Abstract
Patients with gliomas expressing high levels of epidermal growth factor receptor (EGFR) and plasminogen activator inhibitor-1 (PAI-1) have a shorter overall survival prognosis. Moreover, EGF enhances PAI-1 expression in glioma cells. Although multiple known signaling cascades are activated by EGF in glioma cells, we show for the first time that EGF enhances expression of PAI-1 via sequential activation of c-Src, protein kinase C delta (PKCδ), and sphingosine kinase 1 (SphK1), the enzyme that produces sphingosine-1-phosphate. EGF induced rapid phosphorylation of c-Src and PKCδ and concomitant translocation of PKCδ as well as SphK1 to the plasma membrane. Down-regulation of PKCδ abolished EGF-induced SphK1 translocation and up-regulation of PAI-1 by EGF; whereas, down-regulation of PKCα had no effect on the EGF-induced PAI-1 activation but enhanced its basal expression. Similarly, inhibition of c-Src activity by PP2 blocked both EGF-induced translocation of SphK1 and PKCδ to the plasma membrane and up-regulation of PAI-1 expression. Furthermore, SphK1 was indispensable for both EGF-induced c-Jun phosphorylation and PAI-1 expression. Collectively, our results provide a functional link between three critical downstream targets of EGF, c-Src, PKCδ, and SphK1 that have all been implicated in regulating motility and invasion of glioma cells.
Keywords: glioma, invasiveness
Gliomas are characterized by rapid growth, intensive vascularization, and an invasive phenotype that results in the diffuse infiltration of tumor cells into the surrounding normal brain tissue (1). Since individual tumor cells can invade as deeply as several centimeters into the brain, gliomas are resistant to the established standard treatments, including surgery and radiotherapy. Deregulation of expression of glioma proteolytic enzymes, which allows them to degrade the extracellular matrix (ECM), migrate, and invade healthy tissue, is likely critical for their invasion (2, 3). The ECM degradation involves the activation/inhibition of proteinases and their corresponding inhibitors in glioma cells, and the subsequent deregulation of the proteolytic balance at the site of invasion. These processes involve at least two major proteolytic systems: the plasminogen activator (PA) system, controlling the release/activity of plasmin, and metalloproteinases and their tissue inhibitors (4–7).
Plasmin is generated from inactive plasminogen by a precisely controlled PA system that includes plasminogen activators [urokinase-type (uPA), tissue-type (tPA)], as well as their inhibitors [plasminogen activator inhibitors (PAI-1, -2, and -3) and the protease nexin 1] (4). In addition, uPA and tPA can bind to their cell surface receptor (uPAR) and activate multiple signaling cascades (8). In the brain, microglia are the predominant source of inactive plasminogen, whereas astrocytes and glioma cells secrete the components of the PA system. The expression of these proteins is regulated by proinflammatory cytokines and growth factors in both astrocytes and invading glioma cells (9–11). Among the growth factors, epidermal growth factor (EGF) has attracted much attention since it is produced in the brain and readily crosses the blood-brain barrier (12). In gliomas, the receptor for EGF (EGFR) is frequently amplified (13, 14), overexpressed (13, 14), or mutated (14, 15). The amplification and overexpression are associated with high-grade progression (16). Although the high expression of EGFR alone is not a prognostic marker in gliomas (17), patients expressing high levels of both EGFR and PAI-1 have a shorter overall survival prognosis (18). Recently, EGF has been shown to regulate expression of PAI-1, uPA, and vascular endothelial growth factor (VEGF) in glioma cells (10, 19), suggesting that it may be critical for the invasive phenotype of gliomas and their intensive vascularization. In addition, PAI-1 has been shown to stimulate VEGF expression and thereby increase vascularization of glioblastomas (20).
To date, both uPA and uPAR have been strongly associated with the invasiveness and metastasis of a variety of cancer cells (21). In gliomas, the knockdown of uPAR resulted in the significant inhibition of cell invasion in both the Matrigel and spheroid invasion assays (22, 23). Furthermore, the knockdown of both uPAR and MMP-9 resulted in total regression of pre-established intracerebral tumors (22). These findings are likely attributed to the diminished membrane-associated proteolytic activity of uPA bound to uPAR, and the diminished signaling capabilities of uPAR. Surprisingly, high expression of the proteinase inhibitor PAI-1 has also been reported in highly invasive high-grade gliomas (18), suggesting that it may also be an important determinant of their invasiveness. These observations parallel the findings that breast cancer patients with high levels of PAI-1 have a poor prognosis for survival due to metastasis (24). Recently, it has been proposed that elevated levels of PAI-1 promote cancer cell detachment from their substratum by causing the internalization of the PAI-1/uPA/uPAR/integrin complex (25, 26). Hence, the enhanced expression of PAI-1 may be critical for the detachment of cancer cells and the subsequent invasion into the healthy tissue. Future therapies may, therefore, involve either targeting the expression of PAI-1, uPA, and uPAR directly, or indirectly by targeting signaling molecules that control their expression.
Increased expression of PAI-1 has been reported in high-grade gliomas in vivo (18) and in cultured glioma cells (10). In vitro, the expression of PAI-1 is regulated by cytokines and growth factors, including EGF (10). However, the molecular mechanisms linking EGF and its receptor to up-regulation of PAI-1 are not well understood. In this study, we identified a novel signaling pathway of EGF-mediated up-regulation of PAI-1 in glioblastoma cells that requires activation of c-Src, PKCδ, and sphingosine kinase 1.
MATERIALS AND METHODS
Cell culture
Human glioblastoma U373-MG cells were obtained from the American Type Culture Collection (Manassas, VA, USA), and human glioblastoma A172 cells were obtained from Dr. Jaharul Haque (Cleveland Clinic Foundation, Cleveland, OH, USA). Cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, antibiotics, sodium pyruvate, and nonessential amino acids.
Cytokines and cell stimulation
Cells were stimulated with 25 ng/ml EGF (Peprotech, Rocky Hill, NJ, USA), 25 ng/ml oncostatin M (OSM; R&D Systems, Inc., Minneapolis, MN, USA), or 10 ng/ml IL-1 (a gift from Immunex Corp., Seattle, WA, USA). For inhibitor studies, cells were pretreated with 1 µM SP600125, 10 µM BAY-117082, 10 µM SB202190, 5 µg/ml actinomycin D (Sigma, St. Louis, MO, USA), 100 nM staurosporin, 5 µM U73343, 5 µM U73122, 10 µM PP2, 5 µM Gö6983 (EMD Biosciences, Inc., San Diego, CA, USA), 10 µM LY294002 or 1 µM U0126 (Cell Signaling Technology, Inc., Beverly, MA, USA), and then stimulated with EGF as described in figure legends.
RNA preparation and Northern blot analysis
Total RNA was prepared by phenol extraction exactly as described previously (27). The filters were prehybridized at 65°C for 3 h in 0.5 M sodium phosphate buffer, pH 7.2; 7% SDS (sodium dodecyl sulfate); and 1 mM EDTA and hybridized in the same solution with cDNA fragments of PAI-1 labeled by random priming (28). After the hybridization, nonspecifically bound radioactivity was removed by four washes in 40 mM phosphate buffer and 1% SDS at 65°C for 20 min each.
Quantitative PCR
SphK1 and PAI-1 mRNA levels were measured using TaqMan technology (Applied Biosystems, Foster City, CA, USA) according to the supplier’s instructions. Briefly, 1 µg of total RNA was reverse-transcribed using the high-capacity cDNA archive kit. Subsequently, the cDNAs were diluted 10-fold (Sphk1 and PAI-1) or 10,000-fold (28S rRNA). For real-time PCR, premixed primer-probe sets and TaqMan Universal PCR Master Mix were purchased from Applied Biosystems, and cDNAs were amplified using ABI 7900HT cycler.
Synthetic oligonucleotides
The following oligonucleotides were synthesized to amplify the −15,025 to −14,320 fragment from the 5′ flanking region of the PAI-1 gene: 5′-GCCATCTAGA GCTATGGCCATTATGGCCT-3′ and 5′-TCATTCTAGAAATCCCAGCAATTTGGGAGG-3′. The Alu element located at −11.7 to −11.4 kb was amplified using the 5′-ACCCTCTAGAAACC AGTGGGCTCAAG-3′ and 5′-TTTGTCTAGACATCGTGGCTGGTG GCGTAC-3′ primers. A double-stranded oligonucleotide containing the ATF binding site derived from the −11.7 to −11.4 kb Alu element (used in gelshifts and construction of the ATF reporter) was generated by annealing the following oligonucleotides: 5′-GATCTCGAACT CCTGACCTCATGTAGTCCA-3′ and 5′-GATC TGCACTACATGAGGTCAGGAGTTCGA-3′. The AP-1 and STAT oligonucleotides were described before (11, 29).
