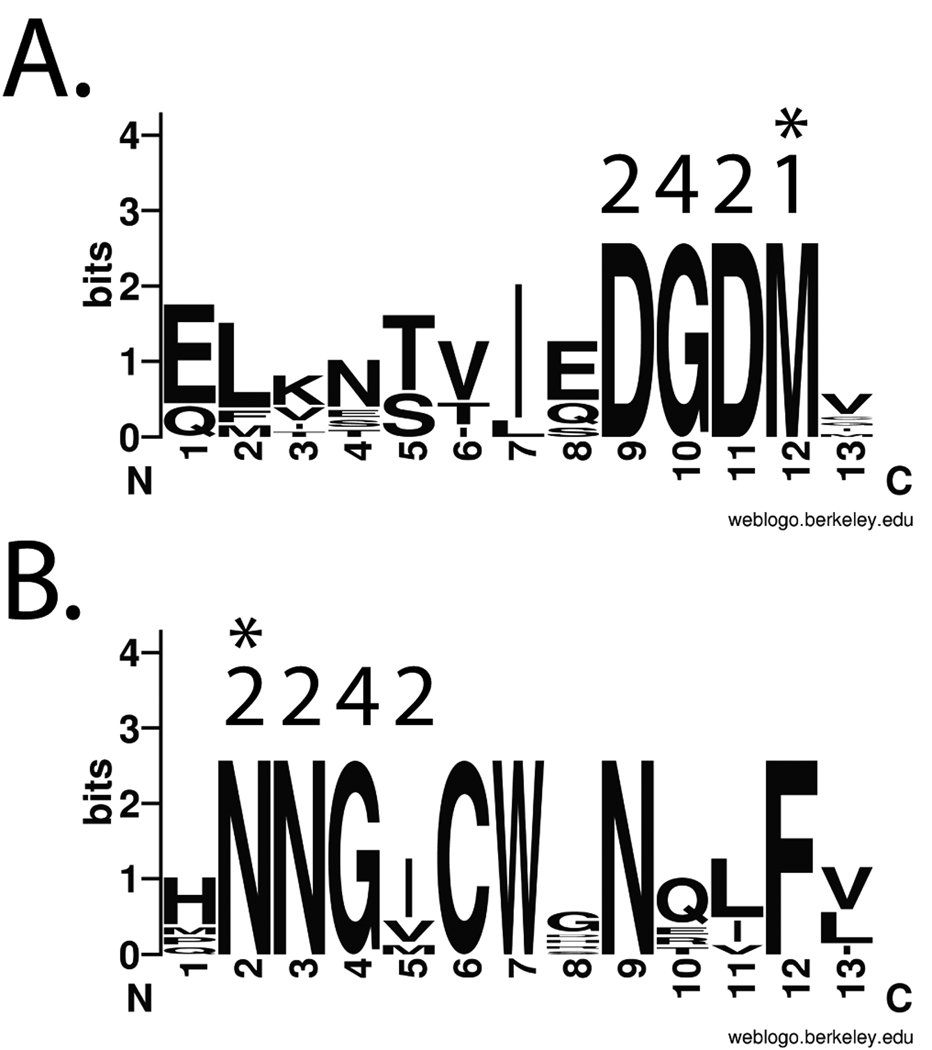Abstract
Consensus-degenerate hybrid oligonucleotide primers (CODEHOPs) have proven to be a powerful tool for the identification of novel genes. CODEHOPs are designed from highly conserved regions of multiply-aligned protein sequences from members of a gene family and are used in PCR amplification to identify distantly-related genes. The CODEHOP approach has been used to identify novel pathogens by targeting amino acid motifs conserved in specific pathogen families. We initiated a program utilizing the CODEHOP approach to develop PCR-based assays targeting a variety of viral families that are pathogens in non-human primates. We have also developed and further improved a computer program and website to facilitate the design of CODEHOP PCR primers. Here, we detail the method for the development of pathogen-specific CODEHOP PCR assays using the papillomavirus family as a target. Papillomaviruses constitute a diverse virus family infecting a wide variety of mammalian species, including humans and non-human primates. We demonstrate that our pan-papillomavirus CODEHOP assay is broadly reactive with all major branches of the virus family and show its utility in identifying a novel non-human primate papillomavirus in cynomolgus macaques.
Keywords: polymerase chain reaction, consensus, degenerate, oligonucleotide, primers, pathogens, virus, non-human primate, papillomavirus
1. Introduction
Traditional methods for detection of viral infections include standard viral cultures, direct fluorescent antibody tests, immunohistochemistry staining and enzyme immunoassays. More recently there has been a shift to molecular detection of viral nucleic acids, including PCR and microarray-based screening. Typically, such molecular assays utilize specific nucleic acid primers derived from known viral sequences to detect identical or closely related viral variants. Especially when based on an amplification step, these assays are highly sensitive and specific for the targeted viral pathogen. To deal with the complex nature of infections, multiplex testing platforms have been developed to screen for a set of different viral variants or species simultaneously. The development of PCR-based assays to identify distantly-related or unknown viral species is more problematic and relies upon mixtures of nucleic acid primers and the ability of primers to hybridize to non-complementary sequences with a required degree of specificity. Pools of related primers carrying known or predicted nucleotide sequence differences throughout the length of the primer have been used with moderate success to amplify unknown or distantly related genes. These are referred to as degenerate primers and can contain hundreds or thousands of individual primers in the pool to cover all possible nucleotide variations in a particular sequence. Alternatively, consensus PCR primers have been utilized to amplify unknown or related virus variants. A consensus primer carries the most common actual or predicted nucleotide variant in each position of a primer sequence and relies on its ability to specifically hybridize to a target sequence with mismatched or unpaired bases. When basing primer design on protein coding sequences, standard degenerate primers will contain most or all of the possible nucleotide sequences encoding a large conserved amino acid motif, while consensus primers will contain the most common nucleotide at each codon position in the targeted motif. While useful with adequate concentrations of closely-related template targets in non-complex mixtures, both standard degenerate- and consensus-primer approaches suffer from a lack of specificity and sensitivity when these conditions are not met.
1.1 CODEHOP PCR primers
We have developed a PCR approach for detecting and identifying unknown and distantly-related viruses using consensus-degenerate hybrid oligonucleotide primers (CODEHOPs)[1]. CODEHOPs are designed from short highly-conserved regions of multiply-aligned protein sequences from members of a gene family and are used in PCR amplification to identify unknown members of the family. Each CODEHOP consists of a short 3’ degenerate core region corresponding to all possible codons specifying 3–4 highly conserved amino acids and a longer 5’ consensus clamp region containing a single “best guess” nucleotide sequence derived from the consensus sequences flanking the target motif. Thus, a CODEHOP PCR primer consists of a pool of primers that are heterogenous at the 3’ end and homogenous at the 5’ end. The CODEHOP primer design strategy overcomes problems of both degenerate and consensus PCR primer methods [1]. The limited degeneracy in the short 3’ core region minimizes the total number of individual primers in the degenerate pool, yet provides a broad specificity during the initial PCR amplification cycles. Hybridization of the 3’ degenerate core is stabilized by the 5’ consensus clamp which allows higher annealing temperatures without increasing the degeneracy of the primer pool. Although mismatches between the 5’ consensus clamp and the target sequence may occur during the initial PCR cycles, they are situated away from the 3’ hydroxyl extension site of the polymerase, thus minimizing their disruptive effects on polymerase priming and extension. Further amplification of primed PCR products during subsequent rounds of primer hybridization and extension is enhanced by the sequence similarity of all primers in the pool. This allows utilization of all primers in the PCR reaction cycles [1]. The CODEHOP PCR approach provides the necessary specificity and sensitivity to allow for the amplification of disparate viral species, at low titer, in complex mixtures of genetic material [2].
1.2 Description of optimal CODEHOP PCR targets
The first step in developing a CODEHOP PCR assay to detect unknown members of a particular pathogen family is to identify amino acid motifs that are highly conserved within the targeted pathogen family and are suitable for the design of CODEHOP PCR primers and assay development. In general, this requires two amino acid motifs of approximately 5–10 amino acids each that are separated from each other by approximately 10–300 amino acids. Thus, a primer derived from the sense-strand encoding the upstream motif coupled (~30 bp) with a primer derived from the anti-sense strand encoding the downstream motif (~30 bp) would yield a PCR product of approximately 90–1000 base pairs. Such a product would provide approximately 30–940 bases of sequence from a novel viral template. Optimal amino acid motifs are those that are highly conserved across disparate members of the targeted pathogen family and contain amino acids with low codon degeneracy. CODEHOP PCR primers are composed of a degenerate 3’ core that contains all possible sequences needed to encode a 3–4 amino acid motif. Therefore, choosing motifs with amino acids that have limited codon degeneracy decreases the total number of primers in the CODEHOP primer pool. Optimal amino acids would include Met and Trp (one codon) and Phe, Tyr, Asp, Glu, Gln, Asn, Arg, and Lys (two codons). The most optimal motifs would contain one of these amino acids in the penultimate position of the motif, thus limiting the degeneracy of the CODEHOP PCR primer at the 3’ hydroxyl end, the site of polymerase extension. The ideal motif would contain a C-terminal amino acid with a codon containing two invariant bases at the first and second position (the third wobble position in this codon is not utilized), ie if the amino acid in the motif was valine with the codon GTN, the two invariant bases would be G and T.
