Abstract
Identifying the requirements for the regulatory HBx protein in hepatitis B virus (HBV) replication is an important goal. A plasmid-based HBV replication assay was used to evaluate whether HBx subcellular localization influences its ability to promote virus replication, as measured by real time PCR quantitation of viral capsid-associated DNA. HBx targeted to the nucleus by a nuclear localization signal (NLS-HBx) was able to restore HBx-deficient HBV replication, while HBx containing a nuclear export signal (NES-HBx) was not. Both NLS-HBx and NES-HBx were expressed at similar levels (by immunoprecipitation and Western blotting), and proper localization of the signal sequence-tagged proteins was confirmed by deconvolution microscopy using HBx, NLS-HBx, and NES-HBx proteins fused to GFP. Importantly, these findings were confirmed in vivo by hydrodynamic injection into mice. Our results demonstrate that in these HBV replication assays, at least one function of HBx requires its localization to the nucleus.
Keywords: Hepatitis B virus, HBx, nuclear-localization, HBV replication
Introduction
Chronic infection with hepatitis B virus (HBV) is a significant risk factor for the development of hepatocellular carcinoma (Beasley et al., 1981). Despite the recognized public health concern of HBV, little progress has been made in defining the critical virus-cell interactions that underlay the initiation of virus infection and maintenance of the chronic state. Such knowledge is necessary for designing rational therapies that will interrupt chronic HBV replication.
HBV encodes a 17-kDa regulatory HBx protein whose role in virus replication remains undefined. Studies of HBx in transfected cells have identified a number of functions that are thought to be important for HBV replication and pathogenesis, including effects on cell cycle, cytoplasmic signaling, DNA repair, and apoptosis [reviewed in (Bouchard and Schneider, 2004)]. However, the link between these properties of HBx in cell culture, and virus replication remains unproven.
It is established that HBx can localize to either the cytoplasm or the nucleus of infected liver cells (Dandri et al., 1998; Doria et al., 1995; Haruna et al., 1991; Henkler et al., 2001; Hoare et al., 2001; Nomura et al., 1999; Su et al., 1998; Wang et al., 1991), and it is reasonable to anticipate that HBx may exert different effects on virus replication and/or pathogenesis in different subcellular compartments. Furthermore, studies reveal that a subset of cytoplasmic HBx may be associated with the mitochondria (Clippinger and Bouchard, 2008; McClain et al., 2007; Rahmani et al., 2000; Takada et al., 1999; Waris et al., 2001). The function of HBx in these subcellular compartments is an important issue.
The recent descriptions of models that recapitulate portions of the HBV replication cycle (Bouchard et al., 2002; Yang et al., 2002) provide an opportunity to directly examine functions of HBx within the context of virus replication. In HepG2 cells, virus replication from an HBx-deficient HBV plasmid (Bouchard et al., 2002; Melegari et al., 1998) is significantly reduced compared to replication from an HBx-proficient HBV plasmid (Bouchard et al., 2002; Keasler et al., 2007; Leupin et al., 2005; Melegari et al., 1998; Tang et al., 2005). The requirement for HBx in virus replication has also been established in hydrodynamically-injected mice, where HBx-deficient replication is reduced to just 1% of HBx-proficient replication (Keasler et al., 2007). HBx provided in trans on a second plasmid restores HBx-deficient replication to wildtype (HBx-proficient) levels in both HepG2 cells and mouse hepatocytes in vivo. Interestingly, there is no difference between HBx-proficient replication and HBx-deficient replication in human liver HuH7 cells, although the mechanism for that observation is not known (Melegari et al., 1998).
Previous studies to address a role of HBx subcellular localization in virus replication have not yielded definitive conclusions. One study reported that NLS-HBx was unable to restore HBx-deficient woodchuck hepatitis virus (WHV) replication, but did not include NES-HBx as a positive control for cytoplasmic HBx (Klein et al., 2001). In addition, that study utilized Chang cells (http://www.atcc.org. search term: CCL-13 cells) that likely do not support HBx-dependent replication. A second study was performed with HepG2 cells, but used plasmid vectors (for HBx, NLS-HBx, and NES-HBx) that were expressed fused to green fluorescence protein (GFP) (Leupin et al., 2005). The effect of the longer-lived GFP protein [half-life, 26 hrs; (Corish and Tyler-Smith, 1999)] on HBx [reported half-life ∼30 min (Schek et al., 1991; Sirma et al., 1998) and 3 hr (Schek et al., 1991)] was not examined, and so it is possible that HBx may have been expressed to high levels. Therefore the goals of the present study were to characterize the HBV replication assay to establish its sensitivity for HBx function, and to examine the ability of HBx targeted to the nucleus or the cytoplasm to complement HBx-deficient pHBVΔX replication, using transfected HepG2 cells and the hydrodynamic injection model.
Results
HBx expression vectors
Recent HBV replication models utilize plasmid DNA encoding a greater-than-unit length (120%) wildtype HBV (pHBV) (Scaglioni et al., 1997) which express HBV proteins, including HBx (McClain et al., 2007) under control of the native HBV enhancers and promoters. Previous studies addressed the ability of nuclear- and/or cytoplasmic localized HBx to restore HBx-deficient (pHBVΔX) replication (Klein et al., 2001; Leupin et al., 2005) utilizing the cytomegalovirus (CMV) early promoter (Brondyk, 1994) and the SRα promoter (Takebe et al., 1988) that are each anticipated to drive HBx expression to high levels (Table 1). High levels of HBx are reported to aggregate within the cytoplasm and to affect cell cycling (Henkler et al., 2001; Hoare et al., 2001; Song et al., 2003), which may influence the interpretation of those results. Therefore, we began our study by re-creating a panel of plasmid DNAs in which expression of HBx (and its signal-tagged derivatives) was driven from the SV40 early promoter/enhance (Brondyk, 1994) (Fig. 1). All HBx sequences are from the adw2 subtype of HBV, and based on relative promoter strengths (Table 1) are predicted to express HBx at lower levels than in previous studies (Klein et al., 2001; Leupin et al., 2005).
Table 1.
Effect of HBx subcellular localization on levels of HBV capsid-associated DNA
| Cell type | HBxa | Promoterb | Promoter strengthc | Rescue of ΔX replication?d | Reference |
|---|---|---|---|---|---|
| Chang livere | NLS-HBx | pCMV | 1.8 to 6.5 | No | Klein et al. 2001 |
| HepG2f | NLS-GFP-HBx | pSRα | 40 | Yes | Leupin et al. 2005 |
| NES-GFP-HBx | pSRα | 40 | No | Leupin et al. 2005 | |
| HepG2 | NLS-HBx | pSI | 1 | Yes | Current study |
| NES-HBx | pSI | 1 | No | Current study | |
| Mouse (viremia)g | NLS-HBx | pSI | 1 | Yes | Current study |
| NES-HBx | pSI | 1 | No | Current study |
HBx expressed from transfected plasmid DNA as an in-frame fusion protein with one or more of the following: NLS, nuclear localization signal; NES, nuclear export signal; GFP, green fluorescence protein.
