Abstract
Jasmonate signaling plays an important role in both plant defense and development. Here, we have identified a subunit of the Mediator complex as a regulator of the jasmonate signaling pathway in Arabidopsis thaliana. The Mediator complex is a conserved multiprotein complex that acts as a universal adaptor between transcription factors and the RNA polymerase II transcriptional machinery. We report that the PHYTOCHROME AND FLOWERING TIME1 (PFT1) gene, which encodes the MEDIATOR25 subunit of Mediator, is required for jasmonate-dependent defense gene expression and resistance to leaf-infecting necrotrophic fungal pathogens. Conversely, PFT1 appears to confer susceptibility to Fusarium oxysporum, a root-infecting hemibiotrophic fungal pathogen known to hijack jasmonate responses for disease development. Consistent with this, jasmonate gene expression was suppressed in the pft1 mutant during infection with F. oxysporum. In addition, a wheat (Triticum aestivum) homolog of PFT1 complemented the defense and the developmental phenotypes of the pft1 mutant, suggesting that the jasmonate signaling functions of PFT1 may be conserved in higher plants. Overall, our results identify an important control point in the regulation of the jasmonate signaling pathway within the transcriptional machinery.
INTRODUCTION
In response to an attempted infection, plants activate an inbuilt system of defense, which results in the production of a variety of pathogenesis-related (PR) proteins and secondary metabolites, as well as low molecular weight defense-related hormones, such as salicylates (SAs) and jasmonates (JAs). These defense-related hormones act both locally and systemically to orchestrate the plant's defense signaling network through the activation of transcription factors. Thus, transcriptional regulation of defense gene expression plays a major role in determining whether a plant is more or less resistant to pathogen attack (McGrath et al., 2005).
Recently, a new component of the plant's transcriptional machinery has been identified with the purification of the Mediator complex in Arabidopsis thaliana (Bäckström et al., 2007). The Mediator complex is a large multiprotein complex that is conserved in all eukaryotes, from yeast to humans, whose characterization had until recently remained elusive in plants. The function of the Mediator complex is to act as a bridge between the RNA polymerase II complex and the myriad of transcription factors present within the cell (Kim et al., 1994; Koleske and Young, 1994). By binding to distal activators/repressors as well as general transcription factors at the promoter site, Mediator fine-tunes diverse regulatory inputs and presents a balanced output to the RNA polymerase II complex to initiate transcription (Malik and Roeder, 2005).
Mediator subunits are organized into three core modules, termed the head, middle, and tail, as well as an additional detachable kinase module. The tail module of Mediator is thought to interact primarily with DNA-bound transcription factors, while the head and middle modules bind to the C-terminal domain of RNA polymerase II (reviewed in Malik and Roeder, 2005). Depending on the organism, the Mediator complex contains ∼20 to 30 subunits. For example, the Mediator from the yeast Saccharomyces cerevisiae is composed of 25 subunits, of which 22 subunits are at least partially conserved among eukaryotes (Boube et al., 2002; Bourbon et al., 2004). Bäckström et al. (2007) identified 21 conserved and six putative plant-specific Mediator subunits in Arabidopsis. It is expected that individual Mediator subunits recognize and respond to a subset of the ∼1500 transcription factors present in the Arabidopsis genome. Therefore, determining which transcription factors each Mediator subunit recognizes would provide a step forward in the understanding of how the Mediator complex manages to integrate the complex transcriptional information in plants.
Prior to its identification as a Mediator subunit, MEDIATOR25 (MED25) was first described as a positive regulator of shade avoidance in Arabidopsis and was termed PHYTOCHROME AND FLOWERING TIME1 (PFT1) (Cerdán and Chory, 2003). Cerdán and Chory (2003) hypothesized that PFT1 might act downstream of phytochrome B to promote flowering in response to shade. However, Wollenberg et al. (2008) have since found that the pft1 mutant did not show an altered flowering response when grown in simulated shade (far-red light–enriched long-day [LD] conditions). As a result, PFT1 is now considered to be a gene that negatively regulates the phytochrome signaling pathway as opposed to being a component of the phytochrome signaling pathway itself.
Here, we report that PFT1 also acts as a positive regulator of JA signaling that regulates plant defense responses during fungal pathogen infection. In a genome-wide analysis of Arabidopsis transcription factor gene expression, we previously reported that PFT1 expression was reduced in response to methyl jasmonate (MeJA) and during the defense response to an incompatible isolate of the necrotrophic fungal pathogen Alternaria brassicicola (McGrath et al., 2005). In this report, we show that PFT1 is required for uncompromised expression of JA-dependent defense genes and resistance to the leaf-infecting necrotrophic pathogens A. brassicicola and Botrytis cinerea. We have also found that PFT1 is necessary for susceptibility to the root-infecting hemibiotrophic fungal pathogen Fusarium oxysporum, which requires intact JA signaling in the host to promote disease symptoms (Thatcher et al., 2009). In addition, through the analysis of several Mediator subunit mutants, we have identified an additional Mediator subunit, MED8, which is also a regulator of both flowering time and disease resistance. These results provide new insights into the regulatory role of plant Mediator subunits in determining fungal disease outcomes as well as the initiation of flowering.
RESULTS
PFT1 Is Required for Basal Resistance to Necrotrophic Fungal Pathogens
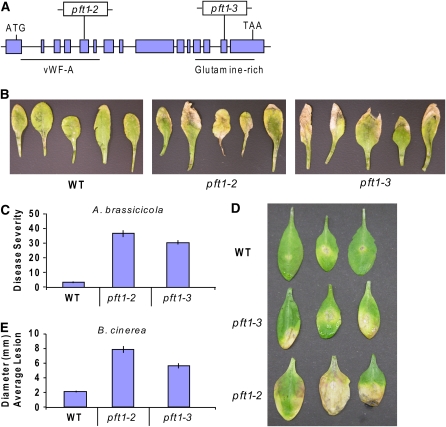
Previous research investigating the responses of the Arabidopsis transcription factor transcriptome during plant defense had demonstrated that PFT1 transcript levels were reduced in Arabidopsis leaves either challenged with A. brassicicola or treated with MeJA (McGrath et al., 2005; see Supplemental Table 1 online). To analyze potential roles of PFT1 in plant defense, we isolated two homozygous lines harboring independent T-DNA insertions in the PFT1 gene (Alonso et al., 2003). These lines were designated as pft1-2 and pft1-3 in sequence after the pft1-1 mutant previously characterized by Cerdán and Chory (2003). Like the pft1-1 mutant, the pft1-2 mutant contains a T-DNA insertion located in the 5th exon of the PFT1 gene. This insertion lies within a genomic region that encodes a von Willebrand Factor A domain (vWF-A) located at the N terminus of the PFT1 protein (Figure 1A). The vWF-A domain is a widely distributed protein–protein interaction domain (Whittaker and Hynes, 2002) and has been shown to be critical for the binding of MED25 to the Mediator complex in human cell lines (Mittler et al., 2003). The T-DNA in the pft1-3 mutant is inserted into the 14th exon of PFT1 (Figure 1A) and disrupts the Gln-rich region predicted to function as a putative transcriptional activation domain (Cerdán and Chory, 2003).
Figure 1.
PFT1 Is Required for Basal Resistance to the Leaf-Infecting Necrotrophic Pathogens A. brassicicola and B. cinerea.
(A) Schematic representation of the PFT1 gene and the locations of two independent T-DNA insertions designated as pft1-2 (SALK_129555) and pft1-3 (SALK_059316). Introns (solid line) and exons (boxes) are indicated.
(B) Symptoms on rosette leaves of 4-week-old soil-grown wild-type, pft1-2, and pft1-3 plants 2 weeks following drop inoculation with freshly harvested spores of A. brassicicola.
(C) The percentages of leaves showing chlorotic regions calculated 2 weeks after inoculations with A. brassicicola.
(D) Symptoms on rosette leaves of 4-week-old soil-grown wild-type, pft1-2, and pft1-3 plants following drop inoculation with freshly harvested spores of B. cinerea.
(E) The average lesion diameter measured 4 d after inoculation with B. cinerea.
Error bars in (C) and (E) represent se from three replicated experiments that contained 20 to 30 plants each.
To determine whether PFT1 is required during plant defense, pft1-2, pft1-3, and wild-type plants were inoculated with the leaf-infecting necrotrophic fungal pathogens A. brassicicola and B. cinerea. The A. brassicicola isolate used here is incompatible on wild-type (Columbia-0 [Col-0]) Arabidopsis (Schenk et al., 2000, 2003) but has been shown to be capable of causing lesions on Arabidopsis mutants with attenuated plant defenses (Trusov et al., 2006). As shown in Figure 1B, inoculations with A. brassicicola resulted in the development of distinct chlorotic regions restricted to older rosette leaves of the pft1 mutant. In these experiments, 35 and 30% of the inoculated pft1-2 and pft1-3 leaves, respectively, showed chlorosis (Figure 1C). Furthermore, the pathogen was often able to sporulate on these chlorotic lesions. By contrast, only 3% of the inoculated leaves from wild-type plants had similar chlorotic regions (Figure 1C). Similarly, B. cinerea, although poorly compatible on wild-type plants, produced larger chlorotic regions on the pft1 mutant leaves than those on wild-type leaves (Figures 1D and 1E). Thus, these pathogen inoculation experiments suggest that PFT1 is an important component of basal resistance to these necrotrophic pathogens in Arabidopsis.
