Abstract
Papillomavirus capsids are composed of 72 pentamers reinforced through inter- and intrapentameric disulfide bonds. Recent research suggests that virus-like particles and pseudovirions (PsV) can undergo a redox-dependent conformational change involving disulfide interactions. We present here evidence that native virions exploit a tissue-spanning redox gradient that facilitates assembly events in the context of the complete papillomavirus life cycle. DNA encapsidation and infectivity titers are redox dependent in that they can be temporally modulated via treatment of organotypic cultures with oxidized glutathione. These data provide evidence that papillomavirus assembly and maturation is redox-dependent, utilizing multiple steps within both suprabasal and cornified layers.
Human papillomaviruses (HPVs) exclusively infect cutaneous or mucosal epithelial tissues (14, 15, 30). HPV types that infect the mucosal epithelia can lead to the development of benign or malignant neoplasms, thus allowing for their categorization into low-risk or high-risk HPV types, respectively (14, 15, 30). A small subset of the more than 200 HPV types now identified are the causative agents of over 75% of all cervical cancers. HPV16 is the most prevalent type worldwide, found in ca. 50 to 62% of squamous cell carcinomas (14, 50).
HPV16 virions contain a single, circular double-stranded DNA genome of ∼8 kb which associates with histones to form a chromatin-like structure. This minichromosome is packaged within a nonenveloped, icosahedral capsid composed of the major capsid protein L1 and the minor capsid protein L2. Similar to polyomaviruses, 72 capsomeres of L1 are geometrically arranged on a T=7 icosahedral lattice (2, 9, 17, 19, 36, 42). Recent cryoelectron microscopy images of HPV16 pseudovirions (PsV) suggest that L2 is arranged near the inner conical hollow of each L1 pentamer, although it is not known whether each L1 pentamer is occupied with a single L2 protein (5, 42).
Due to technical constraints in the production of native HPV virions in organotypic culture, assembly studies of HPV particles have largely been restricted to the utilization of in vitro-derived particles such as virus-like particles (VLPs), PsV, and quasivirions (QV) (6, 12, 25, 40, 43). Recent research suggests that HPV and bovine papillomavirus PsV can undergo a redox-dependent conformational change that takes place over the course of many hours. This conformational change is characterized by resistance to proteolysis and chemical reduction and the appearance of a more orderly capsid structure via transmission electron microscopy (TEM) (7, 20).
We present evidence that native virions, in the context of the complete papillomavirus life cycle, utilize a tissue-spanning redox gradient that facilitates multiple redox-dependent assembly and maturation events over the course of many days. We show that stability and specific infectivity of 20-day virions increases over 10-day virions, 20-day virions are more susceptible to neutralization than 10-day virions, and both viral DNA encapsidation and infectivity of HPV-infected tissues are redox dependent in that they can be manipulated via the treatment of organotypic tissues with oxidized glutathione (GSSG), which is concentration and temporally dependent.
MATERIALS AND METHODS
Mutagenesis.
pBSHPV16(114/B) DNA, a generous gift from M. Dürst, was utilized as the template for site-directed mutagenesis using a QuikChange II XL site-directed mutagenesis kit (Stratagene) (26). To create a full-length, HPV16(114/B) genome with a Cys428Ser substitution, the two complementary oligonucleotides—forward (5′-GTAACCCAGGCAATTGCTTCTCAAAAACATACACCTCC-3′) and reverse (5′-GGAGGTGTATGTTTTTGAGAAGCAATTGCCTGGGTTAC-3′)—were used with the change of G to C at nucleotide 6917. Multiple mutant viral genomic clones containing correctly mutagenized sequences were isolated per mutation and utilized in subsequent experimentation.
Keratinocyte cultures, and electroporation.
Primary human foreskin keratinocytes (HFKs) were isolated and grown from newborn circumcision as described previously (33). For electroporations, 30 μg of wild-type or Cys428Ser pBSHPV16(114/B) DNA was digested with BamHI, linearizing the viral DNA at nucleotide 6151 in L1 and separating it from the vector sequence. HFKs were electroporated with the prepared DNA as described previously (33, 34). Multiple cell lines were obtained for each wild-type and mutant construct.
Southern blot hybridization.
Total cellular DNA was isolated as previously described (34). Briefly, 5 μg of total cellular DNA was digested with BamHI, which linearizes the HPV16 genome. Samples were separated by 0.8% agarose gel electrophoresis and transferred onto a GeneScreen Plus membrane (New England Nuclear Research Products) as previously described (41). Hybridization of the membrane utilized an HPV16-specific, whole-genomic probe as previously described (34).
Organotypic “raft” culture-derived native virion production.
Immortalized HFK lines which stably maintained episomal HPV16 DNA were grown in monolayer culture using E medium in the presence of mitomycin C-treated J2 3T3 feeder cells (34). Raft tissues were grown as previously described (32-34). For GSSG (Sigma) treatment, E-medium was supplemented with the appropriate concentration and fed to organotypic cultures either beginning on day 8, followed by tissue harvest on day 10 (8 to 10 days), or beginning on day 15, followed by tissue harvest on day 20 (15 to 20 days).
293TT cell-based QV production.
HPV16 QV were generated in 293TT cells as previously described (12, 38, 39). Briefly, 293TT cells, plated to 90% confluence in T-150 flasks were cotransfected by using Lipofectamine 2000 with 25 mg of each p16L1h (L1 expression plasmid), p16L2h (L2 expression plasmid), and a cloned HPV16 genome that had been excised from pBS-HPV16 (114/B) (7). Cells were split 1:2 at 24 h posttransfection and harvested 48 h posttransfection. Cell pellets were resuspended in a total of 750 μl of phosphate-buffered saline (PBS). Cells were lysed by Dounce homogenization as in the HPV isolation step described below. MgCl2 was spiked into the lysate at a final concentration of 2 mM. Lysates were then incubated overnight at 37°C for maturation (7). Unpackaged DNA was digested by adding 0.2% benzonase (Sigma) and incubated for 1 h at 37 C°. After digestion, NaCl was spiked into the lysate to a final concentration of 1 M. Cellular debris was removed, and virus-containing supernatant was collected postcentrifugation at 10,500 rpm.
Identification of free sulfhydryls and disulfide bonds.
Frozen sections of HPV16(114/B)-infected HFKs were stained with DACM (N-[7-dimethylamino-4-methyl-3-coumaring]-maleimide) to detect free sulfhydryls and disulfide bond-specific sulfyhydryls as previously described (38). A Nikon fluorescence microscope equipped with an UV filter was used to detect the fluorescence of DACM.
Histology and immunohistochemical staining.
Tissues were fixed in 10% neutral buffered formalin and embedded in paraffin. Sections (4 μm) were cut and stained with hematoxylin and eosin (H&E) as previously described (34). Sections were stained with an anti-HPV16 L1 monoclonal antibody, Camvir-1 (BD Pharmingen), diluted 1:5,000 in PBS. Immunostaining was done by using a Vectastain Elite ABC kit (Vector Laboratories, Burlingame, CA) according to the manufacturer's instructions.
HPV isolation.
For Optiprep (Sigma-Aldrich) fractionation, reverse transcription-PCR (RT-PCR), quantitative RT-PVR (RT-qPCR), and qPCR-based DNA encapsidation assays, three-raft crude viral preps (CVPs) or 293TT cell viral lysates were prepared by Dounce homogenization in 500 μl of phosphate buffer (0.05 M sodium phosphate [pH 8.0], 2 mM MgCl2). Homogenizers were rinsed with 250 μl of phosphate buffer. Then, 1.5 μl (375 U) of benzonase was added to 750 μl of CVPs or 293TT cell viral lysates, followed by incubation at 37°C for 1 h. Samples were brought to 1 M NaCl by adding 130 μl of ice-cold 5 M NaCl. Then, samples were vortex mixed and centrifuged at 4°C for 10 min at 10,500 rpm in a microcentrifuge. The supernatants were stored at −20°C.
Optiprep purification of virions.
Optiprep purification was performed as described previously (6, 18). Briefly, 27, 33, and 39% Optiprep gradients were produced by underlayering. Gradients were allowed to diffuse for 1 to 2 h at room temperature. Then, 600 μl of clarified benzonase-treated virus preps were layered on top of the gradient. Tubes were then centrifuged in a SW55 rotor (Beckman) at 234,000 × g for 3.5 h at 16°C. After centrifugation, 11 500-μl fractions were carefully collected from the top of each tube.
qPCR-based DNA encapsidation assay.
