Abstract
The purpose of this study was to evaluate the potential for use of image volume based registration (IVBaR) to aid in measurement of changes in the tumor during chemotherapy of breast cancer. Successful IVBaR could aid in the detection of such changes in response to neoadjuvant chemotherapy and potentially be useful for routine breast cancer screening and diagnosis. IVBaR was employed in a new method of automated estimation of tumor volume in studies following the radiologist identification of the tumor region in the prechemotherapy scan. The authors have also introduced a new semiautomated method for validation of registration based on Doppler ultrasound (U.S.) signals that are independent of the grayscale signals used for registration. This Institutional Review Board approved study was conducted on 10 patients undergoing chemotherapy and 14 patients with a suspicious∕unknown mass scheduled to undergo biopsy. Reasonably reproducible mammographic positioning and nearly whole breast U.S. imaging were achieved. The image volume was registered offline with a mutual information cost function and global interpolation based on a thin-plate spline using MIAMI FUSE© software developed at the University of Michigan. The success and accuracy of registration of the three dimensional (3D) U.S. image volume were measured by means of mean registration error (MRE). IVBaR was successful with MRE of 4.3±1.7 mm in 9 out of 10 reproducibility automated breast ultrasound (ABU) studies and in 12 out of 17 ABU image pairs collected before, during, or after 115±14 days of chemotherapy. Semiautomated tumor volume estimation was performed on registered image volumes giving 86±8% mean accuracy compared to the radiologist hand-segmented tumor volume on seven cases. Doppler studies yielded fractional volume of color pixels in the region surrounding the lesion and its change with changing breast compression. The Doppler study of patients with detectable blood flow included five patients with suspicious masses and three undergoing chemotherapy. Spatial alignment of the 3D blood vessel data from the Doppler studies provided independent measures for the validation of registration. In 15 Doppler image volume pairs scanned with differing breast compression, the mean centerline separation value was 1.5±0.6 mm, while MRE based on a few identifiable structural points common to the two grayscale image volumes was 1.1±0.6 mm. Another measure, the overlap ratio of blood vessels, was shown to increase from 0.32 to 0.59 (+84%) with IVBaR for pairs at various compression levels. These results show that successful registration of ABU scans may be accomplished for comparison and integration of information.
Keywords: cancer, chemotherapy monitoring, medical imaging, breast ultrasound, diagnosis, image registration, image processing, image fusion, Doppler, validation
INTRODUCTION
Mammography is the accepted technique for breast screening aimed at detecting cancer at early stages. However, exposure to radiation, low positive predictive values, and low sensitivity in dense breasts are concerns.1, 2, 3 Contrast enhanced magnetic resonance imaging (MRI) is very useful, but it would be helpful to have noninvasive, less expensive, and time-consuming procedures.4 Physical palpation is used to study the breast qualitatively for size, shape, firmness, or location of mass. Despite the utility of clinical breast examination or palpation, it has been shown to be dependent on the examiner, interpreter, and other factors that could be relatively inaccurate.5 Ultrasound (U.S.) is used frequently to evaluate breast masses (mainly mammographic soft tissue densities as opposed to focal and clustered calcifications) as well as changes in the breast structure.6, 7 U.S. has been mainly useful in its ability to differentiate simple cysts from solid masses8, 9, 10 notably in mammographically dense breasts.11, 12, 13, 14 Breast U.S. scans are currently performed free hand by radiologists and technologists using U.S. systems providing real time two dimensional (2D) images with relatively small field of view. Conventional U.S. imaging is performed freehand in a different geometry than mammography, which can make it difficult to correlate lesions in the two modalities. Studies have shown that at least 10% of the time, lesions found in the U.S. images do not correspond with those in the mammograms.15
Currently, for tracking response to breast cancer therapy, qualitative and coarse quantitative changes are detected using palpation as the primary method. This is supplemented in some cases by visual comparison of images during the course of therapy. Even more accuracy and precision are required for treatment trials where early prediction of response is attempted. In patients undergoing therapy for malignant tumors, it can be vitally important to gauge the effectiveness of the therapy when there are questions regarding appropriate follow-up therapy. According to a study on the change in tumor volume measured using MRI, volume change estimation could possibly aid in evaluating response to chemotherapy and may act as a detection and diagnostic tool in the future.16 As for U.S. tracking, breast tumor boundaries have been typically poorly defined in image volumes after the onset of chemotherapy or radiation therapy.17 A small study on chemotherapy showed that it may induce inflammatory or fibrotic changes in the tumor and standard methods of assessing tumor size are only moderately successful.18, 19
Spatial registration that aligns temporally separated image volumes should aid in visual and automated detection and characterization of changes in volumetric images that are now available in several modalities.20, 21 We suggest that 3D U.S. registration of periodic screening studies may become highly useful in detecting and discriminating malignant changes. Manual registration for the evaluation of large sets of image volumes may be extremely time consuming and is subject to substantial user variability depending on skill, patience, and experience. Automated or semiautomated registration of two image volumes taken at different times was performed primarily to spatially align the two in the same coordinate system in order to allow better comparison and visualization of changes.22 This is becoming an effective tool in diagnostics, therapy planning, image-guided surgery, and treatment assessment.23, 24, 25 Registration helps in correcting for slight changes in scan positioning during subsequent studies of the patient and resolves ambiguities in positioning and actual location of some of the structures. This may help in detecting subtle changes that may go unnoticed in unregistered scans.
There are many different techniques to compute the geometric transformations that map the coordinates of corresponding points between two image volumes. The transformation model can be a simple rotate-translate model that is applicable for a rigid site, e.g., human skull. A more general linear transform is the affine transform that includes rotations, translations, shearing, and scaling. There are many situations where affine registration techniques are not sufficient to achieve alignment of anatomy. Hence, a nonrigid deformable model is needed to accurately represent the transformation. Some of the commonly used nonrigid models are based on parametrization, including general diffeomorphism (e.g., fluid model), spline based methods [e.g., B-spline and thin-plate spline (TPS)], Fourier based methods (e.g., statistical parametric mapping), etc.
In the “Demons” algorithm based on the fluid model, the image entities or demons push according to local characteristics of the image in a way similar to that proposed by Maxwell in solving for Gibbs free energy in thermodynamics.26 This algorithm, which is remarkably fast, uses the optical flow equation to determine the demons force at each pixel. When the gradient on the reference image is low, the model may not be efficient. The strength of the force that gets adjusted in the iterative process has been refined in subsequent studies.27, 28 Some of the salient features of this model include the use of gradient information from a moving image in order to deform it to a static reference image. At the same time, good understanding of the regularization is critical to obtain a useful deformation field.
B-splines are piecewise polynomial of the order of n with compact support and continuous (n−1)th derivative. Due to compact local support, B-splines can be used to model localized deformations and have advantages of reduced complexity and low computational time.29 The displacement at any point is given by the weighted sum of basis functions defined over a limited region. The mapping function in a B-spline based transformation is modeled based on translations of a regularly spaced grid of control points. In another spline based model—TPS, the transformation parameters are determined by minimizing the bending energy of a thin, hypothetical 3D metal plate with edges clamped at infinity.30 The spline coefficients are calculated by the least-squares method. In the TPS-based deformation model, control points are iteratively moved to maximize the similarity metric between the two image volumes. TPS can be used even if the control points are irregularly spaced, and changing the position of one control point changes the deformation of the overall image. Using more control points reduces this influence but leads to higher computation cost.31, 32 Selection of a few control points results in a rough match, whereas selection of a larger number of control points results in large local oscillations in the deformation model. Both these spline based methods suffer because the ideal number of spline parameters (number of control points) is unknown. In order to maintain the focus of this preliminary study on clinical application of image registration, we have applied a single nonrigid deformation model based on TPS.
Earlier registration work was applied to small volumes of tissue usually containing a central mass such as a tumor. In this study, we aim to evaluate the accuracy of a more difficult problem, image-based spatial registration of compressed, nearly whole breast image volumes. Our 3D U.S. system has been augmented with a motorized transducer carriage above a special compression plate developed in order to improve the coverage over the whole breast.33, 34, 35, 36, 37, 38
U.S. imaging, along with other coherent imaging systems, suffers from speckle noise caused by interference effects between overlapping echoes from randomly distributed structure scatterers that are too small to be resolved. Speckle degrades the signal-to-noise ratio of the image and this could lead to reduced performance in registration compared to most other imaging modalities.22 Registration of whole breast U.S. studies to each other is somewhat difficult due to noisy images, artifacts, heterogeneity, and high mobility of breast structures under mammographic-style compression. Hence, some of the U.S. studies were performed in the same session with patient repositioning to assess image registration accuracy under nearly ideal conditions. The accuracy was estimated by the evaluation of displacement of visually selected corresponding points (referred to as “fiducials”) in the two image volumes by registration. For increased accuracy in tumor change evaluation, three studies on each malignant tumor case (prior, during, and after chemotherapy) were performed.
