Abstract
Used for decades for biological warfare, Bacillus anthracis (category A agent) has proven to be highly stable and lethal. Quantitative risk assessment modeling requires descriptive statistics of the limit of detection to assist in defining the exposure. Furthermore, the sensitivities of various detection methods in environmental matrices are vital information for first responders. A literature review of peer-reviewed journal articles related to methods for detection of B. anthracis was undertaken. Articles focused on the development or evaluation of various detection approaches, such as PCR, real-time PCR, immunoassay, etc. Real-time PCR and PCR were the most sensitive methods for the detection of B. anthracis, with median instrument limits of detection of 430 and 440 cells/ml, respectively. There were very few peer-reviewed articles on the detection methods for B. anthracis in the environment. The most sensitive limits of detection for the environmental samples were 0.1 CFU/g for soil using PCR-enzyme-linked immunosorbent assay (ELISA), 17 CFU/liter for air using an ELISA-biochip system, 1 CFU/liter for water using cultivation, and 1 CFU/cm2 for stainless steel fomites using cultivation. An exponential dose-response model for the inhalation of B. anthracis estimates of risk at concentrations equal to the environmental limit of detection determined the probability of death if untreated to be as high as 0.520. Though more data on the environmental limit of detection would improve the assumptions made for the risk assessment, this study's quantification of the risk posed by current limitations in the knowledge of detection methods should be considered when employing those methods in environmental monitoring and cleanup strategies.
According to the Centers for Disease Control (CDC), a category A agent is an organism that poses a risk to national security because it can be easily disseminated or transmitted from person to person, results in high mortality rates, has the potential for major public health impact, might cause public panic and social disruption, and requires special action for public health preparedness (http://emergency.cdc.gov/agent/agentlist-category.asp). Quantitative information on category A agents in environmental matrices (soil, air, fomite, and water) is very limited (62). However, from the literature, it has been concluded that Bacillus anthracis is the most environmentally stable category A agent overall (62).
After the release of B. anthracis through mail envelopes in 2001, assessment of the decontamination process revealed an important question: could the detection methods effectively determine if the environment is clean? An evaluation of the effectiveness of sampling methods at a U.S. postal facility in Washington, DC, that was contaminated with B. anthracis spores concluded that neither of the sampling methods used (HEPA vacuum or wipes) were sensitive enough to ensure that spores had been removed completely. In addition, the event exposed the necessity of quantifying recovery and extraction efficiency during sample collection and processing to improve the method limit of detection (61, 67).
In this literature review, the limit of detection of methods for B. anthracis is characterized as either an instrument limit of detection or an environmental limit of detection. An instrument limit of detection is generally evaluated with pure cultures. An environmental limit of detection is evaluated with cultures/cells spiked into an environmental matrix (soil, air, fomites, water), which then undergoes various recovery and concentration procedures (i.e., filtration and extraction or direct extraction) before detection (see Fig. S1 in the supplemental material).
Compared to an instrument limit of detection, the establishment of an environmental limit of detection poses more challenges, including dilute target concentrations, environmental impurities, background inhibitors, organisms in a viable but not cultivable state, and overall processing efficiency. There are many steps for processing environmental samples prior to detection. At each process step, there can be a loss of the initial target organism, and thus, each step has a recovery efficiency, which could be interpreted as a set number, distribution, or range (see Fig. S1 in the supplemental material). Since recovery efficiency directly affects the limit of detection, improving recovery efficiency would result in a more sensitive detection method.
In determining if an environmental site is “clean,” another component that should be evaluated is the quantification and characterization of the potential health risk. Quantitative microbial risk assessment (QMRA) is a method used to assess the likelihood of infection based on specific exposures to hazardous pathogenic organisms. QMRA risk modeling has been used with water and food and could be useful for management decisions during a disease outbreak or a bioterrorism attack (35). Environmental monitoring is used to inform the exposure assessment and the efficiency of disinfection. The limit of detection is a critical criterion for any method, which dictates the application and usefulness of demonstrating a “zero” during environmental monitoring. The limit of detection of a chosen analytical method is also an input variable for the QMRA model; a statistical distribution quantifying the variability in limit of detection is preferred for realistic modeling.
The objectives of this study were to review, in the literature, the instrument limit of detection and the environmental limit of detection for methods to detect B. anthracis and to compare the estimated risk at the instrument limit of detection and the environmental limit of detection. Though the number of articles on B. anthracis was extensive, there was a paucity of articles that specifically included environmental limits of detection. This information is essential for a QMRA of B. anthracis in the establishment of future environmental monitoring strategies and cleanup goals.
MATERIALS AND METHODS
Journal articles were searched on the ISI Web of Science database for B. anthracis and the following keywords: method, sensitivity, limit of detection, detection limit, limit, water, air, soil, fomite, surface, specificity, PCR sensor, environmental, rapid, assay, diagnostic, immunoassay, antibody, real time, real-time PCR, microfluidic, polymerase, quantitative, bioaerosol, aerosol, microarrays, biosensor, electrochemiluminescence (ECL), Raman spectrometry, and mass spectrometry. Approximately 1,700 references (and abstracts, when available) were retrieved and were saved in an EndNote file. Though the search defaults were set for the years 1900 through 2007, the oldest article used to evaluate the limit of detection was published in 1994. Abstracts were manually screened for information on the detection of B. anthracis. Some studies used a surrogate for B. anthracis to determine the limit of detection. It was assumed that B. anthracis would behave as the surrogate and was included in this review. If the abstract pertained to a detection method, then the full article was downloaded, saved in another database, and reviewed for quantitative data describing the limit of detection. The remaining references and abstracts that were not used in this literature review either did not indicate information about detection methods or were not retrievable. At the end, 71 articles were retrieved and analyzed to obtain the instrument limit of detection or the environmental limit of detection.
Instrument limit of detection.
The instrument limit of detection was extracted from the articles describing a method that detected B. anthracis in a pure culture without spiking B. anthracis into an environmental matrix (soil, air, fomite, or water). Raw data that were extracted were recorded in numbers of units of cells, spores, DNA, CFU, protective antigens, and genomic copies in volumes that ranged from liters to microliters. Articles that used units of protective antigens were not used in this literature review due to the unknown conversion factor from antigens to cells. All data were converted into standard units of cells per milliliter of reaction solution, and the data by method were graphed and compared.
Environmental limit of detection.
In studies reporting the environmental limit of detection, B. anthracis spores were spiked into the matrix, extracted, and detected using various detection methods. The articles that reported the environmental limit of detection of B. anthracis were categorized according to the matrix in which B. anthracis was detected (soil, air, fomite, or water). Additional parameters extracted from the articles varied with the matrix (see Fig. S2 in the supplemental material). These included the following parameters. (i) For soil, they include the amount of soil, sample concentration, extraction volume, volume of extracted sample added to the reaction, and total volume. In addition, the type of pretreatment or extraction method and the soil type or location were noted (Table 1). (ii) For air, they include the sample volume, airflow rate, duration, sample concentration, extraction volume, volume of extracted sample added to the reaction, and total volume. (iii) For fomites, they include the surface area, sample concentration, surface seeding method, extraction volume, and total volume. In some cases, recovery efficiency and extraction efficiency were available and noted. In addition, the type of fomite, sampling method, extraction method, and culturing method were noted (Table 2). (iv) For water, they included the sample volume, sample concentration, extraction volume, volume of extracted sample added to the reaction, and total volume. In addition, the condition of the water was noted.
