Abstract
Pathogen/microbe-associated molecular patterns (PAMPs/MAMPs) trigger plant immunity that forms the first line inducible defenses in plants. The regulatory mechanism of MAMP-triggered immunity, however, is poorly understood. Here, we show that Arabidopsis thaliana transcription factors ETHYLENE INSENSITIVE3 (EIN3) and ETHYLENE INSENSITIVE3-LIKE1 (EIL1), previously known to mediate ethylene signaling, also negatively regulate PAMP-triggered immunity. Plants lacking EIN3 and EIL1 display enhanced PAMP defenses and heightened resistance to Pseudomonas syringae bacteria. Conversely, plants overaccumulating EIN3 are compromised in PAMP defenses and exhibit enhanced disease susceptibility to Pseudomonas syringae. Microarray analysis revealed that EIN3 and EIL1 negatively control PAMP response genes. Further analyses indicated that SALICYLIC ACID INDUCTION DEFICIENT2 (SID2), which encodes isochorismate synthase required for pathogen-induced biosynthesis of salicylic acid (SA), is a key target of EIN3 and EIL1. Consistent with this, the ein3-1 eil1-1 double mutant constitutively accumulates SA in the absence of pathogen attack, and a mutation in SID2 restores normal susceptibility in the ein3 eil1 double mutant. EIN3 can specifically bind SID2 promoter sequence in vitro and in vivo. Taken together, our data provide evidence that EIN3/EIL1 directly target SID2 to downregulate PAMP defenses.
INTRODUCTION
Plant innate immunity is activated upon the recognition of pathogen/microbe-associated molecular patterns (PAMPs/MAMPs) by surface-localized immune receptors or stimulation of cytoplasmic immune receptors by pathogen effector proteins (Ausubel, 2005; Jones and Dangl, 2006). PAMP-triggered immunity (PTI) is central to plant resistance to numerous potential pathogens and is thus indispensable for plant survival in the environment (Chisholm et al., 2006).
The PTI signal transduction pathway is not well understood. The best understood PTI pathway is mediated by FLS2, the Arabidopsis thaliana receptor for bacterial flagellar peptide flg22 (Schwessinger and Zipfel, 2008). The binding of flg22 induces the association of FLS2 with BAK1, a receptor-like kinase. This ligand-induced oligomerization activates the FLS2 kinase, which subsequently activates cytoplasmic signaling pathways. Downstream, two mitogen-activated protein (MAP) kinase cascades are rapidly activated to regulate defenses (Bittel and Robatzek, 2007). MEKK1, MKK1/MKK2, and MPK4 constitute a MAP kinase cascade that negatively regulates PTI defenses. The mekk1 mutant, mkk1 mkk2 double mutant, and mpk4 mutant all display constitutive defenses (Petersen et al., 2000; Ichimura et al., 2006; Qiu et al., 2008). MPK3 and MPK6, two related MAP kinases, are thought to positively regulate PTI defenses, but genetic demonstration of their function is hampered by the lethality of the mpk3 mpk6 double mutant (Bittel and Robatzek, 2007).
Salicylic acid (SA) is a major plant defense hormone central to the activation of a range of defenses including the induction of pathogenesis-related (PR) genes, systemic acquired resistance, and hypersensitive response (Durrant and Dong, 2004). Recent data indicate that SA is also required for the full activation of PTI (Mishina and Zeier, 2007; Tsuda et al., 2008). Genetic and biochemical studies in the last 15 years have led to a comprehensive understanding of the signaling mechanism underlying SA-mediated disease resistance. NPR1 plays a central role in SA-dependent disease resistance. The conformation of the NPR1 protein is regulated by cellular redox state, enabling SA-induced entry of NPR1 into the nucleus (Mou et al., 2003; Tada et al., 2008). The nuclear entry and function of NPR1 are also regulated by phosphorylation and ubiquitination (Spoel et al., 2009). NPR1 interacts with the TGA class transcription activators and activates the transcription of a number of defense genes. In addition, PAD4 and EDS1 function to amplify the SA defenses by a positive feedback loop (Feys et al., 2001).
Contrary to our extensive knowledge concerning SA-mediated signal transduction, little is known about the control of SA biosynthesis. The biosynthesis of SA is strongly induced upon pathogen infection. This pathogen-induced SA biosynthesis is controlled by SID2, which encodes isochorismate synthase 1 (ICS1; Wildermuth et al., 2001). Arabidopsis sid2 mutants are defective in pathogen-induced SA accumulation and are severely compromised in disease resistance to biotrophic pathogens (Nawrath and Metraux, 1999; Wildermuth et al., 2001). Thus, the regulation of SID2 expression is fundamental to plant immunity.
Here, we show that ETHYLENE INSENSITIVE3 (EIN3) and ETHYLENE INSENSITIVE3-LIKE1 (EIL1), two closely related Arabidopsis transcription factors previously known to regulate the ethylene pathway, negatively regulate SID2 expression and SA biosynthesis to repress plant immunity. The ein3 eil1double mutants constitutively expressed SID2 and a large number of PAMP response genes, overaccumulate SA, and showed increased disease resistance to Pseudomonas syringae bacteria. The enhanced resistance and defense gene expression were abolished in the ein3 eil1 sid2 triple mutant. Conversely, plants that overaccumulate EIN3 protein display enhanced susceptibility to P. syringae bacteria. The SID2 promoter-LUC reporter gene showed greatly increased activity in ein3 eil1 mutant protoplasts. Moreover, the EIN3 protein was capable of binding to the SID2 promoter. These results uncover a role for EIN3 and EIL1 in the crosstalk of ethylene and SA signaling pathways.
RESULTS
The rrb6 Mutant Is a Novel ein3 Allele
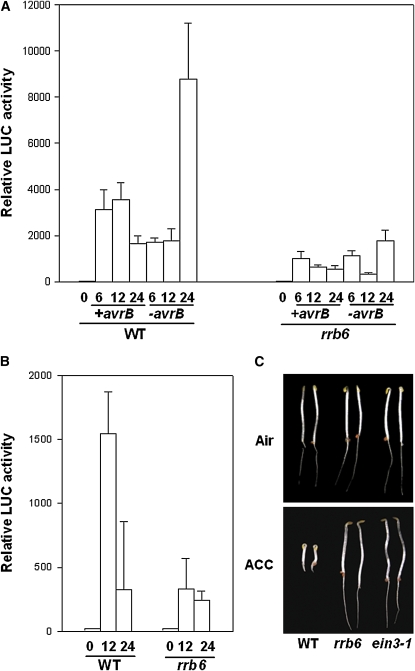
We previously showed that a jasmonate (JA) and ethylene (ET) response gene, RAP2.6, which encodes an ET response factor, is transcriptionally activated by P. syringae effectors, including AvrB (He et al., 2004). To identify Arabidopsis mutants with altered responses to P. syringae bacteria, we developed a RAP2.6-LUC promoter reporter-based mutant population and screened for mutants displaying reduced reporter activity in response to P. syringae DC3000 (avrB) bacterial infection (Shang et al., 2006). One of these mutants, rrb6 (for reduced responsiveness to avrB) displayed reduced reporter gene expression when infiltrated with the virulent strain DC3000, incompatible strain DC3000 (avrB), and water (Figures 1A and 1B), indicating that the mutant was nonspecifically affected in biotic and abiotic responses. Because the RAP2.6 reporter gene is regulated by phytohormones JA and ET, we tested if the rrb6 mutant was affected in ET signaling. Wild-type Arabidopsis seedlings treated with 1-aminocyclopropane-1-carboxylate (ACC), a precursor that can be rapidly converted to ET by plants, showed inhibition of root and hypocotyl elongation, exaggerated tightening of the apical hook, and the swelling of hypocotyl collectively called the triple response (Guzmán and Ecker, 1990). rrb6 showed a uniform insensitive phenotype at 10 μM ACC (Figure 1C). In the absence of ACC, the mutant seedlings were similar to the wild type. The genetic nature of the rrb6 mutation was determined by crossing the homozygous mutant plants with the wild-type transgenic plants carrying RAP2.6-LUC. The F1 seedlings showed a normal ET response, indicating that rrb6 is a recessive mutation. The morphology of rrb6 plants was quite similar to that of the wild type, with the exception that the leaves are slightly larger.
Figure 1.
rrb6 Displays Reduced Expression of RAP2.6-LUC Reporter and Defects in Triple Response.
(A) RAP2.6-LUC reporter activity in wild-type and rrb6 mutant leaves infiltrated with DC3000 bacteria with or without avrB.
