains were transformed with wild type and mutant Arn1-GFP plasmids. Strains were grown in iron-poor medium without FC, and the surface binding of radiolabeled FC was determined (Fig. 7A). In the arn1–4Δ GGA2 strain, none of the wild type Arn1-GFP was detected on the surface, as the FC binding did not rise above the nonspecific binding of the vector-transformed strain. In the arn1–4Δ GGA2 strain expressing the Arn129–31A mutant, very large amounts of FC binding confirmed that the mutant accumulated on the plasma membrane. The amount of Arn129–31A on the cell surface exceeded the amount of wild type Arn1 mislocalized to the surface in the arn1–4Δ gga2Δ strain by more than 4-fold. The lower amount of mis-localized Arn1 in the arn1–4Δ gga2Δ strain may reflect a contribution of Gga1 to the sorting of Arn1, with both Gga1 and Gga2 interactions lost in the Arn129–31A mutant. Consistent with this idea, the amount of surface-mislocalized Arn1 is greater in the gga1Δ gga2Δ strain than in the gga2Δ strain (data not shown). The Arn138–40A mutant in the arn1–4Δ GGA2 strain also exhibited slightly elevated FC binding indicative of surface mislocalization (Fig. 7A), although to a much lower extent, consistent with the much lower level of expression of this mutant.
FIGURE 7.
Plasma membrane mislocalization of amino-terminal mutants of Arn1-GFP. A, elevated surface binding of FC in mutant forms of Arn1-GFP. Congenic arn1-4Δ and arn1-4Δ gga2Δ strains were transformed with vector (pRS316), Arn1-GFP, Arn129–31A-GFP, or Arn138–40A-GFP and were grown overnight in iron-poor medium with no FC. Cells were treated with 20 mm NaN3/KF, harvested, washed, and mixed with 100 mm [55Fe-FC] on ice for 15 min. Cells were again washed, and retained [55Fe-FC] was measured. B, co-sedimentation of mutant forms of Arn1-GFP (Arn129–31A and Anr138–40A) with plasma membrane protein Pma1p. The membrane fraction from YPH499 expressing wild type Arn1-GFP or mutant forms of Arn1-GFP (Arn129–31A and Arn138–40A) or from a congenic gga2Δ strain expressing wild type Arn1-GFP was applied to a 20–60% sucrose gradient. After centrifugation, the gradient was fractionated, and samples from each fraction were analyzed by SDS-PAGE followed by Western blotting. The blots were re-probed with antibodies to marker antigens as follows: plasma membrane, Pma1p, and late Golgi, Vps10p. Extended exposure times were required for visualization of wild type Arn1-GFP and Arn138–40A.
To confirm that both the Arn129–31A mutant and the Arn138–40A mutant were mislocalized to the plasma membrane in the absence of FC, we collected yeast expressing wild type and mutant Arn1-GFP, separated their membranes by sucrose density gradient centrifugation, and examined the distribution of Arn1 by Western blotting (Fig. 7B). Wild type Arn1 was primarily detected in fractions containing lighter membranes that co-migrated with Vps10, which included TGN and endosomal membranes. In contrast, the Arn129–31A mutant and the Arn138–40A mutant were primarily detected in fractions containing heavier membranes that co-migrated with the plasma membrane marker, Pma1, although some mutant Arn1 was also present in lighter fractions. As a control, we also fractionated the membrane from a gga2Δ strain and demonstrated that the majority of Arn1 was present in the plasma membrane fractions, although a substantial amount of Arn1 was also present in intracellular membranes. We attempted to determine whether these tripeptide sequences were directly involved in binding Gga2, but no specific binding between Arn1 and Gga2 could be demonstrated using co-immunoprecipitation, in vitro binding assays, or yeast two-hybrid assays using the Arn1 amino and carboxyl termini (data not shown). Taken as a whole, these data indicated that the tripeptides THN and YGL in the Arn1 amino terminus were required for the genetic interaction of Arn1 with clathrin adaptor proteins in the TGN.