1.3 Web server for design of CODEHOP PCR primers
We have previously developed a software program to predict CODEHOP PCR primers from multiply-aligned protein sequences and have provided this as a web service to the scientific community [1, 3]. The CODEHOP web site was hosted by the Fred Hutchinson Cancer Research Center (Seattle, WA) as an integral part of the BLOCKS protein database developed by Steve and Jorja Henikoff [4]. This web site has been used extensively by researchers world-wide for the development of CODEHOP PCR assays to identify novel genes and pathogens [3]. We have recently made significant revisions to the CODEHOP prediction program and associated web server and now provide an interactive iCODEHOP web server to the scientific community hosted by the Center for Public Health Informatics, University of Washington (Seattle)[5].
1.4 Pathogen detection and characterization using CODEHOP-mediated PCR amplification
The CODEHOP approach is a significant improvement over the existing consensus primer and degenerate primer techniques for detecting distantly-related sequences [1]. It provides a robust and sensitive approach to rapidly isolate unknown and widely-diverse members of a gene family out of a vast background of genomic DNA. One benefit of this technology has been the identification of new organisms, such as viruses, bacteria and fungi, from which the new genes were derived. We have used CODEHOP assays to detect fourteen previously unknown DNA polymerase sequences from members of the alpha, beta and gamma subfamilies of herpesviruses [6]. We have also employed CODEHOPs to identify novel retroviruses targeting conserved motifs within the reverse transcriptase gene [1]. Using reverse-transcriptase targeted CODEHOPs, we have identified and characterized a new lentivirus in Talapoin monkeys [7] and a new endogenous retrovirus in pigs [8]. We have also identified viruses implicated in cancer, such as three novel herpesviruses in retroperitoneal fibrosarcoma, a macaque fibroproliferative malignancy related to the human Kaposi’s sarcoma-associated herpesvirus [9, 10]. Finally, we have used the CODEHOP PCR amplification approach for the characterization of viral genomes [11] and have previously reviewed this approach utilizing the herpesvirus family as a target [2].
1.5 Identification of non-human primate pathogens using CODEHOP PCR assays
As the close evolutionary relationships among primates have become more apparent, the study of non-human primates has become increasingly relevant to understanding human health and well-being. Furthermore, the study of non-human primate pathogens has become increasingly important due to similarities with pathogens and pathogen-related diseases in humans. While many different animal species can be reservoirs of pathogens that can infect humans directly or indirectly via vectors, the close phylogenetic relationship between humans and non-human primates increases the potential for cross-species transmission of some viral agents [12, 13]. Although some non-human primate-specific pathogens have been identified in monkey species, the detection and characterization of viral pathogens infecting non-human primates lags significantly behind studies on human pathogens.
As part of a program to promote the health and well-being of captive macaque colonies, we are currently developing general methods to identify and characterize common pathogens of four non-human primate species maintained at the Washington National Primate Research Center (WaNPRC), including the macaque species: M. nemestrina, M. mulatta, and M. fascicularis, and the baboon species, Papio cynocephalus. Although the long term goal is to target all known viral pathogen families, our approach was initially restricted to viruses belonging to the retrovirus, herpesvirus, adenovirus, and papillomavirus families. The choice of these four virus families was based on their ability to persist in the host in chronic or latent form, cause significant health-related problems within the NPRCs and/or be problematic for on-going research. The detection and characterization of such pathogens not only enhances the health of animals in the primate centers by providing the basis for diagnostic assays, but also leads to surveillance capabilities for zoonotic transmission to humans and insight into pathogen biology by comparative studies with human pathogens.
In this report, we describe the development of a CODEHOP PCR assay for non-human primate pathogens targeting the diverse family of papillomaviruses. We demonstrate the broad specificity of this assay for all branches of the diverse papillomavirus family and show its utility in the identification of a novel papillomavirus in the macaque, M. fascicularis.
2. Development of a broadly reactive pan-papillomavirus CODEHOP PCR assay
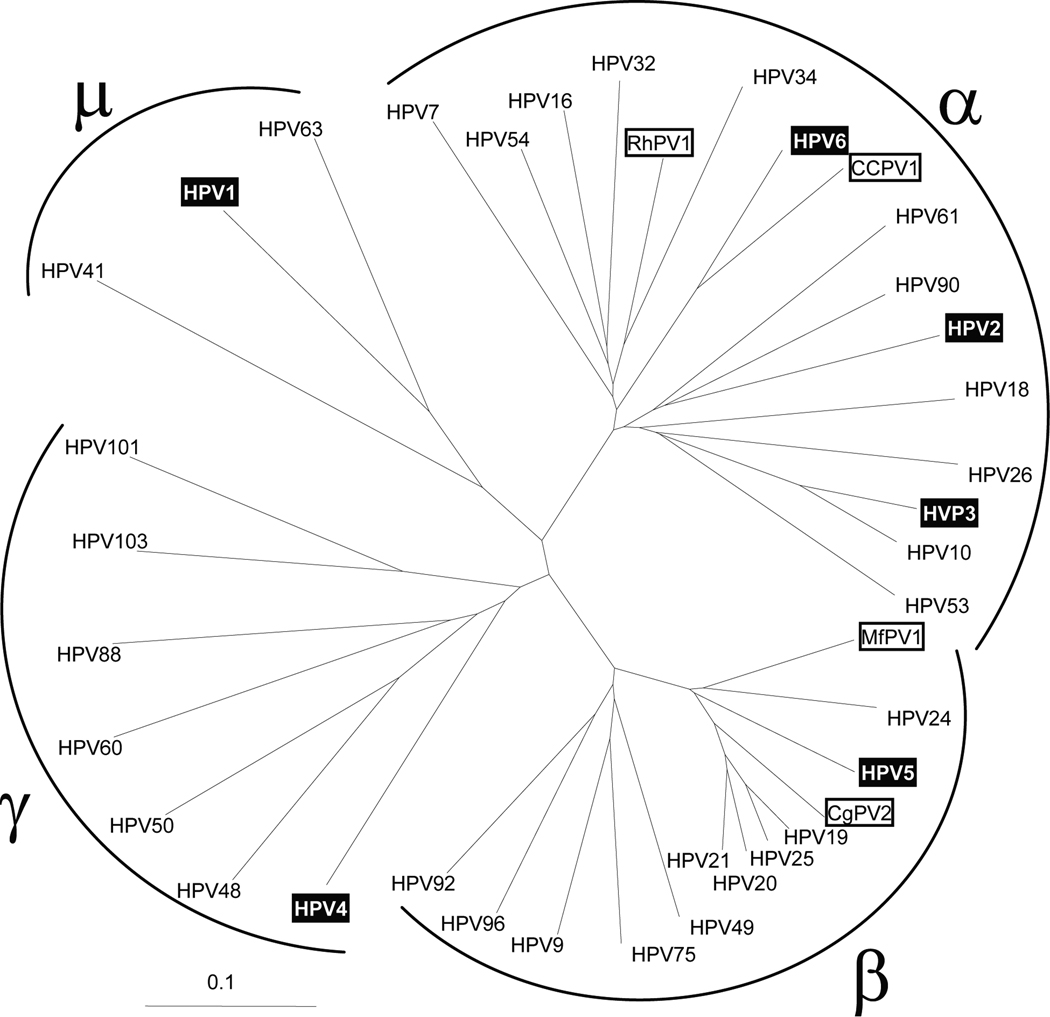
Papillomaviruses, like herpesviruses, have co-evolved with their primate host species. Evolutionary studies have shown that the diversification of papillomaviruses occurred before the evolutionary split between monkeys and apes [14]. Therefore, each primate species, including the different species of macaques and baboons, has its own complement of papillomavirus species. More than 100 different papillomavirus species have been detected in humans. These species cluster within supergroups (genera) that have approximately 50–55% nucleotide similarity [15] (Fig. 1). However, only a few papillomavirus species have been described in primate species used in biomedical research. Twelve papillomavirus isolates have been identified and genotyped in the rhesus macaque (M. mulatta), while twenty-six papillomavirus sequences have been detected in M. fascicularis [16, 17]. No papillomavirus isolates have been reported for M. nemestrina. This indicates that there are a large number of unknown and distinct papillomaviruses in the targeted primate species at the WaNPRC that have yet to be identified. The occurrence of papillomaviruses in cutaneous oral warts, in a squamous cell carcinoma of the penus and in a transmissible cervical cancer in monkeys highlights the importance of discovery and characterization of macaque papillomaviruses [18–20].
Figure 1. Phylogenetic analysis of the L1 open-reading frame sequences of a variety of papillomavirus types.
