Abstract
The cell surface of Trypanosoma cruzi, the etiologic agent of Chagas disease, is covered by a dense layer of glycosylphosphatidylinositol (GPI)-anchored molecules. These molecules are involved in a variety of interactions between this parasite and its mammalian and insect hosts. Here, using the neutral detergent Triton X-114, we obtained fractions rich in GPI-anchored and other membrane proteins from insect developmental stages of T. cruzi. These fractions were analyzed by two-dimensional liquid chromatography coupled to tandem mass spectrometry (2D-LC-MS/MS), resulting in the identification of 98 proteins of metacyclic trypomastigotes and 280 of epimastigotes. Of those, approximately 65% (n=245) had predicted lipid posttranslational modification sites (i.e., GPI-anchor, myristoylation, or prenylation), signal-anchor sequence, or transmembrane domains that could explain their solubility in detergent solution. The identification of some of these modified proteins was also validated by immunoblotting. We also present evidence that, in contrast to the noninfective proliferative epimastigote forms, the infective nonproliferative metacyclic trypomastigote forms express a large repertoire of surface glycoproteins, such as GP90 and GP82, which are involved in adhesion and invasion of host cells. Taken together, our results unequivocally show stage-specific protein profiles that appear to be related to the biology of each T. cruzi insect-derived developmental form.
Keywords: Trypanosoma cruzi, insect-dwelling stages, glycosylphosphatidylinositol (GPI)-anchored proteins, Triton X-114 partition, surface glycoproteins, trans-sialidase superfamily, membrane proteins, host-cell invasion, pathogenesis
1. Introduction
The major cell surface molecules expressed by insect-transmitted protozoan parasites from the order Kinetoplastida (Trypanosoma brucei, Trypanosoma cruzi, and Leishmania spp.) are glycosylphosphatidylinositol (GPI)-anchored glycoconjugates. T. brucei major glycoconjugate is the variant surface glycoprotein (VSG), whereas the T. cruzi glycocalyx is mainly formed by mucins and members of the trans-sialidase (TS) superfamily. The glycocalyx coat in trypanosomatids is also rich in complex GPI-related glycophospholipids, such as lipophosphoglycans (LPG) and glycoinositolphospholipids (GIPLs) (reviewed in 1-5). Considering that GPI-anchored molecules have been shown to be involved in host-cell adhesion and invasion, signaling and activation of the host proinflammatory response, and escape from host immune response,1, 3-6 they are attractive targets for the development of vaccines or drugs against these pathogenic protozoa.
T. cruzi is the etiologic agent of Chagas disease, or American trypanosomiasis, an infection without any effective drug treatment or immunotherapy that affects over 11 million people in the Americas.7 T. cruzi is naturally transmitted to mammalian hosts through the excrements of infected triatomine bugs. When a triatomine blood feeds on an infected animal, bloodstream trypomastigotes are ingested and reach the insect's midgut, where they differentiate into epimastigotes. The epimastigotes proliferate by asexual reproduction, and later in the insect's rectum, they transform into the infective metacyclic trypomastigotes. During a second blood meal by the insect vector, metacyclic forms concomitantly released into the feces may infect a new mammalian host through exposed bite wound or mucosal tissues, and immediately invade a wide range of host cells.
Similarly to the mammalian stages of the parasite, the cell surface of epimastigotes and metacyclic trypomastigotes is extensively covered by GPI-anchored mucins and GIPLs, formerly designated lipopeptidophosphoglycan (LPPG).2, 3, 5, 6 Metacyclic trypomastigotes which establish the initial parasite-host cell interaction, express two major stage-specific GPI-anchored glycoproteins, namely GP90 and GP82, with no counterpart in bloodstream trypomastigotes.8-10 GP82 is a cell adhesion molecule that induces a bidirectional Ca2+ response, an event essential for T. cruzi penetration into the host cell (reviewed in 11). As opposed to GP82, GP90 binds to mammalian cells in a receptor-mediated manner without triggering a Ca2+ signal and functions as a negative modulator of cell invasion.11 Sequence analysis showed that GP90 and GP82 share similarity with members of the T. cruzi TS superfamily, which comprises a large number of genes encoding major surface antigens of parasite infective forms.12-14
The plasma membrane of T. cruzi is likely to contain proteins that could serve as novel drug targets, diagnostic probes, or antigens for vaccines against Chagas disease. These surface proteins are coded by several hundred genes, and it would be essential to know which genes are expressed at each life cycle stage. However, since T. cruzi regulates gene expression primarily by posttranscriptional mechanisms, such as mRNA turnover and translation control, this limits the use of tools based on nucleic acid information (such as RNA microarrays) to study gene expression in the different developmental forms of the parasite. Thus, sequencing proteins by proteomic analysis is particularly an attractive approach. In the present study we undertook a comparative proteomic analysis of GPI-anchored membrane protein-enriched fractions from epimastigotes and metacyclic trypomastigotes. Identifying and characterizing these membrane proteins is a special challenge because of their structural complexity and physicochemical properties. GPI-anchored proteins were extracted using a simple, fast, and sensitive method that employs the neutral detergent Triton X-114 (TX-114), and further identified by immunoblotting and two-dimensional liquid chromatography coupled to tandem mass spectrometry (2D LC-MS/MS). This study highlights the efficiency of an integrative proteomic approach that combines experimental and computational methods to provide the selectivity, specificity, and sensitivity required for characterization of posttranslationally modified membrane proteins.
2. Materials and Methods
2.1. Parasites
T. cruzi G strain was maintained alternately in mice and at 28 °C in liver infusion tryptose (LIT) medium containing 5% fetal calf serum (FCS).15 Epimastigotes were harvested from 3-day cultures, whereas metacyclic trypomastigotes were harvested from cultures in the stationary growth phase (10−12 days) and purified by anion-exchange chromatography on a diethylaminoethyl (DEAE)-cellulose column as described elsewhere.8
2.2. Protein extraction and Triton X-114 partitioning
Proteins were extracted according to a protocol published elsewhere,16 with minimal modifications (Fig. 1A). Parasites from cultures were washed three times with PBS to remove proteins from the medium. A parasite pellet containing 1 × 108 cells was homogenized in 1 mL lysis buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2% TX-114, 1 mM PMSF) on ice for 1 h with periodic agitation. The homogenate was clarified by centrifugation at 8,800 × g for 10 min at 0 °C and the supernatant (S1) was stored at −20 °C for 24 h. The pellet was re-extracted with buffer A (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.06% TX-114, 1 mM PMSF), incubated for 10 min on ice and centrifuged at 8,800 × g at 0 °C to produce the pellet (P1) and buffer-A supernatant (SBA).
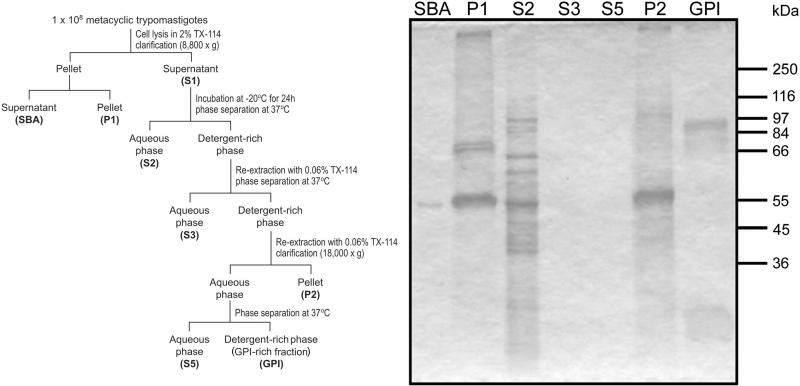
Figure 1.
Panel A) Schematic representation of the extraction protocol for membrane proteins. Metacyclic trypomastigotes (1 × 108) were lysed in TBS containing 2% Triton X-114 (TX-114). The homogenate was clarified by centrifugation and the supernatant stored at −20 °C for 24 h. The supernatant was then submitted to phase separation with TX-114 at 37 °C. The final detergent-rich extract corresponds to the GPI-anchored protein-enriched fraction. Panel B) Analysis by SDS-PAGE and Coomassie staining of molecules obtained in the purification steps of GPI-anchored proteins. SBA, supernatant from extraction step with buffer A; P1, Pellet 1; S2, supernatant 2; S3, supernatant 3; S5, supernatant 5; P2, pellet 2; GPI, GPI-rich fraction. The relative molecular masses are indicated on the right.
After incubation at −20 °C, the first supernatant (S1) was thawed, homogenized and submitted to phase separation by incubation at 37 °C for 10 min. Phases were separated by centrifugation at 3,000 × g for 3 min at room temperature. The upper phase (S2) was collected and the detergent-rich phase (lower phase) re-extracted with 1 mL of buffer A, mixed, and incubated at 0 °C for 10 min, and then submitted to a new phase extraction under the same conditions as above. The upper phase (S3) was collected, and the detergent-rich phase was extracted with 1 mL of buffer A, homogenized, incubated for 10 min at 0 °C, and clarified by centrifugation at 18,000 × g for 10 min at 0 °C. The pellet (P2) was stored and the upper phase (S4) was submitted to a new phase separation as described above. The upper phase (S5) was then collected and the detergent-rich phase (approximately 100 μL) was precipitated with 3 volumes of cold acetone at −20 °C for 30 min. Proteins were recovered by centrifugation at 18,000 × g at 0 °C, and the pellet was dried on the bench at room temperature. This final extract was named GPI-rich fraction (GPI) (Fig. 1A).