(B) RAP2.6-LUC reporter activity in leaves infiltrated with water. Note that the scale is different compared with that in (A) because water treatment induces lower reporter activity than bacteria treatment.
(C) Triple response of 3-d-old wild-type and rrb6 seedlings in the presence or absence of ACC. The ein3-1 mutant was included as a control.
Each data point in (A) and (B) consisted of at least three leaves. Error bars indicate sd.
The rrb6 mutation was initially mapped to the top arm of chromosome III between the simple sequence length polymorphism markers MSJ11 and MLM2. The size of rrb6 F2 population was then increased to fine-map the mutation in a region of ∼100 kb between the markers MQC12 and MOE17 (see Supplemental Table 3 online). Because rrb6 is ET insensitive and was mapped to the region containing EIN3, which is a known ET signaling gene, we tested whether rrb6 is an ein3 allele. Sequence analysis identified a point mutation in rrb6 at nucleotide 1599 of EIN3 (C to T), which resulted in a P216S substitution in the Pro-rich domain. The previously reported allele ein3-1 has a mutation at nucleotide 1598 (G to A) to introduce a stop codon at amino acid 215 (Chao et al., 1997). To further confirm that the ET-insensitive phenotype in rrb6 is due to the mutation in the EIN3 gene, we crossed the rrb6 mutant with ein3-1 plants. As shown in Supplemental Table 1 online, F1 plants were completely insensitive to ET. Several alleles of ein3 have been discovered to date (Chao et al., 1997; Alonso et al., 2003), and we renamed rrb6 as ein3-4.
EIN3 and EIL1 Negatively Regulate Disease Resistance to P. syringae
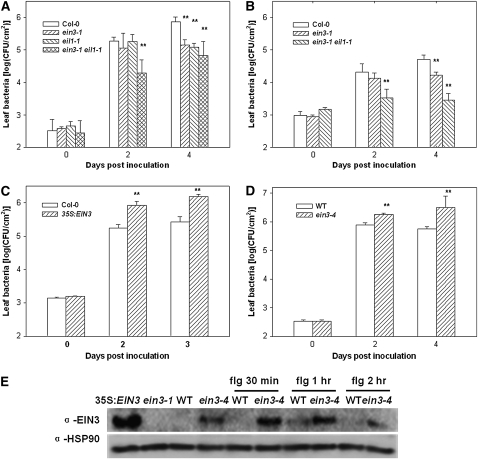
EIN3 and its close homolog EIL1 belong to a family of transcription factors known to regulate the ET response. We sought to determine the role of EIN3 and EIL1 in disease resistance. We inoculated ein3-1, eil1-1, and the ein3-1 eil1-1 double mutant with P. syringae DC3000 bacteria. The ein3-1 eil1-1 double mutant was consistently more resistant than Columbia-0 (Col-0) in multiple experiments and supported ∼10-fold less bacterial growth at 2 and 4 d after inoculation (Figure 2A). The ein3-1 and eil1-1 mutants showed slightly enhanced resistance compared with Col-0 at 4 d after inoculation in this particular experiment, but the results were variable in other experiments. We also inoculated the ein3-1 and ein3-1 eil1-1 mutants with DC3000 carrying avrRpt2, which conditions RPS2-specific resistance in Arabidopsis plants. Figure 2B shows that the ein3-1 eil1-1 double mutant displayed enhanced resistance to this bacterium.
Figure 2.
EIN3 and EIL1 Negatively Regulate Resistance to P. syringae Bacteria.
(A) ein3-1 eil1-1 exhibits enhanced resistance to DC3000. Plants of the indicated genotypes were inoculated with DC3000 bacteria, and bacterial populations in the leaf were determined at the indicated times.
(B) ein3-1 eil1-1 shows enhanced resistance to DC3000 (avrRpt2). Plants of the indicated genotypes were inoculated with DC3000 (avrRpt2) bacteria, and bacterial populations in the leaf were determined at the indicated times.
(C) EIN3 overexpression enhances resistance to DC3000. Plants of the indicated genotypes were inoculated with DC3000 bacteria, and bacterial populations in the leaf were determined at the indicated times.
(D) ein3-4 enhances susceptibility to DC3000. Wild-type RAP2.6-LUC transgenic plants were used as a control.
(E) ein3-4 enhances EIN3 protein stability. Four-day-old etiolated seedlings were treated with 1 μM flg22 for the indicated times and examined by immunoblot using anti-EIN3 antibodies or anti-HSP90 antibodies (for loading control). EIN3 protein exists at a low level in wild-type RAP2.6-LUC transgenic seedlings and was not detected in this blot. The 35S:EIN3 and ein3-1 seedlings were used as positive and negative controls, respectively.
Each data point consisted of at least three samples. Error bars indicate sd. * and ** indicate significant difference at 0.05 and 0.01, respectively, between mutants and Col-0 or wild-type RAP2.6-LUC plants at the same time point (Student's t test). Results shown are a representative of three ([A] to [C]) or five (D) independent experiments.
To further test the role of EIN3 in P. syringae resistance, we inoculated an EIN3 overexpression line with DC3000. This line carried a 35S:EIN3 transgene and displays a dwarf phenotype indicative of constitutive ET response (Solano et al., 1998). Figure 2C shows that the EIN3 overexpression line was more susceptible to DC3000 and supported ∼10-fold more bacterial growth than did the Col-0.
We also inoculated the ein3-4 mutant with DC3000 and determined bacterial growth. To our surprise, the ein3-4 mutant was consistently more susceptible than the wild type to DC3000 in multiple experiments. DC3000 bacteria grew ∼10-fold more in ein3-4 than in the wild type (Figure 2D). The ein3-4 mutant plants also developed more severe disease symptoms than the wild type (see Supplemental Figure 1 online). To further determine the effect of ein3-4 mutation on disease resistance, we inoculated wild-type, ein3-4, and ein3-4 × wild-type F1 plants with DC3000 (avrB) bacteria. Supplemental Figure 2 online shows that the ein3-4 acted semidominantly in the suppression of disease resistance. An examination of the EIN3 protein level showed that the EIN3 protein accumulated to a high level in the ein3-4 mutant plants, indicating that the ein3-4 mutation resulted in greater stability of the EIN3 protein (Figure 2E). Interestingly, flg22 induced the accumulation of EIN3 protein in both Col-0 and ein3-4 seedlings 1 h after treatment. These results indicated that the EIN3P216S mutant protein encoded by ein3-4 was fully functional in repressing plant disease resistance, although it was completely nonfunctional in regulating the ET induced triple response. Together, these results demonstrated that EIN3 and EIL1 negatively regulate resistance to both compatible and incompatible P. syringae bacteria in Arabidopsis.
EIN3 and EIL1 Are Negative Regulators of PTI Defenses
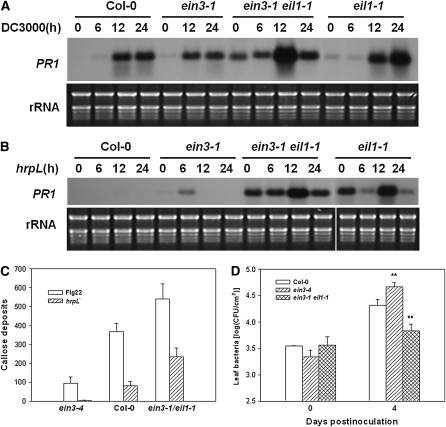
The results described above indicate that EIN3 and EIL1 are involved in basal resistance to P. syringae bacteria, which suggests a role in PTI. Perception of flg22 and other PAMPs activates PR1 gene expression and callose deposition at the cell wall. Indeed, the eil1-1 mutant and ein3-1 eil1-1 double mutant showed constitutive PR1 expression, and the ein3-1 mutant showed elevated PR1 expression when inoculated with the DC3000 (Figure 3A) and hrpL mutant bacteria (Figure 3B). The hrpL mutant lacks the HrpL transcription factor required for expression of type III genes required for effector secretion and is thus thought to be a nonpathogenic bacterium carrying a collection of PAMPs. The elevated PR1 expression in ein3-1 plants in response to hrpL mutant bacteria suggests that EIN3 plays a role in PTI responses. To further determine if EIN3 and EIL1 play a role in PTI defenses, we treated the ein3-4 and ein3-1 eil1-1 mutants with flg22 and quantitatively examined callose deposition. Figure 3C shows that flg22 induced greater callose deposition in the ein3-1 eil1-1 double mutant, whereas the ein3-4 mutant had significantly less callose. Similarly, when treated with the DC3000 hrpL mutant bacteria, greater callose deposition was observed in the ein3-1 eil1-1 double mutant, and reduced callose depositon was seen in the ein3-4 mutant (Figure 3C). Bacterial growth assay showed that the ein3-1 eil1-1 double mutant was slightly more resistant, whereas the ein3-4 mutant was slightly more susceptible to the hrpL mutant bacteria compared with Col-0 (Figure 3D). Together, these results indicate that EIN3 and EIL1 are negative regulators of the PTI defenses.
