Abstract
Flowering plants control energy allocation to their photosystems in response to light quality changes. This includes the phosphorylation and migration of light-harvesting complex II (LHCII) proteins (state transitions or short-term response) as well as long-term alterations in thylakoid composition (long-term response or LTR). Both responses require the thylakoid protein kinase STN7. Here, we show that the signaling pathways triggering state transitions and LTR diverge at, or immediately downstream from, STN7. Both responses require STN7 activity that can be regulated according to the plastoquinone pool redox state. However, LTR signaling does not involve LHCII phosphorylation or any other state transition step. State transitions appear to play a prominent role in flowering plants, and the ability to perform state transitions becomes critical for photosynthesis in Arabidopsis thaliana mutants that are impaired in thylakoid electron transport but retain a functional LTR. Our data imply that STN7-dependent phosphorylation of an as yet unknown thylakoid protein triggers LTR signaling events, whereby an involvement of the TSP9 protein in the signaling pathway could be excluded. The LTR signaling events then ultimately regulate in chloroplasts the expression of photosynthesis-related genes on the transcript level, whereas expression of nuclear-encoded proteins is regulated at multiple levels, as indicated by transcript and protein profiling in LTR mutants.
INTRODUCTION
Changes in light quality result in imbalanced excitation of the two photosystems and decrease the efficiency of the photosynthetic light reactions. Plants counteract such conditions via acclimatory responses that result in modifications of thylakoid proteins and reorganization of the photosynthetic machinery (Kanervo et al., 2005; Walters, 2005). In the short term, this involves the so-called state transitions, which depend on the reversible association of the mobile pool of major light-harvesting complex II (LHCII) proteins with photosystem II (PSII) (state 1; favored under dark, far-red, and high light conditions) or photosystem I (PSI) (state 2; favored under low light conditions). The phosphorylation of LHCII, stimulated in low white light, causes association of LHCII with PSI (state 2) in the stroma lamellae, grana ends, and grana margins, thus directing additional excitation energy to PSI (reviewed in Allen and Forsberg, 2001; Haldrup et al., 2001; Wollman, 2001; Rochaix, 2007; Eberhard et al., 2008).
State transitions occur in the order of minutes and have been proposed to be triggered by conformational changes within LHCII proteins upon phosphorylation (Nilsson et al., 1997), leading to docking of phosphorylated LHCII (Pi-LHCII) to PSI. The docking most likely involves the H, L, and O subunits of PSI, as demonstrated by the impairment of state transitions in the corresponding Arabidopsis thaliana null mutants (Lunde et al., 2000; Jensen et al., 2004). The lateral displacement of Pi-LHCII, possibly favored by electrostatic repulsion due to increases in negative charges upon phosphorylation, is also accompanied by rapid and reversible changes in thylakoid membrane and protein complex organization (Chuartzman et al., 2008; Tikkanen et al., 2008).
The membrane-bound protein kinase responsible for phosphorylating LHCII is activated upon reduction of the cytochrome b6/f complex via the plastoquinone (PQ) pool under low light conditions. PQ oxidizing conditions, such as dark or far-red light conditions, inactivate the LHCII kinase, and Pi-LHCII becomes dephosphorylated by the action of an as yet unknown protein phosphatase, resulting in association of LHCII with PSII (state 1; reviewed in Wollman, 2001; Rochaix, 2007). The LHCII kinase activity, however, is also inactivated under high white light conditions, when the stromal reduction state is very high. In vitro and, more recently, in vivo studies suggest that suppression of LHCII kinase activity might be mediated by reduced thioredoxin (Rintamäki et al., 2000; Lemeille et al., 2009). In this context, the redox state of thioredoxin is thought to fine-tune photosynthetic performance by coordinating the light absorption characteristics of the photosystems and the redox regulation of key enzyme activities of the Benson-Calvin cycle (Buchanan and Balmer, 2005).
LHCII phosphorylation and state transitions have been extensively studied in the green alga Chlamydomonas reinhardtii and the flowering plant Arabidopsis (Wollman, 2001; Allen and Race, 2002; Rochaix, 2007; Eberhard et al., 2008). In C. reinhardtii, the impact of state transitions on energy balancing between photosystems and the promotion of cyclic electron flow around PSI is well established (Wollman, 2001; Eberhard et al., 2008). In flowering plants, however, the physiological significance of state transitions is less clear because their mobile LHCII pools are significantly smaller than those in green algae (Allen, 1992; Delosme et al., 1994). Moreover, plant development and fitness are only marginally affected in A. thaliana mutants impaired in state transitions (Lunde et al., 2000; Bonardi et al., 2005; Tikkanen et al., 2006), even under fluctuating light (Bellafiore et al., 2005) or field conditions (Frenkel et al., 2007) when the ability to adapt to illumination changes should become crucial. Therefore, it has been speculated that state transitions play a less prominent role in land plants than in green algae (Finazzi et al., 2002; Finazzi, 2005), becoming important only under low light conditions (Tikkanen et al., 2006).
Recently, a protein kinase (State Transition-deficient mutant 7 [STT7]) involved in state transitions in C. reinhardtii has been identified using a fluorescence-based mutant screen that took advantage of the distinct fluorescence features associated with states 1 and 2 (Depege et al., 2003). The stt7 mutant was locked in state 1 and failed to phosphorylate LHCII under conditions that favor state 2. The STT7 gene encodes a thylakoid Ser-Thr protein kinase required for LHCII phosphorylation. STT7 is characterized by a transmembrane region that separates its stroma-exposed catalytic kinase domain from its lumen-located N-terminal end with two conserved Cys residues that are critical for its activity and state transitions (Lemeille et al., 2009). Moreover, coimmunoprecipitation assays have revealed that the STT7 kinase interacts with cytochrome b6/f, PSI, and LHCII; however, it is not clarified yet whether STT7 is part of a kinase cascade or whether it directly phosphorylates LHCII. Two STT7 homologs exist in land plants: State Transition 7 (STN7) and STN8 (Bellafiore et al., 2005; Bonardi et al., 2005). Arabidopsis stn7 knockout plants displayed much less LHCII phosphorylation than wild-type plants and were not able to perform state transitions, whereas stn8 mutants failed to phosphorylate PSII core proteins (Bellafiore et al., 2005; Bonardi et al., 2005; Vainonen et al., 2005).
In the long term, imbalances in energy distribution between the photosystems are counteracted by adjusting photosystem stoichiometry, changing the abundance of reaction center and light-harvesting proteins within hours and days (long-term response [LTR]) (Melis, 1991; Fujita, 1997; Pfannschmidt et al., 2003). Like state transitions, the LTR is triggered by the redox state of the PQ pool, and in Arabidopsis it is mainly mediated by changes in the number of PSI complexes that are thought to be caused by altered expression of the corresponding plastid- and nucleus-encoded proteins (Fey et al., 2005a). As a consequence, the chlorophyll a/b ratio and the chlorophyll fluorescence parameter Fs/Fm (reflecting the structural differences in the photosynthetic apparatus as described in Results and Methods) were found to be useful for assessing the ability of plants to perform a proper LTR (Dietzel et al., 2008). Most species investigated indeed exhibit enhanced expression of the PSI reaction center operon psaAB (encoding the P700 apoproteins) upon reduction of the PQ pool or a respective repression upon its oxidation. Additionally, LTR induces an extensive restructuring of the thylakoid membrane system, including increased grana stacking and less accumulation of transitory starch in plastids from plants exposed to PSI light (reviewed in Dietzel et al., 2008). Transcriptomic analyses performed on plants impaired in the LTR process, however, resulted in diverse transcript profiles. This can be explained at least in part by the different types of arrays and growth conditions employed in the experiments (Bonardi et al., 2005; Fey et al., 2005b; Tikkanen et al., 2006).
In Arabidopsis, studies of the stn7 mutant uncovered an intriguing connection between state transitions and the LTR. Analysis of the chlorophyll a/b ratio and Fs/Fm parameters indeed indicated that the stn7 mutant lacks not only state transitions but also the LTR (Bonardi et al., 2005; Tikkanen et al., 2006). Thus, it appears that the STN7 kinase represents a common redox sensor and/or signal transducer for both responses, supporting earlier suggestions that the two processes are subject to regulatory coupling (Allen, 1995; Allen and Pfannschmidt, 2000; Pursiheimo et al., 2001). However, it is not clear whether the signal pathways leading to state transitions and LTR represent two branches that originate at STN7 and then diverge or a hierarchically organized signaling cascade in which state transitions (or aspects of it) are required for the LTR. Because of the inconspicuous phenotype of stn7 plants, metabolic and physiological changes induced by the lack of STN7 do not appear to be responsible for the suppression of the LTR.
Thus, in principle, (1) the phosphorylation state of LHCII itself could directly provide information for signaling, (2) state transitions and associated conformational changes in thylakoids might stimulate signaling, or (3) an unknown protein that is phosphorylated in an STN7-dependent manner could relay the required signal for the STN7-mediated LTR. Concerning the latter hypothesis, the thylakoid soluble phosphoprotein of 9 kD, TSP9, has been suggested as a possible component of the redox-dependent signaling cascade as a consequence of its mobile nature (Carlberg et al., 2003; Zer and Ohad, 2003). Thus, TSP9 is released from the thylakoid membranes into the stroma upon redox-dependent stepwise phosphorylation of three Thr sites. Recently, the Arabidopsis TSP9 protein has been reported to be phosphorylated in an STN7-dependent manner and to be involved in state transitions (Fristedt et al., 2009). Moreover, transcriptomic data suggest a possible role of TSP9 in high light acclimation (Fristedt et al., 2009).
In this work, we have investigated the physiological relevance of state transitions in the flowering plant Arabidopsis and the relationship between the signal pathways leading to state transitions and the LTR. We show that state transitions play a prominent role in energy distribution between photosystems also in flowering plants. Although state transitions per se are not required for the LTR, the presence of the STN7 kinase with a modulatable activity is necessary for this process. Additionally, the LTR signaling pathway appears to be divided in two main branches: one responsible to transcriptionally regulate chloroplast PSI-related gene expression and a second one involved in the regulation of the expression of nuclear photosynthesis-related genes at multiple levels.
RESULTS
Effects of State Transition Mutations on Chlorophyll Fluorescence Quenching
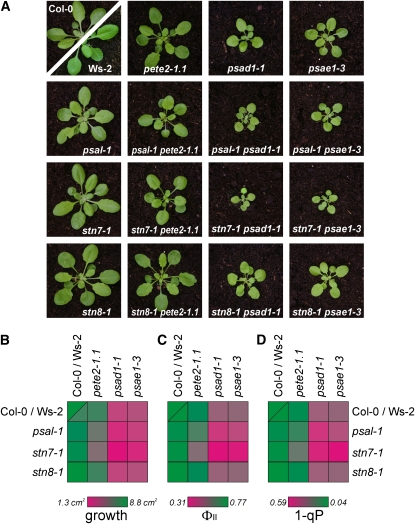
To determine whether state transitions per se or individual steps of this process are essential for the LTR, Arabidopsis lines impaired in the different components necessary for state transitions were tested for their capacity to perform the LTR. The plants analyzed included antisense lines almost completely devoid of LHCII (as LHCB2.1, ecotype Columbia [Col-0]) (Andersson et al., 2003), TSP9 RNA interference (RNAi) lines with a reduced accumulation of the thylakoid phosphoprotein TSP9 (ecotype Col-0) (Carlberg et al., 2003; Fristedt et al., 2009), mutants that lack the docking site for LHCII at PSI (psal-1, ecotype Col-0) (see Supplemental Figure 1 online) (Lunde et al., 2000), and plants that exhibit hyperphosphorylation of LHCII and other thylakoid proteins (psad1-1, ecotype Col-0, and psae1-3, ecotype Wassilewskija [Ws-2]; Ihnatowicz et al., 2008), as well as additional genotypes altered in the accumulation of specific thylakoid proteins (Table 1). State transitions were monitored by measuring chlorophyll fluorescence under light conditions that favored either PSI or PSII.
Table 1.
Overview of Arabidopsis Lines Used in This Study
| Mutant | Effect on State Transitions and Related Photosynthetic Functions | References |
|---|---|---|
| psad1-1 | Lack of PSI subunit D1 (the At4g02770 gene is disrupted by a dSpm insertion +166 bp downstream of the ATG start codon); stable Pi-LHCII-PSI aggregate; drastic increase in Pi-LHCII and in other thylakoid phosphoproteins; 1-qP 10 to 11 times higher than in the wild type; state transitions markedly reduced | Ihnatowicz et al. (2004, 2008); this study |
| psae1-3 | Lack of PSI subunit E1 (the At4g28750 gene is disrupted by a T-DNA insertion in the first exon; germplasm FLAG_486G07); stable Pi-LHCII-PSI aggregate; drastic increase in Pi-LHCII and in other thylakoid phosphoproteins; 1-qP 9 to 10 times higher than in the wild type, state transitions markedly reduced | Varotto et al. (2000); Pesaresi et al. (2002); Ihnatowicz et al. (2007); this study |
| asLHCB2.1 | Downregulation of LHCB1 and 2 (the expression of LHCB1 and LHCB2 gene families is downregulated by an antisense construct in the plant expression vector pSJ10); almost complete lack of LHCII; state transitions markedly reduced | Andersson et al. (2003) |
| psal-1 | Lack of PSI subunit L (the At4g12800 gene is disrupted by a T-DNA insertion in the second exon; germplasm SALK_000637); secondary lack of PSI-H and -O impairing the docking site for LHCII; state transitions markedly reduced | Lunde et al. (2000); Jensen et al. (2004); see Supplemental Figure 1 online |
| chaos | Lack of signal recognition particle 43 (the At2g47450 gene is disrupted by the insertion of the transposable Ds element; germplasm GT14566); impaired LHCII protein targeting; pleiotropic impairment of photosynthetic light reaction including state transitions | Klimyuk et al. (1999) |
| pete2-1.1 | Lack of major isoform of plastocyanin (the At1g20340 gene is disrupted by the insertion of the transposable En element); 1-qP 2 to 3 times higher than in the wild type | Weigel et al. (2003) |
| TSP9 RNAi | Downregulation of thylakoid phosphoprotein TSP9 (the At3g47070 gene is downregulated by RNA interference using the pJawohl8-RNAi vector); impaired state transitions; possible involvement in high light acclimation | Carlberg et al. (2003); Fristedt et al. (2009) |
| stn7-1 | Lack of thylakoid protein kinase STN7 (the At1g68830 gene is disrupted by a T-DNA insertion in the 7th exon; germplasm SALK_073254); drastically reduced LHCII phosphorylation; state transitions markedly reduced | Bellafiore et al. (2005); Bonardi et al. (2005) |
| stn8-1 | Lack of thylakoid protein kinase STN8 (the At5g01920 gene is disrupted by a T-DNA insertion in the 1st exon; germplasm SALK_060869); drastically reduced PSII-core protein phosphorylation; used as control in BN-PAGE analyses (see Figure 3) | Bonardi et al. (2005); Vainonen et al. (2005) |
The gene names are according to the Arabidopsis Genome Initiative. Germplasms are according to T-DNA Express (http://signal.salk.edu/cgi-bin/tdnaexpress). 1-qP refers to the fraction of QA, the primary electron acceptor of PSII, present in the reduced state. Note that except for psae1-3 (Ws-2) all lines are in the genetic background of Col-0.