Promoter used to drive HBx expression: CMV, human cytomegalovirus immediate-early enhancer/promoter region (Brondyk, 1994); pSRα, R-U5 segment from the human T cell leukemia virus I long terminal repeat fused to the SV40 early promoter-enhancer (Takebe et al., 1988); pSI, SV40 early region promoter/enhancer (Brondyk, 1994).
Strength of given promoter relative to the SV40 early promoter/enhancer, as described (Brondyk, 1994; Takebe et al., 1988).
Ability of a specific version of HBx to restore HBx-deficient pHBVΔX replication in trans, compared to wildtype HBV pHBV levels of replication.
Chang liver cells, reported to be cervical cancer cells (http://www.atcc.org, search term: CCL-13).
HepG2, hepatocyte cell line derived from human hepatoblastoma (Aden et al., 1979).
HBV replication in mice hydrodynamically injected as described in Materials and Methods.
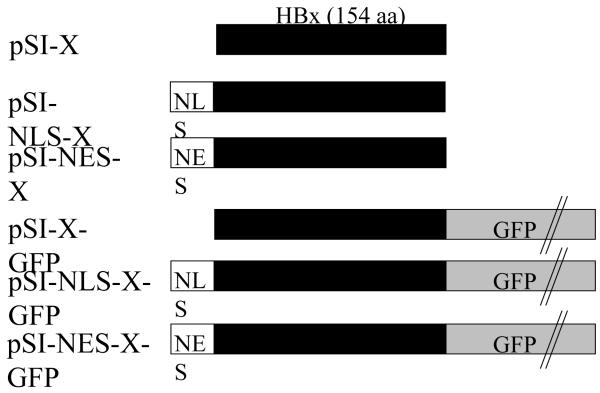
Fig. 1. Schematic representation of the X gene constructs used.
Expression of the 154-aa HBx (subtype adw2; designated as black bar) was from the pSI vector containing the SV40 early region enhancer/promoter (Brondyk, 1994). Variants of HBx were created by cloning one or more of the following in-frame with HBx: NLS, nuclear localization signal; NES, nuclear export signal; GFP, green fluorescence protein. pSI-X plasmids without GFP were used in functional replication assays, while GFP fusion proteins were used to validate the NLS and NES localization signals.
Dose-dependent rescue of HBx-dependent replication in HepG2 cells
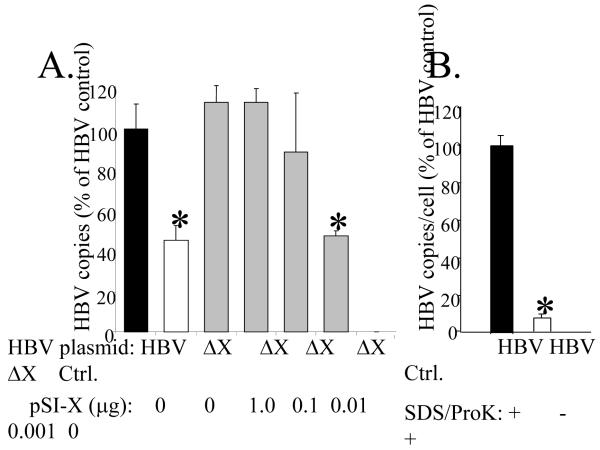
A titration experiment was used to define levels of HBx required for maximal levels of virus replication. HepG2 cells were transfected with either pHBV or pHBVΔX, and plates of cells receiving pHBVΔX were co-transfected with pSI-X diluted over a 1000-fold range. Cells were harvested at 5-days and capsid-associated viral DNA quantitated by real time PCR. Viral copy number in cells transfected with pHBV was set to 100%, and replication from pHBVΔX (alone or with varying amounts of complementing pSI-X) was compared to that level. Replication from pHBVΔX was reduced to 40% of that detected in cells transfected with pHBV (Fig. 2A), a result that confirms our previous findings (Keasler et al., 2007). We show that pSI-X was able to restore HBVΔX replication to wildtype (pHBV) levels when added at 1-, 0.1-, or 0.01-μg per 60-mm plate, but that 0.001-μg pSI-X restored pHBVΔX replication to levels still significantly reduced from the pHBV wildtype levels (Fig. 2A; see asterisk). Importantly, to consistently detect HBx by IP/Western, it is necessary to pool two 60-mm plates of HepG2 cells each transfected with 1 μg pSI-X DNA (Fig. 3B). These results show that HBx can fully restore pHBVΔX replication when expressed at levels below the limit of detection by our sensitive IP/Western blot assay.
FIG. 2. HBx and HBV replication.
Panel A, HepG2 cells were transfected with pHBV, pHBVΔX, pHBVΔX plus pSI-X (1 to 0.001 μg), or a negative control plasmid (Ctrl) and virus replication measured by real time PCR. Viral DNA was normalized to the number of transfected cells (determined by co-transfection of plasmid DNA encoding GFP) and is reported as the average of 3 experiments ± SEM. The statistical significance of reduced HBV expression was determined by comparison to HBV replication measured from the pHBV (wildtype) plasmid, and is noted by an asterisk above the bar (p<0.05). Panel B, Copies of viral DNA measured by real time PCR from core particles disrupted with SDS/Proteinase K (black bar) or left untreated (white bar). Copies of capsid-associated viral DNA detected from cells transfected with pHBV and treated with SDS/proK (black bar) was set to 100% and the relative levels of viral DNA detected in cells transfected with pHBV and no SDS/proK (white bar) or a control plasmid (Ctrl.) are reported ± SEM.
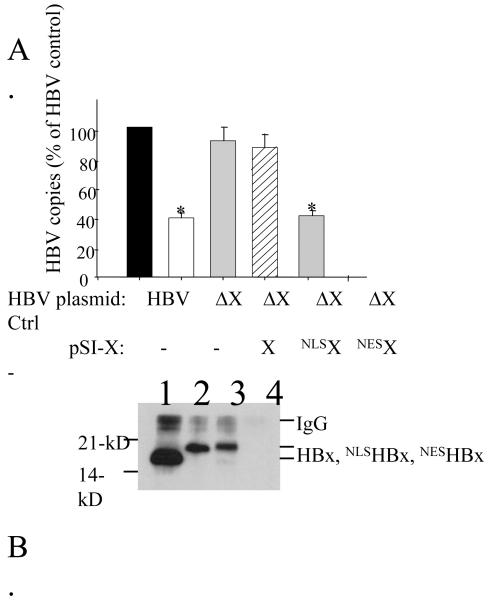
FIG. 3. HBx and NLS-HBX rescue of HBx-deficient replication.
Panel A, HepG2 cells were transfected with pHBV or pHBVΔX, with the latter co-transfected with pSI-X, pSI-NLS-HBX, pSI-NES-HBX, or a negative control plasmid. Virus replication was measured using real time PCR quantitation of capsid-associated DNA. Data is reported as HBV copy number (% of HBV control) from three experiments ± SEM. Statistical significance compared to pHBV is noted by an asterisk above the bar (p<0.05). Panel B, HepG2 cells were transfected with pSI-X (lane 1), pSI-NLS-HBX (lane 2), pSI-NES-HBX (lane 3), or a negative control plasmid (lane 4) and harvested 48 hours post-transfection. Detection of HBx was by IP/Western.