Various L1 sequences from human and non-human primate papillomaviruses were obtained from the NCBI protein database by BLAST analysis using the sequence NP_040309 of HPV1 as query. The sequences were aligned using ClustalW [22] and the resulting guide tree (neighbor-joining) was visualized using TreeView [24]. The clustering of the four different genera of human papillomaviruses is shown. Non-human primate papillomaviruses are indicated (open boxes): rhesus macaque (M. mulatta) papillomavirus type 1 (RhPV1; NP_043338), M. fascicularis papillomavirus type 1 (MfPV1; ABM67070 and this study), Colobus monkey papillomavirus type 2 (CgPV2; AAB39894), common chimpanzee papillomavirus (CCPV1; NP_045018). Representatives of the different human papillomavirus species analyzed in this study are also indicated (shaded boxes): HPV1 (NP_040309); HPV2 (NP_077122); HPV3 (CAA52474); HPV4 (NP_040895); HPV5 (NP_041372); HPV6 (NP_040304). The scale for substitutions per site is provided.
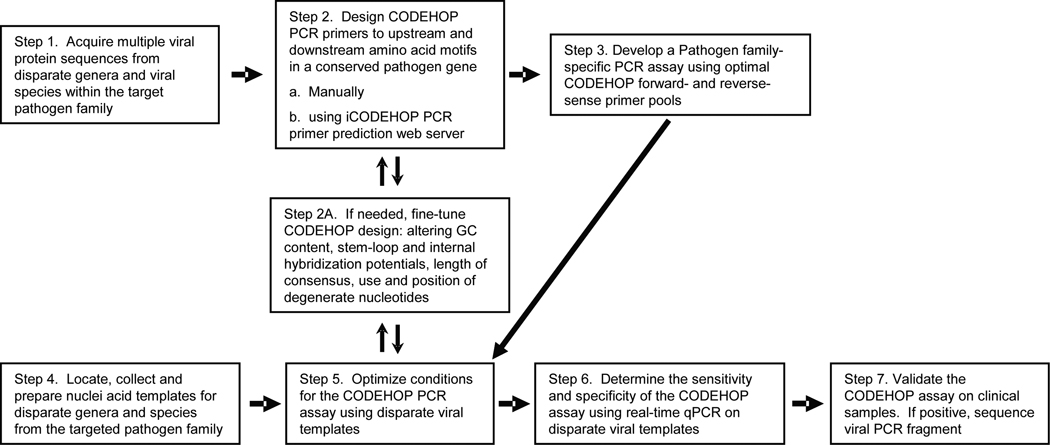
Recently, degenerate primer PCR assays have been developed to detect novel papillomaviruses following the CODEHOP primer design approach [21]. In this study, it was necessary to develop separate PCR assays targeting the different human papillomavirus supergroups. These assays targeted conserved amino acid motifs within the papillomavius L1 protein and utilized degenerate primer pools containing 128–512 individual primers. We set out to develop a more robust CODEHOP assay to identify non-human primate papillomavirus species. Our aim was to limit primer degeneracy and be capable of detecting members of all papillomavirus supergroups in a single assay. The general flow chart for development of a CODEHOP PCR assay is shown in Figure 2.
Figure 2. CODEHOP PCR assay development.
The general flowchart for development of a viral pathogen-specific CODEHOP PCR assay is shown. The different steps are referenced in the text.
2.1 Assembly of sequences of disparate members of the target papillomavirus L1 protein family (Step 1, Fig. 2)
We chose the L1 protein family as a target for our pan-papillomavirus CODEHOP assay due to its ubiquity in the papillomavirus family and its high sequence conservation. The reference sequence for the L1 protein from the human papillomavirus type 1 (HPV1) (NP_040309) was obtained from the NCBI protein database and used in a BLAST search to identify related L1 protein sequences in the database. A set of L1 proteins from the major papillomavirus supergroups was assembled, including a number of non-human mammalian papillomavirus species.
2.2 Design of pan-papillomavirus CODEHOP PCR primers (Step 2, Fig. 2)
2.2.1 Manual approach for design of CODEHOP PCR primers
2.2.1a Identification of CODEHOP target motifs from multiply-aligned L1 protein sequences
An alignment of a set of disparate L1 proteins was performed using ClustalW [22]. Visual examination of the ClustalW output revealed a number of amino acid motifs. Two conserved motifs, “DGDM” and “NNGI/V” that met the optimal criteria described in 1.2 above were identified as possible CODEHOP targets (Figure 3). The codon degeneracy of the “DGDM” motif was 16 while the “NNGI/V” motif was 32 (the wobble position of the I/V codon is not considered, see 2.2.1b, below). These motifs were separated by ~120 amino acids so that PCR amplification between these motifs would generate a product of approximately 420 bp.
Figure 3. LOGOS representation of the “DGDM” and “NNGI” targeted amino acid motifs within the L1 protein family of papillomaviruses.
A) The upstream “DGDM” motif, located at aa193–204 in HPV1 (NP_040309). The C-terminal amino acid which specifies the 3’ codon for the sense-strand CODEHOP is indicated with an asterix (Met-1 codon). B) The downstream “NNGI” motif, located at aa327–338 in HPV1. The N-terminal amino acid which specifies the 3’ codon for the antisense-strand CODEHOP is indicated with an asterix (Asn- 2 codons). The codon degeneracies of the amino acids in the motif are indicated above each residue. The LOGOS representation of amino acid conservation shows the amino acids present in each position of the motif with a size representative of the degree of conservation [25]
2.2.1b Design of the 3’ degenerate core encoding the CODEHOP target motifs
A sense-strand CODEHOP PCR primer was designed from the “DGDM” motif and upstream flanking sequences, while an anti-sense strand CODEHOP PCR primer was designed from the “NNGI/V” motif and downstream flanking sequences. All possible codon sequences for the “DGDM” motif were provided in the 12bp 3’ degenerate core of the DGDMa CODEHOP (the “a” designation indicates sense orientation), yielding a core sequence of 5’-GAYGGNGAYATG-3’ with 16 fold degeneracy. All possible codon sequences for the “NNGI/V” motif were provided in the 11bp 3’degenerate core of the NNGIb CODEHOP (the “b” designation indicates anti-sense orientation), yielding the antisense degenerate core sequence of 5’-AYNACRTTRTT -3’ with 32 fold degeneracy (International code: Y=C or T; R=A or G; N=A,C,G or T).
2.2.1c Design of the 5’ consensus clamp encoding the region flanking the CODEHOP target motifs
To predict the optimal 5’ consensus clamp sequence for these CODEHOPs, the nucleotide sequences encoding the “DGDM” and “NNGI/V” motifs and flanking sequences from the set of genes encoding the L1 proteins from different papillomavirus species were obtained from the NCBI nucleotide database. The nucleotide sequences flanking the codons for each motif were aligned (data not shown) and the consensus nucleotides at each position were chosen. For the DGDMa CODEHOP, a 5’ consensus clamp of 24 nucleotides (5’-GAGCTTATAAACACAGTTATTGAG -3’) was chosen. For the NNGIb CODEHOP, a 5’ consensus clamp of 21 antisense nucleotides (5’-AACAGTTGATTGTCCCAGCAG -3’) was chosen.
2.2.1d Development of a PCR assay based on the DGDMa and NNGIb CODEHOP PCR primers (Step 3, Fig. 2)
Combining the 3’ degenerate core and 5’ consensus clamp regions for the two target protein motifs yielded a 36bp DGDMa CODEHOP (5’-GAGCTTATAAACACAGTTATTGAGgayggngayatg-3’) and a 32bp antisense NNGIb CODEHOP (5’-AACAGTTGATTGTCCCAGCAgaynacrttrtt-3’) PCR primer. The lengths of the 5’ consensus clamps were chosen to have similar melting temperatures, ie. 54 °C. These primers are similar to the ME and MH primers described by Baines et al., [21], except that the degeneracy of the primer pools was significantly less. The DGDMa primer pool was 16 fold degenerate compared to the ME primers, which were 192–768 fold degenerate. The NNGIb primer pool was 32 fold degenerate compared to MH primers, which were 128–1024 fold degenerate. In addition, whereas the ME and MH primer sets contained some degenerate positions in the 5’ clamp region of the primer, the DGDMa and NNGIb CODEHOP primers did not. Amplification of papillomavirus DNA templates with these primers would yield a PCR fragment of ~425 bp (based on the HPV1 sequence).
2.2.2 Automated approach for the design of CODEHOP PCR primers using the iCODEHOP program and web site
2.2.2a – Initiation of the iCODEHOP program
The iCODEHOP web server provides an automated approach to design CODEHOP PCR primers. To design pan-papillomavirus CODEHOP PCR primers, users would access the iCODEHOP program [5] through a web-browser, choose to run either a named session (input and output saved on the web server) or a non-named session and select to enter the “Design Primers” workflow. They would then input either the ClustalW aligned or non-aligned L1 protein sequences assembled in section 2.1 (Step 1) above, select sequences and proceed with the analysis. The following example utilizes the papillomavirus sequences: Rabbit PV (NP_057848); Chimpanzee PV1 (NP_045018); HPV18 (NP_040317); HPV90 (NP_671509); HPV2 (NP_077122); HPV1 (NP_040309); HPV96 (NP_932325); HPV92 (NP_775311).