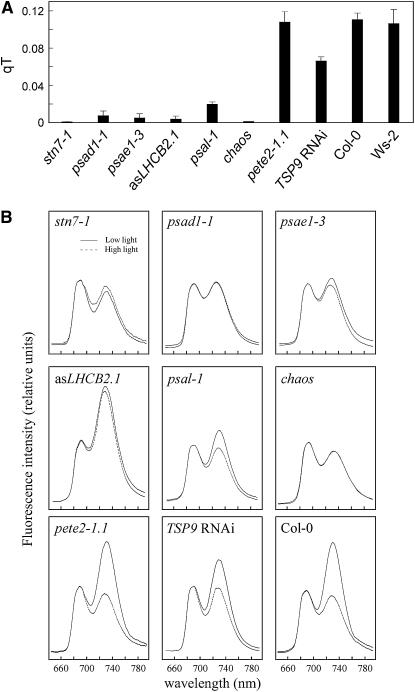
We found that the lines asLHCB2.1, psal-1, TSP9 RNAi, and the chaos (ecotype Col-0) mutant, known to be impaired in LHCII protein targeting (Klimyuk et al., 1999), were defective, although to different extents, in the ability to perform state transitions, similarly to stn7-1 (ecotype Col-0) plants (Figure 1A). The defect in state transitions in these lines was also corroborated by 77 K fluorescence emission measurements (Figure 1B). As a measure of the energy distribution between PSI and PSII, the fluorescence emission ratio at 733 nm (PSI fluorescence emission peak) and 685 nm (PSII fluorescence emission peak) (F733/F685) was calculated (Table 2). Wild-type (ecotypes Col-0 and Ws-2) plants treated with high white light to induce state 1 exhibited a markedly lower ratio than under low white light that induced a transition to state 2. Similar results were obtained for the pete2-1.1 (ecotype Col-0) mutant, implying that state transitions remained unaltered in this genotype. The TSP9 RNAi lines and the psal-1 mutant showed only a limited increase in F733/F685 after exposure to state 2 conditions (Table 2), indicating a partial block of state transitions in state 1, in agreement with previous reports (Lunde et al., 2000; Fristedt et al., 2009). The complete absence of state transitions (i.e., a block in state 1 resulting from the absence of LHCII phosphorylation) occurred in the stn7-1 mutant, in which F733/F685 values of ∼0.9 were obtained, irrespective of whether the plants were adapted to state 1 or state 2 conditions (Table 2). The almost identical F730/F685 ratios in psad1-1 and psae1-3 mutants reflect an absence of state transitions also in these two genotypes. However, because in both psad1-1 and psae1-3 the F730/F685 ratios for state 1 were comparable to the wild-type values, despite a reduction of ∼60% in PSI levels in both mutants (Ihnatowicz et al., 2004; Ihnatowicz et al., 2007), a large fraction of LHCII should be attached to PSI even under state 1 in these genotypes to mimic wild type–like values. A block in state transitions, as a consequence of the marked reduction in amounts of the mobile LHCII pool, was observed in chaos and asLHCB2.1 plants.
Figure 1.
Short-Term Photosynthetic Acclimation in Photosynthetic Mutants.
(A) Quenching of chlorophyll fluorescence due to state transitions (qT). Bars indicate sd using five plants of each genotype.
(B) The 77 K fluorescence emission spectra of the wild type (Col-0) and mutants. Spectra (traces are the average of 10 replicates) were recorded after illumination of plants with low light (80 μmol m−2 s−1, solid lines) and high light (800 μmol m−2 s−1, dotted lines) for 3 h by excitation at 475 nm. The 77 K fluorescence spectra were normalized at 685 nm. Note that, in both psad1-1 and psae1-3, a 3-nm blue shift in the far-red florescence emission peak was observed. The possibility that the light intensities chosen to induce state transitions could elicit photoinhibition was monitored by measurements of Fv/Fm values before and after the light treatment. The identical Fv/Fm values indicated the absence of any photoinibition. Note that wild-type plants of ecotype Ws-2 behaved identically to Col-0.
Table 2.
Energy Distribution between PSI and PSII Measured as F733(F730)/F685, the Fluorescence Emission Ratio at 733 nm (730 nm in the case of psad1-1 and psae1-3) and 685 nm
| F733(F730)/F685
|
||
|---|---|---|
| State 1 | State 2 | |
| Wild type | 0.93 ± 0.02 | 1.63 ± 0.03 |
| pete2-1.1 | 0.89 ± 0.04 | 1.60 ± 0.03 |
| TSP9 RNAi | 0.97 ± 0.03 | 1.39 ± 0.02 |
| psal-1 | 0.96 ± 0.04 | 1.21 ± 0.05 |
| stn7-1 | 0.92 ± 0.02 | 0.87 ± 0.03 |
| psad1-1 | 1.03 ± 0.03 | 1.03 ± 0.02 |
| psae1-3 | 1.02 ± 0.04 | 1.08 ± 0.05 |
| chaos | 0.88 ± 0.03 | 0.88 ± 0.04 |
| asLHCB2.1 | 1.65 ± 0.05 | 1.72 ± 0.04 |
Effects of State Transition Mutations on PSI-LHCI-LHCII Complex Formation
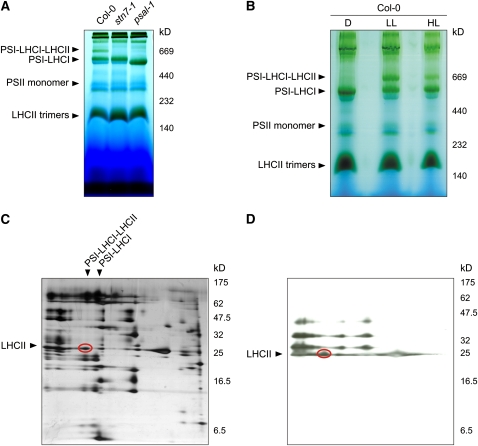
A pigment-protein complex of ∼670 kD appeared in wild-type leaves exposed to low white light conditions that promote attachment of the mobile pool of LHCII to PSI (state 2), whereas it failed to form in stn7-1 and psal-1 thylakoids, as revealed by blue-native PAGE (BN-PAGE) (Figure 2A). The 670-kD complex did not accumulate in dark-adapted wild-type leaves and was markedly reduced in leaves illuminated with high white light levels that promote detachment of LHCII from PSI (state 1) (Figure 2B). The two-dimensional PAGE (2D-PAGE) fractionation showed that the pigment-protein complex consists of PSI and LHCI subunits, together with a portion of Pi-LHCII that associates with PSI upon state 1→2 transition in wild-type plants (Figures 2C and 2D) (Pesaresi et al., 2002; Heinemeyer et al., 2004). The identity of the constituents of the 670-kD pigment-protein complex was also confirmed by mass spectrometry (see Supplemental Table 1 online). Interestingly, the difference of ∼145 kD in the apparent molecular weights of PSI-LHCI-LHCII and PSI-LHCI complexes, matches almost perfectly with the apparent molecular weight of an LHCII trimer, migrating between 140 and 230 kD (Figure 2B), thus confirming electron microscopy analyses where an LHCII trimer attached to a PSI-LHCI complex was shown (Kouril et al., 2005).
Figure 2.
Visualization of the PSI-LHCI-LHCII Complex Associated with State 2–Adapted Thylakoid Membranes.
(A) BN-PAGE of identical amounts of thylakoid proteins isolated from wild-type (Col-0), stn7-1, and psal-1 leaves adapted to low-light conditions (80 μmol m−2 s−1 for 8 h). A pigment-protein complex (PSI-LHCI-LHCII) migrating at ∼670 kD is clearly visible in wild-type (Col-0) thylakoids, whereas it is completely absent in stn7-1 and psal-1 thylakoids; both mutants are blocked in state 1. Note that the PSI-LHCI protein complex from psal-1 thylakoids appears to be smaller than in wild-type (Col-0) and stn7-1 plants, as a consequence of the altered polypeptide composition of PSI (see also Figure 10).
(B) BN-PAGE (as in [A]) of thylakoid proteins isolated from wild-type (Col-0) leaves adapted to darkness for 16 h (D), low light (LL; 80 μmol m−2 s−1 for 3 h), or high light (HL; 800 μmol m−2 s−1 for 3 h).
(C) The wild-type low-light lane from a BN gel was subjected to denaturing PAGE, and the 2D gel was stained with silver. The polypeptide fractionation shows that the ∼670-kD pigment-protein complex consists of PSI and LHCI subunits, together with a portion of LHCII polypeptides (oval).
(D) Thylakoid protein phosphorylation detected by immunoblot analysis with a phosphothreonine-specific antibody. Thylakoid membranes isolated from low light–adapted wild-type (Col-0) leaves were fractionated (as in [C]). The LHCII polypeptides associated with PSI-LHCI complexes are highly phosphorylated (oval).
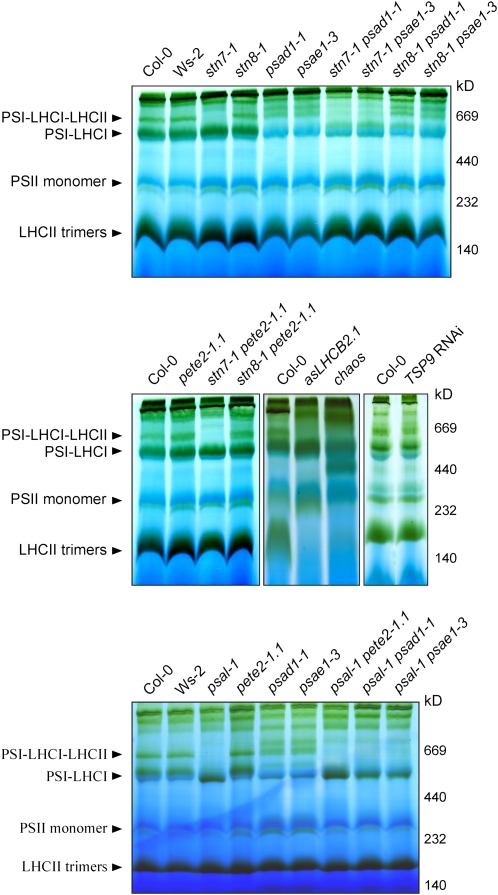
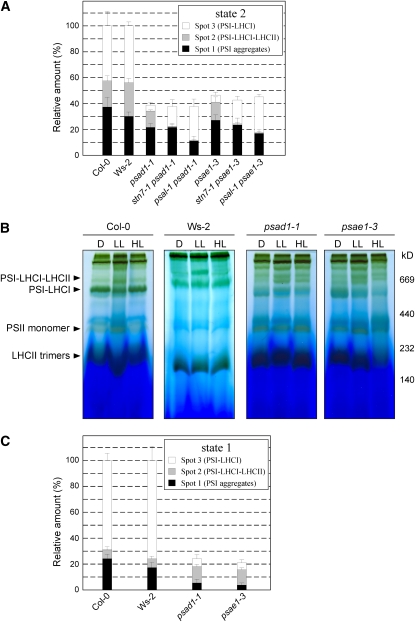
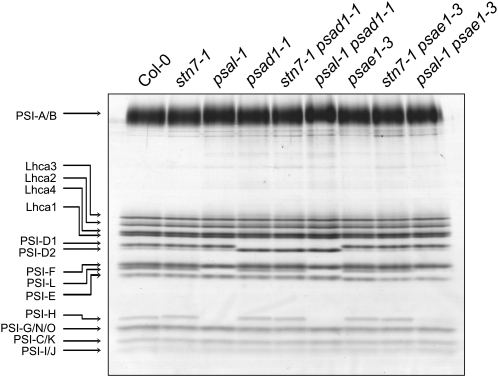
The PSI-LHCI-LHCII complex was not detectable in stn7-1, psal-1, chaos, and asLHCB2.1 plants and was less abundant in TSP9 RNAi lines, as revealed by BN-PAGE fractionation of thylakoid proteins from low light–adapted (state 2) plants (Figure 3). This suggests that in these genotypes state transitions are partially or totally blocked in state 1. Although a drastic impairment in state transitions also occurs in the psad1-1 and psae1-3 mutants (Figure 1A, Table 2), the PSI-LHCI-LHCII complex still accumulated in psad1-1 and psae1-3 thylakoids in conditions inducing state 2, in contrast with the genotypes mentioned above (Figure 3). Indeed, densitometric analyses performed on 2D-PAGE gels revealed that in low light–adapted psad1-1 and psae1-3 thylakoids, the PSI-LHCI-LHCII complex (spot 2 in Figure 4A and Supplemental Figure 2A online) was twofold more abundant than the PSI-LHCI complex (spot 1 in Figure 4A and Supplemental Figure 2A online), whereas the opposite was evident in wild-type (Col-0 and Ws-2) thylakoids. As expected, the PSI-LHCI-LHCII complex disappeared completely in stn7-1 psad1-1, psal-1 psad1-1, stn7-1 psae1-3, and psal-1 psae1-3 double mutants (Figures 3 and 4A; see Supplemental Figure 2A online).
Figure 3.
BN-PAGE Analysis of Single and Double Mutants.
BN-PAGE (as in Figure 2) of thylakoid proteins isolated from wild-type (Col-0 and Ws-2) and single and double mutant leaves adapted to low white light levels (80 μmol m−2 s−1 for 3 h). Note that a highly abundant PSI-LHCI-LHCII complex is present in psad1-1 and psae1-3 leaves, despite the impairment in state transitions (see also Figures 1A and 1B), thus implying a marked shift toward state 2.
Figure 4.
Quantification of PSI-LHCI and PSI-LHCI-LHCII Complexes Associated with Low (State 2) and High Light–Adapted (State 1) Thylakoid Membranes from Wild-Type (Col-0 and Ws-2) and Mutant Plants.
(A) 2D-PAGEs, as in Figure 2, of thylakoid membranes isolated from wild-type (Col-0 and Ws-2) and mutant leaves adapted to low light (state 2 condition, 80 μmol m−2 s−1 for 3 h), were stained with Coomassie blue (see Supplemental Figure 2A online). The densitometric quantification of the spots representing PSI aggregates (spot 1), PSI-LHCI-LHCII (spot 2), and PSI-LHCI (spot 3), shown in Supplemental Figure 2A online, is shown. Values are averages of three independent 2D gels for each genotype. Pools of 20 plants each genotype were used to isolate thylakoids. Bars indicate sd.
(B) BN-PAGE of identical amounts of thylakoid proteins isolated from wild-type (Col-0 and Ws-2), psad1-1, and psae1-3 leaves adapted to darkness for 16 h (D) (state 1 condition), low light (LL; 80 μmol m−2 s−1 for 3 h) (state 2 condition), or high light (HL; 800 μmol m−2 s−1 for 3 h) (state 1 condition).
(C) The BN gel high-light (state 1 conditions, 800 μmol m−2 s−1 for 3 h) lanes from the wild type (Col-0 and Ws-2), psad1-1, and psae1-3 in Figure 4B were subjected to denaturing PAGE, and the 2D gels were stained with Coomassie blue (see Supplemental Figure 2B online). Spots representing PSI aggregates, PSI-LHCI-LHCII and PSI-LHCI, under high light conditions (state 1), displayed in Supplemental Figure 2A online, were densitometrically quantified as in (A).
