Abstract
The mutation rate at fifty-four perfect (uninterrupted) dinucleotide microsatellite loci is estimated by direct genotyping of 96 Arabidopsis thaliana mutation accumulation lines. The estimated rate differs significantly among motif types with the highest rate for AT repeats (2.03 × 10−3 per allele per generation), intermediate for CT (3.31 × 10−4), and lowest for CA (4.96 × 10−5). The average mutation rate per generation for this sample of loci is 8.87 × 10−4 (SE 2.57 × 10−4). There is a strong effect of initial repeat number, particularly for AT repeats, with mutation rate increasing with the length of the microsatellite locus in the progenitor line. Controlling for motif and initial repeat number, chromosome 4 exhibited an elevated mutation rate relative to other chromosomes. A survey of dinucleotide repeats across the entire Arabidopsis genome indicates that AT repeats are most abundant, followed by CT, and CA. The great majority of mutations were gains or losses of a single repeat. Several lines exhibited multiple step changes from the progenitor sequence, although it is unclear whether these are multi-step mutations or multiple single step mutations. Generally, the data are consistent with the stepwise mutation model of microsatellite evolution.
Keywords: Arabidopsis, microsatellite, dinucleotide repeats, mutation rate, mutation accumulation
Introduction
Microsatellites are simple sequence repeats that frequently display length variation within natural populations. These loci can be classified according to the length and type of repeated motif, where the most common lengths are 2, 3, or 4 bases (di-, tri- and tetra- nucleotide repeats, respectively). Because microsatellites are highly polymorphic, they are frequently used as genetic markers in ecological and evolutionary studies (Schlötterer and Pemberton, 1994). The multi-allelic character of microsatellites makes them ideal for paternity analysis (Chase et al.1996; Dow and Ashley, 1998), estimation of parameters in pollination biology (e.g. Kelly and Willis, 2002) and studies of dispersal/spatial-genetic structure (e.g. Sweigart et al., 1999). If one further assumes that microsatellite variation is selectively neutral, they can be used to estimate the effective population size (e.g. Schug et al., 1998).
Polymerase slippage during DNA replication is thought to be the primary source of mutation in microsatellites (Schlötterer et al., 1998). However, much remains unknown about the nature of the mutational process. Most studies suggest that mutations are typically gain or loss of a single repeated unit (Thuillet et al., 2002; Vigouroux et al., 2002), although there are putative examples of multi-repeat gains or losses (Ellegren, 2004). The rate of mutation may depend on allele length, i.e. the number of repeat units (Wierdl et al., 1997; Vigouroux et al., 2002; Thuillet et al., 2004), as can the direction of changes, i.e. the relative likelihood of gain versus loss (see Wierdl et al., 1997). Finally, the mutation rate and other mutational properties may depend on the repeat motif, i.e. AG vs CG (Bachtrog et al., 2000; Kelkar et al., 2008). Most data suggest that dinucleotide microsatellites mutate at a rate that is greater than that of trinucleotide and tetranucleotide microsatellites (Chakraborty et al., 1997 but see Weber and Wong, 1993).
Microsatellites are distributed non-randomly across plant genomes and are associated with non-repetitive DNA (Zhang et al., 2006). In A. thaliana, they are often found in regulatory regions, especially 5’UTRs and 5’flanking regions (Zhang et al., 2006; Grover and Sharma, 2007). A-rich repeats are prominent in introns and intergenic regions. AG is the most common di-nt motif in exons and 5’flanking regions, while AT is most common in introns, intergenic regions, 3’ flanking regions (Zhang et al., 2004).
Microsatellite mutation rates have been estimated for a variety of crop plants (Table 1). Rate estimates range from 0 to 5 × 10–3 per locus per generation. Across these studies, mutations were more frequently observed in loci with long alleles (more repeat units) and most were single repeat changes with gains more frequent than losses. Across all three studies of Table 1, smaller loci (fewer repeats) tended to expand while longer loci (more repeats) tended to lose repeats.
Table 1.
Mutation rates of wheat, corn and chickpea as estimated from mutation accumulation experiments. All mutation rates are haploid (per allele) with a 95% C.I. on the estimate given in parentheses
| Species | Number of loci | Type of repeat | Number of observed mutations | Average mutation rate | Reference |
|---|---|---|---|---|---|
| Wheat | 10 | di-nt | 12 | 2.4 ×10–4 (1.4 ×10–4, 4.2 ×10–4) |
Thuillet et al. 2002 |
| Corn | 88 | di-nt | 73 | 7.7 ×10–4 (5.2 ×10–4, 1.1 ×10–3) |
Vigouroux et al. 2002 |
| Corn | 42 | >2 (compound) | 0 | 0.0 (0.0, 5.1 ×10–5) |
Vigouroux et al. 2002 |
| Chickpea Ghab2 var. | 15 | tri-nt | 167 | 5.0 ×10–3 (4.5 ×10–3, 6.0 ×10–3) |
Udupa and Baum 2001 |
| Chickpea Syrian local var. | 15 | tri-nt | 60 | 1.95 ×10–3 (1.45 ×10–3, 2.5 ×10–3) |
Udupa and Baum 2001 |
Estimates of microsatellite mutation rates are directly relevant to hypotheses about genetic diversity in natural populations. Symonds and Lloyd (2003) found that genetic diversity for 20 microsatellite loci across 126 accessions was positively correlated with the number of contiguous repeats in A. thaliana. This association is predicted by models where mutation rate increases with repeat number. Direct estimates of mutation rate are also essential for evaluating theories of microsatellite evolution. The simplest model is the Infinite Alleles Model (IAM; Kimura and Crow, 1964; Balloux and Lugon-Moulin, 2002) where mutations occur at a constant rate and each mutation creates a novel allele. Seemingly more appropriate for microsatellites is the stepwise mutation model (SMM; Ohta and Kimura, 1973) where mutations occur at a constant rate and involve the gain or loss of a single unit. The two phase model of DiRienzo et al. (1994) is a modification of the SMM with most mutations involving a gain or loss of a single repeat and the remainder of the mutations being multi-step mutations following a geometric distribution. In a survey of variation at five microsatellite loci across 37 populations of A. thaliana, Bakker et al. (2006) found support for both the SMM (2 of the 5 loci) and the IAM (4 of the 5 loci).
