Abstract
The dynamics of chromatin provide the access to DNA within nucleosomes and therefore, this process is critically involved into the regulation of chromatin function. However, our knowledge on the large-range dynamics of nucleosomes is limited. The questions, such as the range of opening of the nucleosome, and the mechanism whereby the opening occurs and propagates, remain unknown. Here we applied single molecule time lapse AFM imaging to directly visualize the dynamics of nucleosomes and identify the mechanism of the large range DNA exposure. With this technique, we are able to observe the process of unwrapping of nucleosomes. The unwrapping of nucleosomes proceeds from the ends of the particles, allowing for the unwrapping of DNA regions as large as dozens of base pairs. This process may lead to a complete unfolding of nucleosomes and dissociation of the histone core from the complex. The unwrapping occurs in the absence of proteins involved in the chromatin remodeling that require ATP hydrolysis for their function, suggesting that the inherent dynamics of nucleosomes can contribute to the chromatin unwrapping process. These finding shed a new light to molecular mechanisms of nucleosome dynamics and provide novel hypotheses to the understanding of the action of remodeling proteins as well as other intracellular systems in the chromatin dynamics.
The dynamics of the nucleosome core particle (NCP), a fundamental unit of chromatin, is a key property of chromatin, providing access to the DNA wrapped around the histone core of the protein. These NCP dynamics are also important in accomplishing the other numerous functions of chromatin. Studies performed over past decade led to the discovery of a class of proteins, the exclusive role of which is to regulate the DNA accessibility within the chromatin (e.g., reviews (1-3). These specialized remodeling machines unwrap the DNA from the histone core to provide the access to the DNA regions inside the nucleosome. This discovery led to the view in which nucleosomes themselves are considered rather stable particles with limited dynamics, so the nucleosome unwrapping process is performed by remodeling protein complexes in an ATP-dependent fashion (3, 4). The reconciliation of this model with the energetic costs required for the unwrapping of DNA from the nucleosome core particle led to the model in which remodeling proteins use the ATP hydrolysis energy to unwrap only a segment of the nucleosomal DNA, creating a bulge and then moving that bulge along the histone like a ratchet (3). At the same time, recent studies performed with the use of various techniques, including single molecule approaches, led to the realization that nucleosomes are quite dynamic rather than static systems. Therefore, the inherent dynamics should be taken into account in the understanding of the nucleosome unwrapping process. Single molecule fluorescence and time resolved techniques showed that nucleosomes undergo local dissociation of DNA in the absence of remodeling proteins (5-11) and this process occurs on the sub-second time scale (8, 10). Earlier AFM imaging was applied to characterization of chromatin structure at the nanoscale level (12-15). The AFM sample preparation is so gentle that AFM enables the study of chromatin structure omitting the rather traditional glutaraldehyde fixation procedure of the sample (16). As a result, the comparison of fixed and unfixed samples led the authors to the conclusion on the high dynamic feature of nucleosomes. However, a number of questions remained unanswered. What is the range of the local dynamics of nucleosomes? Is a non-ATP dependent unwrapping of nucleosomes possible? What are the factors facilitating the large scale opening and unwrapping of nucleosomes? To answer these questions, we used single molecule time lapse AFM approach capable of detecting the dynamics of molecular systems at the single molecule level (17-19). Previously, this capability of time lapse AFM enabled us to follow the dynamics of supercoiled DNA (20), including the dynamics of cruciform structures (21), and to directly test the models of the Holliday junction branch migration (22, 23). The development of high speed AFM, with the ability to perform time lapse AFM imaging at speeds much faster than traditional AFM techniques is also notable (24, 25). Given the capability of time lapse AFM to image the dynamics of molecular systems at nanoscale, we applied this method to study the dynamics of mononucleosome samples. We were able to directly visualize large scale dynamics of nucleosomes, accompanied with unwrapping of long DNA regions, followed by the complete unwrapping of DNA from the histone core, and the dissociation of nucleosome particles. The possibility to unwrap nucleosomes in the absence of the ATP dependent chromatin remodeling proteins in low salt conditions is a feature of nucleosomes, dramatically facilitating the DNA accessibility problem, and prompting new models on the regulation of the nucleosome unwrapping process.
Materials and Methods
Reconstitution of Histone Octamer
Reconstitution of NCP was performed according to the procedure described in (26, 27). Briefly, the human recombinant histones H2A, H2B, H3 and H4 (New England Biolab, Ipswich, MA) were concentrated on Microcon spin column (MW 3,000, Millipore, Billerica, MA), dissolved in unfolding buffer (UB) containing 6M guanidine chloride, 20mM Tris-HCl, pH 7.5, 5mM DTT. Concentration of each histone protein was measured by the absorbance spectra at 276 nm. After one hour incubation at room temperature, all four histone proteins were mixed in equal molar ratio and diluted up to 1mg/ml with refolding buffer (RB), containing 2M NaCl, 10mM Tris-HCl, pH7.5, 1mM EDTA, 5mM 2-mercaptoethanol. After removing the precipitated protein by centrifugation for 10 min at 12,700 g the sample were loaded on Superdex 200 PG column (Pharmacia), which was pre-equilibrated with RB at 40°C. The fraction corresponding to the histone octamer was collected, mixed with 50% (v/v) glycerol and stored at -20°C.
Assembly of nucleosomes
The template for the nucleosome assembly was a 353 bp DNA fragment, containing stable sequence 601 (147 bp) for histone octamer binding (28) flanked by regions of 79 bp and 127 bp. The complete sequence of the DNA fragment is given in Supporting Information (SI). The DNA was obtained by PCR amplification of plasmid pGEM3Z-601 with the following primers:
5′- CGG CCA GTG AAT TGT AAT ACG -3′ and
5′- CGG GAT CCT AAT GAC CAA GG -3′.
