Abstract
Latent infection by Epstein-Barr virus (EBV) requires both replication and maintenance of the viral genome. EBV nuclear antigen 1 (EBNA1) is a virus-encoded protein that is critical for the replication and maintenance of the genome during latency in proliferating cells. We have previously demonstrated that EBNA1 recruits the cellular origin recognition complex (ORC) through an RNA-dependent interaction with EBNA1 linking region 1 (LR1) and LR2. We now show that LR1 and LR2 bind to G-rich RNA that is predicted to form G-quadruplex structures. Several chemically distinct G-quadruplex-interacting drugs disrupted the interaction between EBNA1 and ORC. The G-quadruplex-interacting compound BRACO-19 inhibited EBNA1-dependent stimulation of viral DNA replication and preferentially blocked proliferation of EBV-positive cells relative to EBV-negative cell lines. BRACO-19 treatment also disrupted the ability of EBNA1 to tether to metaphase chromosomes, suggesting that maintenance function is also mediated through G-quadruplex recognition. These findings suggest that the EBNA1 replication and maintenance function uses a common G-quadruplex binding capacity of LR1 and LR2, which may be targetable by small-molecule inhibitors.
Epstein-Barr virus (EBV) is a human gamma herpesvirus that has been implicated in several lymphoid and epithelial cell malignancies (reviewed in reference 60). EBV establishes a long-term latent infection in memory B lymphocytes, where the genome is maintained as a multicopy, chromatinized episome (9). Episome stability in proliferating B cells requires that the viral genome be replicated and faithfully segregated during each cellular division (22, 34). The EBV episomal minichromosome has a genetically defined origin of plasmid replication, referred to as OriP (59). OriP can function as an efficient origin of DNA replication and is thought to be essential for stable maintenance of the viral episome in proliferating cells (58). OriP recruits several components of the cellular replication machinery, including subunits of the origin recognition complex (ORC) and minichromosome maintenance proteins, and replicates indistinguishably from cellular chromosomal origins of replication (8, 13, 42, 43). ORC recruitment to OriP is thought to be essential for replication initiation function but may also be important for other functions, including episome maintenance and chromatin organization (56). ORC recruitment to OriP requires the virus-encoded protein EBV nuclear antigen 1 (EBNA1), but other factors may also contribute to ORC recruitment and function at OriP (2, 11, 12).
EBNA1 is essential for maintaining the latent viral episome during latency. Genetic disruption of EBNA1 from EBV leads to a profound reduction in B-cell transforming activity and an inability to establish episomal latency (28). EBNA1 has several well-characterized functions and protein domains. The EBNA1 C terminus comprises a DNA-binding domain (DBD) with structural similarities to human papillomavirus E2 (5). The DBD binds with high affinity to an ∼18-bp palindromic site that is repeated throughout OriP. OriP contains two distinguishable sites of EBNA1 binding, the family of repeats and the dyad symmetry (DS). The family of repeats is a cluster of 20 EBNA1-binding sites that is important for transcription regulation and episome segregation and maintenance (57). The DS contains two EBNA1 paired-binding sites, the spacing of which is critical for DNA replication initiation function (4).
The replication and plasmid maintenance function of EBNA1 depends on linking region 1 (LR1) and LR2. LR1 and LR2 consist of arginine- and glycine-rich stretches that resemble RGG motifs, which have been described for several RNA-binding proteins (6). LR1 and LR2 have RNA-binding activity, but the significance of this RNA binding has not been clearly established (30, 51). LR1 and LR2 also possess HMGA1a-like AT hooks that confer AT-rich DNA-binding activity (44, 45). HMGA1a AT hook activity has been implicated in origin formation at several cellular origins (53). LR1 and LR2 have been noted to have homotypic binding activity and form higher-order structures in electrophoretic mobility assays (3, 15, 31). LR2 can also interact with cellular proteins P32/TAP (54, 55) and EBP2 (48). In addition, LR1 and LR2 confer the metaphase chromosome-binding activity of EBNA1, which is thought to be a critical component of the episome maintenance function (32, 33, 45). In addition to these functions in DNA replication and plasmid maintenance, EBNA1 is also essential for transcription activation of other viral genes during latent infection in primary B cells (1). The transcription activity was mapped to two regions in the N-terminal domain, referred to as the unique region 1 and LR2 (25, 56). How these various domains of EBNA1 confer transcription, replication, and plasmid maintenance function are not completely understood.