In this paper, we estimate the rate of mutation per allele per generation of dinucleotide repeats in A. thaliana. A large panel of Mutation Accumulation (MA) lines is scored for allele length at fifty-four perfect dinucleotide repeat loci. Perfect repeats are uninterrupted strings of a single motif, e.g. AT. The loci examined in this study are not associated with genes or within intergenic regions of gene clusters. As a consequence, natural selection on allele length within these loci is likely to be much weaker than for gene associated microsatellites. All putative mutations were confirmed by multiple independent PCR amplifications. These results corroborate the effect of allele length on mutation rate. They also indicate an important effect of motif type and possibly also chromosomal location. We also conduct a genomic survey of A. thaliana and interpret our mutation estimates in relation to the full distribution of repeat lengths and motif frequency in the Arabidopsis genome.
METHODS
Plant growth and DNA extraction
Shaw et al. (2002) maintained 118 independent Mutation Accumulation Lines of Arabidopsis thaliana for 30 generations prior to the current study. All lines were initiated from the Columbia accession and each was propagated by single seed descent. We chose a random subset of this population (96 lines) and grew plants to maturity in the University of Kansas greenhouse in February 2008. The soil was equal parts vermiculite and perlite with potting soil sprinkled on top of seeds. Day length was artificially expanded to 18 hours and plants were fertilized every week with Peat-lite (20–10–20 NPK). Tissue was collected for DNA extraction from the basal rosette when each plant was approximately five weeks old.
Tissue was collected into a 96-well plate with a metal bead in each well. 500 µL of CTAB buffer and 1 µL of β-mercaptoethanol was added to each sample. The plate was then sealed and shaken at high speed for 45s in a bead beater. The plate was then incubated for ∼20 min. in 60°C water bath and then centrifuged for ∼10 sec (3980 rpm) to separate solids. We transferred 300 µl liquid from each tube to a new 96-well Costar plate and added 300 µl of chloroform to each sample. This was followed by another round of mixing using the “slanted- vortex technique” and centrifuge for 10 min @ 3980 rpm. Each sample was then fully separated into aqueous (upper) and chloroform (lower) layers. We removed the aqueous layer to a new 96-well plate, added 200 µl isopropanol, and mixed well by inverting the plate repeatedly. The new plate was stored at −20°C overnight and then centrifuged for 10 minutes @ 3980 rpm. This produced a gelatinous pellet in each well. We then poured off the supernatant, added 200 µl 70% ethanol, capped the tubes, and repeated the shake and centrifuge steps. We then poured off the ethanol and air-dried the pellet. Each DNA pellet was resuspended in 50 µL of distilled water. All samples were quantified using a NanoDrop 1000 spectrophotometer (Thermo Scientific) and diluted with distilled H20 to 7–9 ng/µL.
Locus selection for genotyping
Microsatellite loci were identified by searching the Arabidopsis genome sequence via The Arabidopsis Information Resource (TAIR) website (www.arabidopsis.org). Microsatellites were found by searching for each motif in a string of 8 repeats, e.g. ATATATATATATATAT or (AT)8. For coverage of the genome, we divided each of 5 chromosomes into four regions and selected one locus per region per motif type. Not all regions contained a microsatellite satisfying our selection criteria. We eliminated microsatellites that were within 200 bp of start/end of gene, in either a UTR or an intron, had more than 30 repeats, or if the repeat sequence of the microsatellite was interrupted. We found no CG repeats that met these conditions and so our sample consisted entirely of AT, CA, and CT repeats. A number of loci failed to amplify, and as a consequence, we ended up with fewer CA loci (14) than AT or CT loci (20 of each). Primers, described in the Appendix, were designed for the selected loci using the program Primer3 with the default settings (Rozen and Skaletsky, 2000).
For each locus, we genotyped 96 individuals using a 3-primer method for polymerase chain reaction (PCR; Boutin-Ganache et al., 2001). We used one untagged primer for each pair, a second primer with a 5’ tag (CAG sequence: 5’-CAGTCGGGCGTCATCA-3’), and a third CAG-sequence primer with a 5'-6FAM (Applied Biosystems) fluorescent label. The CAG sequence was added to the primer in each pair such that the melting temperature of the tagged primer was approximately 65 ºC. PCRs (15 µl total volume) contained 40ng of template DNA, 0.25 µM untagged primer, 0.025 µM CAG-tagged primer, 0.25 µM 6FAM-labeled CAG primer, 200 µM each dNTP, 0.5 units Taq DNA polymerase (Promega) and 1× PCR buffer (500 mM KCl, 15 mM MgCl2, 100mM Tris-HCl; Promega). For temperature cycling, we implemented a touchdown PCR protocol using an iCycler Thermal Cycler (BioRad): 94 ºC for 1 min, 21 cycles of denaturing at 94 ºC for 30 s, annealing for 20 s, and extension at 72 ºC for 20 s; initial annealing temperature (Ta) = 60 ºC and decreased by 0.5 ºC with each cycle until Ta reached 50 ºC, followed by 9 cycles using this Ta, and a final extension at 67 ºC for 45 min. We detected PCR-amplified fragments on an ABI 3130 Genetic Analyzer (Applied Biosystems), and sized fragments using GeneMapper 4.0 software (Applied Biosystems) calibrated with the ROX500 size standard (Applied Biosystems). Logistic regression and other statistical analyses of the mutation accumulation data were performed in R (www.r-project.org/).