To reconstitute nucleosomes, the histone octamer was mixed with DNA in 1:1 molar ratio in the buffer containing 10mM Tris-HCl, pH7.5, 2M NaCl and incubated for 30 min at room temperature. The mixture after a series of dilutions required by the protocol described in (26, 27). The dialysis with three subsequent changes of the buffer, containing 10mM Tris-HCl, pH 7.5, 0.2 M NaCl was performed at 4°C with one hour incubation time at each step followed by the overnight dialysis against the same buffer. The formation of NCP was controlled by the gel electrophoresis (5% PAGE) and AFM imaging in air.
AFM imaging methodology
The 1-(3-aminopropyl)silatrane (APS-mica) (23, 29, 30) was used as a substrate for AFM imaging. The procedure for mica modification with has been described previously (4). For experiments in air NCP in the buffer, containing 10 mM Tris-HCl, pH 7.5, 4 mM, MgCl2, were deposited on APS mica for 2 min, rinsed with deionized water (Aquamax Lab. Water System, Van Nuys, CA) and dried with argon gas. The images were collected on NanoScope IIId system (Veeco, Santa Barbara, CA) operating in tapping mode in air. Tapping Mode Etched Silicon Probes (TESP; Veeco) with a spring constant 42 N/m and the resonant frequency between 270-320 kHz were used. The scan rate was 1.7Hz. For experiments in liquid, the NCP sample in the buffer, containing in 10 mM Tris- HCl, pH 7.5, 4 mM MgCl2, was injected directly into flow cell avoiding the drying step. Images were acquired by using NanoScope IIID system operating in tapping mode in liquid using the protocols described earlier (20-23). NP Probes (NP, Veeco) with a spring constant 0.06 N/m were used. The resonant peak ca. 8 KHz was optimal for the operating in aqueous in solutions. Before the engage, the initial parameters such as scan size and offset of the microscope were set to 0, the piezo drive amplitude ∼100 mV (varied between 80 and 200 mV corresponding to 2-7 nm of amplitude), the scan rate ∼ 1.5 Hz and the set point voltage ∼0.4 V were typically used. Typically, drive amplitude was 2-3 times smaller than the initially selected value, but the values below 30 mV did yield stable images. The setpoint values were adjusted to the maximal stable values manually during scanning to maintain high-resolution image and minimize the tip sweeping effect (20). The continued scanning over the selected area (about 800×800 nm image size) was performed to follow the dynamics of NCP and images were recorded with 512×512 data density. Note that practically each deposited sample provided the images illustrating high the reproducibility of the APS-mica methodology (31, 32). Additional sample depositions in almost 100% of the cases were due to the need to replace the tip. Image processing for measuring the DNA contour length, arm lengths, angle between DNA arms and protein volume were performed using Femtoscan (Advanced Technologies Center, Moscow, Russia) and described in (30, 33).
AFM data analysis
To estimate the number of DNA turns wrapped around the histone core from the length measurements, we used the following parameters for nucleosomes: 147 bp DNA is wrapped around the histones core making 1.7 turns (34, 35). The following equation for the dependence of the number of DNA turns around histones core on the arms length was used to calculate the number of turns for the NS:
| (1) |
where CL is the contour length of free DNA, Larm1 and Larm2 lengths of short and long DNA arms respectively. The angles between the arms were measured by drawing tangents to the section of the arms comprising 10 nm of the arm starting from the blob (30, 36).
Results
The DNA template designed for this work was the fragment of 353 bp DNA containing 147 bp nucleosome positioning 601 sequence (7), flanked with two regions of different lengths, 79bp and 127bp. Such a design, similar to an early cryo electron microscopy study (37), allowed us to map the nucleosome position and correlate it with the binding of histone core to the central 147 bp region. Depending on the number of DNA turns around the histone core, the complex will adopt different morphologies schematically shown in Fig. 1. The initial design corresponds to the complex with one turn and four other schemes correspond to the complexes with 1.25, 1.5, 1.75 and 2 turns. All designs correspond to the uniform wrapping of both arms starting at the position in the center of the 147 bp region, so the lengths of the arms gradually decrease upon the DNA wrapping while the size of the nucleosome core is increasing. In addition, the DNA wrapping is accompanied by the change of the interarm angle. We assign zero value of the rotation angle to the position of the long arm for the complex with one turn. The design with 1.25 DNA turns is characterized by the rotation angle 90°. Therefore, the complexes with 1.5, 1.75 and 2 turns have the rotation angles 180°, 270° and 360° respectively. These parameters were used in the procedure of assigning of the NCP structure.
Figure 1.
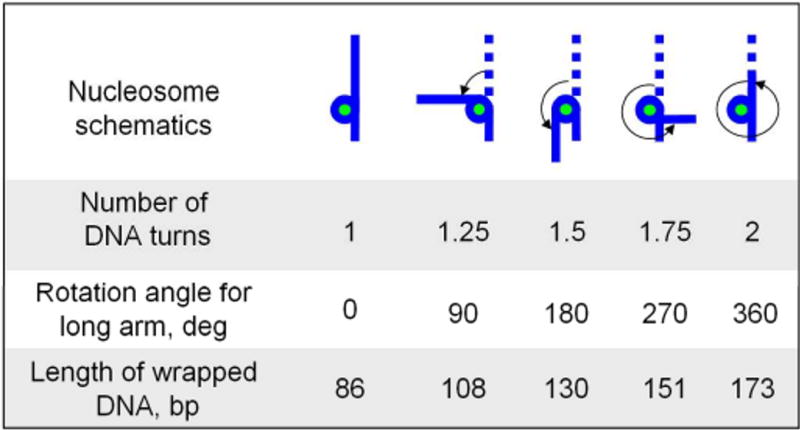
Schematics for nucleosome particles with different number of DNA supercoiling turns around the histone core.
