Abstract
The CREB regulated transcription co-activators (CRTCs) regulate many biological processes by integrating and converting environmental inputs into transcriptional responses. Although the mechanisms by which CRTCs sense cellular signals are characterized, little is known regarding how CRTCs contribute to the regulation of cAMP inducible genes. Here we show that these dynamic regulators, unlike other co-activators, independently direct either pre-mRNA splice-site selection or transcriptional activation depending on the cell type or promoter context. Moreover, in other scenarios, the CRTC co-activators coordinately regulate transcription and splicing. Mutational analyses showed that CRTCs possess distinct functional domains responsible for regulating either pre-mRNA splicing or transcriptional activation. Interestingly, the CRTC1–MAML2 oncoprotein lacks the splicing domain and is incapable of altering splice-site selection despite robustly activating transcription. The differential usage of these distinct domains allows CRTCs to selectively mediate multiple facets of gene regulation, indicating that co-activators are not solely restricted to coordinating alternative splicing with increase in transcriptional activity.
Keywords: alternative splicing, cAMP, CREB, signal transduction, TORC
Introduction
The complexity and diversity of biological responses that occur following receipt of environmental cues far exceeds the absolute number of genes that are transcriptionally induced or repressed by signalling networks. To circumvent this problem, genes are regulated at additional levels, including precursor mRNA (pre-mRNA) processing. One mechanism to generate protein, and therefore functional, diversity is through alternative splicing of pre-mRNAs to include or exclude select coding regions. Remarkably, nearly 60% of the transcripts expressed from human genes are alternatively spliced (Lander et al, 2001; Sharp, 2005; Moore and Silver, 2008).
The splicing process is carried out by the large multi-subunit spliceosome complex and is responsible for catalysing the two transesterification steps that constitute the splicing reaction (Black, 2003). Although the spliceosome is primarily composed of general components that remove intronic sequences in an unregulated manner from the pre-mRNA, it is now evident that several accessory proteins and cis-acting elements in the RNA sequence cooperate with the spliceosome to control splice-site selection (Kornblihtt et al, 2004; Bentley, 2005). Further, there are several instances in which signalling pathways promote alternative mRNA splice patterns by modulating the function of these accessory proteins. For example, signalling directed by c-Jun N-terminal kinase (JNK), calcium/calmodulin-dependent protein kinase IV, the phosphatidylinositol 3′ kinase (PI3K), and mitogen-activated protein kinase/extracellular signal-regulated kinase (MEK) regulate the splicing patterns of several pre-mRNAs (Konig et al, 1998; Lynch and Weiss, 2000; van der Houven van Oordt et al, 2000; Xie and Black, 2001; Matter et al, 2002). In addition, cAMP-dependent protein kinase A (PKA) phosphorylation regulates the nucleo–cytoplasmic localization of the pre-mRNA splicing factor polypyrimidine tract-binding protein (PTB) (Xie et al, 2003). In each of these instances, kinases in these signalling cascades directly mediate RNA processing by phosphorylating key components of the spliceosome.
In addition to regulating pre-mRNA processing through protein phosphorylation, signalling-dependent coupling of transcription and alternative splice-site selection is mediated by recruitment of co-activators by nuclear hormone receptors (Auboeuf et al, 2007). Here, the complement of proteins assembled on promoters can dramatically influence pre-mRNA processing, and in many instances these two processes seam coordinately regulated (Cramer et al, 1997; Monsalve et al, 2000; Auboeuf et al, 2004; Auboeuf et al, 2005). Moreover, transcriptional co-regulators can orchestrate signalling-dependent coupling of transcription and alternative splice-site selection. For example, the nuclear hormone receptor co-activators PGC-1, CoAA and CAPER coordinately regulate transcription and splicing of some target genes through intrinsic RNA recognition motifs (RRM) (Basu et al, 1997; Monsalve et al, 2000; Auboeuf et al, 2002, 2004; Dowhan et al, 2005; Fox et al, 2005).
The CREB regulated transcription co-activators (CRTCs) are signal-dependent transcriptional co-activators of cAMP-responsive promoters and are key regulators of gluconeogenesis (Koo et al, 2005; Dentin et al, 2007), adaptive mitochondrial biogenesis (Wu et al, 2006) and long-term synaptic plasticity (Kovacs et al, 2007). Under basal conditions, CRTCs are phosphorylated by AMP/SNF kinases and are bound by 14-3-3 proteins that sequester CRTCs in the cytoplasm (Screaton et al, 2004; Koo et al, 2005). As cAMP levels rise, PKAc phosphorylates and inhibits AMP/SNF kinases, which prevents CRTC phosphorylation. Dephosphorylated CRTCs are then released from 14-3-3 proteins, translocate to the nucleus and bind to CREB (Bittinger et al, 2004; Screaton et al, 2004). The phosphorylation status of CRTCs and their cytoplasmic retention by 14-3-3 integrates converging cellular signals. For example, hormone and energy-sensing pathways converge on CRTC2 phosphorylation to modulate glucose output through CREB-mediated hepatic gene expression (Koo et al, 2005) and to regulate incretin hormones and glucose to promote β-cell survival (Jansson et al, 2008). Further, CRTC functions as a coincidence detector in excitable cells that funnels cAMP and calcium signalling pathways to CREB-dependent transcription (Screaton et al, 2004; Kovacs et al, 2007).
Here we report that CRTC co-activators, components of the cAMP signalling pathway, are bipartite regulators of gene expression that selectively control pre-mRNA splicing and/or transcriptional activation of CRE-containing genes. Remarkably, unlike other co-activators that contain RNA-binding motifs, we now provide data showing that the CRTC co-activators can induce alternative splicing without increasing transcriptional activation or direct transcriptional activation without altering splice-site selection.
Results
CRTC co-activators regulate alternative splicing
The CRTC co-activators are robust transcriptional co-activators of cAMP-dependent promoters. However, how they direct this function is not understood as they lack identifiable catalytic domains and homology to other proteins. Two previous studies have hinted that unidentified components of the cAMP pathway can mediate pre-mRNA processing, as a small deletion in the fibronectin promoter encompassing a CRE element alters alternative exon usage (Cramer et al, 1997), and a genetic screen for CRTC interactors identified a known regulator of pre-mRNA splicing NONO (p54nrb) (Amelio et al, 2007). Given these connections, we tested whether CRTCs might also contribute to pre-mRNA processing in response to cAMP signalling.
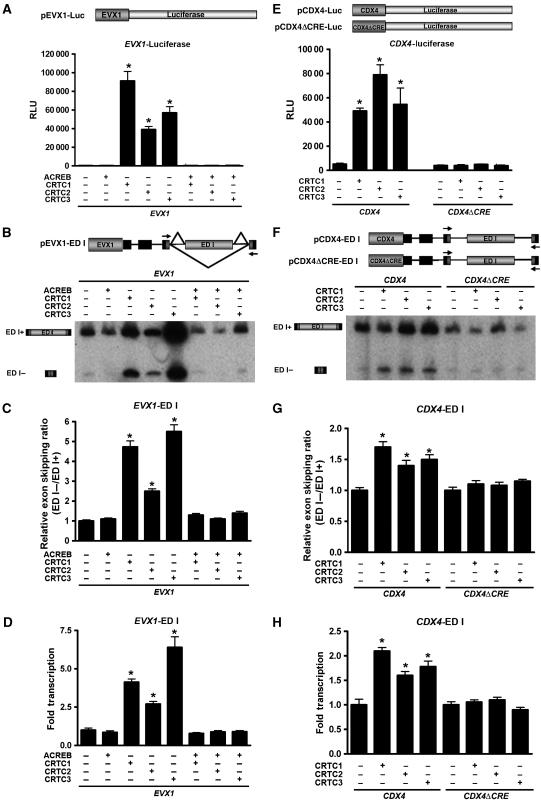
To test the potential dual roles of CRTCs in transcriptional activation and splice-site selection of cAMP-responsive genes, we generated several luciferase reporter genes and minigene splicing reporters with matched promoter fragments containing well-characterized CREB binding sites (Supplementary Figure S1) (Conkright et al, 2003b). Although the luciferase reporters were used to monitor transcriptional activity, the minigene splicing reporters measured inclusion or skipping of the alternatively spliced fibronectin extra domain I (ED I) exon (Caputi et al, 1994; Cramer et al, 1997). Important for these studies, ectopic expression of the CRTC co-activators overrides normal regulatory circuits that sequester CRTCs in the cytoplasm, thereby allowing their nuclear localization. As CRTC co-activators are the limiting components of the CREB transcriptional complex, they trigger robust induction of cAMP-responsive genes and, accordingly, overexpression of each CRTC family member (CRTC1-3) induced robust transcriptional activity of the cAMP-responsive EVX1 promoter (Figure 1A). Further, as expected, the CREB dominant-negative mutant A-CREB, which masks the basic residues of the CREB DNA-binding domain, blocked this response (Figure 1A) (Ahn et al, 1998; Conkright et al, 2003b). Surprisingly, the CRTC co-activators caused a profound alteration in splice-site selection of the ED I minigene construct and these changes were also ablated by addition of A-CREB (Figure 1B). TaqMan primer/probe sets were designed to quantitatively measure alternative splicing or total transcription by detecting sequences that either include or exclude the alternatively spliced ED I exon (Supplementary Figure S2AB). The quantification of minigene alternative splicing transcripts by real-time qPCR showed that co-expression of CRTCs promoted >5-fold increase in skipping of the ED I exon (Figure 1C), a response robust as those of known regulators of pre-mRNA processing (Cramer et al, 1997; Monsalve et al, 2000; Auboeuf et al, 2002). Analysis of RNA transcripts from the ED I minigene by real-time qPCR showed that all CRTC family members activated transcription of the ED I minigene relative to basal conditions similar to findings observed with luciferase enzymatic assays (Figure 1D). Differences between each CRTC family member, on transcriptional activation and alternative splicing, were probably due to differences in their expression levels (Supplementary Figure S3A). Affects on alternative splicing were also blocked by A-CREB, indicating that CRTCs act in conjunction with promoter-bound CREB (Figure 1C). The affects of CRTC on alternative splicing were also observed in cell lines that do not express SV40 large T antigen or adenovirus E1A (Supplementary Figure S3B).