DISCUSSION
Role of Ubiquitination in the Vacuolar Protein Sorting of Arn1
How are cargo proteins sorted into clathrin-coated vesicles at the TGN and what provides the specificity for recognition of these cargos? Here we have shown that in the case of Arn1, ubiquitination is not required for recognition by the clathrin adaptor protein Gga2 at the TGN, but ubiquitination is required for Gga2-mediated sorting into the MVB. Three types of data support this observation. First, in an rsp5-1 mutant, which fails to ubiquitinate Arn1, the transporter traffics from the TGN to the late endosome and accumulates on the limiting membrane of the vacuole (34). Second, lysine mutants of Arn1 that were poorly ubiquitinated were also trafficked from the TGN to the late endosome and accumulated on the vacuolar membrane (Fig. 4). Both findings indicated that, without ubiquitination, Arn1 was efficiently sorted from the TGN to the late endosomal compartment. Third, a mutant allele of Gga2 that exhibited loss of ubiquitin binding was as effective as the wild type allele in rescuing the plasma membrane trafficking defect of Arn1 seen in the gga2Δ strain (Fig. 2). This Gga2-Ubm allele also led to the accumulation of Arn1 on the vacuolar membrane.
Previous studies in yeast have suggested that Gga proteins have roles in trafficking that do not depend on an interaction with ubiquitin. Recently, investigators have found that a mutant Gap1 lacking the ubiquitinated amino-terminal lysine residues traffics from the Golgi to the late endosome in a manner similar to wild type Gap1. Furthermore, substitution of endogenous Gga proteins with a mutant Gga2 lacking the GAT domain did not impair sorting of Gap1 from the Golgi to the endosome, but it did impair sorting into the MVB (31). Strains lacking Gga1, Gga2, and the AP-1 adaptor component Apl2 are inviable (45), but the gga1Δ gga2Δ apl2Δ strain expressing a yGga2/hGga3 chimera with mutations in the ubiquitin-binding residues was viable and exhibited no growth defect (22). These findings and the data presented here indicate that ubiquitin mediates the interaction between Gga2 and cargo proteins in only a specific subset of Gga-mediated trafficking steps.
Because mutations in Rsp5, in ubiquitinated lysine residues of Arn1, and in the ubiquitin-binding domain of Gga2 led to the accumulation of Arn1 on the vacuolar membrane, the ubiquitin binding activity of Gga2 is most likely required at the level of sorting Arn1 into the lumen of the MVB. Previous work supports a role for the Ggas in sorting at the MVB. Yeast Gga1 accumulates in an exaggerated late endosomal/prevacuolar compartment in a vps4 mutant, which is defective in the release of ESCRT components involved in the formation of the MVB (18). Gga1 is similarly mislocalized in a vps27 mutant, which is defective in the early steps of MVB formation (49). Moreover, human Gga3 was shown to bind directly to TSG101, a component of the ESCRT-I complex, and depletion of Gga3 led to a delay in the delivery of internalized epidermal growth factor to lysosomes, a process that depends on the sorting of epidermal growth factor into the MVB (26). Whether yeast Gga2 assists in the recruitment of ESCRT-0 or components of ESCRT-I to ubiquitinated cargo remains to be explored. Our data indicate that in yeast Gga2 is involved in two sequential steps in the trafficking of Arn1 from the TGN to the vacuole. At the TGN-to-endosome step, ubiquitination of Arn1 is not required, although at the late endosome-to-MVB step, ubiquitination of Arn1 is required for Gga2-mediated trafficking.
Role of Enth Domain, Clathrin Adaptor Proteins in Sorting of Arn1
We also observed that deletion of either Ent3 or Ent4 resulted in plasma membrane mislocalization of Arn1 that was phenotypically similar to that observed in the gga2Δ strain. Ent3 and Ent5 function as adaptor proteins at the TGN, and both have been shown to bind Gga2 and clathrin, although Ent5 also binds AP-1 (14, 44). Ent3 and Ent5 can interact directly with cargo proteins, as Ent3 binds the vacuolar SNARE Vti1 and the endosomal SNARE Pep12, and Ent5 associates with Chs3 (14, 50, 51). Cells lacking both Ent3 and Ent5 exhibit delayed processing of Cps1 and mislocalization of Chs3 and Pep12 (14, 44, 50). Localization of Ent3 to the TGN is dependent on the Gga proteins, but deletion of Ent3 and Ent5 does not affect the TGN localization of the Ggas (15). Ent3 appears to cooperate with the Ggas in the sorting of cargo at the TGN. Pep12 is transported from the TGN to the late endosome in a Gga-dependent manner, and in cross-linking experiments, Pep12 co-purified with Gga2 (14). In these experiments, deletion of ENT3 reduced the amount of Pep12 that associated with Gga2. These interactions suggest that Ent3 and Ent5 coordinate with Gga1 and Gga2 to mediate sorting of cargo proteins at the TGN. We observed that deletion of Ent3 alone could disrupt Arn1 trafficking. This, coupled with the observation that deletion of Gga2 alone disrupts Arn1 trafficking, implies that Ent3 and Gga2 may specifically cooperate in the binding of Arn1 at the TGN, and the ubiquitin-independent nature of Arn1 binding may reflect an isoform-specific function of Gga2 and Ent3.