Our previous work has demonstrated that EBNA1 recruits ORC to the DS directly through the RGG-like motifs found in LR1 and LR2 (37). RGG domains were first identified in nucleolin and are known for their RNA-binding ability (26). We found that the interaction between ORC and EBNA1 was RNA dependent. Previous studies, as well as our own, have shown that EBNA1 RNA binding is sequence independent but has a strong preference for G-rich RNA (30, 51). Our previous study indicated that EBNA1 bound preferentially to structured RNA, as demonstrated by the altered mobility of a G-rich RNA probe during gel electrophoresis. Other RGG-like RNA binding proteins are known to bind in a structure-specific, rather than sequence-specific, manner. Both nucleolin and fragile-X mental retardation protein (FMRP) bind to structured G-quadruplex RNA (10, 17). Based on the protein domain similarities between EBNA1 and FMRP, we decided to investigate if EBNA1 also had a preference for G-quadruplex RNA.
G-quadruplex structures occur when four guanine bases interact through noncanonical Hoogstein base pairing to form planar G-quartets, which in turn form stacks (reviewed in reference 20). A minimum of two stacked G-quartets will form a G-quadruplex, but stacks of three or more G-quartets have higher stability. In general, RNA G-quadruplexes are more stable than DNA structures. Many different forms of G-quadruplex structures exist and can be distinguished, for example, by the parallel or antiparallel orientation of the loops that accommodate the G-tetrads. G-quadruplexes, including those that form at the single stranded G-rich DNA overhangs found at telomeres, have been implicated in many different functions (38). G-quadruplex formation at telomeres is thought to limit template accessibility to telomerase and contribute to telomere length regulation (21). G-quadruplex-interacting compounds, such as TMPyP4 and BRACO-19, prevent telomerase function at telomeres, presumably by blocking telomerase access to G-rich telomere DNA (7, 46). Both TMPyP4 and BRACO-19 have been used to selectively inhibit cell growth of telomerase-dependent tumor cells. G-quadruplexes can also form in transcription-regulatory regions of various genes, including the c-Myc gene (49, 50).
In the present study, we investigated the RNA-binding properties of EBNA1. We explored the sequence and structural features of RNA that bind EBNA1 in vitro. We also used G-quadruplex-interacting compounds to determine the role of G-quadruplex RNA in mediating EBNA1 functions in vitro and in vivo. We present evidence that G-quadruplex-interacting compounds may be useful for the pharmacological inhibition of EBNA1-dependent replication and metaphase chromosome attachment.
MATERIALS AND METHODS
Cell lines, constructs, and antibodies.
HeLa cells were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, 2 mM Glutamax, and 100 IU/ml penicillin-streptomycin. DG75, BJAB, Raji, LCL3456, and LCL 3472 cells were cultured in RPMI supplemented with 10% fetal bovine serum, 2 mM Glutamax, and 100 IU/ml penicillin-streptomycin. Glutathione S-transferase (GST)-LR1, GST-LR2, ORC1 peptides, and constructs used for transient replication assay have all been previously described (37). Briefly, GST-LR1 and GST-LR2 were generated with primers that amplified the regions from amino acid (aa) 30 to 53 and 328 to 350, respectively, and cloned into the EcoRI-BamHI sites of pGEX-4T vector. GST-ORC1 aa 1 to 200, GST-ORC1 aa 201 to 511, and GST-ORC1 aa 512 to 861 were generated in the same manner. All proteins were expressed in Bcl Star cells (Invitrogen), purified over glutathione-Sepharose beads (Amersham), and dialyzed into D150 buffer (20 mM HEPES [pH 7.9], 20% glycerol, 1 mM EDTA, 150 mM KCl, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride). For the replication assay, full-length EBNA1 (FL-EBNA1) (lacking Gly-Ala repeats), the DBD (aa 454 to 640), or four tandem copies of LR1 (aa 30 to 56) fused to the DBD (4×LR1-DBD) was cloned into the 3×Flag CMV 24 vector (Sigma). pHeBo was used as the reporter plasmid (52). For metaphase spreads, either the amino terminus of EBNA1 (aa 1 to 440) was cloned into the BglII/EcoRI sites of pmCherry-C1 (Clontech Laboratories, Inc.) or FL-EBNA1 (lacking Gly-Ala repeats) was cloned into the Asp718/BglII sites of pEGFP (Clontech).
For immunoprecipitation, polyclonal rabbit EBNA1 (305/10wk) was used (16). For Western blotting, monoclonal EBNA1 (ABI), ORC2 (MBL), GST (Santa Cruz), β-actin (Sigma), and Flag (Sigma) antibodies were used according to the manufacturer's directions.