Genome scan for dinucleotide microsatellite loci
We downloaded entire chromosome sequences as FASTA files from www.arabidopsis.org and used the program Tandem Repeats Finder v. 4.0 for Windows (TRF; Benson, 1999) to identify microsatellites. We used the following parameter values within TRF for genome analysis: alignment weights +2, −7, −7 (representing match, mismatch and indel penalties); matching probability of 0.80 and an indel probability of 0.10 (pM = 0.80 and pI = 0.10, respectively); a minimum alignment score of 20 and a maximum period size of 10. We extracted the dinucleotide repeats of all motif types from the full TRF output by visual inspection. We statistically analyzed the resulting data in Minitab (v. 14.0) for mean repeat length for each repeat motif category.
Results
For all loci, the majority of lines produced fragments that matched the length of the progenitor sequence: the Col-1 genomic sequence length plus the increment due to the primers. Putative mutations were identified as deviations from this progenitor sequence length. Each putative mutant was subsequently re-amplified and re-genotyped 2–6 times to distinguish real mutations (acquired during mutation accumulation) from those due to PCR error. Approximately 15% (19/124) of all putative mutations identified in the initial screen were determined to be PCR errors.
Across lines and loci, there were 5165 genotypes. Of these, 137 (2.7%) were confirmed mutations (Table 2). If we bin all mutant types in Table 2, the (haploid) mutation rate,μ, can be estimated as the number of mutations divided by the product of the number of lines (L) and the number of generations of mutation accumulation (G). Each line is expected to produce 2µ mutations per locus per generation but only half of these mutations will fix in subsequent generations of propagation. By this method, the estimated μ is 2.03 × 10−3 for the 20 AT repeats, 4.96 × 10−5 for the 14 CA repeats, and 3.31 × 10−4 for the 20 CT repeats. For the entire sample, the estimatedμ= 8.87 × 10−4 with a standard error of 2.57 × 10−4.
Table 2.
The genotypes for all 96 are summarized for each locus Non-mutant genotypes match the progenitor line. Unscored genotypes could not be determined and/or replicated. Het-loss and het-gain denotes lines that were heterozygous for a single repeat mutation and the progenitor allele. The other six categories are homozygous lines that differ from the progenitor by 1, 2, or 3 repeats.
| Locus | Non-Mutant | Unscored | Het-Loss | Het-gain | 1 Loss | 1 Gain | 2 Loss | 2 Gain | 3 Loss | 3 Gain |
|---|---|---|---|---|---|---|---|---|---|---|
| AT.CIW7 | 85 | 1 | 5 | 5 | ||||||
| AT0101 | 96 | |||||||||
| AT0102 | 91 | 1 | 4 | |||||||
| AT0103 | 88 | 1 | 3 | 3 | 1 | |||||
| AT0104 | 96 | |||||||||
| AT0201 | 93 | 1 | 2 | |||||||
| AT0202 | 89 | 3 | 1 | 3 | ||||||
| AT0203 | 95 | 1 | ||||||||
| AT0204 | 93 | 3 | ||||||||
| AT0301 | 82 | 1 | 1 | 6 | 6 | |||||
| AT0302 | 93 | 1 | 1 | 1 | ||||||
| AT0303 | 96 | |||||||||
| AT0304 | 96 | |||||||||
| AT0402 | 66 | 1 | 12 | 11 | 5 | 1 | ||||
| AT0403 | 76 | 2 | 9 | 7 | 1 | 1 | ||||
| AT0404 | 96 | |||||||||
| AT0501 | 89 | 1 | 3 | 3 | ||||||
| AT0502 | 93 | 2 | 1 | |||||||
| AT0503 | 96 | |||||||||
| AT0504 | 83 | 1 | 5 | 7 | ||||||
| CA0101 | 96 | |||||||||
| CA0102 | 95 | 1 | ||||||||
| CA0103 | 96 | |||||||||
| CA0104 | 95 | 1 | ||||||||
| CA0201 | 95 | 1 | ||||||||
| CA0202 | 96 | |||||||||
| CA0301 | 96 | |||||||||
| CA0302 | 96 | |||||||||
| CA0401 | 96 | |||||||||
| CA0501 | 96 | |||||||||
| CA0502 | 95 | 1 | ||||||||
| CA0503 | 95 | 1 | ||||||||
| CA0504 | 96 | |||||||||
| CA72 | 95 | 1 | ||||||||
| CT.nga1145 | 96 | |||||||||
| CT.nga172 | 96 | |||||||||
| CT.nga225 | 95 | 1 | ||||||||
| CT.nga32 | 95 | 1 | ||||||||
| CT.nga59 | 96 | |||||||||
| CT0101 | 96 | |||||||||
| CT0102 | 96 | |||||||||
| CT0103 | 96 | |||||||||
| CT0104 | 94 | 2 | ||||||||
| CT0201 | 96 | |||||||||
| CT0301 | 96 | |||||||||
| CT0302 | 96 | |||||||||
| CT0303 | 95 | 1 | ||||||||
| CT0304 | 96 | |||||||||
| CT0401 | 89 | 3 | 4 | |||||||
| CT0402 | 90 | 1 | 5 | |||||||
| CT0403 | 96 | |||||||||
| CT0501 | 95 | 1 | ||||||||
| CT0502 | 96 | |||||||||
| CT0503 | 93 | 3 |
The preceding calculations are approximate because the number of mutant lines may not exactly match the number of mutant alleles. Counting het-gain and het-loss as full mutations produces a slight upward bias in mutation rate because we expect that half of these lines will revert to the progenitor sequence with random allele loss due to segregation. However, we are likely underestimating mutation rate by single counting the multi-gain and multi-loss lines. These lines might reflect real multi-step mutations but they might also have fixed multiple single repeat mutations. Also, a small fraction of lines are expected to match the progenitor because of canceling of gains and losses.