The previously uncharacterized Ent4 was also found to affect Arn1 localization, although the mislocalization of Arn1 in the ent4Δ strain was less pronounced than in the ent3Δ strain. Although Ent3, Ent4, and Ent5 share an amino-terminal homology domain, Ent4 is shorter than Ent3 and Ent5 by 161 and 164 amino acids, respectively, and the proteins lack significant sequence homology in their carboxyl termini (43). As the clathrin binding domain of Ent1 was mapped to its carboxyl terminus, Ent4 may lack clathrin binding activity. Although Ent4 has not been identified in studies aimed at identifying binding partners for the Ggas or AP-1, our observations suggest that Ent4 may function as an adaptor protein at the TGN. Ent3 and Ent5 are also involved in the sorting of ubiquitinated cargo into the MVB, and they bind to phosphatidylinositol 3,5-bisphosphate, a phospholipid that is required for MVB sorting (16). Whether Ent3, Ent4, or Ent5 cooperate with Ggas to promote sorting into the MVB remains to be investigated.
Amino-terminal Signal Directing Arn1 from the TGN to the VPS Pathway
We found that two tripeptide signals separated by six amino acids in the amino terminus of Arn1 were required for efficient TGN to endosome sorting (Figs. 6 and 7). This THNX6YGL sequence differed from the sequence that was required for the sorting of Pep12 from the TGN to late endosomes, FSDSPEF (45). Although mutations in these two tripeptides produced mis-sorting of Arn1 that was phenotypically similar to that of the GGA2 deletion, the phenotypes were not identical. Mutation of the THN and YGL sequences resulted in mislocalization to both the plasma membrane and to the vacuolar membrane (Fig. 6, 0 FC). As Gga2 is involved in sorting at both the TGN and the MVB, these accumulations may reflect a failure of the mutant Arn1 to interact with Gga2 at both sites. The THN and YGL motifs may be binding sites for Gga2, Ent3, Ent4, or a combination of these. Similar to previous investigators, we were unable to detect a direct, specific interaction between Arn1 and Gga2. Ubiquitin-independent sorting at the TGN may involve a summation of low affinity interactions between Arn1, Gga2, Ent3, Ent4, and perhaps other unidentified adaptor proteins. Although sorting of other cargo proteins at the TGN suggests that Gga1 and -2 and Ent3 and -5 exhibit redundant functions, our data seem to suggest isoform-specific functions for Gga2, Ent3, and Ent4. This may not be the case, however. If Arn1 sorting is dependent on the simultaneous assembly of several low affinity binding partners, partial depletion of the cellular levels of one class of adaptor (Gga1/2 or Ent3/4) may shift the equilibrium of the entire complex to the unbound state and result in a failure to recruit Arn1 to a clathrin-coated vesicle.
Our work with Arn1 indicates that when this transporter reaches the TGN, competing VPS and plasma membrane sorting signals, located in the amino and carboxyl terminus, respectively, are recognized. When no FC is bound to the receptor domain of the transporter, the VPS signals predominate, and the Gga and Ent proteins bind and direct the transporter into clathrin-coated vesicles. When FC is bound to the receptor domain, the plasma membrane signals predominate, and Arn1 is sorted by an unknown mechanism into exocytic vesicles. As long as the interactions with the clathrin adaptor proteins remain weak, FC binding need only produce a small change in the affinity of Arn1 for these adaptors to disrupt their assembly. Thus, this system is poised to respond to small conformational changes in the cargo with altered trafficking patterns. It is likely that other cargo proteins can respond to changes in the intra- or extracellular environment in ways that allow the cell to fine-tune the localization and therefore the activity of integral membrane proteins.
Acknowledgment
We thank G. Payne for helpful discussions and generously sharing strains.
This work was supported, in whole or in part, by National Institutes of Health Intramural Research Programs of the NIDDK (to Y. D., Y. G., M. S.-E., and C. C. P.), the Eunice Kennedy Shriver NICHD (to H. W. and J. S. B.), and the NCI (to W.-C. A. and M. B.).
- FC
- ferrichrome
- TGN
- trans-Golgi network
- MVB
- multivesicular body
- GGA
- Golgi-localized, γ-adaptin ear-containing, Arf-binding protein
- Arf
- ADP-ribosylation factor
- GFP
- green fluorescent protein
- GAT
- Gga and Tom1
- VHS
- Vps27, Hrs, and STAM
- Ub
- ubiquitin
- VPS
- vacuolar protein sorting, BisTris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- HA
- hemagglutinin
- DIC
- differential interference contrast.