2.3. Protein digestion and peptide purification
GPI-rich fractions were resuspended in 20 μL 400 mM NH4HCO3 containing 8 M urea and subjected to trypsin digestion, as described by Stone.17 After reducing the disulfide bonds with 5 mM dithiothreitol at 50 °C for 15 min, the cysteine residues were alkylated with 10 mM iodoacetamide for 15 min at room temperature and protected from light. The reaction mixture was then diluted 8-fold and incubated overnight at 37 °C after adding 2 μg sequencing-grade trypsin (Promega, Madison, WI). The reaction was terminated by adding 0.05% trifluoroacetic acid (TFA) (final concentration) and the digested proteins were desalted using reverse phase (RP) ziptip columns (POROS R2 50, Applied Biosystems, Foster City, CA), as described by Jurado.18
To remove the detergent present in the preparation, the peptides were purified/fractionated in a strong cation-exchange (SCX) ziptip. Peptides eluted from the RP ziptip purification were diluted by addition of an equal volume of SCX buffer [25% acetonitrile (ACN)/0.5% formic acid (FA)]. SCX ziptip columns were manufactured in our laboratory using POROS HS 50 resin (Applied Biosystems) and equilibrated with SCX buffer. After loading the samples, the column was exhaustively washed (30 times the column volume) with SCX buffer to remove any remaining traces of detergent from the preparation. The peptides were eluted with increasing concentrations of NaCl (25, 50, 100, 200, and 500 mM) dissolved in the same buffer. The fractions were dried in a vacuum centrifuge (Vacufuge, Eppendorf) to remove the ACN, desalted in reverse-phase ziptips, and dried again.
2.4. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis
Each fraction was redisolved in 20 μL 0.1% FA and analyzed by LC-MS/MS in an ESI-QTOF-MS (Micromass Q-Tof 1, Waters, Milford, MA) or an ESI-linear ion-trap-MS (LTQ XL, Thermo Fisher Scientific, San Jose, CA). For the Q-Tof 1 analysis, one μL was injected into a PepMap reversed-phase column (15 cm × 75 μm, 3-μm C18, LC Packings, Dionex) coupled to an Ultimate (LC Packings, Dionex) nanoHPLC. The run was performed in a flow rate of 300 nL/min with a gradient from 5 to 35% ACN in 0.1% FA over 100 min. Eluting peptides were directly analyzed in the Q-Tof 1. Spectra were collected in the positive-ion mode in the 400−1800 m/z range for 2 s, and the three most abundant peptides were fragmented with a ramp collision energy (22−60 eV) for 3 s in the 50−2050 m/z range. The dynamic exclusion was set to fragment each peptide only once and then exclude for 2 min.
For the LTQ XL analysis, eight-microliter aliquots of each sample were loaded into a C18 trap column (0.25 μL, C18, OPTI-PAK, Oregon City, OR). The separation was performed on a capillary reverse-phase column (Acclaim, 3-μm C18, 75 μm × 25 cm, LC Packings. Amsterdam, The Netherlands) connected to a nanoHPLC system (nanoLC 1D plus, Eksigent, Dublin, CA). Peptides were eluted using a linear gradient from 5 to 40% ACN in 0.1% FA over 200 min and directly analyzed in the LTQ XL equipped with a nanospray source. MS spectra were collected in centroid mode in a range from 400 to 1700 m/z, and the five most abundant ions were submitted twice to CID (35% normalized collision energy) before dynamic exclusion for 120 s.
2.5. Database search and protein/peptide identification
MS/MS spectra collected in the Q-Tof 1 were converted into peak lists (PKL format) using ProteinLynx 2.0 (Waters). MS peaks were smoothed twice with 5 channels using the Savitzky-Golay method and centered with 4 channels in the top 80%. For the MS/MS peaks a threshold of 5% was set. After smoothing twice with 3 channels using the Savitzky-Golay method, the peaks were centered with 4 channels in the top 80% and converted into monoisotopic ions. MS/MS spectra collected in the LTQ XL from peptides with masses of 600−3500 Da, at least 100 counts, and 15 fragments, were converted into DTA files using Bioworks v.3.3.1 (Thermo Fisher Scientific).
LTQ XL data were submitted to database search using Sequest19 (available in Bioworks), and both Q-Tof 1 and LTQ XL data were searched with Phenyx20 (web interface, version 2.5, GeneBio, Switzerland). The database used in the analyses consisted of the forward and reverse T. cruzi, bovine, human keratin, and porcine trypsin sequences from GenBank (191,762 non-redundant sequences downloaded on Mar 17th, 2008 from http://www.ncbi.nlm.nih.gov/sites/entrez?db=protein&cmd=search&term=).
Database search parameters included i) trypsin cleavage in both peptide termini with one missed cleavage site allowed; ii) carbamidomethylation of cysteine residues as a fixed modification; iii) oxidation of methionine residues as a variable modification; iv) peptide-mass tolerance of 2.0 Da for LTQ XL or 500 ppm for Q-Tof 1 data; and v) 1.0 Da (for both LTQ XL and Q-Tof 1 data) for fragment mass tolerance. In addition, Q-Tof 1 data were submitted to a second round of searching by Phenyx with the same parameters as above, but with a trypsin cleavage in at least one of the peptide termini with two missed cleavage sites allowed, and 1500 ppm for peptide mass tolerance.
To ensure the quality of protein identification, the false-positive rate (FPR) was estimated using the following formula: FPR = Number of proteins matching the reverse sequences / Total number of proteins. The FPR was calculated after applying the following filters in Bioworks (for Sequest analysis): DCn ≥ 0.1; protein probability ≤ 1 × 10−3; and Xcorr ≥ 1.5, 2.0, and 2.5, for singly-, doubly- and triply-charged peptides, respectively. With these parameters we validated all sequences with an FPR < 2%. For Phenyx analysis, a minimum peptide Z-score of 5, p-value of 1 × 10−4, and a minimum of 2 distinct peptides were used. In addition, the protein AC score was set according to each dataset to obtain a maximum of 5% FPR.
2.6. Bioinformatics analysis of identified T. cruzi protein sequences
All identified T. cruzi protein sequences were submitted to GPI-anchor prediction analysis using the FragAnchor algorithm21 (available at http://navet.ics.hawaii.edu/~fraganchor/NNHMM/NNHMM.html). The prediction of transmembrane domains22, myristoylation23 and prenylation24 sites, and signal-anchor sequence25 was performed using the TMHMM, NMT, PrePS, and SignalP algorithms, respectively, available at Expert Protein Analysis System (ExPASy) Proteomics server (http://www.expasy.org).
2.7. Polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting
Polyacrylamide gels (10%) were run for about 4 h at 8 mA/gel, and the resolved proteins were transferred onto nitrocellulose membranes. The membranes were then blocked with 5% skim milk in PBS (PBS-milk) and probed with monoclonal or polyclonal antibodies diluted in PBS-milk. After washing in PBS-milk containing 0.05% Tween 20, the membranes were incubated with anti-mouse IgG conjugated to horseradish peroxidase (Sigma-Aldrich) diluted 1:10,000 in PBS-milk. After extensive washing in PBS containing 0.05% Tween 20, the blot was developed by chemiluminescence using the ECL Western blotting detection reagent and Hyperfilm-MP (Amersham, GE Healthcare Life Sciences). The monoclonal antibodies (mAbs) 1G7, 3F6, and 10D8 are specific for the T. cruzi GPI-anchored surface glycoproteins GP90, GP82, and GP35/50 (i.e., epimastigote and metacyclic trypomastigote mucins), respectively.8 The mAb 25 is specific for the flagellar calcium-binding protein (FCaBP).26, 27
3. Results and Discussion
3.1. SDS-PAGE and immunoblotting analyses of T. cruzi GPI-enriched fractions extracted with Triton X-114
Here, we used a Triton X-114 detergent-based two-phase separation for the extraction of membrane proteins, particularly GPI-anchored proteins from insect-dwelling developmental forms of T. cruzi (Fig. 1A). Triton X-114 is fully water-soluble at 0 °C, forming a single-phase mixture; however, at 37 °C, it partitions into aqueous and detergent-rich phases. After three consecutive phase separation steps, we obtained a putative GPI-rich fraction, which was first analyzed, in parallel with other intermediate fractions, by 10% SDS-PAGE gel stained with Coomassie blue (Fig. 1B). This putative GPI-rich fraction from metacyclic trypomastigotes showed a poor staining profile, yet substantially different from that of the other fractions. When analyzed by SDS-PAGE stained with silver, GPI-rich fraction showed three major bands with relative molecular masses (Mr) of approximately 90, 24, and 14 kDa, and four faint bands of 82, 75, 58, and 20 kDa (Fig. 2). Two broad bands with Mr of approx. 50 and 35 kDa could only be visualized by silver staining (Fig. 2, lane 2). Next, we used mAbs to identify some of these proteins (Fig. 2, lanes 3−6). The mAb 1G7, which is specific for GP90, stained the 90-kDa band (lane 3), and mAb 3F6, directed to GP82, reacted with two bands with Mr of 82 and 30 kDa (lane 4). Metacyclic forms from different T. cruzi isolates expressed GP30, a surface glycoprotein detectable by mAb 3F6.28 The mAb 10D8, which is specific for T. cruzi mucins GP35/50, strongly reacted with the GPI-enriched fraction (lane 5). The 24-kDa protein corresponds to the FCaBP,26 as demonstrated by the reaction with mAb 25 (lane 6). The calpain-like cysteine proteases29 reacted with monospecific polyclonal antibodies against the catalytic domain (Cys Pc) of this enzyme (A. Galetovic and J. F. da Silveira, unpublished) (lane 7), and appeared as two faint bands with Mr of approximately 56 and 66 kDa.