Figure 1.
CRTC co-activators regulate alternative exon splicing and transcriptional activation. (A) Effect of CRTCs on transcriptional activity. Transient transfection assays of EVX1 luciferase reporter co-transfected into HEK293T cells with CRTC 1, 2 or 3 and/or A-CREB were analysed for luciferase activity (n=6 wells; mean±s.e.m.; asterisk denotes P-value ⩽0.05). (B) Effect of CRTCs on alternative exon splicing. Transient transfection assays of EVX1 ED I minigene reporter co-transfected into HEK293T cells with CRTC 1, 2 or 3 and/or A-CREB. RT–PCR products were separated by PAGE and splice variants were visualized by autoradiography to determine the effect of CRTC expression on splice site selection. (C) TaqMan real-time qPCR analysis of CRTCs affects on EVX1 ED I minigene exon skipping. The relative exon skipping levels are expressed as the ratio of skipped versus included transcripts (n=3 experiments; mean±s.e.m.; asterisk denotes P-value ⩽0.05). (D) TaqMan real-time qPCR analysis of CRTCs affects on EVX1 ED I minigene reporter gene expression. Relative amounts of ED I reporter transcripts obtained by each transfection were plotted as folds relative to basal transcription (n=3 experiments; mean±s.e.m.; asterisk denotes P-value ⩽0.05). (E) Effect of CRTCs on CRE-dependent transcriptional activity. Transient transfection assays of CDX4 luciferase or CDXΔCRE reporters co-transfected into HEK293T cells with CRTC 1, 2 or 3 were analysed for luciferase activity (n=6 wells; mean±s.e.m.; asterisk denotes P-value ⩽0.05). (F) Effect of CRTCs on CRE-dependent alternative exon splicing. Transient transfection assays of CDX4 or CDX4ΔCRE ED I minigene reporters co-transfected into HEK293T cells with CRTC 1, 2 or 3. RT–PCR products were separated by PAGE and splice variants analysed to determine the effect of CRTC expression on splice site selection. (G) TaqMan real-time qPCR analysis of CRTCs affects on CDX4 or CDX4ΔCRE ED I minigene exon skipping. The relative exon skipping levels are expressed as the ratio of skipped versus included transcripts (n=3 experiments; mean±s.e.m.; asterisk denotes P-value⩽0.05). (H) TaqMan real-time qPCR analysis of CRTCs affects on CDX4 or CDX4ΔCRE ED I minigene reporter gene expression. Relative amounts of ED I reporter transcripts obtained by each transfection were plotted as folds relative to basal transcription (n=3 experiments; mean±s.e.m.; asterisk denotes P-value ⩽0.05).
A second cAMP-responsive promoter, CDX4 (Conkright et al, 2003b), was tested to establish that whether this was a general property of CRE-containing promoters. Similar to the EVX1-luciferase and -ED I minigenes, CRTCs promoted transcriptional activation and splicing in the CDX4 promoter context (Figure 1E–H). To confirm that CRTCs directly affected mRNA processing and that this required a CREB bound to CRE site, a minigene with a deletion in the essential GC core of the CRE (TGACGTCA → TGATCA)(Montminy et al, 1986) was created within the CDX4 promoter. This CRE-disabling mutation ablates both transcription and mRNA processing induced by CRTCs (Figure 1E–H). Thus, CRTC-mediated induction of transcription and splicing are direct and require functional CRE-containing promoters.
To assess whether the observed effects of CRTCs on alternative splicing could reflect sequestering of splicing factors, assays were conducted using increasing amounts of each reporter substrate (Supplementary Figure S4). Although titration of the EVX1-luciferase reporter resulted in near 10-fold increases in transcriptional activity (Supplementary Figure S4A), this did not translate into a change in the ratio of ED I exon skipping (Supplementary Figure S4B). Therefore, CRTC-mediated alterative splicing is not due to a decrease in pools of available splicing factors and there is not a linear relationship between transcription and splicing induced by CRTCs. Therefore, once recruited to CRE-containing promoters by CREB, CRTC co-activators have bipartite roles in gene regulation, by augmenting transcriptional activation and mediating pre-mRNA processing.
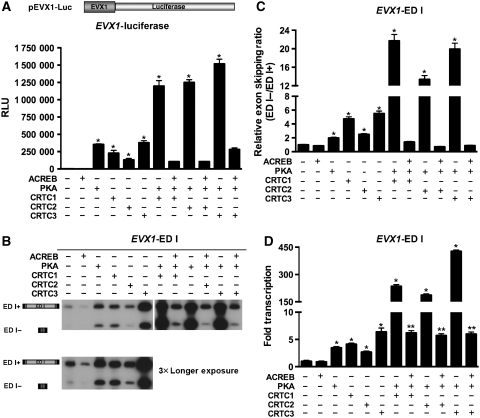
Previous studies have suggested a role for PKA activation in the regulation of alternative pre-mRNA splicing, by directly phosphorylating and altering the subcellular localization of the splicing factor PTB (Xie et al, 2003). As expected, expression of the PKA catalytic subunit (PKAc) induced transcriptional activity of the cAMP-responsive EVX1 promoter and A-CREB blocked this response (Figure 2A). Interestingly, co-expression of each CRTC family member (CRTC1–3) with PKAc led to robust transcriptional response. Real-time qPCR analysis of luciferase transcripts showed that luciferase activity is in agreement with promoter activity (Supplementary Figure S5). Similar to luciferase assays, PKAc augmented splicing through CRTCs, leading to a marked increase in ED I exon skipping (Figure 2B). Real-time qPCR analysis of minigene alternative splicing showed a twofold increase in skipping of the ED I exon induced by PKAc alone, and that co-expression of PKAc with each CRTC family member induced a robust, 10–25-fold increase in exon skipping (Figure 2C). Real-time PCR analysis of minigene transcription showed that co-expression of each CRTC family member with PKAc robustly activated transcription relative to basal conditions (Figure 2D). Notably, co-expression of A-CREB attenuated the shift in splice-site selection mediated by PKA and the CRTC co-activators, indicating that within this setting the role of PKA in alternative splicing is also promoter dependent.
Figure 2.
PKAc activation of the cAMP pathway controls alternative exon splicing. (A) Effect of activating the cAMP pathway on transcriptional activity. Transient transfection assays of EVX1 luciferase reporter co-transfected into HEK293T cells with PKAc in the presence or absence of CRTC 1, 2 or 3 and/or A-CREB were analysed for luciferase activity (n=6 wells; mean±s.e.m.; asterisk denotes P-value ⩽0.05). (B) Effect of activating the cAMP pathway on alternative exon splicing. The schematic illustrates the ED I minigene splicing reporter and alternatively spliced transcripts that either include or skip the ED I exon. Transient transfection assays of EVX1 ED I minigene reporter co-transfected into HEK293T cells with PKAc in the presence or absence of CRTC 1, 2 or 3 and/or A-CREB. RT–PCR products were separated by PAGE and splice variants analysed to determine the effect of CRTC expression on splice-site selection. Identities of spliced and unspliced amplicons are indicated on the right side of the panel. (C) TaqMan real-time qPCR analysis of ED I exon skipping. The relative exon skipping levels are expressed as the ratio of skipped versus included transcripts (n=3 experiments; mean±s.e.m.; asterisk denotes P-value ⩽0.05). (D) TaqMan real-time qPCR analysis of reporter gene expression. Relative amounts of ED I reporter transcripts obtained by each transfection were plotted as folds relative to basal transcription (n=3 experiments; mean±s.e.m.; single asterisk denotes P-value ⩽0.05; double asterisk for A-CREB inhibition denotes P-value ⩽0.05).