Plasmid construction
Plasmids pPAI-1(−1496)CAT and pPAI-1(−115)CAT have been described previously (11). Plasmid pPAI-1(−15 kb-enhancer)-CAT was generated by cloning the XbaI-digested PCR product (covering the −15,025 to −14,320 fragment) into the XbaI site of pPAI-1(−115)CAT. Plasmids p5x(PAI-ATF)CAT and p3x(PAI-ATF)CAT were generated by cloning corresponding double-stranded oligonucleotides into the BamHI site of ptkCATΔEH (29). The XbaI-digested PCR product containing the Alu element located at −11.7 to −11.4 kb was cloned into the XbaI site of pPAI-1(−115)CAT and yielded the plasmid pPAI-1-(Alu)CAT.
Nucleofection
A172 cells (1×106/6-cm dish) were trypsinized, collected by centrifugation, and resuspended in 600 µl of T nucleofactor solution™ (Amaxa Inc., Gaithersburg, MD, USA). The respective plasmids (2 µg) were added to the solution, and transfection was performed using the Nucleofector device (Amaxa Inc.) with the electrical setting of T-20. Warm DMEM medium (1 ml) was added, and cells were incubated at 37°C for 10 min and transferred to 6-cm dishes containing 5 ml of DMEM culture medium. Typical nucleofection efficiencies were greater than 70%, with cell viability close to 100%. One day after transfections, cells were stimulated with the respective cytokines for 18 h and processed as described earlier.
Transient transfections
Cells were transfected in 12-well clusters using FuGENE6 transfection reagent (Roche, Indianapolis, IN, USA) with 400 ng of the reporter CAT plasmid and 100 ng of expression plasmid encoding β-galactosidase. One day after transfection, the cells were stimulated with EGF or cytokines, cultured another 24 h, and harvested. Protein extracts were prepared by freeze thawing, and the protein concentration was determined by the BCA method (Sigma Chemical Co.). Chloramphenicol acetyltransferase (CAT) and β-galactosidase assays were performed as described (29). CAT activities were normalized to β-galactosidase activity and are means + se (3 determinations).
Nuclear extract preparation and electromobility shift assays (EMSA)
Nuclear extracts were prepared as described (30). The oligonucleotides used for EMSA were designed to contain single-stranded 5′ overhangs at each end after annealing. Double-stranded DNA fragments were labeled by filling in 5′ protruding ends with Klenow enzyme using [α32P]dCTP (3000 Ci/mmol). EMSA was carried out according to the published procedures (31). Briefly, 5 µg of nuclear extracts and approx. 10 fmol (10,000 cpm) of probe were used.
Western blotting
Cells were lysed in 10 mM Tris, pH 7.4; 150 mM sodium chloride; 1 mM EDTA; 0.5% Nonidet P-40; 1% Triton X-100; 1 mM sodium orthovanadate; 0.2 mM PMSF; and protease inhibitor cocktail (Roche, Mannheim, Germany). Equal amounts of proteins were resolved using SDS-PAGE and electroblotted onto nitrocellulose membranes (Schleicher & Schuell, Keene, NH, USA). Polyclonal anti-ERK, anti-IkBα, anti-c-fos, anti-c-jun, anti-PKCα, anti-PKCδ, and anti-tubulin antisera were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA), while anti-phospho-p38, anti-phospho-ERK, anti-phospho-ATF-2, anti-phospho-JNK, anti-phospho-Akt, anti-phospho-Src, and anti-phospho-PKCs antisera were purchased from Cell Signaling Technology, Inc. (Beverly, MA, USA). SphK1 (Ser-225 phospho-specific antibodies were purchased from ECM Biosciences (Versailles, KY, USA). Antigen-antibody complexes were visualized by enhanced chemiluminescence according to the manufacturer’s instructions (Pierce, Rockford, IL, USA).
Confocal microscopy
Transfectants grown on glass coverslips were treated as indicated in the figure legends. Cells were washed with PBS, fixed for 20 min at room temperature with 2% paraformaldehyde, and mounted on glass slides using 100% glycerol containing 10 mM n-propylgallate. Alternatively, cells were washed with PBS, fixed for 20 min at room temperature with 3.7% formalin, permeabilized with 0.5% Triton X-100 in PBS for 3 min, and then blocked for 1 h with 4% bovine serum albumin. After washing, cells were incubated for 30–45 min with primary antibodies in PBS containing 1% bovine serum albumin, then for 30 min with the corresponding secondary antibodies. No fluorescence crossover was found between the channels, and images were collected separately with a ×63 oil immersion objective using the appropriate laser excitation.
Quantification of confocal images
Images were quantified using Zeiss LSM Image Examiner (v. 3.2.70). The fluorescence intensity present within 2 µm of the membrane was averaged and divided by the average fluorescence intensity within 2–4 µm from the membrane. This ratio was then normalized to control images.
Fractionation
Cells were collected in 20 mM Tris, pH 7.7; 20% glycerol; 1 mM 2-mercaptoethanol; 1 mM EDTA; 5 mM sodium orthovanadate; 40 mM b-glycerophosphate; 15 mM NaF; 0.5 mM 4-deoxypyridoxine; and protease inhibitor cocktail and homogenized using glass homogenizer. Unbroken cells were removed by centrifugation at 1500 g for 10 min. Cytosol was separated from the membrane fractions by centrifugation at 100,000 g for 60 min at 4°C. Pellets were resuspended in the buffer described above with addition of 1% Triton X-100, and solubilized at 4°C with gentle shaking for 60 min. Triton soluble and triton insoluble fractions were then separated by centrifugation at 100,000 g for 60 min at 4°C. Pellets were resuspended in the buffer described above with addition of 1% Triton X-100 and 0.1% SDS and sonicated.
Down-regulation with siRNA
PKCα and PKCδ expression were down-regulated using siRNAs purchased from Santa Cruz Biotechnology, c-Src SmartPool siRNA was from Dharmacon, Inc. (Lafayette, CO, USA), and SphK1 mRNA was down-regulated with siRNA targeted to a unique hSphK1 sequence as described previously (32). siRNAs was transfected into cells using Oligofectamine (Invitrogen, Carlsbad, CA, USA), Dharmafect 1 (Dharmacon, Inc.), or by nucleofection with the Amaxa Nucleofector (Amaxa).
RESULTS
EGF rapidly activates PAI-1 expression in glioblastoma cells
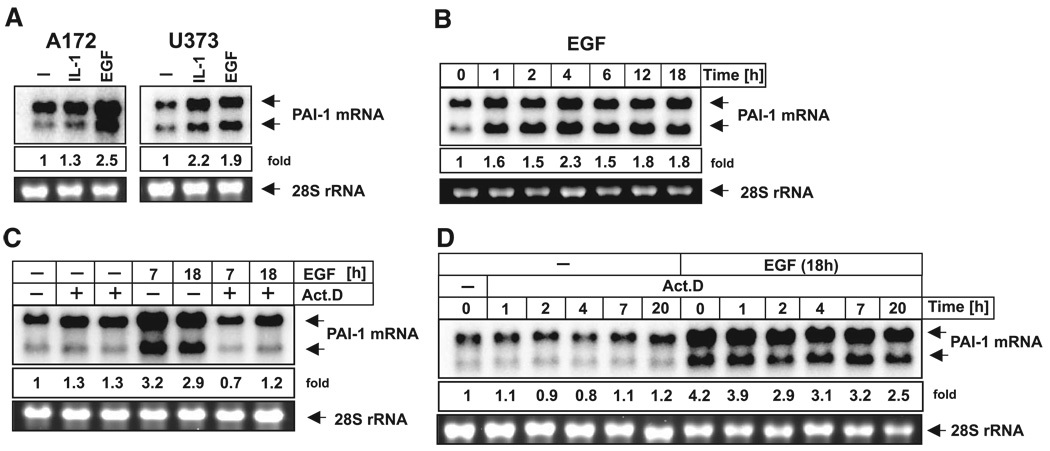
Since EGF signaling appears to be critical for the development and progression of tumors, we analyzed the mechanism of PAI-1 activation by EGF in glioma cells. EGF enhanced PAI-1 expression in U373 glioma cells as previously reported (Fig. 1A and ref. 10). Similar up-regulation was found in A172 glioma cells (Fig. 1A), which were chosen for further analysis due to the higher transfection efficiency of these cells. As expected, PAI-1 expression was also up-regulated by IL-1 in both cell lines (Fig. 1A and ref. 10). The induction of PAI-1 expression by EGF was very rapid (Fig. 1B) and was blocked by pretreatment with actinomycin D, indicating that ongoing transcription is required (Fig. 1C). Interestingly, the half-life of PAI-1 mRNA was greater than 20 h in both the control and EGF-treated cells (Fig. 1D), indicating that mRNA stability was not enhanced by EGF.
Figure 1.
