Abstract
Jmjd3, a JmjC family histone demethylase, is induced by the transcription factor NF-kB in response to microbial stimuli. Jmjd3 erases H3K27me3, a histone mark associated with transcriptional repression and involved in lineage determination. However, the specific contribution of Jmjd3 induction and H3K27me3 demethylation to inflammatory gene expression remains unknown. Using chromatin immunoprecipitation-sequencing we found that Jmjd3 is preferentially recruited to transcription start sites characterized by high levels of H3K4me3, a marker of gene activity, and RNA polymerase II (Pol_II). Moreover, 70% of lipopolysaccharide (LPS)-inducible genes were found to be Jmjd3 targets. Although most Jmjd3 target genes were unaffected by its deletion, a few hundred genes, including inducible inflammatory genes, showed moderately impaired Pol_II recruitment and transcription. Importantly, most Jmjd3 target genes were not associated with detectable levels of H3K27me3, and transcriptional effects of Jmjd3 absence in the window of time analysed were uncoupled from measurable effects on this histone mark. These data show that Jmjd3 fine-tunes the transcriptional output of LPS-activated macrophages in an H3K27 demethylation-independent manner.
Keywords: histone demethylase, JmjC, macrophages, transcription
Introduction
Inflammatory responses require the activation of a complex gene expression program that involves the inducible transcription of hundreds of genes whose products restrain microbial colonization, recruit and activate leukocytes, increase vascular permeability, amplify the response, and protect inflammatory and tissue cells from apoptosis (Medzhitov, 2008). Transcription factors belonging to the NF-kB/Rel, IRF and STAT families (Ivashkiv and Hu, 2004; Hayden et al, 2006; Honda and Taniguchi, 2006) are well-established regulators of inflammatory gene expression. However, knowledge on the essential transcriptional coregulators, chromatin modifiers and transcriptional circuits underlying inflammation is still rather primitive.
We previously reported that the histone demethylase (HDM) Jmjd3 is quickly induced by NF-kB in primary mouse macrophages in response to inflammatory stimuli, whereas its paralog Utx is expressed at low and constant levels (De Santa et al, 2007). With the only exception of LSD1 (Shi et al, 2004), all known HDMs belong to the JmjC family, which includes 27 members in the current human genome annotation (Klose and Zhang, 2007; Shi and Whetstine, 2007; Cloos et al, 2008). The JmjC domain is a variant of a common structural motif found from bacteria to mammals, the 2-histidine-1-carboxylate facial triad (Koehntop et al, 2005), that serves as a platform for binding divalent iron. The metal centre of this motif is used to activate molecular oxygen and transfer an oxygen atom to the substrate. Priming of the metal centre for attack by molecular oxygen depends on binding to the substrate and in some cases to a 2-oxoacid cofactor (most often α-ketoglutarate). Although the spectrum of metabolic transformations carried out by oxygenases with a 2-histidine-1-carboxylate facial triad is extremely broad (Loenarz and Schofield, 2008), JmjC proteins in animals can apparently catalyse only hydroxylation reactions. If the target of hydroxylation is a methyl–lysine, an unstable hydroxy-methyl group is generated that is released as a formaldehyde molecule, thus eventually restoring the unmethylated state of the lysine residue (Tsukada et al, 2006). Overall, JmjC family HDMs are characterized by a high level of specificity both regarding the target lysine in the amino-terminal histone tails, and the level of methylation (mono-, di- and tri-methylation) they can reverse.
The closely related Jmjd3 and Utx specifically demethylate trimethylated lysine 27 in histone H3 (H3K27me3), a chromatin modification associated with transcriptional repression (Agger et al, 2007; De Santa et al, 2007; Lan et al, 2007; Lee et al, 2007). Hence, NF-kB-induced Jmjd3 upregulation links inflammation to the control of a histone modification involved in lineage determination, differentiation and tissue homeostasis (Kirmizis et al, 2004; Boyer et al, 2006; Bracken et al, 2006; Lee et al, 2006; Sparmann and van Lohuizen, 2006; Burgold et al, 2008; Sen et al, 2008), which may provide a mechanistic connection between chronic inflammation and the associated alterations of differentiation (e.g. metaplasia, discussed in De Santa et al, 2007). However, the specific contribution, if any, of Jmjd3 induction to innate immunity and inflammation remains unknown. In this study, we investigated the genomic distribution of Jmjd3, its relationship to H3K27me3, H3K4me3 and RNA polymerase II (Pol_II) occupancy, and finally its role in controlling the gene expression program of lipopolysaccharide (LPS)-activated macrophages. The data we report here show the involvement of Jmjd3 in tuning the transcriptional output of LPS-stimulated macrophages and suggest that this activity is largely independent of H3K27me3 demethylation.
Results
Analysis of Jmjd3 genomic distribution in LPS-activated macrophages
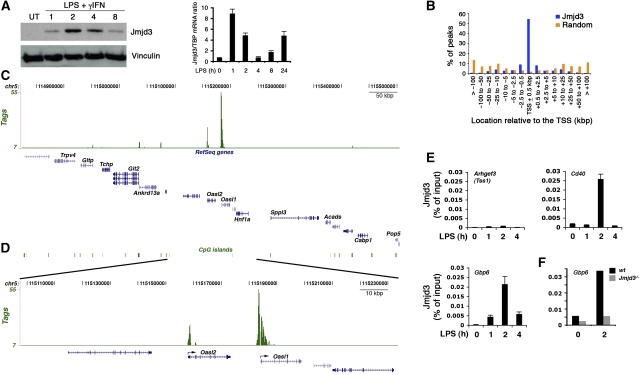
Jmjd3 induction by LPS depends on multiple evolutionary conserved binding sites for the transcription factor NF-kB (De Santa et al, 2007), which map to a region of the gene containing a large CpG island and multiple peaks of H3K4 trimethylation (Supplementary Figure 1), likely representing alternative sites of transcriptional initiation. The presence of multiple conserved binding sites for an inflammatory transcription factor such as NF-kB (Hayden et al, 2006) suggests a possible involvement of Jmjd3 in the control of inflammatory gene expression programs. To address this possibility we analysed the genomic distribution of newly synthesized Jmjd3 in mouse bone marrow-derived macrophages using chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-Seq). Macrophages were stimulated with LPS and interferon gamma (IFNγ) for 2 h, corresponding to the peak of Jmjd3 induction in these cells (Figure 1A). We generated a set of ∼8 million high quality and uniquely aligned reads. Using a very restrictive false discovery rate (FDR)=0.1% (clusters of tags with ⩾9 overlaps) as a cutoff, we identified 4398 peaks, whereas considering clusters of ⩾7 tags 14 013 peaks were detected (Supplementary Table I). Subsequent analyses were carried out considering an FDR=0.1%. Validation by ChIP–qPCR on a representative set of Jmjd3 targets in stimulated versus unstimulated macrophages showed good correlation with peak calling with an overall validation rate of about 93% (Supplementary Figure 2A). Jmjd3 binding preferentially occurred at transcription start sites (TSS), often extending at various distance inside the coding region (Figure 1B; Supplementary Figure 3). Fifty-four per cent of the Jmjd3 peaks were within ±0.5 kb from a mapped TSS, and more than 70% of them were within 2.5 kb, a value much higher than expected for a random distribution.
Figure 1.
