Abstract
Rhodopseudomonas palustris belongs to the group of purple bacteria that have the ability to produce LH2 complexes with unusual absorption spectra when they are grown at low-light intensity. This ability is often related to the presence of multiple genes encoding the antenna apoproteins. Here we report, for the first time to our knowledge, direct evidence that individual low-light LH2 complexes have a heterogeneous αβ-apoprotein composition that modulates the site energies of Bchl a molecules, producing absorption bands at 800, 820, and 850 nm. The arrangement of the Bchl a molecules in the “tightly coupled ring” can be modeled by nine αβ-Bchls dimers, such that the Bchls bound to six αβ-pairs have B820-like site energies and the remaining Bchl a molecules have B850-like site energies. Furthermore, the experimental data can only be satisfactorily modeled when these six αβ-pairs with B820 Bchl a molecules are distributed such that the symmetry of the assembly is reduced to C3. It is also clear from the measured single-molecule spectra that the energies of the electronically excited states in the mixed B820/850 ring are mainly influenced by diagonal disorder.
Introduction
Purple photosynthetic bacteria have evolved an elegant system of modular units that make up the light-harvesting apparatus. These modules consist of pairs of hydrophobic, low-molecular-weight polypeptides, called α and β (usually 50–60 amino acids long), that noncovalently bind a small number of bacteriochlorophyll (Bchl a) and carotenoid (Car) molecules. The modules then oligomerize to produce the native circular, elliptical, or horseshoe complexes (1). Most purple bacteria have two main types of complexes: the core complex (RC-LH1) and the peripheral complex (LH2). In the photosynthetic membranes, LH2 surrounds the RC-LH1 complexes in a two-dimensional network (2,3).
The LH2 complex from Rhodopseudomonas (Rps.) acidophila is a nonameric complex (nine α/β units) that contains 27 Bchl a and nine carotenoid molecules (4,5). The Bchl a molecules are arranged in LH2 into two groups. The first group consists of nine well-separated monomeric Bchl a molecules, one for each αβ-apoprotein pair. The bacteriochlorin rings of this group lie flat within the plane of the membrane. These Bchl a molecules absorb at 800 nm and are collectively called B800. Each αβ-apoprotein apoprotein pair also binds two other Bchl a molecules, whose central Mg2+ are complexed via His residues to the αβ-apoprotein helices. These Bchl a molecules form a closely interacting ring and collectively give rise to the absorption at 850 nm (B850). It is now well established that the spatial arrangement of the pigments determines, to a large extent, the spectroscopic features of the complexes, and that in these systems collective effects (especially in the case of the B850 Bchls) have to be considered to appropriately describe their electronically excited states (6–9). This leads to so-called Frenkel excitons, which arise from the interactions of the transition-dipole moments of the individual pigments and correspond to delocalized electronically excited states (10,11). Since the interaction strength between the individual pigments can be calculated on the basis of the available structural data, information about the pigment arrangement within the LH complexes can be obtained via optical spectroscopy. From this perspective, the LH2 complex from Rps. acidophila has served as a cornerstone for the development of a detailed understanding of structure-function relationships in such antenna systems (12,13).
Of interest, in some species of purple bacteria, such as Rps. acidophila or Rps. palustris, the exact spectral positions of the absorption bands can vary depending on the growth conditions. When grown under low-light (LL) conditions, the light-harvesting apparatus of Rps. acidophila strains 7050 and 7750 features an additional peripheral LH2 complex that absorbs at 820 nm (14–16). The ability to change the type of LH2 in response to growth at different light intensities is related to the presence of multiple αβ-polypeptides, which are (in the case of Rps. acidophila) encoded by at least four different αβ-apoprotein gene pairs (16,17). To distinguish the two types of peripheral LH2 complexes, they are often referred to as B800–850 and B800–820 complexes, or LH2 and LH3. The origin of this spectral variation results from fine-tuning of the electronic energy levels of the B850 molecules by interaction with the protein matrix. In the B800–850 complex, the C3-acetyl group of the α-bound B850 Bchl a molecule is hydrogen (H)-bonded to the αTyr44 residue and the C3-acetyl group of the β-bound B850 Bchl a molecule is H-bonded to the αTrp45 residue (14). In contrast, in the B800–820 complex, the respective residues in these positions, i.e., αPhe44 and αLeu45, are unable to form hydrogen bonds. Instead the C3-acetyl group of the αB820 Bchl a molecules is H-bonded to the αTyr41 residue, and this locks the acetyl group into an out-of-plane position with respect to the bacteriochlorin plane. Similarly, the β-bound B820 Bchl a molecule, lacking any H-bonds, has its acetyl tail in an out-of-plane position with respect to the bacteriochlorin plane. Rotation of the acetyl group of the Bchl a molecule into an out-of-plane position reduces the extent of π-conjugation and results in a blue shift of the BChl a site energies (18). This then is reflected in the shift of the absorption band from 850 nm to 820 nm.
A microscopic understanding of the interplay between pigment organization and its spectral properties requires high-resolution structural information. Unfortunately, this information is only available for very few types of peripheral light-harvesting complexes, two B800–850 complexes (Rps. acidophila 10050 (4,5) and Rhsp. molischianum (19)), and one B800–820 complex (Rps. acidophila 7050 (14)). In each of these cases the LH2 complexes have a well-defined apoprotein composition. When cells of Rps. palustris are grown at HL intensity, they synthesize a standard LH2 complex. However, at LL intensities they produce an LH2 complex with a strikingly different absorption spectrum (20–23), as illustrated in Fig. S1 of the Supporting Material. The LL LH2 complex has been studied before, and indeed a low-resolution model based on crystal diffraction data at 7.5 Å resolution has been described (24). This model suggests that the complex is an αβ-octamer and that each of its αβ-apoprotein pairs binds an extra Bchl a relative to LH2 from Rps. acidophila. It has also been shown that the Rps. palustris genome contains five αβ-apoprotein gene pairs (25,26). Moreover, preparations of both HL and LL LH2 complexes from Rps. palustris contain a complex apoprotein composition, which raises the question as to whether its LH2 complexes consist of rings that each contain a mixture of apoprotein types, as opposed to the homogeneous case of, e.g., Rps. acidophila (22,27). However, it is difficult experimentally to distinguish between a mixture of LH2 complexes where each ring is homogeneous but there are various types of rings present, and where each individual ring has an inhomogeneous apoprotein composition. It is interesting to consider the possible effect that rings with a heterogeneous population of apoproteins might induce, especially since some of the Rps. palustris apoproteins have sequences that would change the site energies of the “B850” Bchls from the standard HL energies to ones more reminiscent of the B820 complex from Rps. acidophila. How would such an energetic heterogeneity affect the absorption properties of LH2? And does such heterogeneity actually exist? One way to address these questions is to use single-molecule spectroscopy.