PFT1 Is a F. oxysporum Susceptibility Gene
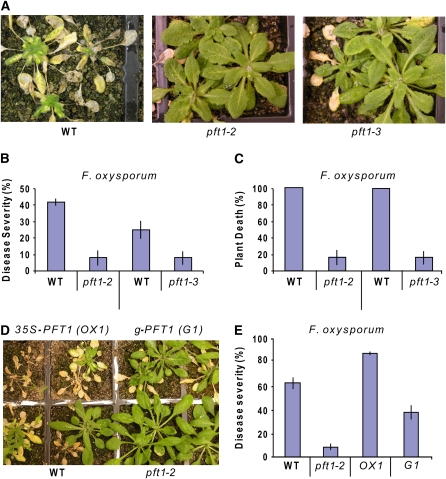
To determine whether PFT1 is also required for resistance to a pathogen with a different lifestyle, we conducted inoculation experiments with the root-infecting hemibiotrophic fungal pathogen F. oxysporum, which causes vascular wilt disease in a wide range of plants, including Arabidopsis (Diener and Ausubel, 2005; Edgar et al., 2006; Berrocal-Lobo and Molina, 2008; Michielse and Rep, 2009). The number of leaves with chlorosis/necrosis and the percentage of plant death after inoculations were substantially lower (P < 0.01) in the pft1 mutants than those in the wild-type plants (Figures 2A to 2C). The increased resistance to F. oxysporum and increased susceptibility against B. cinerea and A. brassicicola in pft1 mutants was reminiscent of the reaction to these pathogens in the coi1 mutant, which has impaired JA signaling (Thatcher et al., 2009).
Figure 2.
PFT1 Is an F. oxysporum Susceptibility Gene.
(A) Typical disease symptoms and plant death observed on wild-type (Col-0) and pft1 mutants 2 weeks after inoculations of the roots of 3- to 4-week-old plants.
(B) The number of leaves showing chlorosis and necrosis 10 d after inoculations and expressed as the percentage of total number of leaves.
(C) Percentages of plant death 2 weeks after the inoculation of roots with F. oxysporum.
(D) Typical disease phenotypes of wild-type (Col-0), pft1, OX1 (35S:PFT1), and G1 (the pft1 mutant complemented with a genomic PFT1) plants at 12 d after F. oxysporum inoculation.
(E) Average percentage of disease severity measured in the same lines shown in (D). Error bars represent se from three replicated experiments that contained 20 to 30 plants each.
To confirm that the increased F. oxysporum resistance was due to a loss-of-function mutation in the PFT1 gene, the Fusarium resistance phenotypes of the pft1-2 mutant, a 35S-PFT1 overexpressing line (OX1) as well as the pft1 mutant transformed with a genomic copy of PFT1 (G1), were analyzed in replicated inoculation experiments. The PFT1-overexpressing line and the PFT1 genomic complement, designated as OX1 and G1, respectively, have been previously characterized in relation to the function of PFT1 in phytochrome signaling (Cerdán and Chory, 2003). Consistent with the previous inoculation experiments, the pft1-2 plants were mostly free of visible symptoms, while typical F. oxysporum disease symptoms (e.g., vein chlorosis) appeared on a high proportion of the wild-type, OX1, and G1 plants (Figure 2D). The OX1 plants were the most affected, with ∼90% of the OX1 leaves showing visible symptoms, while the G1 plants, as expected, showed symptoms comparable to those on wild-type plants (Figures 2D and 2E). Finally, because G1 was generated in the pft1-1 background (Cerdán and Chory, 2003), we tested the response of the pft1-1 mutant to F. oxysporum. As expected, pft1-1 was also resistant to F. oxysporum. Together, these experiments clearly demonstrate that PFT1 increases susceptibility to F. oxysporum.
To determine when PFT1 may be required for susceptibility, F. oxysporum–inoculated roots of pft1, wild-type, and OX1 plants were analyzed by confocal microscopy after staining with a fluorescent wheat germ agglutinin conjugate to differentiate the fungal tissue. These analyses, although qualitative in nature, clearly showed that fungal hyphae were present in the roots of pft1, wild-type, and OX1 plants at the same time periods following inoculations (see Supplemental Figure 1 online). In addition, we could find no evidence of increased callose production or altered structural differences in pft1 lines upon infection with F. oxysporum. This suggests that the pft1 mutation did not alter the sensitivity of pft1 roots to F. oxysporum infection and that resistance was expressed later in infection as symptoms develop.
PFT1 Is a Positive Regulator of Defense Gene Expression in Arabidopsis
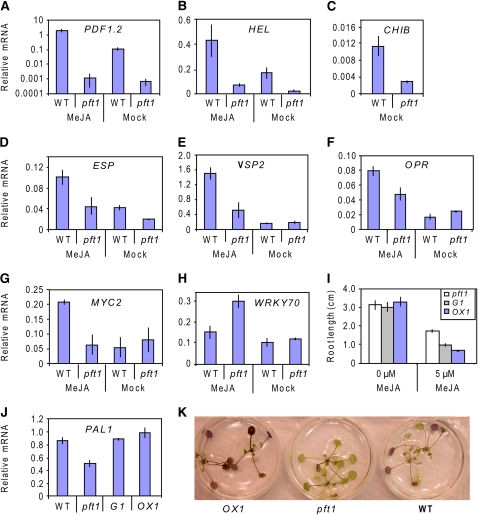
Because the contrasting disease phenotypes displayed by the pft1 mutant against different pathogens resembled those observed in the JA signaling mutant coi1 (Thomma et al., 1998; Thatcher et al., 2009), we analyzed the expression of a number of JA-responsive defense genes in pft1 mutants. As shown in Figures 3A to 3D, the JA-responsive genes PDF1.2, HEL, CHIB, and ESP showed reduced basal expression levels in pft1 mutant plants relative to the wild type. In addition, PDF1.2, HEL, ESP, VSP2, and OPR, although responsive to MeJA, showed reduced expression levels in pft1 mutants (Figures 3A, 3B, and 3D to 3F). CHIB did not show any JA inducibility at this time point; therefore, the expression of CHIB in the treated samples was equivalent to that of the mock samples. MYC2, encoding a regulator of JA signaling, was inducible by MeJA in the wild type but not inducible in pft1 mutants (Figure 3G). By contrast, WRKY70, encoding a negative regulator of JA signaling, showed higher expression levels in pft1 mutants in response to MeJA (Figure 3H). In separate sets of experiments, we further examined the expression of these defense genes in wild-type, G1, and OX1 plants and found increased transcript levels of JA-responsive genes in OX1 plants (see Supplemental Figure 2 online). In addition, pft1 roots were less sensitive to growth inhibition by MeJA than G1 and OX1 roots (Figure 3I). Together, these data suggest a positive regulatory role for PFT1 in JA signaling.
Figure 3.
PFT1 Is a Positive Regulator of JA-Dependent Defenses in Arabidopsis.
(A) to (H) Expression from JA-responsive genes PDF1.2, HEL, CHIB, ESP, VSP, OPR, MYC2, and WRKY70 was examined by quantitative RT-PCR in wild-type (Col-0) and pft1 mutants 6 h following MeJA treatment.
(I) The root lengths of pft1, OX1, and G1 seedlings germinated on half-strength Murashige and Skoog (MS) media containing either 0 or 5 μM of MeJA were measured 6 d after germination. Error bars represent the se from three independent experiments that included ∼10 seedlings each.
(J) Gene expression of PAL1 was examined in 3- to 4-week-old, MeJA-treated wild-type (Col-0), pft1, G1 (genomic complement) , and OX1 (35S:PFT1) plants. The expression data were normalized relative to the expression level of β-ACTIN (See Methods) and presented logarithmically for PDF1.2. Error bars in (A) to (H) and (J) represent se from three independent experiments that contained at least 20 plants each.
(K) To visualize anthocyanin accumulation, 2-week-old soil-grown plants of wild-type, pft1, and OX1 were incubated in water until visible anthocyanin developed in the leaves.
[See online article for color version of this figure.]
We also noted that MYC2 and WRKY70 (differentially regulated in pft1 mutants) encode a positive and a negative regulator of the anthocyanin pathway, respectively (Li et al., 2006; Dombrecht et al., 2007). In addition, the basal transcript levels of PAL1, which encodes a major isoform of the enzyme Phe ammonia lyase involved in phenylpropanoid biosynthesis, was lower in the pft1 mutant than in wild-type, G1, and OX1 plants (Figure 3J). Consistent with these expression data, the pft1 mutant showed a lack of anthocyanin production, while wild-type and OX1 plants produced strong anthocyanin accumulation (Figure 3K).
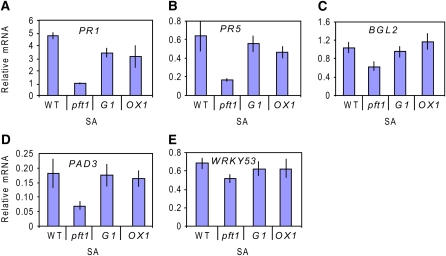
As the pft1 mutant showed attenuated JA defense gene expression, we also investigated whether SA-responsive defense genes showed any differential expression in the mutant. Interestingly, the SA-responsive defense genes PR1, PR5, and BGL2, the phytoalexin biosynthesis gene PAD3, and the transcriptional activator WRKY33 all showed attenuated induction in the pft1 mutant relative to the wild-type, G1, and OX1 plants after treatment with SA (Figures 4A to 4E). Together, these results suggest that PFT1 is a positive regulator of defense gene expression in Arabidopsis.
Figure 4.
PFT1 Is a Positive Regulator of SA-Responsive Defense Gene Expression.
Expression of PR1, PR5, BGL2, PAD3, and WRKY53 was examined by quantitative RT-PCR in wild-type (Col-0), pft1, G1, and OX1 plants 24 h following SA treatment. The expression data were normalized relative to β-ACTIN expression (see Methods). Error bars represent se from three independent experiments that contained ∼20 plants each.