Figure 2.
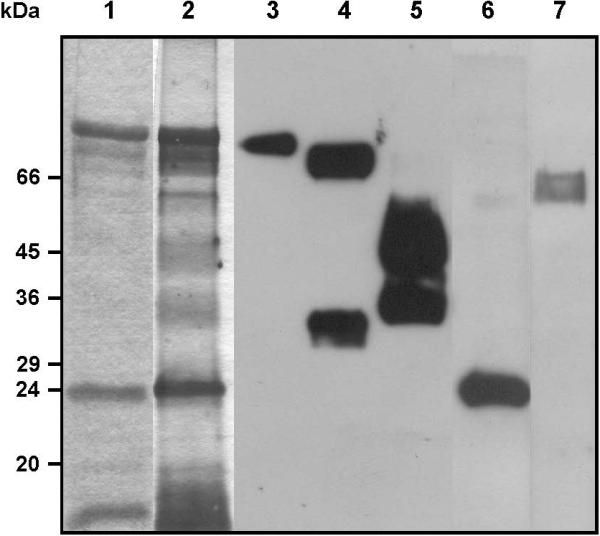
Gel electrophoresis and immunoblotting of Triton X-114-soluble membrane proteins from metacyclic trypomastigotes. Lanes 1 and 2, SDS-PAGE. Proteins were separated by SDS–PAGE on 10%-Laemmli gels and stained with Coomassie blue (lane 1) or silver (lane 2). Lanes 3 to 7, immunoblotting with monoclonal antibodies (mAb) against T. cruzi GPI-anchored surface proteins and flagellar calcium-binding protein (FCaBP) (lanes 3 to 6) and with a monospecific polyclonal antiserum against the catalytic domain of T. cruzi calpain-like cysteine proteinase (lane 7). The mAbs used were: 1G7 against GP90 surface glycoprotein (lane 3); 3F6 against GP82 surface glycoprotein (lane 4); 10D8 against the GP35/50 kDa mucin complex (lane 5); and 25 against the T. cruzi FCaBP (lane 6).
The putative GPI-rich fraction from epimastigotes was also analyzed by SDS-PAGE (data not shown). No proteins could be detected by Coomassie blue staining, suggesting that they were present in small amounts or they did not stain with this particular stain. In fact, the latter was the case two major bands with Mr of 50 and 35 kDa that were detected only by silver staining and identified as GP35/50 mucins with mAb 10D8. Regardless the developmental stage, T. cruzi mucins do not stain with Coomassie blue (I.C. Almeida et al., unpublished observations). Our results differ to some extent from those of Anez-Rojas30, who analyzed the protein profiles of epimastigote detergent extracts from Venezuelan T. cruzi isolates and reported the presence of two other bands with Mr of approximately 30 and 20 kDa. This discrepancy may be due to the fact that different isolates were used in the two studies.
3.2. Identification of metacyclic trypomastigote and epimastigote proteins by mass spectrometry
Next, the GPI-rich fraction was digested with trypsin and analyzed by 2D LC-MS/MS. In total, 378 proteins from epimastigotes (n=280) and metacyclic trypomastigotes (n=98) were unambiguously identified with one or more peptide sequences (Supplementary Table 1; see complete dataset in Suppl. Table 3, and Suppl. Figs. 1 and 2). Of those, 245 (64.8%) were predicted to have lipid posttranslational modifications (PTMs) (i.e., GPI-anchor, prenylation, or myristoylation), or at least one hydrophobic membrane-spanning domain that could account for their presence in the TX-114-rich fraction (Table 1 and Suppl. Table 2). The remaining proteins may represent: (i) soluble proteins that were not completely removed by detergent phase partitioning; (ii) proteins with physicochemical properties that would cause them to partition into the detergent phase; and (iii) peripheral membrane proteins.
Table 1.
Lipid posttranslational modifications (PTMs), hydrophobic membrane-spanning domains, sorting signals, and organellar localization of T. cruzi proteins extracted with Triton X-114 and identified by LC-MS/MS
| Lipid PTM, Hydrophobic Membrane-Spanning Domain, or Sorting Signal | Metacyclic Trypomastigote N (% total) a | Epimastigote N (% total) a |
|---|---|---|
| GPI Anchor | 21 (21.4%) | 1 (0.4%) |
| Myristoylation and Prenylation | 15 (15.3%) | 21 (7.5%) |
| Transmembrane Domain | 4 (4.1%) | 20 (7.1%) |
| Signal Peptide b | 26 (26.5%) | 72 (25.7%) |
|
Total |
66 (67.3%) |
114 (40.7%) |
|
Organelle distribution |
|
|
| Mitochondrial Targeting Sequence | 16 (16.3%) | 49 (17.5%) |
| Reservosomal membrane proteins | N/A c | 145 (51.8%) |
| Total d | 16 (16.3%) | 174 (62.1%) |
Number and percentage of the total identified proteins. Total proteins: metacyclic trypomastigotes, n=98 (100%); epimastigotes, n=280 (100%).
Proteins containing a typical ER signal peptide motif or an uncleaved signal-anchor sequence were included.
N/A, not applicable: reservosomes are not present in metacyclic trypomastigotes.60
Only non-redundant proteins.
We found matching entries in the T. cruzi databases for 98 proteins identified in metacyclic trypomastigotes (Suppl. Tables 1 and 2). Sixty-six (67.3%) of these proteins were predicted to have lipid PTM (i.e., GPI-anchor, prenylation, or myristoylation) (n=36; 36.7%), transmembrane domain (n=4; 4.1%), or endoplasmic reticulum (ER) signal peptide or targeting sequence (n=26; 26.5%) that could explain their presence in the TX-114-rich fraction. Out of the remaining 32 proteins, sixteen (16.3%) had mitochondrial targeting sequences (MTS), and 16 (16.3%) appeared not to have any potential lipid PTM or hydrophobic domain (Table 1 and Supplementary Tables 2 and 3b).
The protein content of epimastigote GPI-rich fraction largely differed from that of metacyclic trypomastigotes (Suppl. Table 1). Among the 280 proteins identified in epimastigotes, 114 (40.7%) contained lipid PTMs (n=22; 7.9%), ER signal peptide (n=72; 25.7%) or transmembrane domain (n=20; 7.1%), whereas 117 proteins (41.8%) did not show any potential lipid PTMs or hydrophobic domains. In contrast to the metacyclic GPI-rich fraction, where 21 of the identified proteins were predicted to be GPI-anchored, the epimastigote counterpart had only one potentially GPI-anchored protein (i.e., Tc85−11) (Table 1 and Supplementary Tables 2 and 3).
Intriguingly, a relatively high proportion (12.7%) (n=48) of proteins identified here contain what seems to be an uncleaved signal anchor sequence at the N-terminus (Table 1 and Supplementary Table 3b).31, 32 Possibly, many T. cruzi surface antigen genes have two or three in-frame ATG initiator codons at their 5’ end. For instance, in several TS protein genes, the predicted translation from the first in-frame ATG encodes a sequence which does not conform to typical ER signal peptide sequences.13, 33 However, after the second or third methionine residue, there is a typical signal anchor sequence (Figure 3). It remains to be determined which is the effective ATG initiator codon used in vivo by the parasite.
Figure 3.
Peptide-mapping coverage of GP90 and GP82. Representative fitting of tryptic peptides (shaded in gray) from 3 independent samples of GPI-rich extracts from metacyclic trypomastigotes that match with the deduced proteins GP90 (AAM47176) and GP82 (ABR19835) are shown. Potential initiator methionine residues are indicated by asterisks. Underlined sequences indicate the following motifs: signal-anchor or uncleaved signal peptide (positions 39−63 in GP90 and GP82); P4 and P8 cell-binding sites of GP82 (positions 462−481 and 502−521, respectively); FLY domain (positions 670−680 and 678−688 in GP90 and GP82, respectively). The hydrophobic C-terminal region of GP82, characteristic of GPI-anchored proteins, is indicated in italics (positions 702−726). The coverage percentage for GP90 and GP82 is 40% and 22%, respectively.