When the registration is based primarily on the mass or on tissue structures surrounding the mass, a nonlinear transform could map the boundaries in the reference image to the boundaries in the homologous image.39, 40 This transform has information about the size change and could be used for the estimation of volume change. Using image volume based registration (IVBaR) and the radiologist identified tumor region in the prechemotherapy image volume, we have devised a new method of automated estimation of tumor volume.
Malignant masses have often been associated with increased vasculature in comparison to benign lesions.41, 42 Studies in the past have shown that the efficacy of U.S. for discriminating breast masses is improved by adding the information from color flow Doppler imaging to the B-mode grayscale information than just from B-mode imaging.43, 44 In this study, color flow Doppler scans were performed at each time point to not only quantify the blood flow in and around the lesion,45 but also to validate IVBaR of the 3D grayscale information in these color flow image volumes by the resulting alignment of the Doppler-imaged blood vessels that were not used in the registration. Previous studies have used information from major blood vessels in Doppler U.S. for fusing with MRI or computed tomography (CT) images.46, 47
The aims were to perform and assess the following:
-
(1)
Image volume based registration of automated U.S. in the mammographic geometry. IVBaR under minimal change in ABU scans was performed before and after repositioning for assessment of registration accuracy.
-
(2)
Tumor volume change estimation. Estimation of chemotherapy induced tumor volume change was performed using IVBaR and validated.
-
(3)
Independent validation using Doppler imaging. Color flow Doppler U.S. imaging was performed on selected subjects with sufficient blood flow to provide an independent measure of the accuracy of B-mode image registration.
METHODS AND MATERIALS
Human studies were conducted at the University of Michigan Comprehensive Cancer Center. An Internal Review Board approved all procedures and informed consents were obtained from patients.
Patient distribution
Subjects selected for this study included 10 women who were scheduled to undergo chemotherapy based on the previously confirmed presence of cancer and 14 women with suspicious∕unknown masses who were scheduled for biopsy with mean age of 46±9 yr. The mean time difference between pre- and postchemotherapy scans was 115±14 days. Among ten tumor cases, only seven were chosen for tumor volume estimation based on tumor visibility as discussed in Sec. 2F. The Doppler study was performed on eight patients (five scheduled for biopsy of a suspicious mass and three scheduled for chemotherapy) who had detectable blood flow (see Sec. 2G) with mean age of 49±8 yr. A subset of patients was repositioned and rescanned for the registration accuracy tests.
Setup
All U.S. scans were performed with a Logiq-9 U.S. system (General Electric Healthcare, Waukesha, WI), modified to acquire individual B-mode or color Doppler images on input trigger pulses. Images were obtained using a 10L linear array transducer at 10 MHz central frequency for grayscale imaging in most cases and in some cases with an M12L transducer at the same frequency using a maximum number of 8 transmit focal zones. Doppler imaging was performed at a central frequency of 6.6 MHz (pulse repetition frequency in the range of 0.6–0.9 kHz depending on flash artifacts).
Each patient scanning session began with a radiologist performing a 3D freehand U.S. scan across the breast to confirm the region of interest and to look for blood flow. Volumetric U.S. data were obtained and stored in the cine loop buffer of the U.S. system by manually sweeping the U.S. transducer across the volume of interest. The patient was then positioned, as shown in Fig. 1, with her breast between the top compression paddle, a TPX plate (polymethyl pentene), and the bottom supporting plate. A biocompatible mild adhesive spray (Got2b glued spiking spray, Advanced Research Laboratories, Costa Mesa, CA) stabilized the compressed breast while providing reasonable acoustic coupling between the top plate and the breast. The patient was seated comfortably throughout the scan to minimize motion artifacts.
Figure 1.
View of the breast under partial compression by mammography-style compression plates with the patient slightly rotated away in order to show the apparatus. Here, the transducer holder was spring loaded in a frame attached to the paddle and moves above the compression plate with the transducer following the paddle surface. Notice that the transducer holder can be rotated about the vertical axis for scans at angles not parallel to the chest wall.
The automated breast ultrasound (ABU) scanning assembly consisted of a two-axis U.S. transducer positioning system above the top compression plate, as shown in Fig. 1. The U.S. transducer was placed in a holder that was attached to computer-controlled positioning system. Spring-loaded vertical motion of the holder allowed the transducer to follow the slightly curved surface of the compression plate. All ABU scans were performed in the cranial-caudal (CC) position with prior assessment of location of the mass and information including pathology and conventional mammograms. A 3D grayscale image volume was obtained by translating the transducer in the elevational direction by 12–13 cm. Proper coupling between the transducer and the TPX plate was achieved with water for CC views and coupling gel for others. In order to reduce the air gap between the breast periphery and the plate, a viscous, bubble-free U.S. gel (LithoClear, Sonotech, Inc., Bellingham, WA) was applied. The 3D U.S. volumes were acquired as a parallel stack of 2D U.S. images taken with elevation separation of 0.4 mm and axial∕lateral voxel dimension of 0.1–0.15 mm depending on the depth of imaging. Imaging a test object using the M12L transducer through the TPX plate in order to determine the line spread function revealed that the elevational, lateral, and axial full width at half maximum resolutions were approximately 1.0, 0.4, and 0.2 mm.35 These methods and the effect of using a compression plate for U.S. scanning were addressed.36 The U.S. system trigger rate was set at 5 Hz resulting in a transducer translation rate of 2 mm∕s. One to three parallel 39 mm wide automated B-mode U.S. sweeps were performed as needed to cover the entire breast, depending on its shape and size under compression and to the extent allowed by the achieved acoustic coupling. Sweeps overlapped by 1 cm. Other hardware and software interfaces developed to perform automated scanning, which included computer triggering of the Logiq-9 system for data acquisition, were discussed elsewhere.33
The region of interest (ROI) was identified in the automated grayscale scans to guide a cardiac-gated Doppler scan that was performed over the ROI in a single sweep of the U.S. transducer. This study was performed on patients with cancers or with undiagnosed suspicious masses having sufficient blood flow to be visible in the freehand U.S. Doppler scan performed by the radiologist. The Logiq-9 image acquisition was triggered at the peak of the R-wave in the ECG to allow blood flow to peak at the breast after an appropriate time delay of 165 ms based on a pilot study of four healthy volunteers. After that the transducer was translated to its next location by the computer-controlled positioning system. A 50 ms time delay was then implemented to let the transducer positioning assembly stabilize and to minimize flash artifacts in the Doppler image before arming the ECG trigger. The process was then repeated for the length of the scan. A Doppler image volume with color information covered approximately 4×4×4 cm3.
In order to evaluate reproducibility, a subset of patients was taken out of compression and given a break, and all the above positioning and scans were repeated within 30 min in the same session.
Image acquisition
A total of 19 3D ABU grayscale scan pairs was acquired in ten women consisting of nine pre- and midchemotherapy scan pairs, five mid- and postchemotherapy scan pairs, and five pre- and postchemotherapy scan pairs. For reproducibility evaluation of image volume by registration in the simplest relevant cases, repositioning and recompressing the breast in the same session within 30 min, ten pairs of grayscale ABU volumes were acquired.
In addition, to evaluate the effects of compression on vascularity measures, automated 3D cardiac-gated Doppler U.S. scans were collected at multiple compressions of the breast in the same session in selected subjects with detectable blood flow (three patients undergoing chemotherapy and five with a suspicious∕unknown mass). The initial Doppler scan was taken at near mammographic compression and the subsequent scans were taken either at the same, relaxed or additional compression as acceptable to the patient. Each Doppler acquisition took less than 4 min.
Image preprocessing
After image acquisition along the slightly curved surface of the compression plate, image data from each sweep were aligned rapidly with those of its neighboring sweep using a 1D cross-correlation technique along the elevation direction in the overlap region of 100×389×383 voxels or about 10×39×153 mm3. This helped in obtaining a self-consistent single image volume covering most of the breast, typically with size of 700×389×383 voxels or about 70×39×153 mm3 for a typical two transducer sweeps. Due to the instability in the transducer holder and stepping motion, a mean misalignment of 1±0.8 mm was detected along the elevation direction between two adjacent sweeps and corrected in all 19 ABU image volume pairs.
Doppler image volumes were acquired by a single sweep of the U.S. transducer over the ROI surrounding the tumor. Every automated 3D color flow image volume was converted into two processed image volumes before registration. First, the grayscale portion of the color flow image was retained, while the color portion was zeroed out (referred to as Doppler image G or DI-G). Second, in every automated 3D color flow image volume, the color portion of each Doppler image was altered to a uniform color while the grayscale portion was zeroed out (referred to as Doppler image C or DI-C). The color portion of the reference Doppler volume set was replaced by uniform red color, shown as dark gray in grayscale figures, and that of the final volume set by uniform green, shown as medium gray in grayscale figures. The utility for these two image types is described in Sec. 2G.