[See online article for color version of this figure.]
We noted that in the experiments described above, defense gene expression was only analyzed in the leaf tissue where disease symptoms were scored. Because F. oxysporum infects the plants through the roots, we also examined the transcript levels of the same defense genes from the shoot by performing quantitative RT-PCR experiments in the roots of F. oxysporum–inoculated pft1 mutant and wild-type plants. For all defense genes tested, with the exception of HEL, no difference in basal expression levels was observed in the pft1 roots. In addition, no significant upregulation of these genes in the roots of either pft1 or the wild type by F. oxysporum was evident (see Supplemental Figure 3 online). HEL, on the other hand, showed increased expression in wild-type roots after F. oxysporum infection, but both the basal and F. oxysporum–responsive expression level was lower in pft1 roots than the basal wild-type expression level (see Supplemental Figure 3 online). We also conducted quantitative RT-PCR experiments using the shoot tissue of F. oxysporum–infected wild-type and pft1 plants. These analyses also showed a significantly lower expression of defense genes in pft1 and confirmed the earlier results from the SA and MeJA quantitative RT-PCR experiments (see Supplemental Figure 3 online). We therefore concluded that the pft1-mediated F. oxysporum resistance occurs independently from elevated defense gene expression in either the roots or the shoots.
F. oxysporum Resistance in pft1 Is Associated with Attenuated Jasmonate Signaling
As defense against leaf-infecting necrotrophic pathogens requires intact JA signaling, the increased susceptibility of the pft1 mutant to A. brassicicola and B. cinerea was most likely due to the attenuated expression of JA-associated defense genes (Thomma et al., 1998, 1999; Zhou et al., 1999). However, the increased F. oxysporum resistance observed in the pft1 mutant despite reduced defense gene expression suggests that an alternative resistance mechanism was operating against this pathogen. To identify the genes that may be contributing to increased F. oxysporum resistance in the pft1 mutant on a genome-wide scale, we performed a microarray experiment using the Affymetrix ATH1 Genome Array, using four independent replicates of pft1 and wild-type Arabidopsis leaves collected 2 d after either a mock treatment or inoculation with F. oxysporum. To analyze the microarray data, we performed a two-way analysis of variance (ANOVA; P < 0.05) on the entire data set with the inclusion of the Benjamini and Hochberg false discovery rate to reduce the number of potential false positives in our results.
This stringent analysis identified 39 genes that were differentially expressed after F. oxysporum treatment, irrespective of the genotype, and 284 genes that were differentially expressed between the genotypes, irrespective of the treatment (see Supplemental Tables 1 and 2 online). Therefore, the genotype had a more substantial effect in this experiment than the F. oxysporum treatment. While the number of differentially expressed genes from the F. oxysporum treatment was quite low, there were several defense genes found to be induced, signifying that the infection was successful (see Supplemental Table 1 online). Of the 39 F. oxysporum differentially expressed genes, 22 genes were also identified as being differentially expressed between pft1 and the wild type (Table 1; see Supplemental Table 3 online). All except for one of the 22 genes were found to be induced by F. oxysporum in both genotypes, but their expression was significantly lower in pft1 than the wild type under both mock and infected conditions (Table 1). Interestingly, a number of these genes were found to have roles in JA biosynthesis and signaling. Included in the list was the JA carboxyl methyltransferase as well as the JA ZIM-domain gene (JAZ9) (Seo et al., 2001; Chini et al., 2007; Farmer, 2007; Thines et al., 2007). Previously mentioned JA-associated genes from the quantitative RT-PCR analysis, such as PDF1.2 and HEL, also appeared in the list as well as other defense-related genes, such as a putative thionin and a lectin family protein.
Table 1.
Genes Identified from the Microarray Experiment That Are Differentially Expressed Both between the Wild-Type and pft1 Genotypes and Also after F. oxysporum Treatment (Two-Way ANOVA, P < 0.05 Adjusted by the False Discovery Rate)
| AGI Locus | P Value | Infected/Mock | pft1/Wild Type | AGI Description | Jasmonate Inducibility |
|---|---|---|---|---|---|
| AT1G19640 | 0.0006 | 2.5266 | 0.6016 | Jasmonic acid carboxyl methyltransferase (JMT) | 10.46 |
| AT4G24340 | 0.0009 | 2.6703 | 0.2733 | Phosphorylase family protein | 11.45 |
| AT4G24350 | 0.0009 | 2.2642 | 0.3178 | Phosphorylase family protein | 11.45 |
| AT3G05730 | 0.0023 | 2.1376 | 0.5933 | Defensin-like protein | 0.92 |
| AT1G70700 | 0.0031 | 1.8562 | 0.5318 | Jasmonate Zim-domain protein 9 (JAZ9) | 28.95 |
| AT1G66100 | 0.0036 | 1.8243 | 0.3275 | Thionin, identical to thionin 2.4 precursor | 14.09 |
| AT3G04720 | 0.0043 | 4.4631 | 0.0841 | Hevein-like protein (HEL) | 1.15 |
| AT5G24420 | 0.0068 | 2.1219 | 0.6044 | Glucosamine/galactosamine-6-phosphate isomerase-related | 33.24 |
| AT1G06830 | 0.0134 | 1.5713 | 0.7332 | Monothiol glutaredoxin-S11 (GRXS11) | 0.67 |
| AT1G11580 | 0.0160 | 1.8307 | 0.6279 | Pectin methylesterase (ATPMECRA) | 1.87 |
| AT5G61160 | 0.0198 | 6.5067 | 0.1410 | Anthocyanin 5-aromatic acyltransferase 1 (AACT1) | 2.62 |
| AT5G44420 | 0.0198 | 32.7145 | 0.0125 | Plant defensin 1.2a (PDF1.2a) | 1.05 |
| AT3G16530 | 0.0198 | 3.7444 | 0.2426 | Legume lectin family protein | 1.03 |
| AT1G19670 | 0.0209 | 2.7618 | 0.3069 | Chlorophyllase 1 (ATCHL1)/coronatine-induced protein 1 | 30.83 |
| AT5G23820 | 0.0209 | 1.3360 | 0.6500 | MD-2–related lipid recognition domain-containing protein | 2.09 |
| AT1G69370 | 0.0210 | 1.4756 | 0.7482 | Chorismate mutase3 (CM3) | 3.59 |
| AT3G62410 | 0.0256 | 1.1571 | 0.8770 | CP12 domain-containing protein (CP12-2) | 0.61 |
| AT5G40610 | 0.0326 | 1.3079 | 0.7687 | Glycerol-3-phosphate dehydrogenase | 1.00 |
| AT4G13410 | 0.0366 | 1.7372 | 0.6285 | Cellulose synthase like protein (ATCSLA15) | 18.97 |
| AT2G03980 | 0.0366 | 1.2918 | 0.6883 | GDSL-motif lipase/hydrolase family protein | 4.24 |
| AT5G02940 | 0.0397 | 1.4524 | 0.7374 | Phosphotransferase-related protein | 4.60 |
| AT4G01450 | 0.0443 | 0.7104 | 0.7634 | Nodulin MtN21 family protein similar to MtN21 | 1.31 |
Values listed under the Infected/Mock and pft1/Wild Type columns are the normalized ratios obtained from the entire microarray data set. Analysis of publicly available microarray data revealed an enrichment of JA-induced genes within the list. Values listed under the Jasmonate Inducibility column indicate the expression ratio after MeJA treatment according to Genevestigator (Zimmermann et al., 2004) (values in bold represent genes >1.5-fold induced). AGI, Arabidopsis Genome Initiative.
Analysis of the list using publicly available microarray data revealed a strong enrichment of JA-induced genes, with 14 out of the 22 genes showing a >1.5-fold induction by MeJA and eight out of the 22 genes showing an induction >10-fold (Zimmermann et al., 2004). However, this is probably an underestimate of the proportion of JA-induced genes identified here, as the expression data from the publicly available arrays is only recorded from plants sampled at an early time point after JA treatment. Therefore, well-known JA-responsive genes, such as PDF1.2 and HEL, which respond more slowly to JA, are not indicated in this comparison as being JA induced. However, one particularly interesting gene that was identified was the At CLH1 gene (also known as CORONATINE INDUCED PROTEIN1), that encodes a pathogen- and JA-inducible chlorophyllase (Kariola et al., 2005). Kariola et al. (2005) found that a reduction in At CLH1 expression, as can be seen in the pft1 mutant, resulted in a decrease in JA defenses and an increased susceptibility to A. brassicicola. The microarray results suggest that infection with F. oxysporum induces a number of JA-associated genes and that these genes have reduced expression in the pft1 mutant. This suggests that an attenuated JA signaling observed in the pft1 mutant might be responsible not only for the reduced F. oxysporum symptom development but also the loss of basal resistance to the leaf-infecting necrotrophic pathogens.