2.2.2b – Sequence alignment and identification of conserved sequence blocks
If non-aligned sequences were input, then the program will align them using ClustalW and present users with a ClustalW alignment (.ALN) and a cladogram of the phylogenetic relationship of the sequences. Users would provide an alignment name and proceed. Alternatively, users can input a set of previously aligned sequeces. In both cases, this will initiate a program to identify conserved protein motifs within the aligned sequences. At this step, the CODEHOP design interface would indicate the protein motifs as “blocks” and provide default values for the design of the 5’ consensus clamp and 3’ degenerate core. These values may be changed under the Advanced Settings page. Users would then initiate the primer design by selecting “Look for primers.”
2.2.2c – Choosing an iCODEHOP PCR primer pair (Steps 2 and 3, Fig. 2)
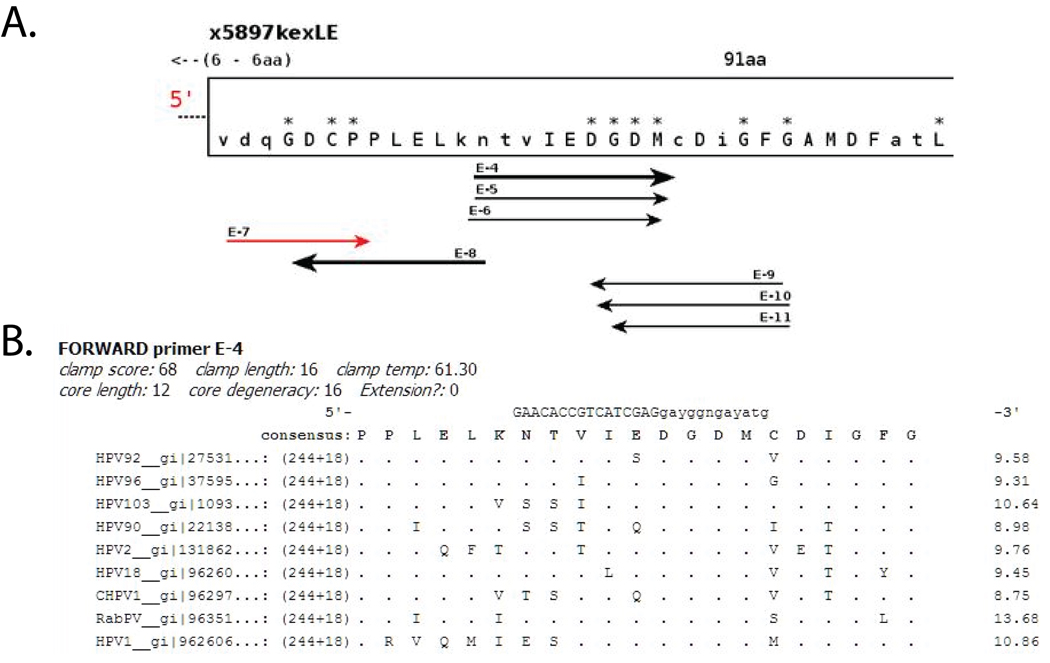
A graphical representation of the conserved motifs (blocks) and predicted CODEHOP PCR primers, in both sense and anti-sense orientations, would be outputted (Fig. 4A). Mousing over the individual blocks would provide an alignment of the amino acid sequences within the block and a consensus sequence. Mousing over the individual primers would show a proposed CODEHOP PCR primer with its associated conserved sequence block (Fig. 4B). The program shows sense-strand primers singly, while anti-sense strand primers (black) are shown in relation to the sense-strand sequence (grayed out) (data not shown). Using the papillomavirus sequences in the example, the fifth block (E; 91aa) contains the “DGDM” protein motif identified visually in 2.2.1a above, with a proposed sense-strand CODEHOP PCR primer E5 derived from that motif: 5’-GAACACCGTGATCGAGgayggngayatg-3’ (16-fold degenerate). Examination of the sixth block (F; 65aa) shows the “NNGI” protein motif identified visually in 2.2.1a, above, with a proposed anti-sense strand CODEHOP PCR primer F8 derived from that motif: 5’ – GTTGCGCCAGCAGaynccrttrtt-3’ (32-fold degenerate) (data not shown).
Figure 4. Output of the automated iCODEHOP primer design web server for the sense-strand DGDMa CODEHOP PCR primer.
A) The L1 protein sequences from a diverse set of papillomaviruses were used in the new interactive iCODEHOP web server [5] to design CODEHOP PCR primers: Rabbit PV (NP_057848); Chimpanzee PV1 (NP_045018); HPV18 (NP_040317); HPV90 (NP_671509); HPV2 (NP_077122); HPV1 (NP_040309); HPV96 (NP_932325); HPV92 (NP_775311). Primer design was performed using the default conditions, including the 60 °C default clamp melting temperature (discussed in the text). The portion of the graphical output of the analysis with the 91 amino acid “E” block (x5897kexLE) containing the DGDM motif is shown (asterices indicate highly conserved residues). The relative position of the “E” block to the preceeding “D” block (not shown) is indicated, ie “6–6aa”. The arrows underneath the block indicate the length and position of possible sense and anti-sense strand CODEHOP primers, labeled with their block and a number. B) The information provided for the sense-strand (forward) E-4 primer, derived from the DGDM motif, includes the length and melting temperature of the consensus clamp region, as well as the length and degeneracy of the core. The predicted E-4 primer sequence (clamp length determined by the 60 °C default temperature setting) is shown above the amino acid consensus derived from the alignment of the L1 proteins used as input (dots indicate identity with the consensus sequence). Further details regarding the output of the program are described on the iCODEHOP website [5].
2.2.2d – Fine-tuning iCODEHOP PCR primer design (Step 2A, Fig. 2)
The 3’ degenerate cores of the E5 and F8 primers designed by iCODEHOP are identical to the 3’ degenerate cores designed manually for DGDMa and NNGIb in 2.2.1c above. The 5’ consensus clamps of the E5 and F8 primers, which were designed using the default melting-point calculation of 60 degrees in the iCODEHOP primer design, were 16 and 13 bp in length, respectively, compared to the 24 and 21 bp 5’ consensus clamps of the manually-designed DGDMa and NNGIb primers. If empirical studies indicate that the initial design needs fine-tuning, then the default values used in the primer design can be altered in the Advanced Settings of the design interface. For example, if the 5’ consensus clamps should be longer to increase the annealing temperature of the PCR reaction in order to obtain a more specific amplification, the iCODEHOP primer design can be redone, altering the temperature in the Advanced Settings. Using a 72 degree temperature setting for the 5’ consensus clamp region, the iCODEHOP proposed E5 (DGDM) and F8 (NNGI) primers are 5’- GAGGCTGAAGAACACCGTGATCGAGgayggngatatg -3’ and 5’- ACATCTGGTTGCGCCAGCAGaynccrttrtt -3’, with similar annealing properties as the manually designed DGDMa and NNGIb CODEHOP primers.
The automated iCODEHOP PCR primer design is performed completely without reference to nucleotide sequences encoding the target protein motifs, by design. The 3’ degenerate core generated by iCODEHOP contains all possible codons for the targeted motif, while the 5’ consensus clamp is based on either the most frequently used codon for the most common amino acid in each position, or the position-specific scoring matrix (PSSM) for the DNA sequences encoding all amino acids at each position. The iCODEHOP-generated PCR primers can be manually modified to include in the design the underlying DNA sequences of the codons used in the cDNA sequences of the target proteins – generating a modified CODEHOP PCR primer that is essentially the same CODEHOP PCR primer as the one designed manually in 2.2.1, above. Additional, fine-tuning of the iCODEHOP generated primer can be done manually, including elimination of stem-loop/self-annealing structures. A new feature of the iCODEHOP program is the ability to optimize the primer design using phylogenetic information to identify and remove sequence outliers which may be masking conserved sequence motifs. Distinct clustering of input sequences can be identified using the phylogenetic analysis, allowing distinct groups of input sequences to be analyzed separately.
Evaluations of secondary structure as well as alignment of the primers with the underlying nucleotide sequence alignments are currently not provided by the iCODEHOP web server and must be performed outside of the program. However, these features will be implemented in a future release. As a final step, the specificity of each primer should be examined by a BLAST search of the non-redundant nucleotide database (GenBank) to detect and exclude primers with strong similarity to cellular sequences.