Image volume based registration (IVBaR)
Registration was performed semiautomatically and with limited user interaction using the well-studied MIAMI FUSE© software developed at the University of Michigan. This algorithm maximizes a similarity measure, the classical Shannon mutual information (MI).22, 48 Registration of two U.S. image volumes depends on the available MI in the image volumes, which, in turn, depends on the amount of true structural information such as specular reflectors and changes in volumetric scatterers as well as shadowing, other artifacts, and deformation of local tissue.
As a first step of registration, a 3D full-affine transformation was performed to take into account changes that are global in nature between the two image volumes including translation, rotation, scaling, and shear.49, 50 Ordered pairs of Nc control points were manually selected at presumed corresponding locations in the two 3D volumes to establish initial correspondence. In the full-affine transform, a mean Nc of four pairs of points (not all in the same plane) was selected. The algorithm utilizes the Nelder∕Mead downhill simplex optimization by iteratively moving the positions of the control points in the homologous image volume with an affine transform. All other voxels were moved and interpolated with the same transform until the MI was maximized.
The nonrigid TPS-based transformation was then performed in order to accommodate elastic deformations of the breast and local changes in the tissues. This needed at least five control points in each 3D image volume pair. The locations of control points in the homologous image volume were moved incrementally by the algorithm and the locations of other voxels were interpolated using TPS. The algorithm maximizes MI as above.51, 52, 53 The TPS mapping has 3Nc degrees of freedom and was globally defined everywhere. The computational time was dependent on the size of the reference image volume and Nc. In nonrigid transforms, additional set(s) of control point pairs were manually selected in the grayscale U.S. image volume, here with mean Nc of 15. In performing the transformations in the sequence of rigid, affine followed by nonrigid, the data volumes were downsampled in a coarse-to-fine approach, referred to as a multiresolution optimization scheme. This strategy assured a smooth global to local deformation and also that the control points did not fall into local minima, keeping the registration process efficient, robust, and less time consuming.54, 55
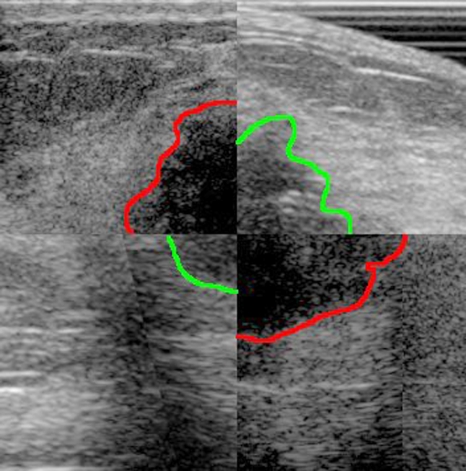
As an example, in Fig. 2, a 2D composite image consisting of registered pair of pre- and postchemotherapy images is shown with a rough hypoechoic boundary. The larger tumor in the quadrants from the pretreatment exam shows that the tumor had reduced in size with therapy. Registration aligned the two image sets in the same coordinate system, thus making comparisons easier.
Figure 2.
Checkerboard (2×2) pattern of a registered pair of a prechemotherapy image (top left and bottom right squares) and a postchemotherapy image (top right and bottom left squares). The hypoechoic boundaries are drawn for convenience [dark gray (red in online figure) for pre-chemotherapy and medium gray (green in online figure) for postchemotherapy images]. The mean registration error was 8.4±2.6 mm. It can be seen from spatial alignment of these images by registration that the tumor had shrunk in size with therapy. Other reasons could include changes due to differences in compression forces and thickness, as well as differences in positioning within the range of normal mammographic repositioning.
Manual selection of fiducial points was a time-consuming procedure that was also prone to inaccuracy. In the absence of a “gold standard,” this constituted a direct way to estimate registration error in vivo without exterior or interior placed landmarks that were impractical in breast imaging. In this preliminary study, a senior radiologist performed the manual selection of fiducials as identical, point-defining structures including branching ducts, vessels, glandular, and connective tissue patterns in the unregistered reference and homologous image volumes. The registration transformation applied to this set of fiducials in the reference image volume gave the location of these reference fiducials in the homologous data space. The mean Euclidean distance between these transformed fiducial points and their corresponding points selected earlier in the homologous image volume gave the mean registration error (MRE). Note that the MRE was dependent on the radiologist’s positioning and distribution of the fiducials in both the image volumes and hence was dependent on the radiologist’s skill and patience. Our estimation of MRE includes the image registration error as well as error in hand-picking fiducials.
Also measured was the mean pixel displacement, which was defined as the Euclidean displacement of every pixel in the U.S. image volume due to registration transformation. Note that this automated measure was not a measure of registration accuracy but overall motion of pixels due to transformation.
Tumor volume estimation using IVBaR in ABU image volumes
For automated estimation of tumor volume in the postchemotherapy ABU image volumes, the tumor in the prechemotherapy image volume was hand segmented by the radiologist using in-house segmentation software programmed in MATLAB®. For validation purposes, the tumor volume in the postchemotherapy image volume was also hand segmented by the radiologist. Using IVBaR and the radiologist identified tumor region in the prechemotherapy scan, we designed a new method of automated estimation of tumor volume in subsequent studies as outlined below.
Typically, boundaries of breast tumors in U.S. image volumes are even more poorly defined after the onset of chemotherapy.19 An assumption in this study was that the only tissues to change in the chemotherapy were those of the tumor and the boundaries of the tumor. The automated estimation of tumor volume of the later scan involved these steps:
-
(1)
Prechemotherapy image volumes were registered with later scan (mid- or postchemotherapy) image volume.
-
(2)
Radiologist hand segmented the tumor region in both image volumes by setting intensity of tumor regions to zero (process of “masking”).
-
(3)
The transformation used in step (1) was applied to the prechemotherapy image volume in step (2) to obtain the automated tumor region in the image space of the later scan.
-
(4)
The automated tumor volume of later scan was obtained by summation of pixels in automated tumor region in step (3).
-
(5)
Radiologist’s estimation of the tumor volume of later scan was obtained by summation of manual segmentation of tumor region, which was then compared to that of step (4).
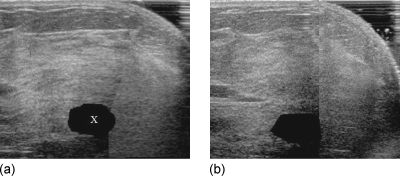
Shown in Fig. 3 are the transformed prechemotherapy tumor volume in Fig. 3a and the hand-segmented postchemotherapy tumor volume in Fig. 3b. In this case, the volume estimate from the registration-based method was within 91% of the radiologist’s measurement of tumor volume.
Figure 3.
(a) Slice from prechemotherapy image volume mapped into the space of postchemotherapy image volume. The blacked out region (marked as “X”) was obtained by applying the transformation of the pre- to postchemotherapy registration to the hand-segmented prechemotherapy tumor volume. (b) The corresponding slice in the hand-segmented postchemotherapy image volume for validation.
Evaluation of registration accuracy using Doppler U.S.
In the 3D color flow Doppler image volume, summation of the number of color pixels gave a value, which, ignoring attenuation effects, was related to the volume of relatively fast moving blood. As blood vessels were widely distributed throughout and hence reflect the deformations of the breast, the movement of blood vessels was used as a surrogate to the movement of muscle, fat tissue, glandular tissue, mammary glands, ducts, and breast masses. Doppler reproducibility studies under similar breast compression and identical system settings were performed in the same session in order to study the overlap of blood vessels using image registration. Various color flow Doppler quantities were also estimated as mentioned in an earlier study.45 Comparing the spatial overlap of color flow pixels between the Doppler image volume pairs before and after registration, three independent and automated measures for validation of B-mode registration were identified in the final aim of this study.
All U.S. system settings were identical between studies on the same individual since Doppler signals are very sensitive to system settings. Color Doppler artifacts were removed and the overlap regions in corresponding color vessel segments in the two registered image volumes were identified and cropped. Three methods were identified for independent validation of B-mode image volume registration using the Doppler image volume:
-
(a)
Blood vessel overlap (BVO) ratio in the region surrounding the mass,
-
(b)
Automated separation of the centerlines of blood vessels, and
-
(c)
Manual separation of radiologist selected landmarks.