REFERENCES
- 1.Hou J. C., Pessin J. E. (2007) Curr. Opin. Cell Biol. 19,466–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takata K., Matsuzaki T., Tajika Y., Ablimit A., Hasegawa T. (2008) Histochem. Cell Biol. 130,197–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.La Fontaine S., Mercer J. F. (2007) Arch. Biochem. Biophys. 463,149–167 [DOI] [PubMed] [Google Scholar]
- 4.Magasanik B., Kaiser C. A. (2002) Gene 290,1–18 [DOI] [PubMed] [Google Scholar]
- 5.Liu X. F., Culotta V. C. (1999) J. Biol. Chem. 274,4863–4868 [DOI] [PubMed] [Google Scholar]
- 6.Stimpson H. E., Lewis M. J., Pelham H. R. (2006) EMBO J. 25,662–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Philpott C. C. (2006) Biochim. Biophys. Acta 1763,636–645 [DOI] [PubMed] [Google Scholar]
- 8.Philpott C. C., Protchenko O. (2008) Eukaryot. Cell 7,20–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erpapazoglou Z., Froissard M., Nondier I., Lesuisse E., Haguenauer-Tsapis R., Belgareh-Touze N. (2008) Traffic 9,1372–1391 [DOI] [PubMed] [Google Scholar]
- 10.Kim Y., Yun C. W., Philpott C. C. (2002) EMBO J. 21,3632–3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim Y., Lampert S. M., Philpott C. C. (2005) EMBO J. 24,952–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Matteis M. A., Luini A. (2008) Nat. Rev. Mol. Cell Biol. 9,273–284 [DOI] [PubMed] [Google Scholar]
- 13.Bonifacino J. S. (2004) Nat. Rev. Mol. Cell Biol. 5,23–32 [DOI] [PubMed] [Google Scholar]
- 14.Copic A., Starr T. L., Schekman R. (2007) Mol. Biol. Cell 18,1803–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costaguta G., Duncan M. C., Fernández G. E., Huang G. H., Payne G. S. (2006) Mol. Biol. Cell 17,3907–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eugster A., Pécheur E. I., Michel F., Winsor B., Letourneur F., Friant S. (2004) Mol. Biol. Cell 15,3031–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J., Sun H. Q., Macia E., Kirchhausen T., Watson H., Bonifacino J. S., Yin H. L. (2007) Mol. Biol. Cell 18,2646–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirst J., Lindsay M. R., Robinson M. S. (2001) Mol. Biol. Cell 12,3573–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pelham H. R. (2004) Curr. Biol. 14,R357–R359 [DOI] [PubMed] [Google Scholar]
- 20.Misra S., Puertollano R., Kato Y., Bonifacino J. S., Hurley J. H. (2002) Nature 415,933–937 [DOI] [PubMed] [Google Scholar]
- 21.Shiba T., Takatsu H., Nogi T., Matsugaki N., Kawasaki M., Igarashi N., Suzuki M., Kato R., Earnest T., Nakayama K., Wakatsuki S. (2002) Nature 415,937–941 [DOI] [PubMed] [Google Scholar]
- 22.Bilodeau P. S., Winistorfer S. C., Allaman M. M., Surendhran K., Kearney W. R., Robertson A. D., Piper R. C. (2004) J. Biol. Chem. 279,54808–54816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawasaki M., Shiba T., Shiba Y., Yamaguchi Y., Matsugaki N., Igarashi N., Suzuki M., Kato R., Kato K., Nakayama K., Wakatsuki S. (2005) Genes Cells 10,639–654 [DOI] [PubMed] [Google Scholar]
- 24.Mattera R., Puertollano R., Smith W. J., Bonifacino J. S. (2004) J. Biol. Chem. 279,31409–31418 [DOI] [PubMed] [Google Scholar]
- 25.Prag G., Lee S., Mattera R., Arighi C. N., Beach B. M., Bonifacino J. S., Hurley J. H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102,2334–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Puertollano R., Bonifacino J. S. (2004) Nat. Cell Biol. 6,244–251 [DOI] [PubMed] [Google Scholar]
- 27.Scott P. M., Bilodeau P. S., Zhdankina O., Winistorfer S. C., Hauglund M. J., Allaman M. M., Kearney W. R., Robertson A. D., Boman A. L., Piper R. C. (2004) Nat. Cell Biol. 6,252–259 [DOI] [PubMed] [Google Scholar]