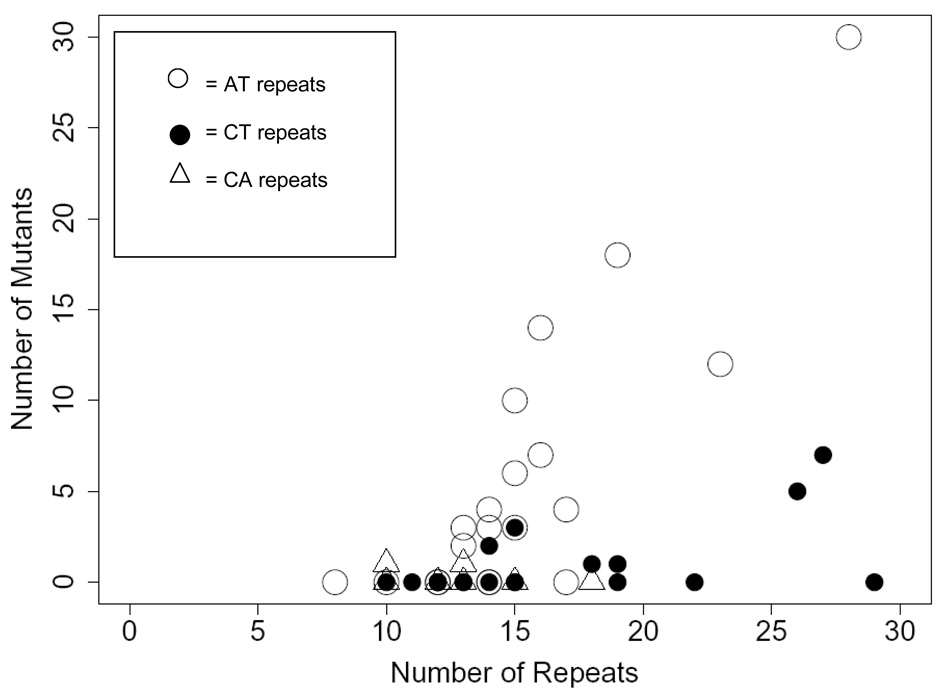
There was a great deal of variability among loci in mutation rate (Table 2). This is partly due to the difference among motif types. However, within both the AT and CT groups, the variance in mutation count substantially exceeds the mean. Much of this variation can be attributed to the strong effect of initial repeat number (Figure 1). For both AT and CT repeats, mutation rate increases substantially with the allele length for that locus in the progenitor line. This is confirmed statistically using a Poisson general linear model with mutant count per locus as the response variable, motif type as a categorical factor, and progenitor repeat number as the covariate. The estimated mutation rate equations for each motif type are:
| (1a) |
| (1b) |
| (1c) |
All coefficients, intercepts and slope, are significantly different from zero (p < 0.001). These equations share the same slope estimate because the test for an interaction between motif type and progenitor repeat number (slope heterogeneity) is non significant.
Figure 1.
The number of lines scored as mutant is given as a function of initial allele length.
Finally, we examined whether the direction of mutation (gain vs. loss) was related to repeat number. In our screen, gains were more frequent than losses. For AT loci, there were an equal number of gains and losses (4 of each), but gains occurred more frequently in shorter alleles (16.5 vs. 20 repeats on average, respectively). For the AC repeat loci, there was equal number of gains and losses (1 of each). The number of repeats in the gain was 10 and the number of repeats in the loss was 13. For the AG repeats, all five mutations were gains. In our second longest locus (AT0402; 28 repeats), 6 of the mutation accumulation lines differed from the progenitor by 2 or more repeats and all were losses. This is consistent with the trend noted in other studies for longer loci to contract with mutation.
The loci were chosen to span all five chromosomes of Arabidopsis. To test for an effect of chromosome on mutation rate, we added it as a factor in the Poisson regression model. Controlling for the effect of initial repeat number and motif type, the chromosomes were indistinguishable except for chromosome 4 which exhibits an elevated mutation rate (Z = 2.876, p<0.005). This is because the most mutable loci within motifs (AT402, AT403 and CT401, CT402) reside on chromosome 4. With chromosome included as a factor in the model, initial repeat number remains the dominant predictor of mutation rate, although the estimated slope is reduced by about 25%.
Results from genome survey
Microsatellites composed of AT repeats were the most frequent followed by AG and then AC microsatellites (Table 3). The scan also identified a small number of short GC repeats, but these were excluded from Table 3. A greater number of perfect microsatellites (uninterrupted repeat strings) were identified than imperfect microsatellites. The latter category included compound microsatellites for all repeat motif types. Compound microsatellites comprise more than one repeat type. Some, but not all, compound microsatellites also have insertions between the multiple repeat types and this is likely to affect the mutational pattern.
Table 3.
The average and standard deviation of repeat length is given for both perfect and imperfect repeats of each motif type.
| Perfect repeats | Imperfect repeats | |||||
|---|---|---|---|---|---|---|
| Average repeat length | SD of repeat length | Number of loci | Average repeat length | SD of repeat length | Number of loci | |
| AT | 7.97 | 4.332 | 9433 | 12.13 | 5.559 | 1737 |
| AC | 6.60 | 2.116 | 2518 | 9.61 | 2.494 | 194 |
| AG | 7.71 | 5.135 | 7258 | 12.22 | 8.066 | 1780 |
Discussion
This survey estimates the rate of mutation at 54 dinucleotide microsatellite loci in A. thaliana. The average estimated rate across loci isµ= 8.87 × 10−4 and the great majority of mutations were gains or losses of a single repeat. The mutation rate is heterogeneous across loci and increases with repeat number. Mutations in longer alleles are more frequently losses than gains (e.g. locus AT0402 in Table 2). These observations are fully consistent with previous mutational studies of plants (Table 1) and other organisms (e.g. Wierdl et al., 1997; Schlötterer et al., 1998; Dieringer and Schlötterer, 2003; Harr and Schlötterer, 2004; Seyfert et al., 2008).