TMPyP2, TMPyP3, and TMPyP4 can be purchased from Frontier Scientific, and BRACO-19 was synthesized by C. Meyers at the Fox Chase Cancer Center.
EMSA and replication assay.
Electrophoretic mobility shift assay (EMSA) conditions have been previously described (37). Briefly, the probe was end labeled with [γ-32P]ATP and T4 polynucleotide kinase. Labeled probe (10 pM) was incubated with purified protein (∼100 nM) in D150 for 30 min at room temperature and then loaded on a 1.5% horizontal agarose gel in 1/2× Tris-borate-EDTA. Gels were dried on DE81 paper and exposed overnight for PhosphorImager analysis. The replication assay has been previously described (11).
Immunoprecipitation, GST pulldown assay, and retention assay.
Immunoprecipitation for Raji cells and GST pulldown assays have been previously described (2). The only modification was that the wash buffer was supplemented with 10 μM TMPyP2, TMPyP3, TMPyP4, or BRACO-19 as indicated. For the retention assay, GST protein was immobilized on glutathione-Sepharose beads as described for the GST pulldown assay. The bead-bound protein was then washed three times in wash buffer supplemented with 10 μM BRACO-19 and 20 μg/ml RNase A (Sigma). The bead-bound protein slurry was spotted onto Whatman paper and allowed to air dry. An image was taken with a digital camera, and BRACO-19 retention was quantified using ImageQuant.
Genome maintenance assay.
Genome copy number was measured by real-time PCR as described previously (62). Cells were either mock treated or treated with 10 μM BRACO-19 and harvested 3 days later. Genome copy number was analyzed by quantitative PCR with primers specific to EBV DS or the cellular β-actin gene.
Viability assay and propidium iodide staining.
DG75, BJAB, Raji, LCL3456, and LCL3472 cells were plated at 2.5 × 105 cells per ml, cultured as described above, and supplemented with 10 μM BRACO-19 as indicated. Cells were collected at day 3 and resuspended at a concentration of 2.5 × 105 cells per ml in medium containing 10 μM BRACO-19 for an additional 3 days. After 6 days of treatment, cells were washed twice in 1× phosphate-buffered saline (PBS), resuspended in 80% ethanol, and incubated at −20°C. Cells were collected by centrifugation and resuspended in 1× PBS supplemented with 20 μg/ml propidium iodide (Sigma) and 6 μg/ml RNase A (Sigma). Cells were then analyzed by flow cytometry.
Metaphase spreads and epifluorescence.
Generation of metaphase spreads and epifluorescence have been previously described (33). Briefly, HeLa cells were transfected with either the empty Cherry or green fluorescent protein (GFP) vector or Cherry-EBNA1 (1 to 440) or GFP-FL-EBNA1 constructs. Cherry-transfected cells were collected at 24 h posttransfection, and cells expressing high levels of fluorescence were collected by fluorescence-activated cell sorting. GFP-positive cells were not sorted since they expressed uniformly high levels after transfection (>60% positive). Cells were replated in 0 or 10 μM BRACO-19 and then at 24 h postsorting were treated with 0.1 μg/ml colcemid (Roche) for 24 h. After 72 h posttransfection, cells were stained with 1 μg/ml Hoechst 33342 for 15 min and immediately collected by gentle pipetting, washed once in 1× PBS, resuspended in 75 mM KCl, and incubated for 20 min at room temperature. Cells were affixed to slides by cytocentrifugation (500 rpm, 3 min). Coverslips were affixed to slides immediately with 20% glycerol in 1× PBS and observed for fluorescence.
RESULTS
EBNA1 LRs bind to G-quadruplex RNA.