Genomic distribution of Jmjd3 in LPS-stimulated macrophages. (A) Jmjd3 induction in primary mouse bone marrow-derived macrophages. A western blot (left) and an RT–qPCR (right) analysis are shown. (B) Genome-wide analysis of Jmjd3 association with TSSs. Macrophages were stimulated with LPS+γIFN for 2 h and distribution of Jmjd3 peaks relative to mapped TSS was determined. (C) Jmjd3 binding to a representative region of mouse chromosome 5. The y axis indicates the number of tags in peaks. (D) A zoomed-in view of the same region shows the association of Jmjd3 with the TSSs of two genes. (E) Kinetics of Jmjd3 recruitment. TSS1 of Arhgef3, which was negative for Jmjd3 in the ChIP-Seq data, was used as a negative control. Guanylate-binding protein 6 (Gbp6) encodes an antiviral GTPase representing one of the most abundant proteins induced by LPS+γIFN. Error bars: s.e.m. from a triplicate experiment. (F) Abrogation of ChIP signals in Jmjd3 knockout macrophages. Anti-Jmjd3 ChIP was carried out in wild type and Jmjd3−/− foetal liver-derived macrophages.
Using ±100 kb around promoters as cutoff, we found 4331 Jmjd3 peaks (98.5%) associated with 3339 genes (based on the annotated TSSs from the DBTSS database; Supplementary Table I). The binding of Jmjd3 to a large (0.85 Mbp) representative region of chr5 is shown as an example in Figure 1C and a zoomed-in view of the same region is shown in Figure 1D. The kinetic profile of Jmjd3 recruitment to individual target genes closely mirrored the behaviour of bulk Jmjd3 protein levels (Figure 1E) and ChIP signals were dependent on the presence of Jmjd3, as indicated by their abrogation in Jmjd3 knockout macrophages (Figure 1F; Supplementary Figure 2B).
In activated macrophages, newly synthesized Jmjd3 is rapidly recruited to the TSSs of thousands of genes (Supplementary Table I) including those encoding LPS-inducible immune response and inflammatory mediators such as chemokines (e.g. Cxcl2, Cxcl11, Ccl5), cytokines (e.g. Tnfα, Il6, Il27), proteins or enzymes involved in microbial recognition and killing (Nos2, Nod1, Nod2, multiple 2′–5′ oligoadenylate synthetase family members, antiviral GTPases such as Gbp2-6), adhesion molecules and immuno-receptors (Sdc4, Icam1, Cd40, Cd83, Cd86), complement components (C3), growth factors (Vegfa, Vegfc), signal transducers and transcription factors (Jak2, Socs1, Socs3, Nfkbie, Nfkbiz, Bcl3, Jun, Junb, Irf2, Irf7), histo-compatibility molecules (H2-Q2) and enzymes involved in prostaglandin biosynthesis (Ptgs2). Thus, widespread Jmjd3 genomic recruitment in activated macrophages seems to provide a satisfactory explanation of the evolutionary link of Jmjd3 gene expression to NF-kB activation in vertebrates.
Jmjd3 distribution correlates with gene activity
To systematically analyse the transcriptional state of Jmjd3-bound TSSs, we first generated genomic maps of H3K4me3, a histone mark associated with the TSSs of genes that are either active or poised for activation (Kouzarides, 2007; Ruthenburg et al, 2007), in both unstimulated and 4 h LPS+IFNγ-stimulated primary macrophages. More than 17 million uniquely aligned sequencing reads were obtained in each condition, corresponding to 21 418 and 19 631 peaks, respectively (FDR 0.1%) (Supplementary Tables II and III); 18 618 (87%) peaks were overlapping between unstimulated and stimulated macrophages and they were used for further analyses (Supplementary Table IV). In agreement with previous gene-specific data (Foster et al, 2007), H3K4me3 was quickly upregulated at several LPS+IFNγ-inducible genes: based on the total tag counts within each H3K4me3 cluster, H3K4me3 intensity increased more than 2-fold at the TSS of 173 genes and more than 1.5-fold at 496 genes (Supplementary Table IV; Supplementary Figure 4), thus indicating a very dynamic behaviour of H3K4me3 in this system (as opposed to what was found during embryonic stem cell differentiation) (Mikkelsen et al, 2007; Zhao et al, 2007). The observation that in yeast H3K4me3 completely turns over completely in less than 2 h (Seward et al, 2007) also supports the dynamic nature of this modification and suggests that steady-state levels measured by ChIP may in fact reflect a dynamic equilibrium.
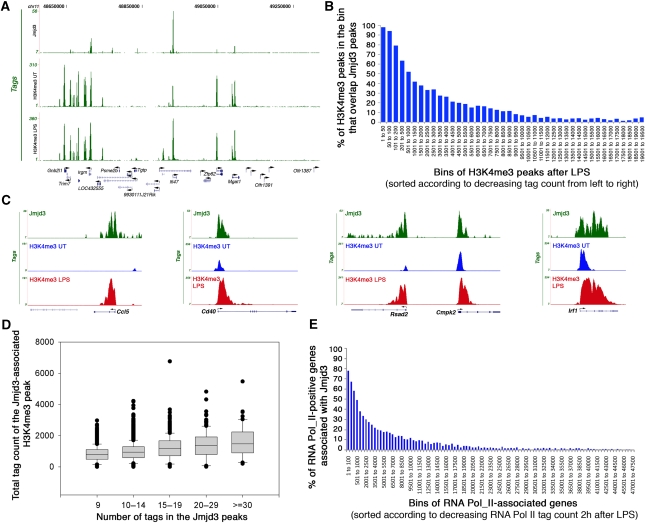
Visual browsing through the data suggested a strong correlation between Jmjd3 binding and H3K4me3 positivity and levels (Figure 2A). To assess the correlation between Jmjd3 and H3K4me3 genome-wide in a quantitative manner, we sorted all 19 631 H3K4me3 peaks in LPS-stimulated cells in the order of their total tag counts (the number of tags that form the peak). Then we calculated the percentage of peaks in each bin that overlap a Jmjd3 peak. Almost 100% of the highest H3K4me3 peaks were found associated with a Jmjd3 peak (Figure 2B; Supplementary Table IV). The association with Jmjd3 decreased steadily in the H3K4me3 peaks with lower tag counts (Figure 2B). The correlation was much weaker when pre-stimulation H3K4me3 levels were considered (data not shown). Importantly, not all strongly positive H3K4me3 genes were bound by Jmjd3 (Figure 2A; Supplementary Table IV) and Jmjd3 seemed to bind preferentially to TSSs with upregulated H3K4me3 levels: out of the 173 genes with ⩾2-fold increase in H3K4me3, 106 genes (61%) were bound by Jmjd3. Some specific examples are shown in Figure 2C and Supplementary Figures 5 and 6.
Figure 2.
Jmjd3 association with transcriptionally active and inducible genes. (A) Jmjd3 association with H3K4me3-positive genes. Jmjd3 peaks and H3K4me3 peaks (in unstimulated and LPS-stimulated macrophages) at a representative region of chr 11 are shown. (B) Correlation between H3K4me3 levels and Jmjd3 binding in LPS-stimulated macrophages. H3K4me3 peaks were grouped in bins of decreasing total tag count from left to right. The y axis indicates the per cent of H3K4me3 peaks overlapping Jmjd3 peaks. (C) Association between Jmjd3 and H3K4me3 at representative genes. (D) Correlation between intensity of Jmjd3 binding and high levels of H3K4me3. (E) Correlation between Pol_II level and Jmjd3 binding at 2 h after LPS stimulation. Genes were grouped in bins of decreasing Pol_II intensity from left to right. The y axis shows the per cent of active, RNA Pol_II-positive genes that are associated with Jmjd3.