To detect endonuclease-resistant genomes in CVPs or Optiprep fractions, only benzonase-treated CVPs were utilized so that all nonencapsidated genomes were digested. For additional DNase I-treatments of Optiprep fractions, 2-μl aliquots from Optiprep-fractionated 10-day CVP fractions 2 and 7 were digested in a total reaction volume of 50 μl containing 4 U of DNase I (NEB), and 5 μl of 10× DNase I reaction buffer. Reactions were incubated for 1 h at 37°C and then heat inactivated for 10 min at 75°C. To release all encapsidated viral genomes, 10 μl of sonicated virus prep or 20 μl of Optiprep fraction was added to 2 μl of proteinase K, 10 μl of 10% sodium dodecyl sulfate, 2 μl of pCMV-GFP (140 ng/μl) carrier DNA, and brought up to 200 μl with Hirt buffer. To harvest cellular DNA, reactions were passed through an 18-gauge needle 10 times prior to the treatment above. Tubes were rotated at 37°C for 2 h. Immediately, an equal amount of phenol-chloroform-isoamyl alcohol (25:24:1) was added, and the mixture was extracted for the aqueous phase. An equal amount of chloroform was added and again extracted for the aqueous phase. DNA was ethanol precipitated overnight at −20°C. After centrifugation, the DNA pellet was washed with 70% ethanol and resuspended in 20 μl of Tris-EDTA overnight. To detect viral genomes or cellular DNA, a Qiagen Quantitect SYBR green PCR kit was utilized. Amplification of the viral target was performed in 0.2 ml, 96-well PCR plates (Bio-Rad) with a total reaction volume of 25 μl. Then, l μl of each endonuclease-resistant viral genome prep was analyzed in triplicate for each independent experiment. Amplification of HPV16 genomes was performed using 0.3 μM 5′-CCATATAGACTATTGGAAACACATGCGCC-3′ as the forward primer (nucleotides [nt] 2839 to 2868) and 0.3 μM 5′-CGTTAGTTGCAGTTCAATTGCTTGTAATGC-3′ as the reverse primer (nt 2960 to 2989). Amplification of human repetitive Alu DNA was performed using 0.3 μM 5′-GTCAGGAGATCGAGACCATCCC-3′ as the forward primer and 0.3 μM 5′-TCCTGCCTCAGCCTCCCAAG-3′ as the reverse primer (37). Oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA). A standard curve was generated by amplifying 1-μl aliquots of 104, 103, 102, and 101 serially diluted pBSHPV16 copy number controls. Acceptable R2 values for standard curves were at or above 0.99. A Bio-Rad iQ5 Multicolor Real-Time qPCR machine and software were utilized for PCR amplifications and subsequent data analysis. The PCR thermocycling profile was as follows: a 15-min hot-start at 95°C, followed by 40 cycles at 15 s at 94°C, 30 s at 52°C, and 30 s at 72°C. The data analysis commenced during the extension phase.
Endpoint RT-PCR infectivity assays.
Nested RT-PCR-based infectivity assays were performed as described previously (31, 33, 35).
RT-qPCR infectivity assays.
HaCaT cells were grown to confluence in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, 2 mM glutamine, 1 mM pyruvate, 100 U of penicillin/ml, and 100 μg of streptomycin/ml and seeded 50,000 cells/well in 24-well plates. HPV16 CVPs were diluted with cell culture medium to a total volume of 0.5 ml. Medium was aspirated from HaCaT cells, and 0.5 ml of diluted CVPs was added per well. As per our DNA encapsidation assay, 30,000 viral genome equivalents (vge) per cell was utilized for infections. For antibody-mediated neutralizations, 30,000 vge of CVP was incubated for 1 h at 37°C prior to infection with a 1:100 dilution of the anti-L1 antibody H16.7E and the anti-L2 antibody RG-1. One well on each plate received 0.5 ml of medium without virus as a negative control. The cells were incubated with the virus for 48 h at 37°C. mRNA was harvested with the SurePrep TrueTotal RNA purification kit (Fisher Scientific). DNA contamination of columns was insignificant in that the optional on-column DNase-I treatment of extracted mRNA had no effect on downstream signal. Amplification of both the viral target and endogenous cellular control target was performed by using a duplex format in 0.2 ml, 96-well PCR plates (Bio-Rad) with a total reaction volume of 25 μl. All reactions containing RNAs from virus-infected cells were performed in duplicate or triplicate. RT-qPCR were performed in the same closed tube with ∼250 ng of total RNA per reaction using the Quantitect probe RT-PCR kit (Qiagen). The HPV16 E1^E4 primers used were the splice-site straddling forward (5′-GCTGATCCTGCAAGCAACGAAGTATC-3′; nt 868 to 3372) and reverse (5′-GGATTGGAGCACTGTCCACTGAG-3′; nt 3535 to 3557) primers at final concentrations of 4 μM. A fluorogenic, dual-labeled, HPV16 E1Ê4 probe (5′-6-FAMCACCGGAAACCCCTGCCACACCACTAAGBHQ-1-3′; nt 3493 to 3520) was utilized at a final concentration of 0.2 μM to detect E1Ê4 cDNA. The HPV18 E1Ê4 primers used were the splice-site straddling forward (5′-GGCTGATCCAGAAACCAGTGAC-3′; nt 916 to 3443) and reverse (5′-CTGGCCGTAGGTCTTTGCGGTG-3′; nt 3522 to 3542) primers at final concentrations of 4 μM. A fluorogenic, dual-labeled, HPV18 E1Ê4 probe (5′- 6-FAM-CCTCACCGTATTCCAGCACCGTGTCCGTGBHQ-1-3′; nt 3490 to 3518) was utilized at a final concentration of 0.2 μM to detect E1Ê4 cDNA. Primers and probes were developed by using the GeneLink Software OligoAnalyzer 1.2 and OligoExplorer 1.2. TATA-binding protein (TBP) amplicons were created using the primers 5′-CACGGCACTGATTTTCAGTTCT-3′ (nt 627 to 648) and 5′-TTCTTGCTGCCAGTCTGGACT-3′ (nt 706 to 686) at final concentrations of 0.125 μM. TBP amplicons were detected by using a fluorogenic TaqMan probe (5′-HEX-TGTGCACAGGAGCCAAGAGTGAAGA-BHQ-1-3′) at 0.2 μM. TBP primer sequences were obtained from those previously described (11). All primers were synthesized by Integrated DNA Technologies (Coralville, IA). All RT-qPCRs were performed using the iQ5 (Bio-Rad). Cycling conditions were 50°C for 30 min (RT) and 95°C for 15 min, followed by 42 cycles of 94°C for 15 s and 54.5°C for 1 min. Amplification efficiencies of each primer set was 93% for E1Ê4 and 97% for TBP. The relative quantities of viral target cDNA were determined by using REST software.
Immunoblot analysis.
Aliquots (40 μl) from Optiprep fractions were boiled for 10 min in 6% 2-mercaptoethanol (2-ME) loading buffer and loaded onto 8 to 10% polyacrylamide gels. To detect HPV16 L1, anti-HPV16 L1 monoclonal antibody (Camvir-1; BD Pharmingen) was utilized at a dilution of 1:4,000 according to the manufacturer's recommendations. To detect cellular keratin within fractions of Optiprep gradients, pan-keratin monoclonal antibody (AE1/AE3; Chemicon) or anti-HPV16 L1 monoclonal antibody (Camvir-1; BD Pharmingen) were utilized according to the manufacturer's recommendations. Cross-reactivity between Camvir-1 and cellular keratin during immunoblot analysis allowed this antibody to be used.
RESULTS
Characterization of an HPV16-infected cell line.
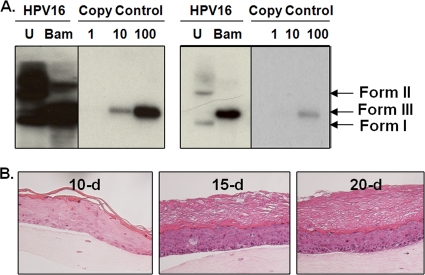
To develop a cell line which can synthesize native virions, primary HFKs were electroporated with linearized HPV16(114/B) DNA (26). The recircularization and maintenance of episomal HPV16 viral genomes within a representative HPV16 cell line can be seen in Fig. 1A. An HPV16 cell line which stably maintains ∼10,000 episomal copies of HPV16 DNA per cell was subsequently allowed to grow in an organotypic culture. To enhance our yield of virions from each organotypic culture, the growth period was extended to 15 and 20 days. H&E staining of organotypic tissues showed that the stratum corneum increased in thickness over time, whereas basal and suprabasal layers retained their original widths, suggesting that virions assembled by day 10 collected in the stratum corneum by days 15 and 20 (Fig. 1B).
FIG. 1.
Establishment of a productive HPV16 cell line. (A) Southern blot of an HPV16(114/B) HFK cell line. Both overexposed and underexposed images of uncut (U) and BamHI-cut (Bam) HPV16(114/B) genomes are shown. The numbers 1, 10, and 100 represent the number of viral genome copies per 5 μg of total cellular DNA that was loaded in lanes U and Bam. Form I, supercoiled viral DNA; form II, nicked viral DNA; form III, linear viral DNA. (B) H&E staining of paraffin-embedded 10-, 15-, and 20-day HPV16-infected organotypic tissues.