PFT1 Function Is Conserved in Plants
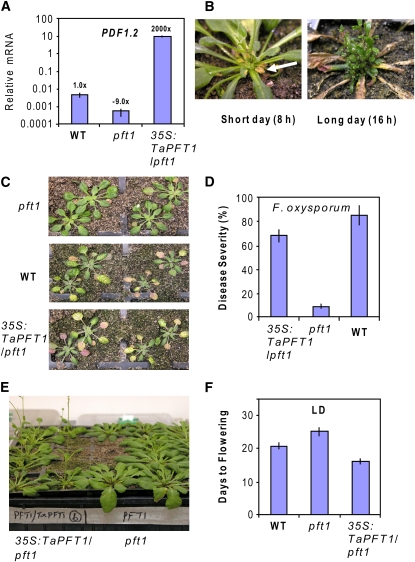
PFT1 is a single-copy gene in Arabidopsis with homologs in diverse plant species (Hecht et al., 2005; Bäckström et al., 2007). The conserved nature of the PFT1 protein indicates the possibility that the role of PFT1 homologs in disease and development might also be conserved in other plants. To test this possibility, we cloned a PFT1 cDNA homolog named here as Ta PFT1 from wheat (Triticum aestivum) and introduced this cDNA under the control of the cauliflower mosaic virus 35S promoter into the Arabidopsis pft1 mutant. As shown in Figure 5A for the PDF1.2 gene, a high level of Ta PFT1 expression in the pft1-2 mutant background has led to a significant increase in JA-dependent defense gene expression. The 35S:TaPFT1-expressing pft1 mutant lines also displayed a lesion-mimic phenotype (Figure 5B), possibly due to constitutive activation of defenses. This lesion-mimic phenotype was particularly visible in the rosette leaves after bolting. In the plants grown under LD conditions (16 h), this lesion mimic phenotype was aggravated, with older leaves dying prematurely in parallel to the development of multiple shoots (Figure 5B). The 35S:TaPFT1/pft1 lines also showed increased F. oxysporum susceptibility (Figures 5C and 5D) and early flowering phenotypes (Figures 5E and 5F), suggesting that Ta PFT1 complements the defense and developmental defects of the pft1 mutant. Although further experiments are required to fully assess the potential roles of PFT1 in wheat, these complementation experiments in Arabidopsis suggest that the role of PFT1 in both plant defense and the control of flowering may be conserved across diverse plant species.
Figure 5.
PFT1 Function Is Conserved in Plants.
(A) Transgenic expression of Ta PFT1, a PFT1 homolog from wheat, in the pft1 mutant background complements the compromised defense gene expression.
(B) The 35S:TaPFT1/pft1 line shows spontaneous lesion development (arrow).
(C) and (D) Expression of Ta PFT1 complements the increased F. oxysporum resistance of the pft1 mutant.
(E) and (F) Expression of Ta PFT1 complements the delayed flowering phenotype of pft1. PDF1.2 transcript levels were examined by quantitative RT-PCR, and the expression data were normalized relative to β-ACTIN expression and presented logarithmically.
Numbers on each bar in (A) represent fold difference in expression of PDF1.2 in untreated plants of pft1 and 35S:TaPFT1/pft1 relative to untreated wild-type plants. Error bars represent se from three independent replicates that contained ∼20 plants each. F. oxysporum inoculation experiments were conducted as described above, and symptom development was scored 10 d after inoculation.
The Role of Mediator Subunits in Plant Defense and Flowering Time Control
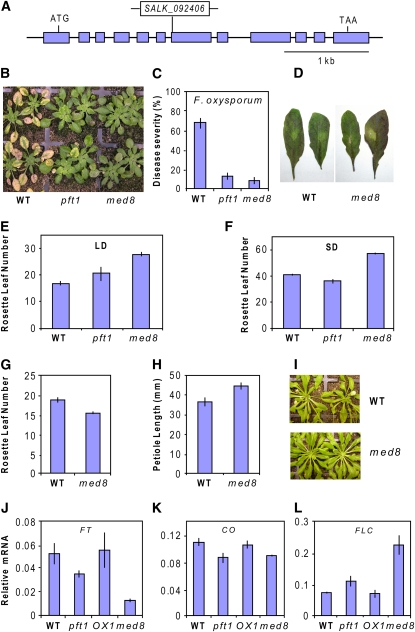
Given that Mediator is a multiprotein complex, the loss of PFT1 function could affect the overall function of Mediator. If this was the case, then inactivation of other Mediator subunits could lead to similar defense and developmental phenotypes observed in the pft1 plants. To test this hypothesis, we isolated homozygous T-DNA insertion lines for 11 individual Arabidopsis Mediator subunit mutants publicly available in the SALK Arabidopsis T-DNA insertion collection (see Methods). Of the Mediator mutants tested, only an insertion in the MED8 subunit (Figure 6A; see Supplemental Figure 4 online) produced an F. oxysporum resistance phenotype that was comparable to that of the pft1 mutant in terms of delayed symptom development (Figures 6B and 6C) and increased survival rates (see Supplemental Figure 5 online). In addition, the med8 mutant had increased susceptibility to A. brassicicola with 42% of the inoculated leaves showing relatively large chlorotic lesions (Figure 6D) as compared with 21% of wild-type leaves that showed some degree of chlorosis in these experiments.
Figure 6.
MED8 Is a Regulator of Plant Defense and Flowering.
(A) Schematic representation of the MED8 gene and the location of the T-DNA insertion (SALK_092406). Introns (solid line) and exons (boxes) are indicated.
(B) and (C) The med8 mutant shows increased resistance to F. oxysporum.
(D) The med8 mutant shows increased susceptibility to A. brassicicola.
(E) and (F) The med8 mutant shows delayed flowering under both LDs and SDs (expressed as the number of rosette leaves at the time of bolting).
(G) to (I) The med8 mutant has reduced petiole lengths and an increased vegetative leaf number compared with the wild type at equivalent growth stages, shown in (I). Measurements for (G) and (H) were taken by counting the total leaves from 10 wild-type and med8 plants that were 8 weeks old. Flowering measurements were taken from two independent experiments with 18 plants per line in each experiment. The rosette leaf number was recorded when the shoot bud had extended 5 mm.
(J) to (L) The transcript levels of FT, CO, and FLC were examined by quantitative RT-PCR and the expression data normalized relative to β-ACTIN. Error bars represent se from three replicates of 30 plants each.
Interestingly, the med8 mutant also demonstrated an altered flowering time with a strong delay in flowering under both short-day (SD) and LD conditions (Figures 6E and 6F). The med8 mutant also possessed an increased number of leaves and shorter petioles under vegetative conditions, giving it a distinctive phenotype (Figures 6G to 6I). In addition to med8, we also measured the flowering time of pft1 as a comparison. Our results confirmed the results of Cerdán and Chory (2003), with a small but statistically significant (P < 0.05) decrease and increase in flowering time under SD and LD, respectively, when determined by rosette leaf number (Figures 6E and 6F), and an increased flowering time under both SD and LD when determined by the number of days to bolting (see Supplemental Figure 6 online).
To explore the molecular mechanism behind the late flowering phenotype of med8, we also quantified the expression of the key flowering genes, FLOWERING LOCUS T (FT) and CONSTANS (CO), which positively regulate flowering, and FLOWERING LOCUS C (FLC), which negatively regulates flowering (reviewed in Farrona et al., 2008) in LD-grown pft1 and med8 plants (Figures 6J to 6L). Cerdán and Chory (2003) previously reported reduced FT and CO expression in pft1, and our results confirmed this finding. We also found reduced expression of the floral promoting genes FT and CO in med8 compared with the wild type. In addition, we found increased expression of the floral repressor FLC in both pft1 and med8. Furthermore, the levels of FT and FLC expression in pft1 and med8 correlated well with the severity of the flowering delay seen in these two mutants, with the later flowering med8 having a noticeably lower and higher level of FT and FLC expression, respectively, than pft1 and the wild type (Figures 6J and 6L). Together, these experiments suggest that MED8, similar to PFT1, is a regulator of both plant defense and flowering time.
Genetic Evidence for the Independent and Additive Functions of PFT1 and MED8
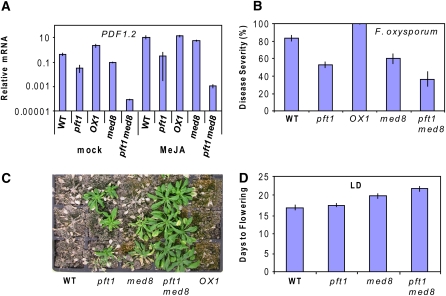
As the pft1 and med8 mutants appear to similarly affect both flowering time and pathogen resistance, we sought to determine whether these two mutations act independently to produce similar phenotypes. To test this possibility genetically, we constructed a pft1 med8 double mutant and analyzed its defense and flowering time phenotypes together with those of the pft1 and med8 single mutants. First, we looked at the expression of PDF1.2 in the double and single mutants after treatment with MeJA. We found that the expression of PDF1.2 was slightly lower in both untreated and 6-h MeJA-treated med8 plants than in similarly treated wild-type plants (Figure 7A). However, in the pft1 med8 double mutant, PDF1.2 expression was greatly reduced, with the expression level in the double mutant being ∼300- and 370-fold less in the MeJA-treated and untreated plants, respectively, than similarly treated pft1 plants (Figure 7A). Inoculation of the pft1 med8 double mutant with F. oxysporum also revealed a relatively small but additive increase in resistance relative to the individual single mutants (Figures 7B and 7C). At 24 d after inoculation, the increased resistance of the pft1 med8 double mutant was more evident, with a noticeable difference in the survival rate and overall vigor of the double mutant compared with the pft1 and med8 lines (Figure 7C).
Figure 7.
The pft1 med8 Double Mutant Shows an Additive Increase in Both the Flowering Time and the Pathogen Defense Phenotypes Compared with the pft1 and med8 Single Mutants.
(A) The expression of PDF1.2 is greatly reduced in the double mutant relative to the wild type and pft1 and med8 single mutants in both untreated and MeJA-treated plants.
(B) and (C) The double mutant also shows reduced symptoms and greater survival after inoculation with F. oxysporum.
