Abstract
MUMmerGPU uses highly-parallel commodity graphics processing units (GPU) to accelerate the data-intensive computation of aligning next generation DNA sequence data to a reference sequence for use in diverse applications such as disease genotyping and personal genomics. MUMmerGPU 2.0 features a new stackless depth-first-search print kernel and is 13× faster than the serial CPU version of the alignment code and nearly 4× faster in total computation time than MUMmerGPU 1.0. We exhaustively examined 128 GPU data layout configurations to improve register footprint and running time and conclude higher occupancy has greater impact than reduced latency. MUMmerGPU is available open-source at http://mummergpu.sourceforge.net.
Keywords: Short read mapping, GPGPU, suffix trees, CUDA
1. Introduction
Graphics Processing Units (GPUs) were originally designed for efficient data-parallel graphics computations, such as in scene rasterization or lighting effects. However, as GPUs have become more powerful with dozens or hundreds of stream processors, researchers have begun using them for general-purpose (GPGPU) computations. Early attempts to exploit GPU’s high level of parallelism for non-graphical tasks required application developers first recast their problem into graphics primitives, and re-interpret graphical results. However, recent toolkits from both nVidia [1] and ATI [2] have enabled developers to write functions called kernels in a restricted variant of C that execute in parallel on the stream processors. High-level toolkits coupled with powerful, low cost hardware have sparked huge interest in developing GPGPU versions of data-parallel applications.
Read mapping is a data-parallel computation essential to genome re-sequencing, a rapidly growing area of research. In this computation, millions of short DNA sequences, called reads, obtained from a donor are individually aligned to a reference genome to find all locations where each read occurs in the reference sequence, with allowance for slight mismatches for biological and technical reasons. Read mapping can be used, for example, to catalog differences in one person’s genome relative to the reference human genome, or compare the genomes of model organisms such as Drosophila melanogaster (fruit fly) or Arabidopsis thaliana (thale cress). Researchers use this information for a wide variety of analyses, since even a single nucleotide difference can have a dramatic effect on health and disease. Next-generation DNA sequencing technologies from Illumina, 454 Life Sciences, and Applied Biosystems have recently become extremely popular because they can create billions of bases of sequence data in a single sequencing run at relatively low cost [3]. The DNA of James Watson, a co-discoverer of the molecule’s structure, was recently sequenced using technology from 454 Life Sciences in just two months. Biotechnology researchers believe that within the next several years, an individual will be able to have his or her DNA sequenced in only a few days and for as little as $1000. [4]. Despite their popularity, the most widely used sequence alignment programs are unable to handle the extreme workload required by the new technology. The MUMmerGPU system uses the highly parallel graphics cards from nVidia and their CUDA GPGPU toolkit to process next generation sequencing reads in a fraction of the time of other programs. [5]
MUMmerGPU 2.0 uses the same suffix tree based match kernel as described in the original version of MUMmerGPU, but we have added several significant improvements to increase performance and capabilities for the overall application. First, we implemented a new query streaming model in which reads are streamed past overlapping segments of the reference, allowing us to compute alignments to Mammalian-sized reference genomes. Second, we implemented a new GPU-based print-kernel that post-processes the tree coordinates from the match kernel into exact alignment coordinates suitable for printing. This computation had previously been the limiting factor in end-to-end application time for commonly used parameters. The print kernel performs the computation via an iterative depth-first-search on the suffix tree using a constant amount of memory and no stack. This non-traditional implementation is required to meet the severe restrictions on kernel code, but is between 1.5-and 4-fold faster than the previous (CPU-based) version of the routine. Popov et al recently reported a different algorithm for traversing trees in a CUDA kernel [6] which requires additional pointers between the leaf nodes in a kd-tree, but our technique is applicable to any tree without additional pointers. Finally, we optimized performance for both kernels by identifying the best organization of the DNA sequencing reads and suffix tree in GPU memory. We explore and report on 128 variations of the data layout policy, and quantify the tradeoffs involved for kernel complexity, cache use, and data placement. We find that optimizing these choices can greatly accelerate performance, and mistuned choices have an equal but negative effect on performance compared to the naïve version. Processor occupancy dominated performance for our data-intensive application, but techniques that reduce GPU memory latency without compromising occupancy were also generally beneficial. We describe several techniques to reduce kernel register footprint and thus improve occupancy that are widely applicable to GPGPU programs. Overall, MUMmerGPU 2.0 is nearly 4× faster in total computation time than the originally published version of the code for the most commonly encountered workloads.
1.1 GPGPU Programming
Recent GPUs from nVidia have up to 256 stream processors running at a core frequency of up to 650 MHz. [7] Each stream processor has an individual arithmetic logic unit (ALU), but the stream processors are grouped into multiprocessors such that all of the stream processors in the same multiprocessor execute the same instruction at the same time (SIMD architecture). The functions that execute on the stream processors are called kernels, and a single instance of a running kernel is called a thread. Threads are launched in groups of 32 called warps that the multiprocessor uses for scheduling, and are further organized in larger groups called thread blocks of user specified size with the guarantee that all threads in the same thread block will execute concurrently. A GPU has up to 1.5 GB of on-board memory, but very small data caches compared to general purpose CPUs (only 8KB per multiprocessor). Cached memory is only available for read-only data and for a small number of word-aligned data types called textures. Non-cached memory has very high latency (400 to 600 clock cycles), but multiprocessors attempt to hide this latency by switching between warps as they stall.
