The importance of protein–protein interactions can never be overemphasized in biology. Important interactions are often weak and/or transient and facilitate cells to turn on and off signals. It still remains a daunting task to study these weak and/or transient protein interactions. Although chemical cross-linking is widely used to study protein–protein interactions, the experiments can be problematic with currently available cross-linking agents, which often suffer from low efficiency and/or poor selectivity. Site-specific protein cross-linking is an attractive option in many studies; however, very few examples of such methods have been demonstrated. Examples include the use of a disulfide bond between two cysteine residues[1] and photo-reactive unnatural amino acids.[2,3] In an effort to provide a better cross-linking method, herein we present a novel, site-specific protein cross-linking assay that utilizes genetically encoded 3,4-dihydroxy-l-phenylalanine (l-DOPA, 1). 
l-DOPA has been demonstrated as a selective chemical cross-linker for a peptide–protein interaction in vitro.[4] Upon oxidation by sodium periodate (NaIO4), l-DOPA forms an ortho-quinone intermediate that is subsequently attacked by nucleophilic side chains of amino acids in proximity on the interacting protein, through either Michael addition or Schiff base formation.[5,6] l-DOPA has also been incorporated into a protein in Escherichia coli using an engineered Methanococcus jannaschii tRNATyr/tyrosyl-tRNA synthetase pair in response to the amber stop codon (TAG).[7,8] Thus, it is becoming feasible to extend the l-DOPA-mediated peptide–protein cross-linking into protein–protein cross-linking (Scheme 1).
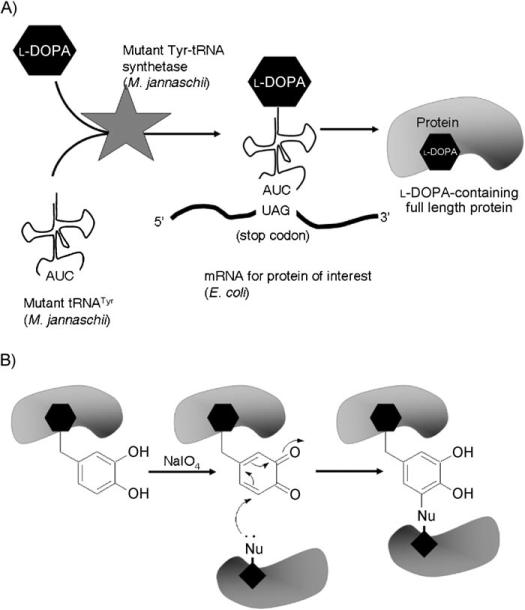
Scheme 1.
l-DOPA as a site-specific protein cross-linker. A) l-DOPA is site-specifically incorporated into proteins in response to an amber codon (TAG) in E. coli. B) Incorporated l-DOPA reacts with the nearby nucleophiles and results in the covalent cross-linking of protein dimer.
To demonstrate the experimental scheme shown above, a therapeutically important Sortase A (SrtA) of Staphylococcus aureus[9] was chosen as a model protein. SrtAΔN59, a truncated SrtA lacking the transmembrane domain, has recently been identified as a homodimer with a modest binding affinity (Kd = 56 μm).[10] The crystal structure of monomeric SrtAΔN59[11] indicates that the surface of this protein is abundant in polar and charged amino acid residues. A point mutation on its surface, K137A, has been shown to disrupt the dimerization of SrtAΔN59 without a significant change in protein conformation.[12] Thus, it is highly likely that this residue is involved in dimerization of SrtAΔN59 by establishing a selective electrostatic interaction with the residue(s) on the surface of the binding SrtAΔN59 counterpart. The cognate residue is a potential nucleophile that can attack the ortho-quinone intermediate formed by the oxidation of l-DOPA. Therefore, mutation of residue K137 of SrtAΔN59 to l-DOPA is likely to result in the cross-linking of the SrtAΔN59 dimer. In contrast, the C-terminal mutant K206A shows no effect in the dimerization state of the SrtAΔN59 protein (Figure S1 and S2 in the Supporting Information); this suggests that this residue is unlikely to be involved in the dimerization. Thus, mutation of K206 to l-DOPA should not result in a positive cross-linking reaction.