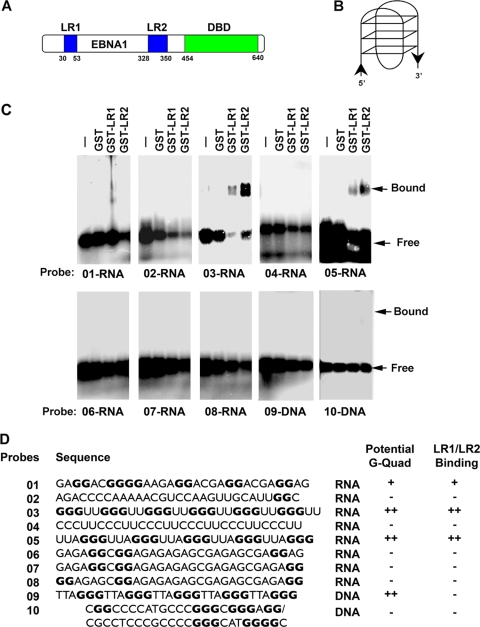
We and others have shown that EBNA1 is able to bind RNA through its LR1 and LR2 domains (30, 37, 51). Our previous studies suggest that EBNA1 binds structured G-rich RNA, but the precise nature of this structure remains unknown. To further investigate the nature of the RNA structures bound by EBNA1, we employed an EMSA using agarose gels to test the abilities of various RNA and DNA probes to bind LR1 or LR2 (Fig. 1). We compared RNA probes that were 30 nucleotides in length and that were predicted to form intramolecular G-quadruplex structures (RNA 01, RNA 03, and RNA 05), were not likely to form intramolecular G-quadruplex structures (RNA 06, RNA 07, and RNA 08), or were G poor (RNA 02 and RNA 04). We also designed a 30-nucleotide G-quadruplex DNA probe (RNA 09) and a G-rich double-stranded DNA probe that could not form an intramolecular G-quadruplex structure (RNA 10). Sequences having at least four separate clusters of two or more guanine nucleotides can form stable intramolecular G-quadruplexes, and stability is increased when the clusters contain three or more guanines and when the loops between the clusters are short (20). We used these considerations to assign G-quadruplex formation potentials for the different probes (Fig. 1B and D).
FIG. 1.
EBNA1 LR1 and LR2 bind to G-quadruplex RNA. (A) Schematic of EBNA1 protein. LR1 and LR2 are shown in blue, and the DBD is represented in green. (B) Representative image of a G-quadruplex structure. (C) The ability of EBNA1 LR1 and LR2 to bind to various RNA and DNA probes was determined by EMSA. Purified GST, GST-LR1, or GST-LR2 was incubated with 32P-labed probes and separated by electrophoresis on agarose gels. Bound and free probes are indicated by arrows. (D) List of probe sequences. All guanines that could potentially contribute to G-quadruplexes are in bold, and the ability of the probe to form a G-quadruplex is indicated. The results of LR1/LR2 EMSA are also indicated.
We found that purified GST-LR1 and GST-LR2 bound preferentially to the RNA probes that have intramolecular G-quadruplex-forming capability (RNA 01, RNA 03, and RNA 05) (Fig. 1C). GST alone did not bind to any of the probes. The binding appears to be better for the canonical G-quadruplex probes RNA 03 and RNA 05, which have three stacks, rather than the less stable two stacks, and shorter loops than the RNA 01 probe. We were unable to detect any binding by GST-LR1 and GST-LR2 to the non-G-quadruplex G-rich RNA (probes RNA 06, RNA 07, and RNA 08), which were scrambled versions of RNA 01, nor do they appear to bind to the G-poor RNA (probes RNA 02 and RNA 04). Furthermore, GST-LR1 and GST-LR2 did not bind to the DNA probes (either the single stranded G-quadruplex DNA [probe DNA 09] or the G-rich double-stranded DNA [probe DNA 10]). This suggests that GST-LR1 and GST-LR2 have a preference for G-rich RNA with predicted G-quadruplex-forming capability.
G-quadruplex-interacting molecules disrupt EBNA1 recruitment of ORC.