Figure 3.
EIN3 and EIL1 Negatively Regulate PTI Responses.
(A) RNA gel blot analysis of PR1 expression in response to DC3000 bacteria. Plants of the indicated genotypes were inoculated with DC3000 bacteria, and RNA was isolated at the indicated times for RNA gel blot analysis.
(B) PR1 expression in response to the hrpL mutant bacteria. Longer exposure was used because the hrpL mutant bacteria induce weaker PR1 expression. Plants of the indicated genotypes were inoculated with hrpL mutant bacteria, and RNA was isolated at the indicated times for RNA gel blot analysis.
(C) Callose deposition of wild-type, ein3-4, and ein3-1 eil1-1 mutant plants in response to flg22 or hrpL mutant bacteria. Leaves were treated with flg22 or hrpL mutant bacteria, stained with aniline blue, and callose deposits/0.1 mm2 were quantified under a fluorescence microscope. The result is a representative of three experiments.
(D) Bacterial growth assay on plants inoculated with the hrpL mutant bacteria. * and ** indicate significant difference at 0.05 and 0.01, respectively, between mutants and Col-0 at the same time point (Student's t test). The result is a representative of three experiments.
For (C) and (D), each data point consisted of at least three samples. Error bars indicate sd.
EIN3 and EIL1 Globally Repress PAMP Response Genes
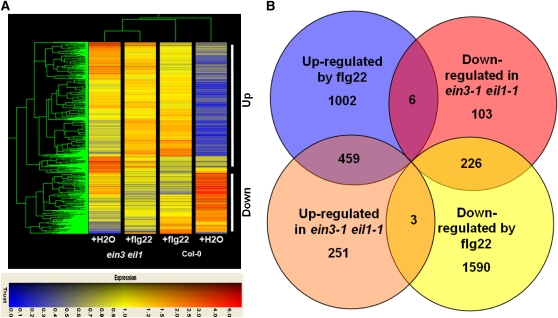
To better understand how EIN3 and EIL1 repress PTI defenses, we conducted microarray analysis on the ein3-1 eil1-1 double mutant using Affimetrix ATH1 oligonucleotide microarray. The ein3-1 eil1-1 double mutant and Col-0 plants were treated with water or flg22 for 3 h, and three biological replicates were examined. As reported previously, a large number of genes were induced or repressed by the flg22 treatment in the Col-0 plants (Zipfel et al., 2004; Navarro et al., 2004). Using a q value < 0.05 and a fold change of >2 as a cutoff, 1467 genes were identified as flg22 induced and 1819 genes as repressed by flg22 in Col-0 plants (see Supplemental Data Sets 1 and 2 online). A total of 713 genes were upregulated and 335 genes were downregulated in water-treated ein3-1 eil1-1 plants compared with Col-0 (Figure 4). Nine genes upregulated in the ein3-1 eil1-1 double mutant were randomly selected and verified by quantitative real-time RT-PCR (see Supplemental Table 2 online). The difference between ein3-1 eil1-1 and Col-0 plants diminished following flg22 treatment. A total of 459 of the 713 upregulated genes are flg22-induced genes (Figure 4B; see Supplemental Data Set 3 online). The overlap is highly significant (P value 1.69e−317). Conversely, 226 of the 335 downregulated genes are normally repressed by flg22 treatment (P value 4.84e−165; see Supplemental Data Set 4 online). These results indicate that EIN3 and EIL1 primarily repress PAMP-triggered transcription programming.
Figure 4.
Microarray Analysis of Genes Regulated by EIN3/EIL1 and flg22.
(A) Hierarchical clustering of genes differentially regulated in Col-0 and ein3-1 eil1-1 plants. Clustering was performed using uncentered Pearson correlation and complete linkage clustering and was visualized with TREEVIEW (Eisen et al., 1998). The 1048 probe sets differentially expressed in water-treated Col-0 and ein3-1 eil1-1 plants were used for the analysis. Genes upregulated and downregulated in water-treated ein3-1 eil1-1 plants are indicated. Colors indicate normalized hybridization signal on a scale of 0 to 5.
(B) Venn diagram of genes commonly regulated by EIN3/EIL1 and flg22.
EIN3 and EIL1 Negatively Regulate the Transcription of SID2 and Repress SA Accumulation
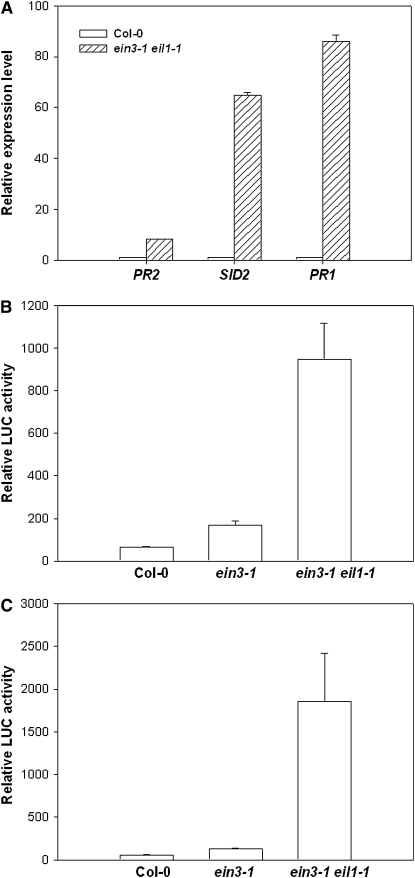
We also compared the genes upregulated in the ein3-1 eil1-1 double mutant with genes induced by SA. An analysis of data from the AtGenExpress consortium (Shimada et al., submission number: ME00364) identified 321 SA-induced genes at 3 h (q value ≤ 0.05 and fold change ≥2; see Supplemental Data Set 5 online). A total of 96 of these genes are upregulated in the ein3-1 eil1-1 mutant plants (P value 3.0e−64) (see Supplemental Data Set 3 online). These include EDS1, PAD4, NPR1/NIM1-interacting1 (NIMIN1), PBS3, and SAG101, which are known to mediate SA-dependent defenses (Loake and Grant, 2007). A close examination showed that genes encoding chorismate synthase and isochorismate synthase 1 (ICS1/SID2) were highly elevated in the ein3-1 eil1-1 double mutant. These two enzymes work in concert downstream of the Shikimate pathway leading to the biosynthesis of isochorismate, which likely is a major precursor for SA biosynthesis (Wildermuth et al., 2001). Indeed, SID2 is known to be required for SA accumulation during pathogen infection (Wildermuth et al., 2001). Consistent with the increased expression of SA pathway genes in microarray results, quantitative real-time RT-PCR showed increased expression of PR1, PR2, and SID2 in ein3-1 eil1-1 double mutant plants (Figure 5A). We therefore tested if EIN3 and EIL1 are required for transcriptional repression of SID2 and the chorismate synthase gene. Promoters of these genes were fused to the firefly luciferase (LUC) reporter gene and transfected into protoplasts prepared from Col-0, ein3-1, and ein3-1 eil1-1 double mutant plants. Consistent with a role in repressing SID2 and the chorismate synthase gene promoters, both reporter genes showed increased expression in the ein3-1 eil1-1 double mutant protoplasts, which was 30- to 100-fold greater than that in the Col-0 protoplasts (Figures 5B and 5C). We also tested if the ein3-4 mutation caused a stronger repression of the SID2 promoter. Because the ein3-4 mutant contained the RAP2.6-LUC reporter gene, we transfected a SID2 promoter-β-glucuronidase (GUS) construct into protoplasts derived from ein3-4 and wild-type RAP2.6-LUC transgenic plants. The SID2-GUS reporter activity in ein3-4 was ∼10% of that in the wild type (see Supplemental Figure 3 online).
Figure 5.
Elevated Expression of SA Pathway Genes in ein3-1 eil1-1 Double Mutant Plants.
(A) Quantitative real-time RT PCR analysis of PR1, PR2, and SID2 gene expression. Expression level for each gene was normalized to that of the wild type (Col-0).
(B) Chorismate synthase promoter-LUC activity in protoplasts isolated from the wild type and ein3-1 eil1-1 double mutant. Relative LUC activity represents arbitrary luminescence units.
(C) SID2 promoter-LUC activity in protoplasts isolated from Col-0 and the ein3-1 eil1-1 double mutant.