CRTC co-activators activate CRE-dependent 3′ splice-site selection
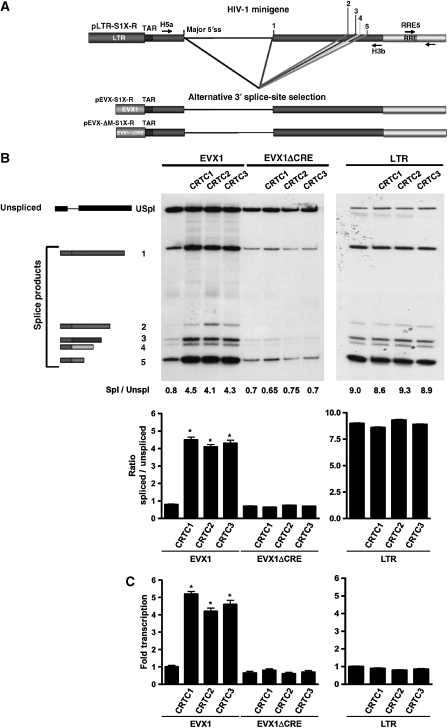
These findings established that CRTC-directed regulation of pre-mRNA splicing is dependent on the promoter. However, the alternatively spliced ED I minigene was derived from the fibronectin gene, which can be induced by cAMP signalling (Dean et al, 1988, 1989). Thus, some intrinsic component of the minigene or its transcript might facilitate CRTC-mediated pre-mRNA splicing. Furthermore, alternative splicing is complex, in which multiple splice sites within an exon can be selected, in addition to complete exon inclusion or exclusion. To address these caveats, a second minigene reporter derived from the HIV-1 LTR was tested, which is not regulated by CREB/CRTC and measures alternative 3′ splice-site selection (Supplementary Figure S6) (Caputi et al, 1999). The effect of the CRTC co-activators on 3′ splice-site selection was examined using minigenes regulated by either the native HIV-1 LTR, wild-type EVX1 or mutated CRE EVX1 promoters (Figure 3A). Each CRTC family member activated transcription and splicing fivefold relative to basal conditions (Figure 3B and C). Moreover, splicing activation is independent of the 3′ splice site chosen, as each of the five 3′ splice sites within the transcript were induced equally, even though each 3′ splice site is activated by different sets of spliceosome factors (Caputi et al, 1999; Zahler et al, 2004). Mutations of the CREB binding sites again effectively blocked this response, confirming the dependency of the CRE-containing promoter for CRTC function on pre-mRNA processing (Figure 3B and C). Importantly, overexpression of CRTCs with a HIV-1 LTR promoter construct devoid of CRE sites did not affect basal transcription or splicing levels, reaffirming that CRTCs do not affect the general transcriptional or splicing machinery independent of promoter recruitment (Figure 3B and C, right panels).
Figure 3.
CRTC co-activators facilitate alternative 3′ splice-site selection. (A) Effect of CRTC co-activators on alternative 3′ splice-site selection. Schematic representation of the pLTR-S1X-R minigene reporter substrates derived from the HIV-1 genome. The major viral 5′ and the alternative 3′ splice sites specific for the tat, rev, env and nef gene products are indicated. Plasmids that substitute the LTR for the EVX1 (pEVX1-S1X-R) and EVX1-mutated CRE (pEVX-ΔM-S1X-R) promoters are also represented. Location of the primers utilized for the detection of alternatively spliced products (H5a, H3b) and qPCR quantification of the transcripts generated by the reporter constructs (RRE5, RRE3) are indicated. (B) HEK293T cells were transfected with the indicated reporter construct together with a CRTC expression plasmid. RT–PCR products were separated by PAGE and splice variants analysed to determine the effect of CRTCs on splicing. Analysis of the ratio of spliced versus unspliced transcripts expressed by the reporter constructs is plotted. The ratio represents the sum of the total amount of spliced transcripts versus unspliced transcripts for each transfection experiment (n=3 experiments; mean±s.e.m.; asterisk denotes P-value ⩽0.05). (C) qPCR analysis of reporter gene expression. Relative amounts of HIV-1 reporter transcripts obtained by each transfection were plotted as fold of total gene products expressed (unspliced+spliced) versus the amount expressed by the pEVX1-S1X-R or pLTR-S1X-R constructs (n=3 experiments; mean±s.e.m.; asterisk denotes P-value ⩽0.05).
To determine whether the observed 3′ splice-site selection is responsive to promoter strength or saturation of splicing factors, the transcription and splicing ratio of each construct was analysed. Comparison of qRT–PCR products expressed from the EVX1 promoter with those expressed from the LTR promoter shows a nearly 40-fold difference in promoter strength (Supplementary Figure S7A). Increasing each minigene substrate resulted in increasing amounts of the HIV-1 reporter transcripts (Supplementary Figure 7B), whereas the ratio between spliced and unspliced mRNA isoforms did not change; therefore, available splicing factors are not being saturated (Supplementary Figure S7C). Thus, despite dramatic differences in promoter strength, the CRTC co-activators are capable of promoting efficient splicing of transcripts originating from the weaker EVX1 promoter. In addition, the ability of the CRTC co-activators to function as mediators of RNA processing is independent of the minigene splicing substrate.
CRTCs direct cell type-specific alternative splicing of endogenous transcripts
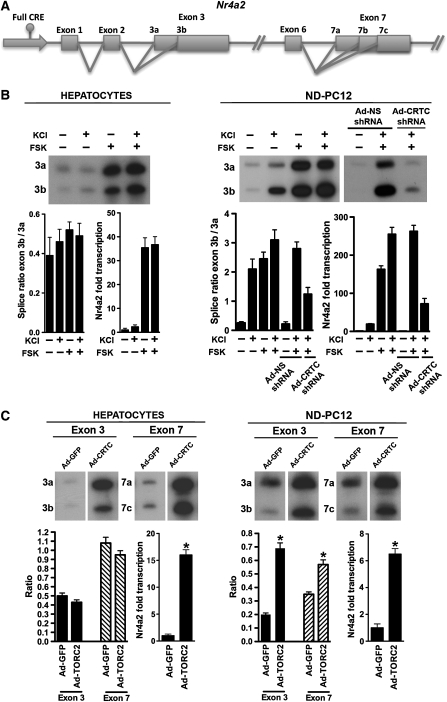
Alternatively spliced isoforms exist for ∼60% of transcripts of the human genome (Lander et al, 2001; Sharp, 2005; Moore and Silver, 2008). As CREB binds to ∼10% of all proximal promoters, it is likely that a subset of CREB/CRTC target genes are also regulated by CRTCs at the level of alternative splicing (Impey et al, 2004; Zhang et al, 2005). To assess whether CRTCs mediate alternative splicing of endogenous CRE-containing genes, we examined the Nr4a2 (Nurr1) gene, which is CRTC responsive (Conkright et al, 2003a). Nr4a2, a member of the structurally related nuclear receptor superfamily, is a transcription factor expressed predominantly in the liver, skeletal muscle and nervous tissues that controls key aspects of metabolism and dopamine biosynthesis (Smits et al, 2003; Pei et al, 2006; Fu et al, 2007). Consequently, alternative splicing in exon 3 and exon 7 of Nr4a2 in nervous tissue (Figure 4A) yields isoforms that encode functionally altered Nr4a2 proteins (Ohkura et al, 1999; Xu and Le, 2004; Michelhaugh et al, 2005).
Figure 4.
CRTC promotes alternative splicing of endogenous Nr4a2. (A) Schematic representation of the Nr4a2 gene. Possible exon 3 and exon 7 products arising from alternative splicing are depicted. (B) RT–PCR analysis of Nr4a2 was carried out using exon-specific primers on cDNA from primary rat hepatocytes or neuronally differentiated PC12 (ND-PC12) cells treated with KCl and/or forskolin (FSK). Analysis of ND-PC12 cells transduced with adenovirus expressing either GFP or CRTC2 and either non-specific (NS) or CRTC2-specific shRNAs. (C) RT–PCR analysis of Nr4a2 was carried out using exon-specific primers on cDNA from primary rat hepatocytes or ND-PC12 cells transduced with adenovirus expressing either GFP or CRTC2. Ratio of alternatively spliced exons are indicated and shown in graphs (n=3 experiments; mean±s.e.m.; asterisk denotes P-value ⩽0.05). Panels of gel images shown here were from the same gel shown in Supplementary Figure 8SB.
Physiologic cues that regulate cAMP or both cAMP and calcium signalling pathways have been shown to converge on the CRTCs in non-excitable and excitable cells, respectively, to regulate CRE-responsive genes (Screaton et al, 2004). Therefore, we analysed RNA isolated from primary rat hepatocytes or neuronally differentiated PC12 (ND-PC12) cells that were treated with forskolin (FSK) or depolarized with KCl (Figure 4B). Primary hepatocytes treated with KCl or FSK did not show significant changes in Nr4a2 exon 3 alternative splice site-selection despite FSK-induced transcription. In contrast, both KCl-mediated depolarization and FSK-induction treatments promote alternative splice-site selection in Nr4a2 exon 3 within ND-PC12 cells, and co-stimulation with both KCl and FSK further potentiates this shift in alternative splicing. This response was attenuated by transduction with adenovirus expressing an shRNA directed against CRTC (Figure 4B, right panel and Supplementary Figure S8A). Induction of the cAMP and calcium signalling pathways may be activating key splicing factors in addition to CRTCs. To show the role of CRTC, we also analysed RNA isolated from either primary rat hepatocytes or ND-PC12 cells that were transduced solely with adenovirus expressing GFP or CRTC (Figure 4C and Supplementary Figure S8B). Alternative splice-site selection in both exon 3 and exon 7 were induced 3.5-fold and 2-fold by CRTC, respectively, in ND-PC12 cells, whereas minimal effects of CRTC on splice-site selection were observed in hepatocytes, despite robust (16-fold) transcriptional activation of the Nr4a2 gene (Figure 4C). Moreover, these effects were reversed by co-transduction with adenovirus expressing an shRNA directed against CRTC (Supplementary Figure S8C). Endogenous CRTC RNA, and protein levels relative to overexpressed adeno-CRTC were monitored by real-time TaqMan PCR and western blot analysis (Supplementary Figures S8DE, respectively).