The usefulness of single-molecule spectroscopy as a tool to unravel the spectroscopic complexities of light-harvesting proteins has been well demonstrated (28–35). (For reviews of single-molecule studies on bacterial light-harvesting complexes, see Cogdell et al. (36) and Berlin et al. (37).) In this study we used fluorescence-excitation spectra (i.e., fluorescence-detected absorption spectra) to investigate individual LL LH2 complexes from Rps. palustris. Our idea was to investigate whether different classes of single complexes exist, and to establish whether there is any evidence for spectral properties that can only be explained by rings with a heterogeneous apoprotein composition.
Materials and Methods
Materials
Rps. palustris strain 2.1.6. was grown anaerobically in C-succinate media at 15 μW m−1 at 30°C. The cells were harvested after 120 h by centrifugation (1248 g, 30 min) and resuspended in 2-[N-Morpholino]ethanesulfonic acid (20 mM, pH 6.8; Sigma-Aldrich, Steinheim, Germany) containing KCl (100 mM). The harvested cells were broken by three passages through a French press (9500 psi) in the presence of a little DNAase and MgCl2. The broken membranes were then sedimented by centrifugation at 184,000 g for 2 h. The pellet was resuspended and homogenized in Tris-HCl (20 mM, pH 8.0; Fisher Scientific, Leicester, UK) and its concentration was adjusted to yield an absorption at B850 of 70 cm−1. The membranes were then solubilized by the addition of 1% v/v lauryldimethylamine N-oxide (LDAO; Fluka, Steinheim, Germany). After the mixture was stirred in the dark at room temperature for 30 min, any unsolubilized material was removed by centrifugation at 16,000 g for 10 min. The supernatant was then layered onto a sucrose step gradient. The gradient consisted of 0.8, 0.6, 0.4, 0.2 M sucrose prepared in Tris-HCl (20 mM, pH 8.0) containing LDAO (0.1% v/v). The gradients were then centrifuged overnight at 149,000 g at 4°C. The upper pigmented band contained the LH2 complexes, and the lower pigmented band contained the LH1-RC complexes. The band containing the LH2 complexes was collected and purified on a DE-52 cellulose column (Whatman, Maidstone, England). The purity of the LH2 complex was determined by calculating the ratio between the absorption maximum at B800 and the maximum at the protein absorption (A260). Fractions with a ratio > 3.0 were collected for spectroscopic analysis.
For single-molecule spectroscopy, the purified LL LH2 complexes were diluted to <10−9 M in Tris-HCl buffer (20 mM, pH 8.0) containing 0.1% LDAO. In the last dilution step, 2% (w/w) polyvinyl alcohol (PVA, mw 30,000–70,000 g mol−1) was added to the LL LH2 and a drop of the solution was spin-coated onto a quartz substrate for 10 s at 500 rpm and 60 s at 2,500 rpm (model P6700; Specialty Coating System, Indianapolis, IN). This produced thin, amorphous polymer films of <1 μm thickness in which the LH2 complexes were embedded. Then the sample was immediately mounted in a helium-bath cryostat and cooled down to 1.4 K.
Low-temperature microscopy
Fluorescence-excitation spectroscopy was performed with the use of an in-house-built microscope that could be operated in either wide-field or confocal mode. The excitation source was a continuous-wave tunable titanium-sapphire laser (3900S; Spectra Physics, Mountain View, CA) pumped by a frequency-doubled continuous-wave neodynium:yttrium-vanadate (Nd:YVO4) laser (Millennia Vs; Spectra Physics). Well-defined changes in the wavelength of the Titanium:Sapphire laser were achieved by rotating the intracavity birefringent filter with a motorized micrometer screw. For calibration purposes, a wavemeter was used, and the accuracy of the laser frequency and a reproducibility of 1 cm−1 were verified. First, a 50 × 50 μm2 wide-field image of the sample was taken by exciting the sample at 800 nm and detecting the fluorescence with a back-illuminated CCD camera (512 SB; Roper Scientific Princeton Instruments, Trenton, NJ) after the light passed through suitable bandpass filters (BP893/21; Dr. Hugo Anders, Nabburg, Germany), which blocked the residual laser light. Subsequently, a spatially well-isolated complex was selected from the wide-field image and a fluorescence-excitation spectrum of this complex was obtained. The setup was switched to the confocal mode and the fluorescence was detected by a single-photon counting avalanche photodiode (APD, SPCM-AQR-16; EG&G Optoelectronics, Vaudreuil, Canada) while the laser was scanned repetitively between 784 and 872 nm. The recorded traces were stored separately in computer memory. With a scan speed of the laser of 3 nm · s−1 (≈50 cm−1 s−1) and an acquisition time of 10 ms per data point, this yields a nominal resolution of 0.5 cm−1, ensuring that the spectral resolution is limited by the spectral bandwidth of the laser (1 cm−1). To examine the polarization dependence of the spectra, a λ/2-plate was put in the confocal excitation path and rotated in steps of 3.2° between two successive scans, changing the angle of polarization of the excitation light by twice that value. The excitation intensity was ∼50 W cm−2.
Results
The fluorescence-excitation spectra of several individual LL LH2 complexes from Rps. palustris are shown in Fig. 1. The top trace shows, for comparison, the fluorescence-excitation spectrum taken from an ensemble of LL LH2 complexes (gray line) together with the spectrum that results from the summation of the spectra of 31 individual LL LH2 complexes (black line). The two spectra are in reasonable agreement, indicating that the selected individual LL LH2 complexes are a fair statistical representation of the ensemble. The ensemble spectrum shows two broad bands centered at 11,684 cm−1 (856 nm) and 12,524 cm−1 (798 nm) with widths (full width at half-maximum (FWHM)) of 353 cm−1 and 174 cm−1, respectively. The peak intensity of the B850 band is about three times lower than that of the B800 band, which agrees with previously published absorption spectra obtained both at room temperature and at 77 K (23).
Figure 1.