We next established that the viral DNA detected by real time PCR was from encapsidated DNA and not from residual input plasmid DNA. HepG2 cells were transfected with pHBV, lysed at day 5, and viral capsids purified and treated with DNaseI to digest non-encapsidated DNA that might stick to the outside of the cores. Viral capsids were then either disrupted by addition of SDS and proteinase K (Fig. 2B, black bar), or left untreated and presumably intact (Figure 2B, white bar) prior to real time PCR quantitation of capsid-associated viral DNA. Virus copy number per cell from SDS/proteinase K-disrupted capsids was set to 100%, and compared to that measured from capsids not disrupted by SDS/proteinase K. There was a 95% reduction in capsid-associated viral DNA detected from samples that were left intact (Figure 2B, white bar). These results indicate that viral capsids must be disrupted in order to measure viral DNA by real time PCR in this assay. Together, the pSI-X titration (Fig. 2A) and the treated/untreated capsid control experiment (Fig. 2B) provide evidence that the role of HBx in virus replication can be measured at very low levels of HBx expression.
Restoration of HBx-deficient replication by nuclear-localized HBx
HBx has been detected in both the cytoplasm and the nucleus of HBV-infected hepatocytes and cultured cells transfected with plasmid DNAs encoding HBx. To determine which subcellular location is critical for virus replication, pSI-HBx, pSI-NLS-HBx, and pSI-NES-HBx expression plasmids (Fig. 1) were tested for their ability to restore pHBVΔX replication. Cells transfected with pHBVΔX had levels of capsid-associated DNA that were 34% of that measured in cells transfected with pHBV (Fig 3A). Capsid-associated viral DNA levels were restored to maximal levels by either pSI-X or pSI-NLS-HBx, but were not restored by pSI-NES-X (Fig. 3A). This result is consistent with a previous report that a GFP-NLS-HBx fusion protein could restore HBx-deficient replication while a similar GFP fusion protein tagged with an NES signal could not (Leupin et al., 2005). Although that study did not examine GFP-HBx expression levels, our present study revealed that HBx (and NLS-HBx) rescue pHBVΔX replication using levels of HBx below the limits of detection of our sensitive IP/Western blot assay.
The failure of NES-HBx to restore pHBVΔX replication was unexpected, given the prevalent idea that HBx function in the cytoplasm is important to virus replication [reviewed in (Ganem and Schneider, 2001)]. We next investigated the possibility that NES-HBx was inherently less stable than NLS-HBx. Plasmid-transfected HepG2 cells were lysed at 48-hr and analyzed for steady-state HBx levels by IP/Western as described in Materials and Methods. Both the NLS-HBx and the NES-HBx proteins migrated at an apparent molecular weight slightly higher than wildtype HBx due to the presence of the added localization signals (Fig. 3B; compare lane 1 with lanes 2 and 3). We consistently found a weaker Western blot signal for the NLS-HBx and NES-HBx proteins, relative to that for wildtype HBx, even though all three proteins are expressed from the same pSI plasmid vector. Importantly, the NLS-HBx and the NES-HBx proteins are expressed at similar levels, indicating that the inability of NES-HBx to restore HBx-deficient replication cannot be explained by reduced protein stability relative to NLS-HBx. We note that other laboratories have found that the addition of the NLS signal, or the Flag epitope tag, to either end of the HBx protein did not effect the ability of HBx to transactivate HBx responsive elements (Nomura et al., 1999) or to induce calcium release (McClain et al., 2007), respectively.
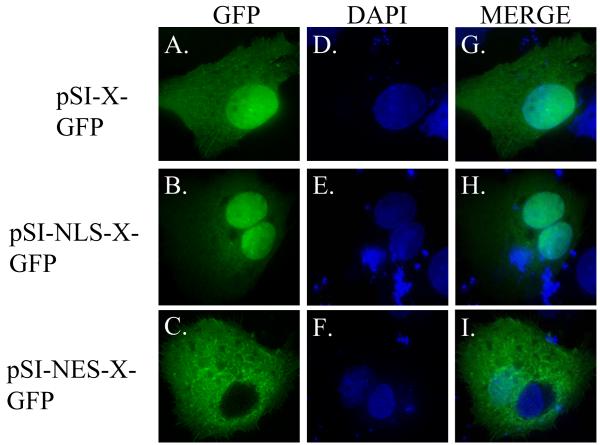
Next we confirmed that the NLS- and NES-targeted HBx proteins were localized to the correct subcellular compartment. For these experiments, HBx, NLS-HBx and NES-HBx were cloned as in-frame fusion proteins with GFP at the C-terminus, expressed by the same SV40 early promoter (Fig. 1). HepG2 cells were then transfected, fixed at 24-hr post-transfection, and processed for analysis by deconvolution fluorescence microscopy. Expression of the wildtype HBx-GFP was detected in both the nucleus and cytoplasm of transfected cells (Fig 4A, D, G). In cells transfected with pSI-NLS-HBx-GFP, localization was primarily to the nucleus, although some cytoplasmic localization was detected (Fig. 4B, E, H), consistent with the report that HBx may possess a nuclear export signal (Forgues et al., 2001). In cells transfected with pSI-NES-HBx-GFP, the majority of fluorescence was localized to the cytoplasm (Fig. 4C, F, I). These data confirm that the X-GFP fusion proteins containing an NLS or NES localization sequence are targeted to the proper subcellular location.
FIG. 4. Subcellular localization of HBx-GFP fusion proteins in HepG2 cells.
HepG2 cells were transfected with pSI-X-GFP, pSI-NLS-X-GFP, or pSI-NES-X-GFP and analyzed by deconvolution microscopy 24 hours post-transfection. Localization of GFP tagged constructs is illustrated in the first column (A-C), staining of the cell nucleus (by DAPI) can be seen in the middle column (D-F), and a merged image can be seen in the right column (G-I).