We next measured the correlation between the levels of Jmjd3 and those of H3K4me3 after LPS stimulation. Figure 2D shows a box plot of the number of overlapping tags in Jmjd3 peaks and the total tag counts of the associated H3K4me3 cluster. It seems that the intensity of the Jmjd3 ChIP signal is positively correlated with H3K4me3 ChIP intensity after LPS treatment, indicating that Jmjd3 binds to active genes in a manner somehow proportional to the intensity of gene activity. As the distribution of H3K4me3 and Jmjd3 often overlaps and because newly synthesized Jmjd3 is transiently incorporated in H3K4 HMT complexes (De Santa et al, 2007), a simple possibility is that Jmjd3 is preferentially recruited to sites of active H3K4me3 deposition or turnover by association with H3K4 histone methyltransferases. Indeed, some of the genes showing the highest levels of Jmjd3 recruitment were those undergoing massive H3K4me3 increase after stimulation (e.g. Ccl5, Nos2, Cd40) (Figure 2C; Supplementary Figure 6; Supplementary Table IV).
To assess the correlation between Jmjd3 recruitment and gene activity in a more direct manner, we generated genomic maps of total Pol_II in unstimulated, 2 and 4 h LPS-stimulated primary macrophages. About 9 million uniquely aligned sequencing reads were obtained in unstimulated macrophages, whereas more than 12 million reads were obtained in stimulated macrophages at both time points. Pol_II transcriptional activity was indicated by the detection of Pol_II signals within the internal regions of several inducible genes including Nos2, Ccl5 and Irf1 (Supplementary Figure 7). Using a high stringency cutoff (FDR=0.1%), we found a total of 55 600 Pol_II peaks in the unstimulated macrophage library and 57 201 and 57 514 peaks in the 2- and 4 h-stimulated libraries, respectively. In each library >70% of the peaks were located ±10 kb of known TSSs, as compared with 26% association with random peaks in simulation experiments. Moreover, >99% of the peaks were associated with gene regions (±100 kb of a gene) whereas less than 1% of Pol_II peaks were found in gene deserts.
Out of the 17 389 genes associated with Pol_II, 3992 genes (23%) showed more than a two-fold increase in the total tag count within the corresponding Pol_II peaks at 2 h after LPS simulation and 1510 of them (1510/3992; 38%) were also bound by Jmjd3. Reciprocally, when the 3339 genes bound by Jmjd3 were considered, 73% of them (2438) showed an increase in Pol_II activity at 2 h after LPS (>25% increase in the total tag count in their peaks), thus indicating that Jmjd3 binding is biased towards a subset of genes whose transcription is induced or increased by LPS. To measure the correlation between transcriptional activity and Jmjd3 binding in a quantitative manner, we first grouped genes in bins of decreasing Pol_II tag count and then we calculated the percentage of genes in each bin that are bound by Jmjd3. Seventy-eight per cent of the genes with the highest total Pol_II tag counts at 2 h after LPS stimulation were Jmjd3 targets (Figure 2E). The association with Jmjd3 decreased steadily in genes with lower tag counts. The correlation was much weaker with the pre-stimulation Pol_II library, and slightly weaker with the 4 h LPS-stimulated library (data not shown). Overall, these data indicate that Jmjd3 is preferentially recruited to sites of high and inducible Pol_II occupancy and gene activity.
H3K27me3 status at Jmjd3 target genes
The only known substrate of Jmjd3 is H3K27me3, and the simplest prediction consistent with its reported biochemical activity as a H3K27 demethylase is that Jmjd3 is recruited to genes associated with basal H3K27me3 levels to reduce them and enable or enhance transcriptional activation.
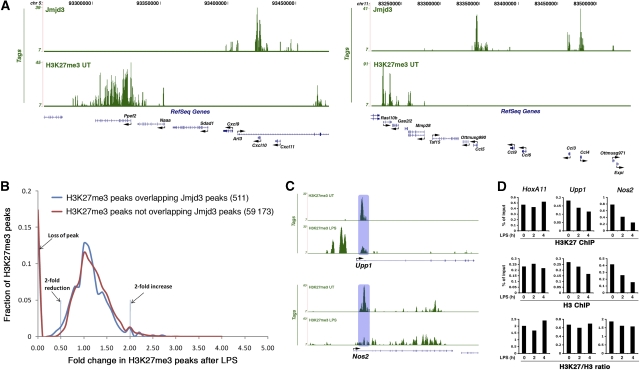
To test this prediction we generated H3K27me3 genomic maps in unstimulated and LPS-stimulated macrophages; 9.7 million and 14 million uniquely aligned sequences were obtained from the anti-H3K27me3 ChIP in untreated and LPS-treated cells, corresponding to 59 684 and 89 093 peaks, respectively, at an FDR of 0.1% (Supplementary Tables V and VI). Similarly to other systems, H3K27me3 peaks were in fact often part of broad regions (previously defined as broad local enrichment, BLOCs) (Pauler et al, 2009) of average size of 21.2 and 27.8 kb (in untreated and LPS-treated macrophages, respectively) (Supplementary Table VII and Supplementary data). The percentage of H3K27me3 peaks contained within the 5733 BLOCs identified was 72.3 and 66.6% in untreated and LPS-treated macrophages, respectively. The behaviour of peaks within and outside these broad regions was however comparable (see Supplementary data) and therefore we will refer to peaks rather than BLOCs in the following section of the manuscript. Jmjd3 target genes were in the majority of cases (2963/3339; 88%) not associated with any H3K27me3 peak within ±1 kb already before LPS stimulation and therefore before induction of Jmjd3 (Figure 3A; Supplementary Table VII). At a few genes with multiple TSSs with differential H3K27me3 association, Jmjd3 seemed to be selectively recruited at the H3K27me3-negative TSS (Supplementary Table VIII). Therefore, Jmjd3 recruitment to target genes does not rely on pre-existing H3K27me3, and at most recruitment sites Jmjd3 will not encounter H3K27me3. Considering all (genic and extragenic) Jmjd3 peaks, only a minority of them (511/4398; 11.6%) was associated with a neighbouring (±500 bp) H3K27me3 peak (Supplementary Table VIII). Of these peaks, only 83 (16.3%) decreased by more than two-fold after a 4 h LPS stimulation. This value is similar to the frequency of a two-fold reduction of H3K27me3 peaks observed elsewhere in the genome after LPS treatment (17.6%, corresponding to 10 385 out of a total of 59 173 peaks). Figure 3B shows the comparison of intensity changes between overall H3K27me3 peaks (shown in red) and peaks overlapping with Jmjd3 (in blue). The change of H3K27me3 patterns on stimulation is extremely similar in the two groups.
Figure 3.
Jmjd3 binding and H3K27me3. (A) Lack of basal H3K27me3 at Jmjd3-bound genes. Jmjd3 and H3K27me3 ChIP-Seq profiles at two representative regions of chr5 and chr11. (B) Reduction in H3K27me3 is statistically similar at Jmjd3-bound and non-bound regions. The x axis indicates fold changes in H3K27me3 levels in response to LPS stimulation. The y axis shows the fraction of H3K27me3 peaks. (C) Reduction in H3K27me3 at Nos2 and Upp1 after LPS stimulation. The H3K27me3 peak downregulated after LPS is indicated by a shaded box. (D) Reduction in H3K27me3 at Upp1 and Nos2 in LPS-stimulated macrophages reflects nucleosome loss. ChIP–qPCRs were carried out with antibodies against H3K27me3 or total H3. H3K27me3/H3 ratios were calculated by dividing H3K27me3 signals by the signal obtained with the anti-H3 antibody. The data refer to one representative experiment out of four with qualitatively similar results.
Taken together, these data show that there is no statistically significant increase in the probability of H3K27me3 reduction at peaks lying close to Jmjd3 as compared with the distant ones.