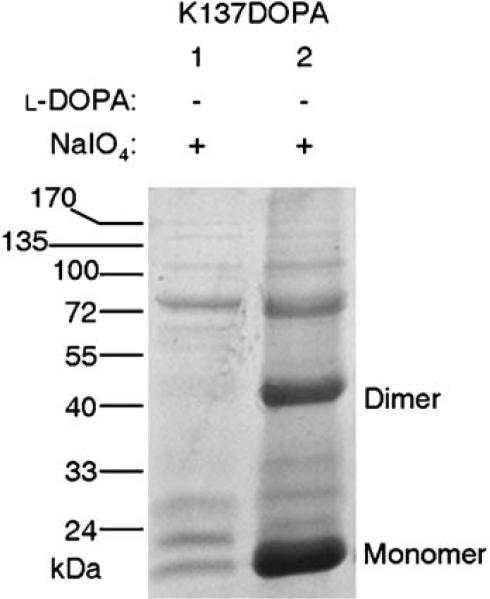
l-DOPA was incorporated into SrtAΔN59 using the established method[7,8] to obtain the C-terminal His6-tagged K137DOPA mutant, and the subsequent total protein analysis revealed the excellent yield of the cross-linking reaction (Figure 1). Following the incorporation, cross-linking of l-DOPA-containing SrtAΔN59 subunits was performed in cell lysate using NaIO4 (see the Experimental Section and Supporting Information). Purified proteins were resolved by SDS-PAGE followed by Coomassie Brilliant Blue staining. The result shows a strong band corresponding to the size of the SrtAΔN59 dimer (35.8 kDa, Figure 1, lane 2, Dimer). Subsequent mass-spectrometric analysis confirmed that this band contains the SrtA protein only (data not shown). In the absence of l-DOPA (Figure 1, lane 1), a trace amount of SrtAΔN59 expression is observed as monomer; this indicates the basal incorporation of endogenous natural amino acids, but no dimer is present. Thus, the cross-linking reaction is caused only by l-DOPA, but not by any endogenous amino acid or external factors present in the experimental process. These data suggest confirm the high yield of the cross-linking reaction that produced the covalent homodimer of SrtAΔN59.
Figure 1.
Cross-linking of the SrtAΔN59 dimer through l-DOPA at K137. l-DOPA incorporation was performed for K137DOPA with (lane 2) or without (lane 1) l-DOPA. Sodium periodate was added prior to the protein purification. Purified proteins were resolved by SDS-PAGE and visualized by Coomassie Brilliant Blue staining.
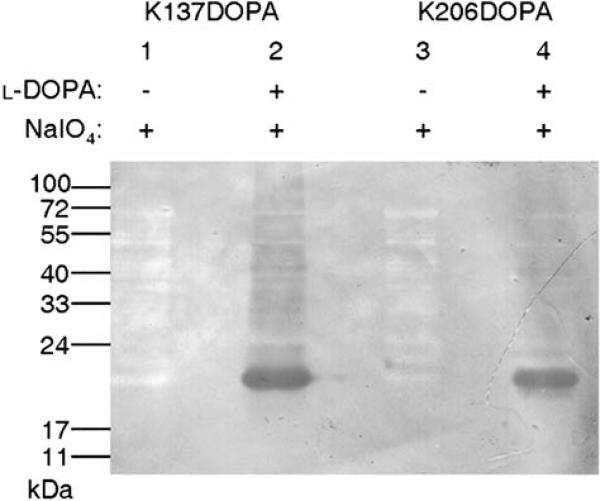
A redox-cycling staining assay that detects quino-proteins[13,14] confirmed the presence of l-DOPA within the mutant proteins K137DOPA and K206DOPA (Figure 2). Monomeric SrtAΔN59 (17.9 kDa) is observed in both K137DOPA and K206DOPA (Figure 2, lanes 2 and 4), indicating the presence of l-DOPA within the mutant SrtAΔN59 proteins. Only a trace amount of dimer can be seen for both K137DOPA and K206DOPA, which might be the noncovalent dimer. The abundant dimer observed in Figure 1 might not be detected by this method since the majority should be the product of the cross-linking reaction which modifies the ortho-quinone originated from l-DOPA.[6] Together with Figure 1, the results in Figure 2 confirm the high fidelity of l-DOPA incorporation at both residues tested and suggest that the covalent modification of the l-DOPA moiety resulted from the cross-linking reaction in the dimer of K137DOPA.