The nucleosome sample prepared as described above was deposited onto APS-mica, rinsed, dried and imaged with AFM in air. Note that the sample was prepared without glurataldehyde crosslinking. A typical AFM image of such samples is shown on Fig. 2. The nucleosomes are clearly seen on these images as bright blobs with the DNA arms at both sides of the particle; however, the morphology of the particles is different. The yield of nucleosomal particle as shown in this image was ca. 60%. We marked the complexes with the identical morphologies with the same numbers. The arms for particles marked as 1 are crossed. The arms are almost parallel for particles labeled as type 2, whereas the angle between the arms for particles labeled as type 3 is 180°. These images can be interpreted in terms of different number of DNA turns around the histone core, and using the schematics in Fig. 1 we assigned groups 1, 2 and 3 to particles with 1.75, 1.5 and 1 DNA turns, respectively. Note as well that blobs of NCP type 3 are less bright compared to those of types 1 and 2, suggesting that type 3 particles have the least number of DNA turns. The AFM provides the height of the topographic images in addition to the lateral sizes, allowing one to calculate the volume values of the particles. The volume measurements were critical for analyses of the protein stoichiometry of various specific protein-DNA complexes (30, 33). The dependence of the proteins' volume on their size obtained for proteins of different sizes is shown as a plot in Fig. S0 The linearity of this plot justifies the use of the AFM volume values to estimate the protein molecular weight and was used for structural characterization of NCP. We interpret the variability of the particle size as an indication of NCP unwrapping, and based on these data, we conclude that nucleosomes in solution are dynamic and capable of large scale unwrapping. Thus, AFM imaging of non-crosslinked NCP samples in air revealed the structural heterogeneity of the particles and we hypothesize that this heterogeneity reflects inherent dynamics of NCP. To test this hypothesis and look directly at the dynamics of NCP in solution, we employed the AFM capability to perform imaging in aqueous solutions using time-lapse imaging of non-dried NCP samples.
Figure 2.
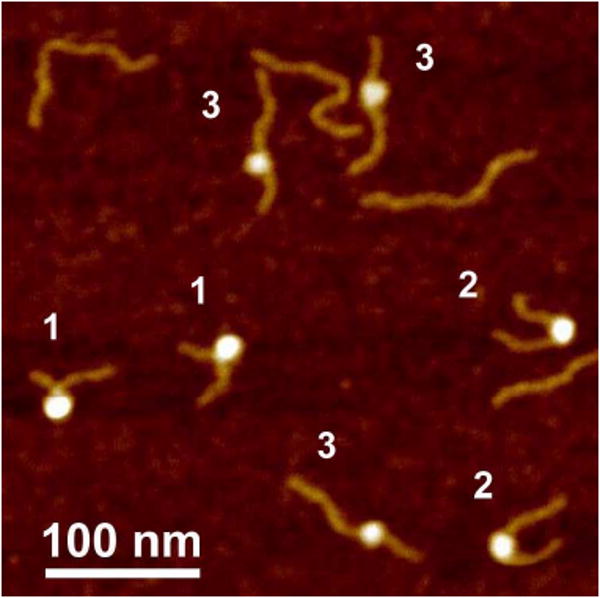
AFM image taken in air of the nucleosome samples. Nucleosomes marked with numbers 1.7, 1.4 and 1.0 correspond to NCPs with ∼1.7, ∼1.4 and ∼1.0 turns of DNA wrapped around the core particle.
In time lapse AFM mode, we performed continuous scanning over a selected area and followed the morphology changes of individual NCPs over time. The sample was deposited on APS mica and imaged omitting the drying step. The use of functionalized APS-mica surface allowed us to perform time lapse experiments without restrictions to the composition of the buffer and this methodology provided almost 100% reproducibility (21-23, 29). Several dozen time-lapse datasets were obtained and analyzed without any bias to the set. These experiments revealed different pathways, and the data below show the examples for each of these pathways. The imaging frames illustrating the dynamics of one selected NCP particle are shown in figures and sets of frames arranged as movie files for each dataset are placed in the supporting information (SI) section.
Figure 3 shows the images and the data for a pathway characterized by two-step changes of the NCP. The frame-by-frame images are shown in Fig. 3A and the set of images assembled as a movie can be seen in the SI section (M1 file). Initially, the NCP with short arms of DNA slightly changes its conformation (frames 1, 2), then unwraps on frame 3. The NCP retains its geometry over 3 frames in a row (frames 3, 4, 5). Then, between frames 5-6, it loosens, unwraps, again on frame 6, stays unchanged on frame 7 and finally undergoes full dissociation on frame 8. This dataset shows also that the NCP dynamics is accompanied by the elongation of the both DNA arms and the decrease of the size of the blob as well the change of the interarm angle. As we mentioned above (see Materials and Methods section), these parameters were used for the characterization of the NCP geometry at each frame.
Figure 3.