TABLE 1.
Parameters for the environmental limit of detection in soil
| Detection methoda | Amt of soil | Sample concn | Pretreatment/extraction method (company) | Time (h) | Difficulty level | Extraction vol (μl) | Vol added to reaction (μl) | Total vol (μl) | Limit of detection (CFU/g soil) | Soil type/location | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PCR-ELISA | 100 g | 1-100 CFU/100 g | Easy DNA kit (Invitrogen) | 2.5 | 2 | 100 | 60 | 60 | 0.1 | Nonsuspicious sites | 10 |
| 100 g | 1-100 CFU/100 g | Easy DNA kit (Invitrogen) | 2.5 | 2 | 100 | 60 | 60 | 1.0 | Contaminated sites with organic compounds and tanning agents | 10 | |
| Nested PCR + 2× cultivation in TSB | 1 g | 0, 1, 10, 102, 103 CFU/g | FastDNA SPIN kit | 36.0 | 4 | 25 | 1.0 | Garden soil with 3% peat | 24 | ||
| Nested PCR + cultivation in TSB | 1 g | 0, 1, 10, 102, 103 CFU/g | FastDNA SPIN kit | 18 | 3 | 25 | 1.0 × 102 | Garden soil with 3% peat | 24 | ||
| Nested PCR | 1 g | 0, 1, 10, 102, 103 CFU/g | FastDNA SPIN kit | 2 | 2 | 25 | 1.0 × 103 | Garden soil with 3% peat | 24 | ||
| 100 mg | 106 CFU/100 mg | Three freeze-thaw cycles/glass beads and glass milk | 3.5 | 5 | 30 | 5 | 25 | 1.0 × 105 | Litter, meadow, cultivated, swamp, and lawn | 63 | |
| PCR | 1 g | 2.5 × 103-2.5 × 107 CFU/g | Hot detergent/bead mill homogenization | 1 | 3 | 100 | 10 | 100 | 2.5 × 103 | Anthony fine sandy loam from New Mexico agriculture fields | 44 |
| Immunofluorescence | 1 g | 103-107 CFU/g | Aqueous polymer two-phase system | 0.75 | 2 | 100 | 20 | 40 | 5.6 × 103 | Sand | 2 |
| 1 g | 103-107 CFU/g | Aqueous polymer two-phase system | 0.75 | 2 | 100 | 20 | 40 | 1.4 × 104 | Garden | 2 | |
| Real-time PCR | 0.1 g | 103-107CFU/g | Heat treatment with 1.22 g/ml sucrose-0.5% Triton X-100 | 0.75 | 3 | 1,000 | 5 | 25 | 1.0 × 104 | National Institute of Health—Korea | 60 |
| Multiplex PCR | 0.1 g | 103-107 CFU/g | Heat treatment with 1.22 g/ml sucrose-0.5% Triton X-100 | 0.75 | 3 | 1,000 | 1 | 25 | 1.0 × 105 | National Institute of Health—Korea | 60 |
| 0.1 g | 103-107 CFU/g | Heat treatment with sterilized water and 10% Triton X-100-PBS | 1.5 | 3 | 1,000 | 1 | 25 | 1.0 × 108 | National Institute of Health—Korea | 60 | |
| IM-ECL | 1 mg | 0-106 CFU/assay | IM separation performed twice and resuspended in PBS | 1.5 | 3 | 1.0 × 105 | Moist, dark brown to black soil and dry, light yellowish sandy soil from diverse military and agriculture fields | 18 | |||
| 1 mg | 0-106 CFU/assay | IM separation performed twice and resuspended in PBS | 1.5 | 3 | 1.0 × 106 | Moist, dark brown to black soil and dry, light yellowish sandy soil from diverse military and agriculture fields | 18 | ||||
| 1 mg | 0-106 CFU/assay | IM separation performed twice and resuspended in PBS | 1.5 | 3 | 1.0 × 107 | Moist, dark brown to black soil and dry, light yellowish sandy soil from diverse military and agriculture fields | 18 | ||||
| Biosensor assay | 1 mg/ml powder in PBS | 3.2 × 103-3.2 × 105 CFU/ml | Washing with 1 ml PBST | 0.25 | 1 | 30 | 5 | 25 | 3.2 × 108 | Talc-based powder, cornstarch, confectioners' sugar, baking soda, and Bacillus thuringiensis-based pesticide | 69 |
TSB, Trypticase soy broth.
TABLE 2.
Parameters for the environmental limit of detection on fomites
| Study authors (reference) | Surface area | Sample concn | Surface seeding | Sampling method | Extraction method | Extraction vol | Total vol | Recovery efficiency (%) | Extraction efficiency (%) | Culture medium | Limit of detection |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Hodges et al. (38) | 10 cm2 | 0.2-3,000 CFU/cm2 | Inoculated with 0.5 ml spore solution | Macrofoam swab | Vortex in 5 ml PBST for 2 min at 10-s intervals | 5 ml | 31.7-49.1 | 93.4 | Sheep blood agar | 12 CFU/cm2 | |
| Rose et al. (59) | 25 cm2 | 2 × 104 CFU/cm2 | Inoculated with 0.5 ml spore solution | Cotton swab, macrofoam swab | Vortex in 5 ml PBST for 2 min at 10-s intervals | 5 ml | 100 μl | 41.7 for cotton swab, 43.6 for macrofoam swab | 93.9 for cotton swab, 93.4 for macrofoam swab | Trypticase soy agar with 5% sheep blood | 20 CFU/cm2a |
| Brown et al. (15) | 25 cm2 | 100-10,000 CFU/cm2 | Dry aerosol deposition | Polyester-rayon blend gauze wipe | Sonication and heat treatment | 30 ml | 1 ml | 35 for stainless steel, 29 for painted wallboard | 93 | Brain heart infusion agar | 90 CFU/cm2 for stainless steel, 105 CFU/cm2 for painted wallboard |
| Brown et al. (16) | 25 cm2 | 100-10,000 CFU/cm2 | Dry aerosol deposition | Rayon swab | Sonication and heat treatment | 10 ml | 1 ml | 41 for stainless steel, 41 for painted wallboard | 76 | Brain heart infusion agar | 1 CFU/cm2 for stainless steel and painted wallboard |
| Brown et al. (14) | 100 cm2 | 100-10,000 CFU/cm2 | Dry aerosol deposition | Vacuum filter sock | Sonication and heat treatment | 30 ml | 1 ml | 29 for stainless steel, 25 for painted wallboard, 28 for carpet, 19 for concrete | Petrifilm aerobic count plate | 105 CFU/m2 for stainless steel and carpet, 102 CFU/m2 for painted wallboard, 160 CFU/m2 for concrete | |
| Buttner et al. (20) | 1 m2 | 105 CFU/m2 | Inoculated with spore solution | BiSKit—wet/dry | Foam compression | 3.3 ml for wet sampling, 16.1 ml for dry sampling | 1 ml | 11.3 for wet sampling, 18.4 for dry sampling | Trypticase soy agar | 42 ± 6 CFU/m2 for wet sampling, 100 ± 10 CFU/m2 for dry sampling |
The limit of detection was not recorded in the article and was calculated to be at least 20 CFU/cm2.
Quantifying limits of risk estimates.