In method (a), nonlinear registration of the two Doppler image volumes was performed. As the first of two steps in registering Doppler image volumes, the original grayscale portion of the Doppler image volume DI-G1 was registered to its later counterpart DI-G2. The resulting transformation was applied to the color-information only DI-C1 image volume, and the registered color image (DI-C) pair was displayed one on top of the other in the same coordinate system. This display shows in red (shown as dark gray in the grayscale figure) the original DI-C1 image volume, in green (shown as medium gray in the grayscale figure) the registered DI-C2 image volume, and in yellow (shown as light gray in the grayscale figure) the overlap region. In this cardiac-gated Doppler study, even with identical system settings as well as breast compression, there were differences in the region of blood vessel imaged in U.S. depending on the time-varying physiological conditions of the subject. As one automated relative measure of registration accuracy, the BVO was defined as the Doppler intersect volume (yellow, or light gray in the grayscale figure) ) divided by the smaller of the two Doppler volumes which is red plus yellow or green plus yellow (dark gray plus light gray or medium gray plus light gray in the grayscale figure). This was chosen to give a scale of 0–1 in all cases and BVO of 0.5 when the segments were of the same size and overlap by half. This measure was evaluated for eight patients at either equal to or at ∼±10% of equal compression levels as measured by plate separation. In addition, changes in the blood volume were evaluated at varying compression levels.56
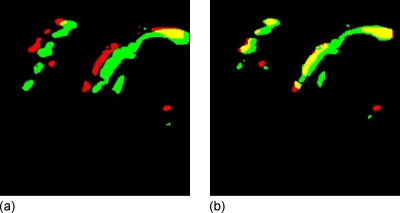
Figure 4 illustrates the use of BVO in color flow data as a method to quantify relative registration error, which is independent of the grayscale data used in the registration process. Breast compression thicknesses were 7.0 and 7.5 cm, and volumes of color flow pixels were measured as 880 and 1220 cm3, giving a fractional change in color pixel density (CPD) of 0.4 and a change in BVO from 0.51 before registration to 0.74 after registration.
Figure 4.
(a) Unregistered pair and (b) registered pair in a slice of 3D Doppler color image volumes with grayscale portion blacked out. Dark gray (red) was the color of blood flow under moderate breast compression, medium gray (green) under relaxed compression, and light gray (yellow) was the overlap region. Notice the increase in flow with relaxation and also the increase in the light gray (yellow) region with registration. The registration was performed on the grayscale portion of the image volume with colored pixels zeroed. Refer to Figs. 56.
In method (b), registration accuracy could be obtained from the displacement of the vessel segments from each other in the registered image volume pair. The appearance of a color pixel in a Doppler image volume depends on Doppler U.S. system settings including PRF, WF, and grayscale-color balance. In a real time 2D Doppler U.S. image, portions of the blood vessel segment were not always seen; the diameter appeared to fluctuate with the cardiac cycle and with subtle variations in attenuation by overlying tissues as the view was changed slightly under identical Doppler settings in the U.S. system. The length of the displayed vessel segment varied even more substantially, and if not tightly curved, contained little measurable information about vessel position along the direction of the vessel. A curved line along the vessel’s geometric center could best represent the position of the blood vessel segment in the 3D Doppler image set, but the information on the location of the curve at any one point was only in the two dimensions normal to the vessel segment at that point. Measurement in 3D of the normal displacement between the two renderings of the vessel gave the registration error at that location in two of the three dimensions.
Centerline separation value (CSV) was defined as the perpendicular distance between the centers of blood vessels measured in the 3D image volume. CSV of blood vessels is based on a curved line passing through the geometric center of the vessels. As totally automatable and independent of the image information used in registration, the CSV is a good alternate indicator of registration error. MRE is based on a few observer-identified points on the structures in grayscale image volume. In order to match the reader study procedure and for convenience in programming, the fitting to the centers of the vessels was performed only within individual 2D images of the 3D Doppler image volume along the maximum length of the vessel. This distance is the registration error vector component in one of the three dimensions in space. Thus, for assumed isotropic error, CSV3D should yield a value that is approximately √3 (1.73) times CSV. If performed in a full 3D fit, CSV3D would give a more accurate 2D estimate of registration error.
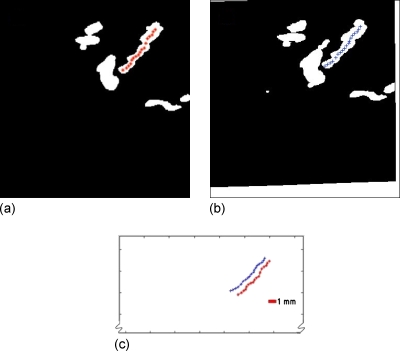
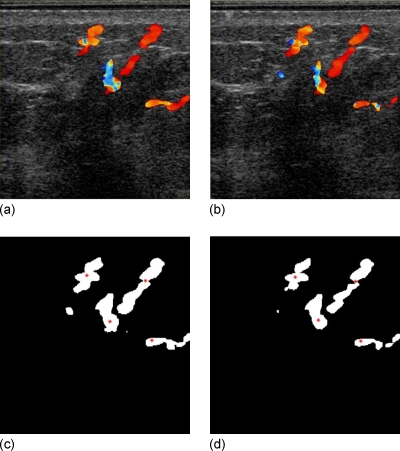
In our software, the semiautomated extraction of the centerline needed two inputs from the user—start and end points of the blood vessel segment. The algorithm worked as follows. The lengths of line segments within the blood vessel passing through the starting point at 0°, 45°, 90°, and 135° were measured in the image plane. The midpoint of the line segment with minimum length among the four was marked as the geometric center of a small region of the blood vessel around that point. Along the line segment of maximum length, another point at a distance d from the location of the previous point was identified as the next center point. The process of identification of center points was repeated until the end point was reached. A value of d=4 pixels was used. As a technique demonstration, two test cases were evaluated with one at equal breast compression and the other at reduced compression, taken in the same session. An example of the method shown in Fig. 5 is a masked Doppler image volume pair acquired before and after relaxation of compression along with the points on the centerline.
Figure 5.
2D automated centerline extraction algorithm has identified points along the centerline on (a) the reference image mask and (b) the registered homologous image mask. (c) Lines of pluses and stars are the centerlines of blood vessel from (a) and (b), respectively. The CSV was estimated to be the mean length of separation between these two line segments. In this compression-relaxation study, CSV was estimated to be 1.0±0.4 mm, while the reference MRE obtained from registration of the corresponding DI-G grayscale image volume pair was 0.8±0.4 mm.
In method (c), the presence of special features such as branches in the blood vessels in the Doppler image volumes could provide identifiable homologous points, which were more valuable than lines for 3D estimate of registration error. The mean separation of the radiologist identified Doppler fiducial (DF) points yields DF separation value (DFSV) that could be compared to the reference grayscale MRE. In order to demonstrate the technique in this study, two cases were selected with sufficient visibly identifiable blood flow patterns, one pair at equal compression levels and the other pair at different compressions. Again an example of the method was shown. Figures 6a, 6b show the original 2D Doppler image pairs of a region around the tumor acquired at equal breast compression, separated by repositioning, in the same session. In Figs. 6c, 6d are shown the registered 2D Doppler image pair with DF points along the center of twisting blood vessel segments or bifurcations.
Figure 6.
A patient with invasive ductal carcinoma showing hypoechoic, irregular shaped mass on the left breast scanned before chemotherapy. (a) and (b) Doppler image pair showing blood flow around the tumor scanned at equal breast compressions in the same session within 30 min of each other. (c) and (d) Registered DI-G image pairs of reference and homologous image volumes, respectively. Radiologist identified Doppler fiducial (DF) points on twisting blood vessel segment are shown for reference as gray (red)-colored stars. Separation between 17 of the DF pairs was 0.3±0.3 mm compared with an MRE of 0.2±0.2 mm from fiducials on the grayscale image volumes.
RESULTS AND SUMMARY
Registration transformations that yielded an MRE less than 10 mm were considered as successful registrations.
IVBaR on reproducibility patients. Registration was successful on nine out of ten ABU reproducibility studies with MRE±SD=3.2±1.2 mm (maximum=5.5 mm). One out of ten registrations of reproducibility study cases failed with MRE±SD=22.4±9.3 mm due to external skin marks from an earlier surgical procedure that made repetition of mammography-style compression very difficult and registration unsuccessful.