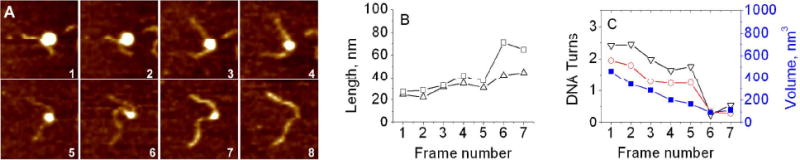
Two step unwrapping process. (A) Consecutive AFM images of nucleosomes with two step unwrapping process, taken during continuous scanning in the buffer. Each frame is 200 nm. (B) Dependence of arm lengths on the frame number. (C) The dependence of number of DNA turns around the core calculated from arm lengths (black) and from the angle between DNA arms (red) on the frame number. The dependence of nucleosome volume on the frame number is shown in blue (right Y-axis). Each frame takes about 170 second to scan.
The values of the arm lengths, depending on the frame number, are plotted in Fig. 3B. The graphs show that, over time, arms increase in length. The arm lengths were used for calculation of the number of the DNA turns within the nucleosome for each frame, and the data are shown in Fig. 3C (triangles, black line). This NCP particle initially has ∼2.5 turns (frame 1), remains unchanged at frame 2, releases ∼ 0.75 turn during frames 3 and 4, stays with 1.75 turns between frames 4 and 5 and then unwraps for 1.2 turns between frames 5 and 6 followed by histone core dissociation on frame 8. The number of turns was also determined using the measurements of the interarm angles and the data are shown with circles (red line) in Fig. 3C. This time-dependence of the DNA turns is qualitatively very similar to the previous one, although the values of the DNA turns measured by this method are slightly different. We consider such pattern of the nucleosome dynamics as a two-step unwrapping process. The DNA wrapping of the histone cores contributes to the overall volume of the NCP particle, so we measured the volumes of NCP on each frame and plotted these values in Fig. 3C (blue squares). This plot shows that the NCP volumes decrease over time (the number of the frame) supporting the idea to use this parameter for the NCP characterization. The dependence is rather smooth and does not show plateaus revealed by the two previous methods indicating to the limited accuracy of the volume measurements for the estimating the number of the DNA turns in the NCP particle.
Figure 4 illustrates another type of the NCP dynamics: one step gradual unfolding. Similar to the previous data set, frames of the images are shown in Fig. 4A and an animated set is appended to the SI section (M2 file). The quantitative analysis of this pathway is presented in Figs. 4B and C. According to the arms lengths measurements (black triangles), the NCP initially with 2.25 turns, is stable for two frames, then loses ∼1 turn between frames 2 and 4 and stays with 1.5 turns for three more frames until finally histone core dissociates (frame 7). Again, the NCP dynamics revealed by the angle measurements (red circles) show a similar pattern. The volume change (blue solid squares) follows the pattern of the DNA unwrapping curves.
Figure 4.
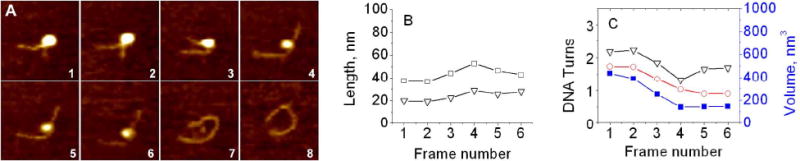
One step unwrapping process. (A) Consecutive AFM images of nucleosomes with one step unwrapping, taken during continuous scanning in the buffer. Scan size is 100 nm. (B) Dependence of arm lengths (in nm) on the frame number. (C) The dependence of number of DNA turns around the core calculated from arm lengths (black) and from the angle between DNA arms (red) on the frame number. The dependence of nucleosome volume on the frame number is shown in blue (right Y-axis). Each frame takes about 170 second to scan.
Figures 5A and B show the results of the analysis of additional sets of the data for one step and two-step unfolding processes, illustrating dynamics of rather stable NCP particles. The images and the data on the arm lengths are shown as figures S1 and S2 respectively along with the corresponding movie files M3 and M4 in SI. The one-step unfolding process of the NCP with ∼2 DNA turns (Fig. 5A and movie M3) starts rather late, so the particle remains stable during four consecutive scanning frames (frames 1-4). The unwrapping occurs between frames 4 and 6 with 1DNA turn left followed by a complete unwrapping of the DNA and dissociation of the histone core detected after frame 7.
Figure 5.
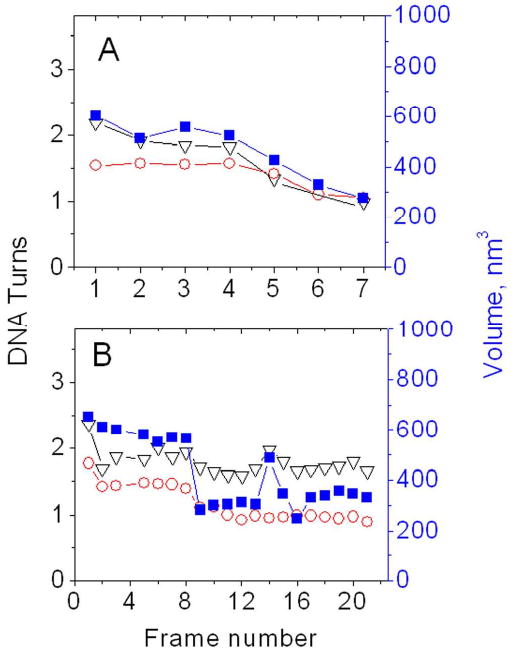
The dependence of the number of DNA turns around the core calculated from the arm lengths (black) and from the angle between the DNA arms (red) on the frame number for two sets of the data shown in Figs. S1 and S2 respectively. The dependence of nucleosome volume on the frame number is shown in blue (right Y-axis).