The risk of mortality by inhalation of B. anthracis spores was estimated for concentrations corresponding to the instrument limit of detection and the environmental limit of detection in the air. For each limit of detection, a distribution of risks was calculated by the Monte Carlo method using 100,000 replicates in Crystal Ball 7.3.1 (2007; Oracle). The number of replicates was chosen at the point where the 90% confidence interval was stable over a range from 1/10 to 10 times the number of replicates used.
A recent evaluation of dose-response data for B. anthracis spores through the inhalation exposure route found that the dose-response relationship could be modeled by the exponential equation (4)
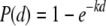 |
where P(d) is the probability of death (P) (when untreated) at dose d, and k is the probability that one organism will survive to initiate the response (4). In this study, a k value generated from a pooled guinea pig and rhesus monkey data set was used. A distribution of 10,000 best-fit k values generated using bootstrap replicates of that data set was provided by Timothy Bartrand of Drexel University and fit to a gamma distribution. The dose was calculated as
 |
where Cair is the number of spores per cubic meter of air (instrument limit of detection or environmental limit of detection), R is the breathing rate (m3/h), and t is the duration of exposure (h). When Cair was evaluated as a range of limits of detection, it was modeled as a lognormal distribution; otherwise, it was evaluated as a point estimate. The breathing rate, R, was modeled as a Pareto distribution fit to the short-term breathing rates of adults (18 years of age and up) of both sexes from rest to moderate activity (71). The exposure time, t, was modeled as a uniform distribution from 1 min to 8 h.
Five risk scenarios were evaluated with this model using different values for Cair. For each risk scenario, either the instrument limit of detection or environmental limit of detection, a sensitivity analysis was generated using Crystal Ball 7.3.1 (2007; Oracle). The median real-time PCR instrument limit of detection and the range of real-time PCR instrument limit of detection were two scenarios used to explore the effect of instrument limit of detection on risk. For the instrument limit of detection scenarios, it was assume that all B. anthracis spores in a cubic meter of air could be collected without any loss and concentrated into 1 ml of solution for analysis. Log-transformed real-time PCR and PCR instrument limits of detection were checked for normality with a Lilliefors test and compared using analysis of variance. Then, the range of PCR instrument limits of detection was combined with the range of real-time PCR instrument limits of detection to increase the data in the distribution.
There were three environmental limit of detection scenarios; Cair was set to the environmental limits of detection reported for B. anthracis detected in the air. There were only two articles on the environmental limit of detection in the air. Due to the lack of data on the environmental limit of detection, the two limits of detection were referred to as the lower and upper environmental limits of detection. These two risk scenarios were evaluated as point estimates. The last risk scenario assumed that the environmental limit of detection for the air fit the same distributions as the lognormal instrument limit of detection, ranging from 17,000 to 50,000 CFU/m3 (this may not be the true range).
RESULTS AND DISCUSSION
Instrument limit of detection.
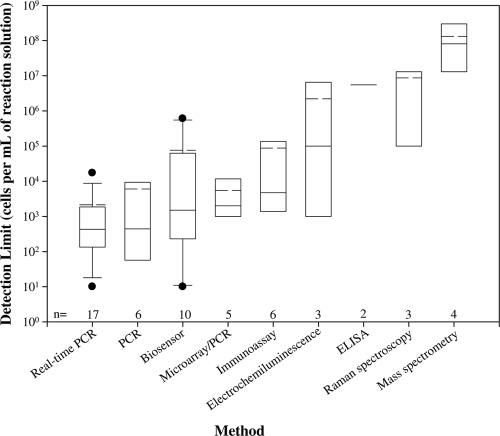
Out of 56 articles on the instrument limit of detection, 17 articles were on real-time PCR (6, 7, 11, 23, 25, 27, 39, 41, 45, 49, 51, 53, 56, 58, 60, 70, 72), 6 were on PCR (13, 31, 48, 57, 76), 10 were on biosensors (1, 3, 21, 22, 33, 36, 40, 52, 73, 74), 5 were on microarray/PCR (5, 19, 50, 64, 75), 6 were on immunoassay (29, 30, 32, 46, 65, 68), 3 were on ECL (17, 34, 77), 2 were on enzyme-linked immunosorbent assay (ELISA) (12, 26), 3 were on Raman spectroscopy (37, 55, 78), and 4 were on mass spectrometry (8, 9, 28, 43) (Fig. 1). Limits of detection ranged from 10 cells/ml (for real-time PCR) to 108 cells/ml (for mass spectrometry). Considering the median instrument limit of detection, real-time PCR and PCR were the most sensitive methods, with median instrument limits of detection of 430 and 440 cells/ml, respectively. It should be noted that there was one instrument limit of detection (4.29 × 106 cells/ml) that was not added to the distribution for real-time PCR because it was a multiplex assay, and the other instrument limits of detection in the distribution were from a singleplex assay (42). The least-sensitive methods were Raman spectroscopy and mass spectrometry, with median instrument limits of detection of approximately 1.0 × 107 and 8.0 × 107 cells/ml, respectively.
FIG. 1.
Distribution of the instrument limits of detection for various methods. On each box plot, the solid line represents the median result, and the dashed line represents the mean result. The box plot whiskers above and below the box indicate the 90th and 10th percentiles, respectively. The solid circles represent the outlying limits of detection, and n represents the number of journal articles available on each detection method for Bacillus anthracis.
The number of journal articles on real-time PCR and biosensors allowed limits of detection to be fit to a statistical distribution. When fewer articles were published, as was true for the other eight methods, assigning distributions was not possible. ECL, ELISA, Raman spectroscopy, and mass spectrometry (having less than four articles) were the methods with the least-sensitive instrument limits of detection. With limited information on these methods, the median instrument limit of detection may not properly represent these detection methods' capabilities for detecting B. anthracis. For example, the instrument limit of detection for ECL had only three published articles, with limits of detection ranging from 102 cells/ml to 106 cells/ml. For some emerging techniques, such as immunomagnetic ECL (IM-ECL) and aptamer-magnetic bead-ECL, limits of detection differed by 4 orders of magnitude. While the instrument limit of detection gives insight into the instruments' capabilities, when evaluating cleanup goals and assessing risk, the environmental limits of detection are needed to understand the challenges and capabilities for addressing the contamination.
Environmental limit of detection.
Out of 15 articles on the environmental limit of detection, 8 articles were on detection in soil (2, 10, 18, 24, 44, 60, 63, 69), 2 were on detection in the air (47, 66), 6 were on detection on fomites (14, 15, 16, 20, 38, 59), and 1 was on detection in water (54). The results for the environmental limit of detection could not be reported as distributions due to the limited number of articles for each matrix. The two most predominant methods used for the environmental limit of detection were cultivation and PCR-based methods.
Soil.
The environmental limit of detection of B. anthracis spiked into soil ranged from 0.1 (reported as 10 CFU/100 g of soil) to 3.2 × 108 CFU/g of soil, with a median limit of detection of 1.2 × 104 CFU/g of soil (Table 1). The median environmental limit of detection for soil should be used with caution, since there is a 9-orders-of-magnitude range due to the many approaches used to evaluate the environmental limit of detection. The approximate time for the extraction method (Table 1) was the time for one sample to be processed based on the information reported. If it was not an automated extraction procedure, then with the increase in samples, there would be an increase in extraction process time. The difficulty level for the extraction process (1 to 5, easy to difficult) was based on the number of steps in the procedure, the preparation time, and the approximated time for the extraction (Table 1). The biosensor assay, the easiest extraction method, resulted in the poorest limit of detection (3.2 × 108 CFU/g of soil). The detection methods with the most-sensitive limits of detection (PCR-ELISA, nested PCR, and PCR) had extraction methods with difficulty levels ranging from 2 to 5 (Table 1).
