Abstract
One major obstacle to membrane protein structure determination is the selection of a detergent micelle that mimics the native lipid bilayer. Currently, detergents are selected by exhaustive screening because the effects of protein-detergent interactions on protein structure are poorly understood. In this study, the structure and dynamics of an integral membrane protein in different detergents is investigated by nuclear magnetic resonance (NMR) and electron paramagnetic resonance (EPR) spectroscopy, and small angle X-ray scattering (SAXS). The results suggest that matching of the micelle dimensions to the protein’s hydrophobic surface avoids exchange processes that reduce the completeness of the NMR observations. Based on these dimensions, several mixed micelles were designed that improved the completeness of NMR observations. These findings provide a basis for the rational design of mixed micelles that may advance membrane protein structure determination by NMR.
Introduction
Integral membrane proteins comprise ≈ 25% of most proteomes and facilitate transport and signaling across cell membranes. Despite their importance, less than 1% of known protein structures are of membrane proteins. One major obstacle to membrane protein structure determination is the selection of detergent that mimics the native lipid bilayer and stabilizes the protein fold.1–5
Detergents are small amphipathic molecules that are used to solubilize membrane proteins for structural and functional investigations. However, unlike phospholipid bilayers, detergents form micelles, which are spheroid and have a core composed of the detergent hydrophobic tails. Micelles have different shapes and sizes depending on the detergent chemical structure. For structural investigations, a multitude of detergents is screened until a condition that provides high quality crystals3 or NMR spectra6 is found. However, a correlation between the physical properties of the detergent micelle and the likelihood of obtaining a membrane protein structure is currently not known.
In this study, we present data on the model polytopic α-helical membrane protein TM0026. TM0026 is a membrane protein of unknown function from the thermophile Thermotoga maritima and was initially characterized as part of the high-throughput structure determination pipeline of the Joint Center for Structural Genomics. 1,7 The data presented demonstrate a correlation between protein conformations, micelle size and thickness, and quality of nuclear magnetic resonance (NMR) spectra. The structure and dynamics of TM0026 in different detergents are investigated by NMR and electron paramagnetic resonance (EPR) spectroscopy, and small angle X-ray scattering (SAXS). The results suggest that matching of the micelle dimensions to the protein’s hydrophobic surface avoids exchange processes that reduce the completeness of the NMR observations. Based on these observations, mixed micelles are designed that improve the completeness of NMR observations. These findings provide a basis for the rational design of mixed micelles that have the potential to advance membrane protein structure determination.
Experimental Section
Cloning, expression, and purification
N-terminal His-tagged TM0026 was cloned as previously published.1 Individual cysteine mutants were produced using standard PCR protocols. Protein expression was performed with LB media containing 1% glycerol (v/v), and 50 mg/mL ampicillin. Expression was induced by the addition of 0.20% arabinose for 3 h. For deuterated, 15N-labeled proteins, published protocols using conventional shakers and minimal media in D2O supplemented with 15NH4Cl were used. TM0026 was purified in each detergent (decyl maltoside, DM; dodecyl maltoside, DDM; decylphosphocholine, FC-10; and dodecylphosphocholine, FC-12 from Anatrace, Inc, Maumee, OH and 1-palmitoyl-2-hydroxy-sn-glycero-3-[phospho-rac-(1-glycerol)], LPPG; and 1,2-dihexanoyl-sn-glycerophosphocholine, DHPC from Avanti Polar Lipids, Inc, Alabaster, AL) as previously described using Co2+-affinty chromatography. For cysteine mutants, the lysis and purification buffers contained 0.2 mM TCEP.
TM0026 in the mixed micelles was prepared differently for the two different mixtures investigated. Since TM0026 was soluble in FC-10, TM0026 was purified in FC-10 as above and DDM was titrated into the NMR tube to produce the desired ratio. The feasibility of this strategy indicates that the disruption of the protein structure in FC-10 is reversible and that protein samples may be “rescued” by titrating an appropriate detergent. TM0026 was previously determined to be insoluble in DHPC and yields were low in LPPG1; therefore, TM0026 was eluted from the Co2+-affinty column with the desired ratio of DHPC/LPPG.
Detergent concentrations were measured by 1D 1H NMR by comparison with samples of known detergent concentrations. Protein concentrations were measured by UV absorbance at 280 nm in 6 M guanidine hydrochloride and by BCA protein assay (Pierce, Rockford, IL). Detergent solutions for SAXS measurements were prepared in 20 mM phosphate buffer (pH 6.2) and 150 mM NaCl.
Spin labeling of TM0026 cysteine mutants
Immediately before spin labeling, TCEP and imidazole were removed using a PD-10 desalting column (GE Healthcare Bio-Sciences Corp., Piscataway, NJ) using a 20 mM phosphate buffer (pH 6.2) with 150 mM NaCl and either 5 mM DM, 5 mM DDM, or 15 mM FC-10. Mutant proteins (typically ≈ 20 µM) were incubated with (1-Oxyl-2,2,5,5-tetramethyl-3-pyrroline-3-methyl)-methanethiosulfonate (MTSL; a gift from Wayne Hubbell, UCLA, and Kalman Hideg, University of Pécs) at a 1:5 molar ratio of protein to label. The reaction was allowed to proceed at room temperature overnight. Protein solutions were then concentrated and unreacted spin label was removed with a PD-10 desalting column using a 20 mM phosphate buffer (pH 6.2) with 150 mM NaCl and either 5 mM DM, 5 mM DDM, or 15 mM FC-10. The protein eluent was concentrated and spectra were recorded.
NMR spectroscopy
NMR samples were prepared as described previously.1 The detergent concentrations ranged between 100 – 150 mM and protein concentrations were ≈ 0.5 mM. All NMR data were recorded at 313 K on a 600 MHz Bruker Avance spectrometer. 2D 15N, 1H-TROSY spectra were recorded with 128 transients per increment, t1max(15N) = 42 ms, t2max(1H) = 285 ms, and a time domain data size of 64(t1) X 2048( t2) complex points. For the sequence-specific resonance assignments of the polypeptide backbone atoms, the following experiments were recorded: 15N,1H-TROSY, TROSYHNCA, and 15N-resolved 1H,1H-NOESY (τm = 200 ms). Using the backbone assignment and the 15Nresolved 1H,1H-NOESY the protein-detergent complex was modeled based on the observed NOEs between the protein amide protons and the alkyl chains of the detergent.