Each data point consisted of at least three replicates. Error bars indicate sd. The experiments were performed twice with similar results.
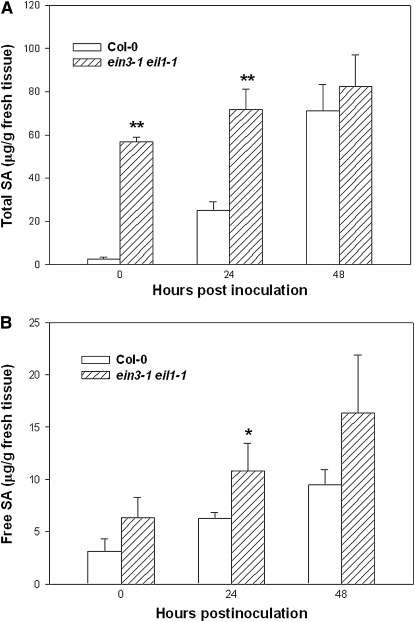
We determined if the increased expression of chorismate synthase and SID2 genes was correlated to SA accumulation in the ein3-1 eil1-1 plants. Much higher levels of total SA were observed in the ein3-1 eil1-1 plants, particularly when uninoculated plants were compared (Figure 6A; see Supplemental Figure 4 online). Inoculation of the ein3-1 eil1-1 double mutant with DC3000 bacteria only modestly elevated the amount of total SA. The ein3-1 eil1-1 plants also contained slightly more free SA compared with Col-0 (Figure 6B). Thus, EIN3 and EIL1 repress SID2 and negatively regulate SA biosynthesis.
Figure 6.
Heightened SA Level in ein3-1 eil1-1 Double Mutant Plants.
(A) Total SA in the ein3-1 eil1-1 and Col-0 plants.
(B) Free SA in the ein3-1 eil1-1 and Col-0 plants.
Plants were inoculated with DC3000 bacteria (106 cfu/mL) for the indicated hours, and tissues were collected for SA extraction. Each data point consisted of three replicates. Error bars indicate sd. * and ** indicate significant difference at 0.05 and 0.01, respectively (Student's t test). The experiment was repeated twice with similar results.
EIN3 Directly Targets the SID2 Promoter
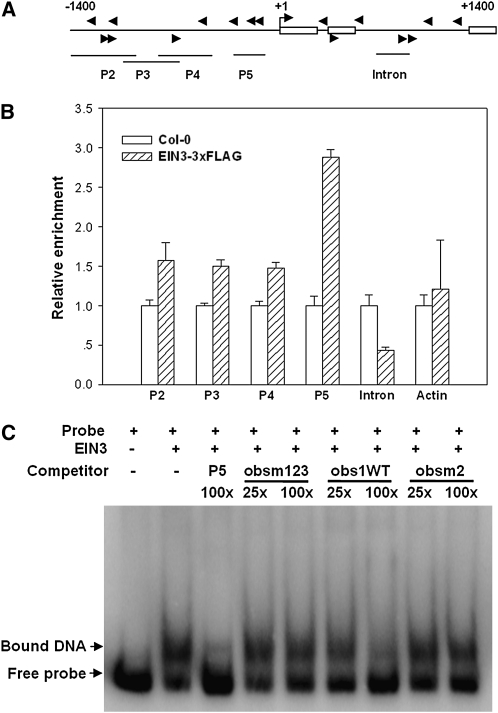
We next tested if EIN3 directly targets the SID2 promoter using chromatin immunoprecipitation (ChIP). Because the EIN3 protein exists at an extremely low level, attempts to test endogenous EIN3-SID2 promoter binding were unsuccessful. We therefore constructed transgenic plants expressing EIN3-3×FLAG fusion protein under an estradiol-inducible promoter (Zuo et al., 2000). Chromatin immunoprecipitated with the anti-FLAG antibody was profoundly enriched in fragment P5 (located −120 to −324 bp upstream of the transcription start site; Figure 7). The EIN3/EIL family protein was reported to bind DNA with a consensus sequence of A(C/T)G(A/T)A(C/T)CT (Kosugi and Ohashi, 2000; Yamasaki et al., 2005). However, only positions 1, 3, and 5 of this sequence were experimentally demonstrated to be important for recognition. P5 contains three relaxed EIN3 binding sites A(C/T)G(A/T)A(C/T). DNA fragments further upstream showed detectable but less enrichment with EIN3-3×FLAG. By contrast, a fragment from the second intron of SID2 and an actin promoter sequence did not show detectable enrichment by EIN3-3×FLAG (Figure 7). These results indicate a specific association of EIN3-3×FLAG with the SID2 promoter sequence, particularly fragment P5.
Figure 7.
EIN3 Binds the SID2 Promoter.
(A) Schematic diagram of potential EIN3 binding sites (arrows) and DNA fragments used for ChIP experiments. Shown are 1.4-kb upstream sequence and part of the coding sequence for SID2. Boxes are exons, and the translational start site (ATG) is shown at position +1.
(B) Enrichment of the indicated DNA fragments following ChIP using anti-FLAG antibody. Chromatin from wild-type and transgenic plants expressing EIN3-3×FLAG was immunoprecipitated with an anti-FLAG antibody, and the presence of the indicated DNA in the immune complex was determined by quantitative real-time PCR. The amounts of DNA amplified from the EIN3-3×FLAG seedlings were normalized to that from Col-0 plants. The Actin promoter fragment was used as a negative control. The experiment was repeated three times with similar results.
(C) EMSA assay for EIN3-P5 DNA fragment binding in vitro. Radiolabeled P5 DNA fragment was incubated with GST-EIN3 protein, and the free and bound DNA (arrows) were separated in an acrylamide gel. Where indicated, the P5 fragment and oligonucleotide primers WTobs1 (5′-AACGATGTACCTGGTCGTATT-3′), obsm2 (5′-GTACATTTACCTGGACCGTGA-3′), and obsm123 (5′-GTACCTTTCCCTGGACCGTGA-3′) were used as competitor DNA.
DNA electrophoresis mobility-shift assay (EMSA) was performed to determine if EIN3 directly binds the SID2 promoter. A truncated EIN3 protein (amino acids 141 to 352) containing the DNA binding domain was expressed as a glutathione S-transferase (GST) fusion protein in Escherichia coli and affinity purified. The GST-EIN3 protein was capable of binding to the radiolabeled P5 DNA fragment in vitro (Figure 7C). The addition of unlabeled P5 DNA fragment blocked the binding. Furthermore, the oligonucleotide obs1 containing the core EIN3 binding sequence (ATGTAC; Kosugi and Ohashi, 2000) was similarly capable of competing with the binding. By contrast, the oligo nucleotides obsm2 and obsm123, in which the core sequence was respectively changed to ATTTAC and CTTTCC, were unable to compete with P5-EIN3 binding. Together, these results demonstrate that EIN3 is capable of binding the conserved EIN3 binding element in the SID2 promoter both in vivo and in vitro.
To determine the importance of EIN3 binding sequence in the SID2 promoter, the P5 sequence was removed from the SID2-GUS construct and transfected into ein3-1 eil1-1 and Col-0 protoplasts. The SID2P5Δ-GUS reporter activity was greatly increased in Col-0 protoplasts but not in ein3-1 eil1-1 protoplasts (see Supplemental Figure 5 online), indicating that the P5 sequence contributes to transcriptional repression of SID2 promoter by EIN3 and EIL1.
SID2 Is Responsible for Enhanced Defenses in the ein3-1 eil1-1 Double Mutant
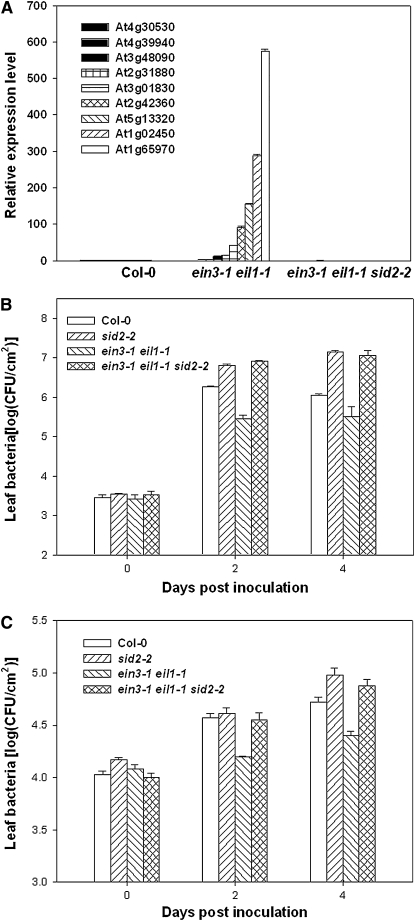
The results described above suggest that EIN3 and EIL1 repress PTI defenses through repressing SID2. We therefore introduced the sid2-2 mutation into the ein3-1 eil1-1 double mutant. The expression of the nine selected genes upregulated in the ein3-1 eil1-1 double mutant was largely restored to wild-type levels in the ein3-1 eil1-1 sid2-2 triple mutant (Figure 8A), indicating that SID2 and possibly other SA biosynthetic genes were responsible for the elevated defense gene expression. To further test if the enhanced SID2 expression contributes to the enhanced disease resistance in the ein3-1 eil1-1 double mutant, we inoculated the ein3-1 eil1-1 sid2-2 triple mutant with DC3000 and the hrpL mutant bacteria. Bacterial growth assays indicated that the triple mutant was completely susceptible to the bacteria compared with the sid2-2 mutant (Figures 8B and 8C), indicating that the SID2-mediated resistance accounted for the elevated basal resistance in the ein3-1 eil1-1 double mutant.