These data, together with the 77 K fluorescence measurements (Figure 1B), suggest that the psad1-1 and psae1-3 mutants are markedly shifted toward state 2. In agreement with that, BN- and 2D-PAGE analyses clearly revealed that a large portion of the PSI-LHCI-LHCII complex persisted in psad1-1 and psae1-3 thylakoids even after exposure to high white light intensities (that normally induce state 1) (i.e., >50% of PSI complexes, excluding PSI aggregates, retained their association with LHCII; Figures 4B and 4C; see Supplemental Figure 2B online). Only a prolonged dark exposure (16 h) of psad1-1 and psae1-3 plants allowed the state 2→1 transition (Figure 4B).
State Transitions Are Not Essential for the LTR
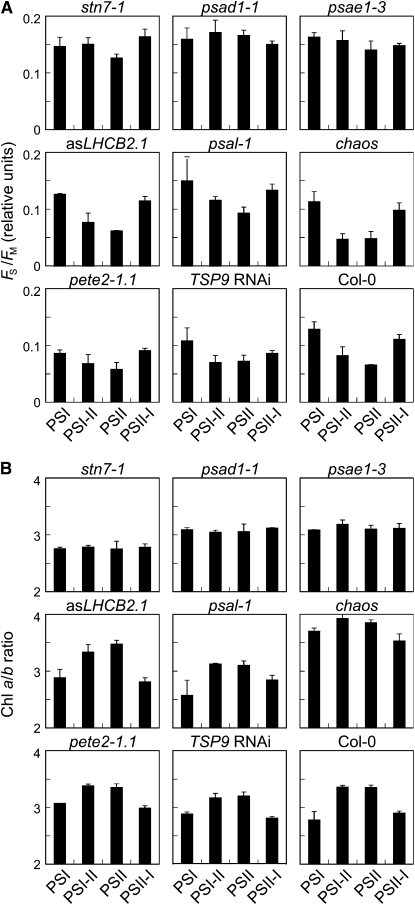
To detect effects on the LTR, wild-type (Col-0 and Ws-2) and mutant plants were exposed to light regimes that favored either PSI (red light, 50% transmittance at 650 nm) or PSII (orange-yellow light, 50% transmittance at 560 nm) (Figure 5). In detail, after growth for 10 d under white light, plants were acclimated to PSI or PSII light for 6 d (PSI or PSII plants), or they were first acclimated to one light source for 2 d followed by 4 d under the respective other light source (PSI-II or PSII-I plants). The development of the LTR was then monitored by measuring not only chlorophyll fluorescence parameters (Fs, the steady state fluorescence; Fm, maximum fluorescence; and its ratio Fs/Fm) and chlorophyll a/b ratios (Figure 5), but also the expression of the plastid genes psaAB (encoding the PSI reaction center, P700) and psbA (encoding the reaction center D1 protein of PSII), as well as of the nuclear genes LHCA3 (encoding LHCA3, a PSI-associated antenna subunit) and LHCB1.1 (encoding LHCB1, a PSII-associated antenna subunit) (Figure 6). The combination of those measurements allows us to define stoichiometric readjustments among the thylakoid complexes (for a review, see Dietzel et al., 2008), as recently confirmed by a detailed biochemical analysis performed on stn7 plants (Tikkanen et al., 2006). In particular, the Fs/Fm parameter reflects the structural differences in the photosynthetic apparatus, and its value typically increases after acclimation of wild-type plants to PSI light and decreases after acclimation to PSII light (Figure 5A). The chlorophyll a/b ratio also reflects the photosystem organization of the photosynthetic complexes and behaved in the opposite manner in wild-type plants; being high after acclimation to PSII light and low in PSI light (Figure 5B). The reversibility of these responses was demonstrated by shifting wild-type plants between the different light regimes (Pfannschmidt et al., 2001; Fey et al., 2005b).
Figure 5.
Long-Term Photosynthetic Acclimation (LTR) in Photosynthetic Mutants.
For LTR measurements, plants were initially grown for 10 d under white light followed by a 6-d acclimation period. Seedlings were acclimated either to PSI or PSII light (see Methods) for 6 d or to PSI light for 2 d followed by 4 d under PSII light (PSI-II plants) or vice versa (PSII-I plants).
(A) Fs/Fm ratios calculated using measurements of the chlorophyll fluorescence parameters Fs (steady state fluorescence) and Fm (maximum fluorescence).
(B) Determination of chlorophyll a/b ratios. All values under the four conditions were calculated as the means of 50 individuals in at least three independent experiments. Bars indicate sd. Note that Ws-2 plants behaved identically to Col-0 with respect to Fs/Fm and chlorophyll a/b ratio changes under the different light conditions.
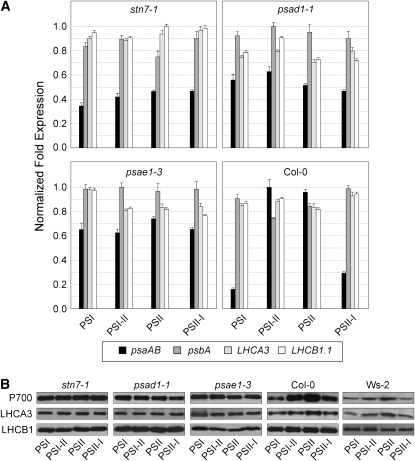
Figure 6.
LTR-Dependent Gene Expression Regulation in Wild-Type (Col-0 and Ws-2) and Mutant (stn7-1, psad1-1, and psae1-3) Plants Adapted to the Different Light Conditions as Described in Figure 5 (See Also Methods).
(A) Transcript amounts of psaAB, psbA, LHCA3, and LHCB1.1 were estimated by real-time PCR analyses (see Methods). Fold expression values were calculated as the means of three biological replicates for each genotype (three pools, five plants each pool) under the four conditions analyzed. Bars indicate sd. Note that Ws-2 plants behaved identically to Col-0.
(B) Levels of protein accumulation under the different light conditions were estimated by immunoblot analyses using antibodies specific for the PSI reaction center (P700) and the antennae proteins LHCA3 and LHCB1.
Characteristic for the adjustment of photosystem stoichiometry during LTR in wild-type plants was the increased accumulation of psaAB transcripts both under PSII light (PSII) and after the shift from PSI to PSII light (PSI-II), whereas no major alteration in the expression of the psbA, LHCA3, and LHCB1.1 genes was observed (Figure 6A). Immunoblot analysis with antisera raised against the PSI reaction center (P700) and the antenna proteins LHCA3 and LHCB1 showed a high abundance of P700 and LHCA3 proteins in wild-type thylakoids exposed to PSI-II and PSII light conditions, whereas no differential accumulation of the LHCB1 antenna protein could be observed (Figure 6B). The stn7-1 mutant, which is incapable of mounting an LTR, exhibited Fs/Fm and chlorophyll a/b values typical of plants acclimated to PSI light under all regimes tested (Figure 5; Bonardi et al., 2005). Additionally, the changes in the accumulation of psaAB transcripts and of the P700 and LHCA3 proteins, which are characteristic for the LTR response in wild-type plants, could not be detected in stn7-1 (Figures 6A and 6B). It is noteworthy that most of the mutants that fail to display state transitions but retain a functional STN7 still manifested the LTR. In particular, asLHCB2.1, psal-1, and chaos lines behaved like the wild type (Figures 5A and 5B). These findings clearly indicate that state transitions per se are not required for LTR.
Modulatable STN7 Activity Is Required for an Efficient LTR
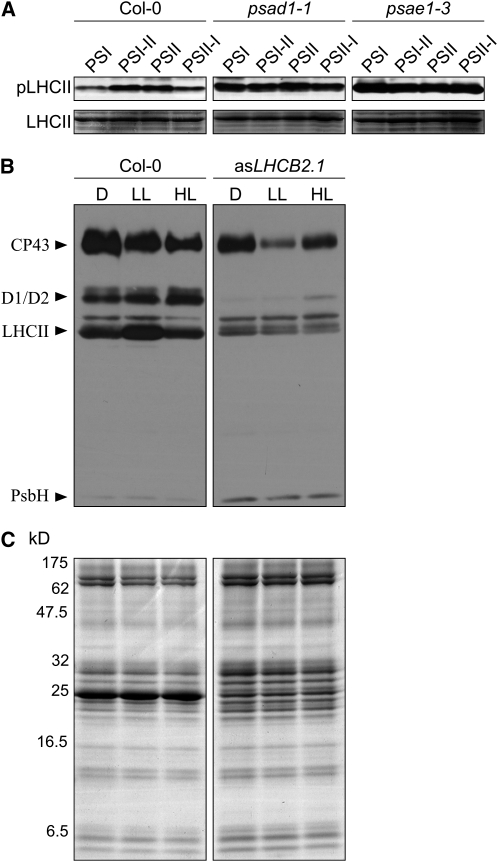
Apart from stn7-1, only the PSI mutants psad1-1 and psae1-3 were incapable of performing the LTR, as monitored by the unchanged values of Fs/Fm and chlorophyll a/b under the different light conditions (Figure 5) and the lack of the characteristic changes in the abundance of psaAB transcripts and of the P700 and LHCA3 proteins (Figure 6). Characteristically, in these two mutants, the fraction of QA, the primary electron acceptor of PSII, present in the reduced state (1-qP; a parameter frequently used for an indirect estimation of the redox state of the PQ pool; Gray et al., 1996; Savitch et al., 1996) is drastically increased even under normal light conditions (low light, 80 μmol m−2 s−1) (see Supplemental Table 1 online; Varotto et al., 2000; Pesaresi et al., 2002; Ihnatowicz et al., 2004, 2007, 2008). As reduction of the PQ pool is known to activate phosphorylation of thylakoid proteins, psad1-1 and psae1-3 leaves also show a marked increase in levels of Pi-LHCII (Ihnatowicz et al., 2008), in complete contrast with stn7-1 (Bonardi et al., 2005). In psad1-1 and psae1-3, unlike the case in wild-type plants, the high level of Pi-LHCII remains the same under all light conditions employed to monitor LTR (Figure 7A). This further implies that no major changes in the PQ redox state take place in these mutants when they are exposed to light conditions that preferentially excite PSI. This, together with the loss of LTR in stn7-1, suggests that the STN7 kinase needs to be functioning but must not be permanently activated to allow an efficient LTR. Interestingly, pete2-1 plants did not show any major perturbation in LTR despite the increased reduction of QA fraction (see Supplemental Table 2 online), implying that a threshold limit exists for QA reduction, above which plants lose their capability to modulate STN7 activity and, therefore, to perform LTR.
Figure 7.
Phosphorylation Patterns in Wild-Type (Col-0) and Mutants (psad1-1, psae1-3, and asLHCB2.1) Acclimated to the Conditions Used to Monitor LTR or State Transitions.
(A) Phosphorylated LHCII (Pi-LHCII; top panels) was detected by immunoblot analysis using a phosphothreonine-specific antibody in wild-type and mutant (psad1-1 and psae1-3) plants adapted to PSI or PSII light or shifted between the two light regimes (PSI-II; PSII-I). Portions of Coomassie-stained SDS-PAGE gels, identical to those used for blotting and corresponding to the LHCII migration region (bottom panels), were used to control for equal loading. Results of one of three independent experiments are shown.
(B) Thylakoid protein phosphorylation in the wild type (Col-0) and asLHCB2.1 plants acclimated to dark (D; 16 h), low light (LL; 80 μmol m−2 s−1, 3 h), and high light (HL; 800 μmol m−2 s−1, 3 h) conditions. Phosphorylated thylakoid proteins were detected by immunoblot analysis using a phosphothreonine-specific antibody. Results of one of three independent experiments are shown.
(C) Coomassie blue–stained SDS-PAGE gels, identical to those used for blotting shown in (B), were used to control for equal loading.
Strikingly, the drastic reduction of LHCII amounts in asLHCB2.1 leaves, together with the basal and constant level of phosphorylation in this genotype independently of light conditions (Figures 7B and 7C), was compatible with a normal LTR (see Figure 5). This allows us to conclude that the structural thylakoid reorganization following the migration of the mobile LHCII pool between the two photosystems is not required for LTR. Together, it appears plausible that the signal transduction pathways associated with the short- and long-term responses must diverge directly at, or immediately downstream from, STN7.
Extreme Changes in STN7-Dependent Phosphorylation Induce Similar Transcriptional Responses of a Distinct Group of Genes
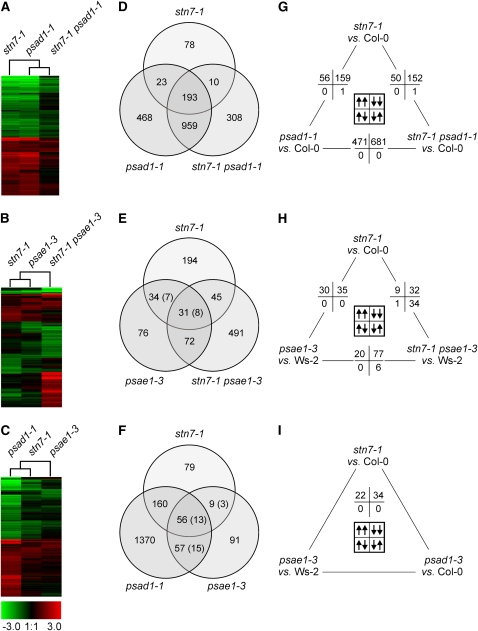
If a modulatable STN7 activity is required for LTR, a common signaling pathway should be perturbed in stn7-1, psad1-1, and psae1-3 mutants, and this should be reflected in their mRNA expression profiles. Whole-genome transcript profiles of leaves of greenhouse-grown stn7-1, psad1-1, and psae1-3 plants, as well as of the double mutants stn7-1 psad1-1 and stn7-1 psae1-3, were obtained by Affymetrix GeneChip analysis, and genes whose transcription levels differed significantly from those in wild-type (Col-0 and Ws-2) plants were identified (Figure 8, Table 3; see Supplemental Tables 3 to 7 online). In addition, for a number of representative genes, the differential expression values obtained by Affymetrix GeneChip analysis were confirmed by real-time PCR analysis (see Supplemental Table 8 online).
Figure 8.
Transcript Profiles of LTR Mutants and Identification of Coregulated Gene Sets.
(A), (D), and (G) stn7-1, psad1-1, and stn7-1 psad1-1 mutants.
(B), (E), and (H) stn7-1, psae1-3, and stn7-1 psae1-3.
(C), (F), and (I) stn7-1, psad1-1, and psae1-3.
(A) to (C) Hierarchical clustering of mRNA expression profiles. The cladogram at the top summarizes the relatedness of transcriptome responses. Colors indicate up- (red) or downregulation (green) of genes, with respect to wild-type (Col-0 or Ws-2) controls.
(D) to (F) Venn diagrams depicting the overlap of genes whose expressions were found in Affymetrix GeneChips analyses to be differentially regulated. The number of genes that are differentially regulated in two different wild-type ecotypes employed in this study (Col-0 and Ws-2) is indicated in parentheses.