Fig. 5B shows the data for a rather interesting two-step dissociation pattern (see images in Figs. S2A, B and movie M4 in SI). This NCP, after an initial rapid loss of about 0.75 of the DNA turn (frame 2), remains stable (7 frames) although the arms move around the core in a rather broad range. Between frames 8 and 9, this NCP loses 0.25 of the turn and remains very stable for another 13 frames. Final dissociation of the histone core is seen on frame 22. Note the increase the number of the DNA turns (∼0.25 turns) between frames 12 and 14 followed by the decrease the number of turns by the same value. This dynamic of NCP is detected by the volume measurements (solid squares).
Discussion
The experiments described above show directly that NCP particles are so dynamic that large DNA segments are capable of unwrapping from the NCP core. This process can lead to full dissociation of nucleosomes. In addition, these data provide insight into the mechanism of the nucleosome dynamics.
Generally, two models are considered for the NCP dynamics. According to the site exposure model, DNA spontaneously dissociates from the core histones via unwrapping from the ends of the nucleosome and the results of the fluorescence studies, including single molecule analysis (8, 10, 11) are in favor of the site exposure model. Moreover, it was shown that unwrapping of DNA is an intrinsic property of NCP, and at physiological conditions individual NCPs are partially unwrapped about 2-10% of the time (2). The alternative model, known as the sliding model, where the core moves along the template was observed mainly in the presence of chaperones or modified histones (e.g., (38, 39)).
Single molecule time lapse AFM data allowed us to distinguish between the two models. Indeed, upon sliding/rolling of the histone core, one of the arms becomes longer whereas another arm becomes shorter. Other structural parameters of NCPs, such as the size of the nucleosome and the angle between the arms should remain unchanged over time. The expectations for the NCP structure of the site exposure model are different. First, the dissociation step is accompanied by the elongation of both arms of the NCP. Second, due to the removal of the DNA from the NCP core, the size of the nucleosome particles decrease. Third, the angle between the arms should vary as well. Our data, regardless of the particular DNA dissociation pathway, reveal the same pattern – both arms of the NCP become longer; this process is accompanied by the decrease of the nucleosome size and the change of the interarm angles. Therefore, the AFM data are in line with the site exposure model. Interestingly, this model is also supported by the imaging in air of dry NCP samples. As it is seen in Fig. 2, there is no same size NCP with different arms lengths that should appear if cores are able to slide along the DNA fragment. Just opposite, two parameters, the blob sizes and the interarm angles are varied. We cannot exclude that the DNA sequence contributes to this particular NCP dynamics pathways as the stability of NCP is DNA sequence dependent (1, 2, 10, 40-42), so the sequences other than 601 type will follow the sliding model. However, for 601 DNA sequence selected for this study, the sliding pattern has not been observed.
There is a common feature for various pathways of the NCP dissociation: the particles with about one DNA turn are rather unstable, so the nucleosome should wrap more DNA or should dissociate. The time-lapse data also provide the information on the core structure at the very last stage of the NCP unwrapping process. Fig. 3A (the two step process) shows that a small blob remains on the full length DNA substrate (frames 6-7), disappearing at the last frame. Similar complexes with small blobs can be seen on Fig.4A (frames 5-6). These are histone cores that remain bound to the DNA template, so we are able to estimate the stoichiometry of the cores based on the molecular weight values obtained from the volume measurements (30, 33). This analysis shows that anticipated volume of histone octamer (108kD) should be in the range of 210 nm3. The size of blob in Fig. 3C (frames 6-7) is smaller than this anticipated value. Interestingly, the volumes of blobs for frames 4-6 of Fig. 4C (one step process) are also lower than 200 nm3, although about 1 DNA turn remains on this particle. These observations suggest that in some NCP complexes, the histone core undergoes partial dissociation prior to the complete DNA unwrapping. At the same time, the volume data in Figs. 5A and B are of a different type: the sizes of blobs prior to the dissociation are larger than 200 nm3, suggesting that no histone dissociation took place. Another example of the dissociation of the full core is shown in SI section (Fig. S3- one step full dissociation and movie file M5), and data analysis for arm lengths, protein volume and number of turns are shown in Figs. S 3B and C respectively. Thus, these data suggest that upon the DNA unwrapping, different pathways with and without partial dissociation of the histones exist and the selection of the pathway may depend on the local environment around each particular NCP. There is biochemical evidence indicating that H2A-H2B dimers can be depleted (see review (43) and also (2, 16)) and this process facilitates the next DNA unwrapping step. The role of H2A-H2B histones on the NCP stability and dynamics has been revealed in the recent single molecule FRET experiments (44, 45) and this important issue has been discussed recently (42).
Thus, the time lapse AFM data presents the picture of the NCP complexes as very dynamic biomolecular systems, in which large DNA segments are capable of dissociating from the NCP particle. Our hypothesis is that the observed unwrapping is due to the thermal motion of nucleosomes rather than the AFM tip sweeping effect. There are a number of major pieces of evidence supporting this assumption. First, AFM images of dried sample show a wide heterogeneity of nucleosome morphologies and essentially the same heterogeneity has been observed in the time lapse single molecule imaging experiments. Second, for the same sample, unwrapping rate for nucleosomes varies in a broad range. It can be relatively fast, so a complete unwrapping occurs during a few frames (6 frames in Figs. 3 and 4) or relatively slow with very subtle changes of the DNA arms lengths (Fig. 5B, frames 1-8). Note that the scanning conditions in these experiments were identical and the same AFM tip was used. Third, we performed experiments in which the scanned and unscanned areas were compared. In these experiments, the large area was scanned once, then a smaller area was selected and continuous scanning over this area was performed until essentially all nucleosomes in the area dissociated. The microscope was unzoomed to the original large area that was rescanned. If the nucleosome unwrapping is caused by the scanning tip only, we would observe NCP unwrapping over the small area with no or little change of the nucleosome morphology in the rest of the image. The data for one of these experiments are shown in Fig. S5; these images show that unwrapped nucleosomes were observed in both areas. Thus, the observed dissociation of nucleosome is not due to the tip sweeping effect. However, we cannot exclude some “pushing” effect of the AFM tips facilitating unwrapping, as a few nucleosomes on the unscanned area remain compared to only one (right edge) on the scanned area.