Temporal increase in virion stability.
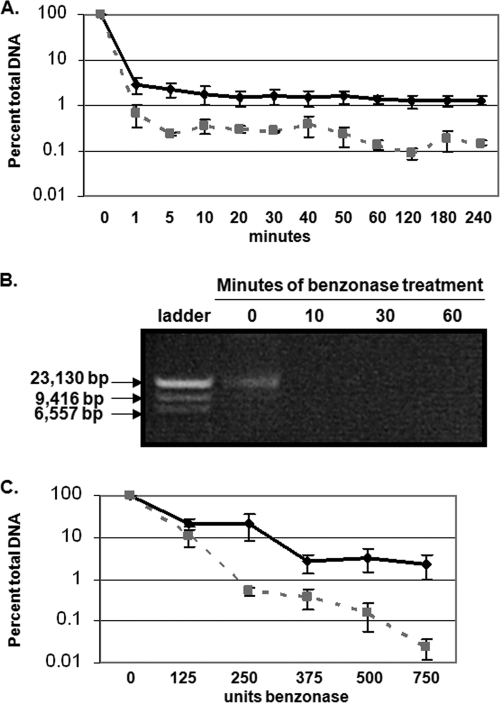
To quantify total virions within 10-, 15-, and 20-day CVPs, we optimized a qPCR-based DNA encapsidation assay to detect the vge (21, 46). Due to the low productivity of organotypic culture, in addition to the high cellular keratin background signal which precludes quantitative Western blot analysis of L1 protein expression directly from CVPs, vge analysis is, at this point, the most quantitative method for normalization (21, 46). Although benzonase has been extensively reported to digest both chromatin-associated and nonassociated DNA, we verified the effectiveness of benzonase to digest chromatin-associated nucleic acid, which is required for complete removal of viral DNA not protected by a capsid (Fig. 2A and B) (1, 24, 29, 49). Aliquots of 20-day CVPs were digested with benzonase for up to 3 h. The viral and cellular DNA was then extracted, followed by SYBR green-based qPCR. Maximum digestion of both viral and cellular DNA occurred by 60 min. However, while 2% of viral DNA remained after benzonase treatment, only 0.1% of cellular DNA remained. Such enhanced protection against benzonase digestion suggests that remaining viral nucleic acid is encapsidated (Fig. 2A and B). The inability of benzonase to completely digest all cellular DNA may also suggest that some cellular DNA is inefficiently encapsidated. Further, efficient digestion of both viral and cellular DNA supports that benzonase digests chromatin-associated and nonassociated DNA. To determine whether the concentration of benzonase was optimal to digest the maximum amount of nonencapsidated genomes, we treated CVPs with increasing concentrations of benzonase (Fig. 2C). After benzonase treatment, viral and cellular DNA was extracted, followed by SYBR green-based qPCR. Although 375 U of benzonase appeared to eliminate as much viral DNA as possible, this concentration was not optimal for complete digestion of cellular DNA. Increasing the benzonase concentration to 500 and 750 U led to increased digestion of cellular DNA, again providing support that the remaining viral DNA is encapsidated and that benzonase efficiently digests both chromatin associated and nonassociated DNA (Fig. 2C).
FIG. 2.
Effectiveness of benzonase in digesting viral and cellular nucleic acids. (A) SYBR green qPCR analysis of viral (solid black line and triangles) and cellular (dashed gray line and squares) DNA extracted from CVPs over a 3-h benzonase digestion time course. (B) Agarose gel analysis of sheared cellular DNA over a 1-h benzonase digestion time course. (C) SYBR green qPCR analysis of viral (solid black line with triangles) and cellular (dashed line and squares) DNA treated with increasing concentrations of benzonase for 1 h at 37°C. The results are means and standard errors for three independent experiments.
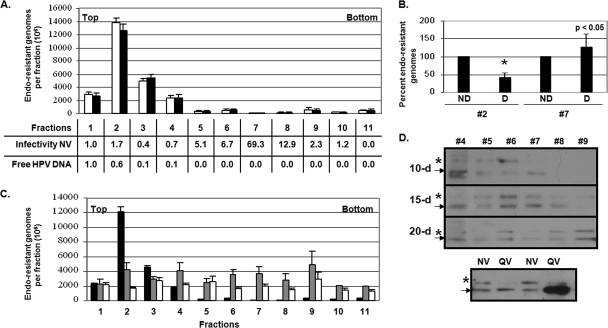
Ten-day CVPs were then treated with benzonase to digest nonencapsidated and endonuclease-susceptible viral genomes, and they were fractionated using a previously described Optiprep step gradient (6). To release viral genomes from capsids, aliquots from each fraction were untreated or treated with 6% 2-ME prior to DNA extraction in order to assess the completeness of encapsidated viral DNA extraction and the necessity of disulfide bonding in capsid stability (Fig. 3A). Figure 3A represents the total number of encapsidated viral genomes detected in each Optiprep fraction by the optimized DNA encapsidation assay. As shown, chemical treatment of fractions with 6% 2-ME prior to DNA extraction did not release viral genomes above that obtained from DNA extraction alone (Fig. 3A). No further increase in reducing reagents (10 and 20% 2-ME) increased the amount of detectable viral genomes beyond 6% 2-ME (data not shown). Surprisingly, the peak of endonuclease-resistant genomes from fractionated 10-day CVPs was detected in fractions 1 to 4 rather than high infectivity fractions 6 to 8, suggesting that high shear forces and hydrostatic pressure associated with ultracentrifugation may have led to the destruction of capsids, as suggested by other studies (7). It should be noted that our top fractions 1 to 4 correspond to a 2-ml fraction that is discarded upon purification of VLPs, PsV, and QV and therefore has not been extensively analyzed (6, 12, 40). To test the possibility that 10-day capsids are destroyed upon ultracentrifugation, thus releasing their encapsidated genomes, SYBR green-based qPCR was used to assay for Optiprep-fractionated free HPV18 DNA. Localization of free viral DNA was found solely in fractions 1 to 4 (Fig. 3A). This idea was further confirmed since viral genomes in fraction 2 were much more susceptible to DNase I digestion than viral genomes in fraction 7, suggesting viral genomes in fraction 2 are free, whereas the genomes in fraction 7 are encapsidated (Fig. 3B). In addition, since specific infectivity (i.e., the vge/infectivity ratio) is enhanced in fractions 6 to 8 compared to fractions 1 to 4, the genomes in fractions 6 to 8 must be in the context of a virion (Fig. 3A). When 15- and 20-day CVPs were Optiprep fractionated and assessed with the DNA encapsidation assay, viral genomes were less prevalent in fractions 1 to 4 and more prevalent in the denser fractions 5 to 11, suggesting that 15- and 20-day virions were more stable than 10-day virions (Fig. 3C). A slight decrease in the detection of 20-day in comparison to 15-day encapsidated viral genomes in multiple fractions may suggest that 20-day virions are more resistant to the used DNA extraction method (Fig. 3C). Western blot analysis of HPV16 L1 from Optiprep-fractionated 10-, 15-, and 20-day CVPs supports that virions are more stable when extracted from 15- and 20-day tissue compared to 10-day tissue since L1 also appears to migrate from the upper fractions to fractions 8 and 9 (Fig. 3D). Analysis of HPV16 L1 was only possible after gradient fractionation in fractions 4 to 9 since we found that its low expression is masked by abundant cellular keratin protein in upper fractions 1 to 3. Such abundant cellular keratin makes detection of L1 from unfractionated CVPs difficult. Similar to a previous report, we found that L1 extracted from organotypic culture exists as two bands (Fig. 3D) (21). Using HPV16 QV L1 as a control for molecular weight, it appears that the faster-migrating L1 species is identical in organotypic culture-derived native virions and QV. The slower-migrating species, however, is not visualized in purified QV even when 10 times as much QV L1 is examined. Since VLP, PsV, and QV L1 is translated solely from the consensus methionine (nt 5638), it is plausible that the slower-migrating species represents a longer translational product which starts at the first methionine (nt 5560) and/or a posttranslationally modified protein product (Fig. 3D) (6, 9, 48). Interestingly, high infectivity fractions of native virions were detected in the same 1.19- to 1.22-g/ml density fraction (fractions 6 to 8) where infectious PsV and QV were reported to exist (Fig. 3A) (6, 12, 18). We attempted to confirm this localization by assessing the specific infectivity in gradient fractions containing 10-day organotypic culture-derived virions and mature HPV16 QV. Although specific infectivity peaked in fractions 6 to 8 for 10- and 20-day native virions (Fig. 3A), specific infectivity was predominant in fractions 10 and 11 for mature HPV16 QV (data not shown).
FIG. 3.