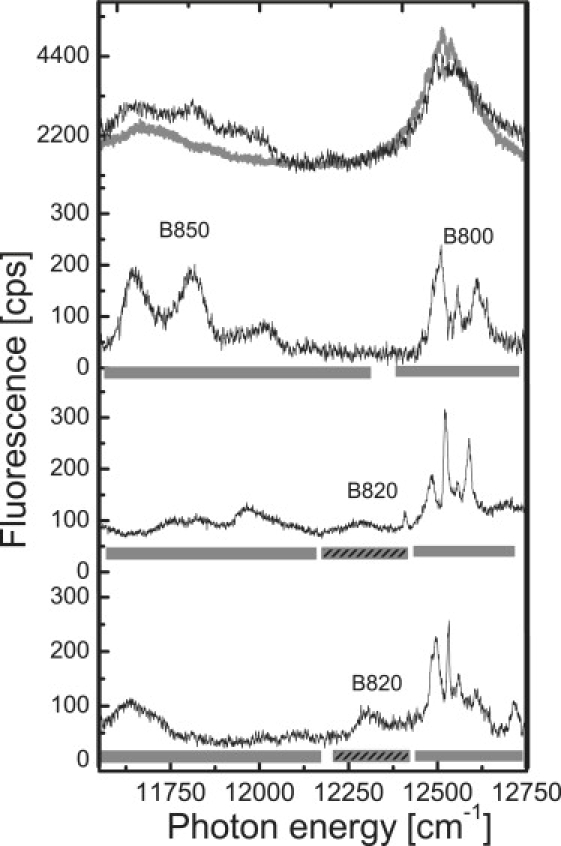
Low-temperature (1.4 K) fluorescence-excitation spectra of LH2 complexes from LL Rps. palustris 2.1.6. The top traces show the spectrum from an ensemble (gray line) and the spectrum that corresponds to the sum of 31 spectra from individual LH2 complexes (black line). The lower three traces display typical fluorescence-excitation spectra from individual LH2 complexes. The spectra have been averaged over all polarizations of the incident laser field. The bars indicate the spectral positions of the 800, 820, and 850 nm bands, respectively. The excitation intensity was 50 W cm−2. The vertical scale is given in counted photons per second (cps).
By measuring the fluorescence-excitation spectra of the individual complexes, as shown for three examples in the lower traces of Fig. 1, we can visualize remarkable features that are obscured in the ensemble average. Around 12,500 cm−1 (800 nm), the spectra show a distribution of narrow absorption bands, with linewidths (FWHM) below 10 cm−1, whereas around 11,750 cm−1 (850 nm) a few broad bands, with linewidths (FWHM) on the order of 160 cm−1, are present. This observation is reminiscent of the situation for LH2 from Rps. acidophila. However, in striking contrast to the single-molecule spectra from LH2 from Rps. acidophila, we find that ∼90% of the individual LL LH2 complexes from Rps. palustris show an additional broad band in the B820 spectral region. The peak position of this band varies between 12,048 cm−1 and 12,288 cm−1 (center of mass at 12,206 cm−1) and its linewidth covers the range between 31 cm−1 and 158 cm−1 (average 79 cm−1).
To analyze the spectral bands from the individual LL LH2 complexes in more detail, we recorded the fluorescence-excitation spectra as a function of the polarization of the incident laser excitation. The excitation spectra were recorded in rapid succession as the polarization of the excitation light was rotated by 6.4° between two consecutive scans. An example of this protocol is shown in the top part of Fig. 2 A in a two-dimensional representation in which 410 individual scans are stacked on top of each other. The horizontal axis corresponds to photon energy, the vertical axis to the individual scans (or equivalently to the polarization of the excitation), and the detected fluorescence intensity is coded by the gray scale. The sum spectrum of these scans is presented at the bottom of Fig. 2 A. The pattern shown at the top of Fig. 2 A clearly reveals the polarization dependence of the absorptions in the B850 region. This becomes even more evident in Fig. 2 B. At the bottom of Fig. 2 B, two individual spectra are shown on an expanded energy scale. The spectra were obtained for mutually orthogonal polarizations of the excitation light. For simplicity, we refer in the following to the absorptions in the B820/850 region as B1, B2, B3, and B4, in order of increasing photon energy, where B stands for band. The angle of the polarization that yields the maximum intensity for the band B1 was set arbitrarily to 0° and provides the reference point. For this particular complex, we find the absorption bands B1, B2, B3, and B4 at the peak positions of 11,562 cm−1, 11,727 cm−1, 11,874 cm−1, and 12,150 cm−1, and the linewidths are 174 cm−1, 125 cm−1, 187 cm−1, and 52 cm−1, respectively. The top part of Fig. 2 B shows the fluorescence intensity of the four bands as a function of the polarization of the excitation light (dots). This variation is consistent with a cos2 dependence (black line).
Figure 2.
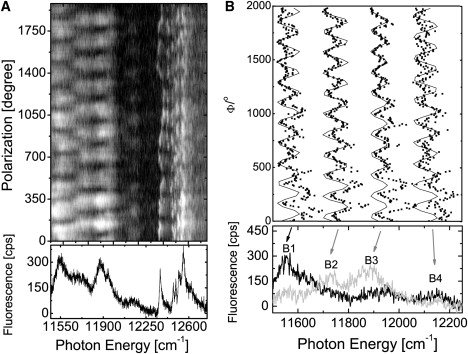
Fluorescence-excitation spectrum from an individual B800–820/850 LH2 complex from Rps. palustris as a function of the polarization of the excitation light. (A) (Top) Stack of 410 individual spectra recorded consecutively. Between two successive spectra, the polarization of the incident radiation has been rotated by 6.4°. The horizontal axis corresponds to the photon energy, the vertical axis corresponds to the scan number (or equivalently to the polarization angle), and the intensity is coded by the gray scale. The excitation intensity was 50 W cm−2. (Bottom) Spectrum that corresponds to the average of the 410 consecutively recorded spectra. (B) (Top) Expanded view of the fluorescence intensity of the four bands (B1–B4) marked by the arrows in the lower part as a function of the polarization of the incident radiation (dots) together with cos2-type functions (black) fitted to the data. (Bottom) Two fluorescence-excitation spectra from the stack that correspond to mutually orthogonal polarization of the excitation light.
From such experiments, we can determine the energetic separation (ΔE) between B1 and the other bands, as well as the relative angle (Δα) between the transition-dipole moments that are associated with them. The distributions of these parameters for the full data set of 28 complexes are shown in histograms (Fig. 3). The energetic separation between the bands B1 and B2 varies between 62 cm−1 and 342 cm−1 and is centered at ∼174 cm−1 (Fig. 3 A). The distribution of the mutual orientation of the transition-dipole moments that are associated with the B1 and B2 bands increases to a maximum at ∼90° (Fig. 3 D). The energetic separation between B1 and B3 is distributed between 230 cm−1 and 491 cm−1, with a maximum at ∼361 cm−1 (Fig. 3 B). The distribution of relative orientations of the transition-dipole moments that is associated with B1 and B3 decreases from a maximum value at ∼10° when going to larger angles (Fig. 3 E). The distribution of the energetic separation between B1 and B4 is between 430 cm−1 and 670 cm−1 and is centered at ∼550 cm−1, whereas the distribution of the relative orientations of the transition-dipole moments associated with B1 and B4 shows a slow decrease from a maximum value at ∼20°.
Figure 3.