(G) to (I) Pairwise comparison of nuclear transcript profiles. Numbers of significantly differentially regulated genes in the same (up/up or down/down) or opposite (up/down or down/up) direction are provided. Note that array data have been validated by monitoring the expression of a subset of genes in the different genotypes using real-time PCR analysis (see Supplemental Table 8 online).
Table 3.
Tentative Target Genes of LTR, Identified as Coregulated Genes in stn7-1, psad1-1, and psae1-3 Leaves
|
stn7-1
|
psad1-1
|
psae1-3
|
||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Description | Subcellular Location | FC | P Value | FC | P Value | FC | P Value |
| Downregulated | ||||||||
| At5g42800 | Dihydroflavonol 4-reductase | Endoplasmatic reticulum | 0.14 | 0.0016 | 0.25 | 0.0099 | 0.07 | 0.0015 |
| At4g22870 | Putative leucoanthocyanidin dioxygenase | Unknown* | 0.15 | 0.0005 | 0.19 | 0.0031 | 0.15 | 0.0025 |
| At1g56650 | MYB75 transcription factor | Nucleus | 0.15 | 0.0003 | 0.06 | 0.0003 | 0.16 | 0.0090 |
| At5g17220 | Putative glutathione S-transferase | Cytoplasm | 0.18 | 0.0050 | 0.22 | 0.0094 | 0.21 | 0.0116 |
| At1g03495 | Transferase family protein | Unknown* | 0.18 | 0.0004 | 0.24 | 0.0030 | 0.08 | 0.0026 |
| At3g16360 | Histidine-containing phosphotransmitter 4 | Cytoplasm, nucleus | 0.18 | 0.0166 | 0.05 | 0.0004 | 0.25 | 0.0019 |
| At4g14090 | UDP-glucosyl transferase family protein | Mitochondrion | 0.19 | 0.0005 | 0.29 | 0.0047 | 0.21 | 0.0108 |
| At5g54060 | UDP-glucose:flavonoid 3-O-glucosyltransferase | Unknown* | 0.20 | 0.0020 | 0.25 | 0.0104 | 0.32 | 0.0216 |
| At1g80130 | Expressed protein | Chloroplast* | 0.23 | 0.0001 | 0.34 | 0.0070 | 0.24 | 0.0007 |
| At1g56600 | Putative galactinol synthase | Unknown* | 0.24 | 0.0010 | 0.10 | 0.0015 | 0.25 | 0.0039 |
| At4g19430 | Expressed protein | Chloroplast | 0.26 | 0.0211 | 0.18 | 0.0008 | 0.23 | 0.0006 |
| At5g52760 | Heavy metal–containing protein | Unknown* | 0.27 | 0.0056 | 0.11 | 0.0051 | 0.45 | 0.0287 |
| At3g12580 | Heat shock protein 70 | Cytosol | 0.29 | 0.0451 | 0.06 | 0.0017 | 0.33 | 0.0010 |
| At4g14365 | Zinc finger (C3HC4-type) protein | Nucleus | 0.30 | 0.0056 | 0.22 | 0.0134 | 0.43 | 0.0210 |
| At5g11930 | Glutaredoxin family protein | Chloroplast* | 0.31 | 0.0035 | 0.25 | 0.0024 | 0.26 | 0.0060 |
| At4g15620 | Integral membrane family protein | Unknown* | 0.32 | 0.0059 | 0.22 | 0.0022 | 0.09 | 0.0001 |
| At2g17040 | No apical meristem (NAM) family protein | Unknown* | 0.34 | 0.0006 | 0.08 | 0.0007 | 0.36 | 0.0086 |
| At5g52750 | Heavy metal–containing protein | Unknown* | 0.35 | 0.0024 | 0.16 | 0.0060 | 0.49 | 0.0359 |
| At1g45145 | Thioredoxin H-type 5 (TRX-H-5) (TOUL) | Cytosol | 0.36 | 0.0044 | 0.22 | 0.0084 | 0.50 | 0.0012 |
| At1g66390 | Putative MYB transcription factor | Nucleus | 0.37 | 0.0003 | 0.30 | 0.0015 | 0.30 | 0.0132 |
| At1g73040 | Jacalin lectin family protein | Unknown* | 0.39 | 0.0028 | 0.21 | 0.0008 | 0.42 | 0.0062 |
| At1g67360 | Rubber elongation factor (REF) | Vacuole | 0.39 | 0.0003 | 0.13 | 0.0003 | 0.43 | 0.0093 |
| At4g32940 | Vacuolar processing enzyme | Vacuole | 0.39 | 0.0006 | 0.28 | 0.0067 | 0.50 | 0.0165 |
| At1g17745 | 3-Phosphoglycerate dehydrogenase | Chloroplast* | 0.40 | 0.0004 | 0.23 | 0.0012 | 0.29 | 0.0002 |
| At5g22460 | Esterase/lipase/thioesterase protein | Cell Wall, vacuole | 0.42 | 0.0317 | 0.49 | 0.0214 | 0.38 | 0.0036 |
| At4g39210 | ADP-glucose pyrophosphorylase | Unknown* | 0.45 | 0.0006 | 0.09 | 0.0010 | 0.44 | 0.0036 |
| At4g33040 | Glutaredoxin family protein | Unknown* | 0.46 | 0.0011 | 0.17 | 0.0011 | 0.42 | 0.0140 |
| At5g55450 | Protease inhibitor | Endomembrane system | 0.46 | 0.0105 | 0.14 | 0.0018 | 0.26 | 0.0106 |
| At5g18130 | Expressed protein | Unknown* | 0.47 | 0.0032 | 0.22 | 0.0019 | 0.43 | 0.0075 |
| At2g39330 | Jacalin lectin family protein | Unknown* | 0.49 | 0.0183 | 0.28 | 0.0049 | 0.25 | 0.0003 |
| At1g29395 | Putative stress-responsive protein | Chloroplast | 0.49 | 0.0019 | 0.14 | 0.0003 | 0.33 | 0.0002 |
| At2g19810 | Zinc finger (CCCH-type) protein | Nucleus | 0.49 | 0.0000 | 0.43 | 0.0013 | 0.45 | 0.0145 |
| At3g15650 | Phospholipase/carboxylesterase family protein | Mitochondrion | 0.50 | 0.0129 | 0.19 | 0.0017 | 0.37 | 0.0055 |
| At2g41410 | Putative calmodulin | Plasma membrane | 0.50 | 0.0047 | 0.27 | 0.0001 | 0.48 | 0.0036 |
| Upregulated | ||||||||
| At3g09580 | Putative oxidoreductase | Chloroplast | 2.02 | 0.0001 | 4.01 | 0.0040 | 2.43 | 0.0008 |
| At5g54180 | Unknown protein | Chloroplast | 2.05 | 0.0007 | 2.61 | 0.0094 | 2.23 | 0.0007 |
| At5g40890 | Anion channel protein | Unknown* | 2.07 | 0.0005 | 5.34 | 0.0002 | 2.19 | 0.0021 |
| At5g24420 | 6-Phospho-gluconolactonase-like protein | Unknown* | 2.08 | 0.0053 | 2.29 | 0.0016 | 3.11 | 0.0006 |
| At5g20250 | Imbitition protein Sip1 | Chloroplast | 2.19 | 0.0179 | 12.88 | 0.0006 | 3.36 | 0.0006 |
| At4g12600 | Ribosomal protein L7Ae-like | Cytosol | 2.20 | 0.0075 | 2.43 | 0.0154 | 2.08 | 0.0162 |
| At3g62950 | Glutaredoxin-C11 | Endomembrane system | 2.23 | 0.0121 | 15.20 | 0.0000 | 6.09 | 0.0006 |
| At3g17170 | Hypothetical protein | Chloroplast* | 2.23 | 0.0009 | 2.12 | 0.0125 | 2.19 | 0.0020 |
| At5g49360 | Xylosidase | Extracellular matrix | 2.28 | 0.0107 | 14.97 | 0.0002 | 2.18 | 0.0245 |
| At1g33340 | Unknown protein | Unknown* | 2.37 | 0.0009 | 2.66 | 0.0013 | 2.43 | 0.0012 |
| At4g04330 | Hypothetical protein | Chloroplast* | 2.39 | 0.0007 | 4.25 | 0.0007 | 2.43 | 0.0011 |
| At4g30610 | Ser carboxypeptidase II-like | Extracellular space | 2.41 | 0.0002 | 2.85 | 0.0071 | 2.82 | 0.0070 |
| At2g40400 | Unknown protein | Chloroplast | 2.50 | 0.0110 | 2.78 | 0.0174 | 3.72 | 0.0003 |
| At1g56050 | GTP binding protein | Chloroplast | 2.62 | 0.0021 | 2.33 | 0.0078 | 2.03 | 0.0058 |
| At5g08610 | RNA helicase-like protein | Chloroplast | 3.02 | 0.0014 | 2.18 | 0.0076 | 2.92 | 0.0001 |
| At1g69200 | pfkB-type carbohydrate kinase family protein | Chloroplast | 3.07 | 0.0001 | 2.13 | 0.0081 | 2.55 | 0.0008 |
| At5g17300 | Unknown protein | Nucleus | 3.14 | 0.0003 | 52.32 | 0.0008 | 2.67 | 0.0021 |
| At3g53460 | RNA binding protein CP29 | Nucleus | 3.16 | 0.0034 | 8.39 | 0.0005 | 2.66 | 0.0020 |
| At3g03630 | O-acetylserine (thiol) lyase | Chloroplast | 3.30 | 0.0009 | 2.71 | 0.0023 | 3.13 | 0.0004 |
| At3g54090 | pfkB-type carbohydrate kinase family protein | Chloroplast | 3.53 | 0.0003 | 2.52 | 0.0084 | 2.53 | 0.0002 |
| At2g44040 | Unknown protein | Chloroplast | 4.02 | 0.0018 | 3.06 | 0.0140 | 2.45 | 0.0055 |
| At5g58310 | Polyneuridine aldehyde esterase-like | Unknown* | 4.66 | 0.0019 | 8.39 | 0.0027 | 3.17 | 0.0007 |
The gene names are according to the Arabidopsis Genome Initiative. FC, the actual fold change in relative expression relative to the wild type. The subcellular location was identified based either on the information available at The Arabidopsis Information Resource database (www.Arabidopsis.org) or by TargetP prediction (asterisks) (www.cbs.dtu.dk/services/TargetP) (Emanuelsson et al., 2000). The expression of the underlined genes was found to be differentially regulated when comparing Col-0 and Ws-2 transcript profiles.
Comparison of the transcript profiles revealed that the transcript levels of 216 of the 304 (71%) differentially regulated genes in stn7-1 were also altered relative to the wild type (Col-0) in psad1-1 leaves, in which a total of 1643 genes showed differential regulation relative to the wild type (Col-0) (Figures 8A, 8D, and 8G). Interestingly, the change in expression of almost all 216 members of this common set showed the same trend in the two mutants, as expected if a common signal transduction pathway is perturbed in both (Figures 8A and 8G). Whereas genes for proteins with unknown function represented the largest group within this 216-gene set, genes coding for proteins involved in gene expression (transcription factors, RNA binding proteins, and ribosomal subunits), signal transduction (two-component responsive regulators and protein kinases), stress responses, and metabolism were also included (see Supplemental Tables 3 and 4 online).
The total number of differentially regulated genes (relative to the wild type [Col-0]) observed in the double mutant stn7-1 psad1-1 was 1470 (Figure 8D). Of these, 1152 also showed changes in expression in psad1-1 plants. Among them, a large fraction of gene products was involved in stress responses, including heat shock proteins, disease resistance protein families, senescence-associated proteins, glutaredoxins, and peroxiredoxins (see Supplemental Tables 4 and 5 online). Additionally, 19 transcription factors belonging to the MYB and AP2 protein families and involved in stress responses were also differentially regulated in both genotypes (see Supplemental Tables 4 and 5 online). Only 203 differentially regulated genes were in common between stn7-1 and the stn7-1 psad1-1 double mutant, implying that the differential regulation of most of the genes in the double mutant is the consequence of the markedly impaired thylakoid electron flow (only in stn7-1 psad1-1; see Supplemental Table 2 online) rather than of suppression of the LTR (in both genotypes).
A similar behavior was observed when comparing the transcript profiles of stn7-1, psae1-3, and stn7-1 psae1-3 mutants (Figures 8B, 8E, and 8H). Sets of 213 and 639 genes were differentially regulated relative to the wild type (Ws-2) in psae1-3 and in the double mutant stn7-1 psae1-3, respectively (see Supplemental Tables 3, 6 and 7 online). Of those, 103 were differentially regulated in both mutants; almost all of them showing again the same trend in expression (Figures 8B and 8H). However, it is noteworthy that, because of the different genetic background of the mutants, the differential regulation of only 95 genes could be specifically attributed to the mutations, whereas the expression of the remaining eight genes was found to be influenced by the different ecotypes (Col-0 versus Ws-2; see Supplemental Table 9 online). Moreover, all 65 genes differentially regulated in both stn7-1 and psae1-3 also showed the same trend. In this case, the expression of 15 of the 65 genes was influenced by the ecotype genetic background. In addition, and similarly to psad1-1, the profile of the double mutant stn7-1 psae1-3 was more closely related to the transcriptional response seen in psae1-3 than to the one observed in stn7-1.
The comparison of the stn7-1, psad1-1, and psae1-3 transcript profiles allowed the identification of 56 genes (13 of which were already differentially regulated when comparing Col-0 and Ws-2 wild-type plants) that were commonly regulated in all three genotypes and showed the same trend in expression, thus representing either tentative nuclear target genes of the LTR signaling pathway or genes regulated to compensate for the lack of LTR signaling under greenhouse conditions (Figures 8C, 8F, and 8I). Interestingly, 12 of the 22 upregulated genes encode proteins targeted to the chloroplast, whereas only 5 of the 34 downregulated genes encode chloroplast proteins. Only two tentative transcription factor genes were found in this gene set, while genes coding for proteins involved in posttranscriptional, translational, or posttranslational regulatory processes (such as RNA binding proteins, RNA helicases, ribosomal proteins, zinc finger proteins, proteases, or glutaredoxins) were highly represented (Table 3). Additionally, besides a few genes coding for photosynthesis-related proteins, such as AGPase (catalyzing the first committed step in starch biosynthesis), several genes for proteins with metabolic functions (for instance, 6-phosphogluconolactonase, pfkB-type carbohydrate kinase family protein, 3-phosphoglycerate dehydrogenase, UDP-glucose:flavonoid 3-O-glucosyltransferase, and dihydroflavonol 4-reductase) could be found in this gene set (Table 3), supporting the view that compensatory effects of the suppressed LTR on nuclear gene expression were monitored in our assay. Considering the protein (Figure 6) and transcript profiling (Figures 6 and 8) data together, we concluded that (1) both the impairment in thylakoid electron flow and the defect in the LTR affect the accumulation of photosynthesis-related proteins in greenhouse-grown plants, and (2) this effect on protein accumulation occurs via posttranscriptional mechanisms.