(D) The flowering time of the double mutant is more delayed than the individual single mutants. PDF1.2 transcript levels were examined by quantitative RT-PCR, and the expression data were normalized relative to β-ACTIN expression and presented logarithmically. The disease symptoms and survival were assessed 14 and 24 d after inoculation, respectively. The error bars from the F. oxysporum inoculation and defense gene experiments are the se from three replicates of ∼30 plants each, whereas the error bars for the flowering time experiment are the se from 18 plants and are representative of two independent replicated experiments.
[See online article for color version of this figure.]
In addition, we compared the flowering time of the double mutant to the wild-type and the single pft1 and med8 mutants. We found that the pft1 med8 double mutant had a similar number of leaves at flowering under LD to the pft1 mutant (see Supplemental Figure 7 online). However, the double mutant flowered later than pft1 and med8 when flowering time was measured by days to flowering under LD, suggesting an additive effect on flowering time (Figure 7D). The stronger effects of the pft1 med8 double mutant on flowering time and defense than either of the single mutants suggest that the pft1 and med8 mutations might be affecting these two phenotypes by independent and additive mechanisms.
DISCUSSION
The Mediator complex was first purified from yeast in the early 1990s (Kim et al., 1994). Subsequent studies have discovered that the Mediator complex is an essential part of the transcriptional machinery in all eukaryotes (Bourbon, 2008). Surprisingly, the existence of the Mediator complex in plants has only recently been shown in Arabidopsis (Bäckström et al., 2007). Using a biochemical purification strategy, Bäckström et al. (2007) identified 21 conserved subunits and six plant-specific Mediator subunits. Two of the Mediator subunits identified by Bäckström et al. (2007), MED14 and MED25, were previously studied plant proteins, SWP1 and PFT1, which are involved in regulating leaf cell number in aerial organs and phytochrome signaling, respectively (Autran et al., 2002; Cerdán and Chory, 2003). More recently, another Mediator subunit, MED21, has been shown to be required for resistance to necrotrophic pathogens in Arabidopsis (Dhawan et al., 2009), although the mechanism of this resistance is currently unknown.
In this article, we show that in addition to its known role in phytochrome signaling, PFT1 is also an important component of the plant's basal defense and is required for uncompromised expression of JA-dependent defenses as well as resistance to necrotrophic fungal pathogens, such as A. brassicicola and B. cinerea (Figures 1 to 4). Interestingly, PFT1 is also essential for susceptibility to the root-infecting fungus F. oxysporum, which is thought to use the host JA pathway to promote host senescence and necrosis (Thatcher et al., 2009).
JA-dependent defense genes and the phytoalexin camalexin have previously been shown to be active against necrotrophic pathogens, such as A. brassicicola and B. cinerea (Thomma et al., 1998, 1999; Zhou et al., 1999). Therefore, it is likely that the attenuated expression of JA-responsive defense genes as well as PAD3 is the cause of increased A. brassicicola and B. cinerea susceptibility in pft1.
The mechanism(s) behind the increased F. oxysporum resistance was initially less apparent. However, as recently reported, the coi1 mutant with compromised JA signaling and JA-dependent gene expression shows a remarkable increase in resistance to this pathogen (Thatcher et al., 2009). Through stringent analysis of the microarray data, we identified that a large proportion of the genes that were differentially expressed after infection by F. oxysporum treatment, as well as between the wild-type and pft1 genotypes, were genes that are also regulated by JA. Importantly, all of the JA-regulated genes that were found to be induced by F. oxysporum in the wild type had significantly reduced expression in the pft1 mutant, under both mock and inoculated conditions (Table 1). Therefore, it is likely that a reduction in JA-responses in pft1 may be providing an increased tolerance to F. oxysporum, as is seen with the coi1 mutant (Thatcher et al., 2009).
Interestingly, we also found a reduction in SA defense gene expression in pft1 from the quantitative RT-PCR experiments. The SA and the JA signaling pathways have been reported to act in a mutually antagonistic manner (Spoel et al., 2003; reviewed in Kazan and Manners, 2008; Koornneef and Pieterse, 2008); therefore, reduced expression from both the JA- and SA-associated defense genes in pft1 was not expected. However, given the fact that Arabidopsis contains >1500 transcription factors and only a limited number of Mediator subunits (Bäckström et al., 2007), it is plausible that PFT1 interacts with several activators/repressors involved in multiple aspects of the disease response pathways. If this was the case, then removal of PFT1 could potentially result in the suppression of both SA and JA defenses. The possibility that PFT1 is a convergence point for the activation of both SA and JA defense signaling within the Mediator complex is an intriguing hypothesis that would justify further investigation. However, we noted that the differential expression of the SA-associated defense genes could not be detected under basal conditions, with a difference seen only after treatment with SA. Therefore, the regulation of JA defense genes in pft1, which occurs under both basal and JA-treated conditions in the pft1 mutant, may be considered to be the primary role of PFT1.
The PFT1 protein is highly conserved across diverse eukaryotes. For example, PFT1 shows sequence similarity to the Drosophila melanogaster and human MED25 proteins, particularly in the N-terminal vWF-A domain but less so in the C-terminal regions of the protein. Interestingly, like the Arabidopsis PFT1, both the Drosophila and human MED25 proteins appear to have a function in host defense. RNA interference–mediated suppression of Drosophila MED25 results in attenuated induction of the AttA gene encoding the antibacterial peptide attacin in response to lipopolysaccharide treatment (Kim et al., 2004). In addition, the human MED25 has been shown to be the cellular target of both VP16, the well-studied activator of herpes simplex virus (Mittler et al., 2003; Yang et al., 2004), as well as the activator IE62, from the closely related Varicella Zoster virus, the virus responsible for chicken pox and shingles (Yang et al., 2008). By functionally complementing the defensive and developmental phenotypes of the Arabidopsis pft1 mutant with a wheat ortholog, we obtained initial evidence that the function of PFT1 is also conserved in plants. In addition to heightened defense gene expression and accelerated flowering relative to the pft1 mutants, the 35S:TaPFT1/pft1 plants displayed a spontaneous lesion phenotype. Transgenic expression of the vWF-A domain of the BONZAI/COPINE1 protein of Arabidopsis in transgenic tobacco (Nicotiana tabacum) activates defense responses and also produces a lesion mimic phenotype (Liu et al., 2005), suggesting that the lesion mimic phenotype we observed in Ta PFT1-expressing pft1 mutant plants could be mediated by the vWF-A domain of Ta PFT1.
In addition to PFT1, our study implicated another Mediator subunit, MED8, in both plant defense and flowering time regulation. The MED8 subunit has been found to be located within the head module of the Mediator complex in other eukaryotes (Kang et al., 2001), while the location of PFT1 within the Mediator complex is currently unknown. However, we noted that the med8 mutant also possesses additional phenotypes that are not present in the pft1 mutant. For instance, the med8 mutant shows a stronger delay in flowering time than pft1 under both LD and SD conditions. Our results also indicated only a slight reduction in PDF1.2 expression in the med8 mutant (Figure 7A). Therefore, it is possible that the effect of the med8 mutation on resistance might also be due to other effects, such as delayed senescence (Schenk et al., 2005; Thatcher et al., 2009), a natural consequence of delayed flowering. However, the effect of the med8 mutation on PDF1.2 expression was more visible in the pft1 mutant background, as the pft1 med8 double mutant showed an additional decrease in PDF1.2 expression over that of the pft1 mutant. The med8 mutation also had an additive effect on both F. oxysporum resistance and flowering time phenotypes of the pft1 mutant. These results suggest that these two Mediator subunits may act independently within the Mediator complex to affect similar developmental and defensive processes as proposed in the model given in Figure 8.
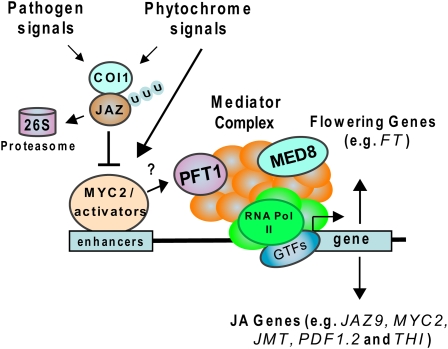
Figure 8.
The Plant Mediator Complex Is a Key Regulator of JA-Dependent Plant Defense and Flowering Time.
The Arabidopsis Mediator subunit PFT1, possibly by acting downstream from a COI1-JAZ-MYC2 complex, integrates JA-dependent defense and developmental (e.g., phytochrome) cues by coordinating information from DNA-bound activators. This enables the RNA polymerase II complex to initiate the transcription of genes involved in flowering time and JA-dependent pathogen defense. GTFs, general transcription factors.
[See online article for color version of this figure.]
Currently, which activator(s) might be involved in relaying JA and phytochrome signals to the Mediator complex through PFT1 is not known. However, recent studies revealed that COI1-dependent ubiquitination of JAZ repressors, which interact with the transcriptional regulator MYC2, modulates downstream transcriptional responses in the JA signaling pathway (Chini et al., 2007). Therefore, one can speculate that MYC2 and/or some of the MYC2-regulated transcriptional activators (Dombrecht et al., 2007) might be involved in relaying JA signals to the transcriptional machinery through PFT1. Because JA signaling is known to act in a positive feedback loop (Kazan and Manners, 2008), inactivation of PFT1 attenuates the JA-dependent expression of a number of JA biosynthesis and signaling genes, including MYC2 and a number of MYC2-regulated genes, such as JAZ9. Therefore, we tentatively place PFT1 downstream from the COI1-JAZ-MYC2 complex in our model given in Figure 8.
