Abstract
Interleukin 17 (IL-17)-producing CD4+ T (TH-17) cells share a developmental relationship with FoxP3+ regulatory T (Treg) cells. Here we show that a TH-17 population differentiates within the thymus in a manner influenced by self-antigen recognition, and by the cytokines IL-6 and transforming growth factor (TGF)-β. Like previously described TH-17 cells, TH-17 cells that develop in the thymus expressed the orphan nuclear receptor RORγt and the IL-23 receptor. These cells also expressed α4β1 integrins and the chemokine receptor CCR6, and were recruited to the lung, gut, and liver. In the liver these cells secreted IL-22 in response to self-antigen and mediated host protection during inflammation. Thus, TH-17 cells, like Treg cells, can be selected by self-antigens in the thymus.
Introduction
CD4+ T helper (TH) cells play a major role in orchestrating immune responses. For nearly 20 years, the TH differentiation paradigm consisted of two mutually exclusive pathways, TH1 and TH2, which were defined by expression of distinct transcription factors and cytokines1. TH1 cells express the transcription factor T-bet and produce interferon (IFN)-γ, which directs cell mediated immunity by promoting clearance of intracellular pathogens2, and is responsible for delayed type hypersensitivity reactions. TH2 cells express the transcription factor GATA-3 and produce IL-4, IL-5 and IL-13, which facilitate host protection against extracellular parasites and are major effectors in atopic disease2. More recently, an IL-17 producing subset (TH-17) was recognized as a third CD4+ T cell effector lineage that plays a critical role during the early innate phase of the immune response3,4. TH-17 cells promote the clearance of extracellular pathogens by recruiting neutrophils, and by inducing production of anti-microbial proteins and inflammatory factors from resident cells.
TH-17 cells express the transcription factors RORγt5 (http://www.signaling-gateway.org/molecule/query?afcsid=A002302) and RORα6, produce IL-227 and IL-17F8 in addition to IL-17 (IL-17A), and express the IL-23 receptor (IL-23R)9. Early work established IL-23 as a survival factor for TH-17 cells4, and showed that in vitro differentiation of TH-17 cells unexpectedly proceeded via a developmental pathway partially shared with the anti-inflammatory FoxP3+ regulatory T cell (Treg) population. Activating naïve T cells in vitro in the presence of transforming growth factor (TGF)-β (http://www.signaling-gateway.org/molecule/query?afcsid=A002271) alone promotes development of Treg cells, and the addition of IL-6 diverts differentiation to the TH-17 lineage10–12. Further supporting a relationship between these subsets, IL-2--a cytokine necessary for Treg survival13--constrains TH-17 cell differentiation; even in the presence of TGFβ and IL-6, IL-2 suppresses TH-17 development and expands Treg cell populations14. Inflammatory conditions, mimicked by the addition of IL-1, rescues the inhibitory effect of IL-2 and restores TH-17 differentiation15. In addition, Treg cells can convert directly to TH-17 producing cells under particular inflammatory conditions16, and retinoic acid produced by dendritic cells (DC) within the gut abrogates inflammation by suppressing TH-17 and enhancing Treg cell differentiation17. More recent work provided definitive data defining a common developmental pathway between these two subsets in that TGFβ operates in a concentration-dependent manner allowing the induction of both FoxP3 and RORγt; differentiation to the TH-17 pathway, for example, is determined by factors such as IL-23 and IL-21, which prevent the direct binding of FoxP3 to RORγt18. FoxP3 and RORγt can both bind to Runx1, and Runx1 promotes RORγt-mediated TH-17 cell induction while facilitating FoxP3 suppression of RORγt19. Further support for a common development of these two lineages comes from the observation that thymocytes which would in wild-type mice become Treg cells, instead express RORγt and produce IL-17 in mice unable to express FoxP3 due to genetic insertion of GFP in the Foxp3 locus20. Thus, in the absence of FoxP3, natural mechanisms selecting the Treg lineage default to the TH-17 lineage.
Self-antigen presentation, important for central tolerance via removal of immature T cells expressing potentially autoreactive T cell receptors (TCR), is also necessary for selection of self-reactive subsets of T cells. These most notably include natural Treg21,22,CD1d-restricted natural killer T cells23, CD8αα intra-epithelial lymphocytes24 and γδ T cells25. Thus, in addition to the production of naïve T cells that migrate to secondary lymphoid organs where they await activation and differentiation, the thymus produces distinct, often smaller, populations of T cells that leave the thymus as differentiated effector populations with the capacity to fine tune the immune response. Here we sought to identify other effector cell subsets that are selected based upon self-reactivity. We specifically focused upon the TH-17 subset given its developmental relationship with Treg cells. We demonstrate that the lineage relationship between Treg and TH-17 cells can be extended to include development in the thymus in response to self-antigen.
RESULTS
Self-reactivity enriches for TH-17 cells
To explore the influence of self-reactivity on the differentiation of distinct T cell subsets, we used a model of natural Treg cell enrichment in which mice express a TCR transgene as well as a transgene encoding a high affinity cognate antigen for this TCR21, 26. We bred B10.BR (H-2k) mice bearing the AND TCR transgene specific for a peptide of pigeon cytochrome c (PCC) with mice expressing PCC under control of an MHC class I promoter27. As expected, AND × PCC double transgenic (DTg) mice showed altered thymocyte development compared to control single transgenic (STg) mice expressing the AND transgene or the PCC transgene (Supplementary Fig. 1a). DTg mice exhibited reduction but not elimination of the peripheral CD4+ T cell population (Fig. 1a). The majority of mature CD4+ T cells in DTg mice expressed CD44 (Fig. 1b), indicating an activated or memory phenotype. Consistent with previous reports21, 26, DTg mice also showed enrichment of Treg cells as determined by FoxP3 expression (Fig. 1c). Treg cells made up only a fraction of the T cell population in DTg mice; we focused our studies on the remaining non- Treg CD4+ T cells.
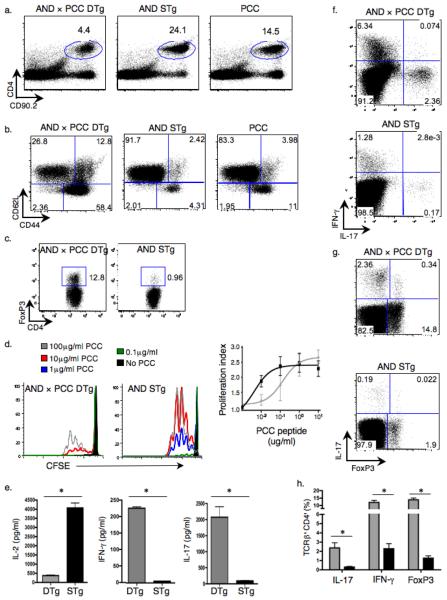
Figure 1.
IL-17+ cells are enriched in peripheral lymphoid organs of AND × PCC DTg mice. (a–c) Splenocytes from indicated mice were analyzed by flow cytometry for percentages of CD4+ T cells (a), activated (CD44hi, CD62Llo) TCRβ+CD4+ cells (b), and CD4+FoxP3+ cells (c). (d) CFSE-labeled CD4+ T cells were cultured with T cell-depleted splenocytes plus the indicated dose of PCC peptide88–104 (left 2 panels). Proliferation profile and proliferation index was measured by CFSE dilution (proliferation index is the average number of divisions of a dividing cell, ignores peak 0). (e) Cytokines in supernatants from cultures of CD4+ T cells and T cell-depleted splenocytes plus 10μg/ml PCC peptide88–104 were analyzed by ELISA (* P ≤ 0.05). (f,g) The percent of IL-17+ compared to IFN-γ+ (f) or IL-17+ versus FoxP3+ splenic TCRβ+CD4+ T cells (g) was assessed by intracellular staining. (h) The percentages of IL-17+, IFN-γ+ and FoxP3+ cells among TCRβ+CD4+ populations from spleens of DTg (gray bars) and STg (black bars) mice were compared (3 mice in each group; * P ≤ 0.05). Data in all panels are representative of at least 3 independent experiments.