Kernel code is written in a restricted variant of C and compiled to GPU specific machine code using the CUDA compiler, NVCC. Developing kernel code can be challenging because commonly used programming features, such as dynamic memory allocation and recursion, are not available. Loops and conditionals are allowed in kernel code, but if different threads in the same warp follow different branches, then the multiprocessor will automatically serialize or stall execution until the threads resynchronize, thus cutting effective parallelism and end-to-end application performance. Furthermore, each multiprocessor has a fixed number of registers available for its stream processors, so the number of threads that can execute concurrently is determined in part by how many registers each thread requires. The percent of stream processors in a multiprocessor that execute concurrently, processor occupancy, is available in discrete levels depending on the number of registers used by each thread, the thread block size, and the physical characteristics of the device including the number of registers present on each multiprocessor, the maximum number of concurrent warps, and the maximum number of concurrent thread blocks Threads are executed in discrete units of the thread block size such that the total number of registers used by all concurrent threads is at most the number available on the device. For example, an nVidia 8800 GTX has 8192 registers per multiprocessor, and can execute at most 8 concurrent thread blocks per multiprocessor and at most 24 concurrent warps of 32 threads per multiprocessor (a maximum of 768 concurrent threads total). If the thread block size is 256 a kernel will have 100% occupancy if it uses at most 10 registers (allowing 3 complete thread blocks), 66% occupancy for at most 16 registers (allowing 2 complete thread blocks), 33% occupancy for at most 32 registers (allowing 1 complete thread block), and fail to launch if each thread requires more than 32 registers because one thread block would require more than 8192 registers Finally, kernel code cannot directly address main memory nor other devices, so inputs to the kernel must be copied to the GPU’s on-board memory prior to execution and outputs must be copied to main memory from on-board memory after execution. The full details of the device capabilities and programming model are described in the CUDA documentation. [1]
GPU accelerated versions of data parallel-applications have been developed for numerous application domains, including molecular dynamics, numerical analysis, meteorology, astrophysics, cryptography, and computational biology. [8–11] The most successful GPGPU applications have generally had high arithmetic intensity, meaning processing time is dominated by arithmetic operations with relatively few memory requests. These applications are well suited to the numerical capabilities of the stream processors. In contrast, data intensive applications requiring fast random access to large data sets have been generally less successful on the GPU, because of the GPUs small data caches and relatively high latency (400–600 clock cycles) for on-board memory accesses.
1.2 DNA Sequence Alignment
DNA is the molecule that encodes the genetic blueprint for the development and traits of an organism. It is composed of a long sequence of four possible nucelotides or ‘base pairs’ (bp): adenine (abbreviated A), cytosine (C), guanine (G) and thymine (T). The sequence of base pairs in biologically active regions called genes determines the amino acid sequence and function of biologically active molecules called proteins. Even a single nucleotide difference in a gene between two individuals can substantially change the function of its protein product and lead to disease. Larger insertion, deletion, or rearrangement events of several nucleotides can have profound effect on development, such as the chromosomal duplication responsible for Down syndrome. Numerous other human diseases and traits have been linked to both small-scale single nucleotide polymorphisms and larger genetic variations, and thus make DNA sequence analysis an extremely active and important field of research. [12]
Until recently, the most widely used protocol for determining the sequence of nucleotides in a genome used Sanger sequencing. Sanger sequencing can determine the order of ~1000 consecutive nucleotides by separating fluorescently tagged molecules based on their charge; each sequence fragment is called a “read.” Longer regions, including full genomes, are sequenced by sequencing random overlapping fragments, and then stitching the reads together computationally into the original full-length sequence. [13] New DNA sequencing protocols from Illumina, 454 Life Sciences, and Applied Biosystems sequence DNA at a much higher rate and dramatically lower cost, but the reads are significantly shorter (30–200bp). Nevertheless, there has been a dramatic shift towards using the cheaper sequencing protocols and placing the burden on computational resources to analyze the result with less information per read.
One of the most widely performed DNA analysis tasks is to align a pair of sequences to find regions that are similar. In the case of short sequencing reads, researchers will generally require that the entire read aligns end-to-end to a reference sequence except for a small number of differences, which may be real polymorphisms or sequencing errors. Modern sequence alignment algorithms use a technique called seed-and-extend to quickly perform the alignment by focusing the search to regions that are reasonably similar. In the first phase, the algorithms find substrings of sufficient length called seeds that are shared between the sequences. In the second phase, the algorithms extend the relatively short exact seeds into longer inexact alignments using a more sensitive dynamic programming algorithm. The widely used BLAST algorithm considers all possible fixed-length substrings called k-mers as seeds. [14] In contrast, the popular MUMmer algorithm and our high-performance variant MUMmerGPU compute variable length maximal exact matches (MEMs) as seeds for alignment. Both algorithms are much faster than using the original Smith-Waterman local sequence alignment algorithm, which requires time that is quadratic with respect to the input sequence sizes. By contrast, MUMmer and MUMmerGPU find MEMs using a suffix tree, which requires linear space and enables substring matching in linear time. [5, 15, 16] MUMmerGPU uses a very similar output format as MUMmer, and thus one can reuse MUMmer’s components for extending the exact seeds into longer inexact alignments.
A suffix tree is a tree that encodes all suffixes of a string on a path from the root node to a leaf. A special character that does not occur in the original string ($) is appended to the reference string to ensure that each suffix ends at a unique leaf node, which is labeled by the starting position of the suffix called the leaf id. Edges of the tree are labeled with substrings of the reference, and internal nodes have at least 2 children representing positions where repeated suffix prefixes diverge. The path string of a node is the concatenation the edge labels along the path from the root to that node. The string depth of a node is the length of its path string. Suffix trees over fixed alphabets, such as for DNA nucleotides, can be constructed in linear time and space using additional pointers called suffix links, that point from node n with path string x∂ to node n′ with path string ∂, where x is a single character and ∂ is a string. [17] Once built, a suffix tree allows one to find occurrences of a query string or substrings of a query string in the reference string in time proportional to the length of the query substring by matching characters of the query along the edges of the tree. Substring matches can be extended into MEMs by walking the suffix tree along the path of the substring matches as described below. For a complete description of suffix tree construction and search algorithms see the comprehensive reference by Gusfield. [18]
2. Alignment Algorithm
MUMmerGPU computes all MEMs that are at least the minimum match length (the parameter l) characters long between a reference sequence and a set of query sequences. The MEM computation is divided into four phases:
Reference Preprocessing – Load the reference from disk and construct a suffix tree of it.