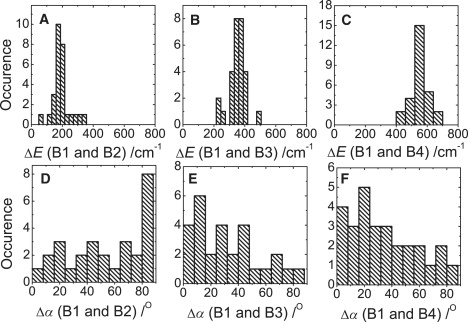
(Top) Distributions of the energetic separations ΔE observed between (A) B1 and B2, (B) B1 and B3, and (C) B1 and B4. (Bottom) Distributions of the mutual angles Δα between the transition-dipole moments that are associated with (D) B1 and B2, (E) B1 and B3, and (F) B1 and B4.
Discussion
When the single-molecule spectra obtained with HL B800–850 complexes from both Rps. palustris and Rps. acidophila are compared with those recorded from LL LH2 complexes from Rps. palustris, both common and strikingly different features can be observed. In the spectral region around 12,500 cm−1 (800 nm), all three types of complexes show narrow bands. These narrow bands have been well explained and arise from essentially monomeric weakly interacting Bchl molecules (B800 manifold) (32,38–40). The single-molecule spectra in the region of ∼11,600–12,250 cm−1 (815–860 nm) differ greatly between the LL and HL complexes. The HL complexes show two and sometimes three broad bands in the 850 nm region. Previous studies have shown that these broad bands arise from excitonic interactions among the strongly coupled B850 Bchls (7,30,41).
All of the LL complexes studied here show broad bands not only in the 11,750 cm−1 (850 nm) region, but also in the 12,200 cm−1 (820 nm) region. They appear to have a mixed composition that gives rise to exciton bands in both the 850 and 820 nm regions. In an effort to explain these differences, we performed a series of simulations to try to understand the structural features that are required to accurately reproduce the experimental data.
The general approach used to describe the electronically excited states of the B850 LH2 ring is based on a model Hamiltonian using the Heitler-London approximation (41,42):
| (1) |
where N refers to the number of Bchl a molecules; and correspond to excitations localized on molecules n and m, respectively; and (E0 + ΔEn) denotes the site energy of pigment n. Due to local variations in the protein environment of the binding site, there is a separation of the individual site energies into an average, E0, and a deviation from this average, ΔEn, which is commonly termed diagonal disorder. This energetic disorder is usually modeled by a Gaussian distribution of site energies. As the simplest approach to evaluate the dependence of the interaction on the distance and the mutual orientation of the pigments, we used a dipole-dipole type function (43):
| (2) |
where κnm denotes the usual orientation factor, and rnm is the distance between pigments n and m. The coupling strength V0 was set to 135,072 cm−1 Å3 (43). Two limiting cases for the dipolar interaction can be distinguished: weak coupling , and strong coupling . In the case of the B850 Bchls, there is strong coupling due to much larger interaction energy (Vnm) compared to the difference in their site energies (ΔEnm). As a result of this, the eigenfunctions of the Hamiltonian (Eq. 1) are given by the combinations of the excited-state wavefunctions of the individual pigments.
In the case of both the 9-mers (HL LH2s from Rps. acidophila 10050 and Rps. palustris) and the 8-mer (LH2 from Rhsp. molischianum), the energy scheme of the excited-state manifold of the B850 ring was constructed by combining symmetric or antisymmetric linear combinations of the localized wave functions of the αβ-dimers in such a way that they formed the basis for the irreducible representations of the C9 and C8 pure-rotational point groups, respectively (7). As previously described for the C9-symmetry case, the symmetric and antisymmetric manifolds consist of one nondegenerate (k = 0) and four doubly degenerate (k = ±1, ±2,…, ±4) exciton states (7). The corresponding C8-symmetry case has the (k = 0, ±1, ±2, ±3, 4) exciton states. To determine which exciton states are involved in giving rise to the measured bands B1, B2, B3, and B4, we calculated the absorption spectra by Monte Carlo simulations, treating the diagonal disorder as a random variable. This simulation was carried out for both 9-mer and 8-mer possibilities. Typical examples of the simulated spectra of individual realizations of the disorder are shown in Fig. 4. The first simulated spectrum of an LL LH2 complex was obtained by introducing diagonal disorder into the circular 9- and 8-mer models (Fig. 4, A and D). For simplicity, the site energies of B850 Bchl a dimers were set according to the previously determined site energies of LH2 from Rps. acidophila (E0 (αB850) = 12,300 cm−1 and E0 (βB850) = 12,060 cm−1) (42). The random diagonal disorder is taken from a Gaussian distribution with a width of Γintra = 160 cm−1, where Γintra stands for intracomplex heterogeneity (41). Within each complex, the 18 or 16 Bchl a molecules will show variations of their site energies with respect to their spectral means. In the case of a 9-mer ring, this simulation produces a sharp band from the k = 0 state, broad bands from the k = ±2 states, and broad bands from the k = ±1 states, which are split in energy and feature mutually orthogonal transition-dipole moments (Fig. 4 A). In the 8-mer ring, only a sharp k = 0 band and the two orthogonally polarized k = ±1 bands are present (Fig. 4 D). In Fig. 4, B and E, this simulation was extended by inserting the expression for the site energies. Here refers to the random diagonal disorder chosen from the Gaussian distribution described above, and introduces an additional correlated modulation of C2-type symmetry into the site energies of the pigments, where and N refers to the number of the pigments in the ring (42). When this modulation is taken into account, the 9-mer model shows a sharp k = 0 band, broad orthogonally polarized k = ±1 bands, and broad k = ±2 bands. Furthermore, although the k = ±3 states gain significant oscillator strength, but they have no observable splitting. Though the k = ±2 bands do contribute, it is apparent that the overall spectrum of the 9-mer model is dominated by the contributions of the k = 0, k = ±1, and k = ±3 states. In the case of the 8-mer model, a sharp k = 0 band, broad orthogonally polarized k = ±1 bands, broad k = ±2 bands, and k = ±3 bands are also present. The splitting of the k = ±3 bands is clear in the 8-mer model. The overall spectrum of the 8-mer model is dominated by the k = 0, k = ±1, and k = ±3 states.
Figure 4.
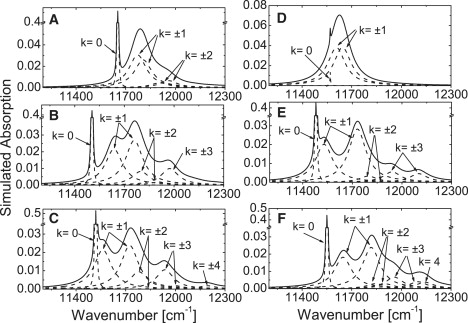
Examples for simulated absorption spectra for an individual LH2 complex (solid line). The spectra correspond to a single realization of the disorder. The calculated energies of the exciton states have been dressed by a Lorentzian with a width of 5 cm−1 for the k = 0 state and a width of 130 cm−1 for all other exciton states (dashed lines). Simulations A–C (left) are based on a nonamaric structure, and simulations D–F (right) are based on an octameric structure. The simulations vary with respect to the type of diagonal disorder: (A and D) random diagonal disorder taken from a Gaussian distribution of width 160 cm−1 (FWHM); (B and E) random diagonal disorder as in A and D, with an additionally correlated diagonal disorder; (C and F) like B and E, additionally using B820-like-pair (E0(αB820) = 12,860 cm−1, ΔE = 260 cm−1) and B850-like-pair (E0(αB850) = 12,300 cm−1, ΔE = 240 cm−1) randomly distributed site energies across the ring assembly. For more details see text.