For a given allele length, mutation rate differed among motif types. Kelkar et al. (2008) review a number of reasons why motifs might differ in mutability. The rate of loss and/or formation of hydrogen bonds can differ among motifs (AT maybe more mutable because fewer H bonds must be broken). The relative mutability of motifs could also depend on the stability of hairpin structures formed (ranked by mutation rate and hairpin stability: ATn > AGn > ACn) or in other secondary structures. Finally, motifs may be recognized differently by DNA repair mechanisms (see Harr and Schlötterer 2000; Schlötterer et al., 2006). We found the AT motif to be most mutable and the CA motif to be least mutable (see difference in intercept estimates in equations 1), which is consistent with each of the first two suggestions (hydrogen bond and hairpin stability). There is also a slight tendency towards greater variability in allele length among A. thaliana lines for AT loci than for other motifs in the surveys of Innan et al. (1997) and Symonds and Lloyd (2003).
Our overall mutation rate estimate is probably less useful than the calibrated functions predicting rate given locus-specific features (equations 1). The strong dependence on motif and initial length implies that the average genome mutation rate depends on the relative frequency of the various motif types and on the distribution of allele sizes currently segregating in the population. The AT motif, which had highest mutation rate, is the most frequent repeat type in the entire genome (Table 3; see also Morgante et al., 2002). The CA motif, which is least mutable, is least frequent. The overall average mutation rate also depends on the distribution of repeat numbers per motif in the genome. We selected loci with allele sizes in the 8–30 range (Figure 1; averages 15.35, 11.86, and 16.35 for AT, CA, and CT, respectively). These average repeat lengths for our sample are higher than the mean for each motif type in our genome survey (Table 3). Since mutation rate increases with repeat number, the average rate across our loci within motifs should be elevated relative to the genomic average. However, this bias is counteracted because the most mutable motif (AT) is more frequent in the genome than in our sample.
Equations (1) use a single slope to describe the linear relationships between mutation rate and repeat length across motifs. This is statistically defensible—the test for slope heterogeneity was not significant—but is unlikely to be literally correct. For example, we see essentially no relationship between allele length and mutation rate in CA repeats of our dataset (Figure 1), although our sample contains few CA loci with large numbers of repeats. Also, the fact that equations (1) have negative intercept estimates is consistent with the idea that there is a minimum size for microsatellite loci to accrue mutations at their typically high rate. According to our linear model, this minimum is identified by where our lines cross the x-axis. However, we caution that the true relationship between mutation rate and repeat length is likely to be non-linear.
Approximately 15% of all putative mutations identified in our initial screen proved to be PCR mutations and were discarded. This proportion is lower than in other studies that have verified putative mutations with multiple rounds of PCR. In their study of corn, Vigouroux et al. (2002) found 166 mutations in their initial screen, but only 72 were confirmed (approximately 43%). Symonds and Lloyd (2003) reported a PCR error rate of 95% for single base pair differences in A. thaliana microsatellites. While replicating PCR eliminates ‘false positives’, it is also possible for PCR to produce false negatives. This occurs if PCR reverts a real mutation back to the allele length of the progenitor. While we did not directly correct for false negatives, this bias should be minimal.
Estimation of the effective population size
There is great interest in estimating Ne, the effective size of natural populations (Frankham, 1995; Leberg, 2005). The neutral theory of molecular evolution predicts that the amount of genetic diversity within a population should be a direct function of the product of Ne and the mutation rate, μ (Kimura, 1983). An independent estimate for μ allows these two variables to be disentangled and permits inference of Ne from genetic diversity.
Symonds and Lloyd (2003) surveyed 126 accessions of A. thaliana for variation at 20 dinucleotide microsatellite loci. The average gene diversity (G) in this survey was 0.76, similar to a previous estimate (0.79) obtained by Innan et al., (1997). Assuming neutrality, the expected G is under the Stepwise Mutation Model (Ohta and Kimura, 1973). Substituting the average G from Symonds and Lloyd (2003) and our average μ across loci, we find that Ne ≈ 2300. With G = 0.79, Ne ≈ 3050. A distinct estimator for Ne is based on V, the variance of allele lengths in a population. The expected value for V is 4 Ne µ, assuming stepwise mutation (Moran 1975). Pooling variance estimates from 20 loci (accounting for differences in sample sizes) in Innan et al. (1997) yields an average V of 25.5. Solving, Ne = 25.5/(4 × 8.87 × 10−4) ≈ 7200.
While reasonable, these Ne estimates are encumbered with a number of notable caveats. First, each is subject to the bias inevitable when substituting point estimates into non-linear functions. Estimation error in either the variation statistics (G or V) or in the mutation rate biases estimation of Ne. Second, these calculations ignore real variation in mutation rate among loci. Finally, and perhaps most importantly, microsatellite allele length may not be selectively neutral. Very weak selection can substantially affect species level polymorphism (Akashi, 1997). The first two issues could be addressed by applying a more elaborate statistical model to the data. A large population survey focused on the same loci for which we have direct mutation rate information could potentially provide a strong test of the neutrality assumption.
The source of mutations
Plants do not have a segregated germ line and as a consequence both mitotic and meiotic mutations will accumulate in MA lines. A few studies have attempted to isolate the mitotic rate by comparing genotypes from ancestral and descendent cells within the same plant. Cloutier et al. (2003) observed no microsatellite mutations in a total of 12 loci of Pinus strobus, allowing the authors to place an upper bound of between 2.3 × 10–7 and 6.9 × 10–8 for the mutation rate per mitotic cell division. Leberg (2005) observed one microsatellite mutation across 8 loci of Thuja plicata and from this estimated 3.13 × 10–4 mitotic mutations per allele per generation.
While our study cannot distinguish between meiotic and mitotic mutations, we suggest that meiotic errors are likely to be more important. Whittle and Johnson (2003) found that a greater proportion of mutations in A. thaliana are transmitted to progeny via pollen than ovule, implying mutation during gametogenesis. Also, our mutation rate estimate and most of the others in Table 1 are much higher than the mitotic rate estimate obtained by Cloutier et al. (2003). However, in long-lived species or those with extensive clonal reproduction, mitotic mutations might contribute a larger fraction of the genetic variation. In the future, application of the molecular tools available for this model plant might provide a quantitative estimate for the contribution of meiotic and mitotic mutation.
Appendix.