Similar to PFT1 and MED8, it has been recently shown that MED21, which is located in the middle module of the Mediator complex (Boube et al., 2002), is also required for resistance to necrotrophic pathogens in Arabidopsis (Dhawan et al., 2009). While med21 shares similar pathogen phenotypes to pft1 and med8, it is unknown whether MED21 possesses altered defense gene expression or flowering time. T-DNA insertions of MED21 show embryo lethality, and an altered flowering time phenotype has not been reported for the MED21 RNA interference lines (Dhawan et al., 2009). Nevertheless, our results together with those of Dhawan et al. (2009) suggest that at least three Mediator subunits are required for necrotrophic pathogen resistance in Arabidopsis.
Our discovery of PFT1 and MED8 as integrators of both plant defense and flowering time raises an intriguing question as to why these two seemingly different but important pathways may be linked. Similarly to what we found in pft1, it has been previously reported that many flowering time mutants also show altered defense gene expression (Wilson et al., 2005). Furthermore, the alteration of flowering times in pathogen-infected plants has been previously reported (Korves and Bergelson, 2003; Veronese et al., 2003). In addition, an intriguing link between phytochrome control and JA responses has recently been discovered (Moreno et al., 2009). The authors found impaired JA responses in the phyB mutant as well as in wild-type plants treated with far-red light. Therefore, the reason why the pft1 mutant possesses reduced JA responses might be due to its involvement in phytochrome regulation. The involvement of MED8 in phytochrome regulation is currently unknown. However, it is plausible that the plant Mediator complex acts as a control panel in integrating both phytochrome and defense-related signals from multiple transcriptional regulators (Figure 8).
At the molecular level, the effect of these two Mediator subunits on defense and flowering time may be due to the involvement of Mediator in chromatin remodeling. In several cases, the Mediator complex has been shown to interact with chromatin modification complexes, such as SWI/SNF and histone deacetylases and acetylases (Sharma and Fondell, 2002; He et al., 2008; Malik and Roeder, 2008). The human MED25 protein has been shown to interact with histone acetylases (Black et al., 2006; Lee et al., 2007). Chromatin remodeling complexes are known to be functionally conserved evolutionarily (Farrona et al., 2008). Thus, if this function of MED25 is conserved between plants and animals, then one would expect that Arabidopsis PFT1 would also play a role in chromatin modification. SWP1/MED14 is known to interact with the transcriptional corepressor LEUNIG, which interacts with the histone deacetylase HDA19 (Gonzalez et al., 2007). Interestingly, a number of genes involved in chromatin modification and remodeling affect both flowering and plant defense (Wagner and Meyerowitz, 2002; He and Amasino, 2005; Zhou et al., 2005; Walley et al., 2008; March-Díaz et al., 2008; Wu et al., 2008).
Similar to our results with the pft1 and med8 mutants, a recent study found that the loss of activity in SPLAYED, one of the SWI/SNF classes of chromatin remodeling ATPases in Arabidopsis, leads to reduced JA-responsive defense gene expression and increased susceptibility to B. cinerea (Walley et al., 2008). The syd mutants also show defects in reproductive development and flowering time (Wagner and Meyerowitz, 2002). Similarly, chromatin modification by the histone deacetylases HDA19 and HDA6 is required for JA-responsive defense gene expression and resistance to necrotrophic pathogens (Zhou et al., 2005; Wu et al., 2008). Again, similarly to pft1 and med8, loss-of-function mutations in the HDA6 gene delays flowering (Wu et al., 2008). HDA6 is also known to interact with COI1 (Devoto et al., 2002), further linking JA responses to chromatin modification.
Finally, the Mediator subunit MED21, required for necrotrophic pathogen resistance, has been shown to interact with HUB1, an E3 ligase responsible for the ubiquination of H2B histones. The hub1 mutant also shows alterations in flowering time (Dhawan et al., 2009). Although further research is required to determine whether perturbation of the Mediator complex in pft1 and med8 would compromise chromatin remodeling, overall, these studies support the view that multiple components of the plant transcriptional machinery are required for the regulation of both plant development, such as flowering time and pathogen defense.
In conclusion, our results reported here link two plant Mediator subunits as integrators of flowering time and JA-dependent defense-related signals to the transcriptional machinery. Future research should reveal new insights into the specific roles of the remaining Mediator subunits and help to advance our understanding of the transcriptional regulation of gene expression in plants.
METHODS
Plant Growth Conditions, Chemical Treatments, and Pathogen Inoculations
Plant growth conditions and MeJA and SA treatments were described previously (Schenk et al., 2000; Campbell et al., 2003; Anderson et al., 2004). Briefly, plants were grown in a controlled environment room with a temperature of 24°C and a light intensity of 150 μmol·m−2·s−1. Photosynthetically active radiation was supplied by high pressure metal halide lamps (Sylvania) and tungsten halogen lamps (Phillips). The red:far-red ratio was ∼1.13, which is within 10% of the observed daylight. All plants used were in the Arabidopsis thaliana Col-0 background. Mutant lines used are listed in the Accession Numbers section at the end of Methods. Homozygous plants of pft1, med8, and the other Mediator subunit mutants were identified using the primer sequences given at http://signal.salk.edu/tdnaprimers.html and used in the experiments described here. The pft1 med8 double mutant was created by pollinating an emasculated med8 floral bud with the pollen from the pft1.2 mutant. All treatments were performed on soil-grown 4- to 5-week-old plants, unless otherwise stated. The Fusarium oxysporum isolate used in this study was strain Fo5176 obtained from Roger Shivas, Queensland Plant Pathology Herbarium, Queensland Department of Primary Industries and Fisheries (QDPIandF), Brisbane, Australia. Inoculations were performed as described by Anderson et al. (2004) and Edgar et al. (2006). The Alternaria brassicicola (UQ4273) infection assays were performed as described previously (Campbell et al., 2003). Botrytis cinerea (BRIP25539) infection assays were performed in a similar manner to the A. brassicicola assays by harvesting spores from half-strength potato dextrose agar plates and pipetting 5-μL droplets (1 × 106 spores/mL) on mature Arabidopsis leaves.
Quantitative RT-PCR Expression Analyses
Quantitative RT-PCR experiments were done as described previously using the Applied Biosystems 7900HT Fast real-time PCR system in conjunction with SYBR Green fluorescence to detect transcript levels (McGrath et al., 2005). Briefly, for all data analysis, the PCR primer efficiency (E-value) of each primer pair in each reaction was calculated from the ΔRn values of each amplification plot using LinReg PCR software (Ramakers et al., 2003). Amplification plots were analyzed using a cycle threshold value of 0.2 across all experiments. Absolute gene expression levels relative to the previously validated reference genes β-ACTIN2, β-ACTIN7, and β-ACTIN8 were used for each cDNA sample using the equation: relative abundance = (Egene^(–Ct gene))/(EACTIN^(–Ct ACTIN)). The mean expression range of the reference gene was found to be within ±1 Ct across all experiments. Three biological replicates of mock and treated samples were used, and the average ratio of these values was used to determine the fold change in transcript level in treatment samples compared with control. The sequences of the primer pairs have been previously published (Anderson et al., 2004; McGrath et al., 2005; Dombrecht et al., 2007).
Flowering Time Measurements
Flowering time measurements were recorded from at least 18 plants per genotype that were grown in soil under either LD or SD conditions. Plants grown in LD conditions had 16 h of light at 28°C and a night period of 21°C. The plants grown in SD conditions had 8 h of light at 24°C and a night period of 21°C. Plants were grown in trays containing 30 (5 × 5 cm) cells and were separated into individual cells once the first true leaves had expanded. The rosette leaf number as well as days to flowering were recorded when the shoot bud had extended 5 mm. For quantitative RT-PCR analysis of flowering control genes, 30 plants of each genotype were grown in LD conditions for 4 weeks before being harvested at the end of the 16-h photoperiod. RNA was extracted, converted to cDNA, and used for quantitative RT-PCR analysis as described above.
Microarray Analysis
Wild-type and pft1 plants were grown and inoculated as described above. Four independent biological replicates consisting of 20 plants each were root dipped in either water or a F. oxysporum spore suspension of 106 spores/mL in water and replanted in soil. The shoot material was harvested 48 h later and total RNA extracted using the RNeasy plant mini kit (Qiagen). The RNA was labeled, hybridized, washed, and scanned by the Australian Genome Research Facility onto 16 ATH1 GeneChip arrays and the resulting data analyzed using GenespringGX 7.3.1 (Agilent) as previously described (Dombrecht et al., 2007). Briefly, the raw CEL files were normalized using the RMA algorithm, and then the resulting expression values were normalized per chip to the median across all chips. A two-way ANOVA was used to investigate differentially expressed genes between both the treatment and genotype. A P value cutoff of 0.05 as well as multiple testing correction using the Benjamini and Hochberg false discovery rate was applied to the data, and the significant genes in both treatment and genotype parameters were obtained. The microarray data have been submitted to the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE15236.
Root Growth Inhibition and Anthocyanin Assays
Surface-sterilized Arabidopsis seeds were plated on half-strength MS medium (supplied with 5% sucrose and 0.7% Bacto Agar, pH 6.0) supplemented with either 0.01% ethanol (mock treatment) or 5 μM MeJA (Aldrich; solubilized in ethanol). Plates were incubated under continuous light at 22°C, and seedlings were monitored 6 d for root growth. For anthocyanin assays, 2-week-old pft1, wild-type, and OX1 plants were detached from their roots and incubated in 6-well microtitre plates (Iwaki) containing distilled water for 3 weeks under SD conditions. Two independent experiments were performed with each containing four plants per genotype.