To determine the functional state of the peripheral CD4+ T cells in the DTg mice, we assessed their proliferation and cytokine production after stimulation with antigen in vitro. In comparison to cells from STg AND mice, those from DTg animals required at least a log more antigen for activation (Fig. 1d), consistent with an antigen-tuning model28. However, having reached the threshold for stimulation, DTg T cells showed similar proliferation kinetics as cells from STg mice, as indicated by an identical proliferation index at higher antigen concentrations (Fig. 1d). In response to antigen, STg T cells secreted significantly more IL-2 than DTg T cells (Fig. 1e). However, DTg T cells produced more IL-17 and IFN-γ than STg T cells (Fig. 1e). Intracellular cytokine staining and analysis of Foxp3 expression established the presence of three distinct and enriched T cell subsets in spleens of DTg mice: Treg, TH-17, and TH1 (Fig. 1f–h).
Thymic selection of self-reactive TH-17 cells
The development of the TH-17 subset in DTg mice could occur intrathymically and/or peripherally, given the widespread tissue expression of the neoself-antigen, PCC. We used intracellular cytokine staining to show that the CD4 single positive (SP) thymocyte pool from DTg mice contained enriched populations of IL-17-producing as well as FoxP3+ and IFN-γ+ cells (Fig. 2a, b); absolute numbers of TH-17 cells were also significantly increased in the thymus and LN of DTg compared to STg mice (Supplementary Fig. 1b). To confirm that these cells were of thymic origin and not re-circulating from peripheral tissues, we sorted HSA+CD4+CD8− SP thymocytes (Supplementary Fig. 2a, b) and cultured them with PCC. Like splenocytes, these thymocytes produced IL-17 intracellularly (Fig. 2c, d). As further confirmation of their immature status, DTg thymocytes producing IL-17 expressed a GFP cassette regulated by the recombinase activating gene-2 (Rag2) promoter29, like CD4+CD8+ double positive (DP) thymocytes from the same mice; in comparison, IFN-γ-producing cells found within the thymus expressed lower amounts of GFP (Fig. 2e). Next, we transferred CFSE-labeled CD4+ T cells from the LN and spleens of DTg mice into PCC STg recipients, to determine if they migrated to the thymus. Forty-eight hours post-transfer, we found that DTg T cells survived, albeit without proliferation (presumably due to TCR tuning)28, and populated the LN, lungs and liver but showed minimal movement to the thymus (Fig. 2f, Supplementary Fig. 2c). Finally, we analyzed AND × TA.TIM transgenic mice, in which PCC expression is restricted to thymic epithelial cells30. We observed enrichment in the percentage and number of IL-17-producing cells within the thymus and LN comparable to that found in DTg mice (Fig. 2g). Together, these results definitively demonstrated that IL-17 producing AND TCR Tg T cells develop in the thymus. In addition, given the thymus restricted self-antigen expression in TA.TIM mice, they indicate that following export from the thymus, these cells are found within secondary lymphoid organs even in the absence of self-antigen.
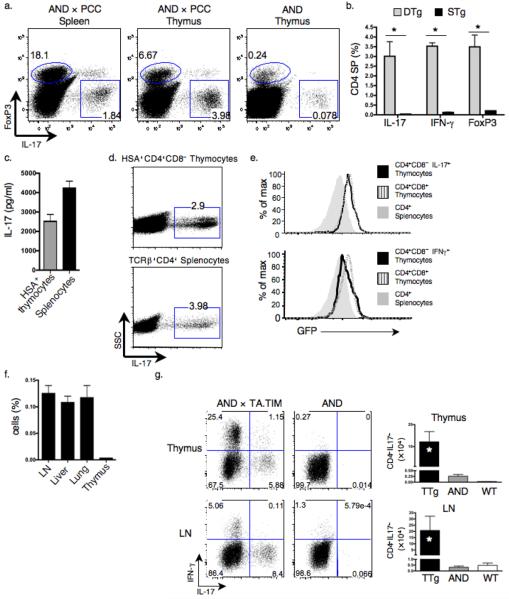
Figure 2.
TH-17 cells develop in the thymus. (a,b) Intracellular staining was used to determine percentages of IL-17+, FoxP3+ and IFN-γ+ cells among TCRβ+CD4+CD8− thymocytes and splenocytes in DTg and STg mice. Left, representative dot plots. Right, graph showing mean ± s.e.m. of 3 mice in each group. (* P ≤ 0.05) (c,d) TCRβ+CD4+CD8−HSA+ thymocytes and TCRβ+CD4+ splenocytes from DTg mice were cultured for 3 d with T cell-depleted splenocytes loaded with 10μg/ml PCC peptide88–104. IL-17 production was analyzed by ELISA (c) and intracellular staining (d). (e) GFP expression in indicated thymocyte subsets from DTg RAG2:GFP mice was measured using flow cytometry. CD4+CD8+ double positive (DP) thymocytes and CD4+ splenocytes were used as positive and negative controls, respectively. (f) Purified CD4+ DTg splenic and LN T cells labeled with CFSE were transferred to wild-type recipients; five days post-transfer, percentages of CFSE+ T cells in the LN, liver, lungs and thymi of recipient mice were determined (3 mice per group). (g) Percentages and absolute numbers of IL-17+, and percentage of IFN-γ+ TCRβ+CD4+ cells within the thymi and LN of AND × TA.TIM (TTg), AND STg and wild-type mice were determined by flow cytometry and cell counting. * P ≤ 0.05 comparing TTg to STg; 6 TTg, 4 STg, and 5 wild-type mice were analyzed. Data are representative of 3 (a,b,g) or 2 (c–f) independent experiments.
We next asked if DTg mice developed an inflammatory or autoimmune phenotype, given their enrichment of TH-17 cells in the setting of global expression of self-antigen. We therefore followed DTg mice up to 1 year of age, and found that their survival curves were identical to those of AND STg and wild-type animals (Supplementary Table 1). Moreover, concentrations of cytokines and chemokines (IL-1α, IL-6, IL-10, IL-17, IFN-γ, IL-12 (p40), tumor necrosis factor (TNF), MIP-1α and RANTES), as well as IgM and autoantibodies (anti-DNA), were equivalent in sera of 12 wk-old DTg and wild-type mice; notably, both groups had reduced concentrations of cytokines and antibodies in comparison to age-matched autoimmune prone MRL/Faslpr mice (Supplementary Fig. 3a, b). Serum concentrations of IL-17 and IFN-γ were similar in DTg and wild-type mice, suggesting that the enriched TH-17 effector populations in DTg mice are present but appropriately suppressed. In addition, upon transfer to DTg recipients, naïve AND STg T cells allotype marked by CD90.1 failed to differentiate into IL-17 producing cells (Supplementary Fig. 3c). Thus spontaneous skewing to TH-17 did not occur in the peripheral environment of DTg mice, further supporting the role of the thymus in TH-17 cell differentiation.