Figure 2.
Redox-cycling staining of l-DOPA-containing SrtAΔN59 proteins. K137DOPA (lanes 1 and 2) and K206DOPA (lanes 3 and 4) were oxidized, purified and resolved by SDS-PAGE, then transferred to a nitrocellulose membrane. DOPA-containing proteins were visualized with a staining solution containing nitroblue tetrazolium.
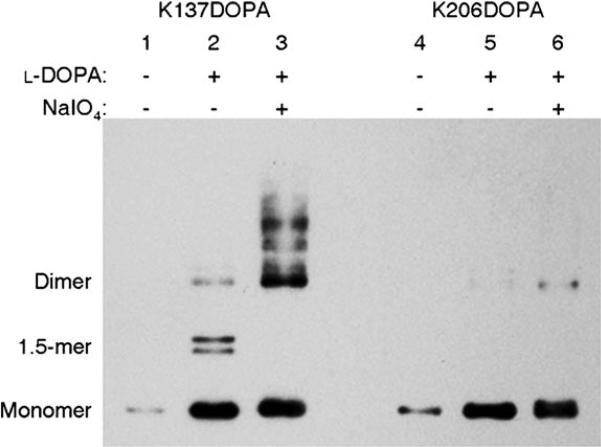
Western blot analysis of both K137DOPA and K206DOPA using an anti-His6 antibody verified the superb site-specific selectivity of the l-DOPA-mediated protein cross-linking assay (Figure 3). K137DOPA shows the covalent dimer in the presence of NaIO4 (Figure 3. lane 3); this is consistent with the total protein analysis (Figure 1). K137DOPA also shows a small amount of dimer in the absence of NaIO4 (Figure 3, lane 2). This band could indicate the trace of noncovalent dimer as seen in Figure 2 and wild type SrtAΔN59,[10] and/or covalent dimer that resulted from the minimal oxididation of l-DOPA by atmospheric oxygen.[6] Interestingly, the same sample also contains the bands that correspond to the size of 1.5-mer (Figure, lane 3, 1.5-mer). The identity of the components of these bands and their biological relevance are currently under investigation. Compared to K137DOPA, K206DOPA shows only a minimal amount of dimer in the presence of NaIO4 (Figure 3, lane 6). The lack of copious cross-linking at residue K206 indicates the absence of nucleophiles that are in the close proximity to this residue and available to react with l-DOPA moiety. The yields of cross-linked dimer from K137DOPA (Figure 3, lane 3) and K206DOPA (Figure 3, lane 6) are consistent with the results from the point-mutation studies[12] (Figures S1 and S2). The data presented here are the first direct evidence to confirm the physical interaction of SrtA at the residue level. Taken together, these results demonstrate the excellent selectivity of l-DOPA-mediated protein–protein cross-linking in the context of protein mixtures as well as its position-dependent specificity.
Figure 3.
Site-specificity of l-DOPA-mediated cross-linking of SrtAΔN59 dimer. K137DOPA (lanes 1−3) and K206DOPA (lanes 4−6) were oxidized and purified, then analyzed by SDS-PAGE followed by western blotting using an anti-His6 antibody.
In conclusion, we have demonstrated an efficient and selective site-specific protein cross-linking methodology by using genetically encoded l-DOPA. Successful cross-linking supports our recent discovery of SrtAΔN59 dimerization.[10,12] Furthermore, we have shown that the efficiency of the cross-linking does not cost its selectivity. The information from serial cross-linking experiments at different residues using the same method would allow us to fairly accurately map the dimerization domain of SrtA. The methodology presented here can also be extended to identify, confirm and/or characterize many protein–protein and protein–small molecule interactions. Highly reactive l-DOPA allows us to capture even weak or transient interactions, in cases in which other methods of characterizations are difficult. l-DOPA-mediated site-specific protein cross-linking can be a valuable addition to the set of tools currently available to study protein–protein and protein–small molecule interactions.