About 35% (n=133) of the total identified proteins (16 in metacyclics and 117 in epimastigotes) had no predicted lipid PTM or transmembrane domain (Suppl. Table 3b, d). However, some of these proteins, such as cytoskeletal proteins (17 identifications in epimastigotes and 4 in metacyclic trypomastigotes), translation elongation factor 1-beta (eEF1B) (two in metacyclic trypomastigotes and one in epimastigotes), enolase and acidic ribosomal proteins (14 identifications), had physicochemical properties that would allow them to be extracted with detergent.34-35
3.3. Functional Classification of Identified Proteins
Functional classification of proteins was based on information from published literature and Swiss-Prot Protein Knowledgebase (www.expasy.org/sprot/). Metacyclic and epimastigote proteins were grouped into functional categories (Table 2 and Suppl. Table 3a, c). Although several categorization schemes could be used, we grouped proteins that share similar functions rather than following a strict biochemical classification. Functions were assigned according to the known or putative involvement of the protein in a cellular process or pathway. Approximately 71% (n=270) of all identifications in epimastigote and metacyclic forms could be classified into seven broad functional categories (Table 2 and Suppl. Table 3a, c). Seventy-five (76.5%) and 195 (69.6%) proteins identified from metacyclic trypomastigotes and epimastigotes, respectively, could be assigned to a cellular function (Table 2 and Suppl. Table 3a, c). The majority of metacyclic proteins fit into the host-parasite category (39.8%) (n=39) and several of them are potentially GPI-anchored (Table 2 and Suppl. Table 3b, d). This group is followed by the categories: intermediate metabolism and bioenergetics (11.2%) (n=11); cellular communication and signal transduction (11.2%) (n=11); protein metabolism (proteolysis/peptidolysis, protein folding, vesicular trafficking) (8.2%) (n=8); structural proteins and cellular dynamics (4.1%) (n=4); and stress response and cell defense (antioxidation and detoxification) (2.0%) (n=2).
Table 2.
Functional Classification of T. cruzi Proteins Extracted with Triton X-114
| Functional Category | Metacyclic Trypomastigote N (%)a | Epimastigote N (%)a |
|---|---|---|
| Host-Parasite Relationship (surface proteins) | 39 (39.8%) | 6 (2.1%) |
| Cellular Communication and Signal Transduction | 11 (11.2%) | 20 (7.1%) |
| Intermediate Metabolism and Bioenergetics | 11 (11.2%) | 71 (25.4%) |
| Bioenergetics and Biological Oxidations | 7 | 19 |
| Lipid Metabolism | 2 | 11 |
| Amino Acid Metabolism | 1 | 18 |
| Carbohydrate Metabolism | 0 | 8 |
| Transporters | 1 | 15 |
| Protein Metabolism | 8 (8.2%) | 48 (17.1%) |
| Proteolysis/Peptidolysis | 6 | 21 |
| Protein Folding | 2 | 20 |
| Vesicular Trafficking | 0 | 7 |
| Structural Proteins and Cellular Dynamics | 4 (4.1%) | 17 (6.1%) |
| Stress Response and Cell Defense | 2 (2.0%) | 10 (3.6%) |
| Transcription, Translation and DNA Repair | 0 | 23 (8.2%) |
| Hypothetical Proteins b | 23 (23.5%) | 85 (30.4%) |
| Total | 98 (100%) | 280 (100%) |
Number and percentage of proteins.
Hypothetical proteins conservedly encoded in the genomes of TriTryps (T. cruzi, T. brucei, and Leishmania major).61
Seventy-one proteins (25.4%) identified in epimastigotes participate in intermediate metabolism: 19 in bioenergetics and biological oxidations, 18 in amino acid metabolism, 11 in lipid metabolism, 8 in carbohydrate metabolism, and 15 are membrane transporters. Proteins involved in protein metabolism (proteolysis/peptidolysis, protein folding, and vesicular trafficking) were in second place, with 48 identifications (17.1%). The other categories were ranked as follows: transcription, translation and DNA repair (23 proteins, 8.2%); cellular communication and signal transduction (20 proteins, 7.1%); structural proteins and cellular dynamics (17 proteins, 6.1%), stress response and cell defense (antioxidation and detoxification) (10 proteins, 3.6%); and host-parasite relationship (6 proteins, 2.1%).
3.4. Metacyclic Proteins
Thirty-nine identifications (39.8%) in metacyclic forms were surface proteins (35 members of the TS superfamily, two procyclic form surface glycoproteins, one mucin, and one GP63 protease) that are involved in host-parasite interplay (evasion from the host immune response, invasion of mammalian cells, and glycosylation of parasite surface molecules) (Tables 2 and 3). From the results of SDS-PAGE and immunoblotting analysis, it was evident that the members of the TS superfamily (i.e., GP90 and GP82 glycoproteins) and mucins were major components of the GPI-enriched fraction (Fig. 2). Although mucins are highly abundant, being easily detected by immunoblotting (with mAb 10D8) and silver staining in detergent fractions from metacyclic and epimastigote forms (data not shown), only one sequence from these glycoconjugates was identified by proteomic analysis of both parasite forms. This is possibly due to the extensive O-glycosylation, peptide sequence variability, and/or fewer proteolytic sites in many T. cruzi mucin sequences, which make their sequencing by conventional proteomic analysis by LC-MS/MS extremely difficult.36 In fact, recent study by Nakayasu et al.37 clearly demonstrated that epimastigote mucins are encoded by the T. cruzi small mucin-like gene (TcSMUG S) family, whose gene products are short polypeptide sequences (56−85 amino acids) with very few trypsin sites located mainly at the N- and C-termini of the protein.
Table 3.
Surface Proteins Identified in T. cruzi Metacyclic Trypomastigotes and Epimastigotes
| Accession Numbera | Protein Nameb | Coverage | Number of Identified Peptides | Developmental Stagec |
|---|---|---|---|---|
| AAM47176.1 | surface glycoprotein GP90 | 40% | 82 | MT |
| EAN87246.1 | trans-sialidase, putative (GP90) | 23% | 14 | MT |
| ABR19835.1 | stage-specific surface protein GP82 | 22% | 31 | MT |
| BAF74647.1 | glycoprotein 82 kDa | 18% | 20 | MT |
| BAF74641.1 | glycoprotein 82 kDa | 14% | 18 | MT |
| EAN81650.1 | trans-sialidase, putative (GP90) | 13% | 24 | MT |
| EAN91385.1 | trans-sialidase, putative (GP90) | 10% | 10 | MT |
| EAN99311.1 | trans-sialidase, putative (GP90) | 10% | 3 | MT |
| ABR19836.1 | stage-specific surface protein GP82 | 9% | 2 | MT |
| EAN95004.1 | trans-sialidase, putative (GP90) | 9% | 20 | MT |
| AAP22091.1 | GP63 group I member b protein | 9% | 3 | MT |
| EAN98599.1 | trans-sialidase, putative (ASP-2) | 9% | 2 | MT |
| EAN82035.1 | procyclic-form surface glycoprotein, putative | 9% | 3 | E |
| EAN95001.1 | trans-sialidase, putative (GP90) | 8% | 14 | MT |
| AAA18827.1 | surface glycoprotein (GP85) | 8% | 6 | MT |
| EAN82322.1 | trans-sialidase, putative (Tc-85/16) | 8% | 15 | MT |
| EAN93322.1 | trans-sialidase, putative (GP85-TSA-E2) | 8% | 5 | MT |
| EAN95642.1 | trans-sialidase, putative (GP85-TSA-E3) | 8% | 16 | MT |
| EAN96370.1 | trans-sialidase, putative (Tc-85/16) | 8% | 16 | MT |
| EAN98005.1 | procyclic-form surface glycoprotein, putative | 7% | 2 | MT, E |
| EAN93266.1 | trans-sialidase, putative (Tc85−11) | 6% | 14 | MT |
| AAB18265.1 | surface protein-1, ASP-1 | 6% | 6 | MT |
| EAN82837.1 | trans-sialidase, putative (GP82) | 6% | 4 | MT |
| AAD10620.1 | surface glycoprotein, GP85 | 6% | 5 | MT |
| AAM47178.1 | surface glycoprotein GP90 | 6% | 1 | MT |
| EAN88221.1 | trans-sialidase, putative (GP90) | 6% | 8 | MT |
| EAN98004.1 | procyclic-form surface glycoprotein, putative | 5% | 1 | E |
| EAN93978.1 | mucin TcMUCII, putative | 5% | 1 | MT |
| EAN88154.1 | trans-sialidase, putative (GP82) | 5% | 6 | MT |
| ABQ53588.1 | surface protein-2, ASP-2 | 5% | 3 | MT |
| EAN82390.1 | trans-sialidase, putative (ASP-2) | 5% | 5 | MT |
| EAN90192.1 | trans-sialidase, putative (GP90) | 5% | 10 | MT |
| BAF74652.1 | glycoprotein 82 kDa | 5% | 3 | MT |
| A48458 | gp85/sialidase homolog | 4% | 1 | MT |
| EAN84247.1 | extracellular receptor, putative | 4% | 1 | E |
| EAN86738.1 | trans-sialidase, putative (GP85-SA85−1.2) | 4% | 4 | MT |
| AAC47720.1 | amastigote surface protein-2, ASP-2 | 3% | 2 | MT |
| AAD13347.1 | surface glycoprotein Tc85−11 | 3% | 1 | E |
| EAN91734.1 | trans-sialidase, putative (GP90) | 3% | 1 | MT |
| EAN93068.1 | trans-sialidase, putative (GP85-TSA-1) | 3% | 3 | MT |
| EAN98006.1 | procyclic-form surface glycoprotein, putative | 2% | 1 | MT |
| EAN82034.1 | procyclic-form surface glycoprotein, putative | 2% | 1 | E |
| ABO28970.1 | trans-sialidase-like protein (GP82) | 2% | 2 | MT |
| EAN85322.1 | trans-sialidase, putative (ASP-2) | 2% | 1 | MT |
GenBank accession number.