The environmental limit of detection depended highly on the pretreatment/extraction process; for instance, in 2003, Ryu et al. (60) used multiplex PCR and reported a difference of 3 orders of magnitude between heat treatment with 1.22 g/ml sucrose-0.5% Triton X-100 and heat treatment with sterilized water and 10% Triton X-100-phosphate-buffered saline (PBS) (Table 1). Similar results were found by Bruno and Yu in 1996 (18) when using IM-ECL as the detection method.
Differences in the environmental limit of detection were also based on the location or the type of soil. In 1999, Beyer et al. (10) reported that the PCR-ELISA method was more sensitive when using soils from nonsuspicious locations compared to using those from former tannery sites. Agarwal et al. (2002) (2) reported that the immunofluorescence assay was more sensitive when spores were spiked into sand (103) rather than into garden soil (104). For the IM-ECL method, Bruno and Yu (1996) (18) reported differences due to different strains, with Sterne (105) being more sensitive in the assay than Ames (106) and Vollum B1 (107).
Air.
There were only two studies of the evaluation of aerosolized B. anthracis spores collected by an air sampler and extracted for detection. The ELISA-biochip system coupled with a portable bioaerosol collection system collected aerosolized spores at an air sampling rate of 150 liters/min for 2 min into 5 ml of PBS. The ELISA-biochip system consisted of an ELISA for antibody-based identification in combination with the biochip detection instrument. The environmental limit of detection of the ELISA-biochip system was 17 CFU/liter. For the ELISA-biochip system, the efficiency of the air sampler was reported as approximately 50%, but the distribution was not fully described (66). The anthrax smoke detector collected aerosolized spores using a bioaerosol collection system at a rate of 15 liters/minute for 1 min onto a glass fiber filter tape. The detection of the spores using the lifetime-gated fluorimeter occurred after a thermal lysis and addition of TbCl3. The environmental limit of detection of the anthrax smoke detector was 50 CFU/liter (47).
Fomites.
Spores were seeded on fomites (stainless steel, plastic, wood, glass, etc.), recovered, extracted, and detected by cultivation. The environmental limit of detection was evaluated from stainless steel fomites ranging in surface area from 10 cm2 to 1 m2 (Table 2). In 2007, Brown et al. (14-16) also evaluated the environmental limit of detection on painted wallboard. In addition, the vacuum filter sock study tested porous fomites, carpet, and concrete (14).
The sampling methods evaluated by the articles were use of a macrofoam swab, cotton swab, polyester-rayon blend gauze wipe, rayon swab, vacuum filter sock, and biological sampling kit (BiSKit) (Table 2). Sampling methods such as use of cotton, macrofoam, polyester, and rayon swabs were all tested by Rose et al. in 2004 (59). It was concluded that the cotton and macrofoam swabs produced the highest recovery when the swabs were premoistened rather than dry. Similarly, in 2004, Buttner et al. (20) tested the BiSKit, cotton swabs, and foam swabs. The BiSKit was designed to do wet and dry sampling of large surfaces for bacteria, viruses, and toxins. The BiSKit resulted in the highest recovery out of the three methods. Using a wetting agent to recover spores from the surfaces enhanced the recovery and environmental limit of detection. Brown et al. (2007) (15, 16) used sterilized deionized water (except when using the vacuum filter sock), Buttner et al. (2004) (20) used potassium phosphate buffer with 0.05% Tween 20, and the other two authors used PBS with 0.04% Tween 80 (PBST). According to the CDC, the recommended wetting agents were sterile water, a sterile saline solution, or a sterile phosphate-buffered solution (http://www.bt.cdc.gov/Agent/Anthrax/environmental-sampling-apr2002.asp).
The detection method used for all fomite studies was cultivation; however, a different agar was used in each study. The focus of the Rose et al. (2004) (59) article was achieving the best recovery, and the study did not determine an environmental limit of detection. From the information given in the article, the environmental limit of detection was calculated by using the initial suspension concentration, the surface area, and the lowest recovery reported. The calculated environmental limit of detection was approximately 20 CFU/cm2.
The recovery efficiencies for all the fomite studies ranged from 10 to 50%, and the extraction efficiencies ranged from 75 to 99%. Recovery of B. anthracis spores from fomites depends on many parameters, such as fomite type, sampling procedure, and sampling processing for detection. The recovery efficiency from the sampling method was primarily the controlling factor in determining the limit of detection and secondarily the efficiency from the extraction method.
Interestingly, in survival studies using cultivation as the detection method on fomites, surface characteristics, relative humidity, and temperature were the most important contributors to viability (62). It was not clear whether recovery and limit of detection changed with time in the environment, as this was difficult to differentiate from survival/degradation of the target. However, this distinction could be made by adding a marker along with the biological agent that does not degrade. For environmental monitoring, the separate time dependence of survival and recovery will be critical to define in future studies. Only the articles from Brown et al. in 2007 (14-16) reported and maintained the relative humidity and temperature in the fomite studies at 30% ± 10% and 25 ± 2°C, respectively. Determining and maintaining the relative humidity and temperature that are most optimal for viability may increase recovery efficiency. In addition, this information could be used at a contaminated site to inform first responders of the possible viability of remaining levels of the biological agent of concern.
Water.
The spores were spiked into a volume of water, filtered through a 0.2- to 0.45-μm-pore-size filter, extracted from the filter, and then detected by various methods. The main challenge for detection of B. anthracis in water was the ability to concentrate the sample. If the sample was too dilute, then the number of B. anthracis cells per liter of water could fall below the environmental limit of detection. When the sample is concentrated, some loss of the initial cells is likely.
There was only one article that evaluated the detection of B. anthracis in water; the lack of articles could be due to this matrix being less likely a vehicle for transmission (62). Perez et al. (2005) (54) spiked B. anthracis spores into tap and source water in volumes ranging from 0.1 to 10 liters. Sample concentrations were detected using sheep red blood cell agar plates, B. anthracis chromogenic agar plates (R&F Laboratories), PCR, or nested PCR.
Cultivation was used to determine the viability of the organisms in the sample, and PCR was used to confirm the identities of any suspect colonies. When using the cultivation approach for the source water samples (Chesapeake Bay and Patuxent River), overgrowth of nontargeted flora occurred in all studies. PCR was only successful for testing source water when the sample concentrations were at least 26 CFU/ml. The environmental limit of detection for tap water was reported as 10 CFU/10 liters using the cultivation methods, while for PCR-based methods, the environmental limit of detection decreased to 534 CFU/liter. Though the PCR-based methods have a rapid detection time compared to that of the cultivation methods (more than 24 h), in this case, PCR was less sensitive. Challenges, such as loss of initial cells, could occur when concentrating large sample volumes (i.e., 10 liters) into 5 to 10 μl for the PCRs.
Quantifying limits of risk estimates.