State Transitions Become Critical for Plant Performance When Linear Electron Flow Is Perturbed
Even under fluctuating light (Bellafiore et al., 2005) or field conditions (Frenkel et al., 2007), only subtle impairments in plant development and fitness have been reported for the stn7-1 mutant, which is affected in both state transitions and the LTR (Bonardi et al., 2005). Moreover, plants specifically impaired in state transitions seem to be capable of compensating to a large degree for their defect (Lunde et al., 2000, 2003). This raises two questions: (1) to what extent do state transitions and the LTR contribute to the conditional phenotype observed in the stn7 mutant, and (2) do state transitions come into play only under extreme light conditions? To clarify whether state transitions and/or the LTR contribute significantly to acclimating photosynthetic performance to perturbations in photosynthetic electron flow, we introduced mutations that affect linear electron flow and are associated with an increased pool of reduced PQ (pete2-1.1, psad1-1, and psae1-3) (Weigel et al., 2003; Ihnatowicz et al., 2004; Ihnatowicz et al., 2007) into the stn7-1 background. In all cases, the three double mutants exhibited a marked decrease in growth rate relative to the parental single mutants (Figures 9A and 9B). With the exception of the maximum quantum yield of PSII (Fv/Fm), which was unchanged (see Supplemental Table 2 online), the exacerbated phenotypes were also evident in the photosynthetic performance of the double mutants: a marked drop in the effective quantum yield of PSII (ΦII) relative to the parental lines was noted, together with an increase in the reduction state of the PQ pool (1-qP) (Figure 9B; see Supplemental Table 2 online). In addition, biochemical analyses indicated that the total abundance of PSI complexes was unchanged in psad1-1, stn7-1 psad1-1, and psal-1 psad1-1 leaves (Figure 4; see Supplemental Figure 2 online), whereas the PSI-LHCI-LHCII complex disappeared in the double mutant genotypes (Figures 3 and 4; see Supplemental Figure 2 online). Identical results were obtained for the psae1-3, stn7-1 psae1-3, and psal-1 psae1-3 plants (Figures 3 and 4; see Supplemental Figure 2 online).
Figure 9.
Exacerbated Phenotypes of Double Mutants with Defects in State Transitions (stn7-1 or psal-1) and in Linear Electron Flow (pete2-1, psad1-1.1, or psae1-3).
As a negative control, the stn8-1 single mutant and the corresponding double mutants are shown.
(A) Four-week-old plants grown in a growth chamber under long-day conditions.
(B) Average leaf areas of 20 plants for each genotype.
(C) Effective quantum yield (ΦII) for each genotype.
(D) 1-qP, the fraction of QA (the primary electron acceptor of PSII) present in the reduced state for each genotype. Detailed values are given in Supplemental Table 2 online.
To exclude a contribution of the LTR to the genetic interaction of stn7-1 with mutations affecting linear electron flow, the stn7-1 mutation was replaced by the psal-1 mutation, which affects state transitions (Lunde et al., 2000, 2003) (see Figures 1A and 1B) but not the LTR (see Figures 5A and 5B). Interestingly, the psal-1 pete2-1.1, psal-1 psad1-1, and psal-1 psae1-3 double mutants resulted in phenotypes that were similar, although more subtly altered, to those described for the double mutant lines carrying the stn7-1 lesion (Figure 9; see Supplemental Table 2 online), although the psal-1 mutation, unlike stn7-1, altered the polypeptide composition of PSI (Figure 10). As a negative control, double mutants resulting from the combination of each of the linear electron flow mutants with stn8-1 (ecotype Col-0), which is defective in PSII core phosphorylation (Bonardi et al., 2005) (see Table 1), were generated, but these did not show exacerbated phenotypes (Figure 9; see Supplemental Table 2 online). As expected, in these lines, the PSI-LHCI-LHCII complex was retained (Figure 3).
Figure 10.
PSI Polypeptide Composition Analysis of the Wild Type (Col-0) and Single and Double Mutants.
SDS-PAGE of PSI complexes isolated from wild-type (Col-0) and mutant leaves (as indicated above the gel) grown under greenhouse conditions.
Further spectroscopic measurements were performed to confirm that the psal-1 and stn7-1 mutations do not destabilize the PSI complexes when introduced into either a pete2-1.1, psad1-1, or psae1-3 background. Pm, the maximum absorption value of P700+ upon transition from the fully reduced to the fully oxidized state, indeed showed no change in the double mutants relative to the values observed in the parental single mutants (pete2-1.1, psad1-1, or psae1-3) (Table 4). As expected from the marked drop in the effective quantum yield of PSII and the increase in the reduction state of the PQ pool of the double mutants (see above), the photochemical quantum yield of PSI, Y(I), was found to be markedly decreased relative to that in the parental single mutants (Table 4). This effect on Y(I) can be attributed to an increased availability of electrons at the PSI donor side, measured as Y(ND), which represents the fraction of total P700 that is oxidized in a given state (see Methods and Table 4), thus leading to the enhancement of the acceptor side limitations, which was measured as Y(NA), the fraction of total P700 that is not oxidized by a saturation pulse in a given state (see Methods and Table 4). In other words, the disappearance of the PSI-LHCI-LHCII complex clearly favors PSII photochemistry to the detriment of PSI activity, causing the overreduction of the electron transport chain.
Table 4.
Parameters of PSI Activity Obtained from P700+ Absorption Measurements on Mutant and Wild-Type (Col-0 and Ws-2) Plants
| Pm | Y(I) | Y(ND) | Y(NA) | |
|---|---|---|---|---|
| Col-0 | 0.27 ± 0.03 | 0.67 ± 0.05 | 0.22 ± 0.03 | 0.11 ± 0.01 |
| Ws-2 | 0.29 ± 0.03 | 0.68 ± 0.06 | 0.20 ± 0.02 | 0.12 ± 0.01 |
| stn7-1 | 0.25 ± 0.03 | 0.66 ± 0.04 | 0.18 ± 0.02 | 0.16 ± 0.02 |
| stn8-1 | 0.27 ± 0.01 | 0.67 ± 0.03 | 0.21 ± 0.01 | 0,12 ± 0.01 |
| psal-1 | 0.25 ± 0.02 | 0.62 ± 0.04 | 0.20 ± 0.02 | 0.18 ± 0.02 |
| pete2-1.1 | 0.25 ± 0.01 | 0.59 ± 0.02 | 0.33 ± 0.01 | 0.08 ± 0.01 |
| psae1-3 | 0.12 ± 0.02 | 0.54 ± 0.03 | 0.07 ± 0,01 | 0.39 ± 0.03 |
| psad1-1 | 0.11 ± 0.01 | 0.51 ± 0.03 | 0.08 ± 0.01 | 0.41 ± 0.03 |
| stn7-1 pete2-1.1 | 0.25 ± 0.03 | 0.53 ± 0.02 | 0.24 ± 0.02 | 0.23 ± 0.01 |
| stn7 psad1-1 | 0.11 ± 0.00 | 0.42 ± 0.03 | 0.04 ± 0.00 | 0.54 ± 0.04 |
| stn7 psae1-3 | 0.12 ± 0.01 | 0.45 ± 0.02 | 0.04 ± 0.00 | 0.51 ± 0.03 |
| psal-1 pete2-1 | 0.26 ± 0.01 | 0.55 ± 0.01 | 0.27 ± 0.00 | 0.18 ± 0.01 |
| psal-1 psad1-1 | 0.10 ± 0.01 | 0.46 ± 0.02 | 0.06 ± 0.01 | 0.48 ± 0.02 |
| psal-1 psae1-3 | 0.13 ± 0.01 | 0.48 ± 0.05 | 0.06 ± 0.00 | 0.46 ± 0.04 |
| stn8 pete2-1 | 0.24 ± 0.00 | 0.58 ± 0.01 | 0.34 ± 0.01 | 0.08 ± 0.01 |
| stn8 psad1-1 | 0.12 ± 0.01 | 0.52 ± 0.03 | 0.08 ± 0.01 | 0.40 ± 0.01 |
| stn8 psae1-3 | 0.12 ± 0.00 | 0.55 ± 0.03 | 0.09 ± 0,01 | 0.36 ± 0.01 |
Measurements were performed as described in Methods. Mean values for five plants (±sd) are shown. Pm, the maximal change of the P700 signal upon quantitative transformation of P700 from the fully reduced to the fully oxidized state; Y(I), the photochemical quantum yield of PSI; Y(ND), the fraction of overall P700 that is oxidized in a given state (provides a measure of PSI donor side limitation); Y(NA), the fraction of overall P700 that cannot be oxidized by a saturation pulse in a given state (provides a measure of PSI acceptor side limitation). For further details, see Methods.
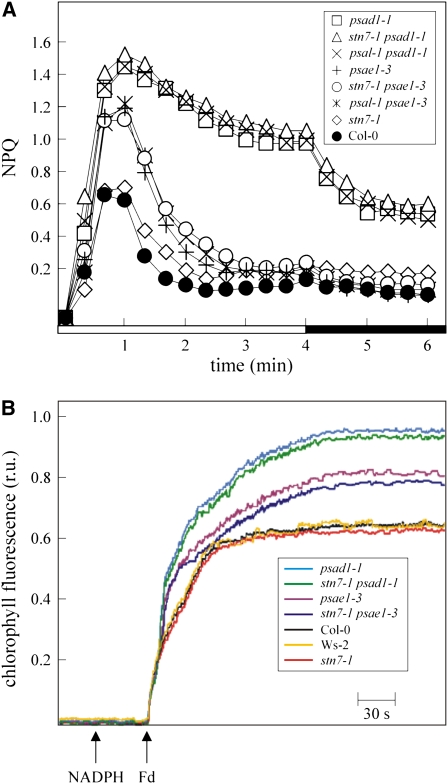
Measurements of nonphotochemical quenching (NPQ) during the dark-to-light transition were performed to monitor the efficiency of cyclic electron flow (Munekage et al., 2002; DalCorso et al., 2008). Identical NPQ inductions occurred in wild-type and stn7-1 plants. Similar NPQ induction kinetics were also observed in psae1-3, psal-1 psae1-3, and stn7-1 psae1-3 plants on the one hand and in psad1-1, psal-1 psad1-1, and stn7-1 psad1-1 plants on the other (Figure 11A). Comparable results were also obtained when cyclic electron transport was measured in ruptured chloroplasts by monitoring the increase in chlorophyll fluorescence under white low light, after addition of NADPH and ferredoxin (Figure 11B), thus implying that, in Arabidopsis, state transitions and cyclic electron transport act independently.
Figure 11.
Measurements of Cyclic Electron Flow.
(A) Time course of induction and relaxation of NPQ monitored during dark-to-light (80 μmol m−2 s−1, white bar) transition in Col-0 and mutant plants. The 4-min light period (white bar) was followed by a 2-min dark period (black bar). Note that NPQ induction during the activation period of photosynthesis is thought to be caused by the transient acidification of the thylakoid lumen when cyclic electron flow activity is higher than the activity of the ATP synthase. Ws-2 plants showed identical NPQ induction kinetics with respect to Col-0.
(B) Quantification of cyclic electron flow in situ. Increases in chlorophyll fluorescence were measured in ruptured chloroplasts under low measuring light (1 μmol m−2 s−1), after the addition of NADPH and ferredoxin (Fd). At these light intensities, the fluorescence level should predominantly reflect the reduction of PQ by cyclic electron transport from ferredoxin, not by PSII photochemistry (Munekage et al., 2002). Average values of five different plants for each genotype are shown. Standard deviation was below 5%.
DISCUSSION
Photosynthetic organisms have developed a variety of molecular mechanisms that serve to acclimate photosynthesis to environmental fluctuations, in particular to changes in the quality or intensity of incident light. Cyanobacteria and unicellular algae, with their short generation times, use mechanisms such as state transitions (Wollman, 2001; Finazzi, 2005; Mullineaux and Emlyn-Jones, 2005; Rochaix, 2007; Eberhard et al., 2008) to respond very dynamically to rapid changes in their environment so as to ensure continuing growth and cell division. Plants exhibit a higher tolerance to rapid environmental changes by virtue of their larger size, which allows for storage of nutrients and energy. However, owing to their sessile lifestyle, plants have evolved long-term acclimatory responses to cope with the environmental rhythms of their habitats to achieve optimal rates of development and reproduction. On the basis of such considerations, it has been speculated that, although the thylakoid protein kinase STN7 functions both in state transitions and the LTR, its major function is to mediate long-term acclimation (Bonardi et al., 2005). Reversible LHCII phosphorylation and state transitions in land plants might then represent an evolutionary relic that plays only a minor role in balancing excitation energy (Finazzi, 2005) and becomes crucial only under special conditions (Tikkanen et al., 2006).
State Transitions Play an Important Role in Flowering Plants
Our data indicate that state transitions play an important role in the photosynthetic acclimation of land plants. In particular, the exacerbated phenotypes of the double mutants psal-1 psad1-1, psal-1 psae1-3, stn7-1 psad1-1, and stn7-1 psae1-3 appear to be the result of a marked increase in the reduction of QA (i.e., the PQ pool; see Supplemental Table 2 online). Such impairments in linear electron flow cannot be attributed to the destabilization of PSI, as indicated by the fact that PSI complexes from stn7-1 psad1-1 and stn7-1 psae1-3 are present in identical amounts and polypeptide composition as in psad1-1 and psae1-3 single mutants (Table 4, Figures 4 and 10; see Supplemental Figure 2 online). Additionally, although the psal-1 mutation alters the PSI polypeptide composition in psal-1 psad1-1 and psal-1 psae1-3 double mutants (Figure 10), their growth rates and phenotypic performance are less altered than in stn7-1 psad-1 and stn7-1 psae1-3 plants. This is most likely a consequence of the residual state transitions activity observed in the psal-1 genetic background (Figure 1; Lunde et al., 2000), together with the fact that psal-1 plants have been reported to compensate for their less efficient PSI by increasing the PSI/PSII ratio, thus preventing, at least in part, the overreduction of the PQ pool (Naver et al., 1999).
Taken together, the data indicate that the PSI-LHCI-LHCII complex plays a crucial role in assisting PSI activity in psad1-1 and psae1-3 mutants, thus meliorating the thylakoid electron flow. Similar, although more subtle, phenotypes were also observed in psal-1 pete2-1.1 and stn7-1 pete2-1.1 double mutants, implying that the PSI-LHCI-LHCII complex plays an important role in balancing excitation energy distribution between photosystems. Mass spectrometry analysis on the PSI-LHCI-LHCII pigment-protein complex (see Supplemental Table 1 online) isolated from wild-type plants did not reveal the presence of two minor monomeric LHCII proteins (CP26 and CP29), unlike the situation reported in C. reinhardtii (Takahashi et al., 2006). However, the association of those minor antenna proteins with the PSI-LHCI-LHCII pigment-protein complex might explain the presence of a larger PSI-LHCI-LHCII complex in the psad1-1 and psae1-3 mutants (see the double PSI-LHCI-LHCII bands in psad1-1 and psae1-3; Figures 3 and 4; see Supplemental Figure 2 online) possibly as a consequence of the increased thylakoid phosphorylation levels in those mutants. Interestingly, and in contrast with the situation in C. reinhardtii (Finazzi et al., 2002), impairment of the ability to undergo state transitions did not affect cyclic electron flow in Arabidopsis, indicating that in higher plants the induction of state 2 is not responsible to promote the switch from linear to cyclic electron transport.