EGF up-regulates PAI-1 gene expression in glioblastoma cells. A172 and U373 cells were treated with EGF or IL-1 for 7 h (A); RNA was isolated and subjected to Northern blot analysis using PAI-1 cDNA as a probe. A172 cells were treated with EGF for indicated time periods (B) or pretreated with 5 µg/ml actinomycin D for 30 min, then treated with EGF for 7 or 18 h (C), and processed as in A. D) A172 cells were left untreated or treated with EGF for 18 h, and then 5 µg/ml actinomycin D was added for indicated time periods and processed as in A. The lower panels show 28S RNA stained with ethidium bromide on the membrane as a loading control.
EGF does not regulate PAI-1 expression via NF-κB, AP-1, or STAT proteins
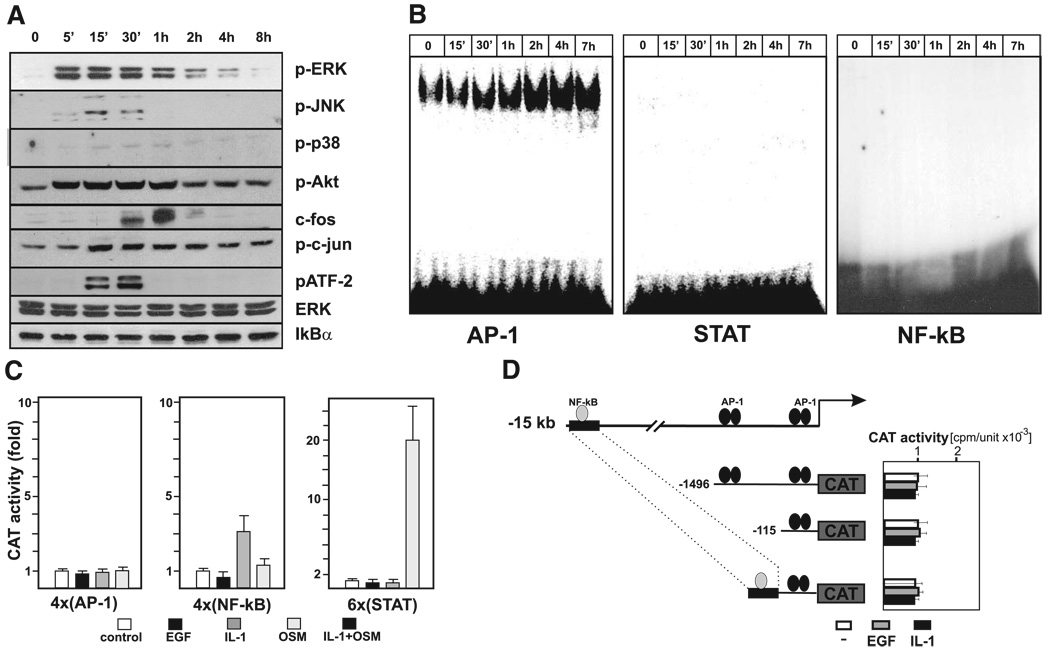
PAI-1 expression has been shown to be enhanced by a broad range of stimuli in a variety of cell types (9). Many of these factors activate PAI-1 transcription via the 1.5 kb 5′ flanking region that includes a critical AP-1 binding element at −58 to −51. Recently, a new TNF-responsive PAI-1 enhancer has been described at −15 kb, which contains a critical putative NF-κB binding element (33). To understand how EGF mediates regulation of PAI-1 expression in glioma cells, we examined activation of relevant signaling molecules and transcription factors, as well as the responsiveness of several reporters. Treatment of glioma cells with EGF resulted in a rapid phosphorylation of ERK1/2, JNK, Akt, c-jun, ATF-2, and also induced expression of c-fos (Fig. 2A). However, EGF did not induce degradation of IκB or alter phosphorylation of p38 kinase (Fig. 2A). In agreement, EGF enhanced AP-1 DNA binding activity, whereas it did not activate binding of NF-κB or STATs (Fig. 2B). Subsequently, we examined the EGF responsiveness of the reporter constructs containing arrays of AP-1, NF-κB, or STAT elements. However, none of these constructs were responsive to EGF, whereas, as expected (11), they were responsive to IL-1 or OSM (Fig. 2C). These results, together with the in vitro binding data, suggest that AP-1, NF-κB, and STAT do not mediate the responsiveness of the PAI-1 gene to EGF. We also tested several reporters containing the PAI-1 5′ flanking region and the newly described enhancer located at −15 kb (33). However, these constructs also did not respond to EGF or IL-1 (Fig. 2D), suggesting that the 1.5 kb 5′ flanking region and the −15 kb enhancer are not necessary. The identity of regulatory element(s) activated by EGF remains unknown. However, the conserved c-jun/ATF binding sites located within the 17 Alu elements found in the 5′ flanking region of the PAI-1 gene may be the targets of EGF-activated complexes that contain c-jun, and ATF family members (Supplemental Fig. 1).
Figure 2.
EGF activates multiple signaling pathways in A172 cells. A) A172 cells were stimulated with EGF for various times, and activation of the indicated signaling molecules was analyzed by Western blotting using specific antibodies. Total ERK is included as a loading control. B) Nuclear extracts were prepared from control and EGF-treated A172 cells as indicated. DNA binding activity of AP-1, STAT, and NF-κB was then analyzed by EMSA using the 32P-labeled oligonucleotide probes. C–D) A172 cells were transiently transfected with the indicated reporter plasmids and a β-galactosidase expression vector. One day after transfection, cells were stimulated with EGF, cultured for an additional 24 h, and harvested. CAT activities were normalized to β-galactosidase activities to account for transfection efficiency. Results are expressed as fold induction.
Both PKCα and PKCδ regulate PAI-1 expression in glioblastoma
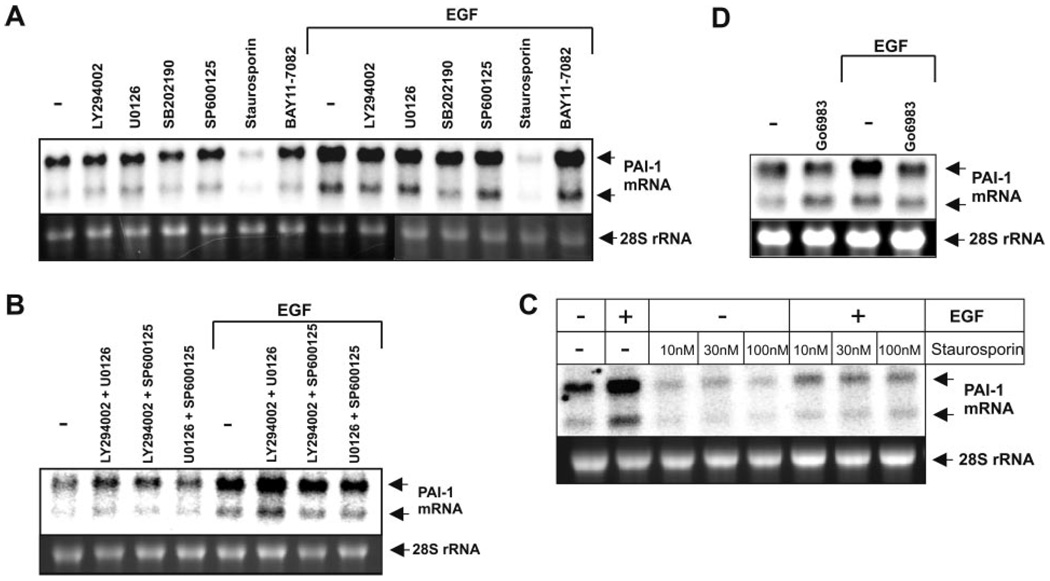
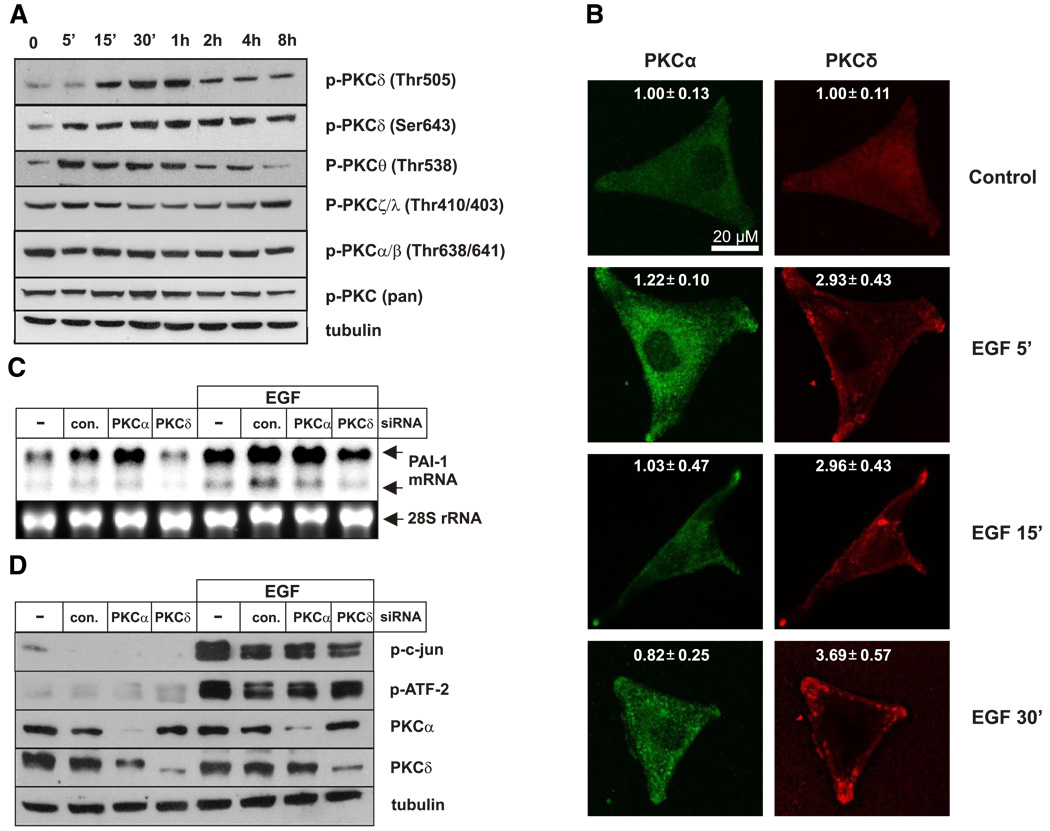
Next, we determined the signaling pathways involved in EGF-induced PAI-1 expression, utilizing pharmacological and molecular approaches. EGF-activated PAI-1 expression was efficiently blocked by staurosporin, a broad-specificity kinase inhibitor and a potent PKC inhibitor at low concentrations, but not by inhibitors of p38 kinase (SB202190), JNK (SP600125), MEK1/2 (U0126), PI3K (LY294002), or IKK (BAY-11–7082), or when used in combinations (Fig. 3A, B). Of note, staurosporin efficiently blocked basal as well as EGF-activated PAI-1 expression, even at a concentration as low as 10 nM (Fig. 3C), suggesting the involvement of PKC(s) in PAI-1 regulation. Indeed, the PKC inhibitor Gö6983 also efficiently blocked EGF-induced PAI-1 expression (Fig. 3D). We then determined which PKC isoforms are activated in response to EGF using isoform- specific phospho-PKC antibodies (Fig. 4A). Increased phosphorylation of PKCδ on both Thr505 and Ser-643, as well as PKCθ on Thr538, was observed within minutes of EGF treatment, while activation of other PKCs was not affected (Fig. 4A). Subsequently, we focused our studies on PKCδ, since EGF did not induce phosphorylation of PKCθ in other glioblastoma cell lines, including U373 cells (data not shown).