Figure 8.
SID2 Is Required for Heightened Defenses and Enhanced Disease Resistance in ein3-1 eil1-1 Plants.
(A) SID2 is required for enhanced expression of flg22 response genes in the ein3-1 eil1-1 double mutant. Expression levels of the indicated genes in untreated wild-type (Col-0), ein3-1 eil1-1 double, and ein3-1 eil1-1 sid2-2 triple mutant plants were determined by quantitative RT-PCR. The value shows normalized expression level for each gene. The experiment was repeated three times with similar results.
(B) SID2 is required for enhanced resistance to DC3000 in the ein3-1 eil1-1 plants. Wild-type (Col-0), sid2-2, ein3-1 eil1-1 double, and ein3-1 eil1-1 sid2-2 triple mutant plants were inoculated with DC3000 bacteria, and bacterial population in the leaf was determined at the indicated times. The experiment was repeated three times with similar results.
(C) SID2 is required for enhanced resistance to the hrpL mutant bacteria in ein3-1 eil1-1 plants. The experiment was repeated twice with similar results. Wild-type (Col-0), sid2-2, ein3-1 eil1-1 double, and ein3-1 eil1-1 sid2-2 triple mutant plants were inoculated with hrpL mutant bacteria, and bacterial population in the leaf was determined at the indicated times.
Each data point consisted of at least three replicates. Error bars indicate sd.
ein2 and ebf Mutants Display Altered Defenses to P. syringae
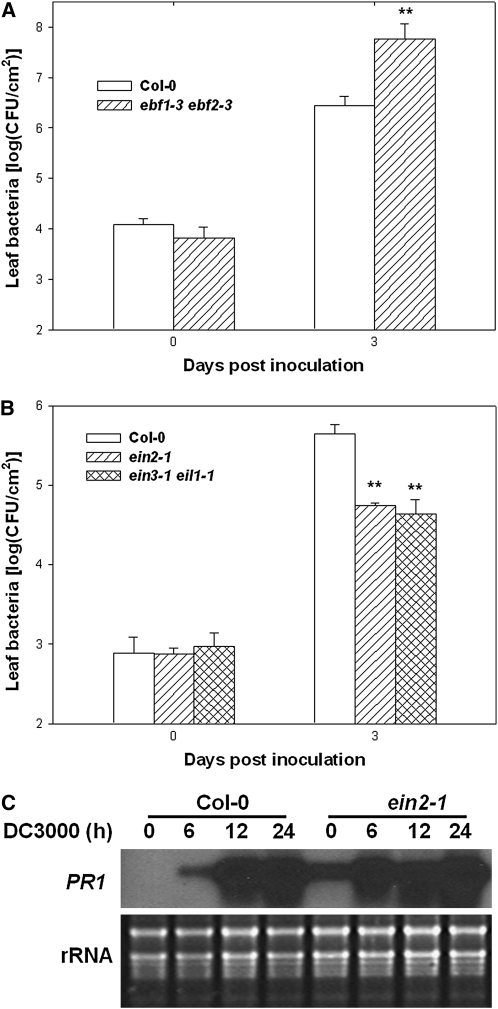
The negative regulation of SA biosynthesis by EIN3 and EIL1 raises a possibility that the ET pathway may negatively regulate the SA pathway through these two transcription factors. We therefore tested if other ET pathway mutants also affected P. syringae bacterial resistance. EBF1 and EBF2, two closely related F-box proteins, direct the ubiquitination and subsequent degradation of EIN3 and EIL1. The ebf1 ebf2 double mutants overaccumulate EIN3 and EIL1 (Guo and Ecker, 2003; Potuschak et al., 2003). We tested if the ebf1-1 ebf2-1 double mutant (Potuschak et al., 2003) was more susceptible to DC3000 bacteria. Indeed, the ebf1-1 ebf2-1 double mutant supported ∼20-fold more bacterial growth than did the wild type (Figure 9A). Another important ET pathway component, EIN2, is an integral membrane protein that acts upstream of EIN3 and EIL1 to regulate the ET pathway (Guo and Ecker, 2004). In contrast with the ebf1-1 ebf2-1 mutant, the ein2-1 mutant is unable to accumulate EIN3 (Guo and Ecker, 2003). In an early report, it was shown that the ein2-5 mutant exhibited reduced disease symptoms when inoculated with virulent P. syringae bacteria (Bent et al., 1992). However, whether this mutant supports less bacterial growth has not been carefully examined. Our repeated bacterial growth assays showed that ein2-1 plants supported 10-fold less DC3000 bacterial growth than did Col-0 plants (Figure 9B). We also examined PR1 expression in the ein2-1 mutant plants. Consistent with a previous report (Lawton et al., 1994), ein2-1 plants showed constitutive PR1 expression (Figure 9C). In addition, greater PR1 expression was observed in ein2-1 plants 6 h after inoculation with DC3000 bacteria (Figure 9C). Together, these results support that EIN3 and EIL1 mediate a crosstalk between the ET pathway and the SA pathway.
Figure 9.
ein2 and ebf Mutants Display Altered Defenses to P. syringae DC3000.
(A) Enhanced susceptibility in ebf1-3 ebf2-3 double mutants. Wild-type (Col-0) and ebf1-3 ebf2-3 plants were inoculated with DC3000 bacteria, and bacterial population in the leaf was determined at the indicated times.
(B) Enhanced disease resistance in ein2-1. Wild-type (Col-0) and ein2-1 plants were inoculated with DC3000 bacteria, and bacterial population in the leaf was determined at the indicated times.
(C) Enhanced PR1 expression in ein2-1 mutant plants in response to DC3000. Plants of the indicated genotypes were inoculated with DC3000 for the indicated times, and RNA was isolated for RNA gel blot analysis.
Each data point consisted of three replicates. Error bars indicate sd. * and ** indicate significant difference at 0.05 and 0.01, respectively, between mutants and Col-0 at the same time point (Student's t test). The experiments were repeated twice with similar results.
DISCUSSION
A major challenge in examining the signal transduction network in PTI defenses is to identify novel regulators in this network. In this study, we demonstrate that EIN3 and EIL1 negatively regulate PTI resistance. The ein3-1 eil1-1 double mutant showed elevated callose deposition and PR1 expression when treated with flg22 or hrpL mutant bacteria. Moreover, plants lacking both EIN3 and EIL1 display enhanced resistance to compatible, incompatible, and nonpathogenic P. syringae strains. By contrast, plants that overaccumulate EIN3 protein (35S-EIN3 transgenic plants and ebf1-3 ebf2-3 double mutants) are compromised in defenses and show enhanced disease susceptibility. Furthermore, microarray analysis uncovered that the majority of genes upregulated in the ein3-1 eil1-1 double mutant are PAMP response genes in wild-type plants. These results demonstrate that EIN3 and EIL1 negatively regulate disease resistance by repressing the PAMP-triggered transcriptional program. The flg22-induced accumulation of EIN3 protein suggests a negative feedback loop in the regulation of PTI defenses.