NLS-HBx restores HBx-deficient HBV replication in vivo
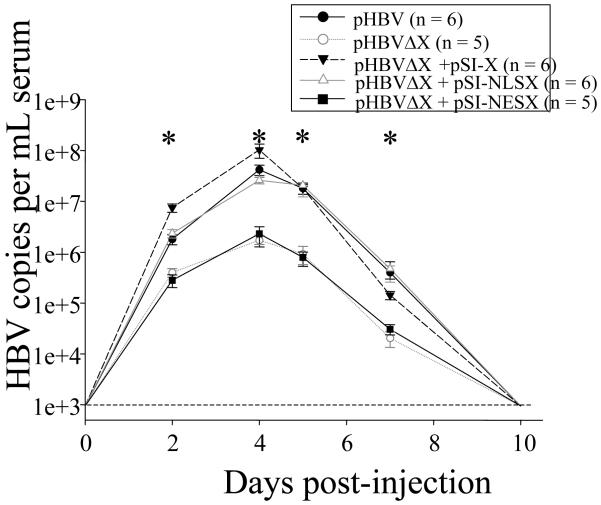
We previously reported that in the hydrodynamic injection model, HBx-deficient pHBVΔX replication was reduced by 99% relative to HBx-proficient replication (Keasler et al., 2007). This model was next used to examine the ability of NLS- and NES-HBx to restore pHBVΔX replication in vivo. Eleven outbred ICR mice were injected with either wildtype pHBV (n = 6) or HBx-deficient pHBVΔX (n=5). Additional mice injected with pHBVΔX were co-injected with pSI-X (n =6), pSI-NLS-X (n = 6), or pSI-NES-X (n = 5). Finally, 4 mice received a negative control plasmid. All 32 mice used in this study were also co-injected with pSI-SEAP (a secretable form of alkaline phosphatase) to normalize injection efficiency among groups. Blood was collected by retroorbital bleeding at days 0, 2, 4, 5, 7, and 10 post-injection and capsid-associated viral DNA from serum was quantitated by real time PCR analysis as described previously (Keasler et al., 2007). Compared to mice injected with pHBV, there was a significant reduction in capsid-associated viral DNA in the serum of mice injected with pHBVΔX at days 2, 4, 5, and 7 post-injection (p<0.05; Fig. 5, see asterisks). The maximum reduction in viremia in the absence of HBx (mice injected with pHBVΔX; 97% reduction) was recorded in sera collected at day 4 post-injection, a result consistent with our previous findings (Keasler et al., 2007). Viremia was restored to maximal pHBV levels in mice injected with pHBVΔX plus pSI-X or pSI-NLS-X. However, pSI-NES-X was unable to restore pHBVΔX replication, with levels of capsid-associated DNA remaining nearly identical to that measured in mice injected with pHBVΔX alone (Fig 5). This result indicates that the requirement for nuclear localized HBx in HBV replication identified in HepG2 cells (Fig. 3A) is also apparent in mouse hepatocytes in vivo.
FIG 5. HBx and NLS-HBX complementation of HBx-deficient replication in vivo.
Mice were injected with pHBV (filled circles; n = 6), pHBVΔX (open circles; n = 5), pHBVΔX plus pSI-X (filled triangles; n = 6), pHBVΔX plus pSI-NLS-X (open triangles; n = 6), pHBVΔX plus pSI-NES-X (filled square; n = 5), or negative control plasmid (n=4). Blood was collected via retroorbital bleeding on days 0, 2, 4, 5, 7, and 10 post-injection. Capsid-associated viral DNA was purified from serum and quantitated by real time PCR. The horizontal dashed line near the bottom of the graph represents the background copies of HBV DNA from the real time PCR reaction (1 copy per μl serum). Error bars represent ± SEM and statistical significance (compared to pHBV levels) is designated with an asterisk.
Discussion
The HBV regulatory X protein is required for virus replication (Bouchard et al., 2002; Chen et al., 1993; Keasler et al., 2007; Zoulim et al., 1994), but the mechanism through which it acts remains unknown. In the present study, we tested the ability of HBx targeted to or excluded from the nucleus to restore HBx-deficient virus replication. We show that in transfected HepG2 cells, the reduced virus replication from the HBx-deficient pHBVΔX plasmid was restored to wildtype levels by nuclear-localized HBx but not by HBx excluded from the nucleus. Deconvolution fluorescence microscopy experiments demonstrated that the NLS and NES signals targeted HBx to the proper subcellular localization, and IP/Western blot was used to verify the similar expression levels of NLS- and NES-HBx. Importantly, dose-response experiments revealed that HBx can restore pHBVΔX replication when HBx is expressed at levels below detection by IP/Western. This study demonstrates that the localization of HBx to the nucleus is essential for its role in virus replication and that this requirement is evident in both human HepG2 cells and in mouse hepatocytes in vivo.
There are at least two interpretations to the data showing that NLS-HBx restores pHBVΔX replication. In the first, HBx is predicted to interact with a cellular target in the nucleus to provide some function critical for HBV replication. This idea is supported by several studies, including that HBx can stimulate transcription from reporter constructs containing the HBV Enhancer I (Doria et al., 1995), leading to a 2- to 3-fold increase in HBV mRNA levels in some (Keasler et al., 2007; Leupin et al., 2005; Melegari et al., 2005; Tang et al., 2005; Zhang et al., 2001) but not all (Bouchard et al., 2002) studies utilizing the HBV replication assay. HBx also stimulates transcription when tethered to a DNA-binding domain (Haviv et al., 1995; Seto et al., 1990), and is reported to interact with the basic leucine zipper region of CREB [cAMP response element-binding; (Barnabas et al., 1997; Palmer et al., 1997; Williams and Andrisani, 1995)] and with binding partner DDB1 (Lee et al., 1995; Leupin et al., 2005). Therefore, the finding that nuclear localized HBx restores HBx-deficient replication may be explained by any or all of the above previous studies.
A second major interpretation of our results, not mutually exclusive from the above explanation of nuclear localization, is that HBx may indeed interact with a critical cytoplasmic target, but additionally require translocation to the nucleus to facilitate replication. Many studies report properties for HBx consistent with its cytoplasmic localization, and so it is somewhat surprising that NES-HBx provided no function in restoring pHBVΔX replication. In the cytoplasm, HBx activates the Ras-Raf-MAP kinase cytoplasmic signaling pathway (Benn et al., 1996; Benn and Schneider, 1994; Cross et al., 1993; Natoli et al., 1994a; Natoli et al., 1994b), and the stress-activated protein kinase/c-Jun NH2-terminal kinase (SAPK-JNK) pathway (Benn et al., 1996; Henkler et al., 1998; Tarn et al., 1999). These properties of HBx have been proposed to facilitate virus replication, although the replication assay was not available for direct testing at the time those studies were published. There are also reports from several laboratories that a subpopulation of cytoplasmic HBx localizes to the mitochondria (Henkler et al., 2001; Huh and Siddiqui, 2002; McClain et al., 2007; Rahmani et al., 2000; Takada et al., 1999; Waris et al., 2001). It is important to recall that these HBV replication assays mimic some, but not all, steps in virus replication. A recent study used the same replication assay to show that a modest mitochondrial release of calcium may be linked to HBV replication (McClain et al., 2007) However, that study did not specifically examine NLS-HBx or NES-HBx effects on calcium release, and so a direct comparison with the present study is not possible. In summary, we cannot eliminate the possibility that there are critical cytoplasmic targets of HBx that must translocate to the nucleus to participate in virus replication.
The present study is the third to investigate the role of HBx subcellular localization in virus replication, and our results both support and differ from those previous studies (Table 1). One study concluded that targeting of HBx to the nucleus had no effect on HBx-deficient replication, but did not examine HBx excluded from the nucleus as a positive control (Klein et al., 2001). Our results do support the findings of another study that utilized HBx-GFP fusion proteins targeted either to the nucleus or the cytoplasm to restore HBx-deficient replication (Leupin et al., 2005). While the author’s use of the strong SRα promoter to drive HBx-GFP expression raised the question of whether HBx expressed at more physiological levels would behave similarly, we now report confirmatory results when HBx is expressed at very low levels, below the limits of detection.