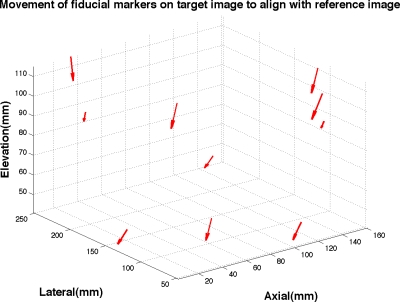
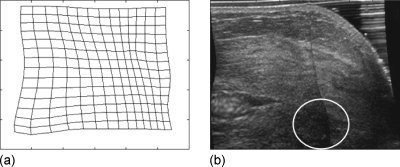
IVBaR on chemotherapy patients. Among the ten subjects undergoing chemotherapy, five were evaluated at all three time points along chemotherapy of which only three were evaluated in the same view (CC) resulting in three pairs each of pre-mid-, mid-post-, and pre-postcombinations. Results on the 17 image volume pairs on chemotherapy cases (as presented in Table 1) includes the successful registration on five out of seven pre- to midchemotherapy, four out of five mid- to postchemotherapy, and three out of five pre- to postchemotherapy scans with an overall MRE±SD=5.2±2.0 mm (maximum=9.2 mm). Shown in Fig. 7 is the displacement of fiducial markers in the transformation of a prechemotherapy to postchemotherapy B-mode ABU image volume. In Fig. 8a, a rectangular grid of deformation in the U.S. imaging plane is shown for a successful registration with MRE of 4.5±2.1 mm. Figure 8b is the corresponding B-mode image from registering the mid- to postchemotherapy image volumes. An indistinct hypoechoic mass is present at the bottom center of the 2D image shown roughly by a white colored oval.
Table 1.
Description of various types of scans with the number of successful registrations. The mean, standard deviation, and maximum registration errors and pixel displacement of registered 3D U.S. grayscale image volumes in the study population going through chemotherapy were shown. 12 out of 17 longitudinal scans and overall 21 out of 27 3D ABU grayscale scan pairs were registered with the MRE=5.2±2 and 4.3±1.7 mm, respectively. Corresponding mean pixel displacements are 12±2.6 and 9.9±2.4 mm.
| Type of scan | No. of registered∕No. of total | Registration error (mm) | Pixel displacement (mm) | ||||
|---|---|---|---|---|---|---|---|
| Mean | SD | Max | Mean | SD | Max | ||
| Reproducibility | 9∕10 | 3.2 | 1.2 | 5.5 | 7.2 | 2.1 | 13.4 |
| Pre- to midchemotherapy | 5∕7 | 4.8 | 1.7 | 7.9 | 13.7 | 3.4 | 24.8 |
| Mid- to postchemotherapy | 4∕5 | 5.5 | 2.5 | 9.2 | 10.5 | 1.9 | 15.6 |
| Pre- to postchemotherapy | 3∕5 | 5.3 | 1.9 | 8.1 | 11.1 | 2.3 | 16.8 |
Figure 7.
Movement of fiducial markers on target postchemotherapy image volume to align with the reference prechemotherapy image volume with MRE=5.5±1.5 mm (maximum=13.2 mm), as estimated by automated registration. (Size of arrows doubled in order to view arrow heads better).
Figure 8.
(a) Rectangular grid of deformation in the U.S. image plane of a successful registration with MRE=4.5±2.1 mm and (b) the corresponding slice in the mid- to postchemotherapy registered image volume with indistinct hypoechoic mass at the bottom center identified roughly by the white oval.
Among these tumor cases, three cases were registered successfully at all stages of chemotherapy and had lower MRE than most of the other registered cases. This might explain the slightly low registration errors as well as pixel displacement in the three pre- to postchemotherapy image volume registration (refer to the last row in Table 1). Tissue deformability, phase aberrations, and refraction artifacts due to inhomogeneities are large in the human breast leading to high registration errors. Reasons for inability to register some of the U.S. scans include necessary changes in compression thickness and force possibly due to patient weight loss and also positioning differences within the range of normal mammographic repositioning. The five unsuccessfully registered image volume pairs of chemotherapy cases have MRE±SD of 30.8±18.4 mm.
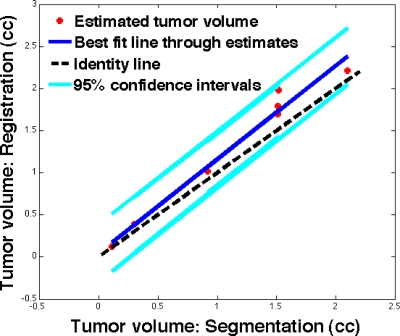
Tumor volume estimation. Using the transformation map obtained in registering the original pre- to later (mid- or post-) chemotherapy grayscale image volumes, later scan-chemotherapy tumor volume was obtained using registration method. Due to unsuccessful registration, partial tumor visibility, dominating shadowing artifacts, and multifocal tumors, only seven out of ten cases were selected for the automated tumor volume estimation. In the seven cases evaluated, the radiologist estimation of tumor volume in prechemotherapy scan ranged from 0.2 to 9.93 cm3 (mean=3.2 cm3). In the later scan, radiologist estimation of tumor volume ranged from 0.1 to 2.1 cm3 (mean=1.3 cm3) as compared to the registration-based automated tumor volume estimate that ranged from 0.1 to 2.2 cm3 (mean=1.5 cm3), as shown in Fig. 9. The correlation coefficient between these estimates was 0.9876 (P<0.0001). The mean percentage change in the ratio of tumor volumes from the tested method and manual method was 86±8%. Thus, prior knowledge on the boundary of the lesion in the reference image volume and successful registration allow the volume of the lesion in the homologous image volume to be obtained with reasonable accuracy. Furthermore, the longest linear tumor dimension from the margins in automated and hand scans were within 20±13% of those from the pathology, where the tumor specimen was often distorted from its shape in vivo. The main advantage of such automated tumor volume estimation was the reduced necessity to manually identify the tumor boundary in subsequent scans. This saved the radiologist’s time and perhaps gave a more accurate fractional volume change.
Figure 9.
Postchemotherapy tumor volume as estimated by automated registration in comparison with that of the radiologist hand-segmentation in seven cases. The reference line (y=1.11x+0.05) derived by least-squares method was shown along with 95% confidence interval lines. The black dotted line was the identity line shown for reference.
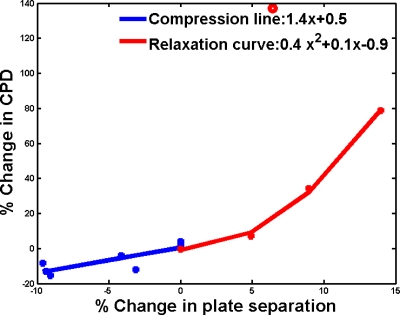
Doppler imaging. The color flow Doppler image pairs were acquired on a small region surrounding the lesion at multiple compressions on three patients undergoing chemotherapy and five patients with suspicious∕unknown masses using identical Doppler settings. From the Doppler studies, the detectable vascularity or CPDs were estimated to change by +1.5%, −9%, and +86%, respectively, for equal compression reproducibility, for 7% increased compression and for 9% relaxation of breast compression. In relaxation of breasts that were under compression, an initial rush of blood suggested that the steady state was not reached in a time frame of 1–2 min and further investigation was needed. For reproducibility and changing compression cases, fractional change in CPD measured as a function of fractional change in plate separation distance is shown in Fig. 10. For additional compressions below 10% of the original compression level, the resulting decrease in CPD gave a linear best-fit estimate of 1.4x+0.5, where x=percentage change in plate separation for increasing compression (x<0). Relaxations up to 15% resulted in a strong increase in CPD, recorded after the initial rush of blood flow, giving a second order polynomial fit to the change in CPD of 0.4x2+0.1x−0.9 for x>0.
Figure 10.
Percentage change in detected vascularity (CPD) given as a function of percentage change (x) in breast compression or plate separation for eight patients. Doppler measurements were performed after subject had been under compression for the approximately 10 min duration of other scans. Additional compression beyond that for the pulsatility resulted in a drop in blood flow, giving a linear fit to CPD change: 1.4x+1.5. Relaxations below 15% resulted in a sudden increase in CPD, which may suggest that measurements should be acquired after a brief relaxation period to avoid the non-steady-state rush of blood from the measured region. Ignoring the obvious outlier (open circle), CPD changed in a nonlinear manner (refer to text).
Doppler registration accuracy. Table 2 indicates that applying the transform obtained via grayscale registration to the color pixels of the Doppler images produces an increase in BVO, showing that the blood vessels, tissues, and other structures overlay better spatially after registration. Increase in BVO was nominal (31%) when registering image volumes acquired at the same compression level, as there was little misalignment between scans. On the other hand, the increase in BVO (213% and 88%) was high in cases with unequal breast compression levels, indicating that there was considerable shift of tissue structures with changing compression. Also summarized in the third column in Table 2, the MRE is substantially lower in registrations of image volumes acquired with changing breast compression than that with the pre- to postchemotherapy scans (refer to the third column in Table 1). In these 15 Doppler image volume pairs, BVO increased from 0.32 to 0.59 or about +84% with IVBaR for various compression levels.
Table 2.