EPR spectroscopy
EPR spectra were recorded on a Varian E-109 spectrometer fitted with a two-loop one-gap resonator.8 Protein samples of 5 µL (≈ 100 µM) were loaded in Pyrex capillaries (0.84 mm o.d. × 0.6 mm i.d.) sealed on one end. All spectra were acquired using a 2 mW incident microwave power. The modulation amplitude at 100 kHz was optimized for each spectrum to avoid spectral distortion. All spectra were normalized to the same area. All labeled mutants lacked spin-spin interaction indicating that the protein is monomeric in all detergent conditions in agreement with previously published results.1
SAXS
SAXS data were recorded on beam line BESSERC CAT 12-ID at the Advanced Photon Source, employing a 2 m sample-detector distance and a CCD detector read out. The measurements were performed at a photon energy of 12 keV using a custom-made cell9. For each data point, a total of three measurements of 0.5 sec integration time were recorded. Data were image-corrected and circularly averaged; the three profiles for each condition were averaged to improve signal quality. Buffer profiles were collected using identical procedures and subtracted for background correction. We tested for possible radiation damage by comparing subsequent exposures of the same sample, and no change was detected.
Modeling of TM0026 in micelles
The protein backbone atoms were assigned using HNCA and HN(CO)CA 3D spectra.10 The DDM/FC-10 mixed micelle condition, where 59 out of 68 residues could be assigned (M1-A5, P31, P46, K57, and K58 were not assigned), allowed assignment of eleven additional residues compared to DM. Using the paramagnetic relaxation enhancement (PRE) measurements from A13R1 (in combination with the backbone assignment) several low resolution models of the protein structure were calculated.11–13 A conformation from one of these calculations was used to model the protein – detergent complex. High-resolution structure determination is in progress and will be published elsewhere. The micelles were approximated using the head group – head group spacing and the aggregation number determined by SAXS and the 3D 15N-editted NOESY spectrum (Figure S9), which provided the amino acids that were within 5 Å of the alkyl chain of the detergent molecules. For the FC-10 micelle, the α-helices were moved as rigid bodies to match the hydrophobic surface area to the micelle dimensions and the termini were exposed to aqueous solvent. Pymol (www.pymol.org) was used to build the protein detergent models and render all protein Figures.14
Results and Discussion
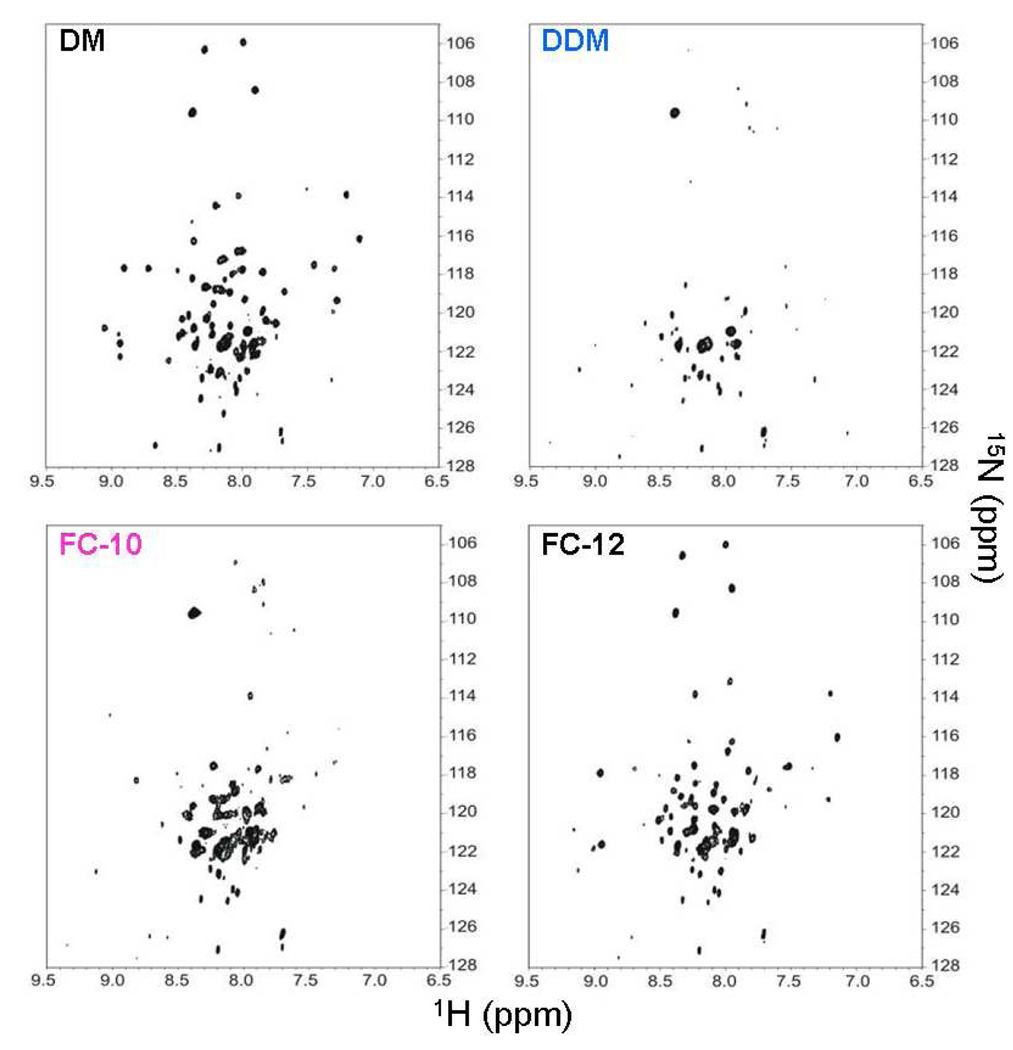
TM0026 contains two transmembrane α-helices and was found to be α-helical, monodisperse, and monomeric in four different detergents: DM, DDM, FC-10, and FC-12.1 Despite these similarities, the NMR signals were drastically different as assessed by the 15N, 1H-TROSY spectra (Figure 1).1 For the detergents DM and FC-12, 51 of 66 expected cross peaks were observed while for the detergents FC-10 and DDM, only 32 and 36 cross peaks were observed, respectively. When observable, the cross peaks have similar chemical shifts in each detergent micelle suggesting that the protein structures are similar in the different detergents (Figure 1). The remainder of the expected cross peaks is not observed in DDM and FC-10 because of extensive line broadening. The observed line broadening, which impedes NMR structure determination, is attributed to conformational heterogeneity and exchange processes and not to aggregation, unfolding, or to the overall size of the protein-detergent complex.1,15
Figure 1.