Five risk scenarios using instrument limits of detection and environmental limits of detection for Cair were evaluated. Log-transformed PCR and real-time PCR instrument limits of detection were normally distributed (Lilliefors test, P of 0.65 for PCR, P of 0.78 for real-time PCR) and were not significantly different (analysis of variance, P of 0.94). Therefore, the PCR and real-time PCR instrument limit of detection distributions were combined to increase the data set for the real-time PCR instrument limit of detection. With the assumption of 100% recovery, the median risk when Cair equaled the median real-time PCR instrument limit of detection was 0.006. When Cair was modeled with a lognormal distribution of real-time PCR instrument limits of detection, the estimated risk was 0.0062. The median risk of death from the inhalation of the entire dose of B. anthracis at the environmental limit of detection in air was 0.22 at the lower reported environmental limit of detection and 0.52 at the upper environmental limit of detection. Assuming that the environmental limit of detection would have a similar distribution as the instrument limit of detection (lognormal) and ranged from 17,000 to 50,000 spores/m3, the median risk of death was 0.32 (Table 3). This assumption should be further evaluated with environmental studies to confirm that the environmental limit of detection would have the same distribution as the instrumental limit of detection. These risk estimates assumed that 100% of the spore sample was inhalable. Risk estimates were also reported for the percentages of 66.5%, 10%, and 1% of spores in the sample that were inhalable or respirable (Table 3). Approximately 70% of inhaled air volume actually contacts alveoli in the lungs, allowing spores to enter the body (71). In addition, the 5th and 95th percentiles of each risk distribution were used to define a 90% confidence interval for each risk estimate (Table 3).
TABLE 3.
Risk estimates using the instrument limit of detection and environmental limit of detection scenarios
| Risk scenario | Analyzed limit of detection | Percentile | Estimates of risk for % of sample inhaled
|
|||
|---|---|---|---|---|---|---|
| 100% | 66.5% | 10% | 1% | |||
| Real-time PCR median instrument limit of detection | 429 cells/ml | 5th | 0.007 | 0.0047 | 0.0007 | 0.00007 |
| Median | 0.006 | 0.0042 | 0.00063 | 0.000063 | ||
| 95th | 0.037 | 0.025 | 0.0038 | 0.00038 | ||
| Real-time PCR instrument limit of detection | 10-34,300 cells/ml | 5th | 0.0001 | 0.000067 | 0.00001 | 0.000001 |
| Median | 0.0062 | 0.0041 | 0.00062 | 0.000062 | ||
| 95th | 0.28 | 0.19 | 0.032 | 0.0032 | ||
| Lower environmental limit of detection in the air | 17,000 CFU/m3 | 5th | 0.026 | 0.017 | 0.0026 | 0.00026 |
| Median | 0.22 | 0.15 | 0.025 | 0.0025 | ||
| 95th | 0.78 | 0.63 | 0.14 | 0.015 | ||
| Upper environmental limit of detection in the air | 50,000 CFU/m3 | 5th | 0.075 | 0.051 | 0.0078 | 0.00078 |
| Median | 0.52 | 0.39 | 0.071 | 0.0073 | ||
| 95th | 0.998 | 0.98 | 0.46 | 0.06 | ||
| Assumed environmental limit of detection in the air | 17,000-50,000 CFU/m3 | 5th | 0.03 | 0.02 | 0.003 | 0.00031 |
| Median | 0.32 | 0.23 | 0.038 | 0.0038 | ||
| 95th | 0.94 | 0.85 | 0.25 | 0.028 | ||
A sensitivity analysis of the risk model was generated by Crystal Ball 7.3.1 (2007; Oracle) for each of the five risk scenarios. For the real-time PCR instrument limit of detection lognormal distribution, the limit of detection (79.4%) was the most sensitive factor in determining risk, followed by the exposure time (11.7%) and breathing rates (8.4%). The dose-response function parameter k (0.5%) had the least impact on the risk estimates. Similarly, for the assumed environmental limit of detection lognormal distribution, the analysis resulted in the exposure time (45%) being the most significant factor, followed by the limit of detection (27.5%), breathing rates (26.1%), and the k parameter (0.9%). The median real-time PCR instrument limit of detection and the two environmental limit of detection (lower and upper) scenarios resulted with the exposure time being the dominant factor in determining risk, followed by breathing rates and the k parameter. Cair values in these scenarios are point estimates rather than a distribution; therefore, the limit of detection was not a measured parameter in the sensitivity analysis.
Even assuming perfect sample collection and processing (no loss in initial concentration), the estimated risk at the instrument limit of detection was far above the commonly used 1:10,000 level. Environmental limits of detection increase due to the imperfect efficiency of sample collection and processing, increasing the risk at these higher detectable concentrations. These risk estimates show that, using current techniques reported in the literature, even allowing for all possible improvements in collection technology, any detectable B. anthracis constitutes an unacceptable risk. Moreover, these estimates define the lowest risk that could be determined from measurement, quantifying the risk that can exist even when no B. anthracis was detected.
Finding significant risk at B. anthracis limits of detection suggests that direct measurement will rarely be adequate for declaring a contaminated site as “clean,” and alternative approaches (e.g., extrapolating from demonstrated log reductions) are needed. For fomites, soil, and water, further work is needed regarding the probability of infection by ingestion and contact before one can adequately address limits of detection and risk estimates. Direct measurement could, at best, reveal a catastrophic failure of decontamination. With respect to preventative monitoring, these estimates showed that significant risk was posed by undetectable concentrations of B. anthracis spores. This means that a low-concentration B. anthracis release would be more likely to be detected by the symptoms in exposed humans rather than by current sampling technology. Where there was danger or suspicion of a B. anthracis release, close monitoring of human health would be needed, in addition to environmental sampling in order to ensure timely medical treatment. Health monitoring alone may be preferred where resources are limited.
The risk assessment approach presented here could be further improved if an experimental probability distribution of the estimated dose was available. However, such a probability distribution was not available even for the most common matrix (soil). To obtain such a distribution, a large number (e.g., 30) of different true doses must be spiked in the environmental matrix of interest, and the sample must be processed through an entire protocol. This time-consuming process has not yet been reported.
Conclusion.
Instrument and environmental limits of detection are necessary for QMRA when evaluating exposure to human pathogens in a contaminated environment. Due to the lack of pertinent data on the detection of B. anthracis, the environmental limit of detection could not be represented as a distribution. These distributions were necessary for estimating the risk at the environmental limit of detection. Even so, it was clear that environmental samples may be expected to have broad distributions due to the many challenges in sample processing that affect the limit of detection. More environmental detection studies need to be conducted in order to produce distributions similar to those of the instrument limit of detection. This will improve the risk assessment and improve the applicability of the information in regard to survival and cleanup goals, providing valuable information for first responders.
Supplementary Material
Acknowledgments
This research has been supported by the Center for Advancing Microbial Risk Assessment, funded by the U.S. Environmental Protection Agency Science to Achieve Results (STAR) program, and U.S. Department of Homeland Security University Programs grant R83236201.