Microarray analysis showed a strong association of genes repressed by EIN3/EIL1 and genes induced by SA. Among the genes highly elevated in the ein3-1 eil1-1 double mutant are EDS1 and PAD4, which are known to amplify the SA defenses through a positive feedback loop (Feys et al., 2001). The SA biosynthetic gene SID2 was also strongly expressed in the ein3-1 eil1-1 double mutant. Reporter assays showed that the promoter of SID2 is hyperactive in ein3-1 eil1-1 double mutant protoplasts, suggesting that EIN3 and EIL1 transcriptionally target SID2 to regulate defenses. ChIP and EMSA experiments showed that the EIN3 protein can specifically bind to the promoter of SID2 in plants and in vitro. Furthermore, elimination of the major EIN3 binding sequence P5 resulted in a promoter less responsive to repression by EIN3. Taken together, these results support that EIN3, and possibly EIL1, directly targets the SID2 promoter to repress plant disease resistance. Importantly, the sid2 mutation completely eliminates the enhanced disease resistance phenotype caused by the ein3-1 and eil1-1 mutations. Moreover, the nine selected genes constitutively expressed at a high level in the ein3-1 eil1-1 double mutant were all reduced to normal levels by the sid2-2 mutation. These results collectively demonstrate that SID2 is a key target gene under transcriptional control of EIN3 and EIL1. Thus, EIN3 and EIL1 negatively regulate PTI defenses primarily by downregulating SID2 transcription. It was recently shown that PAMPs can induce SA biosynthesis in a SID2-dependent manner (Tsuda et al., 2008) and that the SA signaling pathway is required for the full activation of PTI defenses (Mishina and Zeier, 2007; Tsuda et al., 2008). Thus, SID2 is subjected to both positive and negative regulation during PTI defenses.
EIN3 and EIL1 have been shown to activate transcription of ET response genes, such as ERF1 (Solano et al., 1998). The defense suppression function of EIN3, however, is uncoupled from ET-induced plant development. The ein3-4 allele, which carries a P216S substitution, enhances EIN3 protein stability in plants through an unknown mechanism. Similar to the ein3-1 null mutant, ein3-4 is a recessive mutant defective in ET-induced triple response, indicating that P216 is critical for ET-induced plant development. Contrary to ein3-1, the ein3-4 mutant phenocopies EIN3 overexpression plants and represses disease resistance to P. syringae bacteria. Furthermore, ein3-4 exhibits greater repressor activity toward the SID2 promoter and acts as a semidominant allele to suppress disease resistance to P. syringae (avrB), indicating that the EIN3P216S mutant protein is fully capable of repressing plant defenses. It is clear that the defense suppression activity of EIN3 is independent of ET-induced plant development. Thus, EIN3 possesses both transcriptional activator and repressor activities, depending on the target genes. It has long been recognized that some transcription factors can act as both activator and repressor (Roy et al., 1998). It is conceivable that EIN3 and EIL1 may interact with other transcription factors or cofactors to repress SID2 transcription. For example, WRKY54 and WRKY70 are known to repress SID2 transcription, although it is not clear if they directly bind to the SID2 promoter (Wang et al., 2006).
Plant immunity is actively repressed in the absence of pathogen attack, and unregulated defenses are detrimental to plant growth and development. Thus, negative regulation is an integral part of plant immunity regulatory network (Schwessinger and Zipfel, 2008). An important feature of this network is crosstalk among different hormonal pathways (Spoel and Dong, 2008). The crosstalk allows plants to devote resources for optimum growth in the absence of pathogen attack and choose appropriate defense strategies when attacked by pathogens of different lifestyles. For example, JA and ET play an important role in plant development and mediate defenses primarily against necrotrophs and insects, whereas SA mediates resistance against biotrophic pathogens (Spoel and Dong, 2008). The crosstalk between JA and SA pathways is primarily antagonistic and has been studied extensively; here we show direct crosstalk between the ET and SA pathways. Consistent with this, elevated PR1 expression in an ein2 mutant has been observed in a previous report (Lawton et al., 1994). Our findings that ET signal transduction pathway components EIN2, EBF1, EBF2, EIN3, and EIL1 regulate disease resistance show crosstalk between the ET and SA signaling pathways in which EIN3 and EIL1 act as a regulatory node to fine-tune PTI defenses in plants.
METHODS
Plant Materials and Mutant Screen
Plant materials used in this study include a RAP2.6-LUC line (He et al., 2004), wild-type Col-0, ein3-1 (Chao et al., 1997), eil1-1 (Alonso et al., 2003), ein3-1 eil1-1 (Alonso et al., 2003), ebf1-1 ebf2-1 (Potuschak et al., 2003), sid2-2 (Dewdney et al., 2000), ein2-1 (Roman et al., 1995), and 35S:EIN3 transgenic line (Chao et al., 1997). Plants were grown in a growth room with 75% humidity under 12-h daylight at 20°C (night) and 23°C (day). The RAP2.6-LUC mutant population, mutant screening, and RAP2.6-LUC reporter assay are as described (Shang et al., 2006).
For complementation test, the rrb6 mutant was crossed to ein3-1, and F1 seedlings were scored for triple response in the presence of 10 μM ACC in Murashige and Skoog medium according to Guo and Ecker (2003). The ein3-1 eil1-1 double mutant was crossed to sid2-2 to generate the ein3-1 eil1-1 sid2 triple mutant. The triple mutant was confirmed by PCR. The single nucleotide substitution in ein3-1 and was verified using a cleaved-amplified polymorphic sequence marker (primers 5′-TACCAAGTATCAAGCGGAG-3′ and 5′-AGGCCACCAATCCTCTTTC-3′; HaeIII digest). eil1-1 contains a transposon insertion and was verified by PCR using primers 5′-GGGAATGGTGGAAAGATAAG-3′ and 5′-CTTTCGCCGTCATCTTATCC-3′. sid2-2 carries an ∼50-bp deletion in exon IX and was verified using PCR primers 5′-TTCTTCATGCAGGGGAGGAG-3′, 5′-CAACCACCTGGTGCACCAGC-3′, and 5′-AAGCAAAATGTTTGAGTCAGCA-3′ (Wildermuth et al., 2001).
To construct transgenic plants expressing EIN3-3FLAG, EIN3 cDNA was PCR amplified from reverse transcription product with primers 5′-AGGTTCGAAGAACCATATGGATACATCTTG-3′ and 5′-AAACTCGAGATGATGTTTAATGAGATGGGAATG-3′, digested with XhoI and Csp45I, inserted into a pER8-derived plasmid containing triple FLAG tag (Zuo et al., 2000; Li et al., 2005), introduced into Agrobacterium tumefaciens strain GV3101, and transformed into Arabidopsis thaliana Col-0 (Clough and Bent, 1998). A stable transgenic line expressing EIN3-3FLAG was selected and used in the ChIP experiment.
Bacterial Growth Assay
Bacterial strains used include Pseudomonas syringae pv tomato DC3000 and the hrpL mutant strain derived from DC3000 (Zwiesler-Vollick et al., 2002). Bacteria were infiltrated into 5-week-old Arabidopsis plants at 105 colony-forming units (cfu)/mL or 106 cfu/mL for bacterial growth assay as described (He et al., 2004).
RNA Gel Blot and Quantitative Real-Time RT-PCR Analyses of Gene Expression
Plants were infiltrated with 106 cfu/mL bacteria or water, and leaves were collected at the indicated times for RNA isolation. Ten micrograms of total RNA was loaded in each lane, and the RNA gel blot was hybridized with the indicated radiolabeled probes. For quantitative real-time RT-PCR, RNA was reverse transcribed into cDNA using SuperScript reverse transcriptase (Invitrogen). Real-time PCR was then performed using SYBR Green Mix and specific primers listed in Supplemental Table 3 online. The expression level was normalized to actin control.
Microarray Analysis
Wild-type and ein3-1 eil1-1 plants were treated with 2 μM flg22 or water for 3 h prior to RNA isolation. Each treatment contains three biological replicates for each genotype (each replicate consisting of a pool of RNA from six plants) for a total of 12 array hybridizations. Affymetrix ATH1 arrays were used for hybridization. Experiment data from the AtGen Express consortium were used for analyzing SA-regulated genes (Shimada et al., submission number: ME00364). GCOS software and MAS5 algorithm were used for data collection and normalization. To analyze the differentially expressed genes between control and treatment, fold change and significance analysis of microarrays (two class-paired; Tusher et al., 2001) were applied. A q value ≤0.05 and fold change ≥2 between treatment and control samples were considered as cutoff, and the induced or repressed genes were selected. Contingency tests were conducted for random overlap between two gene lists, and P value was calculated using GeneSpring GX software (http://www.chem.agilent.com/en-US/Support/FAQs/Informatics/GeneSpring%20GX/Lists2/Pages/KB001066.aspx).
Callose Staining
Five-week-old Arabidopsis leaves were infiltrated with 2 μM flg22 or 106 cfu/mL hrpL mutant bacteria for 14 h, cleared, stained with aniline blue (Hauck et al., 2003), and mounted in 50% glycerol, and epifluorescence was visualized with a fluorescence microscope under UV light. The number of callose deposits per microscopic field of 0.1 mm2 was calculated from six leaves using the Image J software (http://www.uhnresearch.ca/wcif).