Generation of 35S:TaPFT1/pft1 Arabidopsis Lines
The PFT1 protein of Arabidopsis (NP_173925.3) was used to search the homologous gene of wheat in The Institute of Genomic Research database using tBLASTn program. The search resulted in identification of the UniGene (Ta.39294) that is 61% identical to the Arabidopsis PFT1 protein. Ta.39294 is named here as Ta PFT1. To clone Ta PFT1, total RNA was isolated from young seedling of the wheat (Triticum aestivum) variety Kennedy using Promega SV total RNA isolation system. cDNA synthesis was done using the cDNA synthesis kit Superscript III (Invitrogen). The Ta PFT1 cDNA was amplified using the following primers: 5′-CCCGGATCCCGGATTCGCGAGGGCGAG-3′ and 5′-CCCGGATCCACTCGCAATGCTCTGTAC-3′. The amplification product was cloned into the pBlunt vector (Invitrogen) and confirmed by sequencing. Ta PFT1 was released by digesting the plasmid with BamHI and cloned into the BamH1-digested binary vector pPCV91 (Strizhov et al., 1996), which was then mobilized into the Agrobacterium tumefaciens strain GV3101. The pft1 mutant plants were transformed using the floral dip method, and the seeds collected from infiltrated plants were grown on half-strength MS medium containing 15 mg/L hygromycin (Sigma-Aldrich) to select the transformants. The presence of Ta PFT1 was confirmed by PCR. Homozygous lines were used in gene expression and phenotypic analyses.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: PFT1 (At1g25540, NP_173925.3), MED8 (At2g03070), PDF1.2 (At5g44420), CHIB (At3g12500), PR1 (At2g14610), PR5 (At1G75040), PAL1 (At2g37040), MYC2 (At1g32640), HEL (At3g04720), ESP (At1g54040), OPR (At1g17990 and At1g18020), BGL2 (At3g57260), PAD3 (At3g26830), WRKY70 (At3g56400), WRKY53 (At4g23810), iASK (At5g26751), FT (At1G65480), CO (At5G15840), FLC (At5G10140), β-ACTIN2 (At3g18780), β-ACTIN7 (At5g09810), β-ACTIN8 (At1g49240), and Ta PFT1 (Unigene Ta.39294). The microarray data have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under the accession number GSE15236. The following mutant lines were used: pft1-2 (At1g25540, SALK_129555), pft1-3 (SALK_059316), med6 (At3g21350, SALK_055723C), med8 (At2g03070, SALK_092406), med9 (At1g55080, SALK_115775), med10a (At5g41910, SALK_115673), med22b (At1g07950, SALK_001024C), med31 (At5g19910, SALK_051025), med19a (At5g12230, SALK_020936), med32 (At1g11760, SALK_028490), med33a (At3g23590, SALK_119561), med33b (At2g48110, SALK_015532), and med34 (At1g31360, CS87663).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Confocal Microscopy and Fungal Quantification of F. oxysporum–Infected Wild-Type, pft1, and OX1 Lines.
Supplemental Figure 2. Quantitative RT-PCR Expression of JA-Associated Genes in the Wild-Type, G1, and OX1 Lines.
Supplemental Figure 3. The Expression of Defense Genes after F. oxysporum Infection in the Roots and Shoots.
Supplemental Figure 4. Disease Severity following the Inoculation of 11 Mediator Subunit Mutant Lines with F. oxysporum.
Supplemental Figure 5. The Survival of Wild-Type, pft1, OX1, med8, and the pft1 med8 Double Mutant after Infection with F. oxysporum.
Supplemental Figure 6. The Flowering Phenotype of pft1 and med8.
Supplemental Figure 7. The Flowering Phenotype of the pft1 med8 Double Mutant.
Supplemental Table 1. The List of Genes Differentially Regulated by the F. oxysporum Treatment from the Microarray.
Supplemental Table 2. The List of Genes Differentially Regulated by the Genotype from the Microarray.
Supplemental Table 3. The List of Genes That Are Differentially Regulated by Both the Genotype and the F. oxysporum Treatment.
Supplementary Material
Acknowledgments
B.N.K. and C.I.E. were supported by postgraduate scholarships from the Cooperative Research Centre for Tropical Plant Protection as well as the Grains Research and Development Corporation. K.K.K. was the recipient of a fellowship from the Department of Biotechnology of the Indian Government. We thank the ABRC for the seeds of Arabidopsis T-DNA insertion lines used in the study; Pablo Cerdán and Joan Chory for kindly providing the pft1-1, OX1, and G1 seeds; Roger Shivas for the F. oxysporum and B. cinerea isolates used in the study; Christina Bakker and Carol Kistler for the isolation of homozygous lines of the PFT1 T-DNA insertion lines; Christina Bakker for optimization of the B. cinerea inoculation assay; Rosemary White for assistance in confocal microscopy; Bruno Dombrecht for useful discussions at the early stages of this work; and Louise Thatcher, Donald Gardiner, and Timothy Fitzgerald for critical manuscript reading and useful discussions.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Kemal Kazan (kemal.kazan@csiro.au).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Anderson, J.P., Badruzsaufari, E., Schenk, P.M., Manners, J.M., Desmond, O.J., Ehlert, C., Maclean, D.J., Ebert, P.R., and Kazan, K. (2004). Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16: 3460–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autran, D., Jonak, C., Belcram, K., Beemster, G.T., Kronenberger, J., Grandjean, O., Inze, D., and Traas, J. (2002). Cell numbers and leaf development in Arabidopsis: A functional analysis of the STRUWWELPETER gene. EMBO J. 21: 6036–6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckström, S., Elfving, N., Nilsson, R., Wingsle, G., and Björklund, S. (2007). Purification of a plant Mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol. Cell 26: 717–729. [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo, M., and Molina, A. (2008). Arabidopsis defense response against Fusarium oxysporum. Trends Plant Sci. 13: 145–150. [DOI] [PubMed] [Google Scholar]
- Black, J.C., Choi, J.E., Lombardo, S.R., and Carey, M. (2006). A Mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol. Cell 23: 809–818. [DOI] [PubMed] [Google Scholar]
- Boube, M., Joulia, L., Cribbs, D.L., and Bourbon, H.M. (2002). Evidence for a Mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell 110: 143–151. [DOI] [PubMed] [Google Scholar]
- Bourbon, H.M. (2008). Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional Mediator complex. Nucleic Acids Res. 36: 3993–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbon, H.M., et al. (2004). A unified nomenclature for protein subunits of Mediator complexes linking transcriptional regulators to RNA polymerase II. Mol. Cell 14: 553–557. [DOI] [PubMed] [Google Scholar]
- Campbell, E.J., Schenk, P.M., Kazan, K., Penninckx, I.A., Anderson, J.P., Maclean, D.J., Cammue, B.P., Ebert, P.R., and Manners, J.M. (2003). Pathogen-responsive expression of a putative ATP-binding cassette transporter gene conferring resistance to the diterpenoid sclareol is regulated by multiple defense signaling pathways in Arabidopsis. Plant Physiol. 133: 1272–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdán, P.D., and Chory, J. (2003). Regulation of flowering time by light quality. Nature 423: 881–885. [DOI] [PubMed] [Google Scholar]
- Chini, A., Fonseca, S., Fernández, G., Adie, B., Chico, J.M., Lorenzo, O., García-Casado, G., López-Vidriero, I., Lozano, F.M., Ponce, M.R., Micol, J.L., and Solano, R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671. [DOI] [PubMed] [Google Scholar]
- Devoto, A., Nieto-Rostro, M., Xie, D., Ellis, C., Harmston, R., Patrick, E., Davis, J., Sherratt, L., Coleman, M., and Turner, J.G. (2002). COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 32: 457–466. [DOI] [PubMed] [Google Scholar]
- Dhawan, R., Luo, H., Foerster, A.M., AbuQamar, S., Du, H.-N., Briggs, S.D., Scheid, O.M., and Mengiste, T. (2009). HISTONE MONOUBIQUITINATION1 interacts with a subunit of the Mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21: 1000–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener, A.C., and Ausubel, F.M. (2005). RESISTANCE TO FUSARIUM OXYSPORUM 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics 171: 305–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht, B., Xue, G.P., Sprague, S.J., Kirkegaard, J.A., Ross, J.J., Reid, J.B., Fitt, G.P., Sewelam, N., Schenk, P.M., Manners, J.M., and Kazan, K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, C.I., McGrath, K.C., Dombrecht, B., Manners, J.M., Schenk, P.M., Maclean, D.J., and Kazan, K. (2006). Salicylic acid mediates resistance to the vascular wilt pathogen Fusarium oxysporum in the model host Arabidopsis thaliana. Australas. Plant Pathol. 35: 581–591. [Google Scholar]
- Farmer, E.E. (2007). Jasmonate perception machines. Nature 448: 659–660. [DOI] [PubMed] [Google Scholar]
- Farrona, S., Coupland, G., and Turck, F. (2008). The impact of chromatin regulation on the floral transition. Semin. Cell Dev. Biol. 19: 560–573. [DOI] [PubMed] [Google Scholar]
- Gonzalez, D., Bowen, A.J., Carroll, T.S., and Conlan, R.S. (2007). The transcription corepressor LEUNIG interacts with the histone deacetylase HDA19 and Mediator components MED14 (SWP) and CDK8 (HEN3) to repress transcription. Mol. Cell. Biol. 15: 5306–5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Q., Battistella, L., and Morse, R.H. (2008). Mediator requirement downstream of chromatin remodeling during transcriptional activation of CHA1 in yeast. J. Biol. Chem. 283: 5276–5286. [DOI] [PubMed] [Google Scholar]
- He, Y., and Amasino, R.M. (2005). Role of chromatin modification in flowering-time control. Trends Plant Sci. 10: 30–35. [DOI] [PubMed] [Google Scholar]