To demonstrate that our observations were not limited to one DTg system or to the B10.BR mouse strain, we also bred mice bearing the OTII TCR transgene specific for a peptide of ovalbumin (OVA) in the context of H-2Kb (C57BL/6) with those expressing OVA under the chicken beta actin promoter (Act-mOVA). In these OT-II × OVA DTg animals, percentages and absolute numbers of TH-17 cells were enriched within the thymus and LN to a degree similar to that found in AND × PCC DTg mice (Supplementary Fig. 4a–d). Finding a similar absolute number of TH-17 cells in two different double transgenic systems of different strains and presumed neoself-ag affinity indicates that other factors such as local or circulating cytokines may contribute to development of this subset, analogous to Treg cell dependence upon IL-213. Self-reactivity is presumably only one step in the pathway of thymic TH-17 cell differentiation.
Confirming the self-antigen specificity of the enriched TH-17 cell populations in DTg mice, stimulation of CD4+ T cells from DTg mice with PCC peptide induced proliferation (Fig. 1d, Supplementary Fig. 5a), and IL-17 production (Fig. 1e, 2c). These experiments indicate that TH-17 thymocytes and splenocytes from DTg mice are neoself-ag specific, even in the setting of possible expression of endogenous TCR rearrangements in the Tg T cells. Nonetheless, we went on to demonstrate that TH-17 enrichment also occurred in the thymi and spleens of Rag1−/− DTg mice (Supplementary Fig. 5b). These data, in aggregate, indicate that in DTg mice TH-17 cells may develop in the thymus upon selection by neoself-antigen, analogous to development of natural Treg cells21, 22.
IL-6 and TGF-β promote thymic TH-17 differentiation
IL-6 and TGFβ are critical for TH-17 differentiation in vitro10–12 and in vivo31, 32. Thus, we asked if the same cytokines were necessary for development of thymic derived TH-17 cells in the absence of overt inflammation. We used DTg mice bred to animals either lacking IL-6 or expressing a dominant negative form of the TGF-β receptor under control of the Cd4 promoter33. In both cases, the TH-17 population was significantly reduced in the thymus and in peripheral lymphoid organs, while the Treg and TH1 populations were either unaffected or slightly increased (Fig. 3a, b)34. Thus, IL-6 and TGFβ play specific and unique roles in thymic TH-17 development. Previous work identified IFN-γ as a negative regulator of IL-17 production35. It is unclear, however, if this cytokine inhibits differentiation of TH-17 cells or regulates IL-17 production following differentiation. To address this issue we bred DTg mice to animals genetically lacking IFN-γ. These mice showed similar TH-17 populations in the thymus or LN as IFN-γ-intact DTg animals (Fig. 3c). On the other hand, LN CD4+ T cells from Ifng−/− DTg mice produced significantly more IL-17 after culture with PCC than did Ifng+/+ DTg LN CD4+ T cells (Fig. 3d). Addition of IFN-γ to such cultures resulted in a dose dependent decrease in IL-17 secretion (Fig. 3e). Thus, whereas IL-6 and TGF-β influence TH-17 differentiation in the thymus, IFN-γ appears to regulate these cells once differentiated and has the potential to down-modulate IL-17 production during the course of an inflammatory response.
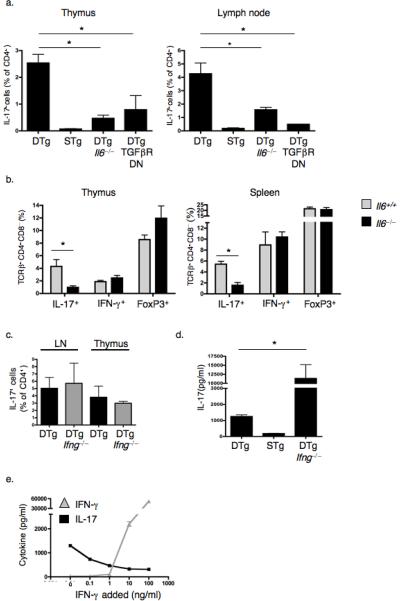
Figure 3.
Thymic TH-17 development depends upon basal IL-6 and TGF-β production, whereas IFN-γ is inhibitory during peripheral activation. (a) Percentages of TCRβ+CD4+CD8−IL-17+ cells in thymi and LN of Il6−/− and CD4dnTGFβRII (TGFβRDN)40 mice were determined by flow cytometry. (b) The percentages of IL-17+, IFN-γ+, and FoxP3+ cells among TCRβ+CD4+CD8− thymocytes and splenocytes were compared between Il6+/+ and Il6−/− DTg mice. (c) Percentages of TCRβ+CD4+CD8−IL-17+ cells were determined in thymi and LN of Ifng−/− DTg mice. Graphs in a–c show mean ± s.e.m. of a minimum of 3 mice of each genotype. * P ≤ 0.05 (d,e) IL-17 in supernatants of splenocytes from DTg, STg and DTg Ifng−/− mice cultured with 10μg/ml PCC peptide88–104 (d, * P ≤ 0.05) or from DTg mice cultured with peptide plus titrated doses of IFN-γ (e). Data are representative of 3 (a,b) or 2 (c–e) independent experiments.
DTg TH-17 cells express ICOS but not PD-1
Compared to STg or wild-type animals, the majority of CD4+ T cells including both the TH-17 and TH1 populations in DTg mice expressed CD44, a marker of activated or memory T cells (Fig. 1b, 4a). However, compared to the TH1 subset, TH-17 cells in DTg mice expressed higher amounts of the inducible costimulator (ICOS), a member of the CD28 superfamily and an important indicator of T cell activation36. This observation suggests a difference in basal activation status of TH1 and TH-17 subsets in DTg mice (Fig. 4b). By contrast, PD-1, an established negative regulator of T cells37, was expressed in lower amounts on TH-17 than on TH1 cells (Fig. 4c). This protein functionally distinguishes these two populations, as in vitro antibody-mediated blockade of PD-1 ligand enhanced PCC-induced production of IFN-γ but had little effect on IL-17 production (Fig. 4d).
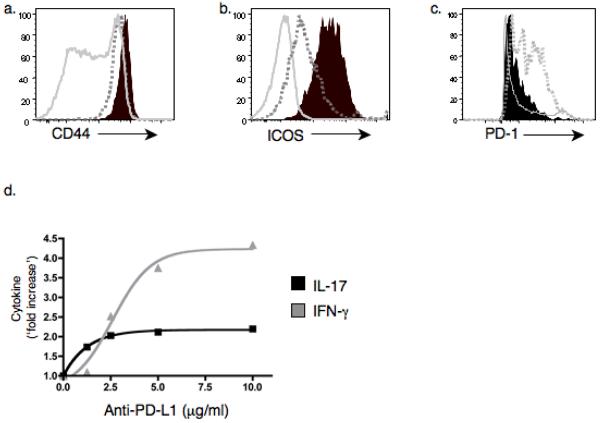
Figure 4.
TH-17 cells from DTg mice express CD44, ICOS but not PD-1. (a–c) CD44, ICOS, and PD-1 were measured by flow cytometry on splenic IL-17+ (black fill), IFN-γ+ (dotted gray) and total CD4+ T cells (solid gray). In b, solid gray represents naïve CD4+CD44− cells. (d) Purified CD4+ T cells from DTg mice were cultured for 3 d with T depleted splenocytes and 10μg/ml PCC peptide88–104 in the presence of a range of concentrations of PD-L1 blocking antibody (clone MIH5). IL-17 and IFN-γ in supernatants was measured by ELISA. Data are representative of 3 (a–c) or 2 (d) independent experiments.