Query Streaming – Load blocks of queries from disk and launch the alignment kernels.
Match Kernel – Match each suffix of each query to the suffix tree to find candidate MEMs.
Print Kernel – Post-processes the candidate MEMs to report all MEMs at least l characters long.
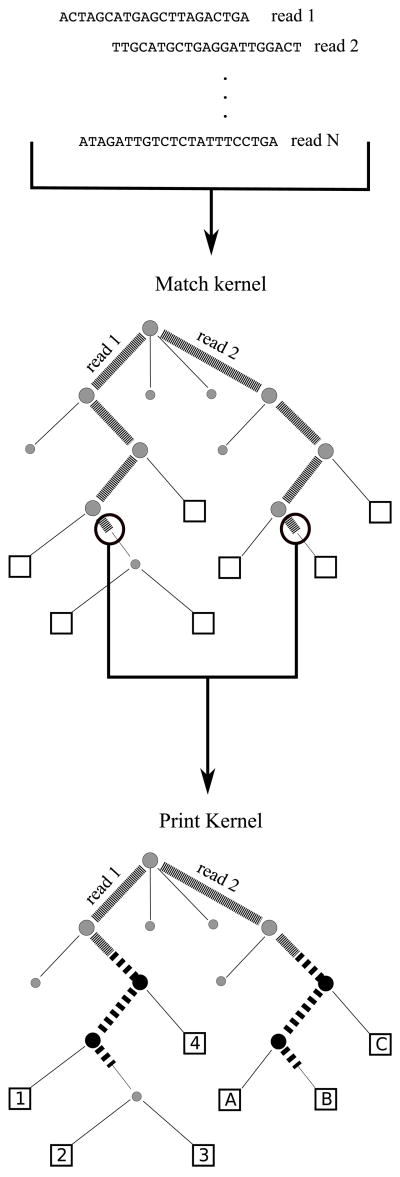
Both the match kernel and the print kernel are executed in parallel on the graphics card, as illustrated in Figure 2. A separate GPU thread running the match kernel processes each query. Then for each matching suffix of each query, a separate instance of the print kernel reports MEMs for that suffix. Suffix tree construction and I/O are executed serially on the CPU and require a small fraction of the overall runtime for large read sets.
Figure 2.
MUMmerGPU constructs suffix trees for overlapping sections of the reference string. Reads are first matched to the suffix tree in the match kernel. Tree coordinates are passed from the match kernel to the print kernel to convert tree coordinates to alignment coordinates in the reference string.
2.1 Reference Preprocessing
Since the reference sequence may be very large, the reference is divided into overlapping 8Mbp segments called pages. For each reference page, the algorithm constructs a suffix tree using Ukkonen’s algorithm in linear time and flattens the tree into a large array suitable for processing on the GPU. Suffix tree construction time is generally a small fraction of the total runtime for typical datasets involving millions of reads. [5] Each suffix tree node requires 32 bytes of data, which is divided into two 16 byte structs called the node and children structs. The node struct contains the coordinates of the reference string for the edge label into that node, the string depth of the node, and the address of the parent and suffix nodes. The children struct contains the address of each of the five children (A,C,G,T,$) and a flag indicating if the node is a leaf. If the node is a leaf, then the leaf id and the character of the reference just prior to that suffix of the reference is stored instead of the children pointers. Node addresses are stored using 24 bit addresses to conserve space but limits the suffix tree to 16 million nodes, and the maximum page size to 8 million base pairs. The nodes of the tree are reordered using the previously described reordering scheme. [5] Briefly, nodes near the top of the tree are numbered according to a breath-first-traversal to maximize locality across threads, while nodes at depth ≥ 16 are assigned using a depth-first-traversal to maximize locality for a particular thread.
2.2 Query Streaming
Unlike previous versions of the code, which processed queries in memory across all reference pages, queries are streamed across the reference, meaning after a query is aligned to a reference page, the alignments are printed immediately and the query is flushed from memory. If the reference is larger than the page size, then it will be necessary to reload the queries multiple times from disk. This tradeoff was necessary to support aligning against very large reference sequence or aligning a large set of reads, either of which required a prohibitively large amount of host RAM in the previous version of the code.
2.3 Match Kernel
The match kernel is essentially the same as described in previous version of the code. Briefly, the kernel finds the longest matching substring of the query starting at each position of the query, i.e. each suffix of the query is considered. Starting with the first character of the query (i=1) and the root node of the reference suffix tree, the characters in the query are matched to the edges of the suffix tree one character at a time until a mismatch or the end of the query is reached. If the number of matching characters is at least l, the match is recorded in the output buffer for position i as the id of the lowest node visited and the length along the edge to that node. The next suffix of the query is then considered by following the suffix link from the parent of lowest node reached. This has effect of removing the first base of the query from consideration, and allows the next suffix to be evaluated without returning to root in the suffix tree.
2.4 Print Kernel
The print kernel post-processes the match kernel results into potentially many MEMs per match (see Figure 3). The match kernel reports the lowest node L in the tree that matches the ith suffix of the query. If the query match to L is longer than l, there are multiple substrings of ith suffix that match the reference and are at least l characters. Call node P the highest ancestor of L that has a string depth at least l characters long. The leaves of the subtree rooted at P determine where in the reference a substring starting at i occurs, and the string depth of the lowest common ancestor of those leaves and L determines the matching substring length. Because the match kernel reports the longest possible match for suffix i, all of the matches at the leaves are guaranteed to be right maximal. However, the print kernel must be careful to not report matches that are fully contained by matches to suffix i−1. That is, the raw matches for suffix i may not be left maximal so the left flanking base must be explicitly checked by comparing the i−1th character of the query to the corresponding character of the reference. The print kernel computes this check for all candidate MEMs via a depth-first-search of the suffix tree to all of the leaves in the subtree rooted at P.