However, some H3K27me3 peaks underwent a rapid reduction following LPS stimulation, and a fraction of these peaks were Jmjd3-associated. Therefore, we sought to understand the molecular basis of this reduction and whether it was due to enzymatic demethylation. Reduced H3K27me3 signals at these peaks after LPS may reflect enzymatic demethylation, histone exchange or nucleosome loss. To discriminate among these possibilities, we analysed two Jmjd3-associated genes among those showing the highest stimulus-induced reduction, Nos2 and Upp1 (Figure 3C; Supplementary Figure 6). In both cases, H3K27me3 downregulation perfectly mirrored the reduction in the total H3 levels that accompanied gene induction (Figure 3D, upper and middle panels). In fact, when H3K27me3 ChIP data were normalized to total H3, no difference was found in untreated and treated cells (Figure 3D, bottom panel), suggesting that nucleosome loss rather than enzymatic demethylation is the mechanism underlying the observed reductions of H3K27me3. Similar data were observed with all the other genes analysed (data not shown). Therefore, nucleosome depletion at inducible genes is a widespread occurrence in LPS-stimulated macrophages, possibly because of the extensive nucleosome displacement linked to massive Pol_II elongation (Supplementary Figure 7); conversely, we could not get evidence supporting the occurrence of H3K27me3 demethylation in the first 4 h after LPS treatment. The Jmjd3-mediated H3K27me3 demethylation, we previously reported at the Bmp2 gene, in fact occurs with much slower kinetics (De Santa et al, 2007). We surmise that the slow rate of the H3K27me3 demethylation reaction, which in vitro requires a high enzyme-to-substrate ratio and a long incubation time (Agger et al, 2007; De Santa et al, 2007; Lan et al, 2007; Lee et al, 2007), combined with the short duration of the encounter with Jmjd3, makes Jmjd3-mediated H3K27me3 demethylation in the very first hours after LPS extremely unlikely. The few kinetic studies published insofar for JmjC-catalysed histone demethylation reactions are compatible with this interpretation, as they reported very slow substrate turnover rates (0.01 min−1) (Culhane and Cole, 2007).
Finally, it should be noticed that increased H3K27 methylation (appearance of new peaks or BLOCs and increased intensity of the pre-existing ones) was more common than its loss or reduction (Figure 3B; Supplementary Table VII). This increase in H3K27 methylation was however strictly gene specific and occurred without any global change in H3K27me3 and Ezh2 levels, which remain both constant after LPS stimulation (De Santa et al, 2007).
Overall, these data suggest that in this system and in this window of time Jmjd3 regulates transcription independently of H3K27me3 demethylation. This possibility is consistent both with the previous observation that most genes downregulated in macrophages depleted of Jmjd3 by retroviral RNA interference were not obvious or reported polycomb targets (De Santa et al, 2007), and with the demethylation-independent control of selected target genes during neural commitment (Burgold et al, 2008).
Jmjd3 contribution to the transcriptional program of activated macrophages
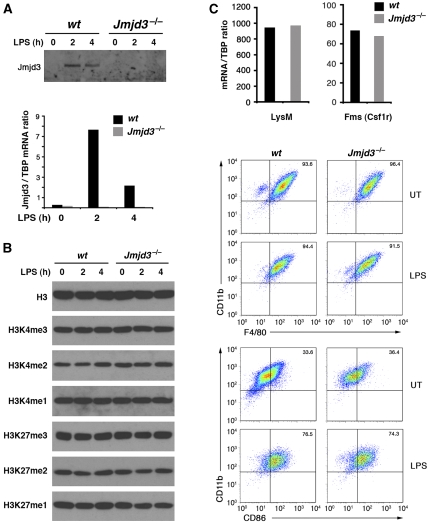
To determine the functional impact of Jmjd3 absence on the gene expression program triggered by LPS stimulation, we derived macrophages from the foetal livers (14.5 dpc) of Jmjd3 knockout mice generated by gene trapping (which will be described elsewhere). Macrophages obtained from these mice lacked Jmjd3 mRNA and protein (Figure 4A). Judging from a western blot analysis, the bulk H3K27me3/2/1 and H3K4me3/2/1 levels were not grossly affected by the absence of Jmjd3 (Figure 4B). Moreover, macrophage differentiation and responsiveness to LPS activation was unperturbed, as indicated by the macrophage markers tested (Figure 4C).
Figure 4.
Analysis of histone methylation and differentiation in Jmjd3−/− macrophages. Macrophages derived from the foetal livers of Jmjd3−/− mice were analysed for Jmjd3 protein and mRNA expression (A) and histone methylation at H3K4 and H3K27 (B). Differentiation and activation markers in Jmjd3−/− macrophages are shown in (C). Upper panel: mRNA levels of macrophages-restricted LysM and Fms genes. Lower panel: FACS analysis of the macrophage markers CD11b and F4/80, and the activation marker CD86 before and after LPS stimulation.
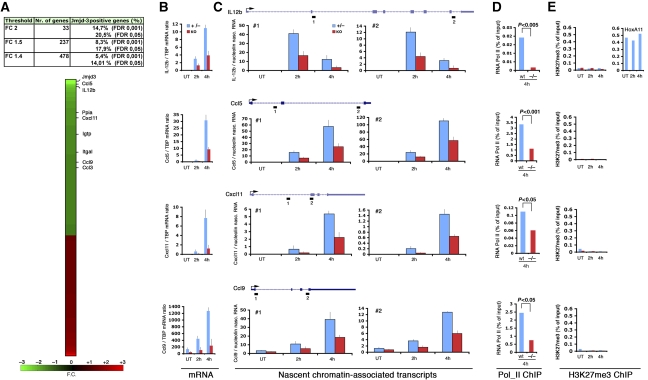
Microarray analyses (Affymetrix 1.0 ST arrays) were performed in triplicate using RNA from biological replicates of LPS+IFNγ-treated (4 h) wt or Jmjd3−/− foetal liver-derived macrophages. Using as cutoff a fold change (FC) of 2, only 33 genes were differentially expressed in Jmjd3−/− cells, of which 20.5% were direct Jmjd3 targets. Considering a FC of 1.5 the expression of 237 genes was affected, whereas 478 genes were influenced by Jmjd3 deletion when a threshold of 1.4 was applied (Figure 5A; Supplementary Table IX). The percentage of direct Jmjd3 targets remained comparable at all thresholds. The ratio of direct versus indirect targets is similar to that previously shown for other coregulators: for instance only 10% of genes that are differentially expressed on depletion of polycomb proteins are their direct targets (Bracken et al, 2006). Data were validated by quantitative RT–PCR on a panel of 20 genes (Figure 5B and data not shown), indicating a high reliability of the results of the microarray. Overall, the fact that the reduction in the selected threshold resulted in large increases in the amount of affected genes, whereas the percentage of direct Jmjd3 targets remained comparable suggests that at most genes only small transcriptional effects are brought about by Jmjd3, consistent with the idea that similarly to other transcriptional coregulators of the JmjC family, Jmjd3 tunes the transcriptional output without being absolutely necessary for the expression of any gene.
Figure 5.
Jmjd3 contribution to gene expression in LPS-activated macrophages. (A) Summary of the microarray results, indicating the number of genes that are differentially expressed in wt versus Jmjd3−/− macrophages at various thresholds and their binding to Jmjd3. The position of selected Jmjd3 targets on the heat plot is indicated. (B) mRNA levels for selected differentially expressed Jmjd3 target genes was quantified by qPCR. (C) Analysis of nascent transcripts for the same genes was carried out using two primer sets for each gene (represented as small rectangles numbered 1 and 2 in the gene diagrams). Data are expressed relative to the nascent transcripts of the housekeeping nucleolin gene. RNA Pol_II ChIP (D) and H3K27me3 ChIP (E) in wt and Jmjd3−/− macrophages. H3K27me3 levels at HoxA11 are shown as a positive ChIP control and used as reference.