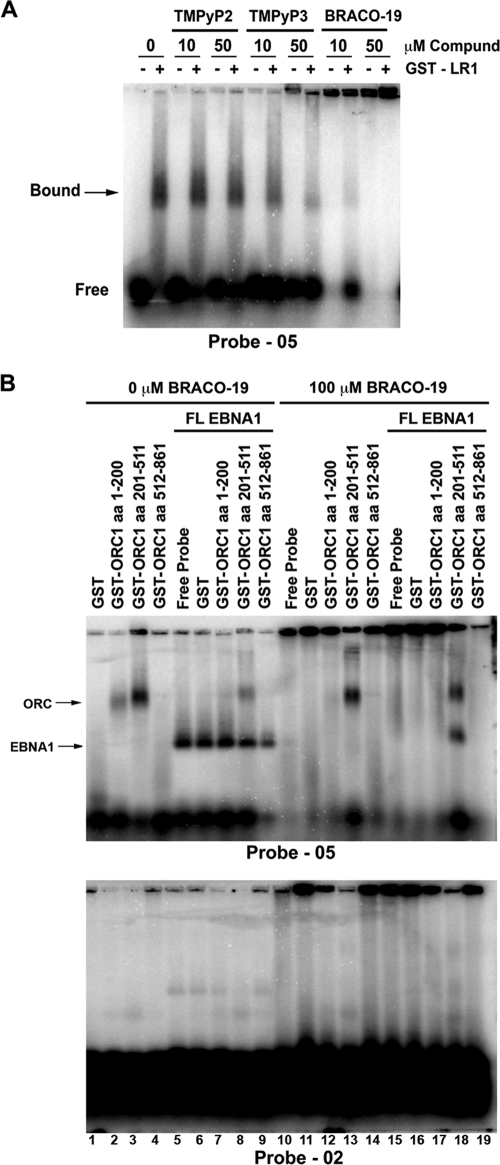
We have previously demonstrated that EBNA1 interaction with ORC can be seen by coimmunoprecipitation from EBV-positive cell lines (37). Furthermore, only the LR1 or LR2 region of EBNA1 is necessary to recruit ORC from HeLa nuclear extract. In both cases, RNA is critical for this interaction, but the role of RNA structure is not fully known. The data in Fig. 1 strongly support the hypothesis that EBNA1 recognizes structured G-quadruplex RNA; however, we wanted to determine if structure was also important in EBNA1 interaction with and recruitment of ORC. Therefore, we used a series of G-quadruplex-specific compounds, TMPyP3, TMPyP4, and BRACO-19, to probe the role of RNA structure in EBNA1 recruitment of ORC. TMPyP3 and TMPyP4 are cationic porphyrins that exhibit specificity for G-quadruplex structures, both DNA and RNA (47). TMPyP4 has been used to study the role of G-quadruplex DNA at both telomeres as well as at the c-Myc promoter. TMPyP2 is a positional isomer of TMPyP3 and TMPyP4 with low specificity for G-quadruplex structures. BRACO-19 is a trisubstituted acridine that also interacts specifically with G-quadruplex structures, both DNA and RNA (41). BRACO-19 has been used to illustrate the role of G-quadruplex DNA at telomeres. These compounds were tested for their ability to interfere with EBNA1 interactions with ORC (Fig. 2).
FIG. 2.
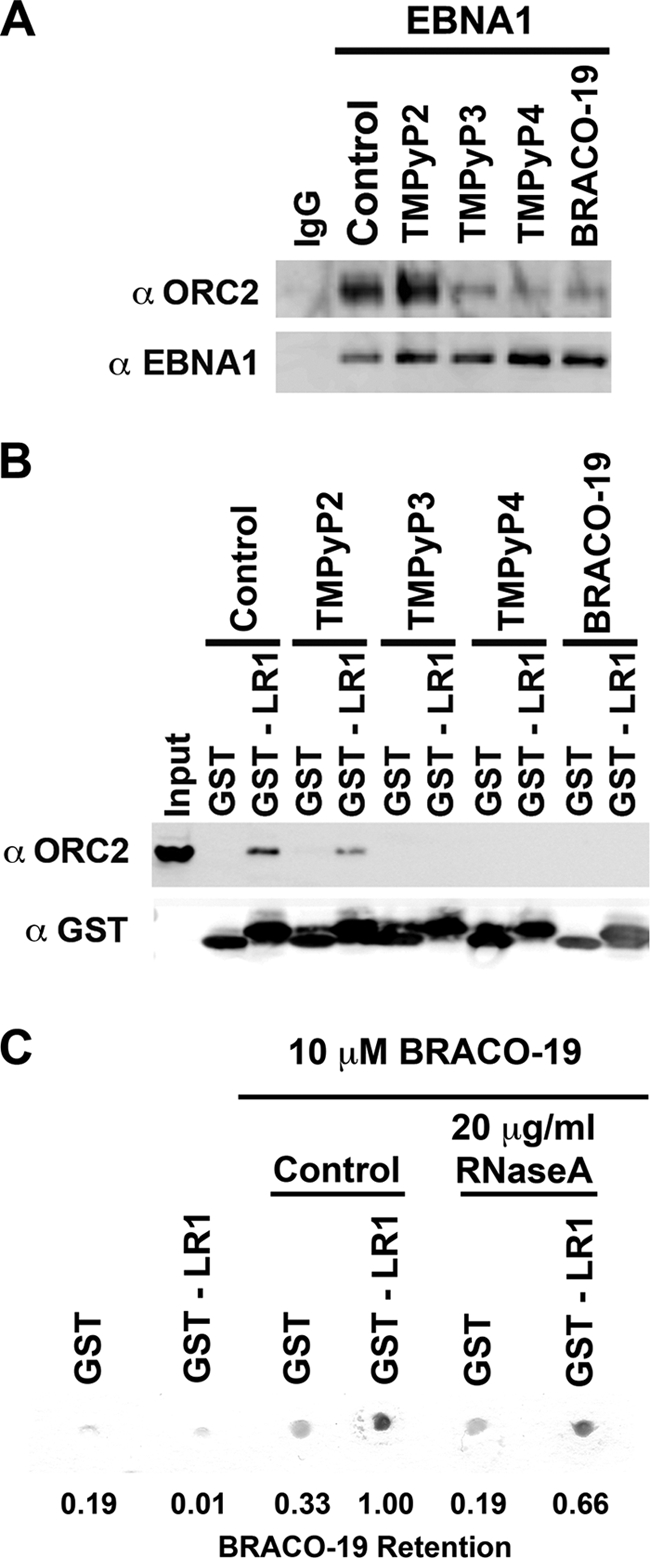
G-quadruplex-interacting compounds disrupt EBNA1 recruitment of ORC. (A) EBNA1 was immunoprecipitated from Raji cells, and the bead-bound complex was washed in the absence or presence of 10 μM G-quadruplex-interacting drugs (TMPyP3, TMPyP4, and BRACO-19) or a noninteracting analogue (TMPyP2) as indicated. Protein was eluted, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and subjected to Western blotting with the indicated antibody. (B) Purified GST or GST-LR1 was bound to glutathione-Sepharose beads and incubated with HeLa nuclear extract. The bead-bound protein complex was washed in the absence or presence of 10 μM G-quadruplex-interacting drugs, as indicated. Associated proteins were determined as for panel A. (C) Purified GST or GST-LR1 was bound to glutathione-Sepharose beads and washed in the absence