There is strong evidence from mass spectroscopic analysis of LL LH2 polypeptides from Rps. palustris that this LL complex contains multiple types of αβ-polypeptides (T. H. P. Brotosudarmo, A. Gall, V. Moulisova, A. T. Gardiner, and R. J. Cogdell, unpublished results). The sequence of some of these polypeptides features changes for key amino acids shown to be involved in H-bonding to the Bchl a macrocycles of the strongly coupled ring of Bchls in LH2. This heterogeneity raises the possibility that some of these Bchl a molecules may have quite different site energies. In other words, some may have site energies characteristic of B850 and some may have ones characteristic of B820. Therefore, the simulation was extended to introduce two different site energies—E0 (αB850) = 12,300 cm−1, E0 (βB850) = 12,060 cm−1 (B850-pair) (42); and E0 (αB820) = 12,860 cm−1, E0 (βB820) = 12,600 cm−1) (B820-pair) (44)—for the Bchl a-pairs in which the B850- and B820-pairs of Bchl a are randomly distributed in the ring (Fig. 4, C and F). In this case the 9-mer model shows the gain of the oscillator strength of all of the exciton states (Fig. 4 C). The bands at 11500 cm−1 (870 nm) and 11700 cm−1 (855 nm) are due to the k = +1 and k = −1 states, respectively. The k = ±2 bands are obscured under the k = −1 band. Although the splitting of the k = ±3 and k = ±4 states are seen, the k = +3 and k = +4 bands are the most intense. The overall features of the simulated spectrum (solid line) from the 9-mer model are dominated by the k = 0, ±1, +3 and +4 states (dashed lines) and is blue-shifted compared to the case shown in Fig. 4 B. In the case of the 8-mer model, the simulated spectrum is identical to that of the 9-mer (Fig. 4 F).
When the absorption spectra of the simulations are compared with the experimental spectrum of the LL LH2 (Fig. 2), it can be seen that the traces in Fig. 4, A, B, and D, are not consistent with the experimental data. The simulations presented in Fig. 4, C, E, and F, however, all are in reasonable agreement with the experimental spectrum. Simulations with a 9-mer model can only account for the experimental spectrum if two different site energies for Bchl a-pairs are considered (Fig. 4 C). If this model describes the situation properly, then the experimentally measured bands B1, B2, B3, and B4 can be assigned to the k = +1, −1, +3, and +4 states, respectively. Simulations with an 8-mer model produce agreement with the experimental data both when different site energies for the pairs of Bchl a molecules are taken into account and when they are not. If the model in Fig. 4 E provides a proper description of the experimentally determined situation, then the B1, B2, B3, and B4 bands in the experimental data can be assigned to the k = +1, −1, −3, and + 3 states, respectively. If, on the other hand, the model that corresponds to the simulation shown in Fig. 4 F is appropriate, then the B1, B2, B3, and B4 bands can be assigned to the k = +1, −1, +3 and 4 states, respectively. Therefore, it is unclear at this stage which of the underlying models that led to the simulations presented in Fig. 4 represents a realistic approximation for describing the structure of the LL LH2.
To distinguish among the three different options presented in Fig. 4, C, E, and F, we considered the extra experimental spectroscopic information that is available (see Fig. 3), i.e., the energetic separation of the spectral bands and the mutual orientation of their transition-dipole moments. Therefore, we compared the experimental histograms with the parameters calculated from the respective simulations for 2000 realizations of the energetic disorder (Fig. 5, circles, triangles, and black squares). Using the 8-mer model represented by Fig. 4 E, it can be seen that the simulated distributions of ΔE and the mutual angle between the transition-dipole moments of the k = ±1 states can be reproduced; however, the simulated distributions of the energetic separations between k = +1 and k = −3, as well as between k = +1 and k = +3, with maxima around 200 cm−1 and 400 cm−1 are not (Fig. 5). Moreover, the simulated distribution for the mutual angle between the transition-dipole moments of the k = +1 and k = −3 states is even worse, with a broad maximum covering the range from 40° to 60°. Simulations based on the 8-mer model represented by Fig. 4 F are able to satisfactorily account for the distribution of the simulated mutual angles between the transition-dipole moments of the k = ±1 states (Fig. 5). However, these simulations also do not satisfactorily reproduce the distributions of the angles between the transition-dipole moments of the k = +1 and k = 4 states. Simulations based on the 9-mer model represented by Fig. 4 C show quite good agreement with the distribution of all three mutual angles (Fig. 5). However, there is a significant though smaller (compared to the two 8-mer models) mismatch between this simulation and the distribution of energetic separations, ΔE (k = +1 and k = +3) and ΔE (k = +1 and k = +4).
Figure 5.
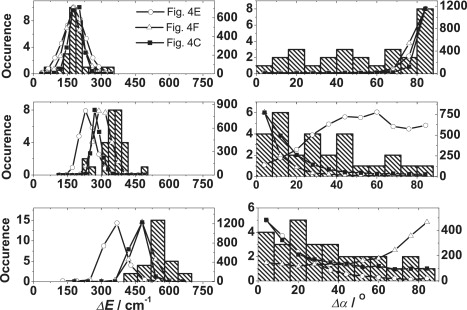
Comparison of the energetic separations of bands B1–B4 with the separations of the respective exciton states (left) and the mutual orientation of the associated transition-dipole moments (right) as predicted from Monte Carlo simulations for 2000 realizations of the disorder for the models shown in Fig. 4E (open circles), Fig. 4F (open triangles), and Fig. 4C (solid squares).