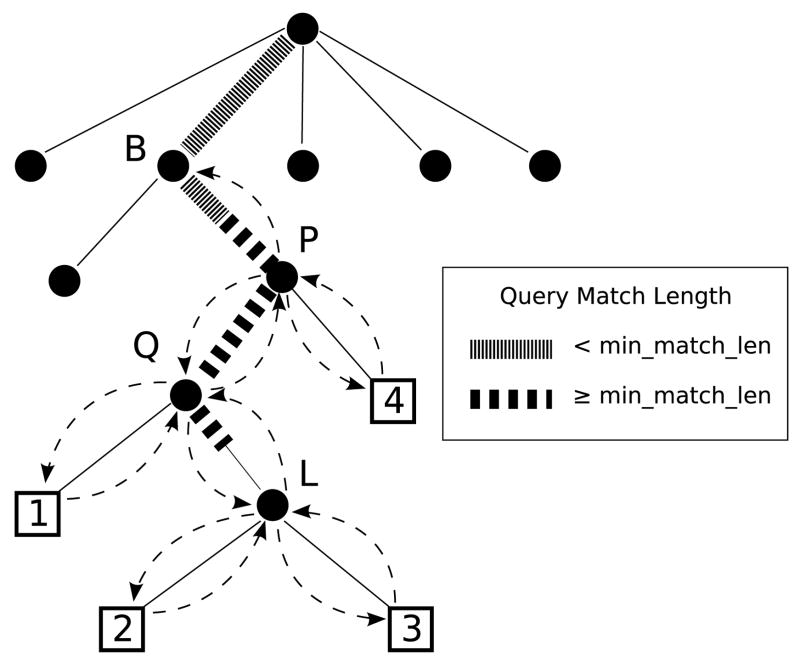
Figure 3.
The query matches to the suffix tree along the bold path from the root to L. The print kernel post-processes this information to consider MEMs at the leaves by a stackless depth-first-search starting at P to leaves 1,2,3 & 4 shown by dashed arrows. The match length at leaf 1 is the string depth of Q, the match length at 2 and 3 is the string depth of Q plus the partial edge match to L, and the match length at leaf 4 is the string depth of P.
The algorithm begins by following parent pointers from L to find node P by following the parent pointer stored in each node, and stopping when the string depth field is < l. It also finds the parent of P called node B. Starting at P, it attempts to traverse to the A child. If the A child is null, it tries the C child and so forth in lexicographical order. It proceeds down the tree in this way to the first (lexicographically smallest) leaf below P where the MEM criterion is evaluated by comparing the i−1th character of the query to the corresponding character in the reference string. This character is the character in the reference that is just before the suffix ending at that leaf, and is stored in the leaf node for efficiency. If the characters are different, the substring is a MEM and the coordinates of the substring are stored to the output buffer, as explained below. After processing the first leaf, the kernel traverses up to the parent of the leaf, and resumes processing with the lexicographically next child. Because leaves are always visited in lexicographic order and because the last child visited can be determined with a pointer comparison, the algorithm does not require a stack to determine where to search next. After processing the $ child, the kernel traverses up the tree and continues processing lexicographically. The algorithm ends when the current node is B.
The coordinates of a MEM depend on the leaf id of the leaf for the start position, and the location of the leaf in the suffix tree relative to L, for the length. The length of the MEM is the string depth of the lowest common ancestor of the leaf node and L in all cases except for the leaves below L because the query may only have a partial match along the edge to L. Call the path of nodes between the parent of L and P the query path. Note the lowest common ancestor of a visited leaf must fall along the query path. When the traversal algorithm begins at P, the substring length is the string depth of P, and by definition P is along the query path. When traversing down the tree and the current node is along the query path, the algorithm checks if the child node is also on the query path. Call the character of the query at the string depth of the current node the query character. The query character determines which child of the current node is also in the query path. If the next node is along the query path, the matching substring length is the string depth of that node. If the next node is not on the query path, the matching substring length is not updated. Instead the kernel records and updates the distance to the query path throughout the traversal. When the distance returns to 0, the current node is once again in the query path and the algorithm resumes checking for the query child as before. Since there may be only a partial match to node L, a special condition checks when this node is visited, and the match length is set to the string depth of L’s parent plus the partial edge match length reported by the match kernel.
3. Data Policies
MUMmerGPU 1.0 organized data on the GPU according to the few “best practices” that existed at the time. However, those best practices, such as whether or not to use texture memory, were developed for arithmetically intensive applications. In MUMmerGPU 2.0, we have revisited our decisions for seven possible boolean data organization policies and exhaustively tested all 128 possible combinations of choices. The policy choices are as follows.
Two-dimensional reference – store the reference string in a two-dimensional array instead of in linear memory.
Query texture – store the query strings in texture memory instead of global memory.
Reference texture – store the reference string in texture memory instead of global memory.
Tree texture – store the tree in a pair of textures instead of global memory.
Two-dimensional tree – store the tree in two-dimensional arrays instead of linear memory.
Tree reordering – reorder the nodes of the tree to improve locality instead of the node numbering determined by the construction algorithm.
Merged tree – for a given node, store the node and children structs adjacent in memory, instead of two parallel arrays.
MUMmerGPU 1.0 stored the reference in a two-dimensional texture, the queries in linear global memory, and the tree in parallel two-dimensional textures after reordering the nodes. The texture cache in G80 series GPUs is described as being optimized for two-dimensional locality, so the node reordering was designed to exploit a two-dimensional cache block. Textures were selected for the reference and tree after preliminary testing suggested this selection had better performance. The tree structs were placed in parallel arrays to simplify addressing. In the following discussion, the naïve control configuration disables all optimization: no query texture, no reference texture, 1D reference, no tree texture, 1D tree, no tree reordering, and parallel arrays for the node and child structs.