2.3 Assembly and preparation of test DNA templates for PCR amplification (Step 4, Fig. 2)
We obtained papillomavirus plasmids containing the complete genomes of separate viral isolates of each of the different human papillomavirus genera from Dr. De Villiers of the World Health Organization Human Papillomavirus DNA International Collaborative Study Group. These plasmids contained the genomes of the human papillomaviruses HPV1, HPV2, HPV3, HPV4, HPV5, and HPV6 (Figure 1, highlighted in black boxes) in a pBR322 plasmid background. Plasmid DNA was purified and quantitated by optical density at 260 nm. For quantitative comparison purposes, we wanted to ensure that all plasmid templates were assayed at identical concentrations. Therefore, we developed a SYBR-green real-time PCR assay to quantitate the papillomavirus plasmid copy number targeting a region of the pBR322 plasmid backbone shared by all of the papillomavirus plasmids. Utilizing the pBR322 qPCR assay, we verified that the concentrations of the different papillomavirus plasmid DNA templates were equivalent (data not shown). For testing, the papillomavirus plasmid DNA was diluted in an excess of human genomic DNA to determine the ability of the CODEHOP assay to amplify papillomavirus templates in a complex genomic mixture. In these assays, we used 50 ng genomic DNA per PCR reaction which corresponds to ca. 5,000 cellular genomes, to mimic the amount of cellular DNA that can be expected in DNA extracts from solid tissue. Standard curves were obtained from a dilution series of plasmid using the optimized pBR322 qPCR assay. Plasmid DNA was assayed in duplicate at concentrations ranging from 104–108 copies per reaction. The assays were linear across this range with a slope of −3.55 (91.3% efficiency) and r2 = 0.992.
2.4 Optimization of the DGDMa/NNGIb CODEHOP PCR assay conditions (Step 5, Fig. 2)
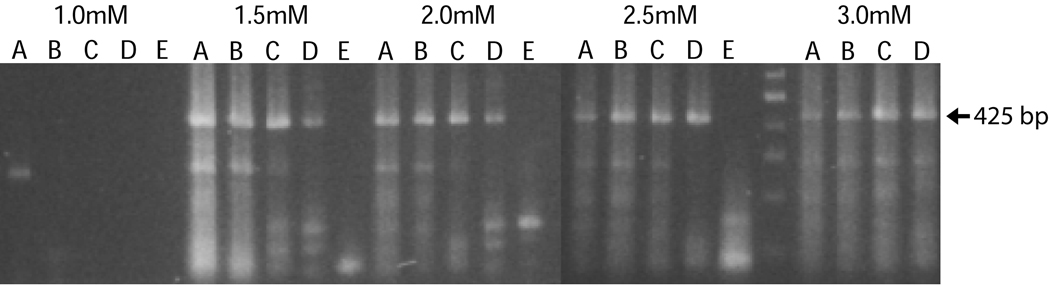
To empirically determine the optimal MgCl2 and annealing conditions for our DGDMa/NNGIb CODEHOP PCR primers, we performed PCR amplification reactions using a thermal gradient of annealing temperatures. We picked the plasmid containing HPV1 as the expected lowest-performing template and the plasmid containing HPV6 as the expected highest-performing template, due to the number of mismatches with the consensus clamps of DGDMa and NNGIb. Initially, we used the HPV6 plasmid as template with the following PCR conditions in a 25 µl reaction volume: 1x PCR buffer (Invitrogen) with 0.2 mM each dNTP, sense-strand primer DGDMa (2 µM), anti-sense strand primer NNGIb (2 µM), fluorescein (10 nM), SYBR green (1:10,000; Invitrogen), and 2.5 units Platinum Taq polymerase (Invitrogen). Different MgCl2 concentrations (1.0–3.0 mM) were assayed. HPV6 plasmid template (106 copies – as determined by SYBR-green PBR322 qPCR) was diluted into 50ng of genomic DNA and samples were assayed in duplicate. Activation of the polymerase and amplification were performed on a BioRad iCycler for 50 cycles of 95 °C for 30 s, 50–63 °C annealing temperature gradient for 30 s and 72°C for 30 s. As shown in Figure 5 (1.0 mM), no PCR product was obtained with 1.0m M MgCl2. The correct-sized PCR product (426 bp) was obtained with reaction conditions of 1.5–3.0 mM MgCl2 at annealing temperatures of 50–58.5 °C. No amplification was seen at the highest annealing temperature tested, 63 °C. The most HPV6 PCR product was detected with 1.5 mM MgCl2 at the lowest annealing temperature tested, 50 °C (Fig. 5: 1.5 mM-A). However, significant non-specific amplification was also detected under these conditions. Increasing the annealing temperature to 58.5 °C at 1.5 mM MgCl2 increased the specificity of the amplification but also decreased the amount of product (Fig. 5: 1.5 mM-D). An increase in the amount of HPV6 PCR product was obtained at higher annealing temperatures (54.7 and 58.5 °C) as the concentration of MgCl2 was increased to 2.5 mM. Conversely, the amount of non-specific PCR products decreased at the higher annealing temperatures and higher concentrations of MgCl2. Optimal amplification (product amount) and specificity (lack of non-specific products) was obtained at 2.0 mM MgCl2/ 54.7 °C and at 2.5 mM MgCl2/ 58.5 °C. Similar results were obtained with the HPV1 plasmid template (data not shown). These results indicate that the DGDMa/NNGIb CODEHOP PCR assay was robust, with good amplification across a broad range of MgCl2 concentrations.
Figure 5. Optimization of MgCl2 concentrations and PCR annealing conditions for the DGDMa/NNGIb CODEHOP PCR assay.
The DGDMa and NNGIb CODEHOP PCR primers were used to amplify the HPV6 plasmid template (106 copies) in a background of 50 ng genomic DNA under different MgCl2 concentrations and annealing temperatures. Gel analysis of the PCR products obtained with HPV6 is shown (expected product size = 425 bp). MgCl2 concentrations ranged from 1.0–3.0 mM and annealing temperatures were (A) 50 °C, (B) 52 °C, (C) 54.7 °C, (D) 58.5 °C, (E) 63 °C).
While the concentration of standard primers used in normal PCR amplification is generally 0.1–0.5 µM, it is not clear what the optimal concentration should be for the pools of degenerate primers used in a CODEHOP PCR assay. We compared the amplification efficiency of the DGDMa/NNGIb CODEHOP PCR assay using increasing primer concentrations. Ten-fold dilutions of HPV1 plasmid templates were prepared in a background of 50ng genomic DNA and a range of 1011 to 104 plasmid copies were used per reaction. PCR was performed in duplicate in 2 mM MgCl2. Activation of the polymerase and amplification was performed on a BioRad iCycler equipped with an optical module for 50 cycles of 95°C for 30 s, a 58 °C annealing temperature for 30 s and 72°C extension temperature for 30 s. Standard curves were obtained from dilution series using SYBR-green qPCR with primer concentrations of 2 µM (1x), 4 µM (2x), or 8 µM (4x). The assay using 2 µM of each primer pool was linear across a four log range of dilutions (107–1011) with a slope of −4.369 (69.4% efficiency) and r2 = 0.979 (data not shown). Doubling the primer concentration to 4 µM yielded a slope of −3.323 (100% efficiency) and r2 =0.993. Further doubling of the primer concentration to 8 µM decreased the efficiency to 74.3% (slope = −4.146) with r2 = 0.999. These results indicate that primer concentration can play an important role in amplification efficiency, but this may depend on the nature of the primer pairs utilized, particularly their overall degeneracy.