The forward and reverse primer sequence is given for each locus in our survey.
| Locus | Repeat composition | Forward primer sequence | Reverse primer sequence | PCR product length (bp) |
|---|---|---|---|---|
| (AT)15 | aatttggagattagctggaat | ccatgttgatgataagcacaa | 144–148 | |
| AT0101 | (AT)14 | ttgtcaaaatgcactcttcattatc | ctagttacccgccaatccaa | 220–222 |
| AT0102 | (AT)16 | cgtgatattgatcactcgtcaga | ggcacatccgttttgaagat | 182–184 |
| AT0103 | (TA)16 | tcaattctacaagaaaaatgctga | gcccatataatgtgcatcacg | 121–127 |
| AT0104 | (AT)10 | aacataaagggcgtgaggtg | tttaaagtaagcattttcattgcat | 237 |
| AT0201 | (AT)13 | gcaaaactgcctaaataacacc | tcgtttgaggtcaatttttgaa | 181–185 |
| AT0202 | (AT)14 | gggttagacaattcaaatgttttt | aaacccaagatcaatattttctttaca | 180–184 |
| AT0203 | (AT)14 | tgcgatatattatgcacggatt | caaaacgtgttcgattttggt | 161 |
| AT0204 | (AT)14 | ttctcaaagtctccaagtatggtg | aaagcttttgttaggcaagca | 215–217 |
| AT0301 | (AT)16 | ttggcctaacctaaccatcaa | ctaaaaacaacaatagaagccaca | 213–217 |
| AT0302 | (AT)12 | catcaatatgatatgttcctattttca | aagccgtattgacaggagaa | 192–196 |
| AT0303 | (AT)12 | ccatgatttcattcacaacca | ttccatgatccaccacttctc | 211 |
| AT0304 | (AT)17 | tgaaatgaacagaagaagaaacca | agaagcaccatgattcaaaga | 165 |
| AT0402 | (AT)28 | acatggttttgctcccaagt | tgcagcccagaactttctct | 198–204 |
| AT0403 | (AT)23 | ttttcccgacagctcgtagt | tctcacatggttagggaaacaa | 182–190 |
| AT0404 | (AT)8 | ggtctctttagtctttaagtttgtcca | tgccgttatagcggtcattt | 178 |
| AT0501 | (AT)15 | aagaaagtgctgaatgttgatga | tgcataagccaaatgaattttt | 168–172 |
| AT0502 | (AT)15 | tgtacgtaaaatataagaaggacgatt | gaatgaaccatttcgcacct | 198–200 |
| AT0503 | (AT)12 | atcctacccgaattccgaac | ccatgccaaaatttacacga | 229 |
| AT0504 | (AT)23 | tttggatcttcaacaaatgctc | ttacccaaaccaagcaaagc | 257–261 |
| CA0101 | (AC)14 | acgaggacttcgcctgtcta | cggaaacacagtactgcttga | 180 |
| CA0102 | (TG)10 | ttatgagactggtcgactgga | catgtcgagaccgatttcaag | 164 |
| CA0103 | (TG)12 | tcacatcaaggtttgctcca | cgtgtttccttatccggtgt | 202 |
| CA0104 | (TG)10 | gacaaacaaaatccgttctgg | tatcgtgacgctctcacctg | 202–204 |
| CA0201 | (TG)10 | ccatgcatgtaaataatgaatagtga | ttgatgcttgtttgttttcca | 190 |
| CA0202 | (TG)12 | aatactgcttcggtggcatc | tggaaatcccgtgttaccat | 222 |
| CA0301 | (TG)10 | tccagcatttctttgccttt | aagctgaaaaatttcccttaatgt | 224 |
| CA0302 | (TG)12 | aatggctggccatcaaact | ttgggtgtcattctcctcgt | 263 |
| CA0401 | (CA)12 | atcacatacgccgtcctaca | tgtagctccgaatcctactcc | 174 |
| CA0501 | (TG)10 | catcgtttctcaattcgatgg | gggtgcacagggatttaaca | 263 |
| CA0502 | (TG)13 | ttcccttcaccgaacttgag | aaagccttcttcaatcaaagc | 165 |
| CA0503 | (TG)10 | tttttctacacattttctctcaatttc | atgaactatctttgatccaatgc | 166 |
| CA0504 | (TG)13 | aaaacgggaaaggtggaagt | gcctcgtgaggagtttggta | 233 |
| CA72 | (CA)18 | aatcccagtaaccaaacacaca | cccagtctaaccacgaccac | 168 |
| CT.nga1145 | (GA)14 | ccttcacatccaaaacccac | gcacatacccacaaccagaa | 229 |
| CT.nga172 | (GA)29 | agctgcttccttatagcgtcc | catccgaatgccattgttc | 175 |
| CT.nga225 | (CT)18 | gaaatccaaatcccagagagg | tctccccactagttttgtgtcc | 134–136 |
| CT.nga32 | (GA)13 | ggagactttttgagattggcc | ccaaaacaattagctcccca | 275 |
| CT.nga59 | (CT)19 | gcatctgtgttcactcgcc | ttaatacattagcccagacccg | 124 |
| CT0101 | (CT)11 | cagagacgaaagaggtgatgg | tcgaagagagagaaaatcccttt | 169 |
| CT0102 | (AG)15 | agacctccacctccaagacc | tcttccacgatccttatcgaa | 228 |
| CT0103 | (CT)10 | caacactgtgaaaccaaaaacc | ccaacctcatgaaacaaagga | 198 |
| CT0104 | (AG)14 | ttgttcggctctgcttcttt | ttgccctccaaacatggtat | 211–213 |
| CT0201 | (AG)12 | tgtgcgtgtaattttgttgct | tcagaaacgtgggtgtgtgt | 223 |
| CT0301 | (AG)12 | gggctctgtgttttgaggaa | ggatttccgcaatcatcatc | 230 |
| CT0302 | (CT)12 | gcactcgcaagtgtgaacat | tcgtttgcttcttctgtttgtc | 266 |
| CT0303 | (CT)15 | caatggtgatgtggcattgt | aaagaagaggagcagcgtgt | 193 |
| CT0304 | (AG)13 | caatttccgatggaggaaga | cccttttctcaatgcccttt | 167–169 |
| CT0401 | (AG)27 | aacaatgaggcgtatgtgagg | tgaaacttttgttgtttgggttt | 193–197 |
| CT0402 | (AG)25 | gccgctgacacttgtcacta | tcagatttccttggctttcg | 229–231 |
| CT0403 | (CT)12 | cttaggggccagctttctct | ccgaggcgtattttgtcatc | 215 |
| CT0501 | (AG)19 | gaagaagcgtgggatatgga | ggcctcacatgaaaccctaa | 204–206 |
| CT0502 | (CT)22 | cccgactcggaattcactaa | ctggcccaaccactactcat | 218 |
| CT0503 | (AG)15 | cttccatttttggcttagca | tgctttttcctcggtaatgaa | 212–214 |
Acknowledgements
We thank J. Gleason, S. Macdonald, L. Hileman, J. Preston, J. Mojica, and M. Holder for comments on this paper. C. Baer provided insightful criticism of an early draft. This research was supported by NIH grant GM073990 and NSF grant DEB-0543052 to J. K. Kelly, NSF grants DEB-9629457 and DEB-9981891 to R. G. Shaw, and NSF DEB-0108242 to M. Orive. M. E. Mort acknowledges DEB-0344883. We thank S. Macdonald and J. Gleason for use of their laboratory equipment and Lisa Darmo for establishing the MA lines.