Figure 3.
Inhibition of PKC prevents up-regulation of PAI-1 expression by EGF. A172 cells were pretreated with 10 µM LY294002, 1 µM U0126, 10 µM SB202190, 1 µM SP600125, 100 nM staurosporin, 5 µM BAY11–7082 (A), the indicated combination of the inhibitors (B), the indicated concentration of staurosporin (C), or 5 µM Gö6983 (D) for 1 h and subsequently stimulated with EGF for 7 h. RNA was isolated, and PAI-1 expression was analyzed by Northern blotting.
Figure 4.
Both PKCα and PKCδ regulate PAI-1 expression in glioblastoma cells. A) A172 cells were stimulated with EGF for indicated times, and phosphorylation of PKC isoforms was analyzed by Western blotting using specific antibodies. B) A172 cells were treated with EGF for indicated times, and localization of PKCα and PKCδ was examined by confocal immunofluorescence microscopy. The numbers represent fluorescent intensity analyzed as described in Materials and Methods. C) A172 cells were transfected with control siRNA (con.) or siRNAs targeted to PKCα or PKCδ for 48 h as indicated, and then stimulated with EGF for 7 h. RNA was isolated after 7 h and subjected to Northern blot analysis using PAI-1 cDNA as a probe. D) Cells were transfected with PKCα siRNA or PKCδ siRNA for 48 h as indicated and stimulated with EGF for 30 min, and indicated proteins were analyzed by Western blotting. Blots were stripped and reprobed with anti-tubulin antibody as a loading control.
Because activation of PKCδ is usually accompanied by translocation to the plasma membrane, EGF-induced changes in cellular localization were examined by confocal microscopy. EGF stimulation resulted in the dramatic translocation of PKCδ into the membrane, whereas it had a negligible effect on PKCα translocation, which was used as a control (Fig. 4B). To address the possible roles of PKCδ and PKCα in PAI-1 regulation, they were down-regulated using specific siRNAs. Surprisingly, down-regulation of PKCα enhanced basal levels of PAI-1 (Fig. 4C), suggesting that PKCα and PKCδ may exert opposing effects on PAI-1 expression. Importantly, knockdown of PKCδ expression, but not PKCα, significantly repressed activation of PAI-1 by EGF (Fig. 4C). Since c-jun and ATF-2 are possible targets of PKC signaling, we examined their phosphorylation in cells depleted of PKCα and PKCδ. Although knockdown of PKCα had no effect, EGF-induced phosphorylation of c-jun but not ATF-2 was diminished when PKCδ was down-regulated (Fig. 4D). Taken together, these results suggest that EGF-induced activation of PAI-1 is mediated via PKCδ and that it involves c-jun.
Sphingosine kinase 1 (SphK1) is required for EGF-activated expression of PAI-1
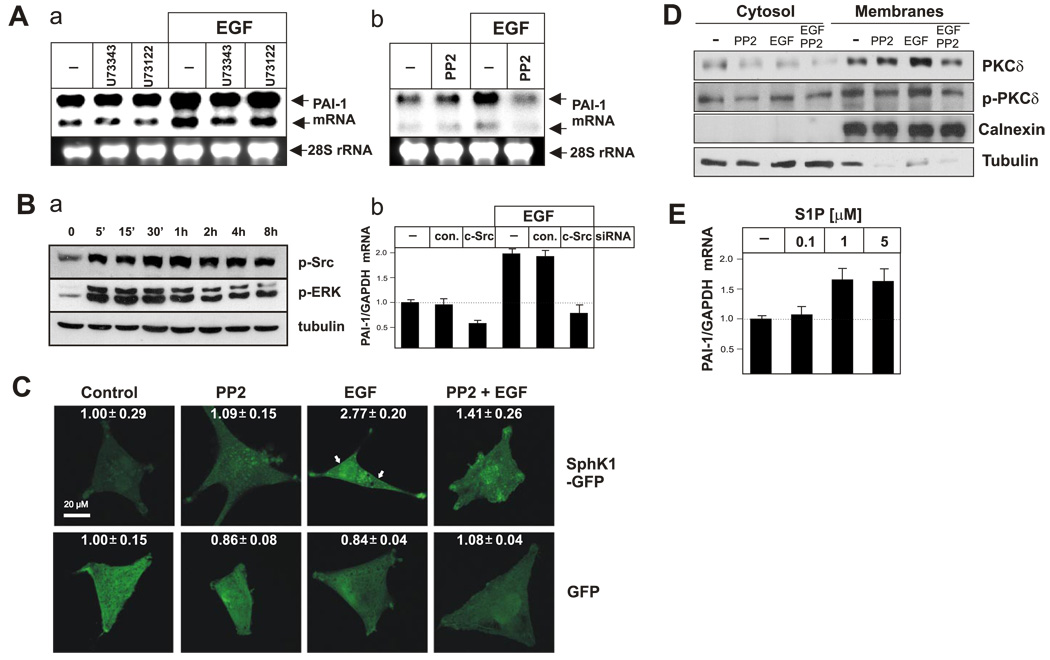
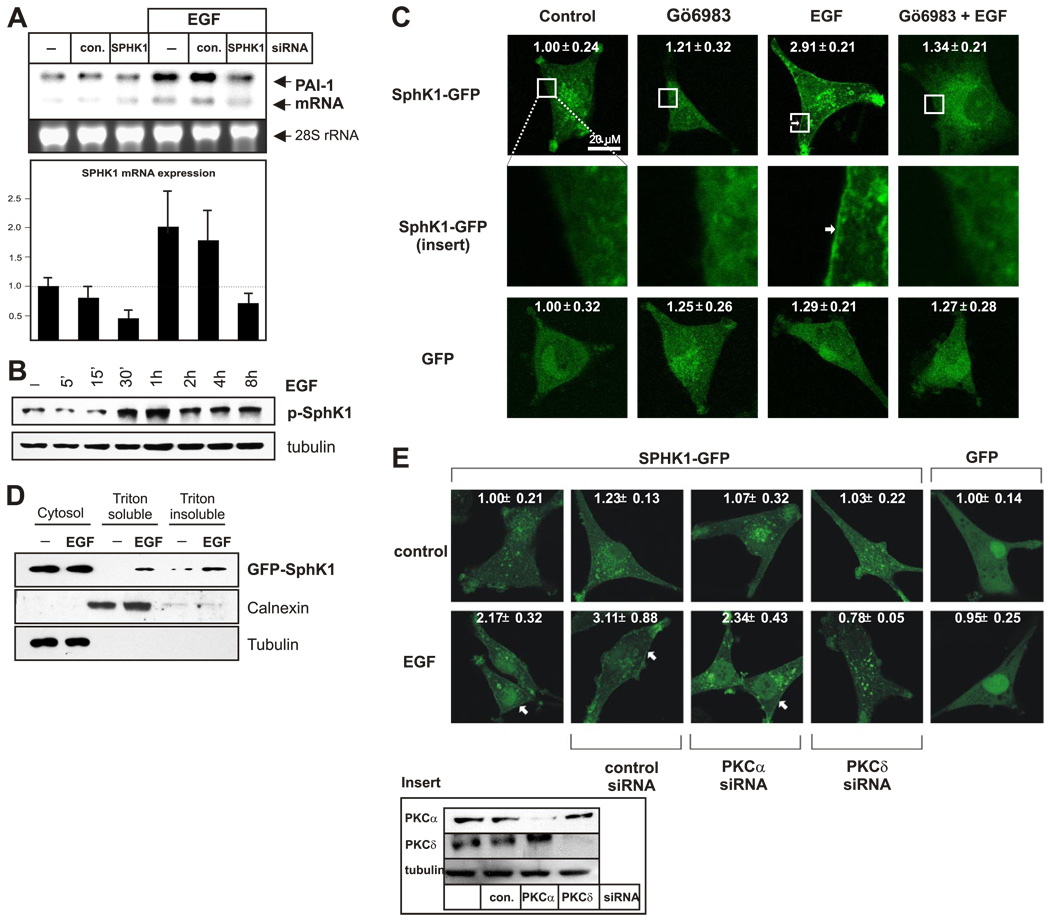
We recently discovered that the bioactive sphingolipid metabolite sphingosine-1-phosphate (S1P) activates PAI-1 expression in several glioma cell lines (Bryan, L., in preparation), including A172 cells (Fig. 6E). In addition, S1P has recently been shown to activate PAI-1 expression in cultured adipocytes (34). Because EGF and PKC activation stimulate sphingosine kinase 1 (SphK1), the enzyme that generates S1P by phosphorylation of sphingosine (35), it was of interest to determine whether SphK1 is involved in EGF-induced PAI-1 expression. Although knockdown of SphK1 expression using siRNA had no effect on basal PAI-1 expression, it essentially abolished its up-regulation by EGF (Fig. 5A). Furthermore, EGF induced efficient phosphorylation of SphK1 on Ser-225 (Fig. 5B), indicating activation of this enzyme (36). Previous studies have shown that activation of SphK1 by EGF or PKC in other types of cells requires its translocation to the plasma membrane where its substrate sphingosine resides (35). In agreement, SphK1-GFP was distributed throughout the cytosol of unstimulated cells and was rapidly translocated to the plasma membrane after EGF stimulation (Fig. 5C, D). Moreover, this translocation was efficiently blocked by pretreatment with a PKC inhibitor (Fig. 5C). As expected, the diffuse localization of GFP was not affected by either EGF or the PKC inhibitor (Fig. 5C). Importantly, similar to their effects on PAI-1 expression, down-regulation of PKCδ prevented EGF-induced translocation of SphK1 to the plasma membrane, whereas down-regulation of PKCα had no effect (Fig. 5E). These results suggest that EGF stimulates PKCδ, which in turn, activates SphK1; SphK1 produces S1P, which is required for EGF-induced PAI-1 activation.
Figure 6.