2.5 Determining the sensitivity of the DGDMa/NNGIb CODEHOP PCR assay (Step 6, Fig. 2)
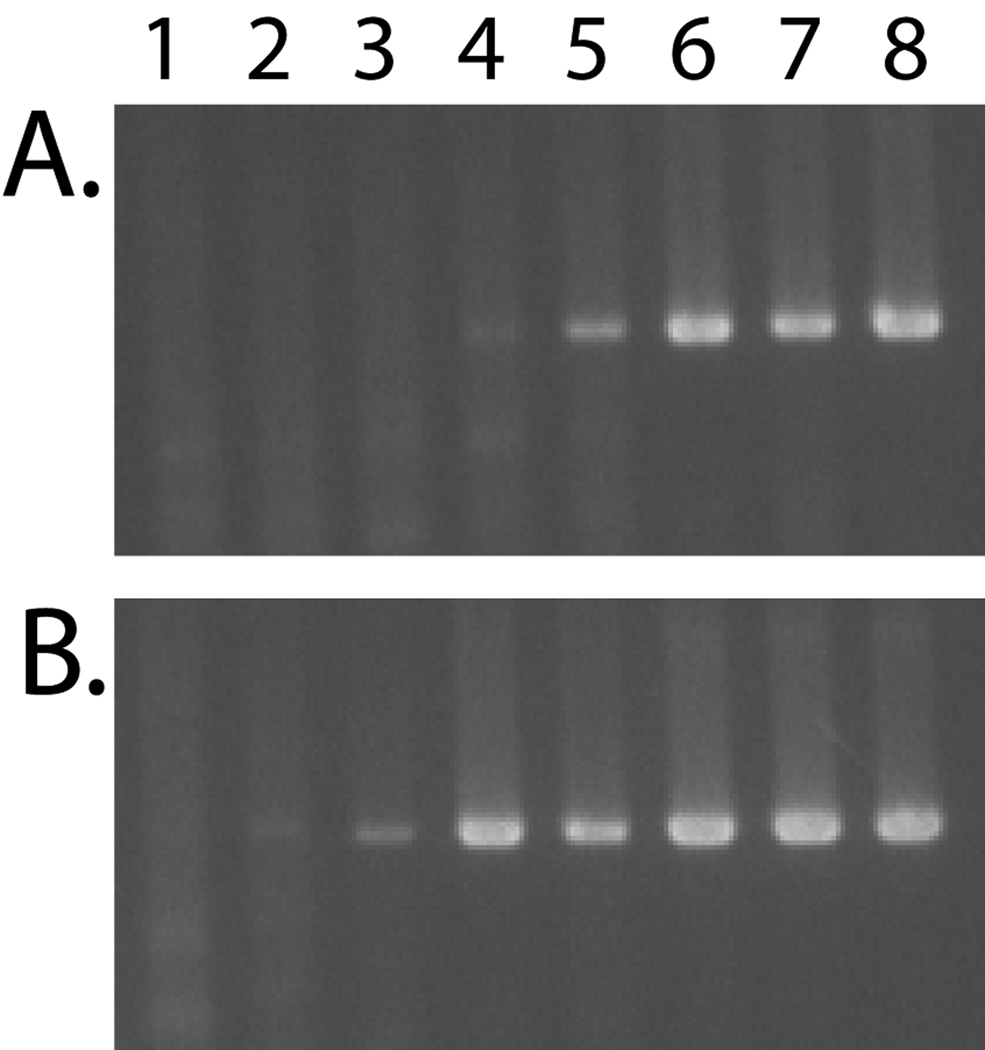
To compare the sensitivity of the DGDMa/NNGIb CODEHOP PCR assay for disparate papillomavirus templates, ten-fold dilution series were prepared for the HPV1 and HPV6 templates ranging from 108 to 10 copies per reaction in a constant background of 50 ng genomic DNA, equivalent to 5,000 cellular genomes per reaction. All reactions were performed in 2 mM MgCl2 at an annealing temperature of 58° C for 50 cycles and the products were analyzed by gel electrophoresis. With the HPV1 template, the expected 426 bp PCR product was detected in the dilution series down to 107 copies per reaction (Fig. 6A). With the HPV6 template, the expected PCR product was detected down to 105 copies per reaction (Fig. 6B).
Figure 6. Comparison of the sensitivity of the DGDMa/NNGIb CODEHOP assay for the HPV1 and HPV6 templates.
The DGDMa and NNGIb CODEHOP PCR primers were used to amplify A) HPV1 and B) HPV6 plasmid templates in a 10-fold dilution series. Lane 1: 104; Lane 2: 105; Lane 3: 106; Lane 4: 107; Lane 5: 108; Lane 6: 109; Lane 7: 1010; Lane 8: 1011 viral plasmid copies per 25 µl reaction. Viral plasmids were analyzed in a background of 50 ng genomic DNA using 2 mM MgCl2 and 58 °C annealing temperature. PCR products were analyzed by gel electrophoresis.
2.6 Determining the broad specificity of the DGDMa/NNGIb CODEHOP PCR assay (Step 6, Fig. 2)
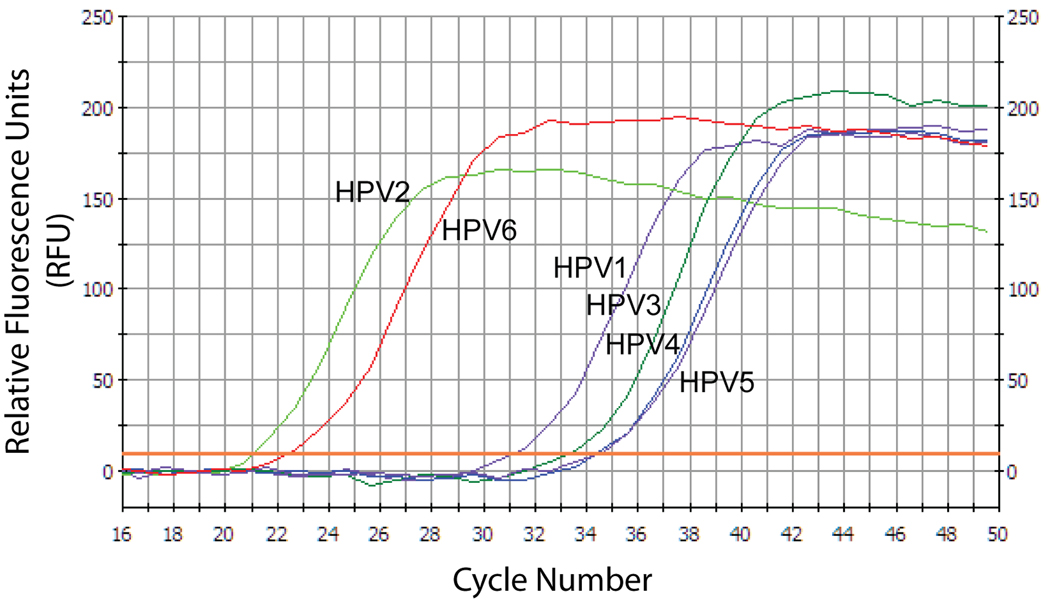
To determine the broad specificity of the DGDMa/NNGIb CODEHOP PCR assay for disparate papillomavirus templates, we performed SYBR-green qPCR assays in 2 mM MgCl2 at an annealing temperature of 58° C using the six plasmids containing genomes for HPV1, HPV2, HPV3, HPV4, HPV5 and HPV6 as templates. PCR was performed for 50 cycles with 108 copies of template per 25 µL reaction in the presence of 50 ng genomic DNA and run in duplicate. The SYBR-green qPCR assay targeting the pBR322 backbone sequence of the templates was run in parallel to validate the concentrations of input DNA template. As shown in Table 1, the CT values obtained for the different HPV plasmid templates using the pBR322 qPCR assay ranged from 16.2–18.3 showing very similar input plasmid template concentrations. The cumulative fluorescence curves for the DGDMa/NNGIb CODEHOP PCR assays on the different HPV plasmid templates showed similar slopes (Fig. 7), although different CT values were obtained, ranging from 20.4–36.1 (Table 1). Amplification of the HPV2 template by the DGDMa/NNGIb assay was only two cycles below that obtained with the pBR322 specific assay. Conversely, amplification of HPV5 was 20 cycles below that obtained with the pBR322 specific assay. Thus, while all HPV species tested were amplified under these conditions, there were significant differences in their ability to be amplified by the DGDMa/NNGIb assay.
Table 1.
Comparison of HPV plasmid templates and amplification results
| HPV plasmid |
HPV Genus/ subgroup/ species |
pBR322- specific assay (CT)1 |
DGDMa/NNGIb CODEHOP assay (CT)2 |
ΔCT3 | Tm 5’ clamp4 |
|---|---|---|---|---|---|
| HPV2 | α3–2 | 18.2 | 20.4 | 2.2 | 18/24 |
| HPV6 | α10–6 | 16.2 | 22.5 | 6.3 | 16/18 |
| HPV1 | μ1–1 | 16.7 | 31.2 | 14.5 | 14/18 |
| HPV3 | α2–3 | 18.3 | 33.2 | 14.9 | 13/21 |
| HPV4 | γ1–4 | 18.2 | 34.2 | 16.0 | 10/20 |
| HPV5 | β1–5 | 17.6 | 34.5 | 16.9 | 10/14 |
The cycle threshold (CT) values obtained from the specific SYBR green qPCR assay targeting the pBR322 sequences common to the different HPV templates using an estimated input of 108 plasmid copies per reaction. Copy number can be back calculated using a CT of 43 for a single copy: 243-CT. ie a CT of 17 = 226 = 6.7 × 107 copies of plasmid.
The CT values obtained using the DGDMa/NNGIb CODEHOP PCR assay on the same plasmid templates as used in the pBR322-specific assay.
The difference in CT values obtained using the degenerate CODEHOP assay and the specific pBR322 assay.
The melting temperature (Tm) was calculated for the mismatched duplex of the 10 bp at the 3’ end of the primer 5’ consensus clamp region (underlined in Figure 8) with the corresponding sequence in the different HPV templates (DGDMa/NNGIb) using 2 degrees Celsius for A-T pairs and 3 degrees Celsius for G-C pairs. The Tm of the DGDMa CODEHOP PCR primer is inversely correlated with the delta CT, showing that HPV templates that closely match the primer in this 10 bp region are amplified the most efficiently (ie. HPV2). HPV templates that have the most mismatches in this region are amplified the least efficiently (ie. HPV5). A similar correlation is noted with the NNGIb primer.
Figure 7. Comparison of the specificity of the DGDMa/NNGIb CODEHOP assay for DNA templates representing HPV species from different papillomavirus genera.