Migration of DTg TH-17 cells
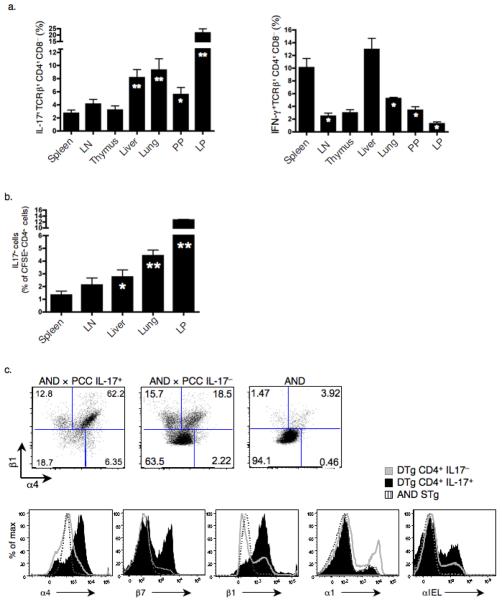
In DTg mice, TH-17 cells were enriched in the liver, lungs, Peyer's patches (PP) and the lamina propria (LP) compared to the spleen (Fig. 5a). In contrast, IFN-γ-producing cells preferentially populated the spleen and liver (Fig. 5a). To confirm that TH-17 cells had specific migration patterns compared to non-IL-17 secreting populations, we purified total CD4+ T cells from the spleen and LN of DTg mice, labeled them with the intracellular fluorescent dye CFSE, and transferred them to wild-type recipients. Two days following transfer, we found that, compared to CD4+ T cells lacking the capacity to produce IL-17, those that made this cytokine had the greatest relative recruitment to the LP followed by the lungs, liver, LN, and then the spleen (Fig. 5b). We noted a different migration pattern of IFN-γ-producing cells (Supplementary Fig. 6a). Of note, the transferred cells did not proliferate at these sites, indicating that their accumulation was due to migration and not local population expansion (Supplementary Fig. 6b).
Figure 5.
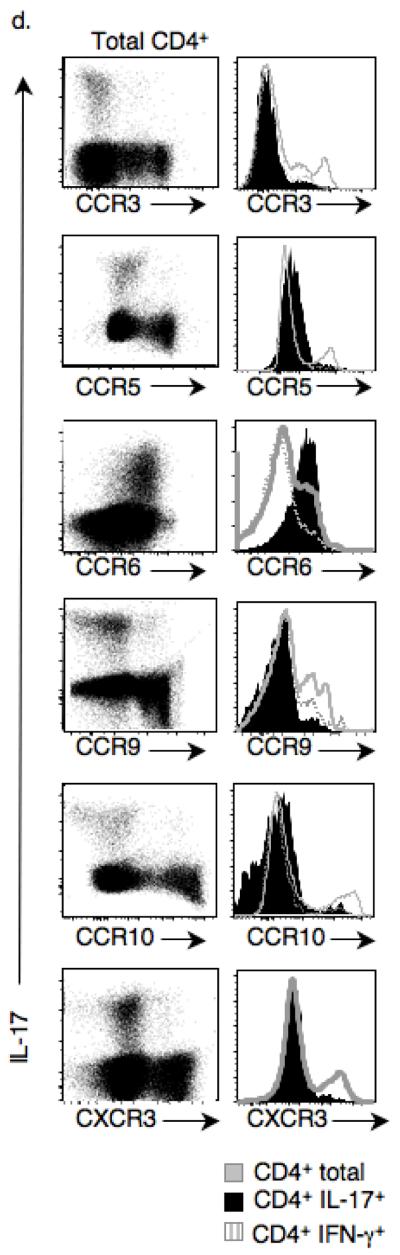
TH-17 cells from DTg mice express CCR6, integrins α4β1, and are enriched in the lamina propria (LP), liver, lung, and Peyer's patches (PP). (a) Percentage of IL-17+or IFN-γ+ cells among TCRβ+CD4+CD8− cells from organs of DTg mice, as measured by intracellular staining (* P ≤ 0.05, ** P ≤ 0.005 compared to spleen). Data represent results from a minimum of 3 mice. (b) Purified splenic and LN CD4+ DTg T cells labeled with CFSE were transferred to wild-type recipients and percentages of IL-17+ cells among CFSE+CD4+ cells were measured in peripheral organs at two days post transfer (* P ≤ 0.05, ** P ≤ 0.005). Data is composite of 3 separate transfer experiments, each using 4 mice per group, with data points combined for statistical analysis. (c) Surface expression of α4, β1, β7, α1 and αIEL integrins on splenic CD4+IL-17+ and CD4+IL-17− DTg cells and on CD4+ STg cells was measured by flow cytometry. (d) Expression of CCR3, CCR5, CCR6, CCR9, CCR10 and CXCR3 on CD4+IL-17+, CD4+IFN-γ+ and total CD4+ T cells taken from DTg mice was measured by flow cytometry. Data are representative of 2 (a) or 3 (c,d) independent experiments.
Given this pattern of migration, we asked if TH-17 cells isolated from spleens of DTg mice displayed unique markers allowing selected peripheral homing38. The majority of DTg TH-17 cells expressed α4β1 integrins, with heterogeneous expression of β7 and αIEL but minimal expression of α1 (Fig. 5c). In addition, chemokine receptor analysis revealed that the T cell compartment of DTg mice contained CD4+ T cells expressing CCR3, CCR5, CCR6 (http://www.signaling-gateway.org/molecule/query?afcsid=A000629), CCR9, CCR10 and CXCR3. Further inspection indicated that DTg TH-17 cells uniquely expressed CCR6, an established TH-17 cell associated chemokine receptor39, but did not express any other inflammatory chemokine receptors tested (Fig. 5d). These findings are consistent with DTg TH-17 cell homing to liver and lung, as these are sites of basal expression of CCL20, the ligand for CCR647.
DTg TH-17 cells express RORγt, IL-23R and IL-22
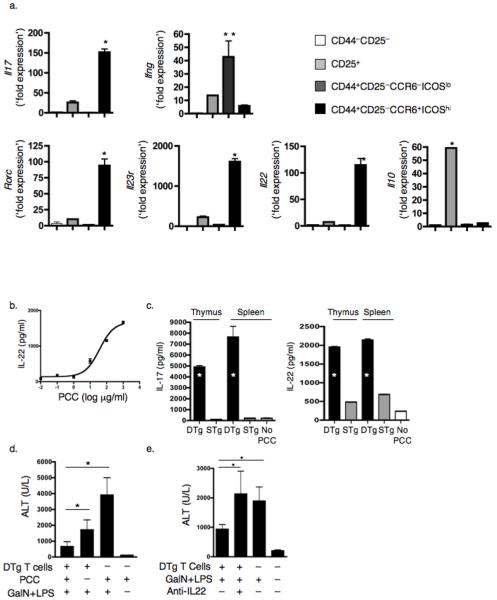
We next asked if self-reactive TH-17 cells that develop intrathymically in the absence of inflammation share features with previously described TH-17 cells5, 9, 41, 42. We sorted CD4+ T cells from LN and spleens of DTg mice into Treg, TH1 and TH-17 populations based upon expression of CD44, CD25, ICOS, and CCR6; we also sorted naïve CD25lo CD44lo cells. Intracellular cytokine staining and RT-PCR for IL-17 and IFN-γ confirmed the validity of this sorting strategy (Supplementary Fig. 7). The TH-17 population uniquely expressed transcripts encoding RORγt (Rorc), IL-23R and IL-22; in contrast Il10 transcripts were expressed only in the Treg subset (Fig. 6a). Using the same sorting strategy for thymocytes from DTg mice (Supplementary Fig. 8a), we found that RORγt, IL-23R and IL-22 were also unique to the thymic TH-17 subset (Supplementary Fig. 8b). This subset also expressed high amounts of Rag1 mRNA, comparable to CD4 SP thymocytes that lacked CD44 expression, and roughly 50-fold higher than LN cells (Supplementary Fig. 8c). Cultures of splenocytes confirmed that DTg T cells produced IL-22 in an antigen-dependent manner (Fig. 6b), and that cells producing this cytokine were also present in the thymus of DTg mice (Fig. 6c). Thus, thymic TH-17 cells obtained from DTg mice express lineage features of TH-17 cells that have been previously found upon in vitro skewing and in inflammatory models.