Inhibition of Src abolishes both EGF-induced activation of SphK1 and expression of PAI-1. A) A172 cells pretreated with 5 µM U73343, 5 µM U73122 (a), or 10 µM PP2 (b) for 1 h and then stimulated with EGF for 7 h. RNA was isolated, and PAI-1 expression was analyzed by Northern blotting. B) A172 cells were treated with EGF for the indicated times and phosphorylation of Src and ERK examined by Western blotting using phospho-specific antibodies. Tubulin was immunoblotted to show equal loading (a). A172 cells were transfected with control siRNA or siRNA targeted to c-Src as indicated. Cells were cultured for 48 h and then stimulated with EGF. RNA was isolated after 7 h, and PAI-1 expression was quantified by real-time PCR using TaqMan technology, normalized to 18S rRNA, and expressed as a ratio to mRNA levels in untreated cells (b). C) A172 cells were transfected with either GFP or SphK1-GFP chimera, and cultured for 24 h. Subsequently, cells were pretreated with 10 µM PP2 for 1 h and then treated with EGF for 5 min and analyzed by confocal microscopy. The numbers represent fluorescent intensity analyzed as described in Materials and Methods. D) A172 cells were pretreated with 10 µM PP2 for 1 h and then treated with EGF for 30 min. Subsequently, cells were fractionated as described in Materials and Methods. Distribution of PKCδ (total protein and its phosphorylated form) was analyzed by Western blotting. Tubulin and calnexin were immunoblotted to show purity of the fractions. E) A172 cells were treated with indicated concentrations of S1P for 7 h, RNA was isolated, and PAI-1 expression was quantified by real-time PCR using TaqMan technology, normalized to GAPDH, and expressed as a ratio to mRNA levels in untreated cells.
Figure 5.
SphK1 is required for EGF-induced PAI activation. A) A172 cells were transfected with control siRNA (con.) or SphK1 siRNA for 48 h as indicated and then stimulated with EGF. RNA was isolated after 7 h, and PAI-1 expression was analyzed by Northern blotting (top panel). The down-regulation of SphK1 mRNA was quantified by real-time PCR using TaqMan technology, normalized to 18S rRNA, and expressed as a ratio to mRNA levels in untreated cells (bottom panel). B) A172 cells were stimulated with EGF for indicated times, and phosphorylation of SphK1 was analyzed by Western blotting using specific antibodies. C) A172 cells were transfected with either GFP or SphK1-GFP chimera, and cultured for 24 h. Subsequently, cells were pretreated with 5 µM Gö6983 for 1 h and then treated with EGF for 5 min and analyzed by confocal microscopy. The numbers represent fluorescent intensity analyzed as described in Materials and Methods. D) A172 cells were transfected with SphK1-GFP chimera and cultured for 24 h. Subsequently, cells were treated with EGF for 30 min and fractionated as described in Materials and Methods. Distribution of SphK1-GFP chimera was analyzed by Western blotting. Tubulin and calnexin were immunoblotted to show purity of the fractions. E) A172 cells were transfected with GFP, SphK1-GFP chimera together with control siRNA, or siRNA targeted to PKCα or PKCδ siRNA as indicated. Cells were cultured for 48 h and then stimulated with EGF for 5 min. Localization of GFP and SphK1-GFP was examined by confocal microscopy. The numbers represent fluorescent intensity analyzed as described in Materials and Methods. PKCα and PKCδ expression was also analyzed by Western blotting (insert). Arrows indicate translocation to the plasma membrane.
EGF-induced c-Src activation is involved in PAI-1 up-regulation
EGFR tyrosine kinase signaling can stimulate PKCδ via activation of phospholipase C (PLC), which generates diacylglycerol that can bind to and activate PKCδ (37). However, neither the PLC inhibitor U73122 nor its inactive analog U73343 had an effect on EGF-induced up-regulation of PAI-1, suggesting that PLC is not required for its activation (Fig. 6Aa). Recently, it has been suggested that PKCδ can also be activated by c-Src, which binds to and phosphorylates CDCP1, a protein that provides a docking site recognized by the C2 domain of PKCδ (38). To test whether c-Src activity is required for activation of PAI-1 by EGF, cells were pretreated with the c-Src inhibitor PP2 and then stimulated with EGF. PP2 effectively blocked PAI-1 expression induced by EGF (Fig. 6Ab). In addition, EGF rapidly activated c-Src as determined by its phosphorylation on Tyr416 (Fig. 6Ba), and the knockdown of c-Src blocked the activation of PAI-1 expression by EGF (Fig. 6Bb). Because activation of c-Src appears to be required for activation of PAI-1, it was important to determine whether it was also required for PKCδ and SphK1 activation/translocation. Indeed, pretreatment with PP2 also reduced translocation of both PKCδ (Fig. 6D) and SphK1-GFP chimera (Fig. 6C) to the plasma membrane in response to EGF. Collectively, these results suggest that c-Src activation is a prerequisite for EGF-induced translocation/activation of both PKCδ and SphK1, and up-regulation of PAI-1. Phosphorylation and translocation of SphK1 in response to EGF suggest that S1P is likely generated and can either be secreted to activate its receptors or alternatively may act via unidentified intracellular mechanisms. In fact, PAI-1 expression is enhanced in cells stimulated with exogenous S1P (Fig. 6E).
