Abstract
Circadian rhythms are modeled as reliable and self-sustained oscillations generated by single cells. The mammalian suprachiasmatic nucleus (SCN) keeps near 24-h time in vivo and in vitro, but the identity of the individual cellular pacemakers is unknown. We tested the hypothesis that circadian cycling is intrinsic to a unique class of SCN neurons by measuring firing rate or Period2 gene expression in single neurons. We found that fully isolated SCN neurons can sustain circadian cycling for at least 1 week. Plating SCN neurons at <100 cells/mm2 eliminated synaptic inputs and revealed circadian neurons that contained arginine vasopressin (AVP) or vasoactive intestinal polypeptide (VIP) or neither. Surprisingly, arrhythmic neurons (nearly 80% of recorded neurons) also expressed these neuropeptides. Furthermore, neurons were observed to lose or gain circadian rhythmicity in these dispersed cell cultures, both spontaneously and in response to forskolin stimulation. In SCN explants treated with tetrodotoxin to block spike-dependent signaling, neurons gained or lost circadian cycling over many days. The rate of PERIOD2 protein accumulation on the previous cycle reliably predicted the spontaneous onset of arrhythmicity. We conclude that individual SCN neurons can generate circadian oscillations; however, there is no evidence for a specialized or anatomically localized class of cell-autonomous pacemakers. Instead, these results indicate that AVP, VIP, and other SCN neurons are intrinsic but unstable circadian oscillators that rely on network interactions to stabilize their otherwise noisy cycling.
Keywords: luciferase, pacemaker, Period gene, suprachiasmatic nucleus, vasoactive intestinal polypeptide
Circadian pacemakers are schematized as intracellular transcription–translation negative feedback loops (1). In mammals, transcription factors including CLOCK and BMAL1 promote the expression of clock genes, including Period 1 (Per1) and 2 (Per2). The protein products of these genes return to the nucleus after a delay of many hours to repress their own transcription. Genetic deletion of these repressors abolishes circadian rhythms in behavior and physiology (2). The strongest evidence for cell-autonomous, circadian rhythm generation in mammals comes from transcriptional rhythms measured from primary and immortalized fibroblasts (3, 4).
The mammalian suprachiasmatic nucleus (SCN) of the anterior hypothalamus coordinates daily rhythms including sleep–wake and hormone release (5). Multielectrode array (MEA) recordings of neuronal firing and luciferase-based reporters of Per1 and Per2 expression showed dissociated SCN neurons in the same culture with different circadian periods (6, 7). Furthermore, Na+-dependent action potentials, vasoactive intestinal polypeptide (VIP), and its receptor, VPAC2, are required for cellular synchrony and maintaining daily oscillations across the SCN (8, 9). Taken together, these results suggest that single SCN neurons are competent circadian oscillators. However, which, if any, SCN neurons are capable of autonomous rhythmicity in the absence of input from other cells is unknown.
Several classes of neurons within the SCN have been proposed to be intrinsically circadian (10). Two neuropeptides, arginine vasopressin (AVP) and VIP, are strong candidates for labeling SCN pacemaking neurons. Approximately 10% of the ≈10,000 neurons in the unilateral SCN are VIP-ergic and found primarily in the ventral SCN; AVP-ergic neurons make up a separate 20% of the population found mostly in the dorsomedial SCN (11). Because AVP and VIP release can have different circadian periods in the same organotypic SCN slice, it was hypothesized that these 2 groups of neurons are separate oscillators (12). Subsequent in vivo (13) and in vitro (8, 14) results continue to support the idea that neurons from the dorsal and ventral SCN maintain daily rhythms. We sought to identify intrinsically circadian neurons within the SCN as AVP- or VIP-ergic.
Here we demonstrate circadian rhythms in gene expression and firing rate from isolated SCN neurons and their subsequent identification by immunocytochemistry. We find intrinsically rhythmic AVP and VIP neurons. However, not all AVP or VIP SCN neurons are circadian. Furthermore, SCN neurons isolated from their network either physically or by tetrodotoxin (TTX) can lose or gain rhythmicity, suggesting that SCN neurons are a heterogeneous population of intrinsic but unstable circadian oscillators.
Results
Circadian PER2 Expression Is Rare in Isolated SCN Neurons.
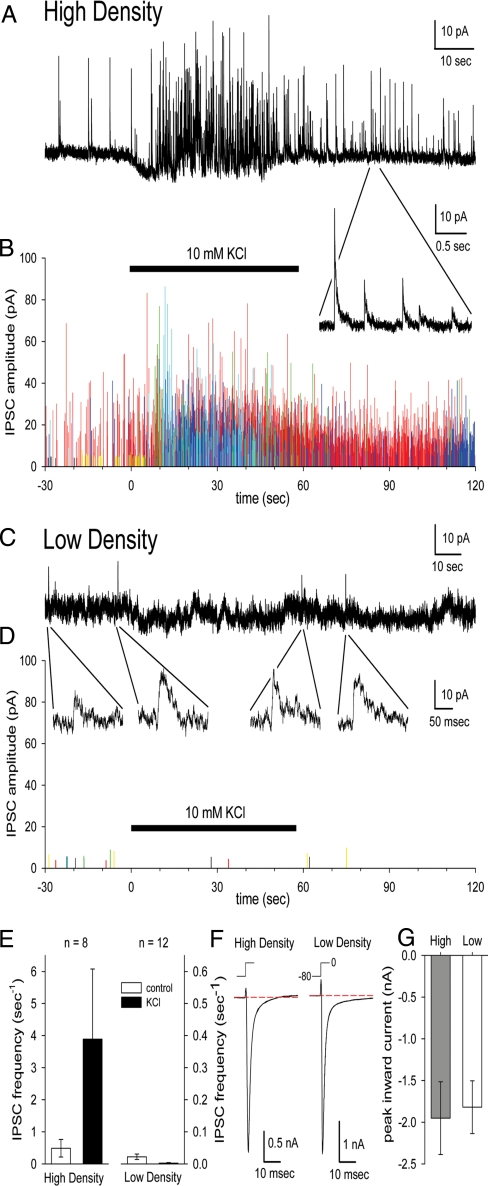
We tested the hypothesis that there is a class of specialized, intrinsically circadian neurons within the SCN. Plating SCN neurons at 10,000 cells/mm2 yields a population containing nearly 80% circadian neurons with a similar period (15), and plating at 3,000 cells/mm2 produces a population of SCN neurons in which ≈50% are circadian with a broad range of periods in the same culture (6, 16, 17). Because inhibitory synaptic currents were still present even in the low-density cultures (6), we tested whether plating at 100 cells/mm2, a density less than 3% of previous methods, would allow simultaneous recordings from cells in which intercellular communication is eliminated. Culture conditions were optimized by growing cells in glial-conditioned medium (Materials and Methods), so that recordings from viable neurons could begin after 3 days in vitro and continue for at least 6 days (Fig. 1A). Using whole-cell patch, we found that these neurons had normal sodium currents but showed few or no postsynaptic currents compared with cultures plated at 10,000 cells/mm2, even in the presence of elevated potassium (Fig. 2). We found anatomic markers suggesting that some neurons synapsed on themselves in very-low-density cultures but no evidence for intercellular communication (supporting information Fig. S1D). These data suggest that, in contrast to previous recordings of dispersed SCN neurons that exhibited synaptic currents (6), these very-low-density cultures lack intercellular communication.
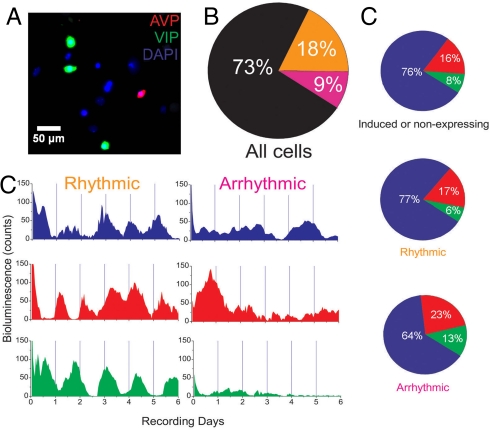
Fig. 1.
Intrinsic circadian rhythmicity is rare and found in diverse classes of SCN neurons. (A) A representative region of a recorded and subsequently immunolabeled very-low-density culture shows SCN neurons containing AVP (red), VIP (green), or neither neuropeptide (blue). (B) Of the neurons recorded over 6 days, 73% (n = 1,027 of 1,413 from 14 cultures) expressed PER2-mediated bioluminescence for <3 days (black), 18% were circadian (orange), and 9% were arrhythmic (pink). Most neurons with sustained PER2 expression (n = 252 of 386 in 14 cultures) were circadian (orange). (C) Representative bioluminescence traces from single identified SCN neurons show circadian or arrhythmic PER2 expression. (D) Neurons that did not express (Top), or showed circadian (Middle) or arrhythmic (Bottom) PER2 profiles were similar in their expression of AVP (red), VIP (green), or neither (blue). Thus, multiple cell types were intrinsically circadian, and neuropeptide expression did not distinguish between rhythmic and arrhythmic neurons.
Fig. 2.
SCN neurons grown at very low density are healthy and functionally isolated. (A) Representative whole-cell recording of spontaneous IPSCs at 0 mV from a neuron 7 days after plating at ≈10,000 cells/mm2. Inset shows IPSCs at higher temporal resolution. (B) IPSC amplitudes and frequencies were high over 2 min in 8 neurons from 1 culture (data from each neuron in a different color) grown at high density. Bar indicates the time of 10 mM KCl exposure in A and B. Note the increase in evoked IPSCs. (C) Representative whole-cell recording of spontaneous IPSCs at 0 mV from a neuron 7 days after plating at ≈100 cells/mm2. Insets show small current events that may reflect rare, spontaneous IPSCs. (D) Plots of IPSC amplitude in 12 neurons combined from 2 cultures grown at low density. (E) KCl elevation boosted IPSC frequency in high-density but not low-density cultures. (F) Inward sodium currents evoked by voltage steps from −80 mV to 0 mV in neurons grown at high and low density. (G) Peak inward sodium current amplitude ± SEM in neurons grown at high density (n = 8) and low density (n = 12).
We recorded bioluminescence for at least 6 days from individual SCN neurons cultured from PERIOD2::Luciferase (PER2::LUC) homozygous knock-in mice (18). Surprisingly, we found that most cells luminesced for <72 h (73%, n = 1,027 of 1,413 neurons; Fig. 1B). This was highly reproducible between cultures, so that the average culture had 71% ± 4% cells (mean ± SD, n = 14 cultures) that expressed PER2 for <3 days. These cells were viable at the end of the recording according to phase contrast imaging, neuropeptide immunolabeling, and nuclear staining. In addition, 10 μM forskolin, a cAMP analog, increased the percentage of PER2-expressing neurons after 6 days of recording to the 70% observed on the first day of recording (Fig. S1B).
Of PER2-expressing neurons, the majority were circadian (65%, n = 252 of 386; Fig. 1 C and D). Rhythmic neurons had periods from 15 to 35 h with no correlation between neighbors (closest neurons were more than 50 μm apart; Fig. S2) and randomly distributed times of peak bioluminescence (Rayleigh test, r = 0.21 ± 0.09, mean ± SD from 14 cultures, P > 0.1). Higher mean bioluminescence, however, did not necessarily equate with rhythmicity (Fig. S3B). Thus, we found that, in contrast to the synchronous rhythms seen in densely packed SCN neurons (15), plating SCN neurons at very low density reveals that intrinsic PER2 expression is lower in all cells and circadian in ≈18% of neurons (252 of 1,413; 16% ± 2%, mean ± SD from 14 cultures).
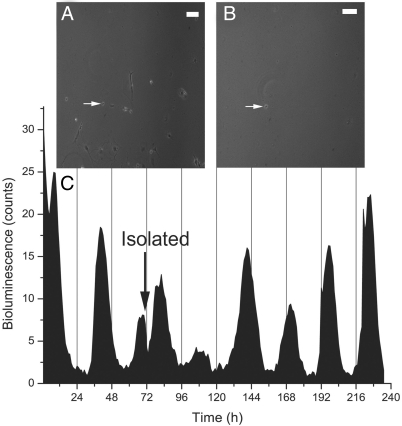
We next physically isolated single, bioluminescent neurons by removing all other cells in the dish. We attempted to isolate and record for 5 days from 12 neurons. We selected for cycling neurons and found that 3 of 5 remained circadian for as long as we recorded (6 days; Fig. 3). An additional 2 neurons were circadian, but at least 1 other cell was discovered in the dish at the end of the recording. The remaining 5 recorded neurons did not survive the isolation procedure for at least 5 days. Thus, ≈60% of PER2-expressing cells were circadian in low-density cultures and when isolated. This suggests that rhythms recorded from SCN neurons plated at very low densities are equivalent to recordings from physically isolated neurons. Together, these results provide the best evidence to date that circadian rhythm generation is intrinsic to only a fraction of SCN neurons at a given time.
Fig. 3.
Circadian rhythms in a fully isolated SCN neuron. A single, representative SCN neuron (white arrow) recorded before (A) and after (B) removal of all other cells. (C) PER2-driven bioluminescence from this neuron had an average period of ≈27.7 h for the last 5 days after isolation (black arrow). This SCN neuron expressed neither AVP nor VIP.
Firing Rate Rhythms in Functionally Isolated SCN Neurons.
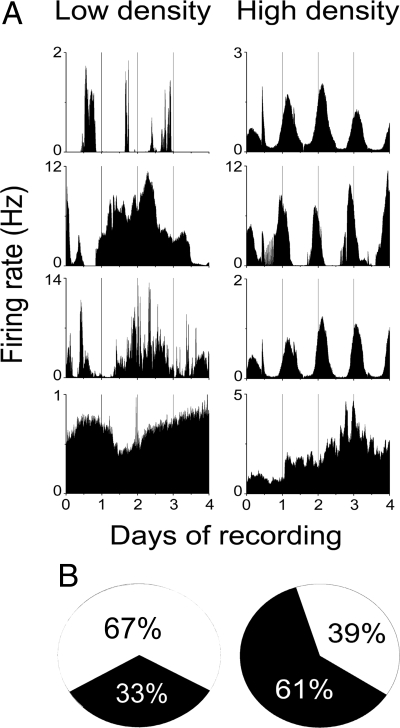
Because we cannot exclude the possibility that PER2 or other cellular processes oscillate under conditions when PER2::LUC fails to report rhythms, we investigated an additional, independent reporter of activity in SCN neurons. Extracellular action potentials were recorded from neurons 1 week after plating at 300–700 cells/mm2 on MEAs. We found that 33% of SCN neurons that discharged action potentials for at least 4 days showed circadian rhythms in firing rate (n = 6 of 18 neurons recorded from 5 cultures; Fig. 4), consistent with the small percentage of SCN neurons that showed intrinsic PER2 rhythms. Our findings that 18–33% of neurons grown in near isolation remain circadian are consistent with the reported 23–45% of circadian SCN neurons in explants and high-density dispersals lacking VIP, VPAC2, or Gi signaling (15, 19). Although the chance remains that circadian rhythms persisted in neurons with no or arrhythmic PER2 or firing, the results indicate that the majority of SCN neurons lose circadian cycling when removed from network interactions. We conclude that single neurons are competent circadian pacemakers that can drive rhythms in firing rate and that intercellular communication dramatically augments the proportion of rhythmic SCN cells.
Fig. 4.
Compared with SCN neurons plated at high density, only a subset of nearly-isolated SCN neurons maintain circadian firing rate rhythms. (A) Firing rate traces over 4 days from representative neurons plated and recorded on multielectrode arrays at extremely low density show unstable circadian rhythms (top 2 traces, Left) compared with the more precise circadian patterns in high-density cultures (top 3 traces, Right). Arrhythmic firing patterns were more common in low-density cultures (bottom 2 traces, Left) than in high-density cultures (bottom trace, Right). (B) The proportion of circadian (black) and arrhythmic (white) SCN neurons from low-density cultures compared with high-density cultures. Note that firing rate, like PER2, rhythms were intrinsic to a small fraction of neurons in low-density cultures.
Both Circadian and Noncircadian SCN Neurons Express AVP or VIP.
To determine the identity of intrinsically rhythmic neurons, we examined the expression of AVP and VIP after PER2::LUC profiling. The percentages of AVP- or VIP-ergic neurons were similar in very-low-density (100 cells/mm2) and control (3,000–5,000 cells/mm2) SCN cultures (20% and 17% of neurons were AVP positive, respectively; 8% and 8% were VIP positive, respectively; 2,608 neurons counted in 5 control cultures and 555 neurons counted in 5 very-low-density cultures). Because these are similar to the proportions reported in the mouse SCN in vivo (11), we conclude that these classes of neuropeptidergic neurons survive in SCN cultures.
We found that 23% of circadian cells (n = 25 of 109 in 5 cultures examined) expressed either AVP or VIP, whereas the majority (n = 84 of 109) expressed neither AVP nor VIP (Fig. 1D). Some of the cells that did not express VIP or AVP in the low-density cultures may not have been neurons. For example, astrocytes can express circadian rhythms in PER2 (20) but were rare in these cultures according to phase contrast imaging. Importantly, the proportion of circadian cells was similar when comparing PER2::LUC and firing rate data, suggesting that the data come from neurons.
The periods of rhythmic AVP and VIP neurons did not differ (AVP: 28.2 ± 0.9 h, mean ± SD; VIP: 28.5 ± 0.3 h; P > 0.05, Student's t test; P > 0.05, Brown-Forsythe's and Levene's tests; Fig. S2A) and were similar to published results from dispersed SCN cultures (7). In addition, rhythmic AVP and VIP neurons did not differ in measures of their rhythm amplitude or expression levels when compared with each other or with rhythmic neurons that were not stained (one-way ANOVA, P > 0.05). Finally, we compared the predicted and measured likelihoods that an isolated SCN neuron was rhythmic and expressed AVP, VIP, or neither (Table S1). The calculated joint probabilities were nearly identical to the observed patterns of neuropeptide and circadian PER2 expression, suggesting that circadian rhythmicity and expression of AVP or VIP are independent in isolated SCN neurons. This demonstrates that single AVP- or VIP-containing neurons exhibit intrinsic circadian rhythms in clock gene expression. We conclude that intrinsic circadian cycling is not restricted to a single class of neuropeptidergic neurons in the SCN.
We also tested whether arrhythmic neurons constituted a unique cell class. Neurons that failed to express PER2::LUC rhythmically were AVP- or VIP-ergic with similar percentages to circadian neurons (Fig. 1D). Arrhythmic AVP and VIP neurons included those that luminesced for <72 h (n = 92 of 382 neurons in 5 cultures) and those whose PER2 expression was noncircadian (n = 23 of 64 neurons). Because AVP and VIP neurons constitute only a fraction of the circadian SCN cells, we asked whether their presence is associated with a circadian phenotype. We found that neither rhythmicity nor arrhythmicity in single SCN neurons is predicted by AVP or VIP expression (P > 0.05, Fisher's exact test). We conclude that both circadian and noncircadian SCN neurons form heterogeneous populations (Fig. S3).
SCN Neurons Can Switch Their Circadian Phenotype.
When we tested the viability of neurons in low-density dispersals (Fig. S1B), we found that application of 10 μM forskolin initiated or ended circadian behavior in 20% of SCN neurons (n = 48 of 241 neurons in 2 cultures). Interestingly, most neurons that changed phenotype became arrhythmic (n = 39). These results, combined with the lack of evidence for a neuropeptide class of pacemaker neurons, led us to hypothesize that a cell's ability to oscillate intrinsically is not deterministic and that circadian cycling becomes random in isolated SCN neurons.
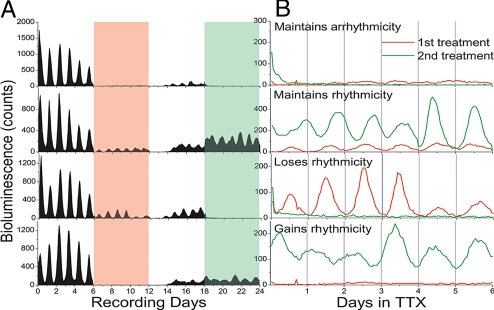
We recorded single-cell bioluminescence under conditions in which cell–cell interactions could be reversibly and repeatedly blocked. Because SCN neurons had been shown to drift out of phase from one another in the presence of TTX and then re-establish their phase relationships when TTX was removed (8), we treated organotypic PER2::LUC SCN explants twice with TTX for 6 days at 6-day intervals. We postulated that if circadian pacemaking is neither unique to, nor a determined property of, a class of SCN neurons, repeated TTX treatments would always reveal circadian cells but not always the same cells. Under baseline conditions, the periods of bioluminescence rhythms in single SCN neurons were tightly clustered (25.7 ± 0.3 h, mean ± SD) with synchronized peak bioluminescence values (Rayleigh test, r = 0.95, P < 0.001). Wash treatment restored rhythmicity to the majority of recorded neurons (n = 162 of 190 neurons in 2 explants). The restored synchrony was significant (Rayleigh test, r = 0.57, P < 0.001) but less than before TTX, suggesting that complete resynchronization requires more than 6 days. We found that 50% of neurons lost and 13% sustained circadian rhythmicity in both TTX treatments (n = 94 arrhythmic and 24 rhythmic of 190 neurons; Fig. 5). In addition, nearly 38% of neurons (n = 72 of 190) gained or lost rhythmicity in the second TTX treatment when compared with the first treatment. Interestingly, the locations of neurons that maintained or increased rhythm amplitude in both TTX applications were scattered throughout the SCN. The locations of arrhythmic cells were also broadly distributed. We then treated a third SCN slice with TTX for 8 days and found that >70% of neurons lost rhythmicity (n = 71 of 98 neurons) and that some rhythmic neurons skipped a circadian peak (n = 18 of 27 rhythmic neurons). When we treated a control SCN explant with fresh medium every 6 days over a 24-day recording, we found that the number of circadian neurons was similar during the first (n = 76 of 100 neurons) and last treatments (n = 62 of 100) and no evidence for switching between rhythmic and arrhythmic states. These observations further support the conclusion that neurons throughout the SCN are capable of cell-autonomous circadian rhythm generation and suggest that network interactions stabilize their intrinsically noisy oscillations.
Fig. 5.
Repeated TTX treatment revealed switching in circadian behavior of SCN neurons. (A) Long-term recordings of PER2::LUC expression from 4 representative SCN cells in an explant before, during, and between 2 6-day TTX treatments (red and green bars). All cells showed drastically reduced PER2::LUC expression, and few cells were circadian in either the first or second TTX administration. (B) Approximately 13% (n = 24 of 190 recorded cells in 2 cultures) remained rhythmic during both treatments, whereas 38% became rhythmic or lost rhythmicity in the second TTX treatment.
PER2 Accumulation Predicts Oscillatory Ability.
We examined PER2 expression for patterns that predict the loss of circadian rhythmicity. When the peak phase or amplitude at the time of TTX application did not correlate with whether a cell remained rhythmic (Fig. S4), we compared 48 h of bioluminescence from neurons that lost (n = 25) or sustained (n = 25) cycling during TTX treatment. We aligned traces to 20% of peak expression in the first 24 h and then averaged the bioluminescence from all neurons in each group in hourly bins (Fig. S5). On the day before losing rhythmicity, neurons had lower peak PER2 expression (Student's t test, P < 0.0002) but similar times to peak compared with rhythmic neurons (P > 0.05). Thus, we found that the rate of PER2 accumulation in one cycle was a better predictor than other measures that a neuron would oscillate in the next cycle (Figs. S3–S5).
Discussion
SCN Neurons Are Weak, Inducible, and Probabilistic Oscillators.
The best examples to date of cell-autonomous circadian rhythm generation are unicellular cyanobacteria (21), dinoflagellates (22), and molluscan retinal neurons (23). Here we have demonstrated that when a single SCN neuron expresses PER2 for multiple days, it is likely to be circadian and can contain AVP, VIP, or another unidentified marker. However, the majority of SCN neurons were arrhythmic and just as likely as rhythmic neurons to express AVP or VIP. In addition, neurons lost or gained daily gene expression rhythms during long recordings in which intercellular communication was blocked (i.e., very low density, isolation, and TTX). It is possible that a small, unique class of self-sustained pacemaker cells exists in the SCN (e.g., the 13% of neurons that remained circadian during both week-long TTX treatments). Because these neurons were not found in a reproducible location from explant to explant and showed unstable circadian periods and amplitudes, we conclude that circadian rhythm generation is not localized to a specialized class of SCN neurons. Instead, it seems that each SCN neuron has approximately a 1 in 4 chance of expressing PER2 rhythmically in the absence of intercellular communication.
The variable nature of daily rhythms in isolated SCN neurons has been predicted by models of the transcription–translation circadian feedback loop in which noise (sometimes associated with low numbers of molecules) was added within a cell (7, 24, 25). Under these conditions, small perturbations (extrinsic or intrinsic to the cell) could push the molecular oscillator on or off a limit cycle. Other modeling efforts have simulated the SCN as a network of damped (26) or sustained, damped, and driven oscillators (27). Bernard et al. (26) found that, compared with self-sustained oscillators, damped oscillators more rapidly synchronized to each other and to environmental cycles, a potential benefit of having cells that change their rhythm amplitude. A third approach modeled the SCN as intrinsic oscillators and nonrhythmic gate cells (28, 29). These models showed that nonrhythmic neurons can contribute by gating input to the SCN depending on feedback from rhythmic cells. Our results support components of each of these models, by revealing that SCN neurons are heterogeneous, intrinsically noisy oscillators that, under some conditions, lose their circadian fluctuations in gene expression and firing rate. We stress, however, that we find no evidence for a specialized or anatomically localized class of cell-autonomous oscillators (or nonoscillatory gate cells). Instead, our results suggest that a single SCN neuron has the potential to move between these regimens and that the network may contribute to overall rhythmicity.
Noisy Gene Expression May Lend Plasticity to the SCN.
Randomness in events like transcription has been implicated in generating cell–cell variability and network plasticity (30). In addition to the known differences in amplitude and period, we find that SCN neurons also differ in their stability of circadian rhythmicity when isolated from their neighbors. This changeable rhythmicity is reminiscent of the circadian properties of Drosophila neurons, which vary according to experience (31), and of Per1-, Per2-, or Cry1-mutant SCN neurons, which show “stuttering” circadian rhythms (7). The present results indicate that intercellular interactions acting on noisy gene expression within SCN neurons stabilize cycling, which may contribute to both the improved precision (32) and adaptable coding of, for example, day length by a population of heterogeneous oscillators (33, 34).
With each neuron having the ability to generate 24-h oscillations or to be driven, the SCN is a multipotential system. In contrast, fibroblasts have been reported to all oscillate independently without damping in vitro (4). It is interesting to speculate that SCN neurons rely on signals from other SCN cells for robust daily cycling while allowing for flexibility with changes in environmental conditions. This is consistent with electrophysiologic recordings and computational modeling in other neural networks, which have shown neurons that transition between oscillatory regimens depending upon a variety of cellular states (35). For example, homeostatic changes in specific ionic conductances can switch neurons in the lobster stomatogastric ganglion from bursting to tonic firing. This kind of tuning provides multiple solutions for individual nodes within a network to produce a system with the desired output. Having a range of solutions lends plasticity, as well as stability, to the system. Individual SCN neurons are variable oscillators when uncoupled but when connected with each other generate a coherent circadian output.
Previous studies have shown that when intercellular signaling is blocked by one of a variety of mechanisms, the amplitude of PER2 expression diminishes (8, 9, 15, 19, 36). Here we reveal the exciting possibility that network interactions act on the rate of PER2 accumulation in single cells (Fig. S5) to determine their ability to generate and sustain daily rhythms. Slower accumulation of PER2 protein, not the amplitude or time of peak expression, is best correlated with the loss of rhythmicity in SCN neurons, strongly suggesting that the processes that modulate the accumulation of PER2 or other clock proteins play a critical role in sustaining circadian cycling. This is consistent, for example, with the observation that phosphorylation sites on the PER2 protein play a critical role in both its accumulation and circadian rhythmicity (37–39). Future studies will likely pursue how intercellular signals impact timekeeping processes, including rates of translation and degradation of clock proteins. It remains to be seen how changes in intracellular state, changes in connectivity within the SCN, or their combined effects mediate some of the developmental or seasonal adaptations in mammalian circadian rhythms (33, 40–43).
Materials and Methods
Cell Culture.
SCN were obtained from 1–7-day-old homozygous PER2::LUC mice (founders generously provided by J. Takahashi, Northwestern University) housed under a 12 h/12 h light/dark schedule. For slice cultures, bilateral SCN from 300-μm coronal sections of hypothalamus were cultured on Millicell-CM membranes (Millipore) for a minimum of 3 days (methods modified from ref. 19). SCN explants were then inverted onto polylysine/laminin-coated glass coverslips and maintained at 37 °C in 200 μL CO2-buffered medium supplemented with 10% newborn calf serum (Invitrogen) for at least 2 days before recording. Dispersed neuron cultures were generated as previously described (16). Viable cells were plated onto polylysine/laminin-coated coverslips attached to 35-mm culture dishes and maintained in 1 mL of CO2-buffered medium that had been conditioned by primary cultures of cortical astrocytes (44) supplemented with 10% newborn calf serum (Invitrogen) at 37 °C. All bioluminescence recordings from dispersed single cells began on in vitro day 4 and continued for a minimum of 6 days.
Whole-Cell Patch Clamp Recording.
We perfused cultures at 1 to 2 mL/min with Tyrode's solution containing 150 mM NaCl, 4 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 10 mM glucose, and 10 mM Hepes (pH adjusted to 7.4 with NaOH). Whole-cell electrodes were filled with 140 mM cesium glucuronate, 5 mM CsCl, 5 mM MgCl2, 10 mM EGTA, 5 mM ATP, 1 mM GTP, and 10 mM Hepes (pH adjusted to 7.4 with CsOH). The reference electrode was in a well with internal solution connected to the bath by an agar bridge equilibrated with 4 M KCl. Once whole-cell recordings were established, we perfused with external solution: 120 mM NaCl, 3 mM KCl, 1 mM NaH2PO4, 4 mM NaHCO3, 10 mM glucose, and 5 mM Hepes (pH adjusted to 7.4 with NaOH). Spontaneous inhibitory postsynaptic currents (IPSCs) were recorded at 0 mV (Axopatch 200A amplifier; Molecular Devices; filtered at 1 kHz, 10 kHz sample rate). After recording IPSCs for 30–60 s in control external solution the superfusion was switched to external solution containing 10 mM KCl. IPSC frequency was quantified using p-Clamp software (Molecular Devices).
Single-Cell Bioluminescence Recording.
We imaged (Versarray 1024 cooled-CCD camera; Princeton Instruments) low-density dispersals in 1 mL of air-buffered medium supplemented with 0.1 mM beetle luciferin (Xenogen) and SCN slices in 500 μL at 37 °C in sealed Petri dishes. Photon counts were integrated over 1 h with 4 × 4 binning using WinView and quantified using ImageJ software (National Institutes of Health). To confirm cell health at the end of recording, we added forskolin (10 μM; Sigma) to the medium and recorded for at least 3 more days. In some SCN slice recordings, we replaced 500 μL of the recording medium with medium containing 0.5 μM TTX (Sigma) and 0.1 mM beetle luciferin. After 6 days, TTX-containing medium was removed and washed with 1 to 2 full exchanges of recording medium.
MEA Recording.
Long-term firing rate patterns were recorded from cells dispersed from the SCN of C57BL/6 mice (Harlan). The SCN from pups (postnatal age 3–5 days) were pooled, dispersed, and plated at ≈300–700 cells/mm2 or 10,000 cells/mm2 onto multielectrode arrays (Multichannel Systems) according to published methods (15, 16). Every 10 min, the number of spikes with identical amplitude and duration was totaled and stored. Using off-line analysis similar to published methods (45), we counted action potentials of similar amplitude and duration from individual neurons. The shape of the spike and presence of a clear refractory period were used as criteria for single-unit activity.
Isolation of Single Cells.
Individual neurons in very-low-density cultures were identified for isolation after 2 to 3 days of initial bioluminescence recording. All other cells were removed by scraping them from the surface of the culture using a glass micropipette and micromanipulator. Cellular debris was rinsed away using medium in the culture.
Immunocytochemistry.
After 6 days of bioluminescence recording, we treated low-density dispersals with colchicine (50 μg/mL; Sigma) for 8–12 h to maximize neuropeptide content. Cultures were then fixed overnight in 4% paraformaldehyde and double-labeled for AVP and VIP using published methods (15). We stained cultures with anti-AVP (mouse monoclonal, 1:50; gift from Dr. H. Gainer) and anti-VIP (rabbit, 1:2,000; Immunostar) followed by donkey antimouse IgG conjugated to Texas Red (1:100; Jackson ImmunoResearch) and donkey antirabbit IgG conjugated to Cy2 (1:50; Jackson ImmunoResearch). Nuclei were visualized using DAPI staining. Immunopositive cells were counted by 2 independent observers. An observer blind to the PER2 expression in each cell scored its neuropeptide content from superimposed immunofluorescence and bioluminescence images.
Very-low-density and control SCN cultures were stained with anatomic markers of neuronal processes and synaptic densities, respectively, anti-β-3-tubulin (rabbit, 1:500; Covance) and anti-SV2 (mouse monoclonal, 1:50; gift from Dr. K. Buckley) followed by donkey antimouse IgG conjugated to Texas Red (1:100; Jackson ImmunoResearch) and donkey antirabbit conjugated to Cy2 (1:50; Jackson ImmunoResearch). For all antibodies tested, fluorescence was imaged across cultures with identical exposure time and gain.
Analysis of Rhythmicity.
We measured the period of single-neuron PER2::LUC expression and firing rate by FFT-NLLS (46) and χ2 periodogram (47). Cells with a statistically significant period between 14 and 36 h by both methods were scored as circadian. Confidence intervals were set to 95% and 99% for FFT-NLLS and χ2 periodogram, respectively.
Receiver Operating Characteristic Curve Analysis.
We used signal detection theory to determine the probability that an ideal observer could predict the cycling behavior of a neuron according to the slope of PER2 accumulation in the previous cycle. For a given slope criterion, we plotted the proportion of circadian and arrhythmic neurons with greater slopes, generating a receiver operating characteristic (ROC) curve. The area under the ROC curve is equivalent to the odds of correct detection by an ideal observer (48).
Supplementary Material
Acknowledgments.
The authors thank Tatiana Simon for expert technical assistance; Sungwon An, Mark Freeman, and Drs. Christian Beaulé, Daniel Granados-Fuentes, and Luciano Marpegan for helpful discussions; and Drs. Sara Aton, Paul Stein, and Kurt Thoroughman for comments on preliminary drafts of the manuscript. This work was supported by National Institutes of Health Grants MH63104 (to E.D.H.) and NS30888 (to J.E.H.), a Beckman Foundation undergraduate fellowship (to N.A.), a National Science Foundation graduate research fellowship (to A.B.W.), and a Washington University Vision Sciences Training grant (to A.B.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902768106/DCSupplemental.
References
- 1.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 2.Bae K, et al. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30:525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 3.Nagoshi E, et al. Circadian gene expression in individual fibroblasts: Cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 4.Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol. 2004;14:2289–2295. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ralph MR, Foster RG, Davis FC, Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science. 1990;247:975–978. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- 6.Welsh DK, Logothetis DE, Meister M, Reppert SM. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron. 1995;14:697–706. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 7.Liu AC, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaguchi S, et al. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science. 2003;302:1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- 9.Maywood ES, et al. Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr Biol. 2006;16:599–605. doi: 10.1016/j.cub.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 10.Antle MC, Silver R. Orchestrating time: Arrangements of the brain circadian clock. Trends Neurosci. 2005;28:145–151. doi: 10.1016/j.tins.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Abrahamson EE, Moore RY. Suprachiasmatic nucleus in the mouse: Retinal innervation, intrinsic organization and efferent projections. Brain Res. 2001;916:172–191. doi: 10.1016/s0006-8993(01)02890-6. [DOI] [PubMed] [Google Scholar]
- 12.Shinohara K, Honma S, Katsuno Y, Abe H, Honma KI. Two distinct oscillators in the rat suprachiasmatic nucleus in vitro. Proc Natl Acad Sci USA. 1995;92:7396–7400. doi: 10.1073/pnas.92.16.7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De la Iglesia HO, Cambras T, Schwartz WJ, Diez-Noguera A. Forced desynchronization of dual circadian oscillators within the rat suprachiasmatic nucleus. Curr Biol. 2004;14:796–800. doi: 10.1016/j.cub.2004.04.034. [DOI] [PubMed] [Google Scholar]
- 14.Noguchi T, Watanabe K, Ogura A, Yamaoka S. The clock in the dorsal suprachiasmatic nucleus runs faster than that in the ventral. Eur J Neurosci. 2004;20:3199–3202. doi: 10.1111/j.1460-9568.2004.03784.x. [DOI] [PubMed] [Google Scholar]
- 15.Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci. 2005;8:476–483. doi: 10.1038/nn1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herzog ED, Takahashi JS, Block GD. Clock controls circadian period in isolated suprachiasmatic nucleus neurons. Nat Neurosci. 1998;1:708–713. doi: 10.1038/3708. [DOI] [PubMed] [Google Scholar]
- 17.Honma S, Shirakawa T, Katsuno Y, Namihira M, Honma KI. Circadian periods of single suprachiasmatic neurons in rats. Neurosci Lett. 1998;250:157–160. doi: 10.1016/s0304-3940(98)00464-9. [DOI] [PubMed] [Google Scholar]
- 18.Yoo SH, et al. Period2::luciferase real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;102:1–8. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aton SJ, Huettner JE, Straume M, Herzog ED. GABA and Gi/o differentially control circadian rhythms and synchrony in clock neurons. Proc Natl Acad Sci USA. 2006;103:19188–19193. doi: 10.1073/pnas.0607466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prolo LM, Takahashi JS, Herzog ED. Circadian rhythm generation and entrainment in astrocytes. J Neurosci. 2005;25:404–408. doi: 10.1523/JNEUROSCI.4133-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mihalcescu I, Hsing W, Leibler S. Resilient circadian oscillator revealed in individual cyanobacteria. Nature. 2004;430:81–85. doi: 10.1038/nature02533. [DOI] [PubMed] [Google Scholar]
- 22.Roenneberg T, Morse D. Two circadian oscillators in one cell. Nature. 1993;362:362–364. doi: 10.1038/362362a0. [DOI] [PubMed] [Google Scholar]
- 23.Michel S, Geusz ME, Zaritsky JJ, Block GD. Circadian rhythm in membrane conductance expressed in isolated neurons. Science. 1993;259:239–241. doi: 10.1126/science.8421785. [DOI] [PubMed] [Google Scholar]
- 24.Forger DB, Peskin CS. Stochastic simulation of the mammalian circadian clock. Proc Natl Acad Sci USA. 2004;102:321–324. doi: 10.1073/pnas.0408465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonze D, Goldbeter A. Circadian rhythms and molecular noise. Chaos. 2006;16:26110–26111. doi: 10.1063/1.2211767. [DOI] [PubMed] [Google Scholar]
- 26.Bernard S, Gonze D, Cajavec B, Herzel H, Kramer A. Synchronization-induced rhythmicity of circadian oscillators in the suprachiasmatic nucleus. PLoS Comput Biol. 2007;3:e68. doi: 10.1371/journal.pcbi.0030068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.To TL, Henson MA, Herzog ED, Doyle FJ., III A molecular model for intercellular synchronization in the mammalian circadian clock. Biophys J. 2007;92:3792–3803. doi: 10.1529/biophysj.106.094086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antle MC, Foley DK, Foley NC, Silver R. Gates and oscillators: A network model of the brain clock. J Biol Rhythms. 2003;18:339–350. doi: 10.1177/0748730403253840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Antle MC, Foley NC, Foley DK, Silver R. Gates and oscillators II: Zeitgebers and the network model of the brain clock. J Biol Rhythms. 2007;22:14–25. doi: 10.1177/0748730406296319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raj A, van Oudenaarden A. Nature, nurture, or chance: Stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wulbeck C, Grieshaber E, Helfrich-Forster C. Pigment-dispersing factor (PDF) has different effects on Drosophila's circadian clocks in the accessory medulla and in the dorsal brain. J Biol Rhythms. 2008;23:409–424. doi: 10.1177/0748730408322699. [DOI] [PubMed] [Google Scholar]
- 32.Herzog ED, Aton SJ, Numano R, Sakaki Y, Tei H. Temporal precision in the mammalian circadian system: A reliable clock from less reliable neurons. J Biol Rhythms. 2004;19:35–46. doi: 10.1177/0748730403260776. [DOI] [PubMed] [Google Scholar]
- 33.Inagaki N, Honma S, Ono D, Tanahashi Y, Honma KI. Separate oscillating cell groups in mouse suprachiasmatic nucleus couple photoperiodically to the onset and end of daily activity. Proc Natl Acad Sci USA. 2007;104:7664–7669. doi: 10.1073/pnas.0607713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ciarleglio CM, et al. Population encoding by circadian clock neurons organizes circadian behavior. J Neurosci. 2009;29:1670–1676. doi: 10.1523/JNEUROSCI.3801-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marder E, Goaillard JM. Variability, compensation and homeostasis in neuron and network function. Nat Rev Neurosci. 2006;7:563–574. doi: 10.1038/nrn1949. [DOI] [PubMed] [Google Scholar]
- 36.Colwell CS, et al. Disrupted circadian rhythms in VIP and PHI deficient mice. Am J Physiol Regul Integr Comp Physiol. 2003;285:R939–R949. doi: 10.1152/ajpregu.00200.2003. [DOI] [PubMed] [Google Scholar]
- 37.Gallego M, Eide EJ, Woolf MF, Virshup DM, Forger DB. An opposite role for tau in circadian rhythms revealed by mathematical modeling. Proc Natl Acad Sci USA. 2006;103:10616–10623. doi: 10.1073/pnas.0604511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Y, et al. Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell. 2007;128:59–70. doi: 10.1016/j.cell.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meng QJ, et al. Setting clock speed in mammals: The CK1epsilontau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron. 2008;58:78–88. doi: 10.1016/j.neuron.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schaap J, et al. Heterogeneity of rhythmic suprachiasmatic nucleus neurons: Implications for circadian waveform and photoperiodic encoding. Proc Natl Acad Sci USA. 2003;100:15994–15999. doi: 10.1073/pnas.2436298100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sladek M, et al. Insight into molecular core clock mechanism of embryonic and early postnatal rat suprachiasmatic nucleus. Proc Natl Acad Sci USA. 2004;101:6231–6236. doi: 10.1073/pnas.0401149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vanderleest HT, et al. Seasonal encoding by the circadian pacemaker of the SCN. Curr Biol. 2007;17:468–473. doi: 10.1016/j.cub.2007.01.048. [DOI] [PubMed] [Google Scholar]
- 43.Sumova A, Kovacikova Z, Illnerova H. Dynamics of the adjustment of clock gene expression in the rat suprachiasmatic nucleus to an asymmetrical change from a long to a short photoperiod. J Biol Rhythms. 2007;22:259–267. doi: 10.1177/0748730407301052. [DOI] [PubMed] [Google Scholar]
- 44.Banker G, Goslin K. In: Culturing Nerve Cells. Banker G, Goslin K, editors. Cambridge: MIT Press; 1991. pp. 75–118. [Google Scholar]
- 45.Meister M, Pine J, Baylor DA. Multi-neuronal signals from the retina: Acquisition and analysis. J Neurosci Methods. 1994;51:95–106. doi: 10.1016/0165-0270(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 46.Plautz JD, et al. Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms. 1997;12:204–217. doi: 10.1177/074873049701200302. [DOI] [PubMed] [Google Scholar]
- 47.Sokolove PG, Bushell WN. The chi square periodogram: Its utility for analysis of circadian rhythms. J Theor Biol. 1978;72:131–160. doi: 10.1016/0022-5193(78)90022-x. [DOI] [PubMed] [Google Scholar]
- 48.Green DM, Swets JA. Signal Detection Theory and Psychophysics. New York: Wiley; 1966. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.