We evaluated MUMmerGPU under each of the 128 possible combinations of policy choices on several workloads. Each workload constitutes a small slice of the input a life sciences researcher would provide to MUMmerGPU when mapping reads to a reference genome. The first workload, HSILL, represents a human resequencing project using next generation Illumina technology. The second workload, CBRIGG, represents a large eukaryotic resequencing project using traditional Sanger sequencing technology. The last two workloads, which we call SSUIS and LMONO, represent typical inputs for resequencing bacteria using next generation sequencing technologies from Illumina and 454 Life Sciences. All four workloads are comprised of genuine (non-simulated) sequencing reads, and is large enough to constitute a representative slice of work from the project, but terminate quickly enough to permit testing all 128 configurations. Table 1 presents the additional details about the reference sequences and read sets for each workload.
Table 1.
Description of the workloads used for testing.
| Workload/Organism | Reference size (bp) | Minimum match Length (bp) | Read type | # of reads |
|---|---|---|---|---|
| HSILL H. sapiens | 16,000,000 | 14 | Illumina (29bp) | 500,000 |
| CBRIGG C. briggsae | 13,000,000 | 100 | Sanger (~700bp) | 500,000 |
| LMONO L. monocytogenes | 2,944,528 | 20 | 454 (~120bp) | 1,000,000 |
| SSUIS S. suis | 2,007,491 | 10 | Illumina (36bp) | 1,000,000 |
In a resequencing project, reads from a donor organism are aligned to a reference genome. Errors in the sequencing reads along with genuine variations between the donor and the reference genome will prevent some reads from aligning end-to-end without error. MUMmerGPU allows users to control the amount of error to tolerate by allowing users to specify the minimum match length (l) to report. The full details for choosing a proper value for l are beyond the scope of this paper, but the choice of l can have a dramatic impact the running time, including determining which CUDA kernel dominates the computation, since smaller values of l produce more MEMs for the print kernel to report. For HSILL, we have chosen l to allow at most 1 difference in an alignment between a read and the reference, 2 differences for SSUIS, and numerous differences for LMONO and CBRIGG.
4. Policy Analysis
For MUMmerGPU 2.0, we looked for a set of policy choices that universally reduced running time, as opposed to workload specific improvement. Ideally, a single configuration would be optimal for all workloads, otherwise, we desired configurations that improve HSILL, since we expect human resequencing projects using short reads will constitute the majority of the read alignment workloads in the near future. To this end, we executed MUMmerGPU with all 128 possible policy combinations on all 4 workloads. The test machine was a 3.0 GHz dual-core Intel Xeon 5160 with 2 GB of RAM, running Red Hat Enterprise Linux release 4 update 5 (32 bit). The GPU was an nVidia GeForce 8800 GTX, using CUDA 1.1. The 8800 GTX has 16 multiprocessors, each with 8 stream processors (128 stream processors total), and 768 MB of on-board RAM, with 8 KB of texture cache per multiprocessor. The data that follows is for the HSILL workload and excludes time spent reading from or writing to disk, as this time was identical within the workload, and only obscures the impact of different policy choices. In the figures, we have isolated the policy choice in question, and each bar represents the percent change in running time for enabling that policy while keeping the policy configuration otherwise the same. A positive value indicates the running time increased after enabling that policy, and negative values indicate the running times decreased. The bars are clustered by which textures are enabled, and are labeled by their non-texture policy choices. The label control indicates the default configuration without any non-texture policies enabled. The full results table for all workloads is available online at http://mummergpu.sourceforge.net.
4.1. Two-dimensional reference
Storing the reference string in a two-dimensional layout instead of a one-dimensional string consistently increased the total computation time, but only by an insignificant. 4% on average (data not shown). We suspect the extra instructions necessary for addressing 2D memory slowed the overall performance relative to any potential gains by 2D locality. Consequently, only configurations that use a one-dimensional layout for the reference string were considered further. The following sections describe the remaining 64 policy configurations.
4.2. Query Texture
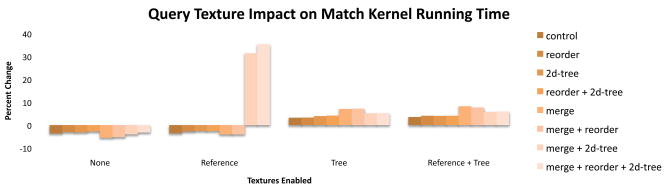
Configurations that placed the queries in a texture increased the print kernel time by 6.% and the match kernel time by 3% on average. When the tree texture is not used, the match runtime consistently decreased except in 2 exception cases with dramatically increased running time due to increased register footprint and decreased occupancy (Figure 4). When the tree texture is enabled, we observe what appears to be a small amount of cache competition in the match kernel which tends to slow down those configurations. The print kernel had similar results.
Figure 4.
Using the query texture with the match kernel generally improves the runtime, except when there is cache competition from the tree texture.
4.3. Reference texture
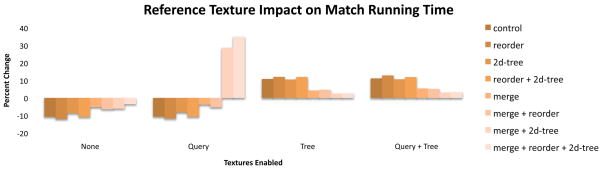
Configurations that placed the reference string in texture memory instead of global memory had significantly different match kernel running times (−12% to +35% change), but had essentially identical print kernel running times. This is as expected, since the print kernel does not access the reference string. Placing the reference string in a texture improved running time for the match kernel by up to 12% (without changing register usage), but only when the tree was not in texture memory as well (Figure 5). We speculate that the tree, queries, and reference negatively compete for the texture cache in those cases, leading to overall lower performance. The two large increases (35%) in match kernel running time observed when the query texture was also used is a result of an increase in register usage and corresponding decrease in processor occupancy.
Figure 5.