Footnotes
Published ahead of print on 31 July 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Acharya, G., D. D. Doorneweerd, C. L. Chang, W. A. Henne, P. S. Low, and C. A. Savran. 2007. Label-free optical detection of anthrax-causing spores. J. Am. Chem. Soc. 129:732-733. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal, G. S., D. V. Kamboj, S. I. Alam, M. Dixit, and L. Singh. 2002. Environmental detection of Bacillus anthracis spores. Curr. Sci. 83:697-699. [Google Scholar]
- 3.Baeumner, A. J., J. Pretz, and S. Fang. 2004. A universal nucleic acid sequence biosensor with nanomolar detection limits. Anal. Chem. 76:888-894. [DOI] [PubMed] [Google Scholar]
- 4.Bartrand, T. A., M. H. Weir, and C. N. Haas. 2008. Dose-response models for inhalation of Bacillus anthracis spores: interspecies comparisons. Risk Anal. 28:1115-1124. [DOI] [PubMed] [Google Scholar]
- 5.Belgrader, P., W. Benett, D. Hadley, G. Long, R. Mariella, F. Milanovich, S. Nasarabadi, W. Nelson, J. Richards, and P. Stratton. 1998. Rapid pathogen detection using a microchip PCR array instrument. Clin. Chem. 44:2191-2194. [PubMed] [Google Scholar]
- 6.Belgrader, P., C. J. Elkin, S. B. Brown, S. N. Nasarabadi, R. G. Langlois, F. P. Milanovich, B. W. Colston, and G. D. Marshall. 2003. A reusable flow-through polymerase chain reaction instrument for the continuous monitoring of infectious biological agents. Anal. Chem. 75:3446-3450. [DOI] [PubMed] [Google Scholar]
- 7.Bell, C. A., J. R. Uhl, T. L. Hadfield, J. C. David, R. F. Meyer, T. F. Smith, and F. R. Cockerill. 2002. Detection of Bacillus anthracis DNA by LightCycler PCR. J. Clin. Microbiol. 40:2897-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beverly, M. B., F. Basile, K. J. Voorhees, and T. L. Hadfield. 1996. A rapid approach for the detection of dipicolinic acid in bacterial spores using pyrolysis mass spectrometry. Rapid Commun. Mass Spectrom. 10:455-458. [DOI] [PubMed] [Google Scholar]
- 9.Beverly, M. B., K. J. Voorhees, and T. L. Hadfield. 1999. Direct mass spectrometric analysis of Bacillus spores. Rapid Commun. Mass Spectrom. 13:2320-2326. [DOI] [PubMed] [Google Scholar]
- 10.Beyer, W., S. Pocivalsek, and R. Bohm. 1999. Polymerase chain reaction-ELISA to detect Bacillus anthracis from soil samples—limitations of present published primers. J. Appl. Microbiol. 87:229-236. [DOI] [PubMed] [Google Scholar]
- 11.Bode, E., W. Hurtle, and D. Norwood. 2004. Real-time PCR assay for a unique chromosomal sequence of Bacillus anthracis. J. Clin. Microbiol. 42:5825-5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borthwick, K. A. J., T. E. Love, M. B. McDonnell, and W. T. Coakley. 2005. Improvement of immunodetection of bacterial spore antigen by ultrasonic cavitation. Anal. Chem. 77:7242-7245. [DOI] [PubMed] [Google Scholar]
- 13.Brightwell, C., M. Pearce, and D. Leslie. 1998. Development of internal controls for PCR detection of Bacillus anthracis. Mol. Cell. Probes 12:367-377. [DOI] [PubMed] [Google Scholar]
- 14.Brown, G. S., R. G. Betty, J. E. Brockmann, D. A. Lucero, C. A. Souza, K. S. Walsh, R. M. Boucher, M. Tezak, and M. C. Wilson. 2007. Evaluation of vacuum filter sock surface sample collection method for Bacillus spores from porous and non-porous surfaces. J. Environ. Monit. 9:666-671. [DOI] [PubMed] [Google Scholar]
- 15.Brown, G. S., R. G. Betty, J. E. Brockmann, D. A. Lucero, C. A. Souza, K. S. Walsh, R. M. Boucher, M. Tezak, M. C. Wilson, and T. Rudolph. 2007. Evaluation of a wipe surface sample method for collection of Bacillus spores from nonporous surfaces. Appl. Environ. Microbiol. 73:706-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown, G. S., R. G. Betty, J. E. Brockmann, D. A. Lucero, C. A. Souza, K. S. Walsh, R. M. Boucher, M. Tezak, M. C. Wilson, T. Rudolph, H. D. A. Lindquist, and K. F. Martinez. 2007. Evaluation of rayon swab surface sample collection method for Bacillus spores from nonporous surfaces. J. Appl. Microbiol. 103:1074-1080. [DOI] [PubMed] [Google Scholar]
- 17.Bruno, J. G., and J. L. Kiel. 1999. In vitro selection of DNA aptamers to anthrax spores with electrochemiluminescence detection. Biosens. Bioelectron. 14:457-464. [DOI] [PubMed] [Google Scholar]
- 18.Bruno, J. G., and H. Yu. 1996. Immunomagnetic-electrochemiluminescent detection of Bacillus anthracis spores in soil matrices. Appl. Environ. Microbiol. 62:3474-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burton, J. E., J. Oshota, E. North, M. J. Hudson, N. Polyanskaya, J. Brehm, G. Lloyd, and N. J. Silman. 2005. Development of a multipathogen oligonucleotide microarray for detection of Bacillus anthracis. Mol. Cell. Probes 19:349-357. [DOI] [PubMed] [Google Scholar]
- 20.Buttner, M. P., P. Cruz, L. D. Stetzenbach, A. K. Klima-Comba, V. L. Stevens, and P. A. Emanuel. 2004. Evaluation of the biological sampling kit (BiSKit) for large-area surface sampling. Appl. Environ. Microbiol. 70:7040-7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campbell, G. A., and R. Mutharasan. 2007. Method of measuring Bacillus anthracis spores in the presence of copious amounts of Bacillus thuringiensis and Bacillus cereus. Anal. Chem. 79:1145-1152. [DOI] [PubMed] [Google Scholar]
- 22.Campbell, G. A., and R. Mutharasan. 2006. Piezoelectric-excited millimeter-sized cantilever (PEMC) sensors detect Bacillus anthracis at 300 spores/mL. Biosens. Bioelectron. 21:1684-1692. [DOI] [PubMed] [Google Scholar]
- 23.Charrel, R. N., B. La Scola, and D. Raoult. 2004. Multi-pathogens sequence containing plasmids as positive controls for universal detection of potential agents of bioterrorism. BMC Microbiol. 4:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheun, H. I., S. I. Makino, M. Watarai, J. Erdenebaatar, K. Kawamoto, and I. Uchida. 2003. Rapid and effective detection of anthrax spores in soil by PCR. J. Appl. Microbiol. 95:728-733. [DOI] [PubMed] [Google Scholar]
- 25.Christensen, D. R., L. J. Hartman, B. M. Loveless, M. S. Frye, M. A. Shipley, D. L. Bridge, M. J. Richards, R. S. Kaplan, J. Garrison, C. D. Baldwin, D. A. Kulesh, and D. A. Norwood. 2006. Detection of biological threat agents by real-time PCR: comparison of assay performance on the RAPID, the LightCycler, and the smart cycler platforms. Clin. Chem. 52:141-145. [DOI] [PubMed] [Google Scholar]
- 26.Dang, J. L., K. Heroux, J. Kearney, A. Arasteh, M. Gostomski, and P. A. Emanuel. 2001. Bacillus spore inactivation methods affect detection assays. Appl. Environ. Microbiol. 67:3665-3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellerbrok, H., H. Nattermann, M. Ozel, L. Beutin, B. Appel, and G. Pauli. 2002. Rapid and sensitive identification of pathogenic and apathogenic Bacillus anthracis by real-time PCR. FEMS Microbiol. Lett. 214:51-59. [DOI] [PubMed] [Google Scholar]
- 28.English, R. D., B. Warscheid, C. Fenselau, and R. J. Cotter. 2003. Bacillus spore identification via proteolytic peptide mapping with a miniaturized MALDI TOF mass spectrometer. Anal. Chem. 75:6886-6893. [DOI] [PubMed] [Google Scholar]
- 29.Farrell, S., H. B. Halsall, and W. R. Heineman. 2005. Bacillus globigii bugbeads: a model simulant of a bacterial spore. Anal. Chem. 77:549-555. [DOI] [PubMed] [Google Scholar]
- 30.Farrell, S., H. B. Halsall, and W. R. Heineman. 2005. Immunoassay for B-globigii spores as a model for detecting B-anthracis spores in finished water. Analyst 130:489-497. [DOI] [PubMed] [Google Scholar]
- 31.Fasanella, A., S. Losito, R. Adone, F. Ciuchini, T. Trotta, S. A. Altamura, D. Chiocco, and G. Ippolito. 2003. PCR assay to detect Bacillus anthracis spores in heat-treated specimens. J. Clin. Microbiol. 41:896-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Floriano, P. N., N. Christodoulides, D. Romanovicz, B. Bernard, G. W. Simmons, M. Cavell, and J. T. McDevitt. 2005. Membrane-based on-line optical analysis system for rapid detection of bacteria and spores. Biosens. Bioelectron. 20:2079-2088. [DOI] [PubMed] [Google Scholar]
- 33.Fujinami, Y., Y. Hirai, I. Sakai, M. Yoshino, and J. Yasuda. 2007. Sensitive detection of Bacillus anthracis using a binding protein originating from gamma-phage. Microbiol. Immunol. 51:163-169. [DOI] [PubMed] [Google Scholar]
- 34.Gatto-Menking, D. L., H. Yu, J. G. Bruno, M. T. Goode, M. Miller, and A. W. Zulich. 1995. Sensitive detection of biotoxoids and bacterial spores using an immunomagnetic electrochemiluminescence sensor. Biosens. Bioelectron. 10:501-507. [DOI] [PubMed] [Google Scholar]
- 35.Haas, C. N., J. B. Rose, and C. P. Gerba. 1999. Quantitative microbial risk assessment. John Wiley & Sons Inc., New York, NY.