State Transitions and LTR Share the STN7 Kinase
The regulatory coupling of state transitions to the LTR (Allen and Pfannschmidt, 2000; Pursiheimo et al., 2001) and the dependence of both processes on STN7 activity are compatible with the view that the signal pathways leading to state transitions and LTR represent a hierarchically organized signaling cascade (Fey et al., 2005a), where changes in PQ redox state trigger first state transitions and then the LTR via a STN7-dependent phosphorylation cascade (and/or reversible relocation of protein complexes). Indeed, the inability of psad1-1 and psae1-3 to adapt to changes in light conditions appears to be the direct consequence of the permanent reduction of the PQ pool, independently from the light conditions. However, neither LHCII phosphorylation nor the thylakoid conformational changes associated to state transitions appear to play any role in LTR (Figure 5). In fact, plants either devoid of the LHCII phosphorylatable fraction (asLHCB2.1 mutant) or altered in the PSI docking site for LHCII (psal-1 mutant) did not show any impairment in the LTR. In conclusion, changes in the PQ pool redox state appear to be required for the reversible STN7-dependent phosphorylation of a specific thylakoid-associated protein, which then triggers signaling events leading to the LTR. The thylakoid phosphoprotein TSP9, thought to dissociate partially from the thylakoid membrane upon phosphorylation (Carlberg et al., 2003), is an obvious candidate for such a signaling component (Zer and Ohad, 2003). However, TSP9 RNAi lines exhibit a normal LTR (Figure 5), indicating that an as yet unknown protein in/on the thylakoid membrane, or temporarily associated with it, might be involved in the LTR signaling cascade. Recently, a transcriptomic analysis suggested a possible involvement of TSP9 in plant acclimation to high light conditions (Fristedt et al., 2009), thus supporting the notion that Arabidopsis displays separate responses to low- (LTR) and high-light intensities (Bailey et al., 2001).
The LTR Process Influences the Expression of a Distinct Group of Chloroplast and Nuclear Genes
A comprehensive picture of modulations of Arabidopsis nuclear gene expression by LTR under greenhouse conditions was obtained by comparing the transcriptome profiles of wild-type (Col-0 and Ws-2 ecotypes) and mutant plants impaired in the LTR process, such as stn7-1, psad1-1, psae1-3, stn7-1 psad1-1, and stn7-1 psae1-3 The different ecotypes analyzed did not appear to markedly influence the LTR process, as indicated by the identical behavior of Col-0 and Ws-2 under the different light treatments, as well as by the very similar effects of the psad-1 (Col-0) and psae1-3 (Ws-2) mutations on the LTR.
Despite the large number of misregulated genes in the different mutants, pairwise comparison of nuclear transcript profiles allowed us to identify a limited set of 56 nuclear genes coregulated in stn7-1, psad1-1, and psae1-3 mutant plants (the expression of 13 of them was also differentially regulated between the Col-0 and Ws-2 ecotypes; see Table 3 and Figure 8). These 56 nuclear genes possibly represent either nuclear target genes of LTR or genes differentially expressed to compensate for the effects of a suppressed LTR under greenhouse conditions. Interestingly, genes involved in different processes, including stress responses, posttranscriptional, translational, and posttranslational gene expression regulation, as well as metabolism, could be identified within this set; however, only few of them appeared to be directly involved in the photosynthetic process. Moreover, none of the 56 LTR-dependent genes were found to be differentially regulated in the tsp9 mutant (Fristedt et al., 2009), thus supporting the notion that TSP9 is not involved in LTR. Additionally, in agreement with our data, two previous independent transcriptomic studies have shown that the stn7 mutant does not display major differences in photosynthetic transcript accumulation when compared with the wild type (Bonardi et al., 2005; Tikkanen et al., 2006).
Furthermore, a detailed transcriptomic analysis combining light shift experiments with application of a thylakoid electron transport inhibitor (Fey et al., 2005b) showed that 286 nuclear genes encoding for plastid proteins were regulated directly by the redox state of the photosynthetic electron transport chain, and only few of them were directly involved in photosynthesis. The effects of the PQ pool redox state on the regulation of nuclear photosynthesis-related genes were also investigated in barley (Hordeum vulgare) by comparing the transcription and accumulation of light-harvesting complexes of the wild type and the mutant viridis zb63, which is depleted of PSI and exhibits a constitutively reduced PQ pool (Frigerio et al., 2007). Indeed, although the antenna size was strongly reduced in viridis zb63, the transcript levels of nuclear photosynthesis-related genes were unaffected. However, a more detailed study in Arabidopsis, extending the approach chosen by Fey et al. (2005b), indicates that, at least under light (shift) conditions that specifically excite PSI or PSII, nuclear photosynthesis genes are also LTR-dependently regulated at the transcript level (T. Pfannschmidt, unpublished data).
In contrast with the LTR-dependent gene regulation in the nucleus, expression analysis of the plastid genes psaAB and psbA encoding the PSI and PSII reaction center subunits, respectively, revealed that the psaAB operon responds robustly to changes in the PQ redox state (Figure 6A). Similar results have been obtained in different plant species: in Arabidopsis, the redox-dependent chloroplast transcriptional regulation appears to be limited to the psaAB operon, whereas in mustard (Sinapis alba), transcriptional regulation is also extended to the psbA gene (Pfannschmidt et al., 1999). Immunoblot analysis confirmed the increased accumulation of the products of psaAB (P700) under PSII light condition and following the shift from PSI to PSII light (Figure 6B). Interestingly, the PSI antenna subunit LHCA3 also showed higher protein levels under PSII light and following the PSI-to-PSII light shift, although no differential LHCA3 transcript accumulation was observed under the same light conditions, again supporting the view that LTR-dependent regulation of nuclear photosynthesis-related gene expression occurs after transcription.
Conclusions
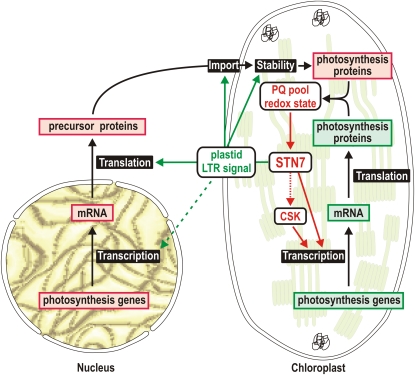
In this work, we have provided data that allow us to conclude that state transitions play an important role in flowering plants. The photosynthetic impairments observed in psal-1 psae1-3, stn7-1 psae1-3, psal-1 psad1-1, and stn7-1 psad1-1 double mutants have allowed us to attribute a physiological relevance to the PSI-LHCI-LHCII pigment-protein complex during state 2. Additionally, we have clarified that the STN7 kinase is the only enzyme known so far that is common to both state transitions and LTR (Figure 12). Indeed, our mutant analyses allowed to us exclude the participation of any other state transitions step in the LTR. Changes in the redox state of the PQ pool are essential to modulate the STN7 activity and are required to trigger both short- and long-term responses. It seems likely that the STN7 kinase is positioned at the beginning of a phosphorylation cascade that communicates the photosynthetic needs to the chloroplast and nuclear genomes, resulting in the expression regulation of a distinct set of genes. Intriguingly, in Arabidopsis, the LTR-associated regulation of thylakoid composition appears to operate at the transcriptional and posttranscriptional level in the two compartments (Figure 12): for instance, of the genes encoding subunits of PSI and its antenna complex, the plastid-located psaAB operon is regulated at the transcriptional level, whereas accumulation of the nucleus-encoded LHCA3 protein is subject to a posttranscriptional regulatory process (see Figure 6).
Figure 12.
A Model Illustrating the Possible Role of STN7 in LTR Signaling.
The LTR signaling pathway is triggered by the STN7 kinase, the activity of which is modulated by the redox state of the PQ pool. The signaling pathway can be divided into two main branches. One branch concerns chloroplast gene expression (chloroplast branch) and is responsible for the transcriptional regulation of PSI-related genes, such as the psaAB operon. The chloroplast sensor kinase (CSK) (Puthiyaveetil et al., 2008), reported to regulate the transcription of psaAB operon, might be a component of the chloroplast branch. A second branch might be responsible for the regulation of the accumulation of products of nuclear photosynthesis-related genes (nucleus-cytosol branch). These regulatory processes seem to occur mainly after transcription and might take place at the level of protein translation or degradation, or protein import efficiency and specificity. Note that the differential expression of genes for metabolism-related proteins could also represent a compensatory reaction in response to suppressed LTR signaling under greenhouse conditions. In this context, it cannot be excluded that nuclear photosynthesis genes are initially regulated at the transcript level upon changes in light quality (dotted line).
Currently, the identity of other components of the STN7-dependent signaling cascade is unknown. Our analyses allowed to exclude the TSP9 protein as a possible component of the LTR signaling pathway. A possible candidate might be the chloroplast sensor kinase, shown to couple the redox state of the PQ pool with the expression of chloroplast photosynthesis-related genes (Puthiyaveetil et al., 2008; Figure 12). In the future, phosphoproteomic studies of chloroplast proteins, comparing wild-type and stn7-1 mutant plants, exposed to light conditions favoring PSI or PSII complexes, appears to be the strategy of choice to identify as yet unknown proteins that, upon phosphorylation, might relay the signal to other regions of the chloroplast and to the nucleus.
METHODS
Plant Material, Propagation, and Growth Measurement
The Arabidopsis thaliana psal-1 (SALK_000637) mutant was identified in the SIGnAL database (Alonso et al., 2003) (see Supplemental Figure 1 online). Lines lacking TSP9 were generated by RNAi (see below). The origins of all other lines are described in Table 1. All plants were grown as reported previously (Bonardi et al., 2005), and plant growth was quantified as before (Leister et al., 1999).
Generation of the TSP9 RNAi Construct and Transformation of Arabidopsis
To silence the TSP9 gene by RNAi, a 287-bp fragment spanning the exon from position 114 in the 5′-untranslated region to position +97 bp was amplified using the primers TSP9_RNAi_s (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCTTGGGTTTATGACAAAACAAG-3′) and TSP9_RNAi_as (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTTAAGAAAGAGTAGCTTATATATAAGT-3′). The PCR product was purified, and BP and LR Clonase reactions (GATEWAY Cloning; Invitrogen) were performed according to the manufacturer's instructions. The final construct (pJAWOHL8-TSP9i; pJawohl8-RNAi: EMBO database ID AF408413) contained an intron between the inverted repeat of part of the TSP9 gene. Expression of the RNAi cassette was driven by the cauliflower mosaic virus 35S promoter. pJAWOHL8-TSP9i was transferred into Agrobacterium tumefaciens, and Arabidopsis (ecotype Col-0) plants were transformed by the floral dip method (Clough and Bent, 1998). T1 seeds were sown on soil, sprayed with 0.0287% (v/v) BASTA (Bayer Crop Science) after 10 and 15 d, and grown under long-day conditions (16 h light/8 h dark, 23°C). T2 and T3 seedlings were tested for TSP9 silencing using real-time PCR experiments with primers TSP91_rt_s (5′-GGTTTCTTCGCT TCTTATGTC-3′) and TSP9i_rt_as (5′-GGTGGTGGTGCCTTCTTTC-3′). Two independent lines in which the TSP9 transcript levels were reduced to 10% of the wild-type level were used for subsequent experiments.
Chlorophyll Fluorescence Measurements
Five plants of each genotype were analyzed, and average values and standard deviations were calculated. In vivo chlorophyll a fluorescence of single leaves was measured using the Dual-PAM 100 (Walz). Pulses (0.5 s) of red light (5000 μmol m−2 s−1) were used to determine the maximum fluorescence and the ratio (Fm − F0)/Fm = Fv/Fm. A 15-min exposure to red light (37 μmol m−2 s−1) was used to drive electron transport before measuring the effective quantum yield of PSII, ΦII (Genty et al., 1989), and 1-qP. Quenching of chlorophyll fluorescence due to state transitions (qT) was determined by illuminating dark-adapted leaves with blue light (35 μmol m−2 s−1, 20 min) and then measuring the maximum fluorescence in state 2 (Fm2). Next, state 1 was induced by superimposing far-red light (255 μmol m−2 s−1, 20 min) and recording Fm1. qT was calculated as described (Ihnatowicz et al., 2004). Electron transport rate (JF parameter) was estimated as previously described (Krall and Edwards, 1992).
For 77 K fluorescence emission spectroscopy, the fluorescence spectra of thylakoids were recorded after illumination of plants with low (80 μmol m−2 s−1) and high white light (800 μmol m−2 s−1) for 3 h as described (Tikkanen et al., 2006). Thylakoids were isolated in the presence of 10 mM NaF, and 77 K fluorescence spectra were obtained by excitation at 475 nm using a Spex Fluorolog mod.1 fluorometer (Spex Industries). The emission between 600 and 800 nm was recorded, spectra were normalized at 685 nm, and the fluorescence emission ratio F733/F685 (or F730/F685 in the case of psad1-1 and psae1-3) was calculated as an indication of energy distribution between PSI and PSII.
PSI activity was monitored by far-red preillumination of leaves and application of a saturation pulse (5000 μmol m−2 s−1) of red light to determine Pm, the maximum P700+ absorption. Actinic light (80 μmol m−2 s−1) was subsequently switched on, and a saturation pulse of red light (5000 μmol m−2 s−1) was applied to determine Pm′, the maximum P700+ absorption under illumination (Klughammer and Schreiber, 1994). The photochemical quantum yield of PSI, Y(I), defined as the fraction of total P700 that is reduced in a given state and is not limited by the acceptor side, was calculated as: Y(I) = 1 − Y(ND) − Y(NA), where Y(ND) represents the fraction of total P700 that is oxidized in a given state (as a measure of donor side limitation), and Y(NA) is the fraction of total P700 that is not oxidized by a saturation pulse in a given state, calculated as (Pm − Pm')/Pm, which provides a measure of acceptor side limitation (Klughammer and Schreiber, 1994).
Photosynthetic Acclimation Analysis
The development of the LTR was monitored on the basis of measurements of steady state fluorescence (Fs) and chlorophyll a/b ratios as described before (Bonardi et al., 2005; Fey et al., 2005b; Wagner et al., 2008). In detail, after growth for 10 d under white light, plants were acclimated to PSI (40-W incandescent light bulbs filtered through a red filter giving 50% transmittance at 650 nm; Lee Filters, 027 Medium Red) or PSII light (30-W cool-white fluorescent strip lamps filtered through an orange-yellow filter giving 50% transmittance at 560 nm; Strand Lightning Filters, 405 Orange) for 6 d (PSI or PSII plants) or they were first acclimated to one light source for 2 d followed by 4 d under the respective other light source (PSI-II or PSII-I plants). Chlorophyll fluorescence determination was performed using the PAM 101 device. Plants dark-adapted for 15 min were used to determine the minimal (F0) and maximal (Fm) chlorophyll fluorescence. Illumination with either PSI or PSII actinic light sources were used to determine the steady state fluorescence Ft, whereas F′0 (minimal fluorescence of light adapted plants) was determined after incubation with far-red light. The steady state fluorescence Fs was calculated as Fs = Ft − F′0. Chlorophyll a/b ratios were determined by harvesting plants under the respective growth light and determining chlorophyll concentrations after grinding the material in liquid nitrogen and extracting the pigments with 80% buffered acetone. Chlorophyll a and b concentrations were calculated according to Porra et al. (1989).