The reference string competes with the suffix tree for the texture cache. Configurations only benefit from placing the reference in texture memory when the tree is not also in texture memory.
4.4. Suffix tree texture
Storing the tree in a texture instead of global memory improves print kernel performance in almost all configurations and by 8% on average. The impact of this policy is more complicated for the match kernel (Figure 6). On average, using a texture for the tree improves match kernel performance by 11%, presumably because the cache lowers effective memory latency. In some configurations, though, the tree competes for the cache with the queries or reference and those configurations are generally slower than the equivalent configuration that uses global memory for the tree.
Figure 6.
Placing the suffix tree in texture memory is not universally beneficial, presumably due to cache competition in some configurations.
Interestingly, cache competition does not always result in an overall slowdown, especially when the register footprint was improved. In the match kernel, two configurations with multiple data types in texture requires 18 registers, yielding 33% occupancy. However, if these configurations are altered such that the tree is placed in a texture, the match kernel requires only 16 registers, achieving 66% occupancy and an overall speedup. In the print kernel, the control configuration uses 17 registers, and only 16 registers when the tree is placed in a texture and thus has improved occupancy and cache use. We also observed the opposite effect: for some configurations, placing the tree in a texture increased the print kernel register footprint, dropped occupancy, and slowed the overall computation.
4.5. Two-dimensional tree
Configurations that placed the suffix tree in two-dimensional arrays were on average 15% slower than the configurations that used one-dimensional arrays for total computation time (Figure 7). Placing the tree in a texture appears to mitigate some of the negative impact of using a two-dimensional array for the tree, but not using the 2D layout was faster overall. In addition, some configurations using a two-dimensional tree array increased the register footprint in the print kernel across the threshold from 66% to 33% occupancy and had drastically reduced performance (52% worse).
Figure 7.
Placing the suffix tree in a two-dimensional array universally slows overall computation time.
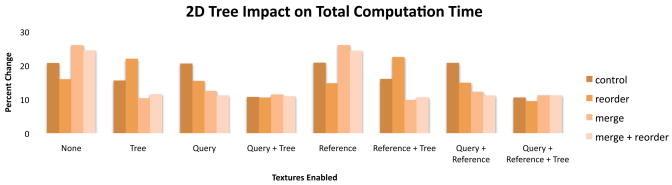
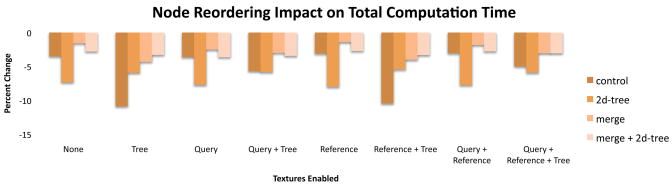
4.6. Tree reordering
In HSILL, configurations that reordered the suffix tree nodes in the GPU memory run significantly and universally faster than the equivalent configurations that do not (between 1% and 11% faster, 5% on average) (Figure 8). This is perhaps the most surprising finding from our benchmark tests, since the reordering is only supposed to improve running time for configurations that use (cached) texture memory for the suffix tree. Furthermore, the node reordering is entirely performed on the CPU, and the GPU kernels are bit-for-bit identical when reordering is enabled over the equivalent configuration with the reordering disabled. However, the actual number of instructions executed by a multiprocessor can vary between invocations based on the access pattern of the kernel. A G80 multiprocessor may serialize execution of threads in a warp if the memory accesses made by threads have significantly different latencies. nVidia offers a profiler that counts events during a kernel execution such as the number of global memory loads and the number of instructions executed. For HSILL, profiling shows a decrease in total instruction count and in the number of instructions due to warp serialization when reordering is enabled.
Figure 8.
Reordering the tree nodes in GPU memory improves running time not only for configurations that place the tree in cached texture memory, but all configurations.
4.7. Merged tree
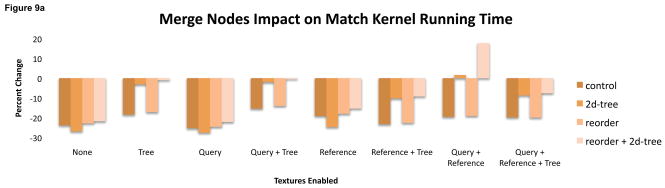
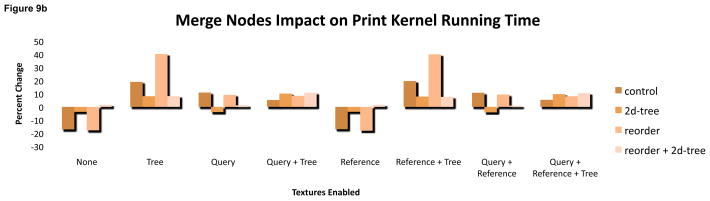
Merging the two suffix tree arrays into a single array places the two halves of a single tree node adjacent in GPU memory. This policy was originally conceived to exploit a common access pattern in the match kernel where the halves are sequentially accessed. This should have improved cache performance from the increased spatial locality. However, merging the arrays also required slightly more complicated kernel code for addressing nodes. This had a more significant impact by altering the register footprint of both the match and print kernel.
In the print kernel, using a merged array increased running time by 6% on average, including some extreme changes in caused by increasing or decreasing the register footprint. (Figure 9). Configurations that placed the suffix tree in a texture or used a two-dimensional array generally suffered when using a merged array. The other configurations saw a reduced footprint, and for a few configurations that reduction boosted the occupancy to 66%. The impact on match kernel time was less dramatic though occupancy differed for some configurations when merged arrays were enabled.
Figure 9.
(top) The print kernel’s register footprint (y axis) depends on whether a configuration uses merged arrays for the suffix tree. In some configurations, only 16 registers are used, increasing occupancy to 66% and dramatically reducing running time (bottom).