At this stage, it appears that the 9-mer model is able to more closely account for the experimental data. The LL LH2 complexes from Rps. palustris have at least four types of α-polypeptides (T. H. P. Brotosudarmo, A. Gall, V. Moulisova, A. T. Gardiner, and R. J. Cogdell, unpublished results). One, the αd-polypeptide, has phenylalanine and methionine residues at positions 44 and 45 (25,45). The other polypeptides have tyrosine and tryptophan residues at these positions. The presence of H-bonding residues at positions 44 and 45, such as tyrosine and tryptophan, correlates with B850-type site energies, whereas if non-H-bonding residues, such as phenylalanine and methionine, are present, it correlates with B820-like site energies. Shifting in the site energy of one α-bound Bchl a will also affect the nearest-neighbor β-bound Bchl a. Therefore, we extended the simulations using this additional information. The parameters of the coordinate arrangement of the Bchls for this simulation were taken from the crystal structures of the B800–850 and B800–820 LH2 complexes (see Table S1 and Fig. S2). The site energies of the α-bound-B820-like Bchl a molecules E0 (αB820) was set to be 12,640 cm−1, whereas the β-bound-B820-like Bchl a molecule E0 (βB820) was 12,400 cm−1. The position of the B820-like pairs was randomly distributed (Fig. 6). The simulation based on this model, though not perfect, shows closer agreement with the experimental distribution of energetic separations ΔE (k = +1 and k = +3). Again, the match to the distributions of the mutual angles of the respective transition-dipole moments between (k = +1 and k = +3) and (k = +1 and k = +4) is good. We extended this simulation further by systematically setting fewer B820-like site energy pairs. However, none of these simulations yielded an improved reproduction of the experimental data (data not shown).
Figure 6.
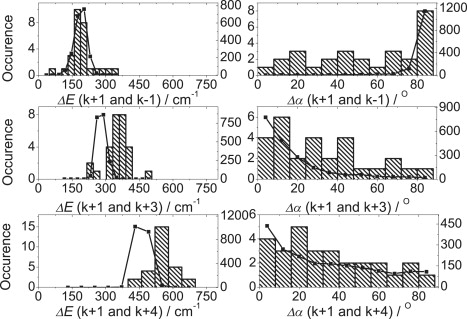
Extended simulations based on the model shown in Fig. 4C, i.e., a nonameric structure with random and correlated diagonal disorder and multipolypeptide composition. The B820-like-pair site energies of E0(αB820) = 12,640 cm−1 with ΔE = 250 cm−1 and the B850-pair site energy of E0(αB850) = 12,300 cm−1 with ΔE = 240 cm−1. The positions of the B820-like pairs are randomly distributed.
Next, the effect of a symmetrical arrangement of these B820-like site energies was tested. Six B820-like site energies were distributed symmetrically with respect to the threefold axis in the ring. The other 12 Bchl a molecules were given B850-site energies. This simulation yielded the best agreement between the simulations and the experimental data that we found (Fig. 7), but only when we introduced a rather high intracomplex heterogeneity, Γintra, at 320 cm−1. This value is significant higher than that previously reported for the B800–850 LH2 of Rps. acidophila (Γintra = 250 cm−1) (41,42). This may be a consequence of having individual rings with a heterogeneous polypeptide composition.
Figure 7.
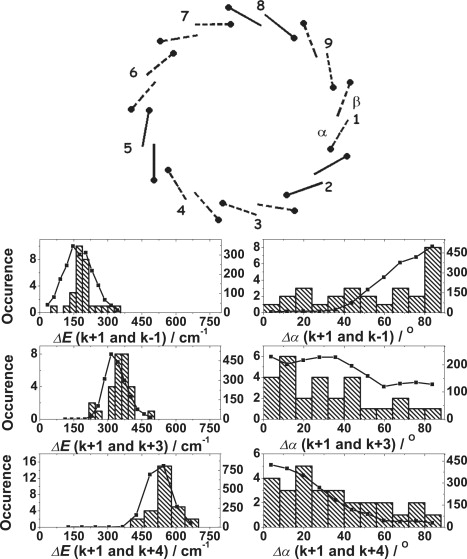
(Top) Model structure featuring six 820-like-pair Bchl a molecules (dashed lines) distributed in C3 symmetry around the ring. (Bottom) Comparison of the experimental data with the results from Monte Carlo simulations (2000 realizations) for the nonameric structure as shown in the top part, taking random and correlated diagonal disorder into account (black squares). The site energies of the pigments were chosen as detailed in Fig. 6. The intracomplex heterogeneity (Γintra) is 320 cm−1. The full and dashed lines in the top part of the figure indicate the B820-like (dashed) and B850 (full) site energies.
It has been suggested that some species of purple bacteria may contain LH2 complexes in which rings of multiple sizes coexist (3). To test whether various mixtures of rings of different sizes could satisfactorily account for our experimental data, we added together simulations for the best 8-mer, 9-mer, and 10-mer models in a number of different combinations. None of these “addition” simulations produced satisfactory agreement with the experimental data (data not shown). Though it is not possible to claim that our simulations can provide definitive proof of the structure of the LL LH2 complex from Rps. palustris, they do strongly favor both a nonameric model and one in which the assembly has multiple apoprotein types within a single LH ring. It is also worth pointing out that the absorption spectrum of the native LL LH2 complex from Rps. palustris studied in this work is not the same as that of the B800-only LH2 complex used in the structural studies mentioned above (46). We do not understand why this is so, but we have never been able to isolate an LL LH2 complex from Rps. palustris that has the spectrum described by Hartigan et al. (24). Photosynthetic bacteria such as Rps. palustris have a very complicated set of control mechanisms that can modify which of the multigene family of LH2 αβ-gene pairs are expressed (25,26,45). These differences, therefore, may reflect variations in the growth conditions and laboratories in which the bacteria were cultured.
Conclusions
The single-molecule spectra reported here can only be satisfactorily explained by assuming that the individual LL LH2 complexes from Rps. palustris contain a heterogeneous polypeptide composition. Multiple types of αβ apoproteins coexist in the same individual complex, producing individual rings, with Bchl molecules having both B820-like and B850-site energies. The simulations described above strongly reinforce this conclusion. The experimental data can best be modeled by taking an arrangement of the Bchl a molecules in the mixed B820/850 ring of nine αβ Bchls dimers, where in six αβ-pairs the Bchl a molecules have B820-like site energies and are distributed around the ring in such a way that the symmetry of the nonameric assembly is reduced to C3. The high intracomplex disorder reflects the heterogeneity in the polypeptide composition.
Supporting Material
A table and two figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(09)01205-3.
Supporting Material
Acknowledgments
This work was supported by the Biotechnology and Biological Science Research Council, the Deutsche Forschungsgemeinschaft, and the Bavarian Elite Network. T.H.P.B. and V.M. received fellowships from the European Commission through the Human Potential Program (Marie-Curie RTN BIMORE, grant MRTN-CT-2006-035859).
Contributor Information
Richard J. Cogdell, Email: gbta18@udcf.gla.ac.uk.
Jürgen Köhler, Email: juergen.koehler@uni-bayreuth.de.