The CRTC-mediated shift in transcript splicing of the Nr4a2 transcription factor generates three distinct open reading frames whose products are either transcriptionally compromised or completely inactive (Ohkura et al, 1999; Xu and Le, 2004; Michelhaugh et al, 2005). Transient transfection of a luciferase reporter containing multimerized Nr4a2 binding sites (3xNBRE) into PC12 or HEK293T cells with each respective Nr4a2 isoform showed a dominant-negative affect of each isoform on full-length Nr4a2 (Supplementary Figures S9A and B, respectively). Therefore, although CRTCs induce endogenous Nr4a2 transcription in multiple cell types, their control of splice-site selection of the Nr4a2 locus is cell type specific. Further, these data suggested that CRTC-mediated splice-site selection is not obligatory to transcriptional activation and is dependent on the cellular context.
CRTC-directed transcription and alternative splicing are independent functions
The activation of gene expression by the CREB/CRTC complex is dictated by promoter context (Conkright et al, 2003a, 2003b). Specifically, in contrast to genes with core promoters containing TATA box elements, TATA-less core promoters such as those possessing GC-rich or initiator (INR) elements do not respond to cAMP stimuli or to CRTC overexpression despite containing a CRE that is bound in vivo by CREB and CRTCs (Conkright et al, 2003a, 2003b). Moreover, mutation of the CRE motif results in significant loss of basal transcriptional activity indicating that this site is regulatory (Conkright et al, 2003b).
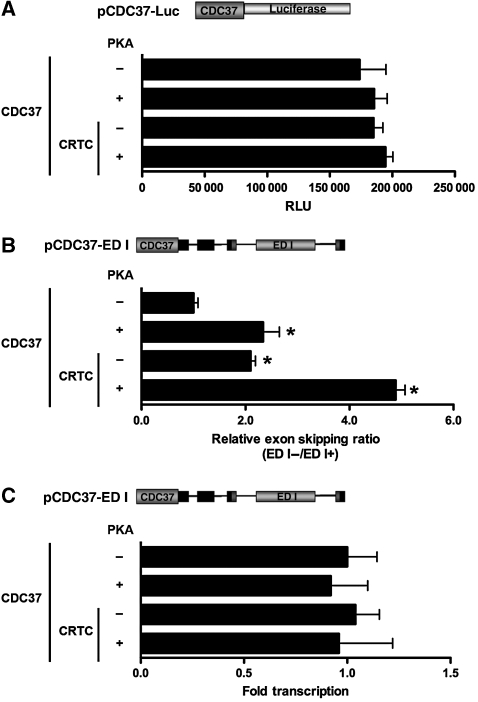
Our data showing that CRTC co-activators augment splicing in a cell context-specific manner suggested that transcriptional activation and splice-site selection might be independent functions of these co-activators. As CREB and CRTCs cannot induce transcription when recruited to CRE-containing TATA-less promoters, we tested whether CRTCs could still mediate splice-site selection in this context. To test this idea, we compared the effects of promoter context on CRTC-mediated transcription and pre-mRNA processing on a natural promoter and in a completely defined system (Figure 5 and Supplementary Figure S10). We generated luciferase and splicing reporters possessing the TATA-less CRE-containing CDC37 promoter, or having promoter fragments derived from previously described constructs containing multimerized GAL4 UAS binding sites upstream of either an INR element or a consensus TATA box (Emami et al, 1995). As expected, PKA or CRTC expression does not direct transcriptional induction of the TATA-less CDC37 promoter (Figure 5A). However, the TATA-less CDC37 promoter is alternatively spliced in response to PKA or CRTC, and co-expression of PKA and CRTC further potentiates this affect, despite a lack of transcriptional activation (Figure 5B and C). Similarly, CRTC had only marginal affects on the INR-containing GAL4 UAS promoter, yet robustly induced transcription of the TATA-containing GAL4 UAS promoter when co-transfected with full-length CREB fused to the GAL4 DBD (Supplementary Figure S10A). In contrast to CRTCs core promoter-dependent affects on transcription, analysis of the ED I minigene reporters showed similar exon skipping ratios (Supplementary Figures S10B and C). Thus, core promoter context does not affect CRTC regulation of pre-mRNA splicing, suggesting that CRTC-mediated transcriptional activation and splicing are separable functions.
Figure 5.
CRTCs elicit alternative ED I splicing independent of core promoter context. (A) Effect of PKA and CRTCs on transcriptional activity of a TATA-less CRE-containing promoter. Transient transfection assays of CDC37 luciferase reporter co-transfected into HEK293T cells in the presence or absence of PKA or CRTC were analysed for luciferase activity (n=6 wells; mean±s.e.m.; asterisk denotes P-value ⩽0.05). (B) TaqMan real-time qPCR analysis of PKA and CRTCs effects on CDC37 ED I minigene exon skipping. The relative exon skipping levels are expressed as the ratio of skipped versus included transcripts (n=3 experiments; mean±s.e.m.; asterisk denotes P-value ⩽0.05). (C) TaqMan real-time qPCR analysis of PKA and CRTCs affects on CDC37 ED I minigene reporter gene expression. Relative amounts of ED I reporter transcripts obtained by each transfection were plotted as folds relative to basal transcription (n=3 experiments; mean±s.e.m.; asterisk denotes P-value ⩽0.05).
The observed effects of CRTC on pre-mRNA processing are also specific for this co-activator, as overexpression of the CBP co-activator had no effect (Supplementary Figures S10B and C). Furthermore, the phospho (Ser 133) CREB mutant, defective for CBP recruitment (Gonzalez and Montminy, 1989), did not block CRTC-mediated splice-site selection, indicating a CBP-independent mechanism for splice-site selection. To minimize the possible recruitment of splicing factors by other regions of CREB, we tested a GAL4–DBD fusion containing only the CREB basic leucine zipper (bZIP) domain. This 70-residue peptide is sufficient to bind the CRTC co-activators (Conkright et al, 2003a; Screaton et al, 2004) and indeed CRTC directed robust transcriptional activation of the TATA-containing construct (Supplementary Figure S10D), and this was blocked by a single amino-acid substitution previously shown to be critical for CREB–CRTC interaction (R314A; relative to full-length CREB)(Screaton et al, 2004). Importantly, alternative ED I exon splicing is also CRTC specific, as it is also blocked by the R314A mutation (Supplementary Figures S10E and F). Direct recruitment of GAL4–CRTC not only resulted in a robust transcription from the TATA-containing construct, but also yielded similar effects on alternative splicing for both promoter configurations (Supplementary Figures 10G–I). Collectively, these data show that the ability of the CRTC co-activators to regulate pre-mRNA processing is not a linear relationship with increases in transcriptional activity, suggesting these events may not be mechanistically linked.
CRTC recruitment to non-cAMP responsive promoters is sufficient to augment alternative splicing but not transcription
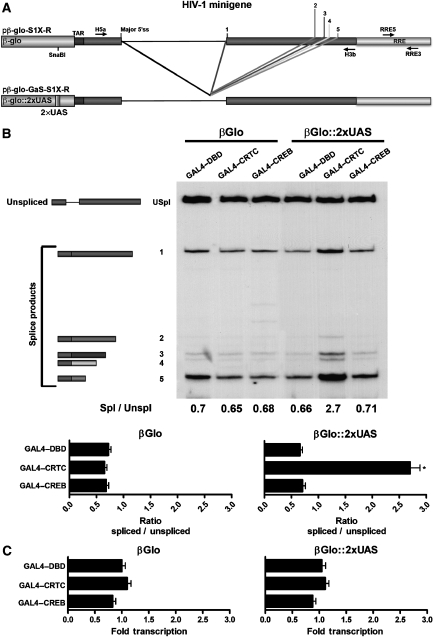
The finding that CRTCs can selectively regulate splicing independent of transcription activation led us to question whether these effects were specific to the configuration of the core promoter and/or to the splicing substrate. To address these alternatives, we tested whether tethering CRTC to a non-cAMP responsive promoter would affect the processing of pre-mRNA (Figure 6A). Neither CRTC nor CREB affect splicing of the HIV-1 minigene when regulated by the β-globin promoter (Figure 6B). However, inserting two tandem GAL4 UAS sites at position −275 in the β-globin promoter and ectopically expressing heterologous GAL4–CRTC was sufficient to provoke a fourfold increase in spliced mRNAs relative to expression of the control GAL4–DBD. In contrast, GAL4–CREB failed to affect splicing confirming that CRTCs role in pre-mRNA processing is independent of CREB. Finally, although CRTC promotes alternative splicing, analysis of transcript levels by qPCR indicates that tethering CRTC to the β-globin promoter does not augment transcription (Figure 6C). CRTC recruitment to promoters is sufficient to augment splicing but not transcriptional activation.
Figure 6.