Soluble protein-detergent complexes differ in conformational heterogeneity. The NMR 15N,1H-TROSY spectra of 2H,15N-labeled TM0026 in DM, DDM, FC-10, and FC-12 are shown.
Structural changes of TM0026 in different detergents
To investigate the physical origin of the line broadening, site-directed spin labeling (SDSL) was employed to study the structure and dynamics of TM0026 in the different detergent conditions. A nitroxide probe (R1, Figure S1) was introduced at four sequential sites (Figure 2). These four residues were chosen because they compose a full α-helical turn at the center of the first hydrophobic sequence and at least one of the residues is likely to be involved in tertiary interactions with the second α-helix. The introduction of the nitroxide side chain does not significantly perturb the overall structure of TM0026 as the 15N, 1H-TROSY spectra of the R1’ (a diamagnetic analog) labeled protein is identical to wild type (Figure S2). The spectral parameters, ΔHpp and 2Azz (Figure 2), provide an assessment of mobility (the central line width, ΔHpp, is determined by the g-tensor anisotropy; that is, nitroxide mobility modulates the averaging of the g-tensor elements and reduction in mobility resolves the anisotropies and the line width is broadened) and can be used to determine the topology16 and backbone dynamics of the α-helix17.
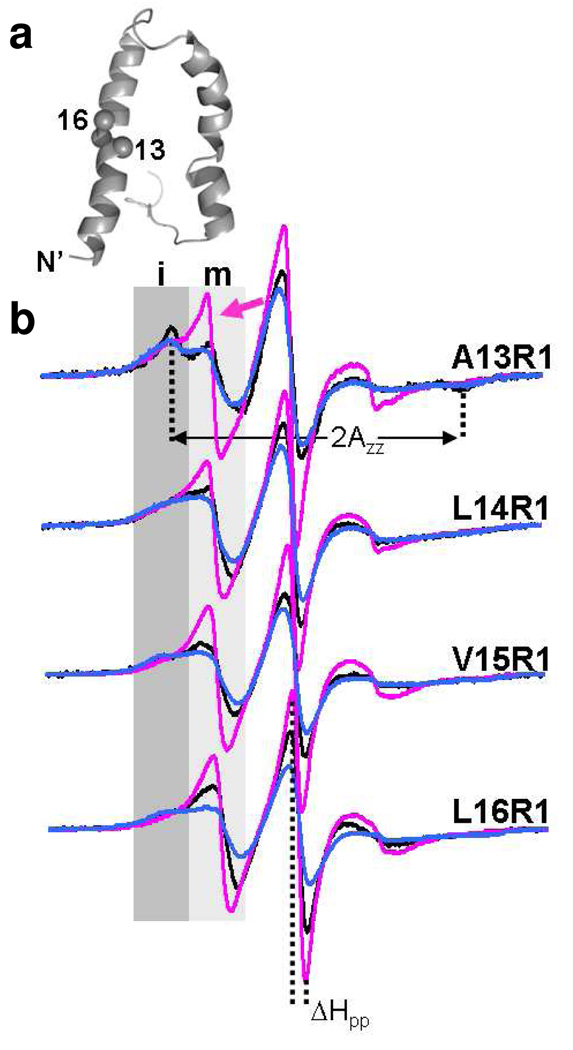
Figure 2.
Effects of the detergent on protein structure and dynamics. (a) A ribbon model of TM0026 with the spin labeled residues A13 – L16 represented by spheres (see Methods section for modeling). (b) The EPR spectra of R1 at residues A13 – L16 in DM, DDM, and FC-10 are shown and colored according to the labels in Figure 1. Spectral intensities in regions labeled i (dark gray) and m (light gray) identify relatively immobile and mobile components, respectively. The arrow indicates a dynamic population observed in FC-10. The semi-quantitative measurements, ΔHpp and 2Azz, are indicated.
In DM, the EPR spectra of residues L14 – L16 are very similar to those observed for R1 at lipid/detergent exposed sites.18,19 In contrast, the EPR spectral lineshape of A13R1 represents a highly restricted nitroxide side chain, based on the evaluation of ΔHpp and 2Azz (Figure 2 and Table S1), indicating a direct interaction with the second transmembrane α-helix. In contrast, in FC-10 the EPR spectrum of A13R1 has two components indicating that R1 samples two conformations. One spectral component is similar to that observed in DM and the other represents a more mobile R1 (Figure 2) and is similar to spectra observed in highly dynamic sequences. 17,20 An increase in mobility for a fraction of the protein population is observed at each labeled position throughout the α-helical turn. These data suggest that the tertiary interaction between the two α-helices is lost in a substantial fraction (≈ 10–20%) of the protein population. The structural heterogeneity and potential exchange between the two populations provide a possible explanation for the observed NMR line broadening in FC-10 detergent micelles. In DDM, the tertiary contact at A13R1 is maintained (Figure 2). However, compared to DM, the EPR spectra for the lipid exposed residues exhibit extensive line broadening as assessed by ΔHpp (the most dramatic difference is observed at L16R1) (Table S1). These data demonstrate that the structure of TM0026 is perturbed in FC-10 and DDM; in order to understand how the detergent influences the protein structure, the characteristics of the detergents were investigated.