References
- 1.Cogdell R.J., Gardiner A.T., Roszak A.W., Law C.J., Southall J. Rings, ellipses and horseshoes: how purple bacteria harvest solar energy. Photosynth. Res. 2004;81:207–214. doi: 10.1023/B:PRES.0000036883.56959.a9. [DOI] [PubMed] [Google Scholar]
- 2.Blankenship R.E. Blackwell Science; Oxford, UK: 2002. Molecular Mechanisms of Photosynthesis. [Google Scholar]
- 3.Scheuring S., Sturgis J.N. Chromatic adaptation of photosynthetic membranes. Science. 2005;309:484–487. doi: 10.1126/science.1110879. [DOI] [PubMed] [Google Scholar]
- 4.McDermott G., Prince S.M., Freer A.A., Hawthornthwaite-Lawless A.M., Papiz M.Z. Crystal structure of an integral membrane light-harvesting complex from photosynthetic bacteria. Nature. 1995;374:517–521. [Google Scholar]
- 5.Papiz M.Z., Prince S.M., Howard T., Cogdell R.J., Isaacs N.W. The structure and thermal motion of the B800–850 LH2 complex from Rps. acidophila at 2.0Å resolution and 100K: new structural features and functionally relevant motions. J. Mol. Biol. 2003;326:1523–1538. doi: 10.1016/s0022-2836(03)00024-x. [DOI] [PubMed] [Google Scholar]
- 6.Hu X.C., Ritz T., Damjanovic A., Autenrieth F., Schulten K. Photosynthetic apparatus of purple bacteria. Q. Rev. Biophys. 2002;35:1–62. doi: 10.1017/s0033583501003754. [DOI] [PubMed] [Google Scholar]
- 7.Matsushita M., Ketelaars M., van Oijen A.M., Köhler J., Aartsma T.J. Spectroscopy on the B850 band of individual light-harvesting 2 complexes of Rhodopseudomonas acidophila. II. Exciton states of an elliptically deformed ring aggregate. Biophys. J. 2001;80:1604–1614. doi: 10.1016/S0006-3495(01)76133-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zigmantas D., Read E.L., Mancal T., Brixner T., Gardiner A.T. Two-dimensional electronic spectroscopy of the B800–B820 light-harvesting complex. Proc. Natl. Acad. Sci. USA. 2006;103:12672–12677. doi: 10.1073/pnas.0602961103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Damjanovic A., Kosztin I., Kleinekathöfer U., Schulten K. Excitons in a photosynthetic light-harvesting system: a combined molecular dynamics, quantum chemistry, and polaron model study. Phys. Rev. E. 2002;65:031919. doi: 10.1103/PhysRevE.65.031919. [DOI] [PubMed] [Google Scholar]
- 10.Davydov A.S. Plenum Press; New York: 1971. Theory of Molecular Excitons. [Google Scholar]
- 11.Knox R.S. Academic Press; New York/London: 1964. Theory of Excitons. [Google Scholar]
- 12.Cogdell R.J., Isaacs N.W., Freer A.A., Arrelano J., Howard T.D. The structure and function of the LH2 (B800–850) complex from the purple photosynthetic bacterium Rhodopseudomonas acidophila strain 10050. Prog. Biophys. Mol. Biol. 1997;68:1–27. doi: 10.1016/s0079-6107(97)00010-2. [DOI] [PubMed] [Google Scholar]
- 13.Georgakopoulou S., Frese R.N., Johnson E., Koolhaas C., Cogdell R.J. Absorption and CD spectroscopy and modeling of various LH2 complexes from purple bacteria. Biophys. J. 2002;82:2184–2197. doi: 10.1016/S0006-3495(02)75565-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McLuskey K., Prince S.M., Cogdell R.J., Isaacs N.W. The crystallographic structure of the B800–820 LH3 light-harvesting complex from the purple bacteria Rhodopseudomonas acidophila strain 7050. Biochemistry. 2001;40:8783–8789. doi: 10.1021/bi010309a. [DOI] [PubMed] [Google Scholar]
- 15.Cogdell R.J., Scheer H. Circular-dichroism of light-harvesting complexes from purple photosynthetic bacteria. Photochem. Photobiol. 1985;42:669–678. [Google Scholar]
- 16.Bissig I., Brunisholz R.A., Suter F., Cogdell R.J., Zuber H. The complete amino acid sequences of the B800–850 antenna polypeptides from Rhodopseudomonas acidophila strain 7750. Z. Naturforsch. [C] 1988;43:77–83. doi: 10.1515/znc-1988-1-216. [DOI] [PubMed] [Google Scholar]
- 17.Gardiner A.T., Takaichi S., Cogdell R.J. The effect of changes in light intensity and temperature on the peripheral antenna of Rhodopseudomonas acidophila. Biochem. Soc. Trans. 1993;21:6S. doi: 10.1042/bst021006s. [DOI] [PubMed] [Google Scholar]
- 18.Sturgis J.N., Jirsakova V., Reiss-Husson F., Cogdell R.J., Robert B. Structure and properties of the bacteriochlorophyll binding site in peripheral light-harvesting complexes of purple bacteria. Biochemistry. 1995;34:517–523. doi: 10.1021/bi00002a016. [DOI] [PubMed] [Google Scholar]
- 19.Koepke J., Hu X., Muenke C., Schulten K., Michel H. The crystal structure of the light-harvesting complex II (B800–850) from Rhodospirillum molischianum. Structure. 1996;4:581–597. doi: 10.1016/s0969-2126(96)00063-9. [DOI] [PubMed] [Google Scholar]
- 20.Hayashi H., Miyao M., Morita S. Absorption and fluorescence spectra of light-harvesting bacteriochlorophyll-protein complexes from Rhodopseudomonas palustris in the near-infrared region. J Biochem. 1982;91:1017–1027. doi: 10.1093/oxfordjournals.jbchem.a133751. [DOI] [PubMed] [Google Scholar]
- 21.Hayashi H., Nakano M., Morita S. Comparative studies of protein properties and bacteriochlorophyll contents of bacteriochlorophyll-protein complexes from spectrally different types of Rhodopseudomonas palustris. J Biochem. 1982;92:1805–1811. doi: 10.1093/oxfordjournals.jbchem.a134110. [DOI] [PubMed] [Google Scholar]
- 22.Gall A., Robert B. Characterization of the different peripheral light-harvesting complexes from high- and low-light grown cells from Rhodopseudomonas palustris. Biochemistry. 1999;38:5185–5190. doi: 10.1021/bi982486q. [DOI] [PubMed] [Google Scholar]
- 23.van Mourik F., Hawthornthwaite A.M., Vonk C., Evans M.B., Cogdell R.J., Sundström V., van Grondelle R. Spectroscopic characterization of the low-light B800–850 light-harvesting complex of Rhodopseudomonas palustris strain 2.1.6. Biochim. Biophys. Acta. 1992;1140:85–93. [Google Scholar]