Literature cited
- Akashi H. Distinguishing the effects of mutational biases and natural selection on DNA sequence variation. Genetics. 1997;147:1989–1991. doi: 10.1093/genetics/147.4.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, Agis M, Imhof M, Schlötterer C. Microsatellite variability differs between dinucleotide repeat motifs- Evidence from Drosophila melanogaster . Mol Biol Evol. 2000;17:1277–1285. doi: 10.1093/oxfordjournals.molbev.a026411. [DOI] [PubMed] [Google Scholar]
- Bakker EG, Stahl A, Toomajian C, Nordborg M, Kreitman M, Bergelson J. Distribution of genetic variation within and among local populations of Arabidopsis thaliana over its species range. Mol Ecol. 2006;15:1405–1418. doi: 10.1111/j.1365-294X.2006.02884.x. [DOI] [PubMed] [Google Scholar]
- Balloux F, Lugon-Moulin N. The estimation of population differentiation with microsatellite markers. Mol Ecol. 2002;11:155–165. doi: 10.1046/j.0962-1083.2001.01436.x. [DOI] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: A progroam to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin-Ganache IRM, Raymond M, Deschepper CF. M13-tailed primers improve the readability and usability of microsatellite analyses performed with two different allele-sizing methods. BioTechniques. 2001;31:24–27. [PubMed] [Google Scholar]
- Chakraborty R, Kimmel M, Stivers DN, Davison LJ, Deka R. Relative mutation rates at di-, tri-, and tetranucleotide microsatellite loci Proc Natl Acad Sci USA 1997. 94 1041 1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase M, Kesseli R, Bawa K. Microsatellite markers for population and conservation genetics of tropical trees. Am J Bot. 1996;83:51–57. [Google Scholar]
- Cloutier D, Rioux D, Beaulieu J, Schoen DJ. Somatic stability of microsatellite loci in Eastern white pine, Pinus strobus L. Heredity. 2003;90:247–252. doi: 10.1038/sj.hdy.6800221. [DOI] [PubMed] [Google Scholar]
- Dieringer D, Schlötterer C. Two distinct modes of microsatellite mutation processes: Evidence from the complete genomic sequences of nine species. Genome Res. 2003;13:2242–2251. doi: 10.1101/gr.1416703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiRienzo A, Peterson AC, Garza JC, Valdes AM, Slatkin M, Freimer NB. Mutational processes of simple-sequence repeat loci in human populations. Proc Natl Acad Sci USA. 1994;91:3166–3170. doi: 10.1073/pnas.91.8.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow BD, Ashley VM. High levels of gene flow in bur oak revealed by paternity analysis using microsatellites. J Hered. 1998;89:62–70. [Google Scholar]
- Ellegren H. Microsatellites: Simple sequences with complex evolution. Nat Rev Genet. 2004;5:435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- Frankhan R. Effective population size/adult population size rations in wildlife: A review. Genet Res. 1995;66:95–107. doi: 10.1017/S0016672308009695. [DOI] [PubMed] [Google Scholar]
- Grover A, Sharma P. Microsatellite motifs with moderate GC content are clustered around genes on Arabidopsis thaliana Chromosome 2. In Silico Biol. 2007;7:201–213. [PubMed] [Google Scholar]
- Harr B, Schlötterer C. Long microsatellite alleles in Drosophila melanogaster have a downward mutation bias and short persistence times, which cause their genome-wide underrepresentation. Genetics. 2000;155:1213–1220. doi: 10.1093/genetics/155.3.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harr B, Schlötterer C. Patterns of microsatellite variability in the Drosophila melanogaster complex. Genetica. 2004;120:71–77. doi: 10.1023/b:gene.0000017631.00820.49. [DOI] [PubMed] [Google Scholar]
- Innan H, Terauchi R, Miyashita NT. Microsatellite Polymorphism in Natural Populations of the Wild Plant Arabidopsis thaliana. Genetics. 1997;146:1441–1452. doi: 10.1093/genetics/146.4.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelkar YD, Tyekucheva S, Chiaromonte F, Makova KD. The genome-wide determinants of human and chimpanzee microsatellite evolution. Genome Res. 2008;18:30–38. doi: 10.1101/gr.7113408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JK, Willis JH. A manipulative experiment to estimate bi-parental inbreeding in Monkeyflowers. Int J Plant Sci. 2002;163:575–579. [Google Scholar]
- Kimura M. The Neutral Theory of Molecular Evolution. New York, Cambridge: University Press; 1983. [Google Scholar]
- Kimura M, Crow JF. The number of alleles that can be maintained in a finite population. Genetics. 1964;49:725–738. doi: 10.1093/genetics/49.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberg P. Genetic approaches for estimating the effective size of populations. J Wildl Manage. 2005;69:1385–1399. [Google Scholar]
- Moran PAP. Wandering distributions and electrophoretic profile. Theor Popul Biol. 1975;8:318–330. doi: 10.1016/0040-5809(75)90049-0. [DOI] [PubMed] [Google Scholar]