Figure 6.
TH-17 cells from DTg mice produce IL-22 that promotes hepatocyte survival during inflammation. (a) Splenic and LN CD4+ T cells were sorted into the indicates subsets, and mRNA transcripts were measured by Q-PCR, with the ratio of gene to β-actin expression determined by the relative quantification method (ΔΔCT) (* P ≤ 0.05, compared to 3 other groups; ** P ≤ 0.05, compared to naive). (b,c) IL-22 was measured by ELISA in supernatants of DTg splenocytes cultured for 3 d with increasing concentrations of PCC peptide88–104 (b) and, along with IL-17, from cultures of CD4+ splenocytes or thymocytes from DTg and STg mice cultured with 10μg/ml PCC peptide88–104 (c). (* P ≤ 0.05, compared to concentrations from STg supernatants) (d,e) Serum aspartate aminotransferase (ALT) was measured in sera taken from mice 5 h post GalN+LPS treatment, with or without 48 h prior administration of CD25+-depleted CD4+ T cells from DTg mice (d; * P ≤ 0.05), and with or without 2 h pre-treatment with anti-IL-22 (e; * P ≤ 0.05; ). Data are representative of 2 (b,c) or a combination of 3 (d,e) experiments, the latter totaling 12 mice per group, with data points combined for statistical analysis.
DTg TH-17 cells suppress hepatic inflammation via IL-22
To determine if AND TCR Tg T cells that produce IL-17 and IL-22 in response to self-antigen were functional, we assessed their effect in a model of toxin-induced hepatitis43. CD4+ T cells were purified from DTg mice, depleted of Treg cells and transferred to PCC-expressing or wild-type recipients. Forty-eight hours later, we exposed recipient mice to the hepatic toxin D-(+)-galactosamine (GalN) in the presence of lipopolysaccharide (LPS). Transfer of DTg T cells protected mice from the toxic effects of GalN and LPS, as evidenced by reduction in hepatic enzyme concentrations; the beneficial effect of DTg cells was enhanced by expression of PCC in the recipient mice (Fig. 6d). An IL-22 blocking antibody abrogated the protective effect of DTg cells during hepatitis induction, whereas IL-22 blockade in the absence of DTg cells had no effect (Fig. 6e, Supplementary Fig. 9). Thus, IL-22 produced by self-reactive DTg cells can suppress inflammation-induced liver toxicity.
Thymic TH-17 cells in wild-type mice
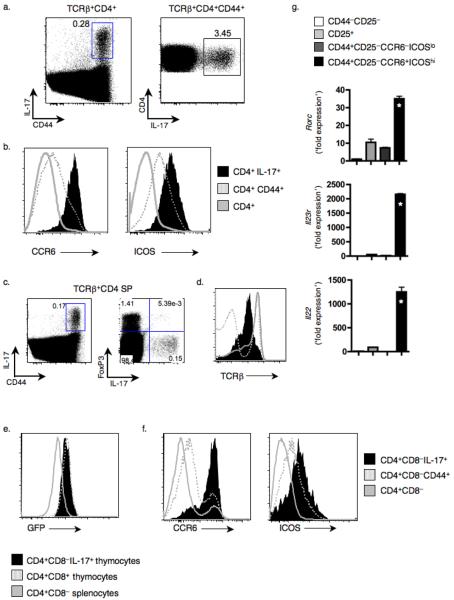
Finally, to demonstrate that our findings were not merely an artifact of double transgenic models, we sought to identify IL-17 producing cells in peripheral lymphoid organs and thymi of wild-type mice. Within the LN, the overall TH-17 population was small but made up over 3% of the CD4+CD44+ T cells (Fig. 7a). These cells were unique in their CCR6 and ICOS expression (Fig. 7b), analogous to the TH-17 population identified in DTg mice. IL-17 producing CD4 SP thymocytes were also identified in wild-type mice (Fig. 7c), and were characterized by reduced TCRβ expression (Fig. 7d), consistent with self-reactivity. Thymic TH-17 cells also expressed GFP regulated by the Rag2 promoter29 (Fig. 7e), consistent with a thymic origin. IL-17+ thymocytes in wild-type mice, like those in DTg animals, expressed ICOS and CCR6 (Fig. 7f, Supplementary Fig. 8a). Using these latter two markers, we were able to employ the same sorting strategy as for DTg thymocytes (Supplementary Fig. 8a) to isolate IL-17-producing thymocytes from wild-type mice and confirm that they selectively expressed Rorc, Il23r, and Il22 mRNA (Fig. 7g). We also observed this population in CD1d-deficient mice, demonstrating that they were not invariant natural killer T cells44 (Supplementary Fig. 10a–d). Thus, IL-17+ thymocytes and CD4+ T cells exist in wild-type mice even in the absence of inflammation.
Figure 7.
TH-17 cells in LN and thymi of wild-type mice have a phenotype identical to TH-17 cells found in DTg mice. (a) Following CD4+ T cell enrichment, TH-17 population size in LN of wild-type B10.BR mice was assessed by intracellular cytokine staining of TCRβ+CD4+ or TCRβ+CD4+CD44+ cells. (b) CCR6 and ICOS expression on the indicated subsets of LN cells of wild-type mice was measured by flow cytometry. (c) CD44 and IL-17 expression by TCRβ+CD4+CD8− thymocytes from wild-type B10.BR mice. (d) Expression of TCRβ on the indicated subsets from wild-type mice was measured by flow cytometry. (e) GFP expression in populations isolated from RAG2:GFP mice was evaluated using flow cytometry. Thymic TCRβ+DP cells and splenic TCRβ+CD4+ cells were used as positive and negative controls, respectively. (f) CCR6 and ICOS expression on the indicated subsets from wild-type mice was measured by flow cytometry. Data (a–f) are representative of 3 independent experiments. (g) Thymocytes from wild-type mice were sorted into the indicated populations and mRNA transcripts were measured using Q-PCR, with the ratio of gene to Hprt1 expression determined by the relative quantification method (ΔΔCT) (* P ≤ 0.05; compared to 3 other groups, with data compiled from 3 replicates using 1 cell sort by flow cytometry).
DISCUSSION
Our work suggests that the current understanding of TH-17 cell differentiation needs to be expanded to include development in the thymus. We found IL-17+ thymocytes in two DTg models, in significantly (log fold) higher percentages and numbers than in STg controls. We also detected small populations of IL-17+ thymocytes in wild-type mice. Several experiments excluded the possibility that IL-17+ thymocytes originated in and recirculated from the periphery. In addition, DTg mice did not have an intrinsically heightened inflammatory environment that might spontaneously promote TH-17 differentiation in the periphery.
Thymic TH-17 cells were selected based on increased reactivity to self-antigens in DTg mice, and produced IL-17 and IL-22 upon stimulation with neoself-antigen. Although we are uncertain if endogenous TCR rearrangement plays a role in the development of these cells, the presence of IL-17+ splenocytes and thymocytes in Rag1−/− DTg mice indicates that they can arise in the absence of such rearrangements. Thymic IL-17-producing cells in wild-type mice express lower amounts of surface TCRβ than other thymic populations, also suggestive of a self-reactive phenotype.