The expression level of HBx is an important consideration in any experiment designed to investigate HBx function. HBx expressed from the strong CMV promoter is reported to form cytoplasmic granules in cells expressing the highest levels of the protein (Henkler et al., 2001; Hoare et al., 2001), and cell lines stably expressing HBx have reduced growth rates relative to control cells (Lee et al., 1998). Indeed, the relative promoter strengths of HBx expression constructs used in different studies should be noted. For example, a previous study that expressed HBx as GFP fusion proteins additionally utilized the SRα promoter that is estimated to be 40-fold stronger than the SV40 early promoter used in our study (Table 1). While a direct comparison of HBx protein levels in the three studies is not possible, we demonstrate here that HBx expressed at levels below detection in HepG2 cells is still able to restore pHBVΔX replication. Furthermore, we calculate that the amount of plasmid DNA taken up per cell via hydrodynamic injection method was approximately 1% of that received per transfected HepG2 cell (see Materials and Methods), indicating even lower levels of HBx in vivo are sufficient to restore HBx-deficient replication. For these reasons, we conclude that HBx was not overexpressed in our replication assays.
The finding that NLS-HBx restores HBVΔX replication in the hydrodynamic injection model is an important first step in characterizing the function of HBx, in vivo in the context of virus replication and in the presence of an intact immune system. It is thought that the inability to infect mice with HBV is due to a lack of virus receptors on mouse hepatocytes. It remains unknown whether this is the only difference in virus replication between human and mouse hepatocytes, or whether these two models of HBV replication will eventually diverge at some molecular level.
It is apparent from this study and others that while HBx-deficiency is associated with a reproducible decrease in capsid-associated DNA, virus replication is not completely eliminated (Keasler et al., 2007; Leupin et al., 2005; Melegari et al., 2005; Tang et al., 2005; Zhang et al., 2001). This residual HBx-independent replication is consistently found when real time PCR is used to quantitate virus copy number, and we have considered several possible explanations. First, there may be occasional read-through of the stop codon at HBx amino acid 7 resulting in very low levels of HBx sufficient to restore some replication. In addition, HBx is not required for virus replication in actively dividing cells (Blum et al., 1992), and it is possible that the HepG2 cell cultures used in our replication studies are not completely contact-inhibited. Despite the residual replication in the absence of HBx, the effect of HBx-deficiency is reproducible and can be complemented by the addition of a second plasmid encoding HBx.
After our experiments were completed, a new study was published of a similar analysis in HepG2 cells (Cha et al., 2009). Although the authors concluded that both NLS- and NES-HBx contribute equally to HBV replication, a close examination of the data revealed that NES-HBx activated pHBVΔX replication to significantly lower levels than did either HBx or NLS-HBx. A comparison with wildtype (pHBV) replication was not included. In addition, that study expressed HBx under control of the strong CMV promoter, shown by others to induce the formation of cytoplasmic granules (Henkler et al., 2001; Hoare et al., 2001; Song et al., 2003). This may provide an explanation for why Cha et al found that HBx restores pHBVΔX to 75% of wildtype (pHBV) replication (versus 100% restoration in our study). Never-the-less, our findings are in general agreement with the Cha study.
In summary, we have examined the requirement of HBx to restore HBx-deficient replication when HBx is targeted to, or excluded from, the cell nucleus. While the HBV replication models used in our study are not true infection models, they do provide the opportunity to quantitate virus replication under varying conditions. These assays represent the best experimental model available to test HBx function in a biologically relevant setting. Our results clearly demonstrate that nuclear localization of HBx is critical for maximal levels of capsid-associated viral DNA in transfected HepG2 cells. These results do not exclude the possibility that HBx may have critical cytoplasmic targets, but do suggest that such interactions would require translocation to the nucleus as part of the HBV replication strategy. Our results support a previous report that NLS-HBx expressed as a GFP fusion protein can restore HBx-deficient replication in HepG2 cells (Leupin et al., 2005), and extends those earlier studies by showing that this occurs when HBx is expressed at more physiological levels. Importantly, we further show that this property of HBx also occurs in an in vivo setting using the hydrodynamic mouse model. Future experiments will seek to identify cellular targets that mediate these functions of HBx during virus replication.
Materials and Methods
Plasmids
Plasmid DNAs encoding a greater-than-unit length HBV genome [payw1.2 (Scaglioni et al., 1997)] or the same plasmid containing a stop codon at amino acid 7 [payw1.2*7 (Melegari et al., 1998)] were obtained from the laboratory of Dr. Robert Schneider. For simplicity, we refer to payw1.2 plasmid as pHBV and the HBx-deficient plasmid payw1.2*7 as pHBVΔX. Expression of HBV in these vectors is driven by the native HBV promoter/enhancers (Brondyk, 1994). The parental HBx expression plasmid used in this study, pSI-X, was created using a plasmid containing the simian virus 40 enhancer/early promoter vector (pSI; Promega) and the subtype adw2 HBx, as described previously (Keasler et al., 2007). pSI-NLS-HBX was created by PCR cloning using primers [5′GTAACGCGTATGCCAAAGAAGAAG CGAAAGGCTGCTAGGCTG’3 and 5′GTAGCGGCCGCTTAGGCAGAGG’3] that contain the SV40 T antigen nuclear localization signal (Lanford and Butel, 1984). pSI-NES-HBX was created using PCR primers [5′GTAACGCGTATGAACGAGCTAGCACTAAAGCTAGCAGGTCTAGACATCAACAAGGCTG TCAGGCTG’3 and 5′GTAGCGGCCGCTTAGGCAGAGG’3] that contain the nuclear export signal (NES) identified in the protein kinase inhibitor alpha gene (Fukuda et al., 1997). GFP was added in frame to the carboxy terminus of pSI-X, pSI-NLS-HBX, and pSI-NES-HBX by linearizing a plasmid construct containing the entire GFP open reading frame (pSI-GFP) using double restriction enzyme digest with Mlu I and Sma I. The X sequence (including the NES or NLS described above) was PCR amplified using the forward primers described above (to add the NES and NLS respectively) and the following primer to remove the stop codon from the X open reading frame and to add a Not I restriction site: 5′ATGCCCGGGCAGAGGTGAAAAAG’3. The PCR amplified sequences for NLS-HBX and NES-HBX were then ligated into the pSI-GFP vector. Removal of the stop codon at the 3′ end of the NLS-HBX and NES-HBX constructs allows read-through into the GFP nucleotide sequence creating an NLS-HBX-GFP or NES-HBX-GFP fusion construct. A plasmid encoding a heat-stable, secretable form of alkaline phosphatase (pSI-SEAP) was constructed as previously described (Keasler et al., 2007). A promoterless pSEAP2-basic plasmid (Clontech) was used as a negative control. All plasmid DNAs were transformed into E. coli and plasmid preparations made using an Endofree Plasmid Maxi kit (Qiagen).
Cell culture and plasmid transfection
Human liver HepG2 cells were obtained directly from the ATCC and used at early passages. Cells were plated and maintained in Eagles minimal media plus 10% fetal bovine serum. The replication assay in HepG2 cells was performed as previously described (Keasler et al., 2007). Briefly, 5×105 cells were plated in 60 mm dishes and transfected twenty-four hours later using TransIT (Mirus Bio Corporation) according to the manufacturer’s protocol. For HBV replication experiments, 2×106 untransfected HepG2 cells were added to each 60-mm dish at 2 hours post-transfection to create the confluent monolayer needed for this assay and cells were harvested at 5 days post-transfection.