CRTC recruitment to promoters is sufficient to augment splicing but not transcriptional activation. (A) Schematic representation of the S1X-R minigene reporter substrates that substitute the LTR for the wild-type β-globin promoter or a β-globin promoter-containing tandem GAL4–UAS binding sites. (B) HEK293T cells co-transfected with the indicated reporter construct together with GAL4–CRTC1 or GAL4–CREB expression plasmids. RT–PCR products were separated by PAGE and splice variants analysed to determine the effect of CRTC recruitment to the β-globin promoter on splicing. Ratio of spliced versus unspliced exons are graphically displayed (n=3 experiments; mean±s.e.m.; asterisk denotes P-value ⩽0.05). (C) qPCR analysis of reporter gene expression. Relative amounts of HIV-1 reporter transcripts obtained by each transfection were plotted as fold of total gene products expressed (unspliced+spliced) versus the amount expressed by the pβGlo-S1X-R or pβGlo-GaS-S1X-R constructs (n=3 experiments; mean±s.e.m.; asterisk denotes P-value ⩽0.05).
CRTC-dependent alternative splicing is mediated by a domain separable from transcriptional activation
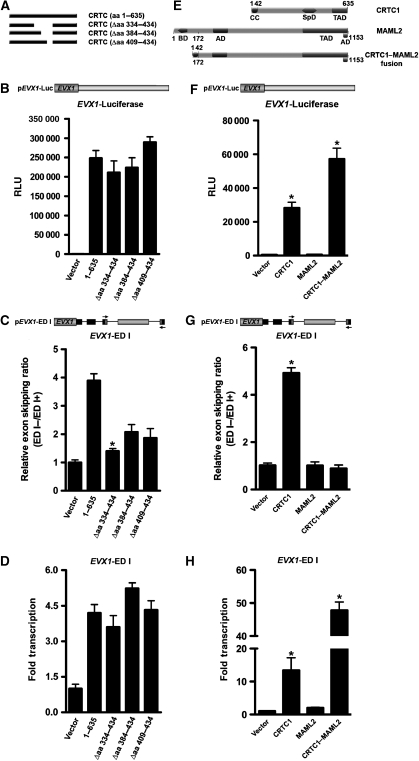
To identify the domain within CRTCs that facilitates splicing and to test whether splicing is separable from transcription, we generated a series of CRTC1 deletion mutants (Figure 7A). Previous experiments defined residues 435–635 as the transactivation domain, whereas a clustal alignment of all three CRTCs showed a conserved proline-rich region, spanning residues 334–434, that shares considerable identity to several proteins including known splicing factors. Analysis of internal deletion mutants encompassing this domain (residues 334–434) ablated alternative splicing activity despite comparable transcriptional output (Figure 7B–D). Analysis of smaller deletions within this domain maintained transcriptional activity and showed attenuated alternative splicing compared with wild-type CRTC1, although the level of attenuation was not as effective as the larger internal deletion (Figure 7B–D).
Figure 7.
Transcriptional activation is not required for CRTC-dependent alternative splicing. (A) Schematic illustration of full-length and internal CRTC deletion proteins with corresponding deleted amino acids indicated in parentheses. (B) Transient co-transfection of CRTC deletion mutants with EVX1 luciferase reporter into HEK293T cells were analysed for luciferase activity (n=6 wells; mean±s.e.m.; asterisk denotes P-value ⩽0.05). (C) TaqMan real-time qPCR analysis of CRTC deletion mutants on EVX1 ED I minigene exon skipping. The relative exon skipping levels are expressed as the ratio of skipped versus included transcripts (n=3 experiments; mean±s.e.m.; asterisk denotes P-value ⩽0.05). (D) TaqMan real-time qPCR analysis of CRTC deletion mutants on EVX1 ED I minigene reporter gene expression. Relative amounts of ED I reporter transcripts obtained by each transfection were plotted as folds relative to basal transcription (n=3 experiments; mean±s.e.m.; asterisk denotes P-value ⩽0.05). (E) Schematic illustration of key CRTC1 and MAML2 protein domains relative to the CRTC1–MAML2 chimeric oncoprotein. Corresponding amino acids derived from CRTC1 are indicated above, whereas those derived from MAML2 are indicated below. Abbreviations are as follows: AD, acidic domain; BD, basic domain; CC, coiled-coil; SpD, splicing domain; TAD, transactivation domain. (F) Transient co-transfection of CRTC1–MAML2 chimeric oncoprotein with EVX1 luciferase reporter into HEK293T cells were analysed for luciferase activity (n=6 wells; mean±s.e.m.; asterisk denotes P-value ⩽0.05). (G) TaqMan real-time qPCR analysis of CRTC1–MAML2 chimeric oncoprotein on EVX1 ED I minigene exon skipping. The relative exon skipping levels are expressed as the ratio of skipped versus included transcripts (n=3 experiments; mean±s.e.m.; asterisk denotes P-value ⩽0.05). (H) TaqMan real-time qPCR analysis of CRTC1–MAML2 chimeric oncoprotein on EVX1 ED I minigene reporter gene expression. Relative amounts of ED I reporter transcripts obtained by each transfection were plotted as folds relative to basal transcription (n=3 experiments; mean±s.e.m.; asterisk denotes P-value ⩽0.05).
Several connections have been made between aberrant pre-mRNA splicing and tumour development (Srebrow and Kornblihtt, 2006; Fackenthal and Godley, 2008). Salivary gland tumours possess a t(11;19) translocation that fuses the 42-amino-acid N-terminal portion of CRTC1 (MECT1) to a 981-amino-acid C-terminal portion of MAML2 to generate the chimeric oncoprotein (Figure 7E) (Tonon et al, 2003; Wu et al, 2005). Although CRTC1 functions as a potent co-activator for CREB, MAML2 (mastermind-like 2) is a key component of the NOTCH signalling pathway and functions as a co-activator for RBPJ (Wu et al, 2002). Consequently, studies suggest that the CRTC1–MAML2 translocation induces the expression of multiple CREB and NOTCH target genes, thereby contributing to transforming activity (Tonon et al, 2003; Wu et al, 2005). Transformation relies on the formation of the chimeric oncoprotein, as overexpression of either parent protein does not promote tumourigenesis. As the CRTC1–MAML2 oncoprotein lacks the identified CRTC splicing domain, we tested whether this fusion oncoprotein leads to aberrant splicing of CREB-dependent targets. Analysis of the CREB-dependent EVX1 luciferase reporter shows robust transcriptional activity with both CRTC1 and CRTC1–MAML2, but not with MAML2 alone (Figure 7F). Notably, TaqMan qPCR analysis of EVX1 ED I alternative splicing shows that the CRTC1–MAML2 oncoprotein is not capable of influencing splice-site selection of the EVX1 minigene reporter (Figure 7G and H). Collectively these data show that the region of CRTCs that recruits factors capable of mediating splice-site selection functions on cAMP-responsive genes is independent of the transcriptional activation domain present.
Discussion
The findings presented herein show that the CRTC co-activators are capable of coupling activated transcription of cAMP-responsive genes to alternative pre-mRNA splicing. Remarkably and unique to the CRTCs, these robust transcriptional co-activators can also direct alternative splicing independent of CRTC-induced transcriptional activation. These autonomous activities of the CRTC co-activators include cell type-specific modes of regulation and support the idea that co-regulators involved in pre-mRNA splicing are not restricted solely to mechanisms requiring transcriptional activation.
Cell type- and developmental stage-specific transcript variations have been documented in several genome-wide mRNA splicing studies (Johnson et al, 2003; Ule et al, 2005; Kwan et al, 2008). These exonomic approaches have posed many questions regarding regulation of gene expression at the exon level and concerted efforts are underway to define the diverse mechanisms that regulate transcript identity (Blencowe, 2006; Moore and Silver, 2008). For example, some transcripts seem to be governed by the promoter-dependent coordination of transcription and alternative splicing (Auboeuf et al, 2007). Our data and a previous study clearly show that promoters containing CRE elements are capable of mediating alternative splicing and here we have shown that the CRTC co-activators regulate this process (Cramer et al, 1997). The idea that the CRTC co-activators perform more than one function is in accord with those of other co-regulators that possess multiple activities (Rosenfeld et al, 2006). For example, the CREB co-activator CBP mediates gene regulation through its intrinsic and associated histone acetyl transferase activity and by promoting the formation of the pre-initiation complex (PIC) through interactions with components of the core transcriptional machinery (Bannister and Kouzarides, 1996; Nakajima et al, 1997). Moreover, the transcriptional co-activators PGC (Monsalve et al, 2000), COAA (Auboeuf et al, 2004) and CAPER (Dowhan et al, 2005) function as transcriptional activators, as well as regulators of transcriptionally coupled pre-mRNA processing. However, in contrast to other co-activators involved in splicing, the CRTCs lack any conserved, definable RNA-binding domain (e.g., an RNA recognition motif (RRM), an arginine–serine (RS) domain, or a K homology (KH) domain). Rather, we hypothesize that CRTCs provide a scaffold for the assembly of a larger complex of proteins that may directly bind the transcript. This idea is supported by our CRTC IP MS/MS experiments that identified several proteins involved in pre-mRNA processing (Amelio et al, 2007). If the CRTCs form the basis for a protein scaffold that mediates RNA processing by recruiting other proteins, then the ability of CRTCs to recruit slightly different complexes (i.e. cell type or promoter specific) may contribute to these independent bipartite functions. Moreover, PKA may contribute to TORC-dependent alternative splicing through phosphorylation of components within the recruited splicing complex (Xie et al, 2003).