Various study groups of population that underwent the different amounts of compression. Note that #C denotes number of patient undergoing chemotherapy and #M denotes number of patients with suspicious or unknown mass who were also scheduled to undergo biopsy. The third and fourth columns give the mean registration error (mm) in registering the grayscale portion of the Doppler image volume (DI-G pair) and the corresponding mean CSV (mm) of registered image volume pair. The fifth column gives the change in BVO with registration.
| Doppler study type | #Scan pairs on #patients | MRE (mm) | CSV (mm) | Change in BVO |
|---|---|---|---|---|
| Reproducibility | 4 on 3C | 0.6±0.4 | 0.7±0.3 | 0.59–0.77 (+31%) |
| More compression | 7 on 2C,5M | 1.2±0.5 | 1.8±0.6 | 0.15–0.47 (+213%) |
| Less compression | 4 on 1C,1M | 1.6±0.9 | 1.8±0.8 | 0.33–0.62 (+88%) |
In the centerline separation method, all the cases shown in Table 2 are evaluated with equal, increased, and relaxed breast compression. In these 15 Doppler image volume pairs, the mean MRE and mean standard deviation were estimated to be 1.1±0.6 mm, while the corresponding mean CSV was 1.5±0.6 mm. In a breast relaxation study shown in Fig. 5, a CSV of 1.0±0.4 mm is obtained from 289 points along the centerlines of 13 blood vessel segments. This is comparable to the reference MRE of 0.8±0.4 mm from registration of DI-G grayscale image volume pair. In the same subject, equal breast compression Doppler image volume pair acquired in the same session yielded a CSV of 0.3±0.1 mm from 299 points along the centerlines of 13 blood vessel segments that is comparable to the reference MRE of 0.2±0.2 mm.
In the radiologist identified fiducial method of evaluating registration accuracy on two test cases, 17 fiducial points were identified on the color flow pairs acquired in the same session with equal compression as shown in Fig. 6 and the DFSV of 0.3±0.3 mm is comparable to the reference grayscale MRE of 0.2±0.2 mm from grayscale image volume. In another Doppler image volume pair with 7% relaxation in breast compression, the DFSV value obtained using 30 fiducial points was 1.3±0.5 mm versus the reference grayscale MRE of 0.8±0.4 mm from registration of DI-G grayscale image volumes.
CONCLUSIONS AND DISCUSSIONS
In the majority of pre- and postchemotherapy ABU scans, it was possible to spatially align the two temporally separated image volumes with a modest alignment error sufficiently small enough to aid identification of tumor remains after half or all of the neoadjuvant chemotherapy treatment. Breast image volume pairs acquired in the same session were registered in order to obtain a baseline registration study with minimal internal change. Almost all the registration on pairs involving immediate repositioning of the breast cases was successful, with registration error within a few millimeters. Furthermore, in this study, automated U.S. scans have fixed slice separation (e.g., 0.4 mm) and thus eliminated the approximation involved in estimating the tumor volume in hand scans. It can be seen from BVO, grayscale MRE, and Doppler MRE in reproducibility and change in compression color flow Doppler studies that spatial registration does align the image volumes. This alignment appears to be enough that it should help with the often difficult identification of the residual tumor location in post-treatment cases with full or substantial regression.
While MIAMI FUSE© is a very versatile algorithm for multimodality registration, it is computationally intensive, limiting the number of control points by working on the entire image volume. Deformations with high spatial frequencies may not be fully recovered. Also, registrations could be improved with better placement of initial control points or by adding control points within the ROI. More work is needed in finding the optimal number of control points for 3D U.S. image volume of a given quality. Clinical registration is expected to become faster with better optimization of algorithms and faster computers. Spatial compounding or alternate registration methods may improve registration results achieved in this study.57 Spatial compound imaging, which is now available in many U.S. systems, increases signal-to-noise ratio by using multiple scan directions and reduces shadowing, motion and refraction artifacts.58 Median filtering59 and speckle reduction imaging60 available in the Logic9 system could also improve registration accuracy. Great care should be taken with image processing techniques as the images may look unfamiliar to clinicians. Thus for practical considerations, it may be therefore be necessary to retain original image and use the processed image as secondary source of information.61
Measuring the accuracy of clinical registrations was a very challenging task in the absence of a gold standard. For such evaluations, multiple observers may be required to mark fiducials as well as the boundary of the tumor. Even though a multireader study would bring out statistically significant results along with inter-reader and intrareader variability comparisons, this could not be performed in this preliminary study and could be a part of a larger clinical trial. A breast phantom with tissues and structures similar to that in breast and which can mimic the chemotherapy induced changes in a breast mass was fairly difficult to make.62 Numerical models have been tested, but it was outside the scope of this study.
Estimation of tumor volume change is considered as a good tool for assessing the effectiveness of therapy. Conventionally, the single longest linear dimension was considered as a measure of the tumor size. In some studies, the three longest orthogonal linear dimensions are considered as a measure of the tumor volume. The effectiveness of therapy was classified as a complete or partial responder depending on the change in these one or three linear dimensions. The methodology used in this manuscript was to estimate the actual tumor volume change using image registration, having measured the initial tumor volume. IVBaR applied on a tumor segmented prechemotherapy image volume provides a means to track the remaining tumor tissues and residues in the postchemotherapy image volume and also to determine change in tumor volume automatically. This has been compared to the radiologist’s hand-segmented estimate of the postchemotherapy tumor volume. In this process, the segmented tumor region in prechemotherapy image volume does not contribute toward the registration. Upon the successful application of this procedure to a larger clinical study, this could have clinical impact by minimal use of radiologist’s time in identifying tumor region in subsequent scans.
Using Doppler image volumes, three independent indicators of accuracy of the registrations of B-mode image volumes (blood vessel overlap, centerline extraction, and radiologist identification of fiducial features) have been explored. The first two of these three measures are automated and minimally dependent on the observer. Among these measures, CSV of blood vessels is based on a best-fit curved line passing through the geometric center of the blood vessels and hence should be less noisy indicator of registration error than MRE and DFSV which are based on a few points on the structures in the grayscale image volumes and blood vessels in the Doppler image volumes, respectively. These methods can be applied to any other easily segmented objects in image volumes from any imaging modality, as those segmented features were not critical for the registration. Doppler scans covered a smaller ROI of breast, generally centered under the compression plate when compared to the whole breast B-mode image volume. This could have contributed to a lower MRE in Doppler image volumes. The Doppler vascularity metric, CPD, can be measured relatively consistently over the range of compressions changes from −10% to +15% of the compression initially judged by the patient as acceptable for a series of scans of up to 10 min.
The cardiac-gated Doppler U.S. study was performed assuming that there was a uniform time delay since the onset of R-wave for blood flow to peak at the breast for all patients and this could be changed. There are various factors that contribute to this time delay including stress-level, emotions, heart rate, and age, etc. Newer U.S. systems have a peak-detect Doppler mode that takes Doppler scan at various time points during a cardiac cycle allowing for postprocessing to detect the peak blood flow in the breast. During the Doppler scan, the patients had been under mammographic compression for a few minutes (as long as it took for getting good coupling in the breast through the compression plate) and this could have contributed toward low blood flow in some subjects. In the future work, care will have to be taken to relax compression for an adequate time period before modest recompression in the Doppler portion of the U.S. study.
ACKNOWLEDGMENTS
This work was supported in part by U.S. Public Health Service Grant Nos. PO1 CA87634 and RO1 CA91713 (a partnership with GE Global Research). The authors would like to thank Dr. Zhi Yang, Dr. Aaron Moskalik, and Dr. Ram Narayanan who worked in various capacities in the Department of Radiology, University of Michigan Health System for guidance, for developing softwares for manual-guided segmentation of breast masses, and for 3D ellipsoid placement around the tumor boundary.