Hydrodynamic injection and retroorbital collection of blood
Approval for all experiments involving animals was obtained from the Institutional Animal Care and Use Committee at Baylor College of Medicine. Hydrodynamic injections were performed on outbred Crl:CD-1 (ICR) female mice at 10-13 weeks of age as previously described (Keasler et al., 2007). Briefly, mice were injected with plasmid DNA diluted in phosphate-buffered saline (PBS) to a volume equivalent to 8% of the total body weight of each animal. Each injected mouse received a total of 18 μg plasmid DNA: 9 μg of HBV plasmid DNA (either pHBV or pHBVΔX), 5 μg pSI-SEAP (encoding secretable, heat-stable alkaline phosphatase), and 4 μg of pSI-X, pSI-NLS-X, pSI-NES-X, or control plasmid. Negative control animals received 5 μg pSI-SEAP and 13 μg of promoterless pSEAP2-Basic plasmid DNA. Approximately 50 μl of blood was collected from each mouse on days 0, 2, 4, 5, 7, and 10 post-injection by retroorbital bleeding to measure viremia as previously described (Keasler et al., 2007).
Measurement of heat-stable alkaline phosphatase
Animal to animal injection variability was controlled by normalizing to levels of heat-stable alkaline phosphatase (expressed from the pSI-SEAP plasmid DNA) detected in the serum at day 4 post-injection (Keasler et al., 2007). Briefly, serum collected by retroorbital bleeding was heated at 65°C for 30 min to inactivate endogenous phosphatases, diluted in sterile saline, and mixed with alkaline phosphatase yellow (pNPP) liquid substrate (Sigma). The optical density at 405 nm was then recorded at 0, 10, 20, and 30 min to determine relative alkaline phosphatase levels among groups of injected mice.
Estimation of plasmid DNA per cell
HepG2 cells (5 × 105 per 60-mm plate) were transfected with 2 μg HBV plasmid DNA. Using an estimated transfection efficiency of 20% (determined by cotransfection with a plasmid DNA encoding GFP), a total of 1×105 HepG2 cells received plasmid for an average of 20 picograms of HBV plasmid DNA per cell (assuming all plasmid DNA was taken up). In comparison, a normal mouse liver contains approximately 108 hepatocytes (Sohlenius-Sternbeck, 2006), and hydrodynamic injection of plasmid DNA encoding beta-galactosidase led to the conclusion that 40% (4 × 107) hepatocytes expressed beta-galactosidase (Liu et al., 1999). In our study, each mouse received 9 μg HBV plasmid DNA, for a total of 0.225 picograms of DNA per transfected hepatocyte. The actual amount per cell is likely less, since plasmid DNA delivered by hydrodynamic injection can also be taken up in other organs (Liu et al., 1999).
Detection of HBx by immunoprecipitation (IP) and Western Blot
Transfected HepG2 cells were harvested and pooled (two 60-mm plates per IP) at 48 hours post-transfection as previously described (Becker et al., 1998). Proteins were separated by 15% SDS-PAGE and transferred to a nitrocellulose membrane. HBx was detected using a rabbit polyclonal antibody (1:1000), an anti-rabbit secondary antibody conjugated to horseradish peroxidase (1:1000; Pierce), and the SuperSignal West Femto detection kit (Pierce).
Deconvolution fluorescence microscopy of HepG2 cells
HepG2 cells were plated on poly-D-lysine coated coverslips in 24-well plates at 5×104 cells per well. Cells were transfected 24 hours later using TransIT (Mirus Bio Corporation) according to the manufacturer’s protocol. Transfected cells were fixed and visualized 24 hours after transfection. Briefly, coverslips were washed with ice-cold PBS (containing Ca++ and Mg++) and fixed with 4% formaldehyde in PEM buffer [80 mM Potassium PIPES (pH 6.8), 5 mM EGTA, (pH 7.0) and 2 mM MgCl2] on ice. Coverslips were washed with PEM buffer, and auto-fluorescence quenched by addition of 0.1M ammonium chloride. Cells were then permeabilized by the addition of 0.5% Triton X-100 in PEM buffer, washed with PEM buffer, and the nuclei stained with DAPI. Finally, coverslips were mounted on slides with Anti-fade mounting media. Transfected cells were visualized using a DeltaVision (Deconvolution) Restoration Microscope equipped with a Plan-APOCHROMAT 63X/1.40 objective and pictures acquired using SoftWoRx software.
Purification of capsid associated viral DNA
Capsid-associated viral DNA was extracted and quantitated from transfected HepG2 cells and from serum of hydrodynamically-injected mice, as previously described (Keasler et al., 2007). Briefly, HepG2 cells were lysed (50 mM Tris, pH 7.5, 0.5% NP-40, 1 mM EDTA, and 100 mM NaCl) and then incubated with 10 μl of 1M MgCl2 and 10 μl of DNaseI (10 mg/ml). Viral cores were next precipitated from solution by the addition of EDTA and polyethylene glycol and the pellet resuspended in 500 μl buffer containing 10 mM Tris, 100 mM NaCl, 1 mM EDTA, 1% sodium dodecyl sulfate (SDS), and 20 μl of 25 mg/ml proteinase K. Viral DNAs released from lysed cores were extracted with phenol and chloroform, precipitated with isopropanol, and resuspended in Tris-EDTA. Purification of capsid-associated viral DNA from serum was nearly identical to that described for transfected HepG2 cells. However, serum was treated with DNaseI overnight and viral DNAs from lysed cores were concentrated using a QIAamp DNA Blood Mini kit (QIAGEN) following the manufacturer’s protocol (instead of phenol and chloroform extraction and isopropanol precipitation).
Real-time PCR detection of HBV DNA
Quantitation of capsid-associated viral DNA was performed by real time PCR using a primer/probe set (Applied Biosystems), as previously described (Keasler et al., 2007). All samples were analyzed in duplicate using a 96-well plate format and the Applied Biosystems 7000 Sequence Detection System with the following cycling parameters: 1 cycle at 50°C for 2 min; 1 cycle at 95°C for 10 min; and 40 cycles at 95°C for 15 s and 60°C for 60 s. Standard curves were included in all real time PCR reactions using the pHBV plasmid DNA diluted over a large dynamic range (107 to 100).
Quantitation and statistics
Statistical significance for all experiments was determined by using the Student’s t test (Microsoft Excel software package). Error bars are reported as standard errors of the means, and significance was assigned for p values of <0.05.