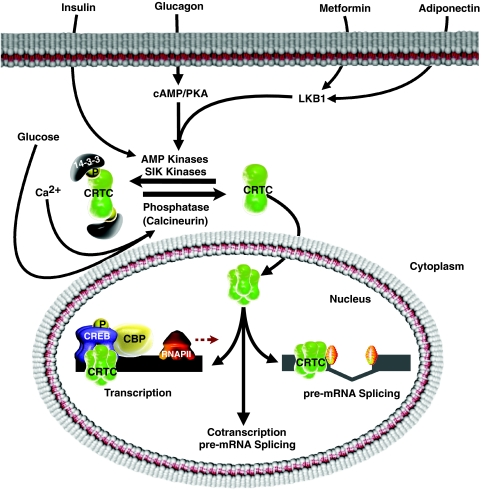
There is now overwhelming evidence that the subcellular localization, and thus function, of the CRTC2 co-activator is mediated by several cues, including glucagon and insulin (Figure 8). CRTC2 increases fasting blood glucose levels and the induction of key genes directly involved in gluconeogenesis (Canettieri et al, 2005; Koo et al, 2005; Dentin et al, 2007). Collectively, these studies showed that CRTC2 is the rate-limiting step of cAMP signalling and that it alone is sufficient to initiate the early stages of the gluconeogenic program. Interestingly, several of the genes within the gluconeogenic program such as Nurr1 (Nr4A2), as well as those encoding proteins that mediate later stages of glucose production such as PGC-1 (Ppargc1a), are alternatively spliced and we show here that CRTC2 can promote alternative splicing of Nr4a2. Thus, CRTCs function as integrators of extracellular signals to influence transcript diversity by acting either as a conduit between components of the transcription and splicing machineries or as autonomous regulators of each process (Figure 8). However, although TORCs can selectively activate splicing in some promoter contexts without activating transcription, these promoters possess basal transcriptional activity, therefore, the effects of the TORCs on pre-mRNA splicing could still formally be considered co-transcriptional despite the absence of co-activator-induced transcription.
Figure 8.
Model of CRTC cytoplasmic and nuclear functions. Schematic representation of environmental signalling cues that converge onto the CRTC co-activators to regulate pre- and post-transcriptional processes.
There are two prevailing models of promoter-dependent coordination of transcription and pre-mRNA processing that could apply to CRTC-mediated alternative splicing. These include splicing factor recruitment to the carboxyl terminal domain (CTD) of RNA polymerase II (RNAP II) (recruitment model) and regulation of RNAP II elongation rates (kinetic model) (Bentley, 2005; Kornblihtt, 2005). These models embody a ‘forward coupling' mechanism that links RNAP II transcription to pre-mRNA splicing. However, ‘reverse coupling' mechanisms have been described, whereby pre-mRNA splicing exerts an influence back on transcription (Brinster et al, 1988; Furger et al, 2002). Indeed, promoter-proximal 5′ splice sites seem to promote reverse coupling by increasing transcription initiation through recruitment of the PIC (Damgaard et al, 2008). CRTCs interact with TAFII130, a component of the PIC (Conkright et al, 2003a). Therefore, the finding that CRTCs can induce splicing independent of their effects on transcription may reflect the failure of CRTCs to recruit components of the PIC to TATA-less promoters. Accordingly, although parts of both models help to explain mechanistically how the CRCT co-activators mediate alterative splicing, neither model can fully account for our observations, suggesting that further refinement to the current working models may be necessary.
Apart from CRTCs mechanistic role in pre-mRNA processing, CRTC-mediated splicing probably has physiological significance. This is supported by the vast number of genes that are alternatively spliced, the large cast of promoters that contain CRE elements (roughly 10%), and by the fact that the CRTC recruitment to promoters (e.g. through interactions with CREB) is sufficient to augment alternative splicing. The inability of the CRTC1–MAML2 oncoprotein to mediate CREB-dependent splice-site selection, despite retaining robust transcriptional activation properties, suggests that this fusion product may also promote cellular transformation through aberrant pre-mRNA splicing of CRE-containing genes. However, transcriptional activators have been shown to promote pre-mRNA splicing, therefore, it is possible that the MAML2 transactivation domain in the CRTC1–MAML2 fusion can mediate splicing in other promoter contexts. Furthermore, although our data clearly show that the CRTC co-activators have the potential to regulate alternative splicing, this does not mandate that transcripts from CRE-containing genes will be alternatively spliced in a CRTC-dependent manner. Indeed, several studies have shown that the significance of the CREB co-activators, CBP/p300, is dictated by gene context and varies among tissues (Cha-Molstad et al, 2004; Zhang et al, 2005; Ravnskjaer et al, 2007; Xu et al, 2007). Similar, cell-type specificity is also observed in CRTC-mediated alternative splicing of the endogenous Nr4a2 transcript. We speculate that the generation of select transcript isoforms may differ between cell types, in part, due to the many diverse signalling events that converge on the CRTC co-activators (Figure 8).
Traditional methods, such as expression arrays or TaqMan PCR focus on gene expression to examine gene regulation in response to signalling cascades. In these technologies, the probes and primer sets intentionally do not account for alternatively spliced products. As a result the canonical analysis of cAMP responsive genes has been restricted to the analysis of transcription induction effectively missing any affects of cAMP activation on pre-mRNA processing. Our findings establish that CRTCs direct alternative splicing independently or in coordination with transcriptional activation, thereby, linking the cAMP signalling cascade with the regulation of pre-mRNA processing. Future experiments will monitor alternative splicing in addition to transcriptional activation when identifying tissue-specific CRTC target promoters.
Materials and methods
Cell culture
Primary rat hepatocytes (Cellz Direct) were maintained according to manufacturer's instructions. Rat pheochromocytoma (PC12) cells were maintained at 37°C in DMEM containing 2 mM GlutaMax (Invitrogen) supplemented with 10% heat-inactivated horse serum (Gibco), 5% FCS (Gemini Bio-Products) and antibiotics (100 units/ml penicillin and 100 mg/ml streptomycin). For NGF-induced differentiation, 5 × 105 cells were plated on 10 cm collagen-coated plates (BD Biosciences) in DMEM supplemented with 100 ng/ml NGF-2.5S (Sigma), 0.05% FCS and antibiotics. Medium was replaced every 2 days. After 4–5 days of exposure to NGF, all cells showed a differentiated morphology and experiments were carried out 6–7 days after the initial plating. HEK293T cells were cultured in DMEM (Invitrogen) supplemented with 10% FCS and antibiotics.
Adenoviruses
The control GFP-expressing or CRTC2-expressing adenoviruses and the non-specific (NS) or CRTC2 RNAi adenoviruses have been described previously by Koo et al (2005). Cells were infected with either GFP-expressing or CRTC2-expressing adenoviruses for 16–18 h. RNAi studies were carried out by co-transducing cells with a 4:1 ratio of adenoviruses expressing either NS or CRTC2 shRNAs relative to CRTC2-expressing adenovirus.
Plasmid construction
ED I minigene. The EVX1 and CDX4 ED I minigene splicing reporters were obtained by PCR amplification of previously described plasmid templates (Conkright et al, 2003b) using primers tagged with ScaI and BssHII restriction enzyme sequences on the forward and reverse primer, respectively, (primer sequences available on request). The pSVEDA_Tot minigene plasmid was digested with ScaI and BssHII, the 7150-bp fragment, used to directionally clone the promoter fragments, and clones were confirmed by sequencing. To obtain the 5 × UAS minigene splicing reporters, a short multiple cloning sequence (MCS) was generated by annealing two oligomers with 5′ ScaI and 3′ BssHII restriction enzyme sequences. The MCS sequence was kinase treated and cloned into the ScaI- and BssHII-digested pSVEDA_Tot minigene. The 5xUAS∷INR and 5xUAS∷TATA promoter sequences were obtained by PCR amplification of previously described plasmid templates (Emami et al, 1995), using primers tagged with AscI and NotI restriction enzyme sequences on the forward and reverse primer, respectively, and subsequently cloned into the new pSVEDA_Tot MCS.
HIV-1 minigene. Splicing reporter plasmid pLTR-S1X-R was obtained by inserting the HIV-1 splicing cassette derived from the plasmid pHS1-X (Amendt et al, 1994) into pcDNA3 (Invitrogen). A PCR-amplified fragment, comprehensive of the viral LTR promoter, was inserted to (Amendt et al, 1994) complete the viral sequence upstream of the major 5′ splice site. Plasmids pEVX1-S1X-R and pEVX-DM-S1X-R were obtained by substituting the LTR promoter in pLTR-S1X-R with a 400-nt fragment containing the wild-type and mutated EVX1 promoter derived from the plasmids pEVX1 and pEVX1DCRE, respectively (Conkright et al, 2003b). The pβglo-S1X-R plasmid was obtained by substituting the LTR promoter in pLTR-S1X for a 700-bp genomic fragment containing the β-globin gene promoter region. The pβglo-GaS-S1X-R plasmid containing the gal4 2 × UAS was generated by annealing, Klenow treating and inserting the following primers into the SnaBI restriction site (−275 nt upstream of +1 nt):
Ga5Ba, GATCC cggagtactgtcctccg cggagtactgtcctccg TACGTA G;
Ga3Ba, GATCC TACGTA cggaggacagtactccgcggaggacagtactccg G.