These findings extend the function of self-antigen presentation during thymic development from one originally thought to be limited to positive selection of functional T cells followed by deletion of autoreactive cells, a concept expanded more recently to include selection of Treg 21, 22, NKT23, CD8αα+24, and γδ T25 cells. These observations also strengthen the existing link between the development of TH-17 cells and natural Treg cells, although it is important to emphasize that the thymic TH-17 cells we describe exhibited effector programming similar to non-classical effectors T cell populations, including γδ and NK T cells. These two lineages offer a paradigm of how inherently self-reactive cells with rearranged TCRs can assist the innate immune system in host responses via recognition of self-ligands, and play a non-redundant role in clearance of pathogens45, 46 and/or down-modulation of inflammatory injury47, 48. Indeed, the thymic TH-17 cells we describe closely resemble γδ T cells with regard to effector function in that they share IL-17 as a proinflammatory mediator49 and have the potential to modulate peripheral inflammatory responses despite numerical inferiority compared to conventional effector T cells. γδ T cells can be activated upon recognition of self-ligands that are upregulated in the periphery during inflammation (e.g. non-classical major histocompatibility complex class I molecules T10 and T2250). Thymic TH-17 cells suppressed hepatic inflammation via IL-22 secretion promoted by neoself-antigen recognition. Thus, effector capability upon self-antigen recognition in peripheral inflammatory responses reflects an already established paradigm.
Thymus-derived TH-17 cells seeded peripheral organs, with enrichment in the LP, liver, lung, and PP in DTg animals. While it is formally possible that such cells arose as a consequence of differentiation in secondary lymphoid organs, as noted above several lines of evidence suggest that this is not the case. In addition, the phenotype of the thymic TH-17 cells (expression of α4β1 integrins and CCR6) favors their migration to such sites as the lung and liver40.
Additional work is needed to determine if thymic TH-17 cells, like previously described TH-17 cells, make IL-17F; this cytokine may exert immunoregulatory functions in allergic responses51. Thus the thymic TH-17 population that migrates to the lung may protect the host against allergic lung injury via IL-17F secretion. The balance between secretion of cytokines with pro- and anti-inflammatory potential, in concert with the tissue-specific expression of their receptors, presumably would resolve the paradox of a self-reactive population that can secrete cytokines having either property. Accordingly, it is likely that IL-17 production is carefully regulated so that its production is restricted to situations demanding inflammation. Simultaneous production of a potentially protective cytokine such as IL-22 may promote survival of cells expressing IL-22R during highly toxic inflammation that thymic TH-17 cells help create52. At the same time, secretion of IL-22 would not necessarily subjugate the pro-inflammatory function of IL-17 given the lack of IL-22R expression on immune cells.
Unexpectedly, basal IL-6 concentrations, in the absence of obvious inflammation in DTg mice, determined the size of the thymic TH-17 population. Although the source(s) of cytokines required for thymic TH-17 cell development is not yet clear, it is intriguing to postulate that during inflammation, IL-6 produced following innate cell activation in the periphery operates in a feedback loop on the thymus to enhance production of TH-17 cells at the expense of natural Treg cells. In an analogous regulatory loop, natural Treg cells in the thymus53 might regulate thymic TH-17 development. Thymic TH-17 cells depend upon TGFβ for optimal development; a possible source of this cytokine is natural Treg cells. We propose that during early T cell development, thymic IL-2 concentrations continue to rise with the expanding mature T cell compartment reaching a critical abundance around day 3, allowing for Treg cell skewing from the self-reactive compartment13. As the Treg population expands, it serves as an IL-2 sink to lift restriction of TH-17 lineage commitment by this cytokine14, and produces TGFβ to promote TH-17 differentiation. Thus, the environment may directly regulate the balance between development of Treg and TH-17 subsets from a common precursor. As such, Treg enrichment of the type we observed in DTg mice may be a necessary component of natural TH-17 development.
Rapid effector capability and self-reactivity may explain why the thymic TH-17 population has been preserved in an immune system that also has induced IL-17 producing cells that recognize foreign antigens. Thymic TH-17 cells in DTg mice have a TCR signaling threshold set at least a log fold above TCR STg cells, suggesting that activation of the former is limited to conditions with elevated self-antigen presentation such as during pathogen challenge with associated tissue injury. If present in wild-type mice, such a phenotype presumably would enable the thymic subset to evaluate the severity of infection by amount of tissue destruction, and upon presentation of self-peptides, resident self-reactive TH-17 cells could promote a robust response, including rapid neutrophil recruitment. Thus, the TH-17 subset would be capable of directing the immune response according to the severity of infection, with host protection mediated by IL-22 whose effects are restricted to non-immune cells52. While the evolutionary retention of the thymic IL-17 producing subset indicates an overall host benefit, its careful regulation can presumably be delinquent given the appropriate environmental trigger in a genetically susceptible individual.
ONLINE METHODS
Mice
AND mice expressing the transgenic Vα11Vβ3 TCR that recognizes PCC88–104, and ePCC mice that express PCC27 were originally provided by S. Hedrick (University of California, San Diego) and were maintained in the B10.BR background. We produced CD90.1 wild-type mice, and AND × PCC DTg, AND STg and ePCC STg animals lacking IL-6 or IFN-γ on the B10.BR background by serial backcross to the C57BL/6J-IghaThy1aGpi1a, B6.129S2-Il6tm1Kopf/J, and B6.129S7-Ifngtm1Ts/J backgrounds, respectively (all from The Jackson Laboratory). OTII (C57BL/6-Tg(TcraTcrb)425Cbn/J), Act-mOVA (C57BL/6-Tg(ACTB-OVA)916Jen/J), B6.129S-Rag1tm1Mom, CD1d-deficient (C.129S2-Cd1tm1Gru/J), BALB/cJ, and C57BL/6 mice were purchased from The Jackson Laboratory. We produced OTII (C57BL/6-Tg(TcraTcrb)425Cbn/J) and Act-mOVA (C57BL/6-Tg(ACTB-OVA)916Jen/J) animals lacking RAG1 by serial backcross to the B6.129S-Rag1tm1Mom background. RAG2:GFP mice29, kindly provided by F. Alt (The Children's Hospital and Harvard Medical School, Boston, MA), and transgenic CD4dnTGFβRII mice33 were backcrossed to B10.BR AND × PCC DTg mice for at least 3 generations, and were selected for H-2k homozygosity. TA.TIM mice30 on the B10.BR background were kindly provided by H. van Santen, D. Mathis, and C. Benoist (Joslin Diabetes Center and Harvard Medical School, Boston, MA). Animals were identically housed in specific pathogen-free facilities at the Yale Animal Resources Center. The Institutional Animal Care and Use Committee at the Yale School of Medicine approved all experiments.