Acknowledgments
This work was supported by NIH grant CA95388 (B.L.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barnabas S, Hai T, Andrisani OM. The hepatitis B virus X protein enhances the DNA binding potential and transcription efficacy of bZip transcription factors. J. Biol. Chem. 1997;272:20684–20690. doi: 10.1074/jbc.272.33.20684. [DOI] [PubMed] [Google Scholar]
- Beasley RP, Hwang LY, Lin CC, Chien CS. Hepatocellular carcinoma and hepatitis B virus. A prospective study of 22 707 men in Taiwan. Lancet. 1981;2:1129–1133. doi: 10.1016/s0140-6736(81)90585-7. [DOI] [PubMed] [Google Scholar]
- Becker SA, Lee TH, Butel JS, Slagle BL. Hepatitis B virus X protein interferes with cellular DNA repair. J. Virol. 1998;72:266–272. doi: 10.1128/jvi.72.1.266-272.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benn J, Schneider RJ. Hepatitis B virus HBx protein activates Ras-GTP complex formation and establishes a Ras, Raf, MAP kinase signaling cascade. Proc. Natl. Acad. Sci. 1994;91:10350–10354. doi: 10.1073/pnas.91.22.10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benn J, Su F, Doria M, Schneider RJ. Hepatitis B virus HBx protein induces transcription factor AP-1 by activation of extracellular signal-regulated and c-Jun N-terminal mitogen-activated protein kinases. J Virol. 1996;70:4978–4985. doi: 10.1128/jvi.70.8.4978-4985.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum HE, Zhang Z-S, Galun E, von Weizsacher F, Garner B, Liang TJ, Wands JR. Hepatitis B virus X protein is not central to the viral life cycle in vitro. J Virol. 1992;66:1223–1227. doi: 10.1128/jvi.66.2.1223-1227.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard MJ, Schneider RJ. The enigmatic X gene of hepatitis B virus. J Virol. 2004;78:12725–12734. doi: 10.1128/JVI.78.23.12725-12734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard MJ, Wang L-H, Schneider RJ. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science. 2002;294:2376–2378. doi: 10.1126/science.294.5550.2376. [DOI] [PubMed] [Google Scholar]
- Brondyk B. pCI and pSI mammalian expression vectors. Promega Notes Magazine. 1994;49:7–12. [Google Scholar]
- Cha MY, Ryu DK, Jung HS, Chang HE, Ryu WS, et al. Stimulation of Hepatitis B Virus Genome Replication by HBx is Linked to Both Nuclear and CYtoplasmic HBx Expression. Journal Of General Virology. 2009 doi: 10.1099/vir.0.009928-0. in press. [DOI] [PubMed] [Google Scholar]
- Chen H-S, Kaneko S, Girones R, Anderson RW, Hornbuckle WE, Tennant BC, Cote PJ, Gerin JL, Purcell RH, Miller RH. The woodchuck hepatitis virus X gene is important for establishment of virus infection in woodchucks. J. Virol. 1993;67:1218–1226. doi: 10.1128/jvi.67.3.1218-1226.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clippinger AJ, Bouchard MJ. The hepatitis B virus HBx protein localizes to mitochondria in primary rat hepatocytes and modeulates mitochondrial membrane potential. J. VIrol. 2008 doi: 10.1128/JVI.00154-08. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corish P, Tyler-Smith C. Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng. 1999;12:1035–1040. doi: 10.1093/protein/12.12.1035. [DOI] [PubMed] [Google Scholar]
- Cross JC, Wen P, Rutter WJ. Transactivation by hepatitis B virus X protein is promiscuous and dependent on mitogen-activated cellular serine/threonine kinases. Proc. Natl. Acad. Sci. 1993;90:8078–8082. doi: 10.1073/pnas.90.17.8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandri M, Petersen J, Stockert RJ, Harris TM, Rogler CE. Metabolic labeling of woodchuck hepatitis B virus X protein in naturally infected hepatocytes reveals a bimodal half-life and association with the nuclear framework. J. Virol. 1998;72:9359–9364. doi: 10.1128/jvi.72.11.9359-9364.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria M, Klein N, Lucito R, Schneider RJ. The hepatitis B virus HBx protein is a dual specificity cytoplasmic activator of Ras and nuclear activator of transcription factors. EMBO J. 1995;19:4747–4757. doi: 10.1002/j.1460-2075.1995.tb00156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forgues M, Marrogi AJ, Spillare EA, Wu CG, Yang Q, Yoshida M, Wang XW. Interaction of the hepatitis B virus X protein with the Crm1-dependent nuclear export pathway. J Biol Chem. 2001;276:22797–22803. doi: 10.1074/jbc.M101259200. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- Ganem D, Schneider RJ. Hepadnaviridae: The Viruses and Their Replication. In: Knipe DM, Howley PM, editors. Virology. Lippincott, Williams, and Wilkins; Philadelphia: 2001. pp. 2923–2969. [Google Scholar]
- Haruna Y, Hayashi N, Katayama K, Yuki N, Kasahara A, Sasaki Y, Fusamoto H, Kamada T. Expression of X protein and hepatitis B virus replication in chronic hepatitis. Hepatology. 1991;13:417–421. [PubMed] [Google Scholar]
- Haviv I, Vaizel D, Shaul Y. The X protein of hepatitis B virus coactivates potent activation domains. Mol Cell Biol. 1995;15:1079–1085. doi: 10.1128/mcb.15.2.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkler F, Hoare J, Waseem N, Goldin RD, McGarvey MJ, Koshy R, King IA. Intracellular localization of the hepatitis B virus HBx protein. J. Gen. Virol. 2001;82:871–882. doi: 10.1099/0022-1317-82-4-871. [DOI] [PubMed] [Google Scholar]
- Henkler F, Lopes AR, Jones M, Koshy R. Erk-independent partial activation of AP-1 sites by the hepatitis B virus HBx protein. J Gen. Virol. 1998;79:2737–2742. doi: 10.1099/0022-1317-79-11-2737. [DOI] [PubMed] [Google Scholar]
- Hoare J, Henkler F, Dowling JJ, Errington W, Goldin RD, Fish D, McGarvey MJ. Subcellular localization of the X protein in HBV infected hepatocytes. J. Med. Virol. 2001;64:419–426. doi: 10.1002/jmv.1067. [DOI] [PubMed] [Google Scholar]
- Huh KW, Siddiqui A. Characterization of the mitochondrial association of hepatitis B virus X protein, HBx. Mitochondrion. 2002;1:349–359. doi: 10.1016/s1567-7249(01)00040-x. [DOI] [PubMed] [Google Scholar]
- Keasler VV, Hodgson AJ, Madden CR, Slagle BL. Enhancement of hepatitis B virus replication by the regulatory X protein in vitro and in vivo. J. Virol. 2007;81:2656–2662. doi: 10.1128/JVI.02020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein NP, Bouchard MJ, Wang LH, Kobarg C, Schneider RJ. Src kinases involved in hepatitis B virus replication. EMBO. 2001;18:5019–5027. doi: 10.1093/emboj/18.18.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford RE, Butel JS. Construction and characterization of an SV40 mutant defective in nuclear transport of T antigen. Cell. 1984;37:801–813. doi: 10.1016/0092-8674(84)90415-x. [DOI] [PubMed] [Google Scholar]
- Lee T-H, Elledge SJ, Butel JS. Hepatitis B virus X protein interacts with a probable cellular DNA repair protein. J. Virol. 1995;69:1107–1114. doi: 10.1128/jvi.69.2.1107-1114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Bong Y, Poo H, Lee Y, Park J, Oh S, Sohn M, Lee S, Park U, Kim N, Hyun S. Establishment and characterization of cell lines constitutively expressing hepatitis B virus X-protein. Gene. 1998;207:111–118. doi: 10.1016/s0378-1119(97)00603-3. [DOI] [PubMed] [Google Scholar]