- 36.Hartley, H. A., and A. J. Baeumner. 2003. Biosensor for the specific detection of a single viable B-anthracis spore. Anal. Bioanal. Chem. 376:319-327. [DOI] [PubMed] [Google Scholar]
- 37.Haynes, C. L., C. R. Yonzon, X. Y. Zhang, and R. P. Van Duyne. 2005. Surface-enhanced Raman sensors: early history and the development of sensors for quantitative biowarfare agent and glucose detection. J. Raman Spectrosc. 36:471-484. [Google Scholar]
- 38.Hodges, L. R., L. J. Rose, A. Peterson, J. Noble-Wang, and M. J. Arduino. 2006. Evaluation of a macrofoam swab protocol for the recovery of Bacillus anthracis spores from a steel surface. Appl. Environ. Microbiol. 72:4429-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoffmaster, A. R., R. F. Meyer, M. P. Bowen, C. K. Marston, R. S. Weyant, K. Thurman, S. L. Messenger, E. E. Minor, J. M. Winchell, M. V. Rassmussen, B. R. Newton, J. T. Parker, W. E. Morrill, N. McKinney, G. A. Barnett, J. J. Sejvar, J. A. Jernigan, B. A. Perkins, and T. Popovic. 2002. Evaluation and validation of a real-time polymerase chain reaction assay for rapid identification of Bacillus anthracis. Emerg. Infect. Dis. 8:1178-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoile, R., M. Yuen, G. James, and G. L. Gilbert. 2007. Evaluation of the rapid analyte measurement platform (RAMP) for the detection of Bacillus anthracis at a crime scene. Forensic Sci. Int. 171:1-4. [DOI] [PubMed] [Google Scholar]
- 41.Hurtle, W., E. Bode, D. A. Kulesh, R. S. Kaplan, J. Garrison, D. Bridge, M. House, M. S. Frye, B. Loveless, and D. Norwood. 2004. Detection of the Bacillus anthracis gyrA gene by using a minor groove binder probe. J. Clin. Microbiol. 42:179-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim, K., J. Seo, K. Wheeler, C. Park, D. Kim, S. Park, W. Kim, S. I. Chung, and T. Leighton. 2005. Rapid genotypic detection of Bacillus anthracis and the Bacillus cereus group by multiplex real-time PCR melting curve analysis. FEMS Immunol. Med. Microbiol. 43:301-310. [DOI] [PubMed] [Google Scholar]
- 43.Krebs, M. D., A. M. Zapata, E. G. Nazarov, R. A. Miller, I. S. Costa, A. L. Sonenshein, and C. E. Davis. 2005. Detection of biological and chemical agents using differential mobility spectrometry (DMS) technology. IEEE Sens. J. 5:696-703. [Google Scholar]
- 44.Kuske, C. R., K. L. Banton, D. L. Adorada, P. C. Stark, K. K. Hill, and P. J. Jackson. 1998. Small-scale DNA sample preparation method for field PCR detection of microbial cells and spores in soil. Appl. Environ. Microbiol. 64:2463-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee, M. A., G. Brightwell, D. Leslie, H. Bird, and A. Hamilton. 1999. Fluorescent detection techniques for real-time multiplex strand specific detection of Bacillus anthracis using rapid PCR. J. Appl. Microbiol. 87:218-223. [DOI] [PubMed] [Google Scholar]
- 46.Lee, S. H., D. D. Stubbs, J. Cairney, and W. D. Hunt. 2005. Rapid detection of bacterial spores using a quartz crystal microbalance (QCM) immunoassay. IEEE Sens. J. 5:737-743. [Google Scholar]
- 47.Lester, E. D., G. Bearman, and A. Ponce. 2004. A second-generation anthrax “smoke detector.” IEEE Eng. Med. Biol. Mag. 23:130-135. [DOI] [PubMed] [Google Scholar]
- 48.Merrill, L., J. Richardson, C. R. Kuske, and J. Dunbar. 2003. Fluorescent heteroduplex assay for monitoring Bacillus anthracis and close relatives in environmental samples. Appl. Environ. Microbiol. 69:3317-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moser, M. J., D. R. Christensen, D. Norwood, and J. R. Prudent. 2006. Multiplexed detection of anthrax-related toxin genes. J. Mol. Diagn. 8:89-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nubel, U., P. M. Schmidt, E. Reiss, F. Bier, W. Beyer, and D. Naumann. 2004. Oligonucleotide microarray for identification of Bacillus anthracis based on intergenic transcribed spacers in ribosomal DNA. FEMS Microbiol. Lett. 240:215-223. [DOI] [PubMed] [Google Scholar]
- 51.Pai, S., A. D. Ellington, and M. Levy. 2005. Proximity ligation assays with peptide conjugate ‘burrs’ for the sensitive detection of spores. Nucleic Acids Res. 33:e162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pal, S., E. C. Alocilja, and F. P. Downes. 2007. Nanowire labeled direct-charge transfer biosensor for detecting Bacillus species. Biosens. Bioelectron. 22:2329-2336. [DOI] [PubMed] [Google Scholar]
- 53.Patra, G., L. E. Williams, Y. Qi, S. Rose, R. Redkar, and V. G. DelVecchio. 2002. Rapid genotyping of Bacillus anthracis strains by real-time polymerase chain reaction. Ann. N. Y. Acad. Sci. 969:106-111. [DOI] [PubMed] [Google Scholar]
- 54.Perez, A., C. Hohn, and J. Higgins. 2005. Filtration methods for recovery of Bacillus anthracis spores spiked into source and finished water. Water Res. 39:5199-5211. [DOI] [PubMed] [Google Scholar]
- 55.Premasiri, W. R., D. T. Moir, M. S. Klempner, N. Krieger, G. Jones, and L. D. Ziegler. 2005. Characterization of the surface enhanced Raman scattering (SERS) of bacteria. J. Phys. Chem. B 109:312-320. [DOI] [PubMed] [Google Scholar]