or presence of 10 μM BRACO-19, with or without 20 μg/ml RNase A, as indicated. The Sepharose beads were then blotted onto Whatman paper and dried. A digital image was taken, and the efficiency of BRACO-19 retention was scored as pigment intensity using ImageQuant. The percent BRACO-19 retention is indicated beneath each lane, normalized to the GST-LR1 control lane in the presence of BRACO-19.
When EBNA1 was immunoprecipitated from EBV-positive Raji cell extracts, ORC subunit ORC2 coimmunoprecipitated (Fig. 2A, control lane). The addition of 10 μM TMPyP2 to the wash buffer did not disrupt ORC2 coimmunoprecipitation by EBNA1. However, the addition of 10 μM G-quadruplex-specific compounds TMPyP3, TMPyP4, and BRACO-19 disrupted the association of ORC2 with EBNA1. We next looked at the ability of GST-LR1 to recruit ORC2 from HeLa nuclear extract in the presence of these compounds. As previously shown, GST-LR1 is able to efficiently recruit ORC2 from HeLa nuclear extract, and 10 μM TMPyP2 did not disrupt this interaction (Fig. 2B). However, the addition of 10 μM TMPyP3, TMPyP4, and BRACO-19 completely disrupted the interaction between EBNA1 LR1 and ORC2. These compounds also disrupted GST-LR2 interaction with ORC2 (data not shown). These findings suggest that EBNA1 interaction with ORC may be mediated by G-quadruplex RNA.
One of the observations during the GST pulldown assay was that the pigmented compounds TMPyP3, TMPyP4, and BRACO-19 appeared to be enriched on the bead-bound LR1 and LR2 peptides in the absence of other cellular proteins. When either GST or GST-LR1 was immobilized on glutathione beads and washed in the presence of 10 μM BRACO-19, there was an enrichment of BRACO-19 compound by GST-LR1 relative to GST alone, as seen by the accumulation of pigmented compound on the beads (Fig. 2C). The addition of 20 μg/ml RNase A partially decreases BRACO-19 association with GST-LR1, by ∼33%. Since the G-quadruplex RNA may be partially resistant to RNases, it may not be surprising that RNase could not eliminate all of the BRACO-19 bound to LR1. These results suggest that LR1 and LR2 bind BRACO-19 through a tight association with G-quadruplex RNA that remained bound to the EBNA1 peptide during the purification from bacterial cells.