Isolation of cells, cell cultures and proliferation assays
Splenocytes, lymphocytes, and thymocytes were obtained by disrupting organs of 6- to 8-week-old mice and were depleted of erythrocytes by hypotonic lysis. Cell cultures were performed in Click's media supplemented with 10% Fetalplex (Gemini Bio-products), 2mM glutamine, 100 Iu/ml penicillin, 0.1 mg/ml streptomycin (Gibco) and 2 μM β-mercaptoethanol. CD4+ T cells were isolated using EasySep mouse CD4+ enrichment kit (Stem Cell Technologies) according to the manufacturer's instructions. T cell-depleted splenocytes were obtained using an EasySep CD90.2 positive selection kit (Stem Cell Technologies) and the unlabelled fraction was used for cultures. CD4+ T cells, labeled via incubation with 2.5μM of the intracellular dye 5,6-carboxyfluorescein diacetate succinimidyl ester (CFSE, Molecular Probes) for 10 min at 37°, and T cell-depleted splenocytes were cultured in a 1:1 ratio at 1 × 106 cells/ml and stimulated with PCC88–104 (10μg/ml) for 3 days. Supernatants were collected at day 3 and cytokines were measured. The proliferation index was calculated as the average number of cell divisions of responding cells (ignores peak 0). For isolation of cells from the lung and liver, mice were sacrificed and their cardiac ventricles perfused with PBS. Organs were diced, incubated in 100 U/ml collagenase (Gibco) at 37° for 1 h and then passed through a 70μm cell strainer and cells separated by 44:56 Percoll gradient.
Hepatitis induction and T cell transfers
Following Treg cell depletion using biotin-conjugated anti-CD25 (PC61), 6–8 × 106 purified splenic and LN CD4+ T cells were injected intravenously (i.v.) into recipient mice. Two days later, animals were given 10mg D-(+)-galactosamine (Sigma) and 300ng LPS from Escherichia coli 055:B5 (Sigma) i.v. in 200μl PBS. When using B10.BR × PCC mice as recipients, an additional 100μg of PCC88–104 peptide was injected with the hepatotoxin. We measured serum ALT concentrations (Teco Diagnostics) five hours later. For IL-22 blockade, 50μg anti-mouse IL-22 polyclonal antibody (R&D Systems) was i.v. injected in 200 μl PBS 2 hours prior to hepatitis induction.
Flow cytometry
For intracellular cytokine staining, cells were stimulated for 2 hours with 50 ng/ml phorbol myristate acetate (PMA) and 1 μg/ml ionomycin (Sigma-Aldrich) in the presence of brefeldin A (GolgiPlug, BD Biosciences). Surface staining with indicated antibodies in the presence of Fc-blocking antibodies was followed by fixation in 1% formaldehyde for 20 min and intracellular staining using the Perm/Wash reagent (BD Biosciences) as directed. All samples were analyzed with FACSCalibur or LSRII (Becton Dickinson) instruments. Flow cytometry data was analyzed with FloJo 8.2 (Tree Star) software. All antibodies were from BD Biosciences, except for those directed against CD25 (PC61.5), CCR9(CW-1.2), FoxP3 (FJK-16s), β1 (HMβ1–1), IL-17 (17B7), PD-L1 (MIH5) (all from eBioscience), CXCR3 (FAB1685P) and CCR10 (248918) (both from R&D Systems), α1 (HMα1) (Abcam), and GFP (600-302-215) (Rockland); for analysis of GFP and IL-17 expression, cells were stimulated as described above with PMA and ionomycin. For analysis of wild-type LN and thymi, single cell suspensions were first enriched for CD4+ T cells and then labeled with antibodies.
Measurement of cytokines, chemokines and antibodies
Supernatants were collected after 3 days of culture and IL-2, IL-17, IFN-γ, and IL-4 concentrations were measured by Bio-Plex (Bio-Rad). IL-22 was detected using ELISA (Antigenix America). Serum cytokines and chemokines were detected by a 9-Plex Bio-Plex consisting of IL-1α, IL-6, IL-10, IL-17, IFN-γ, MIP-1α, IL-12(p40), RANTES, and TNF (Bio-Rad). Total serum IgM was measured by sandwich ELISA using anti-IgM capture antibodies (II-41) (BD Bioscience) and isotype-specific detection Abs conjugated to HRP (Southern Biotechnology Associates). For detection of anti-DNA antibodies, plates were coated with BSA, then dsDNA (Sigma), and developed using isotype-specific detection Abs (F007-1241) (BD Bioscience).
Q-PCR
Following cell sorting, RNA was isolated using an RNeasy mini kit (QIAGEN) and reverse transcribed into cDNA with iScript cDNA synthesis kit (Bio-Rad). Expression was determined using primers listed (Supplementary Table 2) and normalized to Actb or Hprt1.
Statistical analysis
Data are presented as mean ±s.e.m. and were analyzed with Student's t-test using Prism4 (GraphPad Software).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to S. Hedrick and S. Oehan for providing the AND and PCC transgenic mice, F. Alt and D. Schatz for the RAG2:GFP mice, H. van Santen, D. Mathis and C. Benoist for TA.TIM mice, and L. Bockenstedt for supplying the CD1d-deficient animals. We also thank E. Marks and other members of the Craft lab for helpful discussions and L. Zenewicz and E. Espluges for advice. R. Medzhitov and S. Kaech graciously served as critical readers of the manuscript. We also acknowledge those investigators whose work we could not cite because of space limitations. This work was supported by NIH Grants AR40072, AR44076, AI56219, and AR053495, and grants from the Arthritis Foundation, Rheuminations, Inc., and the Connecticut Chapter of the Lupus Foundation. B. Marks was supported by NIH MSTP TG 5T32GM07205.
Footnotes
The authors declare that they have no financial conflicts of interest.
REFERENCES
- 1.Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nat Rev Immunol. 2002;2:933–944. doi: 10.1038/nri954. [DOI] [PubMed] [Google Scholar]
- 2.Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787–793. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- 3.Harrington LE, Mangan PR, Weaver CT. Expanding the effector CD4 T-cell repertoire: the Th17 lineage. Curr Opin Immunol. 2006;18:349–356. doi: 10.1016/j.coi.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 4.Aggarwal S, Ghilardi N, Xie MH, de Sauvage FJ, Gurney AL. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J Biol Chem. 2003;278:1910–1914. doi: 10.1074/jbc.M207577200. [DOI] [PubMed] [Google Scholar]
- 5.Ivanov, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 6.Yang XO, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zenewicz LA, et al. Interleukin-22 but not interleukin-17 provides protection to hepatocytes during acute liver inflammation. Immunity. 2007;27:647–659. doi: 10.1016/j.immuni.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Langrish CL, et al. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowman EP, Chackerian AA, Cua DJ. Rationale and safety of anti-interleukin-23 and anti-interleukin-17A therapy. Curr Opin Infect Dis. 2006;19:245–252. doi: 10.1097/01.qco.0000224818.42729.67. [DOI] [PubMed] [Google Scholar]
- 10.Bettelli E, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 11.Mangan PR, et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 12.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 13.Setoguchi R, Hori S, Takahashi T, Sakaguchi S. Homeostatic maintenance of natural Foxp3(+) CD25(+) CD4(+) regulatory T cells by interleukin (IL)-2 and induction of autoimmune disease by IL-2 neutralization. J Exp Med. 2005;201:723–735. doi: 10.1084/jem.20041982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laurence A, et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26:371–381. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Kryczek I, et al. Cutting edge: opposite effects of IL-1 and IL-2 on the regulation of IL-17+ T cell pool IL-1 subverts IL-2-mediated suppression. J Immunol. 2007;179:1423–1426. doi: 10.4049/jimmunol.179.3.1423. [DOI] [PubMed] [Google Scholar]