Experimental Section
Escherichia coli BL21 was transformed with the plasmid pET28-SrtA59K137TAG or pET28-SrtA59K206TAG and pAC-DHPheRS-6TRN. The culture was grown in glucose minimal media and protein expression was induced in the presence of l-DOPA (1 μm). The cells were harvested and resuspended in lysis buffer (50 μm NaH2PO4, 300 μm NaCl, 10 μm imidazole, pH 8.0). Sodium periodate (1 μm) was added, and the mixtures were incubated on ice for 1 hour. Cells were then lysed and the His6-tagged proteins were purified using Ni-NTA agarose beads. (See the Supporting Information.)
Supplementary Material
Acknowledgements
Authors thank Dr. Peter G. Schultz (The Scripps Research Institute) for the plasmid pAC-DHPheRS-6TRN, and Dr. Thomas J. Kodadek (UT Southwestern Medical Center) for helpful discussions. We also thank Mr. Chintan K. Modi (UT Austin) for assisting in the l-DOPA incorporation, and Dr. Maria D. Person (UT Austin ICMB Mass Spectrometry-Analytical Facility) for protein identification. This work is supported by The Welch Foundation (F1618) and NIH RO1 CA120 168.
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/cbic.200900127.
References
- 1.Bass RB, Butler SL, Chervitz SA, Gloor SL, Falke JJ. Methods Enzymol. 2007;423:25. doi: 10.1016/S0076-6879(07)23002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chin JW, Martin AB, King DS, Wang L, Schultz PG. Proc. Natl. Acad. Sci. USA. 2002;99:11020. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chin JW, Schultz PG. ChemBioChem. 2002;3:1135. doi: 10.1002/1439-7633(20021104)3:11<1135::AID-CBIC1135>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 4.Burdine L, Gillette TG, Lin HJ, Kodadek T. J. Am. Chem. Soc. 2004;126:11442. doi: 10.1021/ja045982c. [DOI] [PubMed] [Google Scholar]
- 5.Burzio LA, Waite JH. Biochemistry. 2000;39:11147. doi: 10.1021/bi0002434. [DOI] [PubMed] [Google Scholar]
- 6.Liu B, Burdine L, Kodadek T. J. Am. Chem. Soc. 2006;128:15228. doi: 10.1021/ja065794h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alfonta L, Zhang Z, Uryu S, Loo JA, Schultz PG. J. Am. Chem. Soc. 2003;125:14662. doi: 10.1021/ja038242x. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Z, Smith BA, Wang L, Brock A, Cho C, Schultz PG. Biochemistry. 2003;42:6735. doi: 10.1021/bi0300231. [DOI] [PubMed] [Google Scholar]
- 9.Ton-That H, Liu G, Mazmanian SK, Faull KF, Schneewind O. Proc. Natl. Acad. Sci. USA. 1999;96:12424. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu C, Zhu J, Wang Y, Umeda A, Cowmeadow RB, Lai E, Moreno GN, Person MD, Zhang Z. Biochemistry. 2007;46:9346. doi: 10.1021/bi700519w. [DOI] [PubMed] [Google Scholar]
- 11.Zong Y, Bice TW, Ton-That H, Schneewind O, Narayana SV. J. Biol. Chem. 2004;279:31383. doi: 10.1074/jbc.M401374200. [DOI] [PubMed] [Google Scholar]
- 12.Zhu J, Lu C, Standland M, Lai E, Moreno GN, Umeda A, Jia X, Zhang Z. Biochemistry. 2008;47:1667. doi: 10.1021/bi7014597. [DOI] [PubMed] [Google Scholar]
- 13.Fluckiger R, Woodtli T, Gallop PM. Biochem. Biophys. Res. Commun. 1988;153:353. doi: 10.1016/s0006-291x(88)81230-0. [DOI] [PubMed] [Google Scholar]
- 14.Paz MA, Gallop PM, Torrelio BM, Fluckiger R. Biochem. Biophys. Res. Commun. 1988;154:1330. doi: 10.1016/0006-291x(88)90285-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.