HPV16 genomes are stabilized over time. (A) DNA encapsidation assay of the absolute numbers of endonuclease-resistant viral DNA from Optiprep-fractionated 10-day WT HPV16 CVPs (fractions 1 to 11) either untreated (white) or treated (black) with 6% 2-ME. In a chart at the bottom of the figure, the relative specific infectivities (i.e., infectivity to vge ratio) of 10-day organotypic culture-derived virions, in addition to the relative vge values of free HPV18 DNA per fraction, are shown. Signal detected in fraction 1 was set to 1.0 in both instances. (B) DNA encapsidation assay of 10-day wild-type HPV16 DNase I-treated (D) and untreated (ND) Optiprep fractions 2 and 7. (C) DNA encapsidation assay of the absolute numbers of endonuclease-resistant viral DNA from Optiprep-fractionated 10-day (black), 15-day (gray), and 20-day (white) wild-type HPV16 CVPs (fractions 1 to 11). (D) Western blot analysis of HPV16 L1 in fractions 4 to 9 from Optiprep-fractionated 10-, 15-, and 20-day CVPs. The arrow and asterisk indicate two forms of observed L1 species in gradient fractions 4 to 9. The lower image shows an equal concentration and a 10-fold increase in concentration of HPV16 QV L1 compared to HPV16 organotypic culture-derived native virus L1. Bar graph results are means and standard errors for three independent experiments. The top and bottom of gradients are also shown.
As a method to quantify the relative stability of particles within CVPs, the ratio of unstably encapsidated viral genomes (fractions 1 to 4) over stably encapsidated viral genomes (fractions 6 to 8) was taken. We detected 27, 1, and 1 unstably encapsidated viral genomes for every stably encapsidated viral genome in 10-, 15-, and 20-day CVPs, respectively. This finding suggests that many more unstably encapsidated genomes exist in 10-day CVPs than in 15- and 20-day CVPs. When Optiprep-fractionated mature HPV16 QV lysates were assessed for the relative stability of particles, we detected 24 unstably encapsidated viral genomes for every stably encapsidated genome, suggesting that the relative stability of mature HPV16 QV resembles that of 10-day organotypic culture-derived native virions.
DNA encapsidation profile is capsid specific.
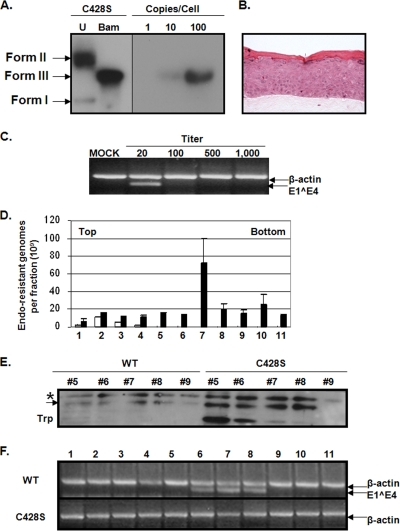
To determine whether modulation of the DNA encapsidation profile was capsid-specific and independent from keratin network formation, 10-day tissues capable of synthesizing virions containing a Cys428Ser substitution were produced (Fig. 4A and B). The Cys428Ser substitution has been shown to be very destabilizing to capsid integrity in both VLPs and PsV (7, 22, 28). Multiple episomal mutant viral DNA-containing cell lines were produced and utilized in experiments to control for PCR fidelity during the mutagenesis protocol. In addition, L1 ORFs from utilized cell lines were sequenced to verify the existence of the intended mutation, and the absence of erroneous mutations in all cases. As seen in Fig. 4C, Cys428Ser virions are infectious as detected via RT-PCR, albeit infectivity was only detected at the lowest threshold of detection. To determine whether the observed low infectivity was due to a decrease in productivity of Cys428Ser organotypic cultures, the total number of encapsidated genomes was quantified and compared to wild-type organotypic cultures (Fig. 4D). Specifically, CVPs made from 10-day tissues were Optiprep fractionated. The DNA encapsidation profile of 10-day Cys428Ser virions depicted a large peak in fraction 7 and smaller peaks in more dense fractions, all of which were higher than wild-type 10-day HPV16 peaks, indicating that Cys428Ser native virions are more susceptible to DNA extraction and/or are more fragile than wild-type and yet still migrate into high-density fractions or that Cys428Ser organotypic cultures are more productive than wild-type organotypic cultures (Fig. 4D). To determine whether the large peak in fraction 7 was due to an increase in productivity of Cys428Ser versus wild-type cultures, equivalent volumes of 20-day CVPs were Optiprep fractionated and Western blot analyses of HPV16 L1 were performed against fractions 5 to 9 (Fig. 4E). Relative densitometric analyses of the slow-migrating L1 species in each fraction were averaged and indicated a fourfold increase in the amount of L1 in Cys428Ser fractions in comparison to wild-type fractions. Since 15- to 80-fold increases were observed in fractions within the Cys428Ser DNA encapsidation profile compared to the wild-type DNA encapsidation profile shown, it did not seem possible that these changes were due to an increase in productivity alone (Fig. 4D). Indeed, Western blot analyses of Cys428Ser fractions 5 to 8 depicted a characteristic 45-kDa tryptic product of L1, which is an indicator of immature and improperly assembled capsids (Fig. 4E) (7, 22, 28).
FIG. 4.
Cys428Ser mutant virions are less infectious and less stable than wild-type virions. (A) Southern blot of a HPV16(114/B) C428S cell line. Images of uncut (U) and BamHI-cut (Bam) HPV16(114/B) genomes are shown. The numbers 1, 10, and 100 represent the number of viral genome copies per 5 μg of total cellular DNA that was loaded in lanes U and Bam. Form I, supercoiled viral DNA; form II, nicked viral DNA; form III, linear viral DNA. (B) H&E staining of paraffin-embedded, HPV16(114/B) C428S-infected organotypic tissue. (C) Endpoint RT-PCR-based infectivity assay detecting E1Ê4 expression from C428S mutant virion-infected HaCaT cells. β-Actin bands were coamplified as a PCR control. Mock samples were prepared from uninfected HaCaT cells, and the titer is the reciprocal dilution of CVP utilized. (D) DNA encapsidation assay of Optiprep-fractionated 10-day wild-type HPV16 (white) and C428S (black) CVPs (fractions 1 to 11). (E) Western blot analysis of HPV16 L1 from Optiprep-fractionated 20-day wild-type (WT) and C428S CVPs (fractions 5 to 9). Species of L1 are discriminated by an arrow and an asterisk. The 45-kDa L1 tryptic product is indicated as Trp. (F) RT-PCR-based stability assay testing 10-day wild-type (WT) HPV16 and C428S CVPs for infectivity after Optiprep fractionation. Bar graph results are means and standard errors for three independent experiments. The top and bottom of gradients are also shown.
To determine whether Cys428Ser virions were more fragile than wild-type virions, approximately 50 × 109 vge of 10-day wild-type and 10-day Cys428Ser virions were Optiprep fractionated (Fig. 4F). Each fraction was assessed for infectivity via RT-PCR and only wild-type virions retained infectivity after ultracentrifugation (Fig. 4F). In all, these results suggest that detection of viral genomes in the DNA encapsidation assay is dependent on capsid architecture and that Cys428 is dispensable for early capsid assembly events yet important in the overall stability of 10- and 20-day virions.
Viral genome amplification and encapsidation kinetics.
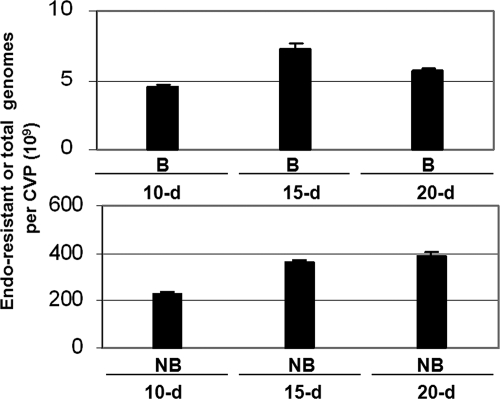
Next, the optimized DNA encapsidation assay was utilized to quantify numbers of DNA-filled virions and total viral DNA within 10-, 15-, and 20-day CVPs (Fig. 5). We detected a 60% increase in endonuclease-resistant genomes in 15-day CVPs in comparison to 10-day CVPs and a slight reduction between 15- and 20-day CVPs, suggesting that DNA encapsidation terminates by 15 days of growth in organotypic culture. These assays detected 4 to 7 × 109 encapsidated genomes per CVP or 1.3 to 2.3 × 109 per raft depending on the time of harvest. To determine the extent that benzonase treatment reduces the background from nonencapsidated viral genomes and to detect total viral genomes, we performed the DNA encapsidation assay on non-benzonase-treated 10-, 15-, and 20-day CVPs and observed that ∼50-fold more genomes were detected in untreated 10-, 15-, and 20-day CVPs than in benzonase-treated CVPs, suggesting that the majority of genomes within a CVP are not protected from endonuclease treatment (Fig. 5). A similar 60% increase in total viral genomes was detected in 15- and 20-day CVPs compared to 10-day CVPs, suggesting that DNA amplification also terminates by 15 days of growth in organotypic culture (Fig. 5).