- Morgante M, Hanafey M, Powell W. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat Genet. 2002;30:194–200. doi: 10.1038/ng822. [DOI] [PubMed] [Google Scholar]
- Ohta T, Kimura M. A model of mutation appropriate to estimate the number of electrophoretically detectable alleles in a finite population. Genet Res. 1973;22:201–204. doi: 10.1017/s0016672300012994. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Schlötterer C, Pemberton J. The use of microsatellites for genetic analysis of natural populations. In: Schierwater B, Streit B, Wagner GP, DeSalle R, editors. Molecular Ecology and Evolution: Approaches and Applications. Basel: Birkhuaser Verlag; 1994. [Google Scholar]
- Schlötterer C, Ritter R, Harr B, Brem G. High mutation rate of a long microsatellite allele in Drosophila melanogaster provides evidence for allele- specific mutation rates. Mol Biol Evol. 1998;15:1269–1274. doi: 10.1093/oxfordjournals.molbev.a025855. [DOI] [PubMed] [Google Scholar]
- Schlötterer C, Imhof M, Wang H, Nolte V, Harr B. Low abundance of Escherichia coli microsatellites is associated with an extremely low mutation rate. J Evol Biol. 2006;19:1671–1676. doi: 10.1111/j.1420-9101.2006.01108.x. [DOI] [PubMed] [Google Scholar]
- Schug M, Hutter C, Wetterstrand K, Gaudette M, Mackay T, Aquadro C. The mutation rates of di-, tri- and tetranucleotide repeats in Drosophila melanogaster. Mol Biol Evol. 1998;15:1751–1760. doi: 10.1093/oxfordjournals.molbev.a025901. [DOI] [PubMed] [Google Scholar]
- Seyfert AL, Cristescu MEA, Frisse L, Schaack S, Thomas WK, Lynch M. The rate and spectrum of microsatellite mutation in Caenorhabditis elegans and Daphnia pulex . Genetics. 2008;178:2113–2121. doi: 10.1534/genetics.107.081927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw FH, Geyer CJ, Shaw RG. A comprehensive model of mutations affecting fitness and inferences for Arabidopsis thaliana. Evolution. 2002;56:453–463. doi: 10.1111/j.0014-3820.2002.tb01358.x. [DOI] [PubMed] [Google Scholar]
- Sweigart A, Karoly K, Jones A, Willis JH. The distribution of individual inbreeding coefficients and pairwise relatedness in a population of Mimulus guttatus. Heredity. 1999;83:625–632. doi: 10.1038/sj.hdy.6886020. [DOI] [PubMed] [Google Scholar]
- Symonds VV, Lloyd AM. An analysis of microsatellite loci in Arabidopsis thaliana: Mutational dynamics and application. Genetics. 2003;165:1475–1488. doi: 10.1093/genetics/165.3.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuillet A-C, Bataillon T, Sourdille P, David JL. Factors affecting polymorphism at microsatellite loci in bread wheat [Triticum aestivum (L.) Thell]: Effects of mutation processes and physical distance from the centromere. Theor Appl Genet. 2004;108:368–377. doi: 10.1007/s00122-003-1443-5. [DOI] [PubMed] [Google Scholar]
- Thuillet A-C, Bru D, David J, Roumet P, Santoni S, Surdille P, Bataillon T. Direct estimation of mutation rate for 10 microsatellite loci in durum wheat, Triticum turgidum (L.) Thell. Ssp durum desf. Mol Biol Evol. 2002;19:122–125. doi: 10.1093/oxfordjournals.molbev.a003977. [DOI] [PubMed] [Google Scholar]
- Udupa SM, Baum M. High mutation rate and mutational bias at (TAA)n microsatellite loci in chickpea (Cicer arietinum L.) Mol Genet Genomics. 2001;265:1097–1103. doi: 10.1007/s004380100508. [DOI] [PubMed] [Google Scholar]
- Vigouroux Y, Jaqueth JS, Yoshihiro M, Smith OS, Beavis WD, Smith JSC, Doebley J. Rate and pattern of mutation at microsatellite loci in maize. Mol Biol Evol. 2002;19:1251–1260. doi: 10.1093/oxfordjournals.molbev.a004186. [DOI] [PubMed] [Google Scholar]
- Weber JL, Wong C. Mutation of human short tandem repeats. Hum Mol Genet. 1993;2:1123–1128. doi: 10.1093/hmg/2.8.1123. [DOI] [PubMed] [Google Scholar]
- Whittle C-A, Johnston MO. Male-biased transmission of deleterious mutations to the progeny in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2003;100:4055–4059. doi: 10.1073/pnas.0730639100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierdl M, M. Dominska M, Petes TD. Microsatellite instability in yeast: Dependence on the length of the microsatellite. Genetics. 1997;146:769–779. doi: 10.1093/genetics/146.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yuan D, Yu S, Li Z, Cao Y, Miao Z, et al. Preference of simple sequence repeats in coding and non-coding regions of Arabidopsis thaliana. Bioinformatics. 2004;20:1081–1086. doi: 10.1093/bioinformatics/bth043. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zuo K, Zhang F, Cao Y, Wang J, Zhang Y, et al. Conservation of noncoding microsatellites in plants: Implication for gene regulation. BMC Genomics. 2006;7:323. doi: 10.1186/1471-2164-7-323. [DOI] [PMC free article] [PubMed] [Google Scholar]