4.8 Comparison to MUMmerGPU 1.0
Based on the above discussion, the new default policy configuration in MUMmerGPU uses a reordered one-dimensional texture for the suffix tree, global linear memory for the queries and reference, and splits the tree into parallel arrays. This configuration is optimal for HSILL and creates a nearly four-fold speedup in total GPU compute time over MUMmerGPU 1.0. For other workloads, it is not optimal, but this configuration consistently outperforms MUMmerGPU 1.0. For example, in LMONO, CBRIGG and SSUIS, reordering suffix tree nodes generally degrades performance, although a few configurations enjoy a modest speedup. Surprisingly, none of the configurations with increased performance placed the tree in texture memory. In general, the impact of reordering appears very sensitive to the specific access pattern of the kernels for a given input and choice of parameters. However, in all workloads, the new configuration speeds up the match kernel by at least 20%, and the new print kernel is between 1.5× and 4× faster than the CPU based print procedure of MUMmerGPU 1.0 (Table 2). To reach as broad a user base as possible, MUMmerGPU also implements tuned, optimized versions of the matching and print procedures that run on the CPU. For HSILL, MUMmerGPU 2.0’s GPU kernels run 13-fold faster than these CPU-based routines.
Table 2.
Comparison of the MUMmerGPU 1.0 and 2.0 runtimes.
| MUMmerGPU 1.0 | MUMmerGPU 2.0 | MUMmerGPU 2.0 vs. 1.0 | ||||
|---|---|---|---|---|---|---|
| Workload | Match time (ms) | Print time (ms) | Match time (ms) | Print time (ms) | Match speedup | Print speedup |
| HSILL | 677 | 56911 | 540 | 14379 | 1.25 | 3.96 |
| LMONO | 6822 | 6518. | 5693 | 4268 | 1.20 | 1.53 |
| CBRIGG | 26419 | 9977 | 22066 | 4640 | 1.20 | 2.15 |
| SSUIS | 1180 | 26265 | 968 | 9383 | 1.22 | 2.80 |
5. Discussion
Our exhaustive policy analysis shows occupancy is the single most important factor for the performance of data-intensive applications. This is because higher occupancy allows for more threads to be executed concurrently. Higher occupancy has the added benefit that memory latency can be better hidden with more threads. As such we attempted to improve occupancy of all configurations by reducing their register footprint. In several configurations, we successfully reduced register use to reach 66% occupancy by making small adjustments to the kernel code, such as moving variable declarations to the tightest possible scope, and using bit masks instead of named fields within structs. We also used more aggressive techniques such as using goto’s to intentionally disable some compiler optimizations with some success. The CUDA compiler NVCC is actively being developed, and as its register allocation and optimization routines improve, it should be easier to achieve higher occupancy.
Our analysis finds proper use of the texture cache is also critical. Haphazardly placing data in texture memory in the hopes of reducing latency is dangerous. The texture cache is limited to 8KB per multiprocessor, so cache competition can easily hurt overall performance. Furthermore, using textures instead of global memory can reduce occupancy and slow execution. Conversely, proper use of cache that places the most important data in textures can greatly improve performance.
Data reordering can also greatly improve performance. Our results show that it universally improved performance when aligning short reads to a large reference, which we expect to be the most common read mapping use case. For this type of workload, data reordering unexpectedly reduced divergence and warp serialization. Normally used to increase locality and reduce latency, data reordering also is a promising avenue for reducing thread divergence. We plan to explore other reordering strategies in future versions of MUMmerGPU. However, reordering should be used with caution and careful measurement. In MUMmerGPU, it was used to improve cache hit rate in two-dimensional textures, but storing the tree in two-dimensions turned out to be a universally bad choice, despite the claims that the texture cache is optimized for 2D locality. Similarly, merging the node and children halves into the same array was supposed to improve data locality and thus cache performance, but this improvement was lost in most configurations to increased register footprint and reduced occupancy from the more complicated addressing.
These conclusions reflect properties and design decisions concerning the current nVidia graphics processing line, and may fail to hold in the future as the hardware and CUDA evolve. A policy analysis such as the one presented here can help identify high performance policy configurations, and can help “future-proof” an application against rapidly evolving hardware. Ideally, MUMmerGPU would be able to self-tune its policies for an individual system, and we are considering such functionality. In the short term, MUMmerGPU’s instrumentation and alternate policy implementations remains in the code, so the application can adapt to new nVidia hardware and CUDA versions as they appear.
6. Conclusions
MUMmerGPU 2.0 is a significant advance over MUMmerGPU 1.0, featuring improved functionality and higher performance over previous versions of the code. With the new query streaming data model, MUMmerGPU 2.0 can map reads to genomes as large or larger than the human genome. The new GPU-based print kernel post-processes the suffix tree matches into full MEMs, and provides a major performance boost, between 1.5× and 4×, over the serial CPU version in MUMmerGPU 1.0. This kernel required a non-traditional stackless implementation of a depth-first-search of the suffix tree. Its tree-walking technique is applicable to essentially any common tree data structure, and thus we expect many data processing tasks could benefit from running on the GPU. In the future, we plan to extend MUMmerGPU with a second post-processing GPU kernel that computes inexact alignments.
Both the match and print kernels benefited from our exhaustive analysis of seven data organization policies. The impact of individual policies is often surprising and counterintuitive, and we encourage other GPGPU developers to carefully measure their applications when making such decisions. Our analysis shows occupancy is the main determining factor of data-intensive kernel performance. We are optimistic that new versions of the CUDA compiler will simplify reaching high occupancy, but currently, it is imperative that developers monitor their register footprint, and reduce it when possible. The next most important factor is proper use of the textures. The texture cache is very small, and haphazard use of textures will quickly overwhelm it. Instead, applications should only use textures for the most important data. Data reordering can be used to improve locality and cache hit rate, but since different workloads may have different access patterns, developers should select a reordering that is appropriate for the most common workload. Reordering also affects thread divergence, and we recommend that developers consider reordering strategies that reduce divergence, even when not using cached memory.