Comparison of DM, FC-10, DDM, and FC-12 micelles
A comparison of the detergent properties (Table 1, Figure S3 and Table S2) indicates that the detergents that produce resolved NMR spectra (DM, FC-12) do not have similar head groups, ionic properties, or alkyl chain lengths. To characterize the micelles formed by the different detergents, we have determined their shape and size using SAXS. In particular, we obtain the aggregation number (N) and hydrophobic core volume (VHC) from fits of a twocomponent spheroid model, and the dominant head group separation (L, the distance between the head groups centers across the short diameter of the spheroid) from the position of the second maximum in the scattering intensity (see reference 21 and Supplementary Material Figure S4). We find that DM and FC-12 form micelles of similar size with respect to N, VHCM, and L (Table 1).21 In contrast, the detergents for which poor NMR spectra were observed form either smaller and thinner (FC-10) or larger and thicker (DDM) micelles compared to DM and FC-12.
Table 1.
Properties of select detergents.
| Detergent (abbreviation) | CMC (mM) | Na | Lb (Å) | rHCc (Å) | VHCd (nm3) |
|---|---|---|---|---|---|
| n-decyl-β-D-maltoside (DM) | 1.8 | 80–90 | 34 | 12.5, 23 | 26 |
| n-dodecyl-β-D-maltoside (DDM) | 0.17 | 135–145 | 39 | 14, 29 | 49 |
| n-decylphosphocholine (FC-10) | 11 | 45–53 | 28 | 21, 13 | 15 |
| n-dodecylphosphocholine (FC-12) | 1.5 | 65–80 | 34 | 26, 16 | 27 |
| 1,2-dihexanoyl-sn-glycerophosphocholine (DHPC) | 15 | 30–35 | 23 | 21, 10 | 8.8 |
| 1-palmitoyl-2-hydoxy-sn-glycero-3-[phospho-rac-(1-glycerol)] (LPPG) | 0.018 | 160–170 | 46 | 19, 30 | 72 |
| DDM:FC-10 (molar ratio 4:7) | NDe | 77 (30/47) | 34 | 14, 21 | 26 |
| DHPC:LPPG (molar ratio 1:1) | NDe | 80 (40/40) | 35 | 17, 20 | 29 |
Aggregation number determined by SAXS21
The dominant distance from head group to head group across the short dimension of the micelle core.
Radii defining spheroid with one axis a and two other axes b.
Hydrophobic volume calculated using rHC
Not determined.
Dimensions of mixed micelles
In order to further probe the relationship between micelle size and thickness and membrane protein conformational homogeneity, it is desirable to be able to systematically influence micelle geometries. Therefore, we explored whether engineering mixed micelles by mixing detergents at different ratios might be a way to systematically change the size and shape of detergent micelles. To this end, we obtained SAXS data for a comprehensive set of mixed micelles, including detergents with varying hydrophobic tails and head groups, including non-ionic, zwitterionic, and ionic species (Figure 3, Figure S4, Figure S5, and Figure S6 and Table S2 and Table S3). The dependence of L (determined from the position of the second peak in the scattering intensity) on the mixed micelle composition for two detergents A and B was fit by the relationship
| (1) |
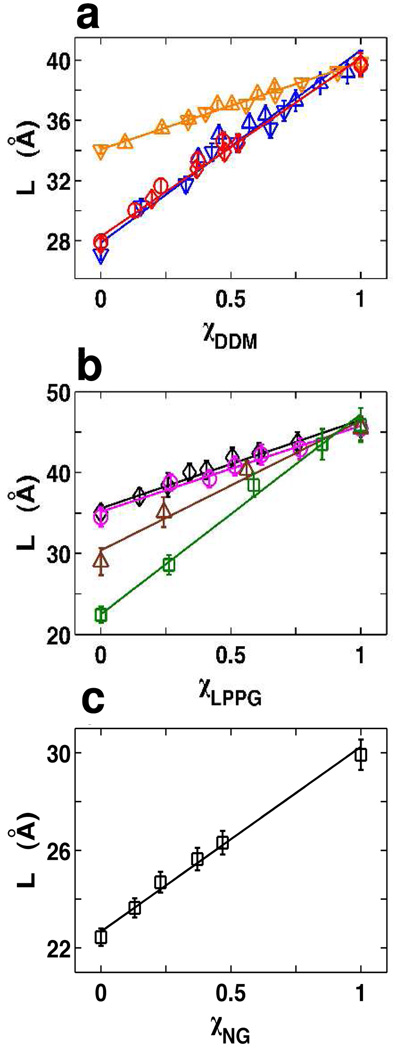
with the mixing ratio χA = ([A])/([A]+[B]) (Figure 3, solid lines). [A], [B] are the detergent concentrations that have been corrected for the monomeric detergent concentration using the relation Xi,monomer ≈ XiCMCi, where Xi and CMCi are the mole fraction and critical micelle concentration of detergent species i.22 The linear dependence of L on the mixing ratio χ appears to hold for a wide range of detergent mixtures (Figure 3). The data set includes mixtures of detergents that only differ by their alkyl chain length (DDM/DM mixtures), that feature different non-ionic head groups (DDM/OG mixtures, a mono- and a disaccharide), and combinations of non-ionic and zwitterionic head groups (DDM/FC-10 and NG/DHPC mixtures), as well as combinations involving an ionic detergent species (LPPG). Furthermore, the relationship of thickness on mixing ratio is largely independent on whether the concentration of detergent A was held fixed and varying concentrations of B were added or whether the reverse strategy was employed. Finally, the relationship seems to be largely independent of the total (absolute) concentration of detergents employed (data not shown), in agreement with the finding that the position of the second peak is independent of detergent concentration for micelles formed by a single detergent species.21
Figure 3.
The micelle thickness L for mixed micelles depends linearly on the mixing ratio. Micelle thickness as a function of mixing ratio determined from SAXS analysis (symbols) and fits to the linear model (equation 1, solid lines). (a) Mixed micelles of DDM mixed with octylglucoside (blue triangles), DM (orange triangles), or FC-10 (red circles). (b) Mixed micelles of LPPG mixed with FC-12 (black diamonds), DM (magneta circles), FC-10 (brown triangles), and DHPC (green squares). (c) Mixed micelles of nonylglucoside and DHPC (black squares).