BN- and 2D-PAGE
Leaves were harvested from plants at the eight-leaf rosette stage, and thylakoids were prepared as described (Bassi et al., 1985) in the presence of 10 mM NaF. For BN-PAGE, thylakoid samples equivalent to 100 μg of chlorophyll were solubilized in solubilization buffer (750 mM 6-aminocaproic acid, 5 mM EDTA, pH 7, and 50 mM NaCl) in the presence of 1.5% digitonin (Serva) for 1 h at 4°C under gentle agitation. Following centrifugation (1 h, 21,000g), the solubilized material was fractionated using a nondenaturing BN-PAGE at 4°C as described (Schägger and von Jagow, 1987). For 2D-PAGE, samples were subsequently fractionated by denaturing SDS-PAGE as described (Pesaresi et al., 2001).
Immunoblot Analysis and PSI Complex Isolation
Leaves from 18-d-old plants used to monitor LTR were harvested, and thylakoids were prepared in the presence of 10 mM NaF, fractionated by SDS-PAGE, and transferred to poly(vinylidene difluoride) membranes (Bonardi et al., 2005). Membranes were then probed with antibodies specific for phosphothreonine-residues (Cell Signaling), PSI reaction center (P700), and LHCA3 and LHCB1 subunits (Agrisera). Signals were detected by enhanced chemiluminescence (Amersham Biosciences). To control for equivalent loading, replicate gels were stained with Coomassie Brilliant Blue (Roth). PSI complex isolation was performed as previously described (Ihnatowicz et al., 2007). PSI protein fractionation was performed using a 16 to 23% gradient SDS-PAGE as described by Jensen et al. (2000), and proteins were visualized by Coomassie Brilliant Blue staining.
Mass Spectrometry Analysis
Proteins were proteolytically digested with trypsin (sequencing grade; Promega). Peptides were separated using a MudPIT system according to Washburn et al. (2001), consisting of a custom-made biphasic column within a fused-silica electrospray ionization emitter tip coupled online to an LTQ XL mass spectrometer (Thermo Fisher Scientific). One full mass spectrometry scan with a scan range from 400 to 2000 was followed by tandem mass spectrometry spectra of the three most intensive ions with normalized collision energy of 35%. Dynamic exclusion was set to 30 s with a range of ±1.5 D as previously described (Lohrig et al., 2009).
cDNA Synthesis and Real-Time Analysis
Total RNA was extracted with the RNeasy plant mini kit (Qiagen) according to the manufacturer's instructions. RNA quality and concentration and the A260 to A280 ratio were assessed by agarose gel electrophoresis and spectrophotometry. cDNA was prepared from 1 μg of total RNA using the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer's instructions. cDNA was diluted 10-fold, and 3 μL of the diluted cDNA was used in a 20-μL iQ SYBR Green Supermix reaction (Bio-Rad). All reactions were performed in duplicate using at least two biological replicates. The following primers were used: 3-phosphoglycerate dehydrogenase (PGDH, At1g17745), forward primer, 5′-AGAAGCTCAGGAAGGTGTAGC-3′, reverse primer, 5′-TGAGATCGGTTCAATGATTCCC-3′; heat shock protein 70 (HSP70, At3g12580), forward primer, 5′-GATCTCGGTACAACCTACTC-3′, reverse primer, 5′-GATTAGACGCTTAGCATCGA-3′; UDP-glucose:flavonoid 3-O-glucosyltransferase (At5g54060, UF3GT), forward primer, 5′-TCCCAAGAAAGCACTAAACC-3′, reverse primer, 5′-GCAAATGTGTCAAGAAGAAAGG-3′; Glutaredoxin-C11 (At3g62950, GRXC11), forward primer, 5′-AAGACGCTATTCTACGAACTAGG -3′, reverse primer, 5′- CTGATCCGATGTACCTTCCTC-3′; histidine-containing phosphotransmitter 4 (At3g16360, AHP4), forward primer, 5′-TGACCAAGCTTTGGAAAGAG-3′, reverse primer, 5′-AAGTCCTCAAGCATCCTTCC-3′ and as a housekeeping gene ubiquitin-protein ligase-like protein (At4g36800), forward primer, 5′-CTGTTCACGGAACCCAATTC-3′, reverse primer, 5′-GGAAAAAGGTCTGACCGACA-3′; psaAB, forward primer, 5′-AACCAATTTCTAAACGCTGG-3′, reverse primer, 5′-TGATGATGTGCTATATCGGT-3′; psbA, forward primer, 5′-GTGCCATTATTCCTACTTCTG-3′, reverse primer, 5′-AGAGATTCCTAGAGGCATACC-3′; LHCA3, forward primer, 5′-GGGTTAGAGAAGGGTTTGGC-3′, reverse primer, 5′-GAGGATGGCGAGCATAGC-3′; LHCB1.1, forward primer, 5′-AGAGTCGCAGGAAATGGG-3′, reverse primer, 5′-AAGCCTCTGGGTCGGTAG-3′. Whenever possible, primers were designed to flank intron sites to make it possible to discriminate amplification of genomic DNA. Thermal cycling consisted of an initial step at 95°C for 3 min, followed by 40 cycles of 10 s at 95°C, 30 s at 55°C, and 10 s at 72°C, and after that a melting curve was performed. RT-PCR was monitored using the iQ5 Multi Color real-time PCR detection system (Bio-Rad). The adjustment of baseline and threshold was done according to the manufacturer's instructions. The relative abundance of PGDH, HSP70, UF3GT, GRXC11, and AHP4 transcripts was normalized to the constitutive expression level of ubiquitin-protein ligase-like protein mRNA. The data were analyzed using LinRegPCR (Ramakers et al., 2003) and according to Pfaffl (2001).
cRNA Synthesis and Hybridization to Affymetrix GeneChips
Five micrograms of total RNA of two to three biological replicates of the different pools (10 plants each pool) of rosette leaves of 4-week-old Col-0, Ws-2, stn7-1, psad1-1, stn7-1 psad1-1, psae1-3, and stn7-1 psae1-3 plants was processed and hybridized to a GeneChip Arabidopsis ATH1 Genome Array using the One-Cycle Target Labeling and Control Reagents according to the manufacturer's instructions (Affymetrix). In brief, reverse transcription was employed to generate first-strand cDNA. After second-strand synthesis, double-stranded cDNA was used in an in vitro transcription reaction to generate biotinylated cRNA. The fragmented, biotinylated cRNA was used for hybridization. Hybridization, washing, staining, and scanning procedures were performed as described in the Affymetrix technical manual. A Hybridization Oven 640, a Fluidics Station, and a GeneChip Scanner 3000 were used. MIAME information describing the samples, as well as raw microarray data, including Affymetrix CEL files, is available at the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE15939 and GSM399895 to GSM399911.
Transcriptome Data Analysis
CEL files were imported into FlexArray (http://genomequebec.mcgill.ca/FlexArray/) for further analysis. Raw intensity data were normalized using the robust multiarray average algorithm (Irizarry et al., 2003). Values of the Pearson correlation coefficients examined between biological replicates were in all comparisons ≥ 0.995. The data were log transformed, and a list of differentially expressed genes was generated using the significance analysis of microarrays algorithm (Tusher et al., 2001) implemented in FlexArray. The significance of differential gene expression was estimated by comparing the observed and expected d values. A threshold of 2 for Δ (the difference between the observed and expected d values) and a threshold of 0.05 for unadjusted P values (rawp < 0.05), together with the twofold change filter, were applied to identify differentially expressed genes. All differentially regulated genes had false discovery rate values < 0.2. Hierarchical clustering was done using the Genesis program (Sturn et al., 2002).
Cyclic Electron Flow Measurements
Time courses of induction and relaxation of NPQ were monitored during dark-to-light (80 μmol m−2 s−1) transition, and the 4-min light period was followed by a 2-min dark period as described (Munekage et al., 2002). In the in vitro assay, ferredoxin-dependent PQ reduction activity was measured in ruptured chloroplasts as described (Munekage et al., 2002) using 5 μM spinach ferredoxin (Sigma-Aldrich) and 0.25 mM NADPH.
Accession Numbers
Complete expression profile data have been deposited at the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE15939 and GSM399895 to GSM399911. Arabidopsis Genome Initiative locus identifiers and germplasm identification information for insertion lines are provided in Table 1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Tagging of the PSAL Gene (At4g12800) Coding for the PSI Subunit L.
Supplemental Figure 2. Quantification of the PSI-LHCI-LHCII Complex in Wild-Type and Mutant Plants Adapted to State 2 (Low White Light) or State 1 (High White Light) Conditions.
Supplemental Table 1. List of Proteins Identified within the 670-kD Pigment-Protein Complex by Mass Spectrometry.
Supplemental Table 2. Parameters of Photosynthetic Electron Flow Based on Chlorophyll Fluorescence Measurements of Mutant and Wild-Type Plants.
Supplemental Table 3. List of Genes Whose Transcript Levels Are at Least Twofold Differentially Regulated in the stn7-1 Mutant Compared with the Wild Type (Col-0).
Supplemental Table 4. List of Genes Whose Transcript Levels Are at Least Twofold Differentially Regulated in the psad1-1 Mutant Compared with the Wild Type (Col-0).
Supplemental Table 5. List of Genes Whose Transcript Levels Are at Least Twofold Differentially Regulated in the stn7-1 psad1-1 Mutant Compared with the Wild Type (Col-0).
Supplemental Table 6. List of Genes Whose Transcript Levels Are at Least Twofold Differentially Regulated in the psae1-3 Mutant Compared with the Wild Type (Ws-2).
Supplemental Table 7. List of Genes Whose Transcript Levels Are at Least Twofold Differentially Regulated in the stn7-1 psae1-3 Mutant Compared with the Wild Type (Ws-2).
Supplemental Table 8. Real-Time PCR Analysis of Exemplary Genes Differentially Regulated in the stn7-1, psad1-1, stn7-1 psad1-1, psae1-3, and stn7-1 psae1-3 Mutants.
Supplemental Table 9. List of Genes Whose Transcript Levels Are at Least Twofold Differentially Regulated in Ws-2 Compared with Col-0.
Supplementary Material
Acknowledgments
We thank Bernd Müller for mass spectrometry analysis. We also thank Bob Buchanan and Paul Hardy for critical comments on the manuscript and the Salk Institute for making T-DNA insertion lines publicly available. Funding was provided by the Deutsche Forschungsgemeinschaft (Grants DL 1265/9, SFB-TR1 TP-B8, FOR 804, and TP 323/3).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Dario Leister (leister@lrz.uni-muenchen.de).
Online version contains Web-only data.