- Leupin O, Bontron S, Schaeffer C, Strubin M. Hepatitis B virus X protein stimulates viral genome replication via a DDB1-dependent pathway distinct from that leading to cell death. J. Virol. 2005;79:4238–4245. doi: 10.1128/JVI.79.7.4238-4245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Song YK, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Therapy. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- McClain SL, Clippinger AJ, Lizzano R, Bouchard MJ. Hepatitis B virus replication is associated with an HBx-dependent mitrochondrion-regulated increase in cytosolic calcium levels. J. Virol. 2007;81:12061–12065. doi: 10.1128/JVI.00740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melegari M, Scaglioni PP, Wands JR. Cloning and characterization of a novel hepatitis B virus X binding protein that inhibits viral replication. J Virol. 1998;72:1737–1743. doi: 10.1128/jvi.72.3.1737-1743.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melegari M, Wolf SK, Schneider RJ. Hepatitis B virus DNA replication is coordinated by core protein serine phosphorylation and HBx expression. J. VIrol. 2005;79:9810–9820. doi: 10.1128/JVI.79.15.9810-9820.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G, Avantaggiati ML, Chirillo P, Costanzo A, Artini M, Balsano C, Levrero M. Induction of the DNA-binding activity of c-jun/c-fos heterodimers by the hepatitis B virus transactivator pX. Mol Cell Biol. 1994a;14:989–998. doi: 10.1128/mcb.14.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G, Avantaggiati ML, Chirillo P, Puri PL, Ianni A, Balsano C, Levrero M. Ras- and Raf-dependent activation of c-Jun transcriptional activity by the hepatitis B virus transactivator pX. Oncogene. 1994b;9:2837–2843. [PubMed] [Google Scholar]
- Nomura T, Lin Y, Dorjsuren D, Ohno S, Yamashita T, Murakami S. Human hepatitis B virus X protein is detectable in nuclei of transfected cells, and is active for transactivation. Biochim. Biophys. Acta. 1999;1453:330–340. doi: 10.1016/s0925-4439(99)00004-6. [DOI] [PubMed] [Google Scholar]
- Palmer CR, Gegnas LD, Schepartz A. Mechanism of DNA binding enhancement by hepatitis B virus protein pX. Biochemistry. 1997;36:15349–15355. doi: 10.1021/bi972076m. [DOI] [PubMed] [Google Scholar]
- Rahmani Z, Huh KW, Lasher R, Siddiqui A. Hepatitis B virus X protein colocalizes to mitochondria with a human voltage-dependent anion channel and alters its transmembrane potential. J. Virol. 2000;74:2840–2846. doi: 10.1128/jvi.74.6.2840-2846.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaglioni PP, Melegari M, Wands JR. Posttranscriptional regulation of hepatitis B virus replication by the precore protein. J. Virol. 1997;71:345–353. doi: 10.1128/jvi.71.1.345-353.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schek N, Bartenschlager R, Kuhn C, Schaller H. Phosphorylation and rapid turnover of hepatitis B virus X-protein expressed in HepG2 cells from a recombinant vaccinia virus. Oncogene. 1991;6:1735–1744. [PubMed] [Google Scholar]
- Seto E, Mitchell PJ, Yen TS. Transactivation by the hepatitis B virus X protein depends on AP-2 and other transcription factors. Nature. 1990;344:72–74. doi: 10.1038/344072a0. [DOI] [PubMed] [Google Scholar]
- Sirma H, Weil R, Rosmorduc O, Urban S, Israël A, Kremsdorf D, Bréchot C. Cytosol is the prime compartment of hepatitis B virus X protein where it colocalizes with the proteasome. Oncogene. 1998;16:2051–2063. doi: 10.1038/sj.onc.1201737. [DOI] [PubMed] [Google Scholar]
- Sohlenius-Sternbeck AK. Determination of the hepatocellularity number for human, dog, rabbit, rat and mouse livers from protein concentration measurements. Toxicol. In Vitro. 2006;20:1582–1586. doi: 10.1016/j.tiv.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Song CZ, Bai ZL, Song CC, Wang QW. Aggregate formation of hepatitis B virus X protein affects cell cycle and apoptosis. World J Gastroenterol. 2003;9:1521–1524. doi: 10.3748/wjg.v9.i7.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Q, Schröder CH, Hofmann WJ, Otto G, Pichlmayr R, Bannasch P. Expression of hepatitis B virus X protein in HBV-infected human livers and hepatocellular carcinomas. Hepatol. 1998;27:1109–1120. doi: 10.1002/hep.510270428. [DOI] [PubMed] [Google Scholar]
- Takada S, Shirakata Y, Kaneniwa N, Koike K. Association of hepatitis B virus X protein with mitochondria causes mitochondrial aggregation at the nuclear periphery, leading to cell death. Oncogene. 1999;18:6965–6973. doi: 10.1038/sj.onc.1203188. [DOI] [PubMed] [Google Scholar]
- Takebe Y, Seiki M, Fujisawa JI, Hoy P, Yokota K, Arai KI, Yoshida M, Arai N. Sr-Alpha Promoter - An Efficient and Versatile Mammalian Cdna Expression System Composed of the Simian Virus-40 Early Promoter and the R-U5 Segment of Human T-Cell Leukemia-Virus Type-1 Long Terminal Repeat. Molecular and Cellular Biology. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Delgermaa L, Huang FJ, Oishi N, Liu L, He F, Zhao LS, Murakami S. The transcriptional transactivation function of HBx protein is important for its augmentation role in hepatitis B virus replication. J. Virol. 2005;79:5548–5556. doi: 10.1128/JVI.79.9.5548-5556.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn C, Bilodeau ML, Hullinger RL, Andrisani OM. Differential immediate early gene expression in conditional hepatitis B virus pX-transforming versus nontransforming hepatocyte cell lines. J Biol Chem. 1999;274:2327–2336. doi: 10.1074/jbc.274.4.2327. [DOI] [PubMed] [Google Scholar]
- Wang WL, London WT, Lega L, Feitelson MA. HBxAg in the liver from carrier patients with chronic hepatitis and cirrhosis. Hepatol. 1991;14:29–37. doi: 10.1002/hep.1840140106. [DOI] [PubMed] [Google Scholar]
- Waris G, Huh KW, Siddiqui A. Mitochondrially associated hepatitis B virus X protein consitutively activates transcription factors STAT-3 and NF-KB via oxidative stress. Mol Cell Biol. 2001;21:7721–7730. doi: 10.1128/MCB.21.22.7721-7730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Andrisani OM. The hepatitis B virus X protein targets the basic region-leucine zipper domain of CREB. Proc Natl Acad Sci U S A. 1995;92:3819–3823. doi: 10.1073/pnas.92.9.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang PL, Althage A, Chung J, Chisari FV. Hydrodynamic injection of viral DNA: A mouse model of acute hepatitis B virus infection. Proc. Natl. Acad. Sci. U. S. A. 2002;99:13825–13830. doi: 10.1073/pnas.202398599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z-S, Torii N, Hu Z, Jacob J, Liang TJ. X-deficient woodchuck hepatitis virus mutants behave like attenuated viruses and induce protective immunity in vivo. J. Clin. Invest. 2001;108:1523–1531. doi: 10.1172/JCI13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoulim F, Saputelli J, Seeger C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J Virol. 1994;68:2026–2030. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]