FIG. 5.
Viral genome amplification and encapsidation kinetics. DNA encapsidation assay quantification of 10-, 15-, and 20-day wild-type HPV16 CVPs either not benzonase treated (NB) or benzonase treated (B) detected total (bottom) or encapsidated (top) viral genomes. The results are means and standard errors for three independent experiments.
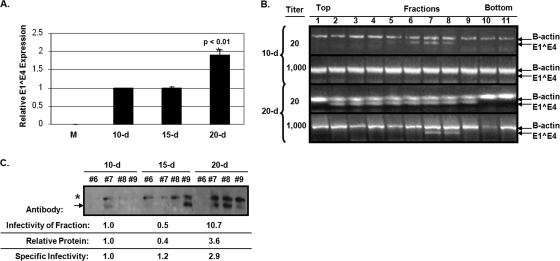
The specific infectivity of 20-day virions is enhanced.
Because of the limited resolution of our RT-PCR infectivity assay, we utilized RT-qPCR to compare relative specific infectivity of virions extracted from 10-, 15-, and 20-day tissues (Fig. 6A). The vge value was utilized to normalize for infections since the productivity of organotypic culture is too low and the cellular keratin background is too high for direct analysis of total L1 expression from CVPs. We consistently detected an ∼2-fold increase in specific infectivity from 20-day virions compared to 10- and 15-day virions when infecting with ∼30,000 vge per cell. The specific infectivity of 10-day and 15-day CVPs remained statistically similar. The infectivity of 20-day virions also appears superior to that of 10-day virions when we analyzed the infectivity of Optiprep-fractionated 10- and 20-day CVPs. Approximately 20 × 109 vge from 10- and 20-day CVPs were fractionated on Optiprep gradients and used to infect HaCaT cells with 1:20 and 1:1,000 dilutions of each fraction, and the infectivity was assessed by RT-PCR (Fig. 6B). Although E1Ê4 amplification can be detected in fractions 5 to 8 from Optiprep-fractionated 10-day CVPs when infected with a dilution of 1:20, E1Ê4 from Optiprep-fractionated 20-day CVPs can be detected in fractions 2 to 9. When infected with a dilution of 1:1,000, E1Ê4 is no longer detected in 10-day gradient fractions, whereas E1Ê4 can still be observed in 20-day gradient fractions 7 and 8. The increased ability to detect E1Ê4 amplification from 20-day compared to 10-day gradient fractions may be due to an increase in the observed specific infectivity of 20-day virions and/or to an increase in stability of 20-day virions post-Optiprep fractionation.
FIG. 6.
Specific infectivity increase in 20-day virions. (A) Relative RT-qPCR infectivity assay of 10-, 15-, and 20-day wild-type HPV16 virions. A total of 30,000 vge per cell was utilized for each infection. M is the mock-infected negative control. (B) Endpoint RT-PCR infectivity assays of Optiprep-fractionated 10- and 20-day wild-type HPV16 CVPs infecting at dilutions of 1:20 and 1:1,000. (C) A Western blot of L1 protein expression using Camvir-1 is shown of Optiprep fractions 6 to 9 from Optiprep-fractionated 10-, 15-, and 20-day CVPs. Below, a table shows the relative infectivity of fractions via RT-qPCR, the relative total protein in fractions via densitometry, and the specific infectivity (i.e., protein to infectivity ratio) of 10-, 15-, and 20-day virions from fraction 7. Bar graph results are means and standard errors for three independent experiments. The top and bottom of gradients are also shown.
To determine whether stability of virions played a role in the observed increase in specific infectivity, Western blot analysis against L1 from Optiprep-fractionated 10-, 15-, and 20-day CVPs was performed (Fig. 6C). The relative densitometric analysis of L1 protein expression in 10-, 15-, and 20-day gradient fraction 7 was used to normalize for relative infectivity of these same fractions. Defining the “protein-to-infectivity ratio” as a second gauge for specific infectivity, 20-day virions were still 2.9-fold more infectious than 10-day virions, which suggests that there is an intrinsic difference between 10- and 20-day virions taken from fraction 7 rather than a difference in the overall stability of particles within 10-, 15-, and 20-day CVPs (Fig. 6C).
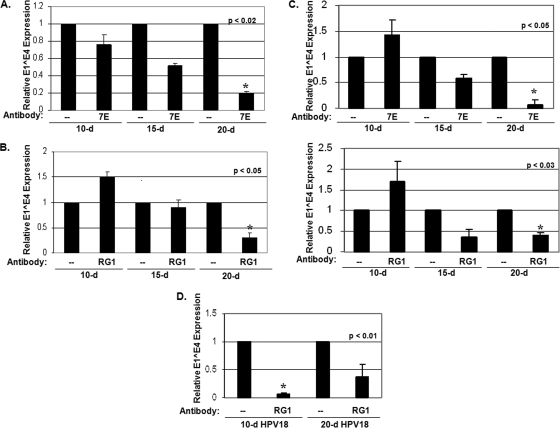
Twenty-day HPV16 virions are more susceptible to neutralization.
Since infections can be neutralized with anti-L1 and anti-L2 neutralizing antibodies, we detected particle-mediated infections. However, neutralization of 30,000 vge from 10-, 15-, and 20-day CVPs with the anti-HPV16 L1 conformation-dependent, monoclonal antibody H16.7E (amino acids 38 to 65) revealed a differential susceptibility to neutralization in that 10-day virions are not neutralized, 15-day virions are partially neutralized, and 20-day virions are effectively neutralized by a 1:100 dilution of H16.7E (Fig. 7A) (10). A similar differential susceptibility to neutralization was shown using the anti-HPV16 L2 mouse monoclonal antibody RG-1 (amino acids 17 to 36) (Fig. 7B) (18). Both antibodies used here have been extensively analyzed in our lab and in others, and a 1:100 dilution of each tends to be the highest concentration tested against PsV, QV, and organotypic culture-derived native virions (10, 13, 18, 27, 31, 40). These results contrast with those obtained through the neutralization of immature and mature HPV16 PsV, which displayed essentially no difference in their 50% neutralization cutoffs (7). Since the utilization of vge to normalize neutralizations may not account for varied ratios of empty to full capsids in CVPs, we performed neutralization assays with virions taken from Optiprep fraction 7 from Optiprep-fractionated 10-, 15-, and 20-day CVPs that had previously been screened for relative L1 concentration (Fig. 6C). These neutralization experiments verified that H16.7E and RG-1 neutralize 20-day virions more effectively than 10-day virions even when more protein was present in the 20-day fraction 7 (Fig. 7C). Surprisingly, we also noticed significant increases in infectivity when 10-day virions were incubated with RG-1 (Fig. 7B and C). In addition, HPV18 native virions exhibited the opposite trend, with 20-day virions less effectively neutralized by RG-1 than 10-day virions, suggesting that conformational alterations of HPV are type specific (Fig. 7D).
FIG. 7.
Antibody-mediated neutralization is enhanced in 20-day virions. (A) Neutralization of 30,000 vge per cell of wild-type HPV16 virions from 10-, 15-, and 20-day CVPs using the anti-L1 antibody, H16.7E, at a dilution of 1:100 (10). (B) Neutralization of 30,000 vge per cell of wild-type HPV16 virions from 10-, 15-, and 20-day CVPs using the anti-L2 antibody, RG-1, at a dilution of 1:100 (18). (C) Neutralization of equal volumes of Optiprep fraction 7, with relative protein levels depicted in Fig. 6C, from Optiprep-fractionated 10-, 15-, and 20-day CVPs using the anti-L1 antibody, H16.7E (top), and the anti-L2 antibody, RG-1 (bottom), both at a dilution of 1:100. (D) Neutralization of 4,000 vge per cell of wild-type HPV18 virions from 10- and 20-day CVPs using the anti-L2 antibody, RG-1, at a dilution of 1:100. The results are means and standard errors for three independent experiments.
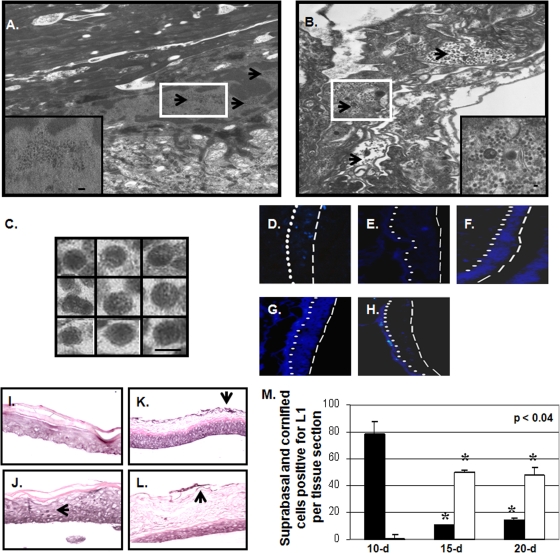
Different temporospatial location of 10-day versus 20-day virions in human tissue.