Design of mixed micelles for NMR structure determination
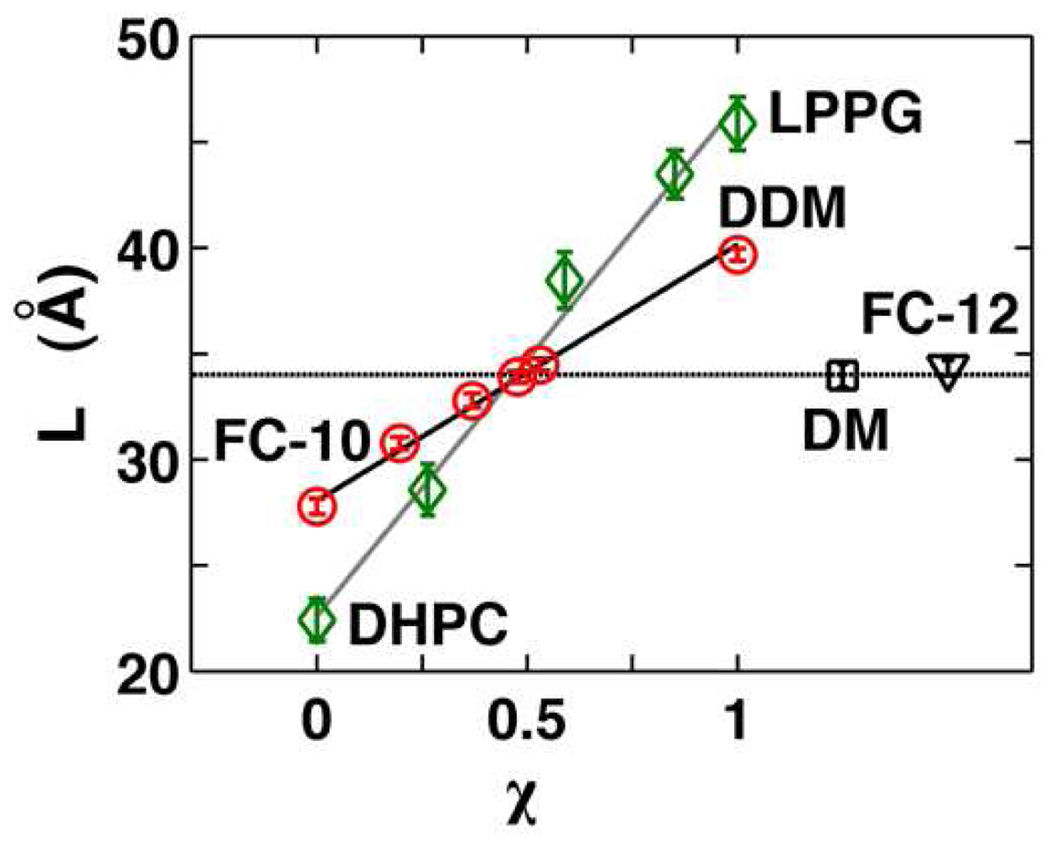
The linear dependence of the characteristic micelle thickness on detergent mixing ratios (Equation 1) provides a straight forward method to engineer micelles of a particular thickness by mixing detergents. We explored this strategy to test how micelle size and thickness influence protein structure in the context of TM0026. Two mixtures were pursued further for NMR structure determination: DDM/FC-10 and LPPG/DHPC. The DDM/FC-10 pair was chosen as an extension of the four detergents investigated with NMR and SDSL. The LPPG/DHPC mixture was chosen because TM0026 in the individual pure micelles was either insoluble (DHPC) or yielded very poor NMR spectra (LPPG)1 and, therefore, was a test of the general applicability of the method. Both DDM/FC-10 and LPPG/DHPC micelles have dimensions L, as well as, VHC and N, similar to DM and FC-12 at mixing ratios of χDDM or χLLPG ≈ 0.4–0.5 (Figure 4, Table 1 and Table S3).
Figure 4.
Optimization of mixed micelles for TM0026 NMR measurements. Characteristic micelle thickness L as a function of the mixing ratio of the detergents χ for DDM/FC-10 (red circles) and LPPG/DHPC (green squares) mixed micelles. A dashed line is drawn at the L value measured for DM and FC-12 (34 Å).
TM0026 in mixed micelles with a matched L parameter
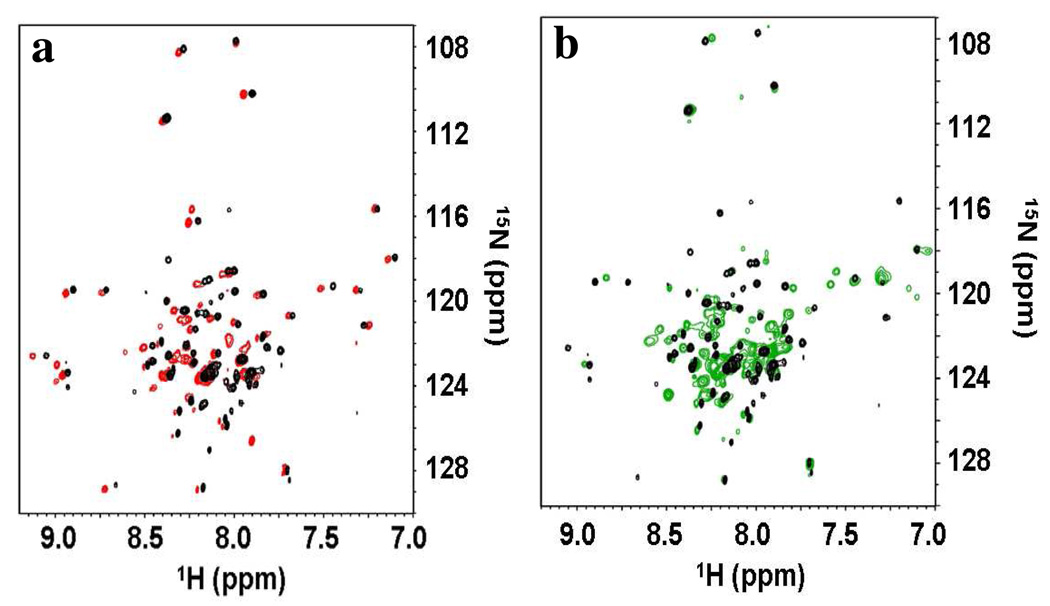
Figure 5 shows 15N, 1H-TROSY spectra of TM0026 in DDM/FC-10 and LPPG/DHPC mixed micelles with thicknesses engineered to match the thickness of pure FC-12 or DM micelles, 34 Å. The 15N, 1H-TROSY spectra of TM0026 in these mixed micelles are similar (with respect to both chemical shift and line broadening) to the spectra in DM and FC-12, indicative of an identical global fold (Figure 5). The EPR spectra of A13R1– L16R1 in the DDM/FC-10 mixed micelle were identical to those observed in DM suggesting similar tertiary interactions (Figure S7). In fact, the quality of the NMR spectra in DDM/FC-10 mixed micelles was such that several additional cross peaks were observed, which facilitated the assignment of eleven additional residues compared to DM, providing a more complete backbone assignment.