- 24.Hartigan N., Tharia H.A., Sweeney F., Lawless A.M., Papiz M.Z. The 7.5-Å electron density and spectroscopic properties of a novel low-light B800 LH2 from Rhodopseudomonas palustris. Biophys. J. 2002;82:963–977. doi: 10.1016/S0006-3495(02)75456-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tadros M.H., Waterkamp K. Multiple copies of the coding regions for the light-harvesting B800–850 α- and β-polypeptides are present in the Rhodopseudomonas palustris genome. EMBO J. 1989;8:1303–1308. doi: 10.1002/j.1460-2075.1989.tb03509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tadros M.H., Katsiou E., Hoon M.A., Yurkova N., Ramji D.P. Cloning of a new antenna gene cluster and expression analysis of the antenna gene family of Rhodopseudomonas palustris. Eur. J. Biochem. 1993;217:867–875. doi: 10.1111/j.1432-1033.1993.tb18315.x. [DOI] [PubMed] [Google Scholar]
- 27.Nishimura Y., Shimada K., Yamazaki I., Mimuro M. Energy transfer processes in Rhodopseudomonas palustris grown under low-light conditions. Heterogeneous composition of LH 2 complexes and parallel energy flow pathways. FEBS Lett. 1993;329:319–323. doi: 10.1016/0014-5793(93)80245-p. [DOI] [PubMed] [Google Scholar]
- 28.Wörmke S., Mackowski S., Brotosudarmo T.H.P., Jung C., Zumbusch A. Monitoring fluorescence of individual chromophores in peridininchlorophyll-protein complex using single molecule spectroscopy. Biochim. Biophys. Acta. 2007;1767:956–964. doi: 10.1016/j.bbabio.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 29.Rutkauskas D., Novoderezhkin V., Cogdell R.J., van Grondelle R. Fluorescence spectroscopy of conformational changes of single LH2 complexes. Biophys. J. 2005;88:422–435. doi: 10.1529/biophysj.104.048629. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.van Oijen A.M., Ketelaars M., Köhler J., Aartsma T.J., Schmidt J. Unraveling the electronic structure of individual photosynthetic pigment-protein complexes. Science. 1999;285:400–402. doi: 10.1126/science.285.5426.400. [DOI] [PubMed] [Google Scholar]
- 31.Hofmann C., Michel H., van Heel M., Köhler J. Multivariate analysis of single-molecule spectra: surpassing spectral diffusion. Phys. Rev. Lett. 2005;94:195501. doi: 10.1103/PhysRevLett.94.195501. [DOI] [PubMed] [Google Scholar]
- 32.van Oijen A.M., Ketelaars M., Köhler J., Aartsma T.J., Schmidt J. Spectroscopy of individual light-harvesting 2 complexes of Rhodopseudomonas acidophila: diagonal disorder, intercomplex heterogeneity, spectral diffusion, and energy transfer in the B800 band. Biophys. J. 2000;78:1570–1577. doi: 10.1016/S0006-3495(00)76709-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tietz C., Chekhlov O., Dräbenstedt A., Schuster J., Wrachtrup J. Spectroscopy on single light-harvesting complexes at low temperature. J. Phys. Chem. B. 1999;103:6328–6333. [Google Scholar]
- 34.Bopp M.A., Jia Y.W., Li L.Q., Cogdell R.J., Hochstrasser R.M. Fluorescence and photobleaching dynamics of single light-harvesting complexes. Proc. Natl. Acad. Sci. USA. 1997;94:10630–10635. doi: 10.1073/pnas.94.20.10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ketelaars M., Hofmann C., Köhler J., Howard T.D., Cogdell R.J. Spectroscopy on individual light-harvesting 1 complexes of Rhodopseudomonas acidophila. Biophys. J. 2002;83:1701–1715. doi: 10.1016/S0006-3495(02)73938-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cogdell R.J., Gall A., Köhler J. The architecture and function of the light-harvesting apparatus of purple bacteria: from single molecules to in vivo membranes. Q. Rev. Biophys. 2006;39:227–324. doi: 10.1017/S0033583506004434. [DOI] [PubMed] [Google Scholar]
- 37.Berlin Y., Burin A., Friedrich J., Köhler J. Low temperature spectroscopy of proteins. Part II: experiments with single protein complexes. Phys. Life Rev. 2007;4:64–89. [Google Scholar]
- 38.van Oijen A.M., Ketelaars M., Köhler J., Aartsma T.J., Schmidt J. Spectroscopy of individual LH2 complexes of Rhodopseudomonas acidophila: localized excitations in the B800 band. Chem. Phys. 1999;247:53–60. [Google Scholar]
- 39.Hofmann C., Ketelaars M., Matsushita M., Michel H., Aartsma T.J. Single-molecule study of the electronic couplings in a circular array of molecules: light-harvesting-2 complex from Rhodospirillum molischianum. Phys. Rev. Lett. 2003;90:013004. doi: 10.1103/PhysRevLett.90.013004. [DOI] [PubMed] [Google Scholar]
- 40.Cheng Y.C., Silbey R.J. Coherence in the B800 ring of purple bacteria LH2. Phys. Rev. Lett. 2006;96:028103. doi: 10.1103/PhysRevLett.96.028103. [DOI] [PubMed] [Google Scholar]
- 41.Ketelaars M., van Oijen A.M., Matsushita M., Köhler J., Schmidt J. Spectroscopy on the B850 band of individual light-harvesting 2 complexes of Rhodopseudomonas acidophila. I. Experiments and Monte Carlo simulations. Biophys. J. 2001;80:1591–1603. doi: 10.1016/S0006-3495(01)76132-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hofmann C., Aartsma T.J., Köhler J. Energetic disorder and the B850-exciton states of individual light-harvesting 2 complexes from Rhodopseudomonas acidophila. Chem. Phys. Lett. 2004;395:373–378. [Google Scholar]
- 43.Sauer K., Cogdell R.J., Prince S.M., Freer A., Isaacs N.W. Structure-based calculations of the optical spectra of the LH2 bacteriochlorophyll-protein complex from Rhodopseudomonas acidophila. Photochem. Photobiol. 1996;64:564–576. [Google Scholar]
- 44.de Ruijter W.P.F., Segura J.M., Cogdell R.J., Gardiner A.T., Oellerich S. Fluorescence-emission spectroscopy of individual LH2 and LH3 complexes. Chem. Phys. 2007;341:320–325. [Google Scholar]
- 45.Evans K., Fordham-Skelton A.P., Mistry H., Reynolds C.D., Lawless A.M. A bacteriophytochrome regulates the synthesis of LH4 complexes in Rhodopseudomonas palustris. Photosynth. Res. 2005;85:169–180. doi: 10.1007/s11120-005-1369-7. [DOI] [PubMed] [Google Scholar]
- 46.de Ruijter W.P., Oellerich S., Segura J.M., Lawless A.M., Papiz M. Observation of the energy-level structure of the low-light adapted B800 LH4 complex by single-molecule spectroscopy. Biophys. J. 2004;87:3413–3420. doi: 10.1529/biophysj.104.044719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