To determine whether gross morphological differences are observed between 10- and 20-day virions, sections of 10- and 20-day tissues were prepared for TEM. Analysis of these tissues depicted a differential localization of virions depending on the day the tissues were harvested (Fig. 8A to C). The results were consistent with previous reports (4, 35, 46). Most virions were ∼50 nm in diameter; however, a minority of irregularly shaped, tube-like capsid structures was apparent within the stratum corneum (Fig. 8C). Full and empty capsids were observed, but none were associated in paracrystalline arrays. Instead, our examination of 10-day tissue (Fig. 8A) depicted large patches of virions within suprabasal nuclei in stark contrast to the 20-day tissue (Fig. 8B), which depicted patches of virions predominantly at the apex of the cornified envelope and not detected in the suprabasal nuclei. This localization was verified by immunohistochemical analysis with the α-HPV16 L1 monoclonal antibody Camvir-1 since L1 signal predominates in the suprabasal nuclei at 10 days, whereas L1 signal is predominantly detected at the apex of the stratum corneum at 15 and 20 days (Fig. 8I to L). Quantification of suprabasal nuclei positive for L1 shows that these nuclei are sparse in 15- and 20-day tissue (Fig. 8M). Quantification of cornified cells positive for L1 shows that L1 signal is increased within the stratum corneum at 15 and 20 days compared to 10 days (Fig. 8M).
FIG. 8.
Differential localization of 10-day versus 20-day virions within wild-type HPV16-infected organotypic cultures. (A to C) TEM examination of 10- and 20-day HPV16-infected organotypic cultures, respectively. (A) A representative suprabasal nucleus with numerous clusters of virions throughout the intranuclear space is shown (black arrowheads point to clusters). (B) A representative cornifed cell with numerous clusters of virions throughout the cell is shown (black arrowheads point to clusters). The insets show enlargements of both the suprabasal nucleus and the cornified cell. Scale bars: 200 nm (A) and 100 nm (B). (C) Depiction of select HPV16 virions visualized within the cornified envelope. Scale bar, 50 nm. (D to H) Detection of disulfide bonds in cornified layer of 15-day HPV16-infected organotypic cultures via DACM staining. (D) DACM-unstained tissue. (E) Sulfhydryls blocked with NEM prior to DACM stain. (F) Free sulfhydryls stained with DACM. (G) Free and disulfide-bonded sulfhydryls stained by treatment with dithiothreitol, followed by DACM stain. (H) Disulfide-bonded sulfhydryls stained by blocking with NEM, followed by treatment with dithiothreitol and then DACM stain. (I to L) Immunohistochemical staining of L1 within 10 (J)-, 15 (K)-, and 20 (L)-day paraffin-embedded raft sections of HPV16-infected organotypic culture and an uninfected 10-day HFK paraffin-embedded raft section (I). L1 signal is shown within suprabasal nuclei at 10 days and at the apex of the cornified envelope at 15 and 20 days (the L1 signal is indicated by black arrowheads). (M) Quantification of the number of L1-positive suprabasal nuclei (black) and cornified cells (white) per paraffin-embedded 10-, 15-, and 20-day raft sections. The results are means and standard errors for three independent experiments.
Ten-day virions are in a reducing environment, and twenty-day virions are in an oxidizing environment.
The highly oxidizing environment of the stratum corneum compared to suprabasal strata allowed us to hypothesize that the temporal phenotypic changes observed were due to the exposure of virions to the oxidizing environment of the stratum corneum (7, 38, 39). To confirm the oxidative potential of the stratum corneum, frozen sections of 15-day HPV16-infected organotypic tissues were stained with the fluorescent, sulfhydryl-alkylating, histochemical staining reagent, DACM (Fig. 8D to H). DACM is not fluorescent until it reacts with sulfhydryl groups, thus producing fluorescent addition products (38, 39). We show that all epithelial strata, except the stratum corneum, contained significant amounts of detectable free sulfhydryl groups when stained with DACM alone (Fig. 8F). When we irreversibly blocked free sulfhydryls with the sulfhydryl-alkylating reagent N-ethylmaleimide (NEM), followed by reduction of disulfide-bonded sulfhydryls with dithiothreitol and then stained with DACM, only the stratum corneum contained significant amounts of detectable disulfide bonds (Fig. 8H). Only faint autofluorescent signals can be detected in unstained samples, or samples blocked with the NEM (Fig. 8D and E). The differential localization of free versus disulfide-bonded sulfhydryls confirms that a natural redox gradient exists within human epithelial tissue whereby the basal and suprabasal compartments can be generally described as reducing, while the cornified compartment is oxidizing.
GSSG-treatment enhances genome encapsidation and infectivity of tissue.
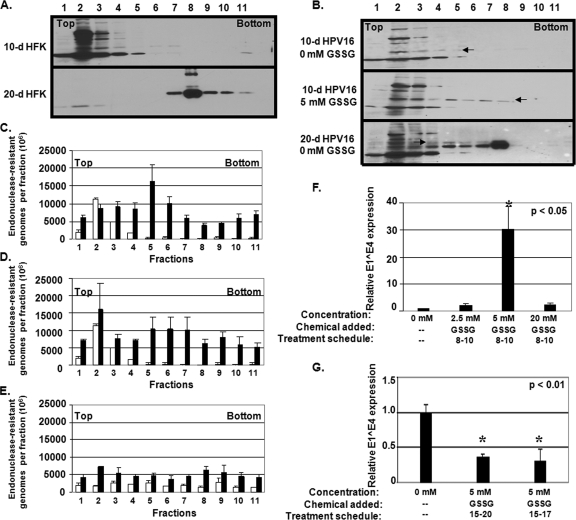
To assess the role of a tissue-spanning redox gradient in the assembly process of virions, we treated HPV16-infected organotypic cultures with GSSG. Concentrations of GSSG were chosen based on previous reports that 5 mM GSSG enhances bovine papillomavirus and HPV16 PsV maturation (7, 20). Treatments were performed beginning on day 8 and terminated on day 10, at which point tissue was harvested. This allowed us to treat virions during initial capsid assembly. Treatments were also performed beginning on day 15 and terminating on day 17 or 20, at which point tissue was harvested. This treatment schedule was chosen to assess whether further GSSG-dependent assembly events take place after the initial assembly. To verify that treatments with GSSG altered the redox potential of organotypic tissues, Western blot analyses were used against cellular keratin and HPV16 L1 from Optiprep-fractionated 10- and 20-day lysates from HFK and HPV16-infected organotypic cultures (Fig. 9A and B). In HFK fractions, cellular keratin network formation can be observed via the complete migration of protein from fractions 1 to 5 to fractions 7 to 11 starting on day 10 and terminating on day 20, respectively (Fig. 9A). Interestingly, keratin from uninfected 20-d HFK tissues are incorporated into dense protein networks much more effectively than in HPV16-infected 20-d HFK tissues, which is reminiscent of E1^E4′s role in keratin network reorganization (Fig. 9A and B) (3, 16, 47). The migration of L1 into dense gradient fractions appears to be redox dependent since HPV16-infected organotypic cultures treated with 5 mM GSSG during days 8 through 10 of growth led to an increase in the localization of L1 in fractions 6 to 8 compared to untreated tissues (Fig. 9B).
FIG. 9.
GSSG treatment alters viral DNA encapsidation and infectivity of tissue. (A) Western analysis of human keratin from Optiprep-fractionated 10- and 20-day HFK lysates, and (B) Western analysis of HPV16 L1 from Optiprep-fractionated 10- and 20-day wild-type HPV16 CVPs. Ten-day organotypic cultures were either untreated or treated with 5 mM GSSG during days 8 through 10. The arrows point to suspected monomeric HPV16 L1. (C to E) DNA encapsidation assay of Optiprep-fractionated wild-type HPV16 CVPs made from either 5 mM GSSG during days 8 through 10 untreated (white) or treated (black), 20 mM GSSG during days 8 through 10, and 5 mM GSSG during days 15 through 20 organotypic cultures, respectively. (F) Relative RT-qPCR infectivity assay of CVPs made from untreated versus 2.5, 5, and 20 mM GSSG-treated wild-type HPV16 organotypic cultures treated during days 8 through 10. (G) Relative RTq-PCR infectivity assay of wild-type HPV16 CVPs made from untreated versus 5 mM GSSG-treated organotypic cultures treated during days 15 through 17 and 15 through 20. Bar graph results are means and standard errors for three independent experiments. The top and bottom of gradients are also shown.