- 16.Xu L, Kitani A, Fuss I, Strober W. Cutting edge: regulatory T cells induce CD4+CD25-Foxp3- T cells or are self-induced to become Th17 cells in the absence of exogenous TGF-beta. J Immunol. 2007;178:6725–6729. doi: 10.4049/jimmunol.178.11.6725. [DOI] [PubMed] [Google Scholar]
- 17.Mucida D, et al. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317:256–260. doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- 18.Zhou L, et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang F, Meng G, Strober W. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat Immunol. 2008;9:1297–1306. doi: 10.1038/ni.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gavin MA, et al. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445:771–775. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- 21.Jordan MS, et al. Thymic selection of CD4+CD25+ regulatory T cells induced by an agonist self-peptide. Nat Immunol. 2001;2:301–306. doi: 10.1038/86302. [DOI] [PubMed] [Google Scholar]
- 22.Hsieh CS, Zheng Y, Liang Y, Fontenot JD, Rudensky AY. An intersection between the self-reactive regulatory and nonregulatory T cell receptor repertoires. Nat Immunol. 2006;7:401–410. doi: 10.1038/ni1318. [DOI] [PubMed] [Google Scholar]
- 23.Kronenberg M, Gapin L. The unconventional lifestyle of NKT cells. Nat Rev Immunol. 2002;2:557–568. doi: 10.1038/nri854. [DOI] [PubMed] [Google Scholar]
- 24.Cheroutre H. Starting at the beginning: new perspectives on the biology of mucosal T cells. Annu Rev Immunol. 2004;22:217–246. doi: 10.1146/annurev.immunol.22.012703.104522. [DOI] [PubMed] [Google Scholar]
- 25.Xiong N, Raulet DH. Development and selection of gammadelta T cells. Immunol Rev. 2007;215:15–31. doi: 10.1111/j.1600-065X.2006.00478.x. [DOI] [PubMed] [Google Scholar]
- 26.Apostolou I, Sarukhan A, Klein L, von Boehmer H. Origin of regulatory T cells with known specificity for antigen. Nat Immunol. 2002;3:756–763. doi: 10.1038/ni816. [DOI] [PubMed] [Google Scholar]
- 27.Oehen S, Feng L, Xia Y, Surh CD, Hedrick SM. Antigen compartmentation and T helper cell tolerance induction. J Exp Med. 1996;183:2617–2626. doi: 10.1084/jem.183.6.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grossman Z, Paul WE. Autoreactivity, dynamic tuning and selectivity. Curr Opin Immunol. 2001;13:687–698. doi: 10.1016/s0952-7915(01)00280-1. [DOI] [PubMed] [Google Scholar]
- 29.Monroe RJ, et al. RAG2:GFP knockin mice reveal novel aspects of RAG2 expression in primary and peripheral lymphoid tissues. Immunity. 1999;11:201–212. doi: 10.1016/s1074-7613(00)80095-3. [DOI] [PubMed] [Google Scholar]
- 30.van Santen HM, Benoist C, Mathis D. Number of T reg cells that differentiate does not increase upon encounter of agonist ligand on thymic epithelial cells. J Exp Med. 2004;200:1221–1230. doi: 10.1084/jem.20041022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li MO, Wan YY, Flavell RA. T cell-produced transforming growth factor-beta1 controls T cell tolerance and regulates Th1- and Th17-cell differentiation. Immunity. 2007;26:579–591. doi: 10.1016/j.immuni.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 32.Veldhoen M, Hocking RJ, Flavell RA, Stockinger B. Signals mediated by transforming growth factor-beta initiate autoimmune encephalomyelitis, but chronic inflammation is needed to sustain disease. Nat Immunol. 2006;7:1151–1156. doi: 10.1038/ni1391. [DOI] [PubMed] [Google Scholar]
- 33.Gorelik L, Flavell RA. Abrogation of TGFbeta signaling in T cells leads to spontaneous T cell differentiation and autoimmune disease. Immunity. 2000;12:171–181. doi: 10.1016/s1074-7613(00)80170-3. [DOI] [PubMed] [Google Scholar]
- 34.Fahlen L, et al. T cells that cannot respond to TGF-beta escape control by CD4(+)CD25(+) regulatory T cells. J Exp Med. 2005;201:737–746. doi: 10.1084/jem.20040685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park H, et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshinaga SK, et al. T-cell co-stimulation through B7RP-1 and ICOS. Nature. 1999;402:827–832. doi: 10.1038/45582. [DOI] [PubMed] [Google Scholar]
- 37.Nishimura H, Nose M, Hiai H, Minato N, Honjo T. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM 38. doi: 10.1016/s1074-7613(00)80089-8. [DOI] [PubMed] [Google Scholar]
- 38.Ley K, Laudanna C, Cybulsky MI, Nourshargh S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat Rev Immunol. 2007;7:678–689. doi: 10.1038/nri2156. [DOI] [PubMed] [Google Scholar]
- 39.Reboldi A, et al. C-C chemokine receptor 6-regulated entry of TH-17 cells into the CNS through the choroid plexus is required for the initiation of EAE. Nat Immunol. 2009;10:514–523. doi: 10.1038/ni.1716. [DOI] [PubMed] [Google Scholar]
- 40.Schutyser E, Struyf S, Van Damme J. The CC chemokine CCL20 and its receptor CCR6. Cytokine Growth Factor Rev. 2003;14:409–426. doi: 10.1016/s1359-6101(03)00049-2. [DOI] [PubMed] [Google Scholar]
- 41.Liang SC, et al. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203:2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McGeachy MJ, et al. TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nat Immunol. 2007;8:1390–1397. doi: 10.1038/ni1539. [DOI] [PubMed] [Google Scholar]
- 43.Arvelo MB, et al. A20 protects mice from D-galactosamine/lipopolysaccharide acute toxic lethal hepatitis. Hepatology. 2002;35:535–543. doi: 10.1053/jhep.2002.31309. [DOI] [PubMed] [Google Scholar]
- 44.Michel ML, et al. Critical role of ROR-gammat in a new thymic pathway leading to IL-17-producing invariant NKT cell differentiation. Proc Natl Acad Sci U S A. 2008;105:19845–19850. doi: 10.1073/pnas.0806472105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mattner J, et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434:525–529. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- 46.Hayday A, Tigelaar R. Immunoregulation in the tissues by gammadelta T cells. Nat Rev Immunol. 2003;3:233–242. doi: 10.1038/nri1030. [DOI] [PubMed] [Google Scholar]
- 47.Lahn M, et al. Negative regulation of airway responsiveness that is dependent on gammadelta T cells and independent of alphabeta T cells. Nat Med. 1999;5:1150–1156. doi: 10.1038/13476. [DOI] [PubMed] [Google Scholar]
- 48.Jahng AW, et al. Activation of natural killer T cells potentiates or prevents experimental autoimmune encephalomyelitis. J. Exp. Med. 2001;194:1789–1799. doi: 10.1084/jem.194.12.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roark CL, Simonian PL, Fontenot AP, Born WK, O'Brien RL. gammadelta T cells: an important source of IL-17. Curr Opin Immunol. 2008;20:353–357. doi: 10.1016/j.coi.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chien YH, Konigshofer Y. Antigen recognition by gammadelta T cells. Immunol Rev. 2007;215:46–58. doi: 10.1111/j.1600-065X.2006.00470.x. [DOI] [PubMed] [Google Scholar]
- 51.Yang XO, et al. Regulation of inflammatory responses by IL-17F. J Exp Med. 2008;205:1063–1075. doi: 10.1084/jem.20071978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aggarwal S, Xie MH, Maruoka M, Foster J, Gurney AL. Acinar cells of the pancreas are a target of interleukin-22. J Interferon Cytokine Res. 2001;21:1047–1053. doi: 10.1089/107999001317205178. [DOI] [PubMed] [Google Scholar]
- 53.Ito T, et al. Two functional subsets of FOXP3+ regulatory T cells in human thymus and periphery. Immunity. 2008;28:870–880. doi: 10.1016/j.immuni.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.