The dynamics of nucleosomes in 50-100 ms time scale corresponds to the fluctuation of the DNA arms at the very edge of the particle (8). Our AFM experiments revealed unwrapping of much larger DNA segments including full unwrapping of nucleosomes. Note recent publications in which large scale dynamics of NCP was detected with single molecule FRET (46, 47). Additional indirect evidence for the large scale dynamics of nucleosome come from the recent paper (9) in which the dynamics of nucleosomes in vivo was probed by the DNA repair with photolyase. These data also revealed the dynamics of nucleosomes at the particle edges in a second time scale, but all lesions within the nucleosome were repaired in two hours suggesting that accessibility of internal regions is limited, and long times are required for exposing remote regions within nucleosomes. If the large scale unwrapping requires for the internal probing of nucleosomes, the obtained data suggest that the time scale for such a process is much larger than the seconds time scale for the small-range fluctuations. It was proposed recently (8) that large range dynamics of nucleosomes may employ a step-wise unwrapping process in which initial unwrapping starts at the edge of the nucleosomal DNA and moves inward. So, the movement of the unwrapped segment inside the nucleosome is a multistep process in which unwrapping of large regions requires long times.
It is tempting to extract the characteristic times from the single molecule AFM data, but these values cannot be obtained due to two major reasons. First, the temporal resolution of the traditional AFM instrument does not allow us to acquire the data with a rate faster than 1 frame per min. According to the data of Li et al (8) unwrapping occurs in the millisecond time scale. Therefore, special AFM techniques with considerably higher temporal resolution are needed. Note, however, in some of the time trajectories for the arms lengths (Figs. S2B and S4B (set of the data in Fig. S4 and movie file M6) we observed the wrapping/unwrapping events suggesting that the use of an instrument with higher temporal resolution may enable us to visualize more details of the NCP dynamics paths. Second, AFM is a topographic technique requiring the sample to be bound to the surface. We used positively charged APS-mica surface that has an elevated affinity for negatively charged DNA filaments compared to NCP particles. However, the APS-mica protocol permits us to control the strength of the sample-surface interaction, allowing us to observe DNA dynamics at the surface liquid-interface. At the same time, the interaction of the NCP samples with the surface is the factor that may change the NCP dynamics. We hypothesize that transiently unwrapped DNA segments can be trapped by electrostatic interactions with the surface increasing the probability for the next unwrapping step. These interactions shift the equilibrium of the unwrapping/wrapping dynamics towards unwrapping that eventually leads to full unwrapping of the nucleosome.
The hypothesis on the effect of electrostatic interactions on the NCP dynamics prompts new ideas to potential mechanisms for the regulation of unwrapping of NCP within chromatin. Large remodeling systems are involved in the regulation of chromatin dynamics; however, the mechanism whereby they perform nucleosome unwrapping is unclear. For example, in the wave-ratchet-wave model for ISWI remodeler, a small internal loop is moved along the DNA within the nucleosome (3). The requirement for a small loop was proposed to meet the energy needs for the unwrapping process. However, the necessary energy deficit can be provided from electrostatics. If electrostatic interactions of free DNA with positively charged surface increase the NCP dynamics, the DNA-surface contacts provided by positively charged groups located on the surfaces of the remodeling proteins facing the chromatin surface will shift the fluctuation towards unwrapping step. Our early estimates for the surface charge density of similar aminopropyl mica provided quite low values for the positive charge density, ca. one unit per an area of several nanometers (20) suggesting that a quite limited number of non-specific electrostatic in nature contacts may shift the equilibrium towards unwrapping. In addition to remodeling protein complexes, positive charges for the contacts with chromatin can be provided by intranuclear surfaces. Therefore, we speculate that the interaction of chromatin with these surfaces can provide additional contribution to the chromatin dynamics facilitating unwrapping of the chromatin. Therefore, APS-mica can be a considered as a model system for understanding the role of electroscopic interactions between the chromatin and intracellular membranes in regulation of the chromatin dynamics and genes activity.
Supplementary Material
Acknowledgments
We thank I. Nazarov for the help in the preparation of nucleosome samples J. Widom and C. Woodcock for critical reading of the manuscript and useful comments and Alex Portillo for proofreading of the manuscript.
The work is supported by grants EPS-0701892 and PHY-0615590 (both NSF) and grant GM062235 (NIH) to YLL.
Abbreviations
- AFM
Atomic Force Microscopy
- NCP
nucleosome core particle
- APS
1-(3-aminopropyl)silatrane
Footnotes
Supporting Information Available: The full sequence of the DNA fragment studied in this paper; the dependence of the protein volume measured with AFM on the protein molecular weight; images and graphs for different modes of NCP dynamics: full dissociation, wrapping-unwrapping, one step full dissociation; large scale AFM images in aqueous solution; movie files for animated data set of time lapse images.