To determine the effect of GSSG on DNA encapsidation, virions were extracted from GSSG-treated organotypic cultures and analyzed with our DNA encapsidation assay, post-Optiprep-fractionation (Fig. 9C to E). Treatment of cultures with 5 mM GSSG during days 8 through 10 of growth predominantly increased the pool of detectable endonuclease-resistant genomes in fractions containing low-density virions (Fig. 9C), whereas treatment with 20 mM GSSG appeared to increase the pool of detectable endonuclease-resistant genomes in both low- and high-density fractions (Fig. 9D). No significant change in detectable endonuclease-resistant genomes was observed in any fraction upon treatment with 5 mM GSSG during days 15 through 20 of growth (Fig. 9E). These data imply that GSSG treatment facilitates the assembly of virions, which is temporospatially dependent in that GSSG treatment facilitates DNA encapsidation only when treating suprabasal layer-localized virions, whereas GSSG treatment has no effect on DNA encapsidation of virions localized within the stratum corneum.
To correlate the effect of GSSG treatments on DNA encapsidation and the production of infectious virus, RT-qPCR-based infectivity analyses were performed using CVPs made from GSSG-treated organotypic cultures (Fig. 9F to G). Treatment of organotypic cultures with 2.5, 5, and 20 mM GSSG during days 8 through 10 of growth increased infectivity of CVPs in comparison to untreated controls 2-, 30-, and 2.5-fold, respectively, suggesting that 5 mM GSSG treatment is optimal for virion assembly within suprabasal nuclei (Fig. 9F). Surprisingly, treatment of cultures with 5 mM GSSG during days 15 through 20 or during days 15 through 17 of growth decreased infectivity ca. 60% (Fig. 9G). Such a decrease in infectivity suggests that an additional post-initial assembly step occurs within the stratum corneum and that GSSG treatment perturbs this step by promoting capsid misfolding (7).
DISCUSSION
The possibility that in vivo papillomavirus capsid assembly and maturation occurs in a redox-dependent fashion has been discussed, but never directly tested (4, 7). These hypotheses suggest that during desquamation, capsids are exposed to an increase in oxidizing conditions (4, 7). Such “treatment” of the capsid to the oxidizing environment would facilitate disulfide interactions that stabilize the capsid prior to release into the environment.
Our ability to grow HPV16 virions in organotypic culture has allowed us to examine the capsid assembly process within native differentiating host tissue. Our original observation was that HPV16 virions reached maximum stability at 15 to 20 days since these genomes were less apt to be released from capsids during ultracentrifugation (Fig. 3A to D). Twenty-day virions also had a twofold higher specific infectivity than 10- and 15-day virions (Fig. 6A and B). Previous studies regarding the maturation step of a variety of virus types have also described a parallel increase in infectivity upon capsid maturation (5, 8, 23, 44, 45). That 20-day virions were more susceptible to antibody-mediated neutralization supports that conformational changes continue to occur in both L1 and L2 surface loops throughout the measured time course or that conformational changes which occur upon binding to the cell surface only efficiently occur in 20-day virions (Fig. 7A to D).
TEM analysis depicted a differential localization of HPV16 virions within 10 and 20-day tissue, showing that 10-day virions are predominantly localized within suprabasal nuclei, whereas 20-day virions are predominantly localized to the apex of the cornified envelope (Fig. 8A to C). This localization overlaps with a natural redox gradient within human tissue, which restricts 10-day virions largely to a reducing environment and 20-day virions to an oxidizing environment (Fig. 8D to G). We hypothesized that this exposure to oxidizing conditions would mediate the observed temporal stability and specific infectivity changes. As such, treatment of organotypic cultures with GSSG was able to enhance viral DNA encapsidation and infectivity of tissues but only if added to cultures before virions reached the oxidizing cornified envelope (Fig. 9C to E and F). Although treatment of organotypic cultures with GSSG had no significant effect on viral DNA encapsidation when virions were in the cornified envelope, GSSG treatment did significantly reduce infectivity of these tissues, suggesting that a postencapsidation step was hindered (Fig. 9E and G). These studies reveal a tissue-spanning redox gradient that is a novel tool unique to papillomavirus assembly. Importantly, these studies provide a means for studying papillomavirus assembly steps within epithelial tissue.
Acknowledgments
We thank Roland Myers for TEM help, Lynn Budgeon for histological expertise, Horng-Shen Chen and Tim Culp for assistance with the quantitative PCR, and Ollie Mikse for computational support.
This study was supported by a PHS grant from the National Institute of Allergy and Infectious Disease (R01AI57988) and in part by grants to R.B.S.R. from the National Institutes of Health (National Cancer Institute, SPORE in Cervical Cancer, P50 CA098252 and CA118790).
Footnotes
Published ahead of print on 5 August 2009.
REFERENCES
- 1.Aygun, O., J. Svejstrup, and Y. Liu. 2008. A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc. Natl. Acad. Sci. USA 105:8580-8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, T. S., W. W. Newcomb, N. H. Olson, L. M. Cowsert, C. Olson, and J. C. Brown. 1991. Structures of bovine and human papillomaviruses: analysis by cryoelectron microscopy and three-dimensional image reconstruction. Biophys. J. 60:1445-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryan, J. T., and D. R. Brown. 2000. Association of the human papillomavirus type 11 E1()E4 protein with cornified cell envelopes derived from infected genital epithelium. Virology 277:262-269. [DOI] [PubMed] [Google Scholar]
- 4.Bryan, J. T., and D. R. Brown. 2001. Transmission of human papillomavirus type 11 infection by desquamated cornified cells. Virology 281:35-42. [DOI] [PubMed] [Google Scholar]
- 5.Buck, C. B., N. Cheng, C. D. Thompson, D. R. Lowy, A. C. Steven, J. T. Schiller, and B. L. Trus. 2008. Arrangement of L2 within the papillomavirus capsid. j. virol. 82:5190-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buck, C. B., D. V. Pastrana, D. R. Lowy, and J. T. Schiller. 2004. Efficient intracellular assembly of papillomaviral vectors. J. Virol. 78:751-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buck, C. B., C. D. Thompson, Y. Y. Pang, D. R. Lowy, and J. T. Schiller. 2005. Maturation of papillomavirus capsids. J. Virol. 79:2839-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Champagne, J., M. E. Laliberte-Gagne, and D. Leclerc. 2007. Phosphorylation of the termini of Cauliflower mosaic virus precapsid protein is important for productive infection. Mol. Plant-Microbe Interact. 20:648-658. [DOI] [PubMed] [Google Scholar]
- 9.Chen, X. S., R. L. Garcea, I. Goldberg, G. Casini, and S. C. Harrison. 2000. Structure of small virus-like particles assembled from the L1 protein of human papillomavirus 16. Mol. Cell 5:557-567. [DOI] [PubMed] [Google Scholar]
- 10.Christensen, N. D., N. M. Cladel, C. A. Reed, L. R. Budgeon, M. E. Embers, D. M. Skulsky, W. L. McClements, S. W. Ludmerer, and K. U. Jansen. 2001. Hybrid papillomavirus L1 molecules assemble into virus-like particles that reconstitute conformational epitopes and induce neutralizing antibodies to distinct HPV types. Virology 291:324-334. [DOI] [PubMed] [Google Scholar]
- 11.Culp, T. D., and N. D. Christensen. 2003. Quantitative RT-PCR assay for HPV infection in cultured cells. J. Virol. Methods 111:135-144. [DOI] [PubMed] [Google Scholar]
- 12.Culp, T. D., N. M. Cladel, K. K. Balogh, L. R. Budgeon, A. F. Mejia, and N. D. Christensen. 2006. Papillomavirus particles assembled in 293TT cells are infectious in vivo. J. Virol. 80:11381-11384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Day, P. M., R. Gambhira, R. B. Roden, D. R. Lowy, and J. T. Schiller. 2008. Mechanisms of human papillomavirus type 16 neutralization by l2 cross-neutralizing and l1 type-specific antibodies. J. Virol. 82:4638-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Villiers, E. M., C. Fauquet, T. R. Broker, H. U. Bernard, and H. zur Hausen. 2004. Classification of papillomaviruses. Virology 324:17-27. [DOI] [PubMed] [Google Scholar]