References
- Allen, J.F. (1992). Protein phosphorylation in regulation of photosynthesis. Biochim. Biophys. Acta 1098: 275–335. [DOI] [PubMed] [Google Scholar]
- Allen, J.F. (1995). Thylakoid protein phosphorylation, state1-state 2 transitions, and photosystem stoichiometry adjustment: Redox control at multiple levels of gene expression. Physiol. Plant. 93: 196–205. [Google Scholar]
- Allen, J.F., and Forsberg, J. (2001). Molecular recognition in thylakoid structure and function. Trends Plant Sci. 6: 317–326. [DOI] [PubMed] [Google Scholar]
- Allen, J.F., and Pfannschmidt, T. (2000). Balancing the two photosystems: photosynthetic electron transfer governs transcription of reaction centre genes in chloroplasts. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, J.F., and Race, H.L. (2002). Will the real LHC II kinase please step forward? Sci. STKE 2002: PE43. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Andersson, J., Wentworth, M., Walters, R.G., Howard, C.A., Ruban, A.V., Horton, P., and Jansson, S. (2003). Absence of the Lhcb1 and Lhcb2 proteins of the light-harvesting complex of photosystem II - Effects on photosynthesis, grana stacking and fitness. Plant J. 35: 350–361. [DOI] [PubMed] [Google Scholar]
- Bailey, S., Walters, R.G., Jansson, S., and Horton, P. (2001). Acclimation of Arabidopsis thaliana to the light environment: The existence of separate low light and high light responses. Planta 213: 794–801. [DOI] [PubMed] [Google Scholar]
- Bassi, R., dal Belin Peruffo, A., Barbato, R., and Ghisi, R. (1985). Differences in chlorophyll-protein complexes and composition of polypeptides between thylakoids from bundle sheaths and mesophyll cells in maize. Eur. J. Biochem. 146: 589–595. [DOI] [PubMed] [Google Scholar]
- Bellafiore, S., Barneche, F., Peltier, G., and Rochaix, J.D. (2005). State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433: 892–895. [DOI] [PubMed] [Google Scholar]
- Bonardi, V., Pesaresi, P., Becker, T., Schleiff, E., Wagner, R., Pfannschmidt, T., Jahns, P., and Leister, D. (2005). Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature 437: 1179–1182. [DOI] [PubMed] [Google Scholar]
- Buchanan, B.B., and Balmer, Y. (2005). Redox regulation: a broadening horizon. Annu. Rev. Plant Biol. 56: 187–220. [DOI] [PubMed] [Google Scholar]
- Carlberg, I., Hansson, M., Kieselbach, T., Schroder, W.P., Andersson, B., and Vener, A.V. (2003). A novel plant protein undergoing light-induced phosphorylation and release from the photosynthetic thylakoid membranes. Proc. Natl. Acad. Sci. USA 100: 757–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuartzman, S.G., Nevo, R., Shimoni, E., Charuvi, D., Kiss, V., Ohad, I., Brumfeld, V., and Reich, Z. (2008). Thylakoid membrane remodeling during state transitions in Arabidopsis. Plant Cell 20: 1029–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- DalCorso, G., Pesaresi, P., Masiero, S., Aseeva, E., Schunemann, D., Finazzi, G., Joliot, P., Barbato, R., and Leister, D. (2008). A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell 132: 273–285. [DOI] [PubMed] [Google Scholar]
- Delosme, R., Beal, D., and Joliot, P. (1994). Photoacoustic detection of flash-induced charge separation in photosynthetic systems - Spectral dependence of the quantum yield. Biochim. Biophys. Acta 1185: 56–64. [Google Scholar]
- Depege, N., Bellafiore, S., and Rochaix, J.D. (2003). Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science 299: 1572–1575. [DOI] [PubMed] [Google Scholar]
- Dietzel, L., Brautigam, K., and Pfannschmidt, T. (2008). Photosynthetic acclimation: State transitions and adjustment of photosystem stoichiometry–functional relationships between short-term and long-term light quality acclimation in plants. FEBS J. 275: 1080–1088. [DOI] [PubMed] [Google Scholar]
- Eberhard, S., Finazzi, G., and Wollman, F.A. (2008). The dynamics of photosynthesis. Annu. Rev. Genet. 42: 463–515. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300: 1005–1016. [DOI] [PubMed] [Google Scholar]
- Fey, V., Wagner, R., Brautigam, K., and Pfannschmidt, T. (2005. a). Photosynthetic redox control of nuclear gene expression. J. Exp. Bot. 56: 1491–1498. [DOI] [PubMed] [Google Scholar]
- Fey, V., Wagner, R., Brautigam, K., Wirtz, M., Hell, R., Dietzmann, A., Leister, D., Oelmuller, R., and Pfannschmidt, T. (2005. b). Retrograde plastid redox signals in the expression of nuclear genes for chloroplast proteins of Arabidopsis thaliana J. Biol. Chem. 280: 5318–5328. [DOI] [PubMed] [Google Scholar]
- Finazzi, G. (2005). The central role of the green alga Chlamydomonas reinhardtii in revealing the mechanism of state transitions. J. Exp. Bot. 56: 383–388. [DOI] [PubMed] [Google Scholar]
- Finazzi, G., Rappaport, F., Furia, A., Fleischmann, M., Rochaix, J.D., Zito, F., and Forti, G. (2002). Involvement of state transitions in the switch between linear and cyclic electron flow in Chlamydomonas reinhardtii EMBO Rep. 3: 280–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel, M., Bellafiore, S., Rochaix, J.D., and Jansson, S. (2007). Hierarchy amongst photosynthetic acclimation responses for plant fitness. Physiol. Plant. 129: 455–459. [Google Scholar]
- Frigerio, S., Campoli, C., Zorzan, S., Fantoni, L.I., Crosatti, C., Drepper, F., Haehnel, W., Cattivelli, L., Morosinotto, T., and Bassi, R. (2007). Photosynthetic antenna size in higher plants is controlled by the plastoquinone redox state at the post-transcriptional rather than transcriptional level. J. Biol. Chem. 282: 29457–29469. [DOI] [PubMed] [Google Scholar]
- Fristedt, R., Carlberg, I., Zygadlo, A., Piippo, M., Nurmi, M., Aro, E.M., Scheller, H.V., and Vener, A.V. (2009). Intrinsically unstructured phosphoprotein TSP9 regulates light harvesting in Arabidopsis thaliana Biochemistry 48: 499–509. [DOI] [PubMed] [Google Scholar]
- Fujita, Y. (1997). A study on the dynamic features of photosystem stoichiometry: Accomplishments and problems for future studies. Photosynth. Res. 53: 83–93. [Google Scholar]
- Gray, G.R., Savitch, L.V., Ivanov, A.G., and Huner, N. (1996). Photosystem II excitation pressure and development of resistance to photoinhibition (II. Adjustment of photosynthetic capacity in winter wheat and winter rye). Plant Physiol. 110: 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genty, B., Briantais, J.M., and Baker, N.R. (1989). The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta 990: 87–92. [Google Scholar]
- Haldrup, A., Jensen, P.E., Lunde, C., and Scheller, H.V. (2001). Balance of power: A view of the mechanism of photosynthetic state transitions. Trends Plant Sci. 6: 301–305. [DOI] [PubMed] [Google Scholar]
- Heinemeyer, J., Eubel, H., Wehmhöner, D., Jänsch, L., and Braun, H.P. (2004). Proteomic approach to characterize the supramolecular organization of photosystems in higher plants. Phytochemistry 65: 1683–1692. [DOI] [PubMed] [Google Scholar]
- Ihnatowicz, A., Pesaresi, P., and Leister, D. (2007). The E subunit of photosystem I is not essential for linear electron flow and photoautotrophic growth in Arabidopsis thaliana Planta 226: 889–895. [DOI] [PubMed] [Google Scholar]
- Ihnatowicz, A., Pesaresi, P., Lohrig, K., Wolters, D., Muller, B., and Leister, D. (2008). Impaired photosystem I oxidation induces STN7-dependent phosphorylation of the light-harvesting complex I protein Lhca4 in Arabidopsis thaliana Planta 227: 717–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihnatowicz, A., Pesaresi, P., Varotto, C., Richly, E., Schneider, A., Jahns, P., Salamini, F., and Leister, D. (2004). Mutants for photosystem I subunit D of Arabidopsis thaliana: Effects on photosynthesis, photosystem I stability and expression of nuclear genes for chloroplast functions. Plant J. 37: 839–852. [DOI] [PubMed] [Google Scholar]
- Irizarry, R.A., Hobbs, B., Collin, F., Beazer-Barclay, Y.D., Antonellis, K.J., Scherf, U., and Speed, T.P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264. [DOI] [PubMed] [Google Scholar]
- Jensen, P.E., Gilpin, M., Knoetzel, J., and Scheller, H.V. (2000). The PSI-K subunit of photosystem I is involved in the interaction between light-harvesting complex I and the photosystem I reaction center core. J. Biol. Chem. 275: 24701–24708. [DOI] [PubMed] [Google Scholar]
- Jensen, P.E., Haldrup, A., Zhang, S., and Scheller, H.V. (2004). The PSI-O subunit of plant photosystem I is involved in balancing the excitation pressure between the two photosystems. J. Biol. Chem. 279: 24212–24217. [DOI] [PubMed] [Google Scholar]
- Kanervo, E., Suorsa, M., and Aro, E.M. (2005). Functional flexibility and acclimation of the thylakoid membrane. Photochem. Photobiol. Sci. 4: 1072–1080. [DOI] [PubMed] [Google Scholar]
- Klimyuk, V.I., Persello-Cartieaux, F., Havaux, M., Contard-David, P., Schuenemann, D., Meiherhoff, K., Gouet, P., Jones, J.D., Hoffman, N.E., and Nussaume, L. (1999). A chromodomain protein encoded by the Arabidopsis CAO gene is a plant-specific component of the chloroplast signal recognition particle pathway that is involved in LHCP targeting. Plant Cell 11: 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klughammer, C., and Schreiber, U. (1994). An improved method, using saturating light-pulses, for the determination of photosystem-I quantum yield via P700+ absorbancychanges at 830 nm. Planta 192: 261–268. [Google Scholar]
- Kouril, R., Zygadlo, A., Arteni, A.A., de Wit, C.D., Dekker, J.P., Jensen, P.E., Scheller, H.V., and Boekema, E.J. (2005). Structural characterization of a complex of photosystem I and light-harvesting complex II of Arabidopsis thaliana Biochemistry 44: 10935–10940. [DOI] [PubMed] [Google Scholar]
- Krall, J.P., and Edwards, G.E. (1992). Relationship between photosystem II activity and CO2 fixation in leaves. Physiol. Plant. 86: 180–187. [Google Scholar]
- Leister, D., Varotto, C., Pesaresi, P., Niwergall, A., and Salamini, F. (1999). Large-scale evaluation of plant growth in Arabidopsis thaliana by non-invasive image analysis. Plant Physiol. Biochem. 37: 671–678. [Google Scholar]
- Lemeille, S., Willig, A., Depege-Fargeix, N., Delessert, C., Bassi, R., and Rochaix, J.D. (2009). Analysis of the chloroplast protein kinase Stt7 during state transitions. PLoS Biol. 7: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrig, K., Muller, B., Davydova, J., Leister, D., and Wolters, D.A. (2009). Phosphorylation site mapping of soluble proteins: Bioinformatical filtering reveals potential plastidic phosphoproteins in Arabidopsis thaliana Planta 229: 1123–1134. [DOI] [PubMed] [Google Scholar]
- Lunde, C., Jensen, P.E., Haldrup, A., Knoetzel, J., and Scheller, H.V. (2000). The PSI-H subunit of photosystem I is essential for state transitions in plant photosynthesis. Nature 408: 613–615. [DOI] [PubMed] [Google Scholar]
- Lunde, C., Jensen, P.E., Rosgaard, L., Haldrup, A., Gilpin, M.J., and Scheller, H.V. (2003). Plants impaired in state transitions can to a large degree compensate for their defect. Plant Cell Physiol. 44: 44–54. [DOI] [PubMed] [Google Scholar]
- Melis, A. (1991). Dynamics of photosynthetic membrane composition and function. Biochim. Biophys. Acta 1058: 87–106. [Google Scholar]
- Mullineaux, C.W., and Emlyn-Jones, D. (2005). State transitions: An example of acclimation to low-light stress. J. Exp. Bot. 56: 389–393. [DOI] [PubMed] [Google Scholar]
- Munekage, Y., Hojo, M., Meurer, J., Endo, T., Tasaka, M., and Shikanai, T. (2002). PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 110: 361–371. [DOI] [PubMed] [Google Scholar]
- Naver, H., Haldrup, A., and Scheller, H.V. (1999). Cosuppression of photosystem I subunit PSI-H in Arabidopsis thaliana. Efficient electron transfer and stability of photosystem I is dependent upon the PSI-H subunit. J. Biol. Chem. 274: 10784–10789. [DOI] [PubMed] [Google Scholar]
- Nilsson, A., Stys, D., Drakenberg, T., Spangfort, M.D., Forsen, S., and Allen, J.F. (1997). Phosphorylation controls the three-dimensional structure of plant light harvesting complex II. J. Biol. Chem. 272: 18350–18357. [DOI] [PubMed] [Google Scholar]
- Pesaresi, P., Lunde, C., Jahns, P., Tarantino, D., Meurer, J., Varotto, C., Hirtz, R.D., Soave, C., Scheller, H.V., Salamini, F., and Leister, D. (2002). A stable LHCII-PSI aggregate and suppression of photosynthetic state transitions in the psae1-1 mutant of Arabidopsis thaliana Planta 215: 940–948. [DOI] [PubMed] [Google Scholar]
- Pesaresi, P., Varotto, C., Meurer, J., Jahns, P., Salamini, F., and Leister, D. (2001). Knock-out of the plastid ribosomal protein L11 in Arabidopsis: Effects on mRNA translation and photosynthesis. Plant J. 27: 179–189. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt, T., Nilsson, A., and Allen, J.F. (1999). Photosynthetic control of chloroplast gene expression. Nature 397: 625–628. [Google Scholar]
- Pfannschmidt, T., Schutze, K., Brost, M., and Oelmuller, R. (2001). A novel mechanism of nuclear photosynthesis gene regulation by redox signals from the chloroplast during photosystem stoichiometry adjustment. J. Biol. Chem. 276: 36125–36130. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt, T., Schutze, K., Fey, V., Sherameti, I., and Oelmuller, R. (2003). Chloroplast redox control of nuclear gene expression – A new class of plastid signals in interorganellar communication. Antioxid. Redox Signal. 5: 95–101. [DOI] [PubMed] [Google Scholar]
- Porra, R.J., Thompson, W.A., and Kriedemann, P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophyll a and chlorophyll b extracted with 4 different solvents - Verification of the concentration of chlorophyll standards by atomic-absorption spectroscopy. Biochim. Biophys. Acta 975: 384–394. [Google Scholar]
- Pursiheimo, S., Mulo, P., Rintamaki, E., and Aro, E.M. (2001). Coregulation of light-harvesting complex II phosphorylation and lhcb mRNA accumulation in winter rye. Plant J. 26: 317–327. [DOI] [PubMed] [Google Scholar]
- Puthiyaveetil, S., Kavanagh, T.A., Cain, P., Sullivan, J.A., Newell, C.A., Gray, J.C., Robinson, C., van der Giezen, M., Rogers, M.B., and Allen, J.F. (2008). The ancestral symbiont sensor kinase CSK links photosynthesis with gene expression in chloroplasts. Proc. Natl. Acad. Sci. USA 105: 10061–10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers, C., Ruijter, J.M., Deprez, R.H., and Moorman, A.F. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339: 62–66. [DOI] [PubMed] [Google Scholar]
- Rintamäki, E., Martinsuo, P., Pursiheimo, S., and Aro, E.M. (2000). Cooperative regulation of light-harvesting complex II phosphorylation via the plastoquinol and ferredoxin-thioredoxin system in chloroplasts. Proc. Natl. Acad. Sci. USA 97: 11644–11649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix, J.D. (2007). Role of thylakoid protein kinases in photosynthetic acclimation. FEBS Lett. 581: 2768–2775. [DOI] [PubMed] [Google Scholar]
- Savitch, L.V., Maxwell, D.P., and Huner, N. (1996). Photosystem II excitation pressure and photosynthetic carbon metabolism in Chlorella vulgaris Plant Physiol. 111: 127–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger, H., and von Jagow, G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166: 368–379. [DOI] [PubMed] [Google Scholar]
- Sturn, A., Quackenbush, J., and Trajanoski, Z. (2002). Genesis: Cluster analysis of microarray data. Bioinformatics 18: 207–208. [DOI] [PubMed] [Google Scholar]
- Takahashi, H., Iwai, M., Takahashi, Y., and Minagawa, J. (2006). Identification of the mobile light-harvesting complex II polypeptides for state transitions in Chlamydomonas reinhardtii Proc. Natl. Acad. Sci. USA 103: 477–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikkanen, M., Nurmi, M., Suorsa, M., Danielsson, R., Mamedov, F., Styring, S., and Aro, E.M. (2008). Phosphorylation-dependent regulation of excitation energy distribution between the two photosystems in higher plants. Biochim. Biophys. Acta 1777: 425–432. [DOI] [PubMed] [Google Scholar]
- Tikkanen, M., Piippo, M., Suorsa, M., Sirpio, S., Mulo, P., Vainonen, J., Vener, A.V., Allahverdiyeva, Y., and Aro, E.M. (2006). State transitions revisited-a buffering system for dynamic low light acclimation of Arabidopsis. Plant Mol. Biol. 62: 779–793. [DOI] [PubMed] [Google Scholar]
- Tusher, V.G., Tibshirani, R., and Chu, G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98: 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainonen, J.P., Hansson, M., and Vener, A.V. (2005). STN8 protein kinase in Arabidopsis thaliana is specific in phosphorylation of photosystem II core proteins. J. Biol. Chem. 280: 33679–33686. [DOI] [PubMed] [Google Scholar]
- Varotto, C., Pesaresi, P., Meurer, J., Oelmuller, R., Steiner-Lange, S., Salamini, F., and Leister, D. (2000). Disruption of the Arabidopsis photosystem I gene psaE1 affects photosynthesis and impairs growth. Plant J. 22: 115–124. [DOI] [PubMed] [Google Scholar]
- Wagner, R., Dietzel, L., Brautigam, K., Fischer, W., and Pfannschmidt, T. (2008). The long-term response to fluctuating light quality is an important and distinct light acclimation mechanism that supports survival of Arabidopsis thaliana under low light conditions. Planta 228: 573–587. [DOI] [PubMed] [Google Scholar]
- Walters, R.G. (2005). Towards an understanding of photosynthetic acclimation. J. Exp. Bot. 56: 435–447. [DOI] [PubMed] [Google Scholar]
- Washburn, M.P., Wolters, D., and Yates III, J.R. (2001). Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19: 242–247. [DOI] [PubMed] [Google Scholar]
- Weigel, M., Varotto, C., Pesaresi, P., Finazzi, G., Rappaport, F., Salamini, F., and Leister, D. (2003). Plastocyanin is indispensable for photosynthetic electron flow in Arabidopsis thaliana J. Biol. Chem. 278: 31286–31289. [DOI] [PubMed] [Google Scholar]
- Wollman, F.A. (2001). State transitions reveal the dynamics and flexibility of the photosynthetic apparatus. EMBO J. 20: 3623–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zer, H., and Ohad, I. (2003). Light, redox state, thylakoid-protein phosphorylation and signaling gene expression. Trends Biochem. Sci. 28: 467–470. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.