Cell transfections and reporter gene analyses
Luciferase assays were carried out as previously described (Amelio et al, 2007). Reverse transfection was carried out in 96-well plates using 50 ng of luciferase reporter per well. Serum-free DMEM (60 ml) containing 25 ng test cDNA constructs and Lipofectamine 2000 (Invitrogen) was allocated into each well. After a 30-min incubation at room temperature, DMEM supplemented with 20% FBS (60 ml) containing 3 × 104 HEK293T cells was dispensed into each well. Cells were cultured for 24 h in a humidified incubator at 37°C in 5% CO2. BrightGlo (Promega) reagent (120 ml) was added to each well and luciferase luminescence was measured with an Acquest plate reader (LJL Biosystems).
Splicing assays were carried out as previously described (Cramer et al, 1997) with minor modifications. HEK293T cells were maintained at below 80% confluence in DMEM (Gibco BRL) supplemented with 5% FCS and gentamicin (0.5 mg/ml). Cells were seeded 2 h before transfection at 70% confluence in fresh media containing 5% FCS. Transfections were carried out with 2 mg of the minigene plasmid and 500 ng of each additional expression plasmid (PKA, A-CREB and/or CRTCs 1–3) using Lipofectamine 2000as the transfection reagent. All transfections were DNA balanced using pSport6 plasmid DNA (Invitrogen) as the control and an equal amount of plasmid pEGFP-N1 (Clontech), expressing EGFP, was added to each transfection mixture as a transfection normalizer. Cells were collected 24 h after transfection and total RNA was isolated. Equal amounts of each total mRNA sample (2 mg) were used in subsequent RT–PCR reactions and reverse transcribed using either oligodT or random pd(N)6 primers. The alternatively spliced ED I isoforms were identified by PCR using the previously described pSV5′j and pSV3′j primers (Cramer et al, 1997), whereas the alternatively spliced HIV-1 minigene mRNAs were detected using the primers H5a (AAGTAGTGTGTGCCCGTCTGTT) and H3b (TCCTGCGTCGATCGATTGGTTT). Primers, H5a and H3b, are located upstream of the major HIV-1 5′ splice site and thus detect all splice products. Quantitative TaqMan real-time PCR was carried out to measure ED I alternative splicing and total transcription. The TaqMan primer/probe sequences are listed in Supplementary figure 1D. Quantitative SYBR green PCR analysis was carried out to measure transcription by amplifying unspliced HIV-1 minigene sequences expressed by the reporter construct with the primers RRE5 and RRE3. Each sample was normalized by quantifying the pEGFP-N1 gene product with the primers E5a (ACCACATGAAGCAGCACGACTTCT) and E5b (TCACCTTGATGCCGTTCTTCTGCT). End-labelled ATPγ32P PCR products were fractionated on polyacrylamide gels, exposed to film respectively, visualized and the band intensities were quantified utilizing either Bio-Rad Quantity One v4.6.2 or Kodak 1D software.
Endogenous gene expression analysis
RNA was extracted from HEK293T cells using TRIzol (Invitrogen) and reverse transcription was conducted with SuperScript III reverse transcriptase (Invitrogen). PCR amplification reactions were run in triplicate in 96-well optical microplates in an ABI Prism 7900 HT SDS instrument (Applied Biosystems). Differences in mRNA expression levels were measured by relative quantification using the comparative ΔΔCt method (Applied Biosystems) and were normalized against GAPDH levels. Probe sequences have been previously described by Amelio et al (2007).
Supplementary Material
Supplementary Material
Review Process File
Acknowledgments
We thank Marc Montminy, Gianluca Canettiere, Robert Screaton, Richard Goodman, Michael Poulos and John Cleveland for their scientific review and/or helpful comments during the preparation of this paper; Alberto Kornblihtt for providing the pSVEDA_Tot ED I minigene construct; Stephen Smale for providing the GAL4 TATA and GAL4 Inr promoters; Marc Montminy for providing the GFP, CRTC2, non-specific and CRTC2 shRNA adenoviruses and Michael Bannon for providing the Nr4a2 isoform cDNAs and 3xNBRE promoter. This study is supported by a Ruth L Kirschstein National Research Service Award from the National Cancer Institute (CA134121) (ALA) and by the National Institutes of Health grant RO1 DK081491 (MDC). Scripps paper number 19697.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ahn S, Olive M, Aggarwal S, Krylov D, Ginty DD, Vinson C (1998) A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol Cell Biol 18: 967–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amelio AL, Miraglia LJ, Conkright JJ, Mercer BA, Batalov S, Cavett V, Orth AP, Busby J, Hogenesch JB, Conkright MD (2007) A coactivator trap identifies NONO (p54nrb) as a component of the cAMP-signaling pathway. Proc Natl Acad Sci USA 104: 20314–20319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amendt BA, Hesslein D, Chang LJ, Stoltzfus CM (1994) Presence of negative and positive cis-acting RNA splicing elements within and flanking the first tat coding exon of human immunodeficiency virus type 1. Mol Cell Biol 14: 3960–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auboeuf D, Batsché E, Dutertre M, Muchardt C, O'Malley BW (2007) Coregulators: transducing signal from transcription to alternative splicing. Trends Endocrinol Metab 18: 122–129 [DOI] [PubMed] [Google Scholar]
- Auboeuf D, Dowhan DH, Dutertre M, Martin N, Berget SM, O'Malley BW (2005) A subset of nuclear receptor coregulators act as coupling proteins during synthesis and maturation of RNA transcripts. Mol Cell Biol 25: 5307–5316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auboeuf D, Dowhan DH, Li X, Larkin K, Ko L, Berget SM, O'Malley BW (2004) CoAA, a nuclear receptor coactivator protein at the interface of transcriptional coactivation and RNA splicing. Mol Cell Biol 24: 442–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auboeuf D, Honig A, Berget SM, O'Malley BW (2002) Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science 298: 416–419 [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T (1996) The CBP co-activator is a histone acetyltransferase. Nature 384: 641–643 [DOI] [PubMed] [Google Scholar]
- Basu A, Dong B, Krainer AR, Howe CC (1997) The intracisternal A-particle proximal enhancer-binding protein activates transcription and is identical to the RNA- and DNA-binding protein p54nrb/NonO. Mol Cell Biol 17: 677–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL (2005) Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol 17: 251–256 [DOI] [PubMed] [Google Scholar]
- Bittinger MA, McWhinnie E, Meltzer J, Iourgenko V, Latario B, Liu X, Chen CH, Song C, Garza D, Labow M (2004) Activation of cAMP response element-mediated gene expression by regulated nuclear transport of TORC proteins. Curr Biol 14: 2156–2161 [DOI] [PubMed] [Google Scholar]
- Black DL (2003) Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72: 291–336 [DOI] [PubMed] [Google Scholar]
- Blencowe BJ (2006) Alternative splicing: new insights from global analyses. Cell 126: 37–47 [DOI] [PubMed] [Google Scholar]
- Brinster RL, Allen JM, Behringer RR, Gelinas RE, Palmiter RD (1988) Introns increase transcriptional efficiency in transgenic mice. Proc Natl Acad Sci USA 85: 836–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canettieri G, Koo SH, Berdeaux R, Heredia J, Hedrick S, Zhang X, Montminy M (2005) Dual role of the coactivator TORC2 in modulating hepatic glucose output and insulin signaling. Cell Metab 2: 331–338 [DOI] [PubMed] [Google Scholar]
- Caputi M, Casari G, Guenzi S, Tagliabue R, Sidoli A, Melo CA, Baralle FE (1994) A novel bipartite splicing enhancer modulates the differential processing of the human fibronectin EDA exon. Nucleic Acids Res 22: 1018–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi M, Mayeda A, Krainer AR, Zahler AM (1999) hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J 18: 4060–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha-Molstad H, Keller DM, Yochum GS, Impey S, Goodman RH (2004) Cell-type-specific binding of the transcription factor CREB to the cAMP-response element. Proc Natl Acad Sci USA 101: 13572–13577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, Montminy M (2003a) TORCs: transducers of regulated CREB activity. Mol Cell 12: 413–423 [DOI] [PubMed] [Google Scholar]
- Conkright MD, Guzman E, Flechner L, Su AI, Hogenesch JB, Montminy M (2003b) Genome-wide analysis of CREB target genes reveals a core promoter requirement for cAMP responsiveness. Mol Cell 11: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Cramer P, Pesce CG, Baralle FE, Kornblihtt AR (1997) Functional association between promoter structure and transcript alternative splicing. Proc Natl Acad Sci USA 94: 11456–11460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard CK, Kahns S, Lykke-Andersen S, Nielsen AL, Jensen TH, Kjems J (2008) A 5′ splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol Cell 29: 271–278 [DOI] [PubMed] [Google Scholar]
- Dean DC, Blakeley MS, Newby RF, Ghazal P, Hennighausen L, Bourgeois S (1989) Forskolin inducibility and tissue-specific expression of the fibronectin promoter. Mol Cell Biol 9: 1498–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean DC, Newby RF, Bourgeois S (1988) Regulation of fibronectin biosynthesis by dexamethasone, transforming growth factor beta, and cAMP in human cell lines. J Cell Biol 106: 2159–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentin R, Liu Y, Koo SH, Hedrick S, Vargas T, Heredia J, Yates J, Montminy M (2007) Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature 449: 366–369 [DOI] [PubMed] [Google Scholar]