References
- Kopans D. B., “The positive predictive values of mammography,” AJR, Am. J. Roentgenol. 158, 521–526 (1992). [DOI] [PubMed] [Google Scholar]
- Berg W. A., Blume J. D., Cormack J. B., Mendelson E. B., Lehrer D., Bohm-Velez M., Pisano E. D., Jong R. A., Evans W. P., Morton M. J., Mahoney M. C., Larsen L. H., Barr R. G., Farria D. M., Marques H. S., and Boparai K., “Combined screening with ultrasound and mammography vs mammography alone in women at elevated risk of breast cancer,” JAMA, J. Am. Med. Assoc. 299, 2151–2163 (2008). 10.1001/jama.299.18.2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg W. A., “Rationale for a trial of screening breast ultrasound: American College of Radiology Imaging Network (ACRIN) 6666,” AJR, Am. J. Roentgenol. 180, 1225–1228 (2003). [DOI] [PubMed] [Google Scholar]
- Padhani A. R. and Husband J. E., “Dynamic contrast-enhanced MRI studies in oncology with an emphasis on quantification, validation and human studies,” Clin. Radiol. 56, 607–620 (2001). 10.1053/crad.2001.0762 [DOI] [PubMed] [Google Scholar]
- Barton M. B., Harris R., and Fletcher S. W., “Does this person have breast cancer? The screening clinical breast examination: Should it be done? How?,” JAMA, J. Am. Med. Assoc. 282, 1270–1280 (1999). 10.1001/jama.282.13.1270 [DOI] [PubMed] [Google Scholar]
- Jackson V. P., “The role of US in breast imaging,” Radiology 177, 305–311 (1990). [DOI] [PubMed] [Google Scholar]
- Durfee S. M., Selland D. L. G., Smith D. N., Lester S. C., Kaelin C. M., and Meyer J. E., “Sonographic evaluation of clinically palpable breast cancers invisible on mammography,” Breast J. 6, 247–251 (2000). 10.1046/j.1524-4741.2000.99111.x [DOI] [PubMed] [Google Scholar]
- Bassett L. W. and Kimme-Smith C., “Breast sonography,” AJR, Am. J. Roentgenol. 156, 449–455 (1991). [DOI] [PubMed] [Google Scholar]
- Hilton S. V., Leopold G. R., Olson L. K., and Willson S. A., “Real-time breast sonography: Application in 300 consecutive patients,” AJR, Am. J. Roentgenol. 147, 479–486 (1986). [DOI] [PubMed] [Google Scholar]
- Jackson V. P., “The current role of US in breast imaging,” Radiol. Clin. North Am. 33, 1167–1170 (1995). [PubMed] [Google Scholar]
- Kolb T. M., Lichy J., and Newhouse J. H., “Occult cancer in women with dense breasts: Detection with screening US- Diagnostic yield and tumour characteristics,” Radiology 207, 191–199 (1998). [DOI] [PubMed] [Google Scholar]
- Taylor K. J. W., Merritt C., and Piccoli C., “Ultrasound as a complement to mammography and breast examination to characterize breast masses,” Ultrasound Med. Biol. 28, 19–26 (2002). 10.1016/S0301-5629(01)00491-4 [DOI] [PubMed] [Google Scholar]
- Moss H. A., Britton P. D., Flower C. D. R., Freeman A. H., Lomas D. J., and Warren R. M. L., “How reliable is modern breast imaging in differentiating benign from malignant breast lesions in the symptomatic population?,” Clin. Radiol. 54, 676–682 (1999). 10.1016/S0009-9260(99)91090-5 [DOI] [PubMed] [Google Scholar]
- Stavros A. T., Thickman D., Rapp C. L., Dennis M. A., Parker S. H., and Sisney G. A., “Solid breast nodules: Use of sonography to distinguish between benign and malignant lesions,” Radiology 196, 123–134 (1995). [DOI] [PubMed] [Google Scholar]
- Conway F. W., Hayes C. W., and Brewer W. H., “Occult breast masses: Use of a mammographic localization grid for US evaluation,” Radiology 181, 143–146 (1991). [DOI] [PubMed] [Google Scholar]
- Partridge S. C., Gibbs J. E., and Lu Y., “MRI measurements of breast tumor volume predict response to neoadjuvant chemotherapy and recurrence-free survival,” AJR, Am. J. Roentgenol. 184, 1774–1781 (2005). [DOI] [PubMed] [Google Scholar]
- Roubidoux M. A., Le Carpentier G. L., Fowles J. B., Bartz B., Pai D., Gordon S. P., Schott A. F., Johnson T. D., and Carson P. L., “Sonographic evaluation of early-stage breast cancers that undergo neoadjuvant chemotherapy,” J. Ultrasound Med. 24, 885–895 (2005). [DOI] [PubMed] [Google Scholar]
- Chagpar A. B., Middleton L. P., Sahin A. A., Dempsey P., Buzdar A. U., Mirza A. N., Ames F. C., Babiera G. V., Feing B. W., Hunt K. K., Kuerer H. M., Meric-Bernstam F., Ross M. I., and Singletary S. E., “Accuracy of physical examination, ultrasonography, and mammography in predicting residual pathologic tumor size in patients treated with neoadjuvant chemotherapy,” Ann. Surg. 243, 257–64 (2006). 10.1097/01.sla.0000197714.14318.6f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Fukutomi T., Tsuda H., Akashi-Tanaka S., Matsuo K., Shimizu C., and Miyakawa K., “Changes in finds of mammography, ultrasonography and contrast-induced computed tomography of three histological complete responders with primary breast cancer before and after neoadjuvant chemotherapy: case reports,” Jpn. J. Clin. Oncol. 30, 453–457 (2000). 10.1093/jjco/hyd119 [DOI] [PubMed] [Google Scholar]
- Brown L. S., “A survey of image registration techniques,” ACM Comput. Surv. 24, 325–356 (1992). 10.1145/146370.146374 [DOI] [Google Scholar]
- Zitova B. and Flusser J., “Image registration methods: A survey,” Image Vis. Comput. 21, 977–1000 (2003). 10.1016/S0262-8856(03)00137-9 [DOI] [Google Scholar]
- Meyer C. R., Boes J. L., Kim B., Bland P. H., LeCarpentier G. L., Fowlkes J. B., Roubidoux M. A., and Carson P. L., “Semiautomatic registration of volumetric ultrasound scans,” Ultrasound Med. Biol. 25, 339–347 (1999). 10.1016/S0301-5629(98)00148-3 [DOI] [PubMed] [Google Scholar]
- Edwards P. J., Hill D. L. G., Little J. A., and Hawkes D. J., “A three-component deformation model for image-guided surgery,” Med. Image Anal. 2, 355–367 (1998). 10.1016/S1361-8415(98)80016-9 [DOI] [PubMed] [Google Scholar]
- Rosenman J. G., Miller E. P., Tracton G., and Cullip T. J., “Image registration: An essential part of radiation therapy treatment planning,” Int. J. Radiat. Oncol., Biol., Phys. 40, 197–205 (1998). 10.1016/S0360-3016(97)00546-4 [DOI] [PubMed] [Google Scholar]
- Weese J., Penney G. P., Desmedt P., Buzug T. M., and Hill D. L. G., “Voxel based 2D∕3D registration of fluoroscopy images and CT scans for image guided surgery,” IEEE Trans. Inf. Technol. Biomed. 1, 284–293 (1997). 10.1109/4233.681173 [DOI] [PubMed] [Google Scholar]
- Thirion J. P., “Image matching as a diffusion process: An analogy with Maxwell’s demons,” Med. Image Anal. 2, 243–260 (1998). 10.1016/S1361-8415(98)80022-4 [DOI] [PubMed] [Google Scholar]
- Pennec X., Cachier P., and Ayache N., “Understanding the Demon’s algorithm: 3D non-rigid registration by gradient descent,” Proceedings of MICCAI, LNCS (Springer, Berlin, 1999), Vol. 1679, pp. 597–606.
- Wang H., Dong L., O’Daniel J., Mohan R., Garden A. S., Ang K. K., Kuban D. A., Bonnen M., Chang J. Y., and Cheung R., “Validation of accelerated ‘demons’ algorithm for deformable image registration in radiation therapy,” Phys. Med. Biol. 50, 2887–2905 (2005). 10.1088/0031-9155/50/12/011 [DOI] [PubMed] [Google Scholar]
- Kashani R., Hub M., Balter J. M., Kessler M. L., Dong L., Zhang L., Xing L., Xie Y., Hawkes D., Schnabel J. A., McClelland J., Joshi S., Chen Q., and Lu W., “Objective assessment of deformable image registration in radiotherapy: A multi-institution study,” Med. Phys. 35, 5944–5953 (2008). 10.1118/1.3013563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookstein F. L., “Principal warps: Thin-plate splines and decompositions of deformation,” IEEE Trans. Pattern Anal. Mach. Intell. 11, 567–585 (1989). 10.1109/34.24792 [DOI] [Google Scholar]
- Kessler M. L., “Image registration and data fusion in radiation therapy,” Br. J. Radiol. 79, S99–S108 (2006). 10.1259/bjr/70617164 [DOI] [PubMed] [Google Scholar]
- Holden M., “A review of geometric transformations for nonrigid body registration,” IEEE Trans. Med. Imaging 27, 111–128 (2008). 10.1109/TMI.2007.904691 [DOI] [PubMed] [Google Scholar]
- Kapur A., Carson P. L., and Eberhard J., “Combination of digital mammography with semi-automated 3D breast ultrasound,” Technol. Cancer Res. Treat. 3, 325–334 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanasamy G., LeCarpentier G. L., Zabuawala S., Fowlkes J. B., Roubidoux M., Sinha S., and Carson P. L., “Non-rigid registration of three-dimensional(3D) grayscale and Doppler ultrasound breast images,” in Proceedings of 29th Annual International IEEE Engineering in Medicine and Biology Society Conference, Lyon, France, 2007, pp. 91–94. [DOI] [PubMed]
- Booi R. C., Krücker J. F., and Goodsitt M. M., O’Donnell M., Kapur A., Lecarpentier G. L., Roubidoux M. A., Fowlkes J. B., and Carson P. L., “Evaluating thin compression paddles for Mammographically compatible ultrasound,” Ultrasound Med. Biol. 33, 472–482 (2007). 10.1016/j.ultrasmedbio.2006.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S. P., Goodsitt M. M., Roubidoux M. A., Booi R. C., LeCarpentier G. L., Lashbrook C. R., Thomenius K. E., Chalek C. L., and Carson P. L., “Automated ultrasound scanning on a dual-modality breast imaging system,” J. Ultrasound Med. 26, 645–655 (2007). [DOI] [PubMed] [Google Scholar]
- Narayanasamy G., Fowlkes J. B., Kripfgans O. D., Jacobson J. A., De Maeseneer M., Schmidt R. M., and Carson P. L., “Ultrasound of the finger for human identification using biometrics,” Ultrasound Med. Biol. 34, 392–399 (2008). 10.1016/j.ultrasmedbio.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Carson P. L., LeCarpentier G. L., Roubidoux M. A., Erkamp R. Q., Fowlkes J. B., and Goodsitt M. M., “Physics and technology of ultrasound breast imaging including automated 3D,” in 2004 Syllabus, Advances in Breast Imaging: Physics, Technology, and Clinical Applications, RSNA Categorical Course in Diagnostic Radiology Physics, edited by Karellas A. and Giger M. L., RSNA (2004), pp. 223–232.