We next examined the effects of G-quadruplex-specific drugs on the RNA-binding properties of EBNA1 LR1 in EMSA (Fig. 3A). The addition of either 10 μM or 50 μM TMPyP2 did not alter GST-LR1 binding to a consensus G-quadruplex-forming RNA probe (RNA 05). The addition of 10 μM TMPyP3 had a small inhibitory effect on RNA binding. Addition of 50 μM TMPyP3 caused a significant loss of binding and an accumulation of probe in the well. Similarly, both 10 μM and 50 μM BRACO-19 caused a disruption of LR1 RNA binding in EMSA. The inhibition of RNA binding correlated with the accumulation of the unbound probe in the well. We propose that both TMPyP3 and BRACO-19 bind to the G-quadruplex RNA, causing the probe to aggregate in the well and reduce its ability to migrate into the gel during electrophoresis. The addition of GST-LR1 appeared to increase the solubility of the unbound probe in the presence of the G-quadruplex drugs, suggesting that the GST-LR1 may alter the aggregation induced by TMPyP3 and BRACO-19 binding. GST-LR2 behaved similar to GST-LR1 in these assays (data not shown).
FIG. 3.
G-quadruplex drugs alter binding between EBNA1 and RNA (A) Binding between purified GST-LR1 and the 32P-labeled G-quadruplex probe RNA 05, in the absence or presence of 10 or 50 μM TMPyP2, TMPyP3, or BRACO-19, was measured by EMSA. The protein-bound probe is indicated by an arrow. (B) The ability of ORC1 peptides to bind to either G-quadruplex RNA or G-poor RNA was measured by EMSA. Purified GST, GST-ORC1 aa 1 to 200, GST-ORC1 aa 201 to 511, or GST-ORC1 aa 512 to 861 was incubated with either the 32P-labeled G-quadruplex probe RNA 05 or the 32P-labeled G-poor probe RNA 02 (lanes 1 to 4). Purified FL-EBNA1 was included with these reactions (lanes 5 to 9). The effect of 100 μM BRACO-19 on binding by these complexes was then tested (lanes 10 to 19). ORC1/RNA and EBNA1/RNA complexes are indicated by arrows.
Our previous work had shown that EBNA1 interacts with a specific subdomain of ORC1 (aa 201 to 511) (37). To determine if the interaction between ORC1 aa 201 to 500 and EBNA1 depended on G-quadruplex RNA, we assayed the effects of BRACO-19 on complex formation in EMSA (Fig. 3B). In the absence of BRACO-19, ORC1 aa 201 to 511 bound to the G-quadruplex RNA probe (RNA 05) but not to the G-poor RNA probe (RNA 02) (Fig. 3B, lane 3, upper and lower panels). The BAH-containing domain of ORC1 (aa 1 to 200) also bound weakly to the structured RNA probe, while that ATPase domain (aa 512 to 861) had no detectable binding activity (Fig. 3B, lanes 2 and 4, upper panel). FL-EBNA1 protein bound specifically to the G-quadruplex RNA probe but not to the control probe, as expected (Fig. 3B, lane 5, upper and lower panels). Addition of ORC1 aa 201 to 511 together with EBNA1 produced two major species but did not produce a clear supershift complex, perhaps due to the limiting size of the 30-nucleotide RNA probe (Fig. 3B, lane 8, upper panel). In the presence of 100 μM BRACO-19, binding of ORC1 aa 201 to 511 was not altered but the weaker binding of ORC1 aa 1 to 200 was disrupted (Fig. 3B, lanes 12 and 13, upper panel). Surprisingly, the addition of BRACO-19 abrogated EBNA1 binding to the G-quadruplex RNA in all cases except when ORC1 aa 201 to 511 was also present (Fig. 3B, lane 18, upper panel). This indicates that ORC1 aa 201 to 511 prevents the BRACO-19 induced disruption of RNA binding by EBNA1. One interpretation of this result is that ORC1 aa 201 to 511 competes with BRACO-19 for binding to G-quadruplex RNA.
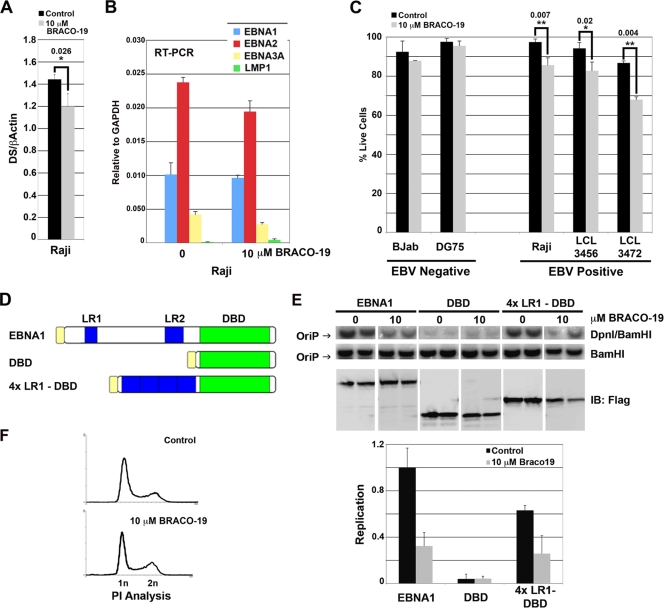
BRACO-19 reduces EBV copy number and inhibits growth of EBV-positive cells.