The DGDMa and NNGIb CODEHOP PCR primers were used to amplify pBR322-based plasmids (~108 copies per reaction) containing the genomes of HPV1, HPV2, HPV3, HPV4, HPV5 or HPV6. PCR amplification was performed in duplicate in a background of 50 ng genomic DNA (5,000 cellular genomes) using 2 mM MgCl2 and an annealing temperature of 58 °C, and was quantitated using SYBR-green real-time qPCR over 50 cycles. Relative fluorescence of representative samples is shown for each of the papillomavirus templates.
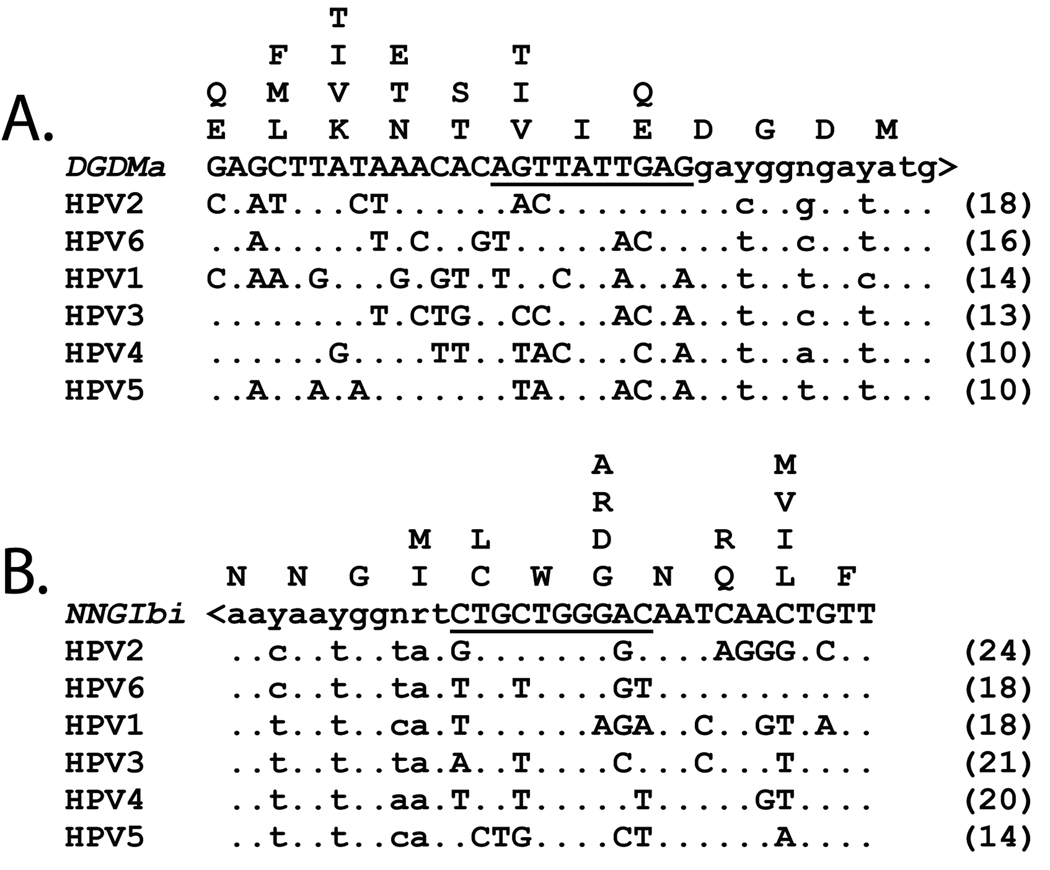
The nucleotide sequences of the different HPV variants were compared to the corresponding sequences of the DGDMa and NNGIb CODEHOP PCR primers in order to determine the extent of mismatches that could explain the differences in amplification. No obvious correlation was detected between the ability of a template to be amplified and the total number of mismatches with the 5’ clamp of the primers. The HPV1 sequence had the most number of mismatches with the DGDMa primer, yet was the third best template for amplification. However, an inverse linear relationship was noted between the amplification efficiency of a template (ΔCT = CT (pBR322)- CT (DGDMa/NNGIb; Table 1) and the calculated melting temperature (Tm) of the 10 bp region at the 3’ end of the consensus clamp of the DGDMa primer (underlined in Figure 8A), ie. the sequence adjacent to the degenerate core. The best amplified template, HPV2, showed a ΔCT of 2.2 and a clamp Tm of 18 degrees, whereas the worst amplified template, HPV5 showed a ΔCT of 19.9 and a clamp Tm of 10 degrees (Table 1). This suggests that modification of the DGDMa clamp region to achieve an equivalent Tm within the 10bp clamp region with all of the HPV templates could result in a PCR assay that would give an equivalent amplification for each template. Experiments to fine-tune this assay are underway.
Figure 8. DGDMa and NNGIb primers aligned with nucleotide sequences of six disparate human papillomavirus species.
The DGDMa (A) and NNGIb (B) primer sequences were aligned with the corresponding sequences in the six human papillomavirus L1 genes HPV1, HPV2, HPV3, HPV4, HPV5, and HPV6. The inverted sequence (ie coding strand) of NNGIb, designated NNGIbi, is shown for comparison purposes. The 3’ degenerate core of each primer is indicated in lower case letters and the 5’ consensus clamp is indicated in capitol letters. The 10 nucleotides immediately upstream of the degenerate core that were used for calculating a Tm value (in parentheses; see Table 1) are underlined. The nucleotide positions in the HPV sequences that differ from the primer sequences are indicated by letters in the alignments. The amino acids in the six HPV templates are aligned vertically above the codons at each position.
3.0 Identification of a novel macaque papillomavirus using the DGDMa/NNGIb CODEHOP PCR assay (Step 7, Fig. 2)
3.1 PCR amplification of DNA extracted from a wart of a cynomolgus macaque
To validate the ability of the DGDMa/NNGIb CODEHOP PCR assay to identify papillomavirus species in tissue samples from non-human primates, we obtained DNA from a wart of a cynomolgus macaque (M. fascicularis), in collaboration with Dr. Albert Jenson (Louisville). Using the PCR protocol developed for the DGDMa – NNGIb CODEHOP PCR assay with 2mM MgCl2, the wart DNA was subjected to 50 cycles of PCR amplification using a gradient of annealing temperatures ranging from 55–65 °C. A strong single PCR amplification product with essentially no background was obtained at each temperature, showing a robust amplification (data not shown). The PCR product was gel-purified and directly submitted for sequencing. The sequence obtained was translated to protein and submitted to a BLAST search against the non-redundant protein database from GenBank. The best scoring hit was obtained with the L1 protein of HPV21 (P50787) with 116 matches out of 132 amino acids (88%) (Fig. 9). Phylogenetic analysis revealed that the DGDMa/NNGIb ORF clustered with HPV21, HPV5 and other papillomaviruses belonging to the β-papillomavirus supergroup (Fig. 1), suggesting that this ORF was derived from a novel β-papillomavirus. This virus has been subsequently characterized and called M. fascicularis papillomavirus 1 (MfPV1) and its sequence has been deposited in the NCBI database (Accession # EF028290.1). Although the HPV5 β-papillomavirus was the most inefficient template in the DGDMa/NNGIb CODEHOP PCR specificity assay (2.7 above), the closely-related MfPV1 gave a strong robust amplification signal suggesting high copy number in the wart tissue.
Figure 9. Sequence alignment of the open reading frame of the DGDMa/NNGIb PCR product obtained from a wart of a cynomolgus macaque.
The DGDMa/NNGIb CODEHOP PCR assay was performed on DNA extracted from a wart of a cynomolgus macaque (M. fascicularis). A single ~462 bp PCR fragment was obtained and the sequence was determined. The open reading frame was identified and used in a BLAST search of the NCBI protein database. The highest scoring hit, the L1 protein of HPV21 was aligned with the DGDMa/NNGIb ORF and dots indicate amino acid identity. Phylogenetic analysis showed that the ORF sequence clustered with HPV21, HPV5 and other papillomaviruses belonging to the β papillomavirus subgroup (see Figure 1). This new macaque papillomavirus has been further characterized (unpublished data, A. Jenson), and is now called M. fascicularis papillomavirus 1 (MfPV1).