- Meyer C. R., Boes J. L., Kim B., Bland P. H., Zasadny K. R., Kison P. V., Koral K., Frey K. A., and Wahl R. L., “Demonstration of accuracy and clinical versatility of mutual information for automatic multimodality image fusion using affine and thin plate spline warped geometric deformations,” Med. Image Anal. 1, 195–206 (1997). 10.1016/S1361-8415(97)85010-4 [DOI] [PubMed] [Google Scholar]
- Krücker J., Ph.D. Thesis, Applied Physics Program, University of Michigan, Ann Arbor, Michigan, 2003. [Google Scholar]
- Folkman J., “Tumor angiogenesis: Therapeutic implications,” N. Engl. J. Med. 285, 1182–1186 (1971). [DOI] [PubMed] [Google Scholar]
- Bergers G. and Benjamin L. E. , “Angiogenesis: Tumorigenesis and the angiogenic switch,” Nat. Rev. Cancer 3, 401–410 (2003). 10.1038/nrc1093 [DOI] [PubMed] [Google Scholar]
- Bhatti P. T., LeCarpentier G. L., Roubidoux M. A., Fowlkes J. B., Helvie M. A., and Carson P. L., “Discrimination of sonographically detected breast masses using frequency shift color Doppler imaging in combination with age and gray scale criteria,” J. Ultrasound Med. 20, 343–350 (2001). [DOI] [PubMed] [Google Scholar]
- LeCarpentier G. L., Roubidoux M. A., Fowlkes J. B., Krücker J. F., Hunt K. A., Paramagul C., Johnson T. D., Thorson N. J., Engle K. D., and Carson P. L., “Suspicious breast lesions: Assessment of 3D Doppler US indexes for classification in a test population and fourfold cross-validation scheme,” Radiology 249, 463–470 (2008). 10.1148/radiol.2492060888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson P. L., Fowlkes J. B., Roubidoux M. A., Moskalik A. P., Govil A., Normolle D., LeCarpentier G. L., Nattakom S., Helvie M. R., and Rubin J. M., “3-D color Doppler image quantification of breast masses,” Ultrasound Med. Biol. 24, 945–952 (1998). 10.1016/S0301-5629(98)00055-6 [DOI] [PubMed] [Google Scholar]
- Porter B. C., Rubens D. J., Strang J. G., Smith J., Totterman S., and Parker K. J., “Three-dimensional registration and fusion of ultrasound and MRI using major vessels as fiducial markers,” IEEE Trans. Med. Imaging 20, 354–359 (2001). 10.1109/42.921484 [DOI] [PubMed] [Google Scholar]
- Nanayakkara N. D., Chiu B., Samani A., Spence J. D., Samarabandu J., Parraga G., and Fenster A., “Nonrigid registration of three-dimensional ultrasound and magnetic resonance images of the carotid arteries,” Med. Phys. 36, 373–385 (2009). 10.1118/1.3056458 [DOI] [PubMed] [Google Scholar]
- Wells III W., Viola P., Atsumi H., Nakajima S., and Kikinis R., “Multi-modal volume registration by maximization of mutual information,” Med. Image Anal. 1, 35–51 (1996). 10.1016/S1361-8415(01)80004-9 [DOI] [PubMed] [Google Scholar]
- Schönemann P. H., “A generalized solution of the orthogonal procrustes problem,” Psychometrika 31, 1–10 (1966). 10.1007/BF02289451 [DOI] [Google Scholar]
- Rueckert D., Sonoda L. I., Hayes C., Hill D. L. G., Leach M. O., and Hawkes D. J., “Nonrigid registration using free-form deformations: Application to breast MR images,” IEEE Trans. Med. Imaging 18, 712–721 (1999). 10.1109/42.796284 [DOI] [PubMed] [Google Scholar]
- Johnson H. and Christensen G., “Landmark and intensity-based, consistent thin plate spline image registration,” in Proceedings of the 17th International Conference on Information Processing and Medical Imaging, Vol. 2082, pp. 329–343 (2001).
- Bookstein F. L., Morphometric Tools for Landmark Data (Cambridge University Press, Cambridge, 1991). [Google Scholar]
- Bookstein F. L., “Shape and information in medical images: A decade of morphometric synthesis,” Comput. Vis. Image Underst. 66, 97–118 (1997). 10.1006/cviu.1997.0607 [DOI] [Google Scholar]
- Shekhar R. and Zagrodsky V., “Mutual information-based rigid and non-rigid registration of ultrasound volumes,” IEEE Trans. Med. Imaging 21, 9–22 (2002). 10.1109/42.981230 [DOI] [PubMed] [Google Scholar]
- Pluim J. P. W., Maintz J. B. A., and Viergever M. A., “Mutual information matching in multiresolution contexts,” Image Vis. Comput. 19, 45–52 (2001). 10.1016/S0262-8856(00)00054-8 [DOI] [Google Scholar]
- Carp S. A., Kauffman T., Fang Q., Rafferty E., Moore R., Kopans D., and Boas D., “Compression-induced changes in the physiological state of the breast as observed through frequency domain photon migration measurements,” J. Biomed. Opt. 11, 064016 (2006). 10.1117/1.2397572 [DOI] [PubMed] [Google Scholar]
- Krücker J. F., Meyer C. R., LeCarpentier G. L., Fowlkes J. B., and Carson P. L., “3D spatial compounding of ultrasound images using image-based non-rigid registration,” Ultrasound Med. Biol. 26, 1475–1488 (2000). 10.1016/S0301-5629(00)00286-6 [DOI] [PubMed] [Google Scholar]
- Moskalik A., Carson P. L., Meyer C. R., Fowlkes J. B., Rubin J. M., and Roubidoux M. A., “Registration of three-dimensional compound ultrasound scans of the breast for refraction and motion correction,” Ultrasound Med. Biol. 21, 769–778 (1995). 10.1016/0301-5629(95)00018-M [DOI] [PubMed] [Google Scholar]
- Czerwinski R. N., Jones D. L., and W. D.O’Brien, Jr., “Ultrasound speckle reduction by directional median filtering,” Proceedings of the International Conference on Image Processing, Washington, DC, Vol. 1, pp. 358–361.
- Milkowski A., Li Y., Becker D., and Ishrak S. O., “Speckle reduction imaging,” Technical White Paper–General Electric Health Care (Ultrasound). Last accessed on July 9, 2009. Available at http://www.gehealthcare.com/usen/ultrasound/education/docs/whitepaper_SRI.pdf.
- Krücker J. F., Lecarpentier G. L., Fowlkes J. B., and Carson P. L., “Rapid elastic image registration for 3-D ultrasound,” IEEE Trans. Med. Imaging 21, 1384–1394 (2002). 10.1109/TMI.2002.806424 [DOI] [PubMed] [Google Scholar]
- Narayanasamy G., Narayanan R., Fowlkes J. B., Roubidoux M. A., and Carson P. L., “Segmentation-free estimation of volume changes in 3D ultrasound of breast phantom lesions,” Proceedings of SPIE Medical Imaging, San Diego, CA, 2007, Vol. 6510, p. 651033.