- 15.Doorbar, J. 2005. The papillomavirus life cycle. J. Clin. Virol. 32(Suppl. 1):S7-S15. [DOI] [PubMed] [Google Scholar]
- 16.Doorbar, J., E. Medcalf, and S. Napthine. 1996. Analysis of HPV1 E4 complexes and their association with keratins in vivo. Virology 218:114-126. [DOI] [PubMed] [Google Scholar]
- 17.Finch, J. T., and A. Klug. 1965. The structure of viruses of the papilloma-polyoma type 3: structure of rabbit papillomavirus, with an appendix on the topography of contrast in negative-staining for electron-microscopy. J. Mol. Biol. 13:1-12. [DOI] [PubMed] [Google Scholar]
- 18.Gambhira, R., B. Karanam, S. Jagu, J. N. Roberts, C. B. Buck, I. Bossis, H. Alphs, T. Culp, N. D. Christensen, and R. B. Roden. 2007. A protective and broadly cross-neutralizing epitope of human papillomavirus L2. J. Virol. 81:13927-13931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagensee, M. E., N. H. Olson, T. S. Baker, and D. A. Galloway. 1994. Three-dimensional structure of vaccinia virus-produced human papillomavirus type 1 capsids. J. Virol. 68:4503-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanslip, S. J., N. R. Zaccai, A. P. Middelberg, and R. J. Falconer. 2008. Intrinsic fluorescence as an analytical probe of virus-like particle assembly and maturation. Biochem. Biophys. Res. Commun. 375:351-355. [DOI] [PubMed] [Google Scholar]
- 21.Holmgren, S. C., N. A. Patterson, M. A. Ozbun, and P. F. Lambert. 2005. The minor capsid protein L2 contributes to two steps in the human papillomavirus type 31 life cycle. J. Virol. 79:3938-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishii, Y., K. Tanaka, and T. Kanda. 2003. Mutational analysis of human papillomavirus type 16 major capsid protein L1: the cysteines affecting the intermolecular bonding and structure of L1-capsids. Virology 308:128-136. [DOI] [PubMed] [Google Scholar]
- 23.Joshi, A., K. Nagashima, and E. O. Freed. 2006. Mutation of dileucine-like motifs in the human immunodeficiency virus type 1 capsid disrupts virus assembly, gag-gag interactions, gag-membrane binding, and virion maturation. J. Virol. 80:7939-7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juodka, B., E. Spiess, A. Angiolillo, G. Joswig, K. Rothbarth, and D. Werner. 1995. High salt- and SDS-stable DNA binding protein complexes with ATPase and protein kinase activity retained in chromatin-depleted nuclei. Nucleic Acids Res. 23:1359-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirnbauer, R., F. Booy, N. Cheng, D. R. Lowy, and J. T. Schiller. 1992. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc. Natl. Acad. Sci. USA 89:12180-12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirnbauer, R., J. Taub, H. Greenstone, R. Roden, M. Durst, L. Gissmann, D. R. Lowy, and J. T. Schiller. 1993. Efficient self-assembly of human papillomavirus type 16 L1 and L1-L2 into virus-like particles. J. Virol. 67:6929-6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kondo, K., Y. Ishii, H. Ochi, T. Matsumoto, H. Yoshikawa, and T. Kanda. 2007. Neutralization of HPV16, 18, 31, and 58 pseudovirions with antisera induced by immunizing rabbits with synthetic peptides representing segments of the HPV16 minor capsid protein L2 surface region. Virology 358:266-272. [DOI] [PubMed] [Google Scholar]
- 28.Li, M., P. Beard, P. A. Estes, M. K. Lyon, and R. L. Garcea. 1998. Intercapsomeric disulfide bonds in papillomavirus assembly and disassembly. J. Virol. 72:2160-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, L., J. H. Choi, H. Yim, J. S. Choi, B. D. Park, S. J. Cho, and S. K. Lee. 2009. ATR (AT mutated Rad3 related) activity stabilizes Cdc6 and delays G2/M-phase entry during hydroxyurea-induced S-phase arrest of HeLa cells. Int. J. Biochem. Cell Biol. 41:1410-1420. [DOI] [PubMed] [Google Scholar]
- 30.Longworth, M. S., and L. A. Laimins. 2004. Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiol. Mol. Biol. Rev. 68:362-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLaughlin-Drubin, M. E., N. D. Christensen, and C. Meyers. 2004. Propagation, infection, and neutralization of authentic HPV16 virus. Virology 322:213-219. [DOI] [PubMed] [Google Scholar]
- 32.McLaughlin-Drubin, M. E., and C. Meyers. 2005. Propagation of infectious, high-risk HPV in organotypic “raft” culture. Methods Mol. Med. 119:171-186. [DOI] [PubMed] [Google Scholar]
- 33.McLaughlin-Drubin, M. E., S. Wilson, B. Mullikin, J. Suzich, and C. Meyers. 2003. Human papillomavirus type 45 propagation, infection, and neutralization. Virology 312:1-7. [DOI] [PubMed] [Google Scholar]
- 34.Meyers, C., M. G. Frattini, J. B. Hudson, and L. A. Laimins. 1992. Biosynthesis of human papillomavirus from a continuous cell line upon epithelial differentiation. Science 257:971-973. [DOI] [PubMed] [Google Scholar]
- 35.Meyers, C., T. J. Mayer, and M. A. Ozbun. 1997. Synthesis of infectious human papillomavirus type 18 in differentiating epithelium transfected with viral DNA. J. Virol. 71:7381-7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Modis, Y., B. L. Trus, and S. C. Harrison. 2002. Atomic model of the papillomavirus capsid. EMBO J. 21:4754-4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicklas, J. A., and E. Buel. 2003. Development of an Alu-based, real-time PCR method for quantitation of human DNA in forensic samples. J. Forensic Sci. 48:936-944. [PubMed] [Google Scholar]
- 38.Ogawa, H., A. Taneda, Y. Kanaoka, and T. Sekine. 1979. The histochemical distribution of protein bound sulfhydryl groups in human epidermis by the new staining method. J. Histochem. Cytochem. 27:942-946. [DOI] [PubMed] [Google Scholar]
- 39.Pang, Y. Y., A. Schermer, J. Yu, and T. T. Sun. 1993. Suprabasal change and subsequent formation of disulfide-stabilized homo- and hetero-dimers of keratins during esophageal epithelial differentiation. J. Cell Sci. 104(Pt. 3):727-740. [DOI] [PubMed] [Google Scholar]
- 40.Pyeon, D., P. F. Lambert, and P. Ahlquist. 2005. Production of infectious human papillomavirus independently of viral replication and epithelial cell differentiation. Proc. Natl. Acad. Sci. USA 102:9311-9316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, J. L., S. K. Campos, and M. A. Ozbun. 2007. Human papillomavirus type 31 uses a caveolin 1- and dynamin 2-mediated entry pathway for infection of human keratinocytes. J. Virol. 81:9922-9931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trus, B. L., R. B. Roden, H. L. Greenstone, M. Vrhel, J. T. Schiller, and F. P. Booy. 1997. Novel structural features of bovine papillomavirus capsid revealed by a three-dimensional reconstruction to 9 Å resolution. Nat. Struct. Biol. 4:413-420. [DOI] [PubMed] [Google Scholar]
- 43.Unckell, F., R. E. Streeck, and M. Sapp. 1997. Generation and neutralization of pseudovirions of human papillomavirus type 33. J. Virol. 71:2934-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Villanueva, R. A., J. L. Galaz, J. A. Valdes, M. M. Jashes, and A. M. Sandino. 2004. Genome assembly and particle maturation of the birnavirus infectious pancreatic necrosis virus. J. Virol. 78:13829-13838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walukiewicz, H. E., M. Banerjee, A. Schneemann, and J. E. Johnson. 2008. Rescue of maturation-defective flock house virus infectivity with noninfectious, mature, viruslike particles. J. Virol. 82:2025-2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, H. K., A. A. Duffy, T. R. Broker, and L. T. Chow. 2009. Robust production and passaging of infectious HPV in squamous epithelium of primary human keratinocytes. Genes Dev. 23:181-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, Q., H. Griffin, S. Southern, D. Jackson, A. Martin, P. McIntosh, C. Davy, P. J. Masterson, P. A. Walker, P. Laskey, M. B. Omary, and J. Doorbar. 2004. Functional analysis of the human papillomavirus type 16 E1=E4 protein provides a mechanism for in vivo and in vitro keratin filament reorganization. J. Virol. 78:821-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Webb, E., J. Cox, and S. Edwards. 2005. Cervical cancer-causing human papillomaviruses have an alternative initiation site for the L1 protein. Virus Genes 30:31-35. [DOI] [PubMed] [Google Scholar]
- 49.Yim, H., I. S. Hwang, J. S. Choi, K. H. Chun, Y. H. Jin, Y. M. Ham, K. Y. Lee, and S. K. Lee. 2006. Cleavage of Cdc6 by caspase-3 promotes ATM/ATR kinase-mediated apoptosis of HeLa cells. J. Cell Biol. 174:77-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.zur Hausen, H. 1996. Papillomavirus infections-a major cause of human cancers. Biochim. Biophys. Acta 1288:F55-F78. [DOI] [PubMed] [Google Scholar]