DISCUSSION
Recent observations demonstrate that breast cancer patients expressing high levels of PAI-1 have a poor prognosis for survival (4, 24, 39). Increased expression of PAI-1 has also been observed in high-grade gliomas, with concurrent high levels of PAI-1 and EGFR being poor prognosis markers for overall survival in glioma patients (18). These findings are in apparent contradiction to the idea that tumor invasion is promoted by increased membrane-associated proteolytic activity of uPA, which is inhibited by PAI-1. High levels of PAI-1 may inhibit cell invasion due to its antiproteolytic activity (4) and also promote cell detachment and invasion by nonproteolytic mechanisms (4, 25, 26). These pro- and antimigratory effects may also depend on whether PAI-1 is present at the leading or trailing edge of the invading tumor cell. Therefore, a correlation between PAI-1 expression and metastatic disease may be related to its effects on the ability of cells to attach to, migrate on, and be detached from the substratum (4). For example, elevated levels of PAI-1 promote breast cancer cell detachment by causing internalization of the PAI-1/uPA/uPAR/integrin complex via interaction with low-density lipoprotein receptor related protein (25, 26). This leads to degradation of both PAI-1 and uPA, whereas uPAR is recycled back to the cell surface. Loss of the PAI-1/uPA/uPAR/ integrin complex from the cell surface promotes breast cancer cell detachment and migration.
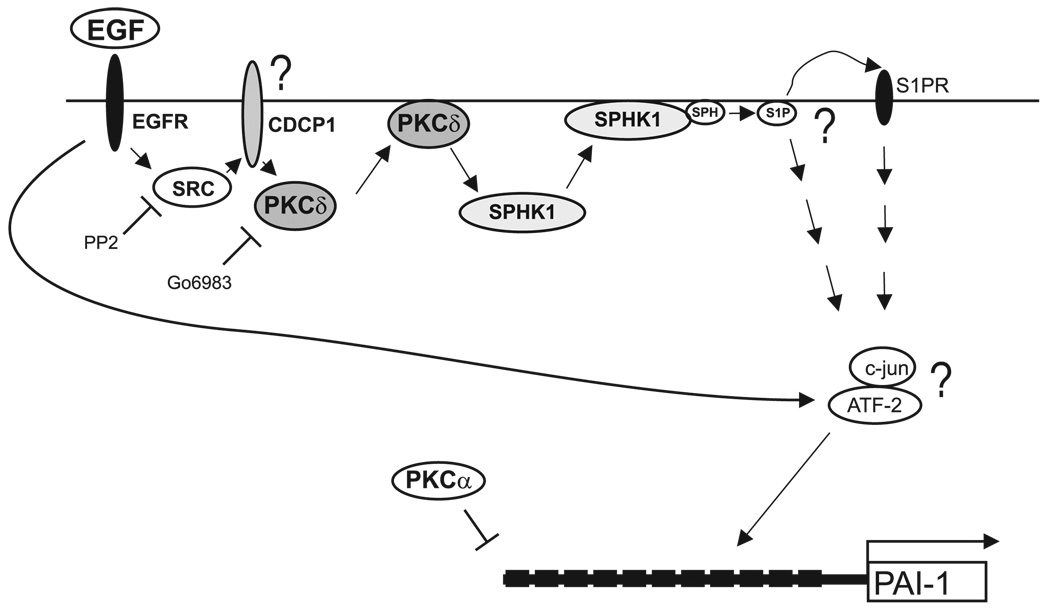
Interestingly, PAI-1 is also an immediate–early gene regulated by a variety of hormones, cytokines, and growth factors, including EGF. EGF enhances PAI-1 expression in a variety of cell types, including gliomas (10), hepatomas (40), trophoblasts (41), microvascular endothelial cells (42), and liver epithelial cells (43). In this study, we have demonstrated that EGF up-regulates the PAI-1 system via a novel signaling pathway involving sequential activation of c-Src, PKCδ, and SphK1 (Fig. 7). In glioma cells, we found that EGF rapidly activated c-Src, and pharmacological inhibition of c-Src blocked both PAI-1 activation and translocation of both PKCδ and SphK1 to the membrane in response to EGF (Fig. 6). Similarly, activation of c-Src by TGFα-induced EGFR has previously been reported to up-regulate PAI-1 expression in smooth muscle cells (44). However, c-Srcdependent activation of PAI-1 in these cells involves the MEK/ERK pathway (44). In contrast, activation of PAI-1 in glioma cells was not blocked by the MEK1/2 inhibitors U0126 and PD98059 (Fig. 3 and data not shown). Likewise phosphorylation of ERK1/2 induced by EGF was not inhibited by the c-Src inhibitor PP2 in breast cancer MCF-7 cells (45), indicating that c-Src may activate target gene expression via MEK/ERK-dependent and -independent mechanisms.
Figure 7.
Scheme for up-regulation of the PAI-1 gene by EGF in glioblastoma. In glioblastoma cells, binding of EGF to EGFR induces rapid phosphorylation of Src, which in turn results in phosphorylation and translocation of PKCδ into the membrane (likely recruited via an adaptor protein CDCP1). Phosphorylated PKCδ activates SphK1, which subsequently translocates to the membrane. The translocation of SphK1 can be efficiently blocked by both Src and PKC inhibitors (PP2 and Gö6983, respectively), and siRNA to PKCδ. Activation of SphK1 is indispensable for both EGFinduced c-jun phosphorylation and PAI-1 activation but not for ATF-2 phosphorylation. Depletion of either SphK1 or PKCδ expression blocks EGFactivated PAI-1 expression and c-jun activation. However, this depletion has no effect on ATF-2 phosphorylation. Phosphorylated c-jun and independently activated ATF-2 may activate PAI-1 expression by binding to the regulatory elements located within 17 Alu elements in the 5′ flanking region of the PAI-1 gene.
Several lines of evidence suggest that PKCδ and SphK1 may be downstream targets of EGF-activated c-Src in glioma cells: i) EGF induced rapid phosphorylation and translocation of PKCδ to the plasma membrane (Fig. 4), which was blocked by PP2 (Fig. 6D); ii) EGF also induced translocation of SphK1 to the plasma membrane, which was blocked by inhibition of c-Src and by down-regulation of PKCδ (Fig. 5 and Fig. 6); and iii) down-regulation of PKCδ and SphK1 abolished EGF-induced PAI-1 expression (Fig. 4 and Fig. 5). Collectively, our data suggest that SphK1 is a downstream target of PKCδ that is indispensable for PAI-1 upregulation by EGF. The activation of PKCδ may involve c-Src-dependent phosphorylation of CDCP1, which may serve as an adapter protein and recruit PKCδ via its C2 domain (38). In contrast to PKCδ, down-regulation of PKCα did not affect translocation of SphK1 nor PAI-1 up-regulation by EGF, yet increased basal expression of PAI-1 (Fig. 4C).
Furthermore, we propose that SphK1 is the downstream target of PKCδ and that it is indispensable for PAI-1 activation by EGF. This observation is supported by the findings that the knockdown of SphK1 expression abolishes the activation of PAI-1 by EGF (Fig. 5A), while the knockdown of PKCδ blocks the translocation of SphK1 to the membrane (Fig. 5E). It is unclear at the moment if the generation of S1P is required for activation of PAI-1 by EGF. However, S1P activates PAI-1 expression when exogenously added to the cells (Fig. 6E), suggesting that S1P is likely generated by EGF-activated SphK1, secreted by the cells, and then activates S1P receptors. It remains to be determined which of the five known S1P receptors are activated and what are their downstream targets. Alternatively, S1P could exert its effects on PAI-1 expression via intracellular mechanisms as proposed for other processes. These mechanisms, however, are not understood.
The transcriptional mechanism of EGF-mediated PAI-1 activation remains elusive. The previously identified regulatory elements located within the 1.5 kb PAI-1 flanking region, and the −15 kb enhancer do not confer responsiveness to EGF (Fig. 2D). However, the down-regulation of PKCδ diminished the phosphorylation of c-jun (Fig. 4D). In addition, EGF rapidly activates ATF-2 (Fig. 2A), suggesting that complexes of c-jun and ATF-2 (or other members of ATF/CREB family) may be involved in PAI-1 regulation. In fact, 17 putative c-jun/ATF binding elements are present within the 17 Alu elements residing in the 5′ flanking region of the PAI-1 gene, and these elements are capable of protein binding in vitro (Supplemental Fig. 1). However, these elements, when integrated into reporters, do not confer responsiveness to EGF in transfection experiments. Since ATF-2 possesses acetyltransferase activity, it is possible that the regulation by EGF occurs on the epigenetic level, which cannot be recapitulated using reporter constructs.