SA Quantitation
Total and free SA was quantitated according to Li et al. (1999). Briefly, frozen leaf tissue (0.1 g) was ground in liquid nitrogen, extracted twice with 90% methanol by vortex and sonication, and the sample (free SA) was dried under vacuum. The free SA sample was treated with β-glucosidase to yield total SA sample. Both free SA and total SA, samples were extracted once with 5% trichloroacetic acid and three times with 100/99/1 (vol) ethylacetate/cyclopentane/isopropanol. The dried SA was then resuspended in 250 μL mobile phase (0.2 M KAc and 0.5 mM EDTA, pH 5) and separated through a 100 × 4.6 sperisorb DDS2 column (Keystone Scientific) with a particle size of 3 μm and a pore size of 80 A° at a mobile-phase flow rate of 1 mL/min. Fluorescent detection was performed on an HPLC spectrofluorescence detector with a Xenon-mercury arc lamp at an excitation/emission wavelength of 295/405 nm.
Immunoblot Assay
Total protein was extracted from 4-d-old etiolated seedlings. EIN3 protein was determined by immunoblot using antibodies raised against recombinant EIN3 protein (Guo and Ecker, 2003). For loading control, the blot was hybridized to anti-HSP90 antibodies (Provided by Gang Zhi, Antibody Core Facility, National Institute of Biological Sciences).
ChIP
ChIP was performed as described previously with minor modifications (Gendrel et al., 2005). Briefly, wild-type and EIN3-3FLAG transgenic seeds were sterilized and grown on half Murashige and Skoog medium in the presence of 10 μM β-estrodial under continuous white light for 8 d. Seedlings (2.5 g) were fixed in 1% formaldehyde for 15 min in vacuum and neutralized with 0.125 M glycine in vacuum for additional 5 min. After washing twice with cold, sterilized water, the tissue was ground in liquid nitrogen. Nuclei were isolated and sonicated. Sonicated chromatin supernatant (300 μL) was diluted to 3 mL, and 20 μL of protein A-agarose bead (Upstate) was added for preclear at 4°C for 1 h. The chromatin was then divided into two 1.5-mL aliquots. Twenty microliters of mouse anti-FLAG M2-agarose beads (Sigma-Aldrich) were added to one tube, 20 μL of protein A-agarose beads were added to the other as “no antibody control.” After incubating at 4°C for overnight, beads were washed with low salt wash buffer, high salt wash buffer, and TE buffer. Elution and reversed cross-linking was done as previously described (Gendrel et al., 2005). Eluates were treated with Proteinase K (10 mg/mL; Sigma-Aldrich) and RNase for 2.5 h at 45°C, phenol/chloroform extracted, and ethanol precipitated with the aid of 20 μg glycogen. The purified DNA was resuspended in 80 μL of water. The enrichment of DNA fragments was determined by quantitative real-time PCR using primers listed in Supplemental Table 3 online.
EMSA
To construct plasmid for the expression of recombinant EIN3 protein (amino acids 141 to 352) in Escheichia coli, the correspond DNA fragment was amplified by PCR using primers 5′-ACTGGATCCAAGGTTAGGTTTGATCGT-3′ and 5′-ACTCTCGAGTCAGAAGAATTCATAACTTTT-3′ and inserted into BamHI and XhoI of the pGEX-6p-1 vector (Pharmacia). For probe, 4 pM PCR-amplified P5 DNA fragment was 32P-labeled using T4-polynucleotide kinase (New England Biolabs) in the presence of 20 μCi [γ-32P]ATP and purified by Sephadex G-50 column. EMSA was performed as described (Kosugi and Ohashi, 2000). Each binding reaction (19 μL) contained 200 ng recombinant protein, 5 ng labeled DNA probe, 0.1 μg poly[d(I-C)] (Sigma-Aldrich), 20 mM HEPES, pH 7.8, 3 mM MgCl2, 1 mM DTT, 1 mM EDTA, 40 mM KCl, and 10% glycerol. After incubation on ice for 30 min, the mixtures were loaded onto 4.5% polyacrylamide gels (29:1) to separate free and bound DNA. For competition experiments, the following primers were used: Obs1, 5′-AACGATGTACCTGGTCGTATT-3′; Obsm2, 5′-GTACATTTACCTGGACCGTGA-3′; and Obsm123, 5′-GTACCTTTCCCTGGACCGTGA-3′ (Kosugi and Ohashi, 2000).
Construct for Promoter-Reporter Assay
A pUC19-GUS-RBS vector was generated from pUC19-35S-FLAG-RBS plasmid (Li et al., 2005), replacing 3×FLAG sequence with GUS sequence between XhoI and PstI. To generate SID2-GUS construct, a 1.6-kb SID2 promoter region was amplified from genomic DNA with primers 5′-CACGAATTCTTCGTAGCATCCACAACAC-3′ and 5′-ACTGGTACCTGCAGAAATTCGTAAAGTG-3′ and inserted between EcoRI and KpnI sites of pUC19-GUS vector. The SID2P5Δ-GUS construct containing a deletion between −324 and −251 (from translational start site) of the SID2 promoter was generated by overlap extension PCR using primers 5′-TAGACCAAGTAAATGAAGTAGGATTAGAAG-3′ and 5′-CTTCTAATCCTACTTCATTTACTTGGTCTA-3′.
To generate 35S-rLUC construct, the coding sequence of Renilla luciferase fragments was amplified from pSP-luc(+) (Promega) using primers 5′-ATAGGTACCATGGCTTCCAAGGTGTACGAC-3′ and 5′-GTACTGCAGTTACTGCTCGTTCTTCAGCAC-3′ and inserted into KpnI and PstI of pUC19-35S-FLAG-RBS plasmid, resulting in pUC19-35S-rLUC-RBS.
To generate SID2-LUC, the 3×FLAG sequence in pUC19-35S-FLAG-RBS plasmid (Li et al., 2005) was first replaced with the coding sequence of firefly luciferase gene LUC between XhoI and PstI, resulting in pUC19-LUC-RBS. The SID2 promoter was PCR amplified with primers 5′- CACCAATTGAATTCAACTAACGTCCTAT-3′ and 5′-ACTGGTACCTGCAGAAATTCGTAAAGTG-3′, digested with MfeI and KpnI, and inserted into pUC19-LUC-RBS vector, resulting in SID2-LUC construct.
Protoplasts were transfected with plasmids as described (Li et al., 2005). For GUS assay, protoplasts were transfected with SID2-GUS or SID2P5D-GUS along with 35S-rLUC and incubated for 12 to 14 h at room temperature before harvested for GUS activity assay. GUS activity was assayed with methyl umbelliferyl glucoronide (Sigma-Aldrich), and renilla luciferase activity was assayed using a luciferase assay kit (Promega) following the manufacturer's instruction. The GUS/rLUC ratio was used to determine the promoter activity. The SID2-LUC reporter activity was determined as described (Li et al., 2005).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Actin (At3g18780), EIN3 (At3g20770), EIL1 (At2g27050), SID2 (At1g74710), PR1 (At2g14610), PR2 (At3g57260), EDS1 (At3g48090), NIMIN1 (At1g02450), PBS3 (At5g13320), a putative defense-related protein (At4g30530), adenosine-5′-phosphosulfate-kinase (AKN2; At4g39940), leucine-rich repeat transmembrane protein (At2g31880), calmodulin-related protein (At3g01830), C3HC4-type RING finger family protein (At2g42360), and thioredoxin-dependent peroxidase 2 (At1g65970).
Supplemental Data
The following materials can be found in the online version of this article.
Supplemental Figure 1. Enhanced Disease Symptoms in ein3-4 Plants.
Supplemental Figure 2. ein3-4 Is a Semidominant Mutation That Enhances Disease Susceptibility.
Supplemental Figure 3. ein3-4 Shows Greater Repression of SID2 Promoter.
Supplemental Figure 4. Total SA Level in ein3-1 eil1-1 Double Mutant Plants after Bacterial Infection.
Supplemental Figure 5. SID2 P5 Sequence Contributes to Transcriptional Repression by EIN3 and EIL1.
Supplemental Table 1. Complementation Test of rrb6 and ein3-1.
Supplemental Table 2. Quantitative Real-Time RT-PCR Analysis of Genes Upregulated in ein3-1 eil1-1.
Supplemental Table 3. Primers Used for Quantitative PCR Analyses.
Supplemental Data Set 1. Genes Induced by flg22.