We next focused on a set of differentially expressed genes, namely Ccl5 (FC -2,7), Il12b (FC -2,3), Cxcl11 (FC -1,63) and Ccl9 (FC -1,48) to analyse in more detail the impact of Jmjd3 deletion on their transcription and the correlation between transcriptional changes and alterations in H3K27me3 and H3K4me3 levels. In the case of Il12b, Jmjd3 binding was detected just upstream of a recently mapped inducible DNAse hypersensitive site (Zhou et al, 2007), whereas the other genes showed signals associated with their TSS. The reduction in mRNA levels (Figure 5B) correlated well with a reduction in elongating RNA Pol_II detected across these genes both at 2 and 4 h poststimulation, as measured by the analysis of nascent (unspliced) chromatin-associated transcripts (Figure 5C). Analysis of Pol_II recruitment to the TSSs of the same genes using ChIP showed a comparable reduction in signals in knockout cells (Figure 5D), although early Pol_II recruitment to Ccl5 was apparently unaffected (data not shown). Therefore, it seems that absence of Jmjd3 results in a defective recruitment of Pol_II followed by a reduction in the amount of Pol_II molecules elongating across target genes. Although the functional relationship and similarities between Jmjd3 and its paralog Utx are unclear at present, it is worth noticing that in Drosophila melanogaster Utx colocalizes with the elongating form of RNA_Pol II, thus suggesting an active role in ongoing transcription (Smith et al, 2008).
Analysis of H3K27me3 at the same genes showed that this histone modification is absent at their promoters and surrounding regions (Figures 3A and 5E). Most importantly, Jmjd3 deletion did not result in any increase whatsoever in the H3K27me3 signals, indicating that Jmjd3 does not act to prevent an increase in H3K27me3 after stimulation (Figure 5E). This was confirmed for several others Jmjd3 target genes (Supplementary Figure 9). We also checked the behaviour of H3K27me3 at Nos2 and Upp1, which are Jmjd3 targets associated with high H3K27me3 levels that rapidly decrease after stimulation (see Figure 3). Consistent with the previous observation that this reduction in H3K27me3 is due to nucleosome loss rather than demethylation, Jmjd3 deletion did not cause any detectable change in the profiles of H3K27me3 at these genes (Supplementary Figure 9). Finally, H3K4me3 signals at target genes were largely unaffected by Jmjd3 deletion (Supplementary Figure 10). Taken together, these data show that the transcriptional effects of Jmjd3 depletion on target genes in macrophages in the early phases of LPS stimulation are independent from measurable effects on H3K27me3.
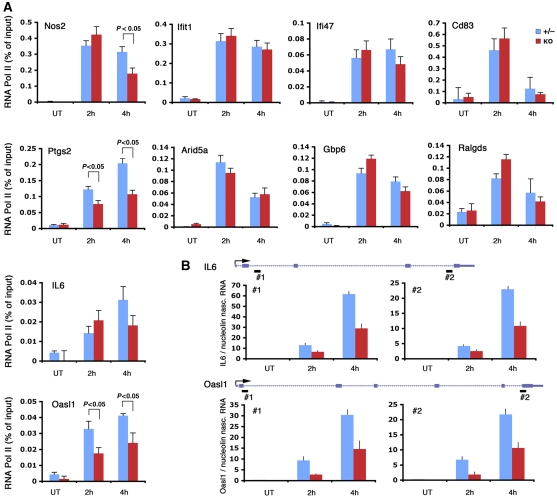
An interesting outcome of this genome-wide analysis is that most genes bound by Jmjd3 do not show obvious transcriptional changes in Jmjd3−/− macrophages. A simple possibility is that functional redundancy provided by other coregulators may prevent the occurrence of transcriptional defects. An alternative (not mutually exclusive) possibility is that the small magnitude of transcriptional changes coupled with the stability of the mature mRNA may mask the transcriptional changes occurring as a consequence of Jmjd3 deletion. Indeed, it has been shown that stabilization of inducible mRNAs by inflammatory cytokines is a major determinant of final mRNA levels, often overriding the consequences of transcriptional changes (Hao and Baltimore, 2009). We therefore analysed Pol_II recruitment and nascent transcripts at a panel of validated Jmjd3 target genes (Il6, Oasl1, Ptgs2, Nos2, CD83, Gbp6, Arid5a, Ifit1) that do not seem to be significantly affected by Jmjd3 deletion in the microarray analysis. We found that at a subset of these genes (Il6, Oasl1, Nos2 and Ptgs2) Pol_II recruitment was reduced in Jmjd3 knockout cells (Figure 6A); for Il6 and Oasl1 we confirmed this effect at the level of nascent transcripts (Figure 6B). However, at the other genes tested, Jmjd3 deletion was devoid of any consequences on Pol_II (Figure 6A), thus indicating either that Jmjd3 does not exert any role in their activation or that the system is equipped with mechanisms providing functional redundancy. Therefore, in some cases small transcriptional impairments because of the absence of Jmjd3 may not be apparent at the mRNA level, most likely because of the effects of mRNA stabilization.
Figure 6.
RNA_Pol II and nascent transcript analyses show transcriptional effects of Jmjd3 that are not apparent at the mRNA level. (A) Pol_II ChIP was carried out at a panel of validated Jmjd3 target genes. P-values are indicated when statistical significance is reached. (B) Nascent transcript analysis at the IL-6 and Oasl1 genes. Two primer sets corresponding to the 5′ and 3′ of each gene were used, as indicated by black rectangles (#1 and #2).
Discussion
Jmjd3-mediated transcriptional control in LPS-activated macrophages
In this study we analysed the contribution of Jmjd3 to the gene expression program of LPS-activated mouse macrophages. The main findings of this study are the following: (i) Jmjd3 showed a widespread association with the TSSs of active (Pol_II- and H3K4me3-positive) and inducible genes; (ii) most Jmjd3 target genes were unaffected by its absence; (iii) a few hundred genes underwent mild (less than two-fold) changes in their mRNA levels in Jmjd3−/− macrophages, whereas only a handful of genes such as Il12b and Ccl5 showed a stronger dependence on Jmjd3 for full activation; (iv) no gene was completely dependent on Jmjd3 for induction or expression; (v) transcriptional effects of Jmjd3 seemed to be largely or entirely independent of H3K27me3 demethylation.
The counterintuitive combination of a widespread distribution and limited transcriptional effects is common to several transcriptional coregulators (and particularly histone-modifying enzymes) associated with the TSSs of protein-coding genes, and specifically components of the MLL/SET H3K4 HMT complexes and polycomb group (PcG) proteins. In budding yeast, deletion of Set1 (the only H3K4 HMT in the genome of Saccharomyces cerevisiae) caused very limited effects on transcription, with only 20 genes affected by more than 1.5-fold (Miller et al, 2001), in spite of its widespread association with the TSS of active genes (Ng et al, 2003). Menin, a component of a subset of MLL complexes (Yokoyama et al, 2004) is broadly associated with the TSSs of active genes; however, in all cell types analysed its depletion did not cause significant changes in transcript levels (Scacheri et al, 2006). Depletion of the MLL and SET complex component Ash2L, which is required for the conversion of H3K4me2 to the trimethylated state, was shown to cause only a mild reduction in splicing rates without any detectable impact on Pol_II recruitment and transcript levels (Sims et al, 2007).
The same behaviour was reported for PcG proteins, which are broadly associated with silent or repressed genes. Depletion of the H3K27 HMT Ezh2 in human fibroblasts altered by more than 1.2-fold the expression of about 350 genes, of which about 10% were direct targets. Most of the 1000 genes bound by Ezh2, however, were not even marginally affected at the transcriptional level (Bracken et al, 2006).
Apart from the intrinsic tendency of several coregulators to bring about small transcriptional changes, in the specific system and response we investigated an additional factor, namely mRNA stabilization, likely contributes to reduce the extent of detectable effects of coregulator depletion on mRNA levels: as it has been recently shown, mRNA stabilization in response to inflammatory cytokines is a major determinant of final transcript levels, in some cases overriding or masking changes in transcription rates (Hao and Baltimore, 2009).
Given these premises, what may be the biological impact of a transcriptional regulator such as Jmjd3 that brings about mainly modest transcriptional effects? It is tempting to speculate that in analogy to other coregulators, Jmjd3 may have a final impact on the system that reflects the combination of a fairly large amount of simultaneous small changes, rather than a limited number of effects of high intensity (like those observed with several sequence-specific transcription factors). A similar concept has been proposed to account for the biological effects of microRNAs, which in most cases change mRNA and protein levels of dozens or hundreds of targets by no more than 10–20% (Baek et al, 2008).
Although a few hundreds of genes are affected only modestly (less than two-fold) by Jmjd3 deletion, a handful of genes show a higher dependence on Jmjd3 for their expression, as exemplified by Ccl5 and IL12b (which mediate recruitment and Th1 polarization of T lymphocytes, respectively). Overall, the list of genes whose expression is directly or indirectly affected by Jmjd3 deletion includes several known players of the inflammatory and immune response such as chemokines (Ccl3, Ccl4, Ccl5, Ccl9, Cxcl11), cytokines (e. g. Il12b, Il6) and antimicrobial molecules (e. g. the GTPase Igtp); however, it is clear that defining the biological impact of Jmjd3 on the response to microbes (and inflammatory responses in general) will require detailed in vivo analyses in mutant mice.
Stability of the H3K27me3 mark in activated macrophages
Although the long-standing dogma that histone methylation is a non-reversible modification has been broken down by the identification of HDMs (Shi et al, 2004; Tsukada et al, 2006), biochemical studies support the concept that the turnover of this modification is overall rather slow (Byvoet et al, 1972). Moreover, in some specific case it was shown that histone methylation is extremely stable: for instance, on nuclear somatic transfer into oocytes, H3K9me3 in the donor nucleus is not erased over several days (Santos et al, 2003).
For some of the 27 JmjC family proteins encoded in the human genome, the concept that methylated lysines in histone tails are the relevant substrates linking these enzymes to their biological effects is supported by several pieces of evidence: the ability to demethylate histone amino-terminal peptides, isolated histones and in some cases assembled nucleosomes in vitro and in vivo; the incorporation in multi-protein complexes mediating chromatin modifications and transitions; and increased gene- and site-specific histone lysine methylation after depletion of individual JmjC proteins (Klose and Zhang, 2007; Shi and Whetstine, 2007; Cloos et al, 2008). In the case of rbr-2, the Caenorhabditis elegans ortholog of the H3K4me3 demethylase Jarid1a/Rbp2, mutant worms expressing a protein lacking the JmjC domain showed increased levels of H3K4me3, suggesting a global role for rbr-2 in balancing the effects of the cognate H3K4 HMTs (Christensen et al, 2007). Similarly, in yeast the H3K4 methyltransferase Set1 and the JmjC protein Jhd1 act as an antagonistic pair regulating global levels of H3K4me3/2 (Seward et al, 2007).
However, the possibility that non-histone substrates and/or non-enzymatic activities (such as structural functions in multi-molecular complexes) of HMTs and HDMs may mediate several biological actions of these enzymes is also slowly emerging (Huang and Berger, 2008). For instance, the methylation state of selected lysine residues in p53 is controlled by the H3K4me2/1-specific demethylase LSD1 (Huang et al, 2007). It is clear that though the definitive identification of the whole complement of substrates of each JmjC protein will represent a complex and long-lasting endeavour, the assumption that histones represent the only substrate on which they act is restrictive and likely to be overall incorrect.
Overall, our data show that H3K27 trimethylation is a stable histone modification in the first hours after LPS activation of macrophages, which is consistent with its main role in the enforcement of differentiation and maintenance of cell type-specific gene expression programs. Specifically, changes in H3K27me3 peaks in the first 4 h after LPS stimulation occurred in both directions, increments being more common than decrements. When considering broad regions of H3K27me3 enrichment, only 3 of them showed a reduction higher than two-fold in response to stimulation, whereas 113 increased by more than two-fold. Although increased H3K27me3 may reflect gene repression and silencing associated with LPS-triggered terminal differentiation, understanding the nature of decrements and the involvement of Jmjd3 in their occurrence requires a careful analysis. More than 10 000 H3K27me3 peaks (out of a total of almost 60 000 detected genome wide) underwent a two-fold (or more) reduction after LPS stimulation. Considering the group of the 511 H3K27me3 peaks associated with Jmjd3, 83 of them showed a more than two-fold decrease. The respective percentage of H3K27me3 decrease in the two groups was 17.6 and 16.3%, indicating that contiguity of H3K27me3 peaks to Jmjd3 peaks does not increase the chances that H3K27me3 is erased. What is even more important is to determine whether the detected decrements in H3K27me3 peaks reflected true enzymatic demethylation events. In all cases we investigated, H3K27me3 reduction was entirely accounted for by nucleosome depletion associated with strong transcriptional induction. Consistent with this interpretation, H3K27me3 reduction at the Jmjd3-associated genes we investigated was entirely unaffected by Jmjd3 deletion. Therefore, currently we do not have any direct evidence that enzymatic H3K27me3 demethylation occurs in this early window of time. These observations strongly suggest that the early transcriptional program unfolding in the first hours after microbial stimulation occurs independently of H3K27me3 demethylation, although more data are required to make this conclusion definitive.
In this context, Jmjd3 is recruited to active TSSs and in the majority of cases it does not have the chance to encounter H3K27me3. Moreover, the genes most affected by its deletion (e.g. Ccl5) are H3K27me3 negative and do not show any increase in H3K27me3 in Jmjd3 ko cells. Overall, given the previous demonstration that Jmjd3 can demethylate H3K27me3, the most parsimonious interpretation of the results described in this study is that though in some specific contexts (such as developmental transitions requiring the derepression of H3K27me3 genes) (Agger et al, 2007; Jepsen et al, 2007; Burgold et al, 2008; Miller et al, 2008; Sen et al, 2008) Jmjd3 may act by erasing the H3K27me3 mark, in activated macrophages (in which Jmjd3 induction is rapid and transient) it exerts actions that are largely independent of H3K27me3 demethylation. At present, we have identified only a single gene (Bmp2) undergoing a Jmjd3-dependent reduction in H3K27me3 demethylation (De Santa et al, 2007), but it is important to notice that in this case the kinetics of the demethylation reaction was relatively slow (between 4 and 8 h after stimulus).
Whether the enzymatic activity of Jmjd3 is required to participate in the control of inflammatory gene expression is still an open question that remains to be experimentally addressed using suitable approaches. Although it is possible to reconstitute Jmjd3−/− macrophages with wt and catalytically inactive Jmjd3 using viral vectors (data not shown), we have been unable until now to reproduce the physiological, highly regulated temporal profile of its expression; moreover, levels of expression achieved this way are much higher than the endogenous ones, thus making such experiments not reliable.
Conclusions
The data shown here show that, in addition to its described roles in development and tissue renewal, Jmjd3 participates in the inflammatory transcriptional response induced by LPS stimulation in macrophages. Overall, Jmjd3 effects on the transcriptional output (which in this system are largely independent of H3K27me3 demethylation) are rather widespread but in most cases of low intensity, suggesting that its main role is to make fine adjustments to the transcription rates of several genes rather than to provide an essential contribution to their induction.
Materials and methods
Cell culture
Bone marrow cells isolated from female Fvb mice were plated in 10 cm plates in 5 ml of BM-medium (high glucose DMEM supplemented with 20% low-endotoxin foetal bovine serum, 30% L929-conditioned medium, 1% glutamine, 1% Pen/Strep, 0.5% sodium pyruvate, 0.1% β-mercaptoethanol). Cultures were fed with 2.5 ml of fresh medium every 2 days. Stimulations were carried out at day 7.
Jmjd3 knockout macrophages were obtained by differentiating foetal liver-derived cells obtained from E14.5 embryos (in a mixed 129SvEv-C57BL/6 background). Cells were first plated on bacterial (non-coated) plates for 2 days and then transferred to standard cell culture plates for additional 5–6 days before stimulation. Knockout mice will be described elsewhere. LPS from Escherichia coli serotype 055:B5 (Sigma) was used at 10 ng/ml; γIFN (R&D) was used at 10 UI/ml.
Chromatin immunoprecipitation
The ChIP-Seq libraries generated in this study are shown in Supplementary Table X. For the Jmjd3 ChIP-Seq experiment 5 × 108 macrophages were stimulated for 2 h with LPS+γIFN. Fixation with formaldehyde and sonication was carried out as described earlier (De Santa et al, 2007). The lysate was divided in five 3 ml aliquots for further processing. For all the other sequencing experiments, lysates (3 ml) were generated from 1 × 108 cells.
Each 1 × 108 aliquot was immunoprecipitated with 10 μg of the following antibodies: antiH3K4me3 (Abcam Ab8580), anti-H3K27me3 (Upstate Biotechnology #07–449), anti-Pol_II (anti-Rbp1, Santa Cruz Biotechnology, sc-899). The anti-Jmjd3 antibody was raised in rabbits against the C-terminus of the mouse protein and affinity purified. Antibodies were pre-bound overnight to 100 μl of G protein-coupled paramagnetic beads (Dynabeads) in PBS/BSA 0.5%. Beads were then added to lysates (the pre-clearing step was omitted) and incubation was let to proceed overnight. Beads were washed six times in a modified RIPA buffer (50 mM Hepes pH 7.6, 500 mM LiCl, 1 mM EDTA, 1% NP-40, 0.7% Na-deoxycholate) and once in TE containing 50 mM NaCl. DNA was eluted in TE containing 2% SDS and crosslinks reversed by incubation overnight at 65°C. DNA was then purified by Qiaquick columns (Qiagen) and quantified using PicoGreen (Invitrogen). Yields were ∼300 ng/108 cells (H3K4me3), ∼10 ng/108 cells (H3K27me3 and Pol_II), ∼2.5 ng/108 cells (Jmjd3). Primers used for validation by ChIP–qPCR are in Supplementary Table XI.
Preparation of ChIP DNA libraries, sequencing and computational analysis
ChIP DNA was prepared for Solexa 1G sequencing by simultaneously blunting, repairing and phosphorylating ends using a mixture of T4 DNA polymerase, DNA polymerase I and T4 kinase according to manufacturer's instruction (Illumina). The DNA was adenylated at 3′ end and recovered by Qiaquick PCR purification kit (Qiagen) according to the manufacturer's recommendations. Adaptors for Genome Analyzer were added by ligation and the ligated fragments were subjected to limited cycles (17 cycles) of PCR amplification and amplified fragments were gel purified by Qiagen columns. The purified DNA was quantified by picogreen (Invitrogen) and diluted to a working concentration of 10 nM. Cluster generation was performed and loaded into individual lanes of flow cell; 36 cycles of base incorporation were carried out on the Illumina 1G analyzer. We routinely generated over 10 million uniquely mapped tags for each ChIP sample and location of each ChIP fragment was inferred by extending 200 bp from the tag mapping location and regions covered by multiple overlapping ChIP fragments were considered as putative binding sites.
The ELAND program provided with the 1G analyzer software package was used to map 36 bp reads to mouse genome (mm8) allowing at most two mismatches. Only reads mapped uniquely in the genome were used for further analysis. The tag mapping coordinates were extended 200 bp to the 3′ direction to obtain ChIP DNA fragment locations on the mm8 genome. Thus a genome-wide intensity profile was obtained. The peak finding algorithm described in Chen et al (2008) was applied to this intensity profile to determine the reliable binding regions. Only peaks above a minimum intensity threshold were selected based on a computationally estimated FDR of 0.1%. Furthermore, FC of intensity against negative control library (anti-GFP) was used to filter out the high-intensity peaks in genomic regions with unusually high tag accumulation, such as satellite repeat regions.
The FDR for a library was determined by a Monte Carlo simulation, in which 200 bp fragments randomly (with a uniform distribution) extracted from the mm8 genome were used to estimate the numbers of random peaks with different intensity values. The number of random fragments is equal to the sequencing depth of the library. The FDR at an intensity p is estimated as the Rp/Op where Rp and Op are the number of random and real peaks, respectively, with Intensities ⩾p. The minimum intensity that satisfies the FDR criterion is selected as the lower cutoff for calling confident peaks.
To filter the peaks in ChIP DNA library against the negative control library (anti-GFP), the ratio of peak intensities in the ChIP DNA and control libraries was compared, and only peaks with an intensity FC ⩾5 were kept. The peak intensities were normalized with respect to sequencing depth in the comparison. Let IChIP(x) Icontrol(x) represent the intensity of a peak centred at genomic locus x in the ChIP DNA and control libraries, respectively. Then the FC for this peak is given by
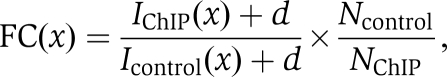 |
where NChIP and Ncontrol are, respectively, the sequencing depths of the ChIP DNA and negative control libraries, and d represents the Dirichlet prior which is set to 1 here. The FC cutoff of 5.0 is relatively stringent.
ChIP_sequencing data were deposited in the GEO public repository (http://www.ncbi.nlm.nih.gov/geo/) and can be retrieved using the accession number GSE17631.
Quantitative RT–PCR and nascent transcript analysis
RNA was extracted from macrophages using Trizol (Invitrogen) and reverse transcribed with random hexamers. For isolation of nascent transcripts, cells were lysed in HB buffer (10% glycerol, 60 mM KCl, 15 mM NaCl, 1.5 mM HEPES pH 7.9, 0.5 mM EDTA) containing 0.3 M sucrose and 0.8% NP40. Nuclei were then pelleted through a 0.9 M sucrose cushion in HB and resuspended in 100 μl of NRB (75 mM NaCl, 20 mM Tris–HCl pH 7.5, 0.5 mM EDTA, 50% glycerol, 100 μg/ml yeast tRNA) and lysed by addition of 750 μl of NLB (0.3 M NaCl, 20 mM HEPES pH 7.6, 0.2 mM EDTA, 7.5 mM MgCl2, 1 M urea, 1% NP-40, 100 μg/ml yeast tRNA). Chromatin was then pelleted in microfuge at 4°C and nascent transcripts extracted in Trizol. As control of the lack of genomic DNA contamination, qPCR was also carried out on RNA that was not reverse transcribed. The sequences of the primers used are in Supplementary Table XII.
Supplementary Material
Supplementary Material
Supplementary Tables I
Supplementary Tables II
Supplementary Tables III
Supplementary Tables IV
Supplementary Tables V
Supplementary Tables VI a
Supplementary Tables VI b
Supplementary Tables VII
Supplementary Tables VIII
Supplementary Tables IX
Supplementary Tables X
Supplementary Tables XI
Supplementary Tables XII
Review Process File
Acknowledgments
We thank Bruno Amati for critically reading the manuscript, and Elena Prosperini for excellent technical support. We also thank the sequencing team at GIS for the generation of high-quality sequencing data. This research was supported by grants from the European Community (Marie Curie Excellence Grant ‘Trans-Tar' to GN), the Italian Association for Research on Cancer (AIRC, to GN and GT), a donation from Manetti and Roberts and the Fondazione Umberto Veronesi (GN), the Armenise-Harvard Foundation (SC), the NIH ENCODE grant 1R01HG003521-01 (CLW) and the Agency for Science, Technology and Research (A*STAR) of Singapore.
Footnotes
GN is a consultant for the CEDD (centre of excellence in drug discovery) in immunoepigenetics of Glaxo Smith Kline.
References
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K (2007) UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449: 731–734 [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP (2008) The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K (2006) Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 20: 1123–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgold T, Spreafico F, De Santa F, Totaro MG, Prosperini E, Natoli G, Testa G (2008) The histone H3 lysine 27-specific demethylase Jmjd3 is required for neural commitment. PLoS ONE 3: e3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byvoet P, Shepherd GR, Hardin JM, Noland BJ (1972) The distribution and turnover of labeled methyl groups in histone fractions of cultured mammalian cells. Arch Biochem Biophys 148: 558–567 [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK et al. (2008) Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 [DOI] [PubMed] [Google Scholar]
- Christensen J, Agger K, Cloos PA, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, Helin K (2007) RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell 128: 1063–1076 [DOI] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Helin K (2008) Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev 22: 1115–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culhane JC, Cole PA (2007) LSD1 and the chemistry of histone demethylation. Curr Opin Chem Biol 11: 561–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G (2007) The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130: 1083–1094 [DOI] [PubMed] [Google Scholar]
- Foster SL, Hargreaves DC, Medzhitov R (2007) Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature 447: 972–978 [DOI] [PubMed] [Google Scholar]
- Hao S, Baltimore D (2009) The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat Immunol 10: 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS, West AP, Ghosh S (2006) NF-kappaB and the immune response. Oncogene 25: 6758–6780 [DOI] [PubMed] [Google Scholar]
- Honda K, Taniguchi T (2006) IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol 6: 644–658 [DOI] [PubMed] [Google Scholar]
- Huang J, Berger SL (2008) The emerging field of dynamic lysine methylation of non-histone proteins. Curr Opin Genet Dev 18: 152–158 [DOI] [PubMed] [Google Scholar]
- Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT, Jenuwein T, Berger SL (2007) p53 is regulated by the lysine demethylase LSD1. Nature 449: 105–108 [DOI] [PubMed] [Google Scholar]
- Ivashkiv LB, Hu X (2004) Signaling by STATs. Arthritis Res Ther 6: 159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepsen K, Solum D, Zhou T, McEvilly RJ, Kim HJ, Glass CK, Hermanson O, Rosenfeld MG (2007) SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature 450: 415–419 [DOI] [PubMed] [Google Scholar]
- Kirmizis A, Bartley SM, Kuzmichev A, Margueron R, Reinberg D, Green R, Farnham PJ (2004) Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev 18: 1592–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y (2007) Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol 8: 307–318 [DOI] [PubMed] [Google Scholar]
- Koehntop KD, Emerson JP, Que L Jr (2005) The 2-His-1-carboxylate facial triad: a versatile platform for dioxygen activation by mononuclear non-heme iron(II) enzymes. J Biol Inorg Chem 10: 87–93 [DOI] [PubMed] [Google Scholar]
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, Roberts TM, Chang HY, Shi Y (2007) A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449: 689–694 [DOI] [PubMed] [Google Scholar]
- Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R (2007) Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 318: 447–450 [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K et al. (2006) Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loenarz C, Schofield CJ (2008) Expanding chemical biology of 2-oxoglutarate oxygenases. Nat Chem Biol 4: 152–156 [DOI] [PubMed] [Google Scholar]
- Medzhitov R (2008) Origin and physiological roles of inflammation. Nature 454: 428–435 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C et al. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Huang AC, Miazgowicz MM, Brassil MM, Weinmann AS (2008) Coordinated but physically separable interaction with H3K27-demethylase and H3K4-methyltransferase activities are required for T-box protein-mediated activation of developmental gene expression. Genes Dev 22: 2980–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A (2001) COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci USA 98: 12902–12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K (2003) Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11: 709–719 [DOI] [PubMed] [Google Scholar]
- Pauler FM, Sloane MA, Huang R, Regha K, Koerner MV, Tamir I, Sommer A, Aszodi A, Jenuwein T, Barlow DP (2009) H3K27me3 forms BLOCs over silent genes and intergenic regions and specifies a histone banding pattern on a mouse autosomal chromosome. Genome Res 19: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J (2007) Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 25: 15–30 [DOI] [PubMed] [Google Scholar]
- Santos F, Zakhartchenko V, Stojkovic M, Peters A, Jenuwein T, Wolf E, Reik W, Dean W (2003) Epigenetic marking correlates with developmental potential in cloned bovine preimplantation embryos. Curr Biol 13: 1116–1121 [DOI] [PubMed] [Google Scholar]
- Scacheri PC, Davis S, Odom DT, Crawford GE, Perkins S, Halawi MJ, Agarwal SK, Marx SJ, Spiegel AM, Meltzer PS, Collins FS (2006) Genome-wide analysis of menin binding provides insights into MEN1 tumorigenesis. PLoS Genet 2: e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA (2008) Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev 22: 1865–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seward DJ, Cubberley G, Kim S, Schonewald M, Zhang L, Tripet B, Bentley DL (2007) Demethylation of trimethylated histone H3 Lys4 in vivo by JARID1 JmjC proteins. Nat Struct Mol Biol 14: 240–242 [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119: 941–953 [DOI] [PubMed] [Google Scholar]
- Shi Y, Whetstine JR (2007) Dynamic regulation of histone lysine methylation by demethylases. Mol Cell 25: 1–14 [DOI] [PubMed] [Google Scholar]
- Sims RJ III, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D (2007) Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell 28: 665–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ER, Lee MG, Winter B, Droz NM, Eissenberg JC, Shiekhattar R, Shilatifard A (2008) Drosophila UTX is a histone H3 Lys27 demethylase that colocalizes with the elongating form of RNA polymerase II. Mol Cell Biol 28: 1041–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M (2006) Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer 6: 846–856 [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y (2006) Histone demethylation by a family of JmjC domain-containing proteins. Nature 439: 811–816 [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, Herr W, Cleary ML (2004) Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol 24: 5639–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung WK, Shahab A, Kuznetsov VA, Bourque G, Oh S, Ruan Y, Ng HH, Wei CL (2007) Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell 1: 286–298 [DOI] [PubMed] [Google Scholar]
- Zhou L, Nazarian AA, Xu J, Tantin D, Corcoran LM, Smale ST (2007) An inducible enhancer required for Il12b promoter activity in an insulated chromatin environment. Mol Cell Biol 27: 2698–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Supplementary Tables I
Supplementary Tables II
Supplementary Tables III
Supplementary Tables IV
Supplementary Tables V
Supplementary Tables VI a
Supplementary Tables VI b
Supplementary Tables VII
Supplementary Tables VIII
Supplementary Tables IX
Supplementary Tables X
Supplementary Tables XI
Supplementary Tables XII
Review Process File