- 56.Qi, Y. A., G. Patra, X. D. Liang, L. E. Williams, S. Rose, R. J. Redkar, and V. G. DelVecchio. 2001. Utilization of the rpoB gene as a specific chromosomal marker for real-time PCR detection of Bacillus anthracis. Appl. Environ. Microbiol. 67:3720-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reif, T. C., M. Johns, S. D. Pillai, and M. Carl. 1994. Identification of capsule-forming Bacillus anthracis spores with the PCR and a novel dual-probe hybridization format. Appl. Environ. Microbiol. 60:1622-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reiman, R. W., D. H. Atchley, and K. J. Voorhees. 2007. Indirect detection of Bacillus anthracis using real-time PCR to detect amplified gamma phage DNA. J. Microbiol. Methods 68:651-653. [DOI] [PubMed] [Google Scholar]
- 59.Rose, L., B. Jensen, A. Peterson, S. N. Banerjee, and M. J. Arduino. 2004. Swab materials and Bacilius anthracis spore recovery from nonporous surfaces. Emerg. Infect. Dis. 10:1023-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ryu, C., K. Lee, C. Yoo, W. K. Seong, and H. B. Oh. 2003. Sensitive and rapid quantitative detection of anthrax spores isolated from soil samples by real-time PCR. Microbiol. Immunol. 47:693-699. [DOI] [PubMed] [Google Scholar]
- 61.Sanderson, W. T., M. J. Hein, L. Taylor, B. D. Curwin, G. M. Kinnes, T. A. Seitz, T. Popovic, H. T. Holmes, M. E. Kellum, S. K. McAllister, D. N. Whaley, E. A. Tupin, T. Walker, J. A. Freed, D. S. Small, B. Klusaritz, and J. H. Bridges. 2002. Surface sampling methods for Bacillus anthracis spore contamination. Emerg. Infect. Dis. 8:1145-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sinclair, R., S. A. Boone, D. Greenberg, P. Keim, and C. P. Gerba. 2008. Persistence of category A select agents in the environment. Appl. Environ. Microbiol. 74:555-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sjostedt, A., U. Eriksson, V. Ramisse, and H. Garrigue. 1997. Detection of Bacillus anthracis spores in soil by PCR. FEMS Microbiol. Ecol. 23:159-168. [Google Scholar]
- 64.Song, L. N., S. Ahn, and D. R. Walt. 2005. Detecting biological warfare agents. Emerg. Infect. Dis. 11:1629-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stopa, P. J. 2000. The flow cytometry of Bacillus anthracis spores revisited. Cytometry 41:237-244. [DOI] [PubMed] [Google Scholar]
- 66.Stratis-Cullum, D. N., G. D. Griffin, J. Mobley, A. A. Vass, and T. Vo-Dinh. 2003. A miniature biochip system for detection of aerosolized Bacillus globigii spores. Anal. Chem. 75:275-280. [DOI] [PubMed] [Google Scholar]
- 67.Subcommittee on Homeland Security, Committee on Appropriations, House of Representatives. 2007. Anthrax detection: DHS cannot ensure that sampling activities will be validated. Statement of Keith Rhodes, Chief Technologist, Center for Technology and Engineering Applied Research and Methods, United States Government Accountability Office (GAO-O-070687T). U.S. Government Accountability Office, Washington, DC.
- 68.Taitt, C. R., G. P. Anderson, B. M. Lingerfelt, M. J. Feldstein, and F. S. Ligler. 2002. Nine-analyte detection using an array-based biosensor. Anal. Chem. 74:6114-6120. [DOI] [PubMed] [Google Scholar]
- 69.Tims, T. B., and D. V. Lim. 2004. Rapid detection of Bacillus anthracis spores directly from powders with an evanescent wave fiber-optic biosensor. J. Microbiol. Methods 59:127-130. [DOI] [PubMed] [Google Scholar]
- 70.Ulrich, M. P., D. R. Christensen, S. R. Coyne, P. D. Craw, E. A. Henchal, S. H. Sakai, D. Swenson, J. Tholath, J. Tsai, A. F. Weir, and D. A. Norwood. 2006. Evaluation of the Cepheid GeneXpert system for detecting Bacillus anthracis. J. Appl. Microbiol. 100:1011-1016. [DOI] [PubMed] [Google Scholar]
- 71.U.S. Environmental Protection Agency (EPA). 1997. Exposure factor handbook. National Center for Environmental Assessment, Washington, DC.
- 72.Van Ert, M. N., W. R. Easterday, T. S. Simonson, J. M. U'Ren, T. Pearson, L. J. Kenefic, J. D. Busch, L. Y. Huynh, M. Dukerich, C. B. Trim, J. Beaudry, A. Welty-Bernard, T. Read, C. M. Fraser, J. Ravel, and P. Keim. 2007. Strain-specific single-nucleotide polymorphism assays for the Bacillus anthracis Ames strain. J. Clin. Microbiol. 45:47-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wan, J. H., B. Fiebor, B. A. Chin, I. H. Chen, J. Brigati, and V. A. Petrenko. 2005. Landscape phage-based magnetostrictive biosensor for detecting Bacillus anthracis spores, p. 1308-1311. In Sensors, IEEE 2005. IEEE, Los Alamitos, CA.
- 74.Wan, J. H., H. H. Shu, S. C. Huang, B. Fiebor, I. H. Chen, V. A. Petrenko, and B. A. Chin. 2007. Phage-based magnetoelastic wireless biosensors for detecting Bacillus anthracis spores. IEEE Sens. J. 7:470-477. [Google Scholar]
- 75.Wang, S. H., J. K. Wen, Y. F. Zhou, Z. P. Zhang, R. F. Yang, J. B. Zhang, J. Chen, and X. E. Zhang. 2004. Identification and characterization of Bacillus anthracis by multiplex PCR on DNA chip. Biosens. Bioelectron 20:807-813. [DOI] [PubMed] [Google Scholar]
- 76.Wilson, W. J., A. M. Erler, S. L. Nasarabadi, E. W. Skowronski, and P. M. Imbro. 2005. A multiplexed PCR-coupled liquid bead array for the simultaneous detection of four biothreat agents. Mol. Cell. Probes 19:137-144. [DOI] [PubMed] [Google Scholar]
- 77.Yu, H., J. W. Raymonda, T. M. McMahon, and A. A. Campagnari. 2000. Detection of biological threat agents by immunomagnetic microsphere-based solid phase fluorogenicand electro-chemiluminescence. Biosens. Bioelectron. 14:829-840. [DOI] [PubMed] [Google Scholar]
- 78.Zhang, X. Y., M. A. Young, O. Lyandres, and R. P. Van Duyne. 2005. Rapid detection of an anthrax biomarker by surface-enhanced Raman spectroscopy. J. Am. Chem. Soc. 127:4484-4489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.