Supplemental Data Set 2. Genes Repressed by flg22
Supplemental Data Set 3. Genes Upregulated in Water-Treated ein3 eil1 Plants.
Supplemental Data Set 4 Genes Downregulated in Water-Treated ein3/eil1 Plants.
Supplemental Data Set 5. Genes Induced by SA.
Supplementary Material
Acknowledgments
We thank Gang Zhi for preparing anti-HSP90 antibodies, Frederick Ausubel for the sid2-2 seeds, Pascal Genschik for ebf1-3 ebf-2-3 seeds, and Mary Wildermuth for sharing sid2-2 genotyping method. J.Z. was supported by grants from Chinese Ministry of Science and Technology (2003-AA210080).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jian-Min Zhou (zhoujianmin@nibs.ac.cn).
Online version contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Alonso, J.M., Stepanova, A.N., Solano, R., Wisman, E., Ferrari, S., Ausubel, F.M., and Ecker, J.R. (2003). Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 2992–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.M. (2005). Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 6: 973–979. [DOI] [PubMed] [Google Scholar]
- Bent, A.F., Innes, R.W., Ecker, J.R., and Staskawicz, B.J. (1992). Disease development in ethylene-insensitive Arabidopsis thaliana infected with virulent and avirulent Pseudomonas and Xanthomonas pathogens. Mol. Plant Microbe Interact. 5: 372–378. [DOI] [PubMed] [Google Scholar]
- Bittel, P., and Robatzek, S. (2007). Microbe-associated molecular patterns (MAMPs) probe plant immunity. Curr. Opin. Plant Biol. 10: 335–341. [DOI] [PubMed] [Google Scholar]
- Chao, Q., Rothenberg, M., Solano, R., Roman, G., Terzaghi, W., and Ecker, J.R. (1997). Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144. [DOI] [PubMed] [Google Scholar]
- Chisholm, S.T., Coaker, G., Day, B., and Staskawicz, B.J. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124: 803–814. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Dewdney, J., Reuber, T.L., Wildermuth, M.C., Devoto, A., Cui, J., Stutius, L.M., Drummond, E.P., and Ausubel, F.M. (2000). Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J. 24: 205–218. [DOI] [PubMed] [Google Scholar]
- Durrant, W.E., and Dong, X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42: 185–209. [DOI] [PubMed] [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95: 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.J., Moisan, L.J., Newman, M.A., and Parker, J.E. (2001). Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20: 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel, A.V., Lippman, Z., Martienssen, R., and Colot, V. (2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2: 213–218. [DOI] [PubMed] [Google Scholar]
- Guo, H., and Ecker, J.R. (2003). Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677. [DOI] [PubMed] [Google Scholar]
- Guo, H., and Ecker, J.R. (2004). The ethylene signaling pathway: New insights. Curr. Opin. Plant Biol. 7: 40–49. [DOI] [PubMed] [Google Scholar]
- Guzmán, P., and Ecker, J.R. (1990). Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck, P., Thilmony, R., and He, S.Y. (2003). A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc. Natl. Acad. Sci. USA 100: 8577–8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, P., Chintamanani, S., Chen, Z., Zhu, L., Kunkel, B.N., Alfano, J.R., Tang, X., and Zhou, J.M. (2004). Activation of a COI1-dependent pathway in Arabidopsis by Pseudomonas syringae type III effectors and coronatine. Plant J. 37: 589–602. [DOI] [PubMed] [Google Scholar]
- Ichimura, K., Casais, C., Peck, S.C., Shinozaki, K., and Shirasu, K. (2006). MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. J. Biol. Chem. 281: 36969–36976. [DOI] [PubMed] [Google Scholar]
- Jones, J.D., and Dangl, J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Kosugi, S., and Ohashi, Y. (2000). Cloning and DNA-binding properties of a tobacco Ethylene-Insensitive3 (EIN3) homolog. Nucleic Acids Res. 28: 960–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Lin, H., Zhang, W., Zou, Y., Zhang, J., Tang, X., and Zhou, J.M. (2005). Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc. Natl. Acad. Sci. USA 102: 12990–12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Zhang, Y., Clarke, J.D., Li, Y., and Dong, X. (1999). Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell 98: 329–339. [DOI] [PubMed] [Google Scholar]
- Loake, G., and Grant, M. (2007). Salicylic acid in plant defence-the players and protagonists. Curr. Opin. Plant Biol. 10: 466–472. [DOI] [PubMed] [Google Scholar]
- Lawton, K., Potter, S., Uknes, S., and Ryals, J. (1994). Acquired resistance signal transduction in Arabidopsis is ethylene independent. Plant Cell 6: 581–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina, T.E., and Zeier, J. (2007). Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J. 50: 500–513. [DOI] [PubMed] [Google Scholar]
- Mou, Z., Fan, W., and Dong, X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113: 935–944. [DOI] [PubMed] [Google Scholar]
- Navarro, L., Zipfel, C., Rowland, O., Keller, I., Robatzek, S., Boller, T., and Jones, J.D.G. (2004). The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 135: 1113–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath, C., and Metraux, J.P. (1999). Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, M., et al. (2000). Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103: 1111–1120. [DOI] [PubMed] [Google Scholar]
- Potuschak, T., Lechner, E., Parmentier, Y., Yanagisawa, S., Grava, S., Koncz, C., and Genschik, P. (2003). EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115: 679–689. [DOI] [PubMed] [Google Scholar]
- Qiu, J.L., Zhou, L., Yun, B.W., Nielsen, H.B., Fiil, B.K., Petersen, K., Mackinlay, J., Loake, G.J., Mundy, J., and Morris, P.C. (2008). Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol. 148: 212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman, G., Lubarsky, B., Kieber, J.J., Rothenberg, M., and Ecker, J.R. (1995). Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: Five novel mutant loci integrated into a stress response pathway. Genetics 139: 1393–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, S., Garges, S., and Adhya, S. (1998). Activation and repression of transcription by differential contact: Two sides of a coin. J. Biol. Chem. 273: 14059–14062. [DOI] [PubMed] [Google Scholar]
- Schwessinger, B., and Zipfel, C. (2008). News from the frontline: Recent insights into PAMP-triggered immunity in plants. Curr. Opin. Plant Biol. 11: 389–395. [DOI] [PubMed] [Google Scholar]
- Shang, Y., Li, X., Cui, H., He, P., Thilmony, R., Chintamanani, S., Zwiesler-Vollick, J., Gopalan, S., Tang, X., and Zhou, J.M. (2006). RAR1, a central player in plant immunity, is targeted by Pseudomonas syringae effector AvrB. Proc. Natl. Acad. Sci. USA 103: 19200–19205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano, R., Stepanova, A., Chao, Q., and Ecker, J.R. (1998). Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12: 3703–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel, S.H., and Dong, X. (2008). Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe 3: 348–351. [DOI] [PubMed] [Google Scholar]
- Spoel, S.H., Mou, Z., Tada, Y., Spivey, N.W., Genschik, P., and Dong, X. (2009). Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137: 860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada, Y., Spoel, S.H., Pajerowska-Mukhtar, K., Mou, Z., Song, J., Wang, C., Zuo, J., and Dong, X. (2008). Plant immunity requires conformational charges of NPR1 via S-nitrosylation and thioredoxins. Science 321: 952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda, K., Sato, M., Glazebrook, J., Cohen, J.D., and Katagiri, F. (2008). Interplay between MAMP-triggered and SA-mediated defense responses. Plant J. 53: 763–775. [DOI] [PubMed] [Google Scholar]
- Tusher, V.G., Tibshirani, R., and Chu, G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98: 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D., Amornsiripanitch, N., and Dong, X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth, M.C., Dewdney, J., Wu, G., and Ausubel, F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565. [DOI] [PubMed] [Google Scholar]
- Yamasaki, K., et al. (2005). Solution structure of the major DNA-binding domain of Arabidopsis thaliana Ethylene-Insensitive3-Like3. J. Mol. Biol. 348: 253–264. [DOI] [PubMed] [Google Scholar]
- Zipfel, C., Robatzek, S., Navarro, L., Oakeley, E.J., Jones, J.D., Felix, G., and Boller, T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767. [DOI] [PubMed] [Google Scholar]
- Zuo, J., Niu, Q.W., and Chua, N.H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273. [DOI] [PubMed] [Google Scholar]
- Zwiesler-Vollick, J., Plovanich-Jones, A.E., Nomura, K., Bandyopadhyay, S., Joardar, V., Kunkel, B.N., and He, S.Y. (2002). Identification of novel hrp-regulated genes through functional genomic analysis of the Pseudomonas syringae pv tomato DC3000 genome. Mol. Microbiol. 45: 1207–1218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.