To investigate the potential role of G-quadruplex RNA on EBNA1 functions in live cells, we first tested the effect of BRACO-19 on EBV genome copy number in latently infected Raji Burkitt lymphoma cells (Fig. 4A). Raji cells were used for this assay because they maintain a high copy number of EBV episomes and are incapable of lytic cycle replication. We found that after 3 days of treatment with 10 μM BRACO-19, there was a small but statistically significant decrease in EBV DNA relative to cellular DNA as measured by quantitative PCR. Longer treatments appeared to reduce Raji cell viability (Fig. 4C). Since EBNA1 can also regulate EBV transcription, we analyzed the effects of BRACO-19 on the mRNA expression for EBNA1, EBNA2, EBNA3A, and LMP1 relative to cellular GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA (Fig. 4B). Quantitative reverse transcription-PCR analysis revealed that BRACO-19 treatment caused a modest decrease (∼20%) in EBNA2 and EBNA3A mRNA expression but no significant change in that of EBNA1 or LMP1 (Fig. 4B).
FIG. 4.
G-quadruplex drugs selectively kill EBV-positive cells and inhibit EBNA1-dependent replication at DS. (A) The copy number of the EBV genome was measured in EBV-positive Raji cells after 3 days of treatment, either mock (control) or 10 μM BRACO-19. Whole-cell DNA was extracted, and quantitative PCR with primers to either DS or β-actin was used to determine the relative ratio of EBV genome to cellular DNA. The unpaired t test was used to determine statistical significance between samples, and * denotes a two-tailed P value of less than 0.026. Error bars indicate standard deviations. (B) Reverse transcription-PCR (RT-PCR) analysis of mRNAs for EBNA1, EBNA2, EBNA3A, and LMP1 relative to cellular GAPDH mRNA in Raji cells treated with 0 or 10 μM BRACO-19 for 3 days. (C) The EBV-negative cell lines BJAB and DG75 and the EBV-positive cell lines Raji, LCL3456, and LCL3472 were cultured in the absence (control) or presence of 10 μM BRACO-19. After 6 days, cells were collected and stained with propidium iodide, and their cell cycle profile was analyzed by flow cytometry. Percentages of live cells are represented by the bar graph. The unpaired t test was used to determine statistical significance between samples and is indicated above the bars. Samples with a two-tailed P value of less than 0.007 are denoted by **, while samples with a two-tailed P value of less than 0.02 are denoted by *. (D) Schematic of constructs used in transient replication assay. The top protein schematic represents Flag-tagged FL-EBNA1. LR1 and LR2 are represented in blue, DBD in green, and the Flag tag in yellow. The middle protein schematic represents Flag-tagged DBD. The bottom protein schematic represents a Flag-tagged fusion peptide with 4×LR1-DBD. (E) Transient-replication assay. The constructs described in panel D were cotransfected into HeLa cells in the absence or presence of 10 μM BRACO-19, and DNA was harvested at 72 h posttransfection. DNA was either linearized with BamHI or digested with DpnI and separated by gel electrophoresis, and a reporter plasmid-specific probe was used to visualize replication. Percent replication was measured by PhosphorImager analysis and quantified by ImageQuant, with replication by FL-EBNA1 set at 100% replication (bottom left panel). (F) Samples of HeLa cells were also stained with propidium iodide (PI) and analyzed by flow cytometry to determine effects of BRACO-19 on cell cycle profile (bottom panel).
We next tested the effect of BRACO-19 on the viability of various EBV-positive and -negative cell lines (Fig. 4C). Cells were cultured for 6 days in the presence of 10 μM BRACO-19 and then analyzed by propidium iodide staining and flow cytometry. We found that there was no significant effect on viability of BJAB and DG75 cells, two EBV-negative B-cell lines (Fig. 4C). However, the three EBV-positive B-cell lines, Raji, LCL3456, and LCL3472, all had decreased viability in the presence of BRACO-19. These findings suggest that BRACO-19 preferentially inhibits viability of a several EBV-positive cell lines.
BRACO-19 inhibits EBNA1-dependent replication of OriP.
We next tested the effects of BRACO-19 on the ability of EBNA1 to stimulate OriP-dependent DNA replication in transient-transfection assays (Fig. 4D to F