Identification of SphK1 as an important mediator of EGF signaling that leads to increased PAI-1 expression is of particular importance. Recently, it has been demonstrated that high expression of SphK1 correlates with poor survival of patients with glioblastoma multiforme (46). In addition, S1P, which is present at substantial levels in the brain, is mitogenic for several glioma cell lines and enhances their motility and invasion (47). Therefore, SphK1 has been proposed as a promising target for therapy for a number of cancers, including gliomas (48). Similarly, c-Src is a promising therapeutic target since it regulates actin dynamics, and c-Src inhibitors decrease glioma cell invasion (49). Furthermore, inhibition of PKCδ decreases glioma cell proliferation (50). To our knowledge, this is the first demonstration that c-Src is an upstream regulator of SphK1 via the activation of PKCδ and that all three are required components of a pathway initiated by EGF, which leads to enhanced PAI-1 expression. Therefore, concurrent inhibition of these enzymes could potentially be a novel therapeutic approach to treat highly invasive gliomas.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (NSO44118 and NSO46280) to T.K., the Pilot Project grant from the Massey Cancer Center, VCU (to T.K.), and in part by R01 CA61774 to S.S. S.M. was supported by the NIMH Intramural Research Program. The imaging cytometry facility was supported in part by the NIH grant P30 CA16059.
REFERENCES
- 1.Bellail AC, Hunter SB, Brat DJ, Tan C, Van Meir EG. Microregional extracellular matrix heterogeneity in brain modulates glioma cell invasion. Int. J. Biochem. Cell Biol. 2004;36:1046–1069. doi: 10.1016/j.biocel.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 2.Mueller MM, Werbowetski T, Del Maestro RF. Soluble factors involved in glioma invasion. Acta Neurochir. (Wien) 2003;145:999–1008. doi: 10.1007/s00701-003-0132-0. [DOI] [PubMed] [Google Scholar]
- 3.Nakada M, Okada Y, Yamashita J. The role of matrix metalloproteinases in glioma invasion. Front. Biosci. 2003;8:e261–e269. doi: 10.2741/1016. [DOI] [PubMed] [Google Scholar]
- 4.Andreasen PA, Egelund R, Petersen HH. The plasminogen activation system in tumor growth, invasion, and metastasis. Cell. Mol. Life Sci. 2000;57:25–40. doi: 10.1007/s000180050497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rooprai HK, McCormick D. Proteases and their inhibitors in human brain tumours: a review. Anticancer Res. 1997;17:4151–4162. [PubMed] [Google Scholar]
- 6.Levicar N, Nuttall RK, Lah TT. Proteases in brain tumour progression. Acta Neurochir. (Wien) 2003;145:825–838. doi: 10.1007/s00701-003-0097-z. [DOI] [PubMed] [Google Scholar]
- 7.Landau BJ, Kwaan HC, Verrusio EN, Brem SS. Elevated levels of urokinase-type plasminogen activator and plasminogen activator inhibitor type-1 in malignant human brain tumors. Cancer Res. 1994;54:1105–1108. [PubMed] [Google Scholar]
- 8.Preissner KT, Kanse SM, May AE. Urokinase receptor: a molecular organizer in cellular communication. Curr. Opin. Cell Biol. 2000;12:621–628. doi: 10.1016/s0955-0674(00)00141-1. [DOI] [PubMed] [Google Scholar]
- 9.Nagamine Y, Medcalf RL, Munoz-Canoves P. Transcriptional and posttranscriptional regulation of the plasminogen activator system. Thromb. Haemost. 2005;93:661–675. doi: 10.1160/TH04-12-0814. [DOI] [PubMed] [Google Scholar]
- 10.Kasza A, Kowanetz M, Poslednik K, Witek B, Kordula T, Koj A. Epidermal growth factor and pro-inflammatory cytokines regulate the expression of components of plasminogen activation system in U373-MG astrocytoma cells. Cytokine. 2001;16:187–190. doi: 10.1006/cyto.2001.0957. [DOI] [PubMed] [Google Scholar]
- 11.Kasza A, Kiss DL, Gopalan S, Xu W, Rydel RE, Koj A, Kordula T. Mechanism of plasminogen activator inhibitor-1 regulation by oncostatin M and interleukin-1 in human astrocytes. J. Neurochem. 2002;83:696–703. doi: 10.1046/j.1471-4159.2002.01163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plata-Salaman CR. Epidermal growth factor and the nervous system. Peptides. 1991;12:653–663. doi: 10.1016/0196-9781(91)90115-6. [DOI] [PubMed] [Google Scholar]
- 13.Wong AJ, Bigner SH, Bigner DD, Kinzler KW, Hamilton SR, Vogelstein B. Increased expression of the epidermal growth factor receptor gene in malignant gliomas is invariably associated with gene amplification. Proc. Natl. Acad. Sci. U. S. A. 1987;84:6899–6903. doi: 10.1073/pnas.84.19.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Libermann TA, Nusbaum HR, Razon N, Kris R, Lax I, Soreq H, Whittle N, Waterfield MD, Ullrich A, Schlessinger J. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumours of glial origin. Nature. 1985;313:144–147. doi: 10.1038/313144a0. [DOI] [PubMed] [Google Scholar]
- 15.Frederick L, Wang XY, Eley G, James CD. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res. 2000;60:1383–1387. [PubMed] [Google Scholar]
- 16.Von Deimling A, Louis DN, von Ammon K, Petersen I, Hoell T, Chung RY, Martuza RL, Schoenfeld DA, Yasargil MG, Wiestler OD, Seizinger BR. Association of epidermal growth factor receptor gene amplification with loss of chromosome 10 in human glioblastoma multiforme. J. Neurosurg. 1992;77:295–301. doi: 10.3171/jns.1992.77.2.0295. [DOI] [PubMed] [Google Scholar]
- 17.Quan AL, Barnett GH, Lee SY, Vogelbaum MA, Toms SA, Staugaitis SM, Prayson RA, Peereboom DM, Stevens GH, Cohen BH, Suh JH. Epidermal growth factor receptor amplification does not have prognostic significance in patients with glioblastoma multiforme. Int. J. Radiat. Oncol. Biol. Phys. 2005;63:695–703. doi: 10.1016/j.ijrobp.2005.03.051. [DOI] [PubMed] [Google Scholar]
- 18.Muracciole X, Romain S, Dufour H, Palmari J, Chinot O, Ouafik L, Grisoli F, Branger DF, Martin PM. PAI-1 and EGFR expression in adult glioma tumors: toward a molecular prognostic classification. Int. J. Radiat. Oncol. Biol. Phys. 2002;52:592–598. doi: 10.1016/s0360-3016(01)02699-2. [DOI] [PubMed] [Google Scholar]
- 19.Valter MM, Wiestler OD, Pietsche T. Differential control of VEGF synthesis and secretion in human glioma cells by IL-1 and EGF. Int. J. Dev. Neurosci. 1999;17:565–577. doi: 10.1016/s0736-5748(99)00048-9. [DOI] [PubMed] [Google Scholar]
- 20.Hjortland GO, Lillehammer T, Somme S, Wang J, Halvorsen T, Juell S, Hirschberg H, Fodstad O, Engebraaten O. Plasminogen activator inhibitor-1 increases the expression of VEGF in human glioma cells. Exp. Cell Res. 2004;294:130–139. doi: 10.1016/j.yexcr.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 21.Mazar AP, Henkin J, Goldfarb RH. The urokinase plasminogen activator system in cancer: implications for tumor angiogenesis and metastasis. Angiogenesis. 1999;3:15–32. doi: 10.1023/a:1009095825561. [DOI] [PubMed] [Google Scholar]
- 22.Lakka SS, Gondi CS, Dinh DH, Olivero WC, Gujrati M, Rao VH, Sioka C, Rao JS. Specific interference of urokinase-type plasminogen activator receptor and matrix metalloproteinase-9 gene expression induced by double-stranded RNA results in decreased invasion, tumor growth, and angiogenesis in gliomas. J. Biol. Chem. 2005;280:21882–21892. doi: 10.1074/jbc.M408520200. [DOI] [PubMed] [Google Scholar]
- 23.Rao JS, Gondi C, Chetty C, Chittivelu S, Joseph PA, Lakka SS. Inhibition of invasion, angiogenesis, tumor growth, and metastasis by adenovirus-mediated transfer of antisense uPAR and MMP-9 in non-small cell lung cancer cells. Mol. Cancer Ther. 2005;4:1399–1408. doi: 10.1158/1535-7163.MCT-05-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foekens JA, Peters HA, Look MP, Portengen H, Schmitt M, Kramer MD, Brunner N, Janicke F, Meijer-van Gelder ME, Henzen-Logmans SC, van Putten WL, Klijn JG. The urokinase system of plasminogen activation and prognosis in 2780 breast cancer patients. Cancer Res. 2000;60:636–643. [PubMed] [Google Scholar]