- Dowhan DH, Hong EP, Auboeuf D, Dennis AP, Wilson MM, Berget SM, O'Malley BW (2005) Steroid hormone receptor coactivation and alternative RNA splicing by U2AF65-related proteins CAPERalpha and CAPERbeta. Mol Cell 17: 429–439 [DOI] [PubMed] [Google Scholar]
- Emami KH, Navarre WW, Smale ST (1995) Core promoter specificities of the Sp1 and VP16 transcriptional activation domains. Mol Cell Biol 15: 5906–5916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fackenthal JD, Godley LA (2008) Aberrant RNA splicing and its functional consequences in cancer cells. Dis Model Mech 1: 37–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Bond CS, Lamond AI (2005) P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol Biol Cell 16: 5304–5315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Luo L, Luo N, Zhu X, Garvey WT (2007) NR4A orphan nuclear receptors modulate insulin action and the glucose transport system: potential role in insulin resistance. J Biol Chem 282: 31525–31533 [DOI] [PubMed] [Google Scholar]
- Furger A, O'Sullivan JM, Binnie A, Lee BA, Proudfoot NJ (2002) Promoter proximal splice sites enhance transcription. Genes Dev 16: 2792–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez GA, Montminy MR (1989) Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell 59: 675–680 [DOI] [PubMed] [Google Scholar]
- Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH (2004) Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell 119: 1041–1054 [DOI] [PubMed] [Google Scholar]
- Jansson D, Ng AC, Fu A, Depatie C, Al Azzabi M, Screaton RA (2008) Glucose controls CREB activity in islet cells via regulated phosphorylation of TORC2. Proc Natl Acad Sci USA 105: 10161–10166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD (2003) Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302: 2141–2144 [DOI] [PubMed] [Google Scholar]
- Konig H, Ponta H, Herrlich P (1998) Coupling of signal transduction to alternative pre-mRNA splicing by a composite splice regulator. EMBO J 17: 2904–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo SH, Flechner L, Qi L, Zhang X, Screaton RA, Jeffries S, Hedrick S, Xu W, Boussouar F, Brindle P, Takemori H, Montminy M (2005) The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature 437: 1109–1111 [DOI] [PubMed] [Google Scholar]
- Kornblihtt AR (2005) Promoter usage and alternative splicing. Curr Opin Cell Biol 17: 262–268 [DOI] [PubMed] [Google Scholar]
- Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G (2004) Multiple links between transcription and splicing. RNA 10: 1489–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs KA, Steullet P, Steinmann M, Do KQ, Magistretti PJ, Halfon O, Cardinaux JR (2007) TORC1 is a calcium- and cAMP-sensitive coincidence detector involved in hippocampal long-term synaptic plasticity. Proc Natl Acad Sci USA 104: 4700–4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan T, Benovoy D, Dias C, Gurd S, Provencher C, Beaulieu P, Hudson T, Sladek R, Majewski J (2008) Genome-wide analysis of transcript isoform variation in humans. Nat Genet 40: 225–231 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K et al. (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Lynch KW, Weiss A (2000) A model system for activation-induced alternative splicing of CD45 pre-mRNA in T cells implicates protein kinase C and Ras. Mol Cell Biol 20: 70–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter N, Herrlich P, Konig H (2002) Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature 420: 691–695 [DOI] [PubMed] [Google Scholar]
- Michelhaugh SK, Vaitkevicius H, Wang J, Bouhamdan M, Krieg AR, Walker JL, Mendiratta V, Bannon MJ (2005) Dopamine neurons express multiple isoforms of the nuclear receptor nurr1 with diminished transcriptional activity. J Neurochem 95: 1342–1350 [DOI] [PubMed] [Google Scholar]
- Monsalve M, Wu Z, Adelmant G, Puigserver P, Fan M, Spiegelman BM (2000) Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol Cell 6: 307–316 [DOI] [PubMed] [Google Scholar]
- Montminy MR, Sevarino KA, Wagner JA, Mandel G, Goodman RH (1986) Identification of a cyclic-AMP-responsive element within the rat somatostatin gene. Proc Natl Acad Sci USA 83: 6682–6686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Silver PA (2008) Global analysis of mRNA splicing. RNA 14: 197–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima T, Uchida C, Anderson SF, Parvin JD, Montminy MR (1997) Analysis of a cAMP-responsive activator reveals a two-component mechanism for transcriptional induction via signal-dependent factors. Genes Dev 11: 738–747 [DOI] [PubMed] [Google Scholar]
- Ohkura N, Hosono T, Maruyama K, Tsukada T, Yamaguchi K (1999) An isoform of Nurr1 functions as a negative inhibitor of the NGFI-B family signaling. Biochim Biophys Acta 1444: 69–79 [DOI] [PubMed] [Google Scholar]
- Pei L, Waki H, Vaitheesvaran B, Wilpitz DC (2006) NR4A orphan nuclear receptors are transcriptional regulators of hepatic glucose metabolism. Nat Med 12: 1048–1055 [DOI] [PubMed] [Google Scholar]
- Ravnskjaer K, Kester H, Liu Y, Zhang X, Lee D, Yates JR, Montminy M (2007) Cooperative interactions between CBP and TORC2 confer selectivity to CREB target gene expression. EMBO J 26: 2880–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld MG, Lunyak VV, Glass CK (2006) Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev 20: 1405–1428 [DOI] [PubMed] [Google Scholar]
- Screaton RA, Conkright MD, Katoh Y, Best JL, Canettieri G, Jeffries S, Guzman E, Niessen S, Yates JR, Takemori H, Okamoto M, Montminy M (2004) The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell 119: 61–74 [DOI] [PubMed] [Google Scholar]
- Sharp PA (2005) The discovery of split genes and RNA splicing. Trends Biochem Sci 30: 279–281 [DOI] [PubMed] [Google Scholar]
- Smits SM, Ponnio T, Conneely OM, Burbach JP, Smidt MP (2003) Involvement of Nurr1 in specifying the neurotransmitter identity of ventral midbrain dopaminergic neurons. Eur J Neurosci 18: 1731–1738 [DOI] [PubMed] [Google Scholar]
- Srebrow A, Kornblihtt AR (2006) The connection between splicing and cancer. J Cell Sci 119: 2635–2641 [DOI] [PubMed] [Google Scholar]
- Tonon G, Modi S, Wu L, Kubo A, Coxon AB, Komiya T, O'Neil K, Stover K, El-Naggar A, Griffin JD, Kirsch IR, Kaye FJ (2003) t(11;19)(q21;p13) translocation in mucoepidermoid carcinoma creates a novel fusion product that disrupts a Notch signaling pathway. Nat Genet 33: 208–213 [DOI] [PubMed] [Google Scholar]
- Ule J, Jensen K, Mele A, Darnell RB (2005) CLIP: a method for identifying protein–RNA interaction sites in living cells. Methods 37: 376–386 [DOI] [PubMed] [Google Scholar]
- van der Houven van Oordt W, Diaz-Meco MT, Lozano J, Krainer AR, Moscat J, Caceres JF (2000) The MKK(3/6)-p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. J Cell Biol 149: 307–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Liu J, Gao P, Nakamura M, Cao Y, Shen H, Griffin JD (2005) Transforming activity of MECT1–MAML2 fusion oncoprotein is mediated by constitutive CREB activation. EMBO J 24: 2391–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Sun T, Kobayashi K, Gao P, Griffin JD (2002) Identification of a family of mastermind-like transcriptional coactivators for mammalian notch receptors. Mol Cell Biol 22: 7688–7700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Huang X, Feng Y, Handschin C, Feng Y, Gullicksen PS, Bare O, Labow M, Spiegelman B, Stevenson SC (2006) Transducer of regulated CREB-binding proteins (TORCs) induce PGC-1alpha transcription and mitochondrial biogenesis in muscle cells. Proc Natl Acad Sci USA 103: 14379–14384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Black DL (2001) A IV CaMK responsive RNA element mediates depolarization-induced alternative splicing of ion channels. Nature 410: 936–939 [DOI] [PubMed] [Google Scholar]
- Xie J, Lee JA, Kress TL, Mowry KL, Black DL (2003) Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proc Natl Acad Sci USA 100: 8776–8781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu PY, Le WD (2004) Novel splicing variant of the human orphan nuclear receptor Nurr1 gene. Chin Med J (Engl) 117: 899–902 [PubMed] [Google Scholar]
- Xu W, Kasper LH, Lerach S, Jeevan T, Brindle PK (2007) Individual CREB-target genes dictate usage of distinct cAMP-responsive coactivation mechanisms. EMBO J 26: 2890–28903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Damgaard CK, Kjems J, Caputi M (2004) SC35 and heterogeneous nuclear ribonucleoprotein A/B proteins bind to a juxtaposed exonic splicing enhancer/exonic splicing silencer element to regulate HIV-1 tat exon 2 splicing. J Biol Chem 279: 10077–10084 [DOI] [PubMed] [Google Scholar]
- Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, Chen H, Jenner R, Herbolsheimer E, Jacobsen E, Kadam S, Ecker JR, Emerson B, Hogenesch JB, Unterman T, Young RA, Montminy M (2005) Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci USA 102: 4459–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Review Process File