Figure 5.
NMR 15N,1H-TROSY spectra of 2H, 15N-labeled TM0026 in (a) FC-10/DDM mixed micelle (χddm ≈ 0.4, red) and (b) DHPC/LPPG mixed micelle (χlppg ≈ 0.5, green). In both panels, the spectrum of TM0026 in DM is shown in black.
Protein – detergent interactions
NMR NOEs and chemical shift perturbation mapping23,24 have been widely used to investigate protein – protein and protein – ligand interactions and can provide information about protein – detergent interactions. The backbone assignments of TM0026 in DM and DDM/FC-10 allow a direct comparison of the chemical shifts in each detergent condition. The chemical shift differences (Δδ = √((ΔδN/5)2 +(ΔδH)2)) between the DM and DDM/FC-10 detergent conditions were calculated and, as expected from the overall similarity of the spectra, the majority of the chemical shift differences (Figure S8) are small (Δδ < 0.4 ppm, the average chemical shift difference). However, two regions of the protein exhibit greater differences in the chemical shifts: L48-R54 and E61-R68 (the protein sequence is shown in Figure S9). The variability in the chemical shifts in the C-terminus (E61-R68) might stem from different interactions of the highly charged C-terminus with the zwitterionic FC-10 headgroups compared to the neutral DM head groups. The residues C-terminal of P46 in the second transmembrane helix (L48-R54) exhibit an increased variability in chemical shift. It is plausible that the proline residue results in a kink that uncouples the motion of these residues from the N-terminal region of the helix. The C-terminal region may adopt slightly different conformations in DM and FC-10/DDM micelles, possibly coupled to the interactions of the C-terminus.
To investigate the protein conformation in the other detergent conditions, the backbone assignments in DM and DDM/FC-10 micelles were compared to the chemical shifts observed in the 15N, 1H-TROSY spectra of detergent conditions in which line broadening impeded obtaining 3D spectra for assignments and structure determination. Assuming that cross-peaks with similar chemical shifts are the same in the different detergent conditions, the observable resonances in DDM and FC-10 are distributed throughout the protein sequence. Cross-peaks in the entire C-terminus and both α-helices were observed in all detergent conditions consistent with previous evidence that TM0026 is α-helical, monomeric, and not aggregated in DDM and FC-101 and that the observed line broadening is due to exchange processes. However, resonances in the loop, at the C-terminal end of the first α-helix and the tertiary contact (A13 determined from the EPR data) were not identified and/or observed in DDM and FC-10 detergent conditions. Therefore, the exchange effects contributing to line broadening do not seem to be localized to one region of TM0026.
In addition, the interactions between the protein and detergents can be mapped using the 15Nresolved 1H,1H-NOESY for the DM and DDM/FC-10 mixed micelles (Figure S9). 25,26 For DDM/FC-10, NOEs with the detergent alkyl chain were observed for both transmembrane α-helices (from L7 – R27 and F34 – V53) and rapid exchange with water was observed for residues in the loop and C-terminus. The cartoon of TM0026 in Figure 6 is colored according to the observed NOEs between the backbone and the alkyl chain of the detergents. A similar trend was observed with DM; however, the assignment was not as comprehensive (see Methods).
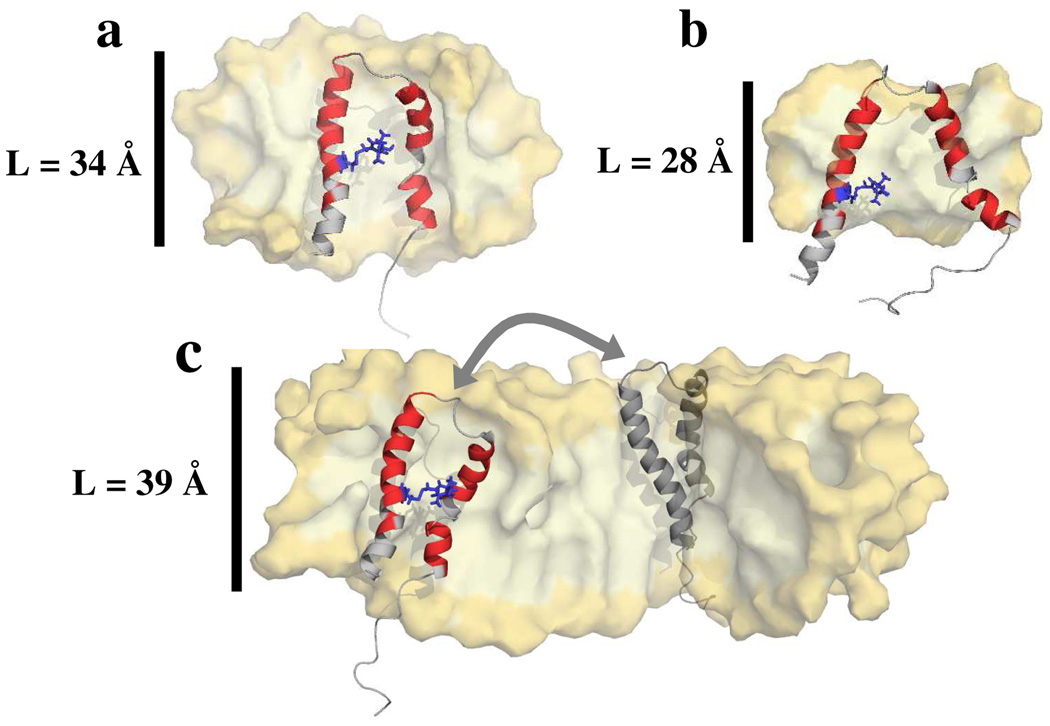
Figure 6.