- 28.Shiba Y., Katoh Y., Shiba T., Yoshino K., Takatsu H., Kobayashi H., Shin H. W., Wakatsuki S., Nakayama K. (2004) J. Biol. Chem. 279,7105–7111 [DOI] [PubMed] [Google Scholar]
- 29.Hein C., Springael J. Y., Volland C., Haguenauer-Tsapis R., André B. (1995) Mol. Microbiol. 18,77–87 [DOI] [PubMed] [Google Scholar]
- 30.Pizzirusso M., Chang A. (2004) Mol. Biol. Cell 15,2401–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lauwers E., Jacob C., André B. (2009) J. Cell Biol. 185,493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hurley J. H., Emr S. D. (2006) Annu. Rev. Biophys. Biomol. Struct. 35,277–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belgareh-Touzé N., Léon S., Erpapazoglou Z., Stawiecka-Mirota M., Urban-Grimal D., Haguenauer-Tsapis R. (2008) Biochem. Soc. Trans. 36,791–796 [DOI] [PubMed] [Google Scholar]
- 34.Kim Y., Deng Y., Philpott C. C. (2007) Mol. Biol. Cell 18,1790–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tong A. H., Evangelista M., Parsons A. B., Xu H., Bader G. D., Pagé N., Robinson M., Raghibizadeh S., Hogue C. W., Bussey H., Andrews B., Tyers M., Boone C. (2001) Science 294,2364–2368 [DOI] [PubMed] [Google Scholar]
- 36.Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Yeast 14,953–961 [DOI] [PubMed] [Google Scholar]
- 37.Sherman F. (1991) in Guide to Yeast Genetics and Molecular Biology ( Guthrie C., Fink G. eds) pp. 3–20, Academic Press, New York [Google Scholar]
- 38.Philpott C. C., Rashford J., Yamaguchi-Iwai Y., Rouault T. A., Dancis A., Klausner R. D. (1998) EMBO J. 17,5026–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore R. E., Kim Y., Philpott C. C. (2003) Proc. Natl. Acad. Sci. U.S.A. 100,5664–5669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaiser C. A., Chen E. J., Losko S. (2002) in Guide to Yeast Genetics and Molecular and Cell Biology ( Guthrie C., Fink G. R. eds) pp. 325–338, Academic Press, San Diego, CA [Google Scholar]
- 41.Heim R., Cubitt A. B., Tsien R. Y. (1995) Nature 373,663–664 [DOI] [PubMed] [Google Scholar]
- 42.Dunn R., Hicke L. (2001) Mol. Biol. Cell 12,421–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wendland B., Steece K. E., Emr S. D. (1999) EMBO J. 18,4383–4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duncan M. C., Costaguta G., Payne G. S. (2003) Nat. Cell Biol. 5,77–81 [DOI] [PubMed] [Google Scholar]
- 45.Black M. W., Pelham H. R. (2000) J. Cell Biol. 151,587–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heymann P., Ernst J. F., Winkelmann G. (2000) Biometals 13,65–72 [DOI] [PubMed] [Google Scholar]
- 47.Yun C. W., Bauler M., Moore R. E., Klebba P. E., Philpott C. C. (2001) J. Biol. Chem. 276,10218–10223 [DOI] [PubMed] [Google Scholar]
- 48.Froissard M., Belgareh-Touze N., Dias M., Buisson N., Camadro J. M., Haguenauer-Tsapis R., Lesuisse E. (2007) Traffic 8,1601–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boman A. L., Salo P. D., Hauglund M. J., Strand N. L., Rensink S. J., Zhdankina O. (2002) Mol. Biol. Cell 13,3078–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chidambaram S., Zimmermann J., von Mollard G. F. (2008) J. Cell Sci. 121,329–338 [DOI] [PubMed] [Google Scholar]
- 51.Chidambaram S., Müllers N., Wiederhold K., Haucke V., von Mollard G. F. (2004) J. Biol. Chem. 279,4175–4179 [DOI] [PubMed] [Google Scholar]
- 52.Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. (2003) Nature 425,686–691 [DOI] [PubMed] [Google Scholar]
- 53.Dell'Angelica E. C., Puertollano R., Mullins C., Aguilar R. C., Vargas J. D., Hartnell L. M., Bonifacino J. S. (2000) J. Cell Biol. 149,81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mullins C., Bonifacino J. S. (2001) Mol. Cell. Biol. 21,7981–7994 [DOI] [PMC free article] [PubMed] [Google Scholar]