This material is available free of charge via the Internet at http://pubs.acs.org”.
References
- 1.Luger K, Hansen JC. Nucleosome and chromatin fiber dynamics. Curr Opin Struct Biol. 2005;15:188–196. doi: 10.1016/j.sbi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Thoma F. Repair of UV lesions in nucleosomes--intrinsic properties and remodeling. DNA Repair (Amst) 2005;4:855–869. doi: 10.1016/j.dnarep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nat Rev Mol Cell Biol. 2006;7:437–447. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- 4.Cairns BR. Chromatin remodeling: insights and intrigue from single-molecule studies. Nat Struct Mol Biol. 2007;14:989–996. doi: 10.1038/nsmb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmad K, Henikoff S. Epigenetic consequences of nucleosome dynamics. Cell. 2002;111:281–284. doi: 10.1016/s0092-8674(02)01081-4. [DOI] [PubMed] [Google Scholar]
- 6.Anderson JD, Thastrom A, Widom J. Spontaneous access of proteins to buried nucleosomal DNA target sites occurs via a mechanism that is distinct from nucleosome translocation. Mol Cell Biol. 2002;22:7147–7157. doi: 10.1128/MCB.22.20.7147-7157.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li G, Widom J. Nucleosomes facilitate their own invasion. Nat Struct Mol Biol. 2004;11:763–769. doi: 10.1038/nsmb801. [DOI] [PubMed] [Google Scholar]
- 8.Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 9.Bucceri A, Kapitza K, Thoma F. Rapid accessibility of nucleosomal DNA in yeast on a second time scale. Embo J. 2006;25:3123–3132. doi: 10.1038/sj.emboj.7601196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tims HS, Widom J. Stopped-flow fluorescence resonance energy transfer for analysis of nucleosome dynamics. Methods. 2007;41:296–303. doi: 10.1016/j.ymeth.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koopmans WJ, Brehm A, Logie C, Schmidt T, van Noort J. Single-pair FRET microscopy reveals mononucleosome dynamics. J Fluoresc. 2007;17:785–795. doi: 10.1007/s10895-007-0218-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leuba SH, Yang G, Robert C, Samori B, van Holde K, Zlatanova J, Bustamante C. Three-dimensional structure of extended chromatin fibers as revealed by tapping-mode scanning force microscopy. Proc Natl Acad Sci U S A. 1994;91:11621–11625. doi: 10.1073/pnas.91.24.11621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leuba SH, Bustamante C, Zlatanova J, van Holde K. Contributions of linker histones and histone H3 to chromatin structure: scanning force microscopy studies on trypsinized fibers. Biophys J. 1998;74:2823–2829. doi: 10.1016/S0006-3495(98)77989-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yodh JG, Lyubchenko YL, Shlyakhtenko LS, Woodbury N, Lohr D. Evidence for nonrandom behavior in 208-12 subsaturated nucleosomal array populations analyzed by AFM. Biochemistry. 1999;38:15756–15763. doi: 10.1021/bi991034q. [DOI] [PubMed] [Google Scholar]
- 15.Yodh JG, Woodbury N, Shlyakhtenko LS, Lyubchenko YL, Lohr D. Mapping nucleosome locations on the 208-12 by AFM provides clear evidence for cooperativity in array occupation. Biochemistry. 2002;41:3565–3574. doi: 10.1021/bi011612e. [DOI] [PubMed] [Google Scholar]
- 16.Nikova DN, Pope LH, Bennink ML, van Leijenhorst-Groener KA, van der Werf K, Greve J. Unexpected binding motifs for subnucleosomal particles revealed by atomic force microscopy. Biophys J. 2004;87:4135–4145. doi: 10.1529/biophysj.104.048983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guthold M, Bezanilla M, Erie DA, Jenkins B, Hansma HG, Bustamante C. Following the assembly of RNA polymerase-DNA complexes in aqueous solutions with the scanning force microscope. Proc Natl Acad Sci U S A. 1994;91:12927–12931. doi: 10.1073/pnas.91.26.12927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansma HG, Bezanilla M, Zenhausern F, Adrian M, Sinsheimer RL. Atomic force microscopy of DNA in aqueous solutions. Nucleic Acids Res. 1993;21:505–512. doi: 10.1093/nar/21.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bustamante C, Rivetti C, Keller DJ. Scanning force microscopy under aqueous solutions. Curr Opin Struct Biol. 1997;7:709–716. doi: 10.1016/s0959-440x(97)80082-6. [DOI] [PubMed] [Google Scholar]
- 20.Lyubchenko YL, Shlyakhtenko LS. Visualization of supercoiled DNA with atomic force microscopy in situ. Proc Natl Acad Sci U S A. 1997;94:496–501. doi: 10.1073/pnas.94.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mikheikin AL, Lushnikov AY, Lyubchenko YL. Effect of DNA supercoiling on the geometry of holliday junctions. Biochemistry. 2006;45:12998–13006. doi: 10.1021/bi061002k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lushnikov AY, Bogdanov A, Lyubchenko YL. DNA recombination: holliday junctions dynamics and branch migration. J Biol Chem. 2003;278:43130–43134. doi: 10.1074/jbc.M308228200. [DOI] [PubMed] [Google Scholar]