Data-intensive applications are believed to be less well suited than arithmetic-intensive applications. Nevertheless, our highly data-intensive application MUMmerGPU achieves significant speedup over the serial CPU-based application. A large part of this speedup is due to tuning techniques that may be used in any GPGPU application. The enormous volume of sequencing reads produced by next generation sequencing technologies demands new computational methods. Our software enables individual life science researchers to analyze genetic variations using the supercomputer hidden within their desktop computer.
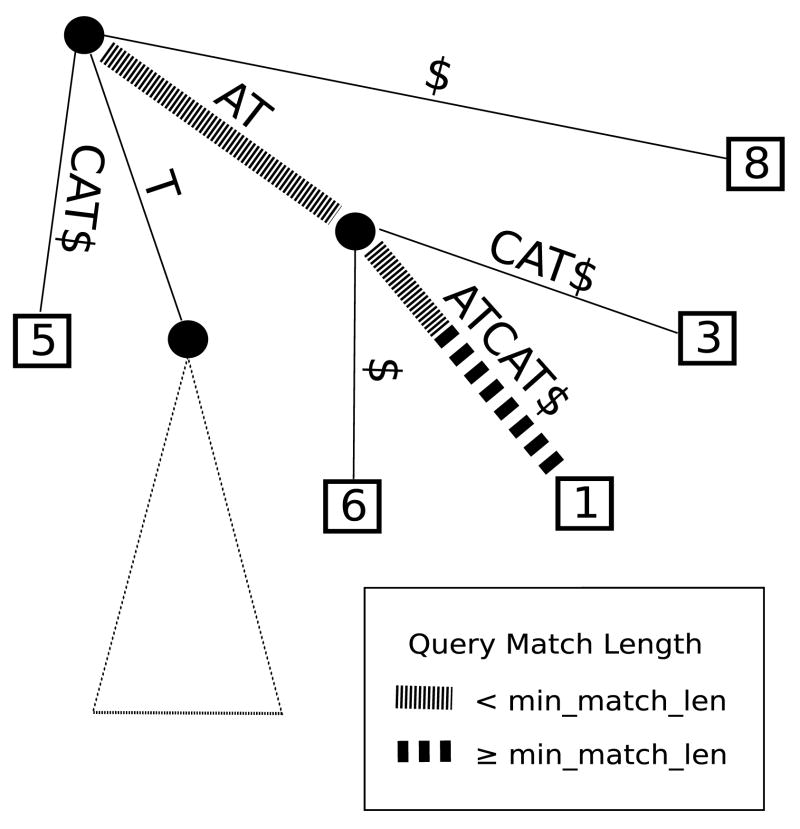
Figure 1.
Aligning the query “ATAT” against the suffix tree for “ATATCAT”. MUMmerGPU will report a match at position 1 in the reference, provided that the minimum match length l ≥ 4.
Acknowledgments
The authors would like to thank Steven Salzberg from the University of Maryland for helpful comments on the manuscript. This work was supported in part by National Institutes of Health grants R01-LM006845 and R01-GM083873.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Cole Trapnell, Email: cole@cs.umd.edu.
Michael C. Schatz, Email: mschatz@umiacs.umd.edu.
References
- 1.nVidia. nVidia Compute Unified Device Architecture (CUDA) Programming Guide, version 1.0. 2007. [Google Scholar]
- 2.AMD. ATI CTM Guide, Technical Reference Manual. [Google Scholar]
- 3.Shaffer C. Next-generation sequencing outpaces expectations. Nat Biotechnol. 2007;25(2):149. doi: 10.1038/nbt0207-149. [DOI] [PubMed] [Google Scholar]
- 4.Chi KR. The year of sequencing. Nat Meth. 2008;5(1):11–14. doi: 10.1038/nmeth1154. [DOI] [PubMed] [Google Scholar]
- 5.Schatz M, et al. High-throughput sequence alignment using Graphics Processing Units. BMC Bioinformatics. 2007;8(1):474. doi: 10.1186/1471-2105-8-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Popov S, et al. Stackless KD-Tree Traversal for High Performance GPU Ray Tracing. Computer Graphics Forum. 2007;26(3):415–424. [Google Scholar]
- 7.nVidia. NVIDIA GeForce 9 Series. 2008. [Google Scholar]
- 8.John Michalakes MV. GPU Acceleration of Numerical Weather Prediction. Workshop on Large Scale Parallel Processing, IEEE International Parellel & Distributed Processing Symposium; 2008. [Google Scholar]
- 9.Stone JE, et al. Accelerating molecular modeling applications with graphics processors. Journal of Computational Chemistry. 2007;28(16):2618–2640. doi: 10.1002/jcc.20829. [DOI] [PubMed] [Google Scholar]
- 10.Liu W, et al. Bio-Sequence Database Scanning on a GPU; 20th IEEE International Parallel & Distributed Processing Symposium (IPDPS 2006) (HICOMB Workshop); Rhode Island, Greece. 2006. [Google Scholar]
- 11.Manavski SA. Cuda compatible GPU as an efficient hardware accelerator for AES cryptography. Proceedings IEEE International Conference on Signal Processing and Communication, ICSPC 2007; 2008. pp. 65–88. [Google Scholar]
- 12.Lewin B. Genes IX. Jones & Bartlett; 2007. [Google Scholar]
- 13.Myers EW, et al. A Whole-Genome Assembly of Drosophila. Science. 2000;287(5461):2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 14.Atschul SF, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 15.Delcher AL, et al. Alignment of whole genomes. Nucleic Acids Res. 1999;27(11):2369–76. doi: 10.1093/nar/27.11.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delcher AL, et al. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30(11):2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ukkonen E. On-line construction of suffix-trees. Algorithmica. 1995;14:249–260. [Google Scholar]
- 18.Gusfield D. Algorithms on Strings, Trees, and Sequences: Computer Science and Computational Biology. New York: Cambridge University Press; 1997. [Google Scholar]