3.2 Characterization of the M. fascicularis DGDMa/NNGIb CODEHOP PCR product
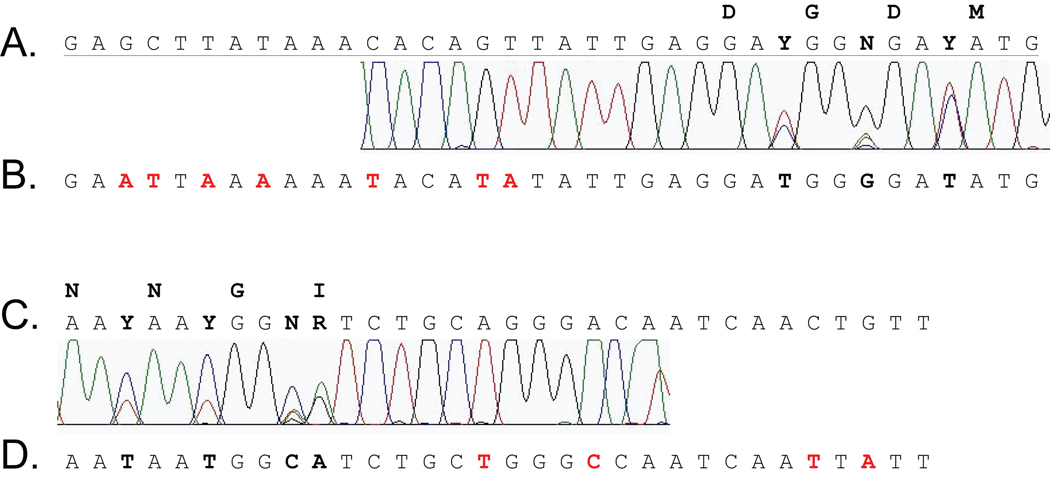
The chromatogram of the ABI sequence obtained from the DGDMa/NNGIb PCR product from the cynomolgus wart showed strong fluorescent signals for each of the nucleotides present in the sequence with the exception of seven positions. These positions corresponded to the degenerate positions within the DGDMa and NNGIb primers that were incorporated into the PCR products (Fig. 10A and C). The relative heights of the fluorescent peaks provides a rough quantitative picture of the primer sequences participating in the PCR amplification at end-point. The sequence of the complete MfPV1 genome was subsequently determined (Accession # EF028290.1) (Albert Jenson, personal communication). The portion of the complete sequence corresponding to the DGDMa and NNGIb primer regions was aligned with the sequence of the PCR product obtained in our assay (Fig. 10B and D). The MfPV1 sequence contained the codons predicted for the “DGDM” and the “NNGI” motifs in our primer design. The presence of multiple nucleotides in the ABI chromatogram at the degenerate codon positions (Y, R and N; in bold, Fig. 10A and C) suggests that the final PCR products contained a wide array of the primers from each of the degenerate pools, as previously demonstrated for other CODEHOP PCR assays[1]. Alignment of the 5’ consensus clamp of each CODEHOP primer with the MfPV1 template sequence revealed numerous mismatched bases, 7/24 (DGDMa) and 4/20 (NNGIb) (red letters in Fig. 10B and D), demonstrating the ability of the CODEHOP primer to amplify distantly-related sequences with a significant number of mismatched bases.
Figure 10. Chromatogram and sequence of the primer region of the DGDMa/NNGIb PCR product.
The sequence chromatograms obtained at each end of the DGDMa/NNGIb PCR product from the amplification of the macaque wart DNA are aligned with the corresponding regions of the DGDMa primer (A) and the NNGIb primer (C) used for amplification. The actual sequence of the underlying MfPV1 genome is aligned below (B and D) with mismatches to the primer sequences indicated in red text. Degenerate positions showing the presence of multiple nucleotides are bolded and the encoded DGDM and NNGI amino acid motifs are indicated. The color code of the chromatogram is as follows: A (green), T (red), G (black), C (blue).
4. Conclusion
We have presented a detailed description of the use of the consensus-degenerate hybrid PCR primer approach for the amplification of unknown and distantly-related sequences. We have developed an assay with broad specificity for the detection of unknown papillomaviruses in humans and non-human primates targeting the L1 gene of the papillomavirus family. This single assay was capable of detecting papillomavirus species belonging to the α, β, γ and μ supergroups of human papillomaviruses. While our assay broadly detected viral species belonging to the major papillomavirus subgroups, it did so with varying sensitivities for individual papillomavirus species. Our data suggests that the 10 base pairs in the 5’ consensus clamp region immediately adjacent to the degenerate core play an important role in the sensitivity of the assay to a particular template. As such, further fine-tuning of the assay could be done to optimize its sensitivity to all the major papillomavirus subgroups. Nevertheless, the new papillomavirus CODEHOP assay was able to amplify an unknown macaque papillomavirus in a robust fashion, even though this virus belonged to the group of viruses that were least sensitive to the assay.
While the DGDMa and NNGIb CODEHOP PCR primers utilized in this study were designed manually utilizing nucleotide sequence information available for different members of the L1 gene family, we have developed an automated interactive software program and web site for designing CODEHOP PCR primers from multiply-aligned protein sequences. This web server quickly identifies conserved amino acid motifs and graphically displays the multiply aligned sequences with the predicted CODEHOP PCR primers. The primers can be utilized, as is, or can be further fine-tuned using the underlying nucleotide sequences encoding the conserved amino acid motifs, as was done with the DGDMa and NNGIb primers. The same general technique can be used to develop CODEHOP PCR assays to detect conserved genes in other pathogen families, including viral, fungal, eukaryotic and bacterial genes, and to identify distantly-related cellular genes, as we have shown previously [1, 23].
Acknowledgement
We would like to thank E.M. de Villiers for the papillomavirus plasmids and A.B. Jenson for the macaque wart sample. The project described was supported by Award Numbers R24-RR021346 and P51-RR00166 from the National Center for Research Resources. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rose TM, Schultz ER, Henikoff JG, Pietrokovski S, McCallum CM, Henikoff S. Nucleic Acids Res. 1998;26:1628–1635. doi: 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rose TM. Virol J. 2005;2:20. doi: 10.1186/1743-422X-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rose TM, Henikoff JG, Henikoff S. Nucleic Acids Res. 2003;31:3763–3766. doi: 10.1093/nar/gkg524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henikoff S, Henikoff JG, Pietrokovski S. Bioinformatics. 1999;15:471–479. doi: 10.1093/bioinformatics/15.6.471. [DOI] [PubMed] [Google Scholar]
- 5.Boyce R, Henikoff J, Henikoff S, Rose TM. 2008 " https://icodehop.cphi.washington.edu/i-codehop-context/Welcome".
- 6.VanDevanter DR, Warrener P, Bennett L, Schultz ER, Coulter S, Garber RL, Rose TM. J Clin Microbiol. 1996;34:1666–1671. doi: 10.1128/jcm.34.7.1666-1671.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Osterhaus AD, Pedersen N, van Amerongen G, Frankenhuis MT, Marthas M, Reay E, Rose TM, Pamungkas J, Bosch ML. Virology. 1999;260:116–124. doi: 10.1006/viro.1999.9794. [DOI] [PubMed] [Google Scholar]
- 8.Wilson CA, Wong S, Muller J, Davidson CE, Rose TM, Burd P. J Virol. 1998;72:3082–3087. doi: 10.1128/jvi.72.4.3082-3087.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rose TM, Strand KB, Schultz ER, Schaefer G, Rankin GW, Jr, Thouless ME, Tsai CC, Bosch ML. J Virol. 1997;71:4138–4144. doi: 10.1128/jvi.71.5.4138-4144.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schultz ER, Rankin GW, Jr, Blanc MP, Raden BW, Tsai CC, Rose TM. J Virol. 2000;74:4919–4928. doi: 10.1128/jvi.74.10.4919-4928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rose TM, Ryan JT, Schultz ER, Raden BW, Tsai C-C. J Virol. 2003;77:5084–5097. doi: 10.1128/JVI.77.9.5084-5097.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiss RA. Nat Med. 1998;4:391–392. doi: 10.1038/nm0498-391. [DOI] [PubMed] [Google Scholar]
- 13.Chomel BB, Belotto A, Meslin FX. Emerg Infect Dis. 2007;13:6–11. doi: 10.3201/eid1301.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan SY, Bernard HU, Ratterree M, Birkebak TA, Faras AJ, Ostrow RS. J Virol. 1997;71:4938–4943. doi: 10.1128/jvi.71.7.4938-4943.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Virology. 2004;324:17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 16.Chan SY, Ostrow RS, Faras AJ, Bernard HU. Virology. 1997;228:213–217. doi: 10.1006/viro.1996.8400. [DOI] [PubMed] [Google Scholar]
- 17.Antonsson A, Hansson BG. J Virol. 2002;76:12537–12542. doi: 10.1128/JVI.76.24.12537-12542.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wood CE, Chen Z, Cline JM, Miller BE, Burk RD. J Virol. 2007;81:6339–6345. doi: 10.1128/JVI.00233-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patterson MM, Rogers AB, Mansfield KG, Schrenzel MD. Comp Med. 2005;55:75–79. [PubMed] [Google Scholar]
- 20.Kloster BE, Manias DA, Ostrow RS, Shaver MK, McPherson SW, Rangen SR, Uno H, Faras AJ. Virology. 1988;166:30–40. doi: 10.1016/0042-6822(88)90143-2. [DOI] [PubMed] [Google Scholar]
- 21.Baines JE, McGovern RM, Persing D, Gostout BS. J Virol Methods. 2005;123:81–87. doi: 10.1016/j.jviromet.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 22.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 23.Rose TM, Schultz ER, Todaro GJ. Proc Natl Acad Sci U S A. 1992;89:11287–11291. doi: 10.1073/pnas.89.23.11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Page RD. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 25.Schneider TD, Stephens RM. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]