- Hecht, V., Foucher, F., Ferrándiz, C., Macknight, R., Navarro, C., Morin, J., Vardy, M.E., Ellis, N., Beltrán, J.P., Rameau, C., and Weller, J.L. (2005). Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol. 137: 1420–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J.S., Kim, S.H., Hwang, M.S., Han, S.J., Lee, Y.C., and Kim, Y.J. (2001). The structural and functional organization of the yeast mediator complex. J. Biol. Chem. 276: 42003–42010. [DOI] [PubMed] [Google Scholar]
- Kariola, T., Brader, G., Li, J., and Palva, E.T. (2005). Chlorophyllase 1, a damage control enzyme, affects the balance between defense pathways in plants. Plant Cell 17: 282–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan, K., and Manners, J.M. (2008). Jasmonate signaling: toward an integrated view. Plant Physiol. 146: 1459–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T.W., Kwon, Y.J., Kim, J.M., Song, Y.H., Kim, S.N., and Kim, Y.J. (2004). MED16 and MED23 of Mediator are coactivators of lipopolysaccharide- and heat-shock-induced transcriptional activators. Proc. Natl. Acad. Sci. USA 101: 12153–12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.J., Bjorklund, S., Li, Y., Sayre, M.H., and Kornberg, R.D. (1994). A multiprotein Mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 4: 599–608. [DOI] [PubMed] [Google Scholar]
- Koleske, A.J., and Young, R.A. (1994). An RNA polymerase II holoenzyme responsive to activators. Nature 368: 466–469. [DOI] [PubMed] [Google Scholar]
- Koornneef, A., and Pieterse, C.M. (2008). Cross talk in defense signaling. Plant Physiol. 146: 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korves, T.M., and Bergelson, J. (2003). A developmental response to pathogen infection in Arabidopsis. Plant Physiol. 133: 339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.K., Park, U.H., Kim, E.J., and Um, S.J. (2007). MED25 is distinct from TRAP220/MED1 in cooperating with CBP for retinoid receptor activation. EMBO J. 26: 3545–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Brader, G., Kariola, T., and Palva, E.T. (2006). WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 46: 477–491. [DOI] [PubMed] [Google Scholar]
- Liu, J., Jambunathan, N., and McNellis, T.W. (2005). Transgenic expression of the von Willebrand A domain of the BONZAI 1/COPINE 1 protein triggers a lesion-mimic phenotype in Arabidopsis. Planta 221: 85–94. [DOI] [PubMed] [Google Scholar]
- Malik, S., and Roeder, R.G. (2005). Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 30: 256–263. [DOI] [PubMed] [Google Scholar]
- Malik, S., and Roeder, R.G. (2008). Epigenetics? Mediator does that too! Mol. Cell 31: 305–306. [DOI] [PubMed] [Google Scholar]
- March-Díaz, R., García-Domínguez, M., Lozano-Juste, J., León, J., Florencio, F.J., and Reyes, J.C. (2008). Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant J. 53: 475–487. [DOI] [PubMed] [Google Scholar]
- McGrath, K.C., Dombrecht, B., Manners, J.M., Schenk, P.M., Edgar, C.I., Maclean, D.J., Scheible, W.R., Udvardi, M.K., and Kazan, K. (2005). Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol. 139: 949–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michielse, C.B., and Rep, M. (2009). Pathogen profile update: Fusarium oxysporum. Mol. Plant Pathol. 10: 311–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler, G., Stuhler, T., Santolin, L., Uhlmann, T., Kremmer, E., Lottspeich, F., Berti, L., and Meisterernst, M. (2003). A novel docking site on Mediator is critical for activation by VP16 in mammalian cells. EMBO J. 22: 6494–6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, J.E., Tao, Y., Chory, J., and Ballaré, C.L. (2009). Ecological modulation of plant defense via phytochrome control of jasmonate sensitivity. Proc. Natl. Acad. Sci. USA 106: 4935–4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers, C., Ruijter, J.M., Deprez, R.H.L., and Moorman, A.F.M. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339: 62–66. [DOI] [PubMed] [Google Scholar]
- Schenk, P.M., Kazan, K., Manners, J.M., Anderson, J.P., Simpson, R.S., Wilson, I.W., Somerville, S.C., and Maclean, D.J. (2003). Systemic gene expression in Arabidopsis during an incompatible interaction with Alternaria brassicicola. Plant Physiol. 132: 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk, P.M., Kazan, K., Rusu, A.G., Manners, J.M., and Maclean, D.J. (2005). The SEN1 gene of Arabidopsis is regulated by signals that link plant defense responses and senescence. Plant Physiol. Biochem. 43: 997–1005. [DOI] [PubMed] [Google Scholar]
- Schenk, P.M., Kazan, K., Wilson, I., Anderson, J.P., Richmond, T., Somerville, S.C., and Manners, J.M. (2000). Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 97: 11655–11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, H.S., Song, J.T., Cheong, J.J., Lee, Y.H., Lee, Y.W., Hwang, I., Lee, J.S., and Choi, Y.D. (2001). Jasmonic acid carboxyl methyltransferase: A key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. USA 98: 4788–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, D., and Fondell, J.D. (2002). Ordered recruitment of histone acetyltransferases and the TRAP/Mediator complex to thyroid hormone-responsive promoters in vivo. Proc. Natl. Acad. Sci. USA 99: 7934–7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel, S.H., et al. (2003). NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15: 760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strizhov, N., Keller, M., Mathur, J., Koncz-Kalman, Z., Bosch, D., Prudovsky, E., Schell, J., Sneh, B., Koncz, C., and Zilberstein, A. (1996). A synthetic cryIC gene, encoding a Bacillus thuringiensis delta-endotoxin, confers Spodoptera resistance in alfalfa and tobacco. Proc. Natl. Acad. Sci. USA 93: 15012–15017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher, L.F., Manners, J.M., and Kazan, K. (2009). Fusarium oxysporum hijacks COI1-regulated jasmonate signaling to promote disease in Arabidopsis. Plant J. 58: 927–939. [DOI] [PubMed] [Google Scholar]
- Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G.H., Nomura, K., He, S.Y., Howe, G.A., and Browse, J. (2007). JAZ repressor proteins are targets of the SCFCO11 complex during jasmonate signalling. Nature 448: 661–665. [DOI] [PubMed] [Google Scholar]
- Thomma, B., Eggermont, K., Penninckx, I., Mauch-Mani, B., Vogelsang, R., Cammue, B.P.A., and Broekaert, W.F. (1998). Separate jasmonate-dependent and salicylate-dependent defense response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 95: 15107–15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P., Nelissen, I., Eggermont, K., and Broekaert, W.F. (1999). Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J. 19: 163–171. [DOI] [PubMed] [Google Scholar]
- Trusov, Y., Rookes, J.E., Chakravorty, D., Armour, D., Schenk, P.M., and Botella, J.R. (2006). Heterotrimeric G proteins facilitate Arabidopsis resistance to necrotrophic pathogens and are involved in jasmonate signaling. Plant Physiol. 140: 210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronese, P., Narasimhan, M.L., Stevenson, R.A., Zhu, J.K., Weller, S.C., Subbarao, K.V., and Bressan, R.A. (2003). Identification of a locus controlling Verticillium disease symptom response in Arabidopsis thaliana. Plant J. 35: 574–587. [DOI] [PubMed] [Google Scholar]
- Wagner, D., and Meyerowitz, E.M. (2002). SPLAYED, a novel SWI/SNF ATPase homolog, controls reproductive development in Arabidopsis. Curr. Biol. 12: 85–94. [DOI] [PubMed] [Google Scholar]
- Walley, J.W., Rowe, H.C., Xiao, Y., Chehab, E.W., Kliebenstein, D.J., Wagner, D., and Dehesh, K. (2008). The chromatin remodeler SPLAYED regulates specific stress signaling pathways. PLoS Pathog. 4: e1000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker, C.A., and Hynes, R.O. (2002). Distribution and evolution of von Willebrand/integrin A domains: Widely dispersed domains with roles in cell adhesion and elsewhere. Mol. Biol. Cell 13: 3369–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, I.W., Kennedy, G.C., Peacock, J.W., and Dennis, E.S. (2005). Microarray analysis reveals vegetative molecular phenotypes of Arabidopsis flowering-time mutants. Plant Cell Physiol. 46: 1190–1201. [DOI] [PubMed] [Google Scholar]
- Wollenberg, A.C., Strasser, B., Cerdán, P.D., and Amasino, R.M. (2008). Acceleration of flowering during shade-avoidance in Arabidopsis thaliana alters the balance between FLOWERING LOCUS C-mediated repression and photoperiodic induction of flowering. Plant Physiol. 148: 1681–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, K., Zhang, L., Zhou, C., Yu, C.W., and Chaikam, V. (2008). HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J. Exp. Bot. 59: 225–234. [DOI] [PubMed] [Google Scholar]
- Yang, F., DeBeaumont, R., Zhou, S., and Naar, A.M. (2004). The activator-recruited cofactor/Mediator coactivator subunit ARC92 is a functionally important target of the VP16 transcriptional activator. Proc. Natl. Acad. Sci. USA 101: 2339–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, M., Hay, J., and Ruyechan, W.T. (2008). The Varicella Zoster Virus IE62 protein utilizes the human Mediator complex in promoter activation. J. Virol. 82: 12154–12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, C., Zhang, L., Duan, J., Miki, B., and Wu, K. (2005). HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell 17: 1196–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, N., Tootle, T.L., and Glazebrook, J. (1999). Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell 11: 2419–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.