- 25.Czekay RP, Aertgeerts K, Curriden SA, Loskutoff DJ. Plasminogen activator inhibitor-1 detaches cells from extracellular matrices by inactivating integrins. J. Cell Biol. 2003;160:781–791. doi: 10.1083/jcb.200208117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Czekay RP, Loskutoff DJ. Unexpected role of plasminogen activator inhibitor 1 in cell adhesion and detachment. Exp. Biol. Med. (Maywood) 2004;229:1090–1096. doi: 10.1177/153537020422901102. [DOI] [PubMed] [Google Scholar]
- 27.Gopalan S, Kasza A, Xu W, Kiss DL, Wilczynska KM, Rydel RE, Kordula T. Astrocyte- and hepatocyte-specific expression of genes from the distal serpin subcluster at 14q32.1 associates with tissue-specific chromatin structures. J. Neurochem. 2005;94:763–773. doi: 10.1111/j.1471-4159.2005.03204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 29.Kordula T, Rydel RE, Brigham EF, Horn F, Heinrich PC, Travis J. Oncostatin M and the interleukin-6 and soluble interleukin-6 receptor complex regulate alpha1-antichymotrypsin expression in human cortical astrocytes. J. Biol. Chem. 1998;273:4112–4118. doi: 10.1074/jbc.273.7.4112. [DOI] [PubMed] [Google Scholar]
- 30.Baeuerle PA, Baltimore D. I kappa B: a specific inhibitor of the NF-kappa B transcription factor. Science. 1988;242:540–546. doi: 10.1126/science.3140380. [DOI] [PubMed] [Google Scholar]
- 31.Sawadogo M, Van Dyke MW, Gregor PD, Roeder RG. Multiple forms of the human gene-specific transcription factor USF. I. Complete purification and identification of USF from HeLa cell nuclei. J. Biol. Chem. 1988;263:11985–11993. [PubMed] [Google Scholar]
- 32.Bektas M, Jolly PS, Muller C, Eberle J, Spiegel S, Geilen CC. Sphingosine kinase activity counteracts ceramide-mediated cell death in human melanoma cells: role of Bcl-2 expression. Oncogene. 2005;24:178–187. doi: 10.1038/sj.onc.1208019. [DOI] [PubMed] [Google Scholar]
- 33.Hou B, Eren M, Painter CA, Covington JW, Dixon JD, Schoenhard JA, Vaughan DE. Tumor necrosis factor alpha activates the human plasminogen activator inhibitor-1 gene through a distal nuclear factor kappaB site. J. Biol. Chem. 2004;279:18127–18136. doi: 10.1074/jbc.M310438200. [DOI] [PubMed] [Google Scholar]
- 34.Samad F, Hester KD, Yang G, Hannun YA, Bielawski J. Altered adipose and plasma sphingolipid metabolism in obesity: a potential mechanism for cardiovascular and metabolic risk. Diabetes. 2006;55:2579–2587. doi: 10.2337/db06-0330. [DOI] [PubMed] [Google Scholar]
- 35.Spiegel S, Milstien S. Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat. Rev. Mol. Cell Biol. 2003;4:397–407. doi: 10.1038/nrm1103. [DOI] [PubMed] [Google Scholar]
- 36.Pitson SM, Xia P, Leclercq TM, Moretti PA, Zebol JR, Lynn HE, Wattenberg BW, Vadas MA. Phosphorylation-dependent translocation of sphingosine kinase to the plasma membrane drives its oncogenic signalling. J. Exp. Med. 2005;201:49–54. doi: 10.1084/jem.20040559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizuno K, Kubo K, Saido TC, Akita Y, Osada S, Kuroki T, Ohno S, Suzuki K. Structure and properties of a ubiquitously expressed protein kinase C, nPKC delta. Eur. J. Biochem. 1991;202:931–940. doi: 10.1111/j.1432-1033.1991.tb16453.x. [DOI] [PubMed] [Google Scholar]
- 38.Benes CH, Wu N, Elia AE, Dharia T, Cantley LC, Soltoff SP. The C2 domain of PKCdelta is a phosphotyrosine binding domain. Cell. 2005;121:271–280. doi: 10.1016/j.cell.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 39.Janicke F, Schmitt M, Graeff H. Clinical relevance of the urokinase-type and tissue-type plasminogen activators and of their type 1 inhibitor in breast cancer. Semin. Thromb. Hemost. 1991;17:303–312. doi: 10.1055/s-2007-1002624. [DOI] [PubMed] [Google Scholar]
- 40.Hopkins WE, Westerhausen DR, Jr, Sobel BE, Billadello JJ. Transcriptional regulation of plasminogen activator inhibitor type-1 mRNA in Hep G2 cells by epidermal growth factor. Nucleic Acids Res. 1991;19:163–168. doi: 10.1093/nar/19.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anteby EY, Greenfield C, Natanson-Yaron S, Goldman-Wohl D, Hamani Y, Khudyak V, Ariel I, Yagel S. Vascular endothelial growth factor, epidermal growth factor and fibroblast growth factor-4 and -10 stimulate trophoblast plasminogen activator system and metalloproteinase-9. Mol. Hum. Reprod. 2004;10:229–235. doi: 10.1093/molehr/gah031. [DOI] [PubMed] [Google Scholar]
- 42.Mawatari M, Okamura K, Matsuda T, Hamanaka R, Mizoguchi H, Higashio K, Kohno K, Kuwano M. Tumor necrosis factor and epidermal growth factor modulate migration of human microvascular endothelial cells and production of tissue-type plasminogen activator and its inhibitor. Exp. Cell Res. 1991;192:574–580. doi: 10.1016/0014-4827(91)90078-9. [DOI] [PubMed] [Google Scholar]
- 43.Seebacher T, Manske M, Zoller J, Crabb J, Bade EG. The EGF-inducible protein EIP-1 of migrating normal and malignant rat liver epithelial cells is identical to plasminogen activator inhibitor 1 and is a component of the ECM migration tracks. Exp. Cell Res. 1992;203:504–507. doi: 10.1016/0014-4827(92)90029-8. [DOI] [PubMed] [Google Scholar]
- 44.Samarakoon R, Higgins CE, Higgins SP, Kutz SM, Higgins PJ. Plasminogen activator inhibitor type-1 gene expression and induced migration in TGF-beta1-stimulated smooth muscle cells is pp60(c-src)/MEK-dependent. J. Cell Physiol. 2005;204:236–246. doi: 10.1002/jcp.20279. [DOI] [PubMed] [Google Scholar]
- 45.Guerrero J, Santibanez JF, Gonzalez A, Martinez J. EGF receptor transactivation by urokinase receptor stimulus through a mechanism involving Src and matrix metalloproteinases. Exp. Cell Res. 2004;292:201–208. doi: 10.1016/j.yexcr.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 46.Van Brocklyn JR, Jackson CA, Pearl DK, Kotur MS, Snyder PJ, Prior TW. Sphingosine kinase-1 expression correlates with poor survival of patients with glioblastoma multiforme: roles of sphingosine kinase isoforms in growth of glioblastoma cell lines. J. Neuropathol. Exp. Neurol. 2005;64:695–705. doi: 10.1097/01.jnen.0000175329.59092.2c. [DOI] [PubMed] [Google Scholar]
- 47.Van Brocklyn JR, Young N, Roof R. Sphingosine-1-phosphate stimulates motility and invasiveness of human glioblastoma multiforme cells. Cancer Lett. 2003;199:53–60. doi: 10.1016/s0304-3835(03)00334-3. [DOI] [PubMed] [Google Scholar]
- 48.Van Brocklyn JR, Lee MJ, Menzeleev R, Olivera A, Edsall L, Cuvillier O, Thomas DM, Coopman PJ, Thangada S, Liu CH, Hla T, Spiegel S. Dual actions of sphingosine-1-phosphate: extracellular through the Gi-coupled receptor Edg-1 and intracellular to regulate proliferation and survival. J. Cell Biol. 1998;142:229–240. doi: 10.1083/jcb.142.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Angers-Loustau A, Hering R, Werbowetski TE, Kaplan DR, Del Maestro RF. SRC regulates actin dynamics and invasion of malignant glial cells in three dimensions. Mol. Cancer Res. 2004;2:595–605. [PubMed] [Google Scholar]
- 50.Yamaguchi K, Richardson MD, Bigner DD, Kwatra MM. Signal transduction through substance P receptor in human glioblastoma cells: roles for Src and PKCdelta. Cancer Chemother. Pharmacol. 2005;56:585–593. doi: 10.1007/s00280-005-1030-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.