Sequences annotated as putative trans-sialidases (TS) in the T. cruzi genome-sequencing project were assigned to the families (indicated in the parentheses) of TS superfamily according to Azuaje.29
MT, metacyclic trypomastigote; E, epimastigote.
Analysis of metacyclic GPI-rich fraction resulted in the identification of 390 peptides mapped to 35 proteins of the TS superfamily (Table 3 and Supplementary Tables 1-3), which members can be classified into four groups according to sequence similarity and function.38, 39 All the TS sequences (i.e., GP90, GP82, ASP-1, ASP-2, GP85, and Tc85) found in our study belong to group II, which comprises genes encoding a number of related surface glycoproteins expressed in trypomastigotes and intracellular amastigotes.38, 39 During the genome project, 1430 sequences (737 genes and 693 pseudogenes) were annotated as putative TS in the clone CL Brener.14 In our proteomic analysis, twenty-one (21.4%) proteins identified in metacyclic forms were annotated as putative TS and further classified as GP82, GP90, GP85, Tc85, ASP-1, and ASP-2, by their similarity with other previously characterized sequences (Table 3 and Supplementary Table 3).40 GP82, GP90, and Tc85 are adhesins that bind to specific mammalian cell receptors and/or to extracellular proteins. In metacyclic forms, GP90 and GP82 were among the top ten proteins with higher coverage in our study (Table 2). TS glycoproteins, previously reported as specific for amastigotes (ASP-1 and ASP-2)33, 41 or bloodstream trypomastigotes (Tc85−11 and GP85)42 were also present in metacyclic forms. It is possible that these proteins are synthesized in very small quantities in metacyclic forms and/or undergo PTM (e.g., glycosylation), which makes their detection difficult by conventional methods, such as SDS-PAGE and Western blotting.
While metacyclic forms appear to express subsets of the TS superfamily, only one TS protein (i.e., AAD13347.1 surface glycoprotein Tc85−11) was detected in epimastigotes (Table 3 and Suppl. Tables 1-3). A large number of variants of GP90 and GP82 were identified in metacyclic trypomastigotes (Table 3 and Suppl. Tables 1-3), confirming that this developmental form expresses a large repertoire of surface glycoproteins involved in the host-cell adhesion and invasion. In contrast to African trypanosomes, T. cruzi needs to invade host cells in order to survive and proliferate, and maybe for this reason it expresses simultaneously many highly diverse surface antigens. The presence of many variants of surface proteins could allow the parasite to adhere to different molecules on host cell membranes and extracellular matrix. Although GP90 and GP82 are highly abundant surface glycoproteins in metacyclic trypomastigotes, only one GP90 (AAM47176) and one GP82 (AAF02477) were previously identified in proteomic studies of metacyclic trypomastigotes whole-cell lysates.43, 44
The peptide mapping coverage of GP90 and GP82 metacyclic proteins, from a combination of tryptic peptides from 3 independent samples, was 40% and 22%, respectively (Figure 3). These proteins are coded by several isoforms and not all of them seem to have host-cell binding sites. Thus, we examined the GP82 sequence, looking for domains involved in the parasite binding to mammalian cells or extracellular matrix proteins. The host cell-binding site of GP82 is formed by the juxtaposition of two sequences, namely P4 and P8, which contain several charged residues and are separated by a more hydrophobic stretch.45 We identified the peptide LVGLLSNSASGDAWIDDYR that covers 65% of peptide P8 (NSASGDAWIDDYRSVNAKVM) (Fig. 3). The peptide NVFLYNRPL, identified in GP90 and GP82, accounts for approx. 64% of the FLY domain. This domain (VTVXNVFLYNR) localizes at the C-terminal portion of TS proteins from group II and was reported to bind to cytokeratin 18 on the surface of LLC-MK2 epithelial cells, and enhance the infectivity of tissue culture-derived trypomastigotes.42
Here, we also present the first evidence of expression of the procyclic-form surface glycoprotein in T. cruzi. This family of putative genes was identified in the T. cruzi genome sequencing project.14 We found five of these proteins (Table 3 and Suppl. Tables 1-3) that share approximately 40% identity with their T. brucei counterparts.46 T. brucei procyclic-form surface glycoprotein is a stage-specific antigen with features of a typical transmembrane glycoprotein but with unusual cytoplasmic tail composed of proline-rich tandem repeats.46 The putative procyclic-form surface glycoproteins identified in our work have 2−3 transmembrane helices. In addition, three of these glycoproteins (i.e., EAN98004, EAN98005, and EAN98006) seem to contain an uncleaved signal anchor sequence,31 suggesting that they are located on the cell surface.
Metacyclic trypomastigotes experience a shift in environmental conditions when they move from triatomines to the vertebrate host, consequently infecting a range of nucleated cells and transforming into amastigotes. Metacyclic trypomastigotes have a complex cell envelope consisting of different glycoproteins as a form of protection. Many surface proteins that are directly exposed to these extreme conditions are predicted to attach to the plasma membrane via a GPI-anchor, be heavily glycosylated, and in some cases, have a polypeptide sequence with few proteolytic sites.3, 5 This could eventually prevent their degradation by hydrolytic enzymes present in the insect midgut and vertebrate host. The presence of surface glycoproteins could also allow the parasite to adhere to different molecules on the host cell membrane and the extracellular matrix.
About 11% (n=11) and 7% (n=20) of metacyclic and epimastigote identifications, respectively, showed similarities to proteins involved in signal transduction, including multiple rab proteins, phosphatases, ras-related proteins, and calcium-binding proteins (Table 4 and Suppl. Table 3). Most of these proteins identified in epimastigote and metacyclic proteomes can be directed to the cell surface through signal peptides, transmembrane domain(s), or other lipid PTM such as myristoylation (Suppl. Table 3). One typical example is the flagellar calcium-binding protein (FCaBP), which is a Ca2+ sensor located in the flagellum of trypanosomes, a unique organelle involved in motility, chemotaxis, and cell signaling.47 Although typical tyrosine-kinase genes were not identified in the T. cruzi genome,48 protein-tyrosine phosphatases control cell differentiation in trypanosomes.49, 50 Here, we found two protein-tyrosine phosphatases in both metacyclic and epimastigote forms, whereas a regulatory subunit of protein kinase was found only in epimastigotes. The importance of protein phosphorylation/dephosphorylation events was corroborated by the presence of different protein phosphatases (Table 4).
Table 4.
Cellular Communication and Signal Transduction Proteins Identified in Metacyclic Trypomastigotes and Epimastigotes
| Accession Numbera | Protein Name | Coverage | Number of Identified Peptides | Developmental Stageb |
|---|---|---|---|---|
| CAA90898.1 | F29 (flagellar calcium-binding protein) | 46% | 34 | MT, E |
| EAN94827.1 | protein tyrosine phosphatase, putative | 35% | 9 | MT, E |
| AAA99985.1 | calcium-binding protein | 34% | 26 | MT, E |
| EAN92885.1 | membrane-bound acid phosphatase, putative | 29% | 6 | E |
| AAB08762.1 | 24 kDa flagellar calcium-binding protein | 28% | 20 | MT |
| BAA13411.1 | calcium-binding protein | 28% | 8 | MT, E |
| EAN85763.1 | small GTP-binding protein Rab1, putative | 27% | 9 | MT, E |
| AAT77755.1 | 14−3−3 protein | 17% | 3 | E |
| AAT77756.1 | 14−3−3 protein; Tcf2p | 16% | 4 | MT, E |
| EAN99708.1 | protein phosphatase, putative | 15% | 6 | E |
| EAN94492.1 | protein tyrosine phosphatase, putative | 14% | 3 | MT, E |
| EAN95123.1 | 14−3−3 protein, putative | 13% | 2 | E |
| EAN85068.1 | membrane-bound acid phosphatase, putative | 10% | 2 | E |
| EAN94164.1 | 14−3−3 protein, putative | 10% | 3 | E |
| EAN99768.1 | rab7 GTP binding protein, putative | 10% | 3 | MT |
| EAN86478.1 | membrane-bound acid phosphatase, putative | 9% | 2 | E |
| EAN86455.1 | calcium-binding protein, putative | 8% | 30 | MT |
| EAN88612.1 | small GTP-binding protein Rab11, putative | 7% | 2 | E |
| EAN92627.1 | 14−3−3 protein, putative | 7% | 5 | E |
| EAN92863.1 | phosphatidic acid phosphatase protein, putative | 7% | 2 | E |
| AAM91818.1 | flagellar calcium binding protein 3 | 7% | 1 | MT |
| EAN87884.1 | ras-related protein rab-2a, putative | 5% | 1 | E |
| EAN93719.1 | membrane-bound acid phosphatase 2, putative | 4% | 3 | E |
| EAN88194.1 | regulatory subunit of protein kinase a-like protein | 2% | 1 | E |
GenBank accession number.
MT, metacyclic trypomastigote; E, epimastigote.
Recently, Atwood et al.51 reported a glycoproteomic analysis of plasma membrane and organelle-enriched fractions from T. cruzi tissue culture-derived trypomastigotes. Nineteen of the proteins identified in that study were also present in our analysis, which consist of 5 structural proteins, 4 heat shock proteins, 3 hypothetical proteins (two of them having signal peptides), 2 mitochondrial proteins (ATPase beta subunit and phosphate transporter), 2 vesicular trafficking proteins, 1 cysteine proteinase, 1 glycosomal phosphoenolpyruvate carboxykinase, and 1 TS protein (EAN90192) (Suppl. Table 1-3). It is noteworthy that only one TS protein was common to both studies, suggesting that metacyclic and tissue culture-derived trypomastigotes express distinct repertoires of TS gene superfamily.
3.5. Epimastigote Proteins
There are clear qualitative differences between the protein profiles of epimastigotes and metacyclic trypomastigotes identified on this study. Epimastigotes undergo biochemical and morphological changes in the midgut of insect vector. They multiply by binary fission and differentiate into metacyclic trypomastigotes upon migration to the hindgut. Proteins from intermediate metabolic pathways, including membrane transporters, are abundant in epimastigotes. Here, we found that 71 (25.4%) of the identifications in epimastigotes were enzymes related to energy metabolism, including those involved in the uptake and degradation of glucose via glycolysis, pentose-phosphate shunt, or tricarboxylic acid (TCA) cycle (Table 2). Several glycosomal and mitochondrial proteins were detected, including a hexose transporter. A glycosomal malate dehydrogenase and pyruvate phosphate dikinase were only detected in epimastigotes. Several members of the respiratory chain complexes were also detected in epimastigotes (Table 2 and Suppl. Table 3).
Epimastigotes degrade proteins and amino acids to produce NH3 and glucose to reduce the level of toxic catabolites (mainly succinate and l-alanine).52 In this study we identified 19 enzymes and one membrane transporter involved in the metabolism of amino acids, confirming that epimastigotes are particularly adapted to take advantage of free amino acids available in the gut of the insect vector (Suppl. Table 3).43 Among the enzymes involved in amino acid catabolism, four were glutamate dehydrogenases (one NADP-dependent) and seven aminotransferases, including four tyrosine aminotransferases involved in the catabolism of aromatic amino acids. In agreement with our data, three glutamate dehydrogenase isoforms and one aminotransferase were identified in epimastigotes submitted to nutritional stress and in metacyclic trypomastigotes.44 Glutamate dehydrogenase provides a link between carbohydrate and amino acid metabolism in T. cruzi.53 This enzyme may play an important role during the transit of parasites in the insect gut, where carbohydrates are limited and free amino acids derived from hemoglobin degradation are available. The presence of urocanate hydratase, an enzyme that participates in the conversion of histidine to glutamate, is noteworthy. The enzymes of this pathway are abundant in the T. cruzi insect stages, but nearly undetectable in the mammalian stages.43 Interestingly, the genes encoding these enzymes were not found in T. brucei or L. major, suggesting that they are species-specific of T. cruzi.
As expected, proteins involved in stress response and cell defense were detected in epimastigotes: cytosol-localized tryparedoxin peroxidase, tryparedoxin (substrate for tryparedoxin peroxidase), thiol transferase Tc52, GPR1/FUN34/yaaH family, carbonic anhydrase-like protein and TcOYE dehydrogenase (old yellow enzyme) (Suppl. Table 3). The expression of these proteins by epimastigotes is consistent with the preadaptation of metacyclic forms to resist the potential respiratory burst of phagocytic cells in mammalian host. Thiol transferase Tc52 and tryparedoxin peroxidase have been detected in nutritionally stressed and differentiating (adherent) epimastigotes and tissue culture-derived trypomastigotes.35, 44 It has been suggested that in vitro-induced benznidazole resistance in T. cruzi is correlated with deletion of copies of the TcOYE gene.54, 55
Forty-eight (17.1%) epimastigote identifications were associated with protein metabolism (Table 2). Several heat shock proteins (HSP 85, 70, and 60; glucose-regulated protein 78; mitochondrial chaperonin HSP60) and other chaperones (7.1% of total identifications) were found in epimastigotes (Suppl. Table 3). They were also identified in whole extract and organelle-enriched fractions from tissue culture-derived trypomastigotes.35, 51 Proteinases and peptidases represented 7.5% (n=21) of the identified proteins in epimastigotes, whereas proteins involved in transporting, targeting, sorting, and translocation represented 2.5%. We have confirmed the presence of a large family of calpain-related proteins, including a calpain-like cytoskeleton-associated protein (CAP5.5) (EAN92788.1) (Suppl. Table 3). The presence of calpain-like proteins in trypanosomes was first suggested by analysis of a cytoskeleton-associated protein in T. brucei (CAP5.5) that showed similarity to the catalytic region of calpain-type proteases.56 CAP5.5 has been shown to be both myristoylated and palmitoylated, suggesting a stable interaction with the cell membrane.56 Bioinformatic analysis of the T. cruzi genome database29 confirmed the presence of calpain-like genes in this parasite, most of them containing a well-conserved protease domain and a novel N-terminal sequence motif unique to kinetoplastids. Furthermore, they contain N-terminal fatty acid acylation motifs, indicating the association of these proteins with cellular membranes. Proteins involved in the transport machinery and cellular trafficking were also identified and can be expected to facilitate studies of these less-defined processes in T. cruzi.
Our group has recently performed a proteomic analysis of reservosomes,57 membrane-bound organelles exclusive of epimastigote forms of the parasite, which were proposed to store nutrients uptaken through endocytosis.58 Since reservosomes were shown to be rich in membrane and metabolic proteins, we hypothesized that many of the proteins found in our analysis were derived from these organelles. Indeed, one hundred and forty-five (51.8%) epimastigote proteins identified here were also present in the reservosomal membrane fraction (Table 1).57
3.6. Hypothetical proteins
Of the 378 T. cruzi proteins identified in our analysis, one hundred and eight (28.6%) were from genes annotated as hypothetical, confirming these as bona fide genes (Table 2 and Suppl. Table 3). On the other hand, genes annotated as hypothetical in T. cruzi were difficult to assess, since no functional insights could be gained from our findings. However, most hypothetical genes have homologues in Leishmania major and T. brucei, suggesting a common function for the given genes in closely related trypanosomatids.
3.7. Conclusion
Proteomic analysis by conventional approaches is biased against hydrophobic and GPI- anchored membrane proteins, many of which are expressed in low abundance. Here, we have identified a large number of membrane proteins from insect developmental forms of T. cruzi using a simple, fast, and sensitive method for the isolation of these proteins by extraction with the detergent Triton X-114 followed by 2D LC-MS/MS analysis. In summary, we have shown in this paper that each of the insect stages of T. cruzi reveals distinct protein datasets when solubilized with neutral detergent. The metacyclic trypomastigotes have a higher number of potential GPI-anchored glycoproteins that might be involved in the process of adhesion and invasion of host cells. On the other hand, the majority of proteins found in epimastigote samples were related to metabolism, which is in agreement with the high energy requirements of this proliferative stage of the parasite. In addition, we speculate that the identified epimastigote proteins are present in metabolism-related organelles, such as mitochondria, glycosomes, and reservosomes. We also present evidence for the expression in T. cruzi of procyclic-form surface glycoprotein, previously described in T. brucei. We envisage that the results of this study may contribute to the design of more rational therapies against Chagas disease.
Supplementary Material
Acknowledgments
This work was supported by NIH grants # 2S06GM008012-38 (to ICA) and 5G12RR008124 (to BBRC/Biology/UTEP); and by grants from FAPESP and CNPq (Brazil) to JFS. ECV and LGG were awarded postdoctoral and doctoral fellowships, respectively, by CNPq. ESN was partially supported by the Georges A. Krutilek memorial scholarship from the Graduate School/UTEP. We would like to thank A. Galetovic for the generous gift of the polyclonal antiserum against T. cruzi calpain, and Dr. Sergio Schenkman (UNIFESP, Brazil) and Dr. David Engman (Northwestern University, Chicago) for the monoclonal antibody to the T. cruzi FCaBP. We thank the Biomolecule Analysis Core Facility at the BBRC/Biology/UTEP (NIH grant # 5G12RR008124), for the full access to the LC-MS instruments.
Footnotes
Supporting Information Available: Supplementary tables and figures are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ferguson MA. The structure, biosynthesis and functions of glycosylphosphatidylinositol anchors, and the contributions of trypanosome research. J Cell Sci. 1999;112(Pt 17):2799–809. doi: 10.1242/jcs.112.17.2799. [DOI] [PubMed] [Google Scholar]
- 2.Colli W, Alves MJ. Relevant glycoconjugates on the surface of Trypanosoma cruzi. Mem Inst Oswaldo Cruz. 1999;94(Suppl 1):37–49. doi: 10.1590/s0074-02761999000700004. [DOI] [PubMed] [Google Scholar]
- 3.Buscaglia CA, Campo VA, Frasch AC, Di Noia JM. Trypanosoma cruzi surface mucins: host-dependent coat diversity. Nat Rev Microbiol. 2006;4(3):229–36. doi: 10.1038/nrmicro1351. [DOI] [PubMed] [Google Scholar]
- 4.Guha-Niyogi A, Sullivan DR, Turco SJ. Glycoconjugate structures of parasitic protozoa. Glycobiology. 2001;11(4):45R–59R. doi: 10.1093/glycob/11.4.45r. [DOI] [PubMed] [Google Scholar]
- 5.Acosta-Serrano A, Hutchinson C, Nakayasu ES, Almeida IC, Carrington M. Comparison and evolution of the surface architecture of trypanosomatid parasites. In: Barry JD, Mottram JC, McCulloch R, Acosta-Serrano A, editors. Trypanosomes: After the genome. Horizon Scientific Press; Norwich, UK: 2007. pp. 319–337. [Google Scholar]
- 6.Almeida IC, Gazzinelli RT. Proinflammatory activity of glycosylphosphatidylinositol anchors derived from Trypanosoma cruzi: structural and functional analyses. J Leukoc Biol. 2001;70(4):467–77. [PubMed] [Google Scholar]
- 7.Dias JC, Silveira AC, Schofield CJ. The impact of Chagas disease control in Latin America: a review. Mem Inst Oswaldo Cruz. 2002;97(5):603–12. doi: 10.1590/s0074-02762002000500002. [DOI] [PubMed] [Google Scholar]
- 8.Teixeira MM, Yoshida N. Stage-specific surface antigens of metacyclic trypomastigotes of Trypanosoma cruzi identified by monoclonal antibodies. Mol Biochem Parasitol. 1986;18(3):271–82. doi: 10.1016/0166-6851(86)90085-x. [DOI] [PubMed] [Google Scholar]
- 9.Güther ML, de Almeida ML, Yoshida N, Ferguson MA. Structural studies on the glycosylphosphatidylinositol membrane anchor of Trypanosoma cruzi 1G7-antigen. The structure of the glycan core. J Biol Chem. 1992;267(10):6820–8. [PubMed] [Google Scholar]
- 10.de Almeida ML, Heise N. Proteins anchored via glycosylphosphatidylinositol and solubilizing phospholipases in Trypanosoma cruzi. Biol Res. 1993;26(1−2):285–312. [PubMed] [Google Scholar]
- 11.Yoshida N. Molecular basis of mammalian cell invasion by Trypanosoma cruzi. An Acad Bras Cienc. 2006;78(1):87–111. doi: 10.1590/s0001-37652006000100010. [DOI] [PubMed] [Google Scholar]
- 12.Araya JE, Cano MI, Yoshida N, da Silveira JF. Cloning and characterization of a gene for the stage-specific 82-kDa surface antigen of metacyclic trypomastigotes of Trypanosoma cruzi. Mol Biochem Parasitol. 1994;65(1):161–9. doi: 10.1016/0166-6851(94)90124-4. [DOI] [PubMed] [Google Scholar]
- 13.do Carmo MS, dos Santos MR, Cano MI, Araya JE, Yoshida N, da Silveira JF. Expression and genome-wide distribution of the gene family encoding a 90 kDa surface glycoprotein of metacyclic trypomastigotes of Trypanosoma cruzi. Mol Biochem Parasitol. 2002;125(1−2):201–6. doi: 10.1016/s0166-6851(02)00212-8. [DOI] [PubMed] [Google Scholar]
- 14.El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran AN, Ghedin E, Worthey EA, Delcher AL, Blandin G, Westenberger SJ, Caler E, Cerqueira GC, Branche C, Haas B, Anupama A, Arner E, Aslund L, Attipoe P, Bontempi E, Bringaud F, Burton P, Cadag E, Campbell DA, Carrington M, Crabtree J, Darban H, da Silveira JF, de Jong P, Edwards K, Englund PT, Fazelina G, Feldblyum T, Ferella M, Frasch AC, Gull K, Horn D, Hou L, Huang Y, Kindlund E, Klingbeil M, Kluge S, Koo H, Lacerda D, Levin MJ, Lorenzi H, Louie T, Machado CR, McCulloch R, McKenna A, Mizuno Y, Mottram JC, Nelson S, Ochaya S, Osoegawa K, Pai G, Parsons M, Pentony M, Pettersson U, Pop M, Ramirez JL, Rinta J, Robertson L, Salzberg SL, Sanchez DO, Seyler A, Sharma R, Shetty J, Simpson AJ, Sisk E, Tammi MT, Tarleton R, Teixeira S, Van Aken S, Vogt C, Ward PN, Wickstead B, Wortman J, White O, Fraser CM, Stuart KD, Andersson B. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309(5733):409–15. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- 15.Camargo EP. Growth and differentiation In Trypanosoma cruzi. I. Origin of metacyclic trypanosomes in liquid media. Rev Inst Med Trop Sao Paulo. 1964;12:93–100. [PubMed] [Google Scholar]
- 16.Ko YG, Thompson GA., Jr. Purification of glycosylphosphatidylinositol-anchored proteins by modified triton X-114 partitioning and preparative gel electrophoresis. Anal Biochem. 1995;224(1):166–72. doi: 10.1006/abio.1995.1024. [DOI] [PubMed] [Google Scholar]
- 17.Stone KL, Williams KR. Enzymatic digestion of proteins in solution and in SDS polyacrilamide gel. In: Walker JM, editor. The protein protocol handbook. Humana Press Inc.; Totowa, NJ: 1996. [Google Scholar]
- 18.Jurado JD, Rael ED, Lieb CS, Nakayasu E, Hayes WK, Bush SP, Ross JA. Complement inactivating proteins and intraspecies venom variation in Crotalus oreganus helleri. Toxicon. 2007;49(3):339–50. doi: 10.1016/j.toxicon.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 19.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 20.Colinge J, Masselot A, Giron M, Dessingy T, Magnin J. OLAV: towards high-throughput tandem mass spectrometry data identification. Proteomics. 2003;3(8):1454–63. doi: 10.1002/pmic.200300485. [DOI] [PubMed] [Google Scholar]
- 21.Poisson G, Chauve C, Chen X, Bergeron A. FragAnchor: a large-scale predictor of glycosylphosphatidylinositol anchors in eukaryote protein sequences by qualitative scoring. Genomics Proteomics Bioinformatics. 2007;5(2):121–30. doi: 10.1016/S1672-0229(07)60022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305(3):567–80. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 23.Maurer-Stroh S, Eisenhaber B, Eisenhaber F. N-terminal N-myristoylation of proteins: prediction of substrate proteins from amino acid sequence. J Mol Biol. 2002;317(4):541–57. doi: 10.1006/jmbi.2002.5426. [DOI] [PubMed] [Google Scholar]
- 24.Maurer-Stroh S, Eisenhaber F. Refinement and prediction of protein prenylation motifs. Genome Biol. 2005;6(6):R55. doi: 10.1186/gb-2005-6-6-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340(4):783–95. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 26.Engman DM, Krause KH, Blumin JH, Kim KS, Kirchhoff LV, Donelson JE. A novel flagellar Ca2+-binding protein in trypanosomes. J Biol Chem. 1989;264(31):18627–31. [PubMed] [Google Scholar]
- 27.Elias MC, da Cunha JP, de Faria FP, Mortara RA, Freymuller E, Schenkman S. Morphological events during the Trypanosoma cruzi cell cycle. Protist. 2007;158(2):147–57. doi: 10.1016/j.protis.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 28.Cortez M, Neira I, Ferreira D, Luquetti AO, Rassi A, Atayde VD, Yoshida N. Infection by Trypanosoma cruzi metacyclic forms deficient in gp82 but expressing a related surface molecule, gp30. Infect Immun. 2003;71(11):6184–91. doi: 10.1128/IAI.71.11.6184-6191.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ersfeld K, Barraclough H, Gull K. Evolutionary relationships and protein domain architecture in an expanded calpain superfamily in kinetoplastid parasites. J Mol Evol. 2005;61(6):742–57. doi: 10.1007/s00239-004-0272-8. [DOI] [PubMed] [Google Scholar]
- 30.Añez-Rojas N, García-Lugo P, Crisante G, Rojas A, Añez N. Isolation, purification and characterization of GPI-anchored membrane proteins from Trypanosoma rangeli and Trypanosoma cruzi. Acta Trop. 2006;97(2):140–5. doi: 10.1016/j.actatropica.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 31.von Heijne G. Transcending the impenetrable: how proteins come to terms with membranes. Biochim Biophys Acta. 1988;947(2):307–33. doi: 10.1016/0304-4157(88)90013-5. [DOI] [PubMed] [Google Scholar]
- 32.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2(4):953–71. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 33.Low HP, Tarleton RL. Molecular cloning of the gene encoding the 83 kDa amastigote surface protein and its identification as a member of the Trypanosoma cruzi sialidase superfamily. Mol Biochem Parasitol. 1997;88(1−2):137–49. doi: 10.1016/s0166-6851(97)00088-1. [DOI] [PubMed] [Google Scholar]
- 34.Pereira NM, Timm SL, da Costa SC, Rebello MA, de Souza W. Trypanosoma cruzi: isolation and characterization of membrane and flagellar fractions. Exp Parasitol. 1978;46(2):225–34. doi: 10.1016/0014-4894(78)90135-2. [DOI] [PubMed] [Google Scholar]
- 35.Paba J, Ricart CA, Fontes W, Santana JM, Teixeira AR, Marchese J, Williamson B, Hunt T, Karger BL, Sousa MV. Proteomic analysis of Trypanosoma cruzi developmental stages using isotope-coded affinity tag reagents. J Proteome Res. 2004;3(3):517–24. doi: 10.1021/pr034075o. [DOI] [PubMed] [Google Scholar]
- 36.Buscaglia CA, Campo VA, Di Noia JM, Torrecilhas AC, De Marchi CR, Ferguson MA, Frasch AC, Almeida IC. The surface coat of the mammal-dwelling infective trypomastigote stage of Trypanosoma cruzi is formed by highly diverse immunogenic mucins. J Biol Chem. 2004;279(16):15860–9. doi: 10.1074/jbc.M314051200. [DOI] [PubMed] [Google Scholar]
- 37.Nakayasu ES, Yashunsky DV, Nohara LL, Torrecilhas ACT, Nikolaev AV, Almeida IC. GPIomics: Global Analysis of Glycosylphosphatidylinositol-Anchored Molecules of Trypanosoma cruzi. Molecular Systems Biology. 2009 doi: 10.1038/msb.2009.13. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colli W. Trans-sialidase: a unique enzyme activity discovered in the protozoan Trypanosoma cruzi. Faseb J. 1993;7(13):1257–64. doi: 10.1096/fasebj.7.13.8405811. [DOI] [PubMed] [Google Scholar]
- 39.Frasch AC. Functional diversity in the trans-sialidase and mucin families in Trypanosoma cruzi. Parasitol Today. 2000;16(7):282–6. doi: 10.1016/s0169-4758(00)01698-7. [DOI] [PubMed] [Google Scholar]
- 40.Azuaje F, Ramirez JL, Da Silveira JF. An exploration of the genetic robustness landscape of surface protein families in the human protozoan parasite Trypanosoma cruzi. IEEE Trans Nanobioscience. 2007;6(3):223–8. doi: 10.1109/tnb.2007.903482. [DOI] [PubMed] [Google Scholar]
- 41.Santos MA, Garg N, Tarleton RL. The identification and molecular characterization of Trypanosoma cruzi amastigote surface protein-1, a member of the trans-sialidase gene super-family. Mol Biochem Parasitol. 1997;86(1):1–11. [PubMed] [Google Scholar]
- 42.Magdesian MH, Giordano R, Ulrich H, Juliano MA, Juliano L, Schumacher RI, Colli W, Alves MJ. Infection by Trypanosoma cruzi. Identification of a parasite ligand and its host cell receptor. J Biol Chem. 2001;276(22):19382–9. doi: 10.1074/jbc.M011474200. [DOI] [PubMed] [Google Scholar]
- 43.Atwood JA, 3rd, Weatherly DB, Minning TA, Bundy B, Cavola C, Opperdoes FR, Orlando R, Tarleton RL. The Trypanosoma cruzi proteome. Science. 2005;309(5733):473–6. doi: 10.1126/science.1110289. [DOI] [PubMed] [Google Scholar]
- 44.Parodi-Talice A, Monteiro-Goes V, Arrambide N, Avila AR, Duran R, Correa A, Dallagiovanna B, Cayota A, Krieger M, Goldenberg S, Robello C. Proteomic analysis of metacyclic trypomastigotes undergoing Trypanosoma cruzi metacyclogenesis. J Mass Spectrom. 2007;42(11):1422–32. doi: 10.1002/jms.1267. [DOI] [PubMed] [Google Scholar]
- 45.Manque PM, Eichinger D, Juliano MA, Juliano L, Araya JE, Yoshida N. Characterization of the cell adhesion site of Trypanosoma cruzi metacyclic stage surface glycoprotein gp82. Infect Immun. 2000;68(2):478–84. doi: 10.1128/iai.68.2.478-484.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jackson DG, Smith DK, Luo C, Elliott JF. Cloning of a novel surface antigen from the insect stages of Trypanosoma brucei by expression in COS cells. J Biol Chem. 1993;268(3):1894–900. [PubMed] [Google Scholar]
- 47.Buchanan KT, Ames JB, Asfaw SH, Wingard JN, Olson CL, Campana PT, Araujo AP, Engman DM. A flagellum-specific calcium sensor. J Biol Chem. 2005;280(48):40104–11. doi: 10.1074/jbc.M505777200. [DOI] [PubMed] [Google Scholar]
- 48.Parsons M, Worthey EA, Ward PN, Mottram JC. Comparative analysis of the kinomes of three pathogenic trypanosomatids: Leishmania major, Trypanosoma brucei and Trypanosoma cruzi. BMC Genomics. 2005;6:127. doi: 10.1186/1471-2164-6-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cuevas IC, Rohloff P, Sanchez DO, Docampo R. Characterization of farnesylated protein tyrosine phosphatase TcPRL-1 from Trypanosoma cruzi. Eukaryot Cell. 2005;4(9):1550–61. doi: 10.1128/EC.4.9.1550-1561.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szöor B, Wilson J, McElhinney H, Tabernero L, Matthews KR. Protein tyrosine phosphatase TbPTP1: A molecular switch controlling life cycle differentiation in trypanosomes. J Cell Biol. 2006;175(2):293–303. doi: 10.1083/jcb.200605090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Atwood JA, 3rd, Minning T, Ludolf F, Nuccio A, Weatherly DB, Alvarez-Manilla G, Tarleton R, Orlando R. Glycoproteomics of Trypanosoma cruzi trypomastigotes using subcellular fractionation, lectin affinity, and stable isotope labeling. J Proteome Res. 2006;5(12):3376–84. doi: 10.1021/pr060364b. [DOI] [PubMed] [Google Scholar]
- 52.Cazzulo JJ. Intermediate metabolism in Trypanosoma cruzi. J Bioenerg Biomembr. 1994;26(2):157–65. doi: 10.1007/BF00763064. [DOI] [PubMed] [Google Scholar]
- 53.Barderi P, Campetella O, Frasch AC, Santome JA, Hellman U, Pettersson U, Cazzulo JJ. The NADP+-linked glutamate dehydrogenase from Trypanosoma cruzi: sequence, genomic organization and expression. Biochem J. 1998;330(Pt 2):951–8. doi: 10.1042/bj3300951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murta SM, Krieger MA, Montenegro LR, Campos FF, Probst CM, Avila AR, Muto NH, de Oliveira RC, Nunes LR, Nirde P, Bruna-Romero O, Goldenberg S, Romanha AJ. Deletion of copies of the gene encoding old yellow enzyme (TcOYE), a NAD(P)H flavin oxidoreductase, associates with in vitro-induced benznidazole resistance in Trypanosoma cruzi. Mol Biochem Parasitol. 2006;146(2):151–62. doi: 10.1016/j.molbiopara.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 55.Andrade HM, Murta SM, Chapeaurouge A, Perales J, Nirde P, Romanha AJ. Proteomic analysis of Trypanosoma cruzi resistance to Benznidazole. J Proteome Res. 2008;7(6):2357–67. doi: 10.1021/pr700659m. [DOI] [PubMed] [Google Scholar]
- 56.Hertz-Fowler C, Ersfeld K, Gull K. CAP5.5, a life-cycle-regulated, cytoskeleton-associated protein is a member of a novel family of calpain-related proteins in Trypanosoma brucei. Mol Biochem Parasitol. 2001;116(1):25–34. doi: 10.1016/s0166-6851(01)00296-1. [DOI] [PubMed] [Google Scholar]
- 57.Sant'anna C, Nakayasu ES, Pereira MG, Lourenco D, de Souza W, Almeida IC, Cunha ESNL. Subcellular proteomics of Trypanosoma cruzi reservosomes. Proteomics. 2009;9(7):1782–1794. doi: 10.1002/pmic.200800730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cunha-e-Silva N, Sant'Anna C, Pereira MG, Porto-Carreiro I, Jeovanio AL, de Souza W. Reservosomes: multipurpose organelles? Parasitol Res. 2006;99(4):325–7. doi: 10.1007/s00436-006-0190-3. [DOI] [PubMed] [Google Scholar]
- 59.Sant'Anna C, Nakayasu ES, Pereira MG, Lourenco D, de Souza W, Almeida IC, Cunha-e-Silva N. Subcellular proteomics of Trypanosoma cruzi reservosomes. Proteomics. doi: 10.1002/pmic.200800730. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sant'Anna C, Pereira MG, Lemgruber L, de Souza W, Cunha e Silva NL. New insights into the morphology of Trypanosoma cruzi reservosome. Microsc Res Tech. 2008;71(8):599–605. doi: 10.1002/jemt.20592. [DOI] [PubMed] [Google Scholar]
- 61.Kissinger JC. A tale of three genomes: the kinetoplastids have arrived. Trends Parasitol. 2006;22(6):240–3. doi: 10.1016/j.pt.2006.04.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.