Models of matching micelle size and shape to the hydrophobic surface of TM0026. (a) A model demonstrating the hydrophobic matching between the hydrophobic dimensions of the protein (hydrophobic length of the α-helices is ≈ 32 Å) and the detergent alkyl chains such as in the case of DM, FC-12, and the 4:7 DDM/FC-10 mixed micelle. (b) A model demonstrating a hydrophobic mismatch between the surface area of the protein and the micelles which are too small, such as in the case of FC-10, causing the α-helices to separate in order to bury more surface area in the interior of the micelle. (c) A possible model for the heterogeneity observed in the DDM larger micelle. The tertiary fold of the protein is maintained but, there are many regions of the micelle which may accommodate the protein. Another protein molecule is shown in gray for which the hydrophobic surface area of the protein is buried within the micelle and an arrow indicates exchange between the two conformers. Approximately 25% of the detergent micelle is removed in order to view the protein and the interior of the micelle. The micelle is rendered as a surface and colored yellow. The protein is displayed as a cartoon model with the residues for which NOEs between the amide proton and the alkyl chain of detergent molecules are observed are colored red and the residues that were unassigned or lacked NOEs with the detergent are colored gray. The A13R1 side chain is rendered as blue sticks. The dominant head group separation L is labeled in each panel.
The influence of micelle dimensions on protein conformations
The results for TM0026 proteindetergent complexes suggest that the protein conformations are strongly influenced by the size and thickness of the detergent micelle. In interpreting these results, we need to consider that the aggregation number N can be different for “empty” micelles and micelles in protein-detergent complexes.15 Similarly, we expect the micelle thickness L to show some variability in the presence of a protein due to the fluid nature of detergents micelles. Nonetheless, the characteristic head group – head group dimension L measured for micelles in the absence of proteins appears to reflect an intrinsic packing preference of the detergent and we observe a similar thickness in TM0026 complexes that yield well resolved NMR spectra (Figure S6).
Taken together, the results suggest that well resolved NMR spectra are observed when the hydrophobic component of L (which is equal to L minus the thickness of the head group, ≈ 29–31 Å; see Table S3 and Figure S5c) matches the length (≈ 30–33 Å) of the hydrophobic part of the α-helices of TM0026 (Figure 6a). We note that this hydrophobic dimension matches the average hydrophobic thickness of the T. maritima bilayer (≈ 30 Å27,28). In the case of the FC-10 micelle, L is too small (Table 1). A possible explanation for the observed loss of tertiary interaction and structural heterogeneity in FC-10 is that the mismatch between the hydrophobic surface and the micelle dimensions may result in a perturbation of the protein structure to avoid hydration of the hydrophobic α-helices (Figure 6b). This structural perturbation is reminiscent of that observed in the case of “hydrophobic mismatch” observed in lipid bilayers29 and inter-helical packing disruption observed in detergents30–32. For the case of DDM, the dominant head group separation is larger than the length of the hydrophobic α-helices, but there is sufficient hydrophobic core volume to accommodate the protein hydrophobic surface area. The protein tertiary structure is maintained in DDM, but line broadening in both the NMR and EPR suggest structural heterogeneity and/or conformational exchange. One of many possible explanations is exchange between protein conformers (for which the tertiary structure is maintained) within a micelle (Figure 6c). For instance, assuming the protein does not perturb the micelle, the protein cannot transverse the center of the micelle. Instead, the protein may pack in regions of the large micelle that better match the hydrophobic regions of the protein. These allowed environments may be heterogenous and/or two or more conformers could be in exchange, which would give rise to the observed line broadening in the NMR and EPR spectra. More experiments need to be performed to understand the effects of the detergent on the protein structure that give rise to the observed line broadening in the DDM micelle.
Conclusion
Using NMR, EPR, and SAXS, we demonstrate a correlation between the dimensions of detergent micelles and the structure of a membrane protein, which leads to the rational design of mixed micelles that facilitates NMR structure determination. In particular, we found that the micelle thickness of mixed micelles depends linearly on the mixing ratio for a significant range of detergent mixtures and that this thickness appears to correlate strongly with the quality of NMR observations.
The generality of the dependence of tertiary contact stability on micelle thickness is difficult to assess at this point, as only very few polytopic membrane protein NMR structures have been determined (7 structures, out of which only 1 is α-helical).33 However, our observations are in qualitative agreement with previous findings. The structure of the Glycophorin A dimer (with a similar hydrophobic thickness as TM0026) was solved in FC-12.34 In sodium dodecyl sulfate (SDS), which has a slightly larger L values of ≈ 36 Å (J.L. and S.D., unpublished results), dimerization is still possible, but significantly reduced compared to FC-12.31 Therien and Deber found that a helix-loop-helix construct from cystic fibrosis transmembrane conductance regulator, which has a shorter hydrophobic thickness compared to Glycophorin A and TM0026, dimerizes in micelles with short alkyl chains, while the dimer interaction is predominantly lost in SDS and other micelles of similar thickness.30
Future work will explore whether similar relationships can be observed for different membrane proteins varying by size (in particular larger helical bundles with larger hydrophobic surface area), fold, and origin. Likely, other micelle properties (such as the total micelle volume) will have to be taken into account to fully understand protein-detergent interactions.
Nonetheless, our data suggests that, rather than exhaustively screening a multitude of detergents, it might be possible to rationally engineer appropriate mixed micelles for NMR structure determination following simple principles and from a limited set of detergents.
Supplementary Material
Acknowledgments
We thank Prof. Wayne L. Hubbell for the use of a Varian EPR spectrometer, Prof. Kurt Wüthrich for support, Drs. Gerard Kroon, Bernhard Geierstanger, and David Jones for helpful discussion and technical NMR assistance, Dr. Cameron Mura for useful discussions, and Vincent Chu, Dmitri Pavlichin, and Dr. Sönke Seifert for data collection assistance at the APS. Support for this research was provided by the National Institutes of Health grants: 1F32GM068286 and The Jeffress Memorial Trust (LC), PO1 GM0066275 (SD) and, Protein Structure Initiative grants, P50 GM62411 and U54 GM074898 (SL). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. A.Y.L.S. is supported by A*STAR, Singapore.
Footnotes
Supporting Information Available. Further details of the SDSL, SAXS, and NMR results. The material is available free of charge via the Internet at http://pubs.acs.org/.
REFERENCES
- 1.Columbus L, Lipfert J, Klock H, Millett I, Doniach S, Lesley SA. Protein Sci. 2006;15:961–975. doi: 10.1110/ps.051874706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vinogradova O, Sonnichsen F, Sanders CR., 2nd J Biomol NMR. 1998;11:381–386. doi: 10.1023/a:1008289624496. [DOI] [PubMed] [Google Scholar]
- 3.Wiener MC. Methods. 2004;34:364–372. doi: 10.1016/j.ymeth.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 4.Bowie JU. Curr Opin Struct Biol. 2001;11:397–402. doi: 10.1016/s0959-440x(00)00223-2. [DOI] [PubMed] [Google Scholar]
- 5.Poget SF, Girvin ME. Biochim Biophys Acta. 2007;1768:3098–3106. doi: 10.1016/j.bbamem.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanders CR, Sönnichsen F. Magn. Reson. Chem. 2006;44:S24–S40. doi: 10.1002/mrc.1816. [DOI] [PubMed] [Google Scholar]
- 7.Lesley SA, Kuhn P, Godzik A, Deacon AM, Mathews I, Kreusch A, Spraggon G, Klock HE, McMullan D, Shin T, Vincent J, Robb A, Brinen LS, Miller MD, McPhillips TM, Miller MA, Scheibe D, Canaves JM, Guda C, Jaroszewski L, Selby TL, Elsliger MA, Wooley J, Taylor SS, Hodgson KO, Wilson IA, Schultz PG, Stevens RC. Proc Natl Acad Sci U S A. 2002;99:11664–11669. doi: 10.1073/pnas.142413399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hubbell WH, Froncisz W, Hyde JS. Rev. Sci. Instrum. 1987;58:1879–1886. [Google Scholar]
- 9.Lipfert J, Millett IS, Soenke S, Doniach S. Rev. Sci. Instrum. 2006;77 046108. [Google Scholar]
- 10.Bax A, Grzesiek S. Acc. Chem. Res. 1993;26:131–138. [Google Scholar]
- 11.Battiste JL, Wagner G. Biochemistry. 2000;39:5355–5365. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- 12.Schwieters CD, Kuszewski JJ, Clore GM. Prog. Nucl. Magn. Reson. Spectrosc. 2006;48:47–62. [Google Scholar]
- 13.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 14.DeLano WL, DeLano Scientific: Palo Alto. 2002 [Google Scholar]
- 15.Lipfert J, Columbus L, Chu VB, Doniach S. J. Appl. Crystallogr. 2007;40:229–234. [Google Scholar]
- 16.Hubbell WL, Gross A, Langen R, Lietzow MA. Curr. Opin. Struct. Biol. 1998;8:649–656. doi: 10.1016/s0959-440x(98)80158-9. [DOI] [PubMed] [Google Scholar]
- 17.Columbus L, Hubbell WL. Trends Biochem. Sci. 2002;27:288–295. doi: 10.1016/s0968-0004(02)02095-9. [DOI] [PubMed] [Google Scholar]
- 18.Altenbach C, Marti T, Khorana HG, Hubbell WL. Science. 1990;248:1088–1092. doi: 10.1126/science.2160734. [DOI] [PubMed] [Google Scholar]
- 19.Gross A, Columbus L, Hideg K, Altenbach C, Hubbell WL. Biochemistry. 1999;38:10324–10335. doi: 10.1021/bi990856k. [DOI] [PubMed] [Google Scholar]
- 20.Columbus L, Hubbell WL. Biochemistry. 2004;43:7273–7287. doi: 10.1021/bi0497906. [DOI] [PubMed] [Google Scholar]
- 21.Lipfert J, Columbus L, Chu VB, Lesley SA, Doniach S. J Phys Chem B. 2007;111:12427–12438. doi: 10.1021/jp073016l. [DOI] [PubMed] [Google Scholar]
- 22.Tanford C. The hydrophobic effect: formation of micelles and biological membranes. 2nd ed. New York: Wiley; 1980. [Google Scholar]
- 23.Gao G, Williams JG, Campbell SL. Methods Mol Biol. 2004;261:79–92. doi: 10.1385/1-59259-762-9:079. [DOI] [PubMed] [Google Scholar]
- 24.Zuiderweg ER. Biochemistry. 2002;41:1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]
- 25.Fernandez C, Hilty C, Wider G, Wüthrich K. Proc Natl Acad Sci U S A. 2002;99:13533–13537. doi: 10.1073/pnas.212515099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee S, Kim Y. FEBS Lett. 1999;460:263–269. doi: 10.1016/s0014-5793(99)01346-0. [DOI] [PubMed] [Google Scholar]
- 27.Eshaghi S, Niegowski D, Kohl A, Martinez Molina D, Lesley SA, Nordlund P. Science. 2006;313:354–357. doi: 10.1126/science.1127121. [DOI] [PubMed] [Google Scholar]
- 28.Lunin VV, Dobrovetsky E, Khutoreskaya G, Zhang R, Joachimiak A, Doyle DA, Bochkarev A, Maguire ME, Edwards AM, Koth CM. Nature. 2006;440:833–837. doi: 10.1038/nature04642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Killian JA. Biochim Biophys Acta. 1998;1376:401–415. doi: 10.1016/s0304-4157(98)00017-3. [DOI] [PubMed] [Google Scholar]
- 30.Therien AG, Deber CM. J Biol Chem. 2002;277:6067–6072. doi: 10.1074/jbc.M110264200. [DOI] [PubMed] [Google Scholar]
- 31.Fisher LE, Engelman DM, Sturgis JN. J Mol Biol. 1999;293:639–651. doi: 10.1006/jmbi.1999.3126. [DOI] [PubMed] [Google Scholar]
- 32.Bowie JU. Nature. 2005;438:581–589. doi: 10.1038/nature04395. [DOI] [PubMed] [Google Scholar]
- 33.Raman P, Cherezov V, Caffrey M. Cell. Mol. Life Sci. 2006;63:36–51. doi: 10.1007/s00018-005-5350-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacKenzie KR, Engelman DM. Proc Natl Acad Sci U S A. 1998;95:3583–3590. doi: 10.1073/pnas.95.7.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.