- 23.Lyubchenko YL. DNA structure and dynamics: an atomic force microscopy study. Cell Biochem Biophys. 2004;41:75–98. doi: 10.1385/CBB:41:1:075. [DOI] [PubMed] [Google Scholar]
- 24.Ando T, Uchihashi T, Kodera N, Yamamoto D, Taniguchi M, Miyagi A, Yamashita H. High-speed atomic force microscopy for observing dynamic biomolecular processes. J Mol Recognit. 2007;20:448–458. doi: 10.1002/jmr.843. [DOI] [PubMed] [Google Scholar]
- 25.Crampton N, Roes S, Dryden DT, Rao DN, Edwardson JM, Henderson RM. DNA looping and translocation provide an optimal cleavage mechanism for the type III restriction enzymes. EMBO J. 2007;26:3815–3825. doi: 10.1038/sj.emboj.7601807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 27.Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 28.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 29.Shlyakhtenko LS, Gall AA, Filonov A, Cerovac Z, Lushnikov A, Lyubchenko YL. Silatrane-based surface chemistry for immobilization of DNA, protein-DNA complexes and other biological materials. Ultramicroscopy. 2003;97:279–287. doi: 10.1016/S0304-3991(03)00053-6. [DOI] [PubMed] [Google Scholar]
- 30.Lushnikov AY, Potaman VN, Oussatcheva EA, Sinden RR, Lyubchenko YL. DNA strand arrangement within the SfiI-DNA complex: atomic force microscopy analysis. Biochemistry. 2006;45:152–158. doi: 10.1021/bi051767c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyubchenko YL, Shlyakhtenko LS. AFM for analysis of structure and dynamics of DNA and protein-DNA complexes. Methods. 2009;47:206–213. doi: 10.1016/j.ymeth.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyubchenko YL, Shlyakhtenko LS, Gall AA. Atomic force microscopy imaging and probing of DNA, proteins, and protein DNA complexes: silatrane surface chemistry. Methods Mol Biol. 2009;543:337–351. doi: 10.1007/978-1-60327-015-1_21. [DOI] [PubMed] [Google Scholar]
- 33.Shlyakhtenko LS, Gilmore J, Portillo A, Tamulaitis G, Siksnys V, Lyubchenko YL. Direct visualization of the EcoRII-DNA triple synaptic complex by atomic force microscopy. Biochemistry. 2007;46:11128–11136. doi: 10.1021/bi701123u. [DOI] [PubMed] [Google Scholar]
- 34.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 35.Feng J, Chun-Cheng Z. Thermodynamics of nucleosomal core particles. Biochemistry. 2007;46:2594–2598. doi: 10.1021/bi061138i. [DOI] [PubMed] [Google Scholar]
- 36.Pavlicek JW, Oussatcheva EA, Sinden RR, Potaman VN, Sankey OF, Lyubchenko YL. Supercoiling-induced DNA bending. Biochemistry. 2004;43:10664–10668. doi: 10.1021/bi0362572. [DOI] [PubMed] [Google Scholar]
- 37.Furrer P, Bednar J, Dubochet J, Hamiche A, Prunell A. DNA at the entry-exit of the nucleosome observed by cryoelectron microscopy. J Struct Biol. 1995;114:177–183. doi: 10.1006/jsbi.1995.1017. [DOI] [PubMed] [Google Scholar]
- 38.Chakravarthy S, Park YJ, Chodaparambil J, Edayathumangalam RS, Luger K. Structure and dynamic properties of nucleosome core particles. FEBS Lett. 2005;579:895–898. doi: 10.1016/j.febslet.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 39.Park YJ, Chodaparambil JV, Bao Y, McBryant SJ, Luger K. Nucleosome assembly protein 1 exchanges histone H2A-H2B dimers and assists nucleosome sliding. J Biol Chem. 2005;280:1817–1825. doi: 10.1074/jbc.M411347200. [DOI] [PubMed] [Google Scholar]
- 40.Luger K. Dynamic nucleosomes. Chromosome Res. 2006;14:5–16. doi: 10.1007/s10577-005-1026-1. [DOI] [PubMed] [Google Scholar]
- 41.van Holde K, Zlatanova J. Scanning chromatin: a new paradigm? J Biol Chem. 2006;281:12197–12200. doi: 10.1074/jbc.R500025200. [DOI] [PubMed] [Google Scholar]
- 42.Sharma S, Dokholyan NV. DNA sequence mediates nucleosome structure and stability. Biophys J. 2008;94:1–3. doi: 10.1529/biophysj.107.119172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kimura H. Histone dynamics in living cells revealed by photobleaching. DNA Repair (Amst) 2005;4:939–950. doi: 10.1016/j.dnarep.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 44.Kelbauskas L, Chan N, Bash R, Yodh J, Woodbury N, Lohr D. Sequence-dependent nucleosome structure and stability variations detected by Forster resonance energy transfer. Biochemistry. 2007;46:2239–2248. doi: 10.1021/bi061289l. [DOI] [PubMed] [Google Scholar]
- 45.Kelbauskas L, Chan N, Bash R, DeBartolo P, Sun J, Woodbury N, Lohr D. Sequence-dependent variations associated with H2A/H2B depletion of nucleosomes. Biophys J. 2008;94:147–158. doi: 10.1529/biophysj.107.111906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomschik M, van Holde K, Zlatanova J. Nucleosome dynamics as studied by single-pair fluorescence resonance energy transfer: a reevaluation. J Fluoresc. 2009;19:53–62. doi: 10.1007/s10895-008-0379-1. [DOI] [PubMed] [Google Scholar]
- 47.Tomschik M, Zheng H, van Holde K, Zlatanova J, Leuba SH. Fast, long-range, reversible conformational fluctuations in nucleosomes revealed by single-pair fluorescence resonance energy transfer. Proc Natl Acad Sci U S A. 2005;102:3278–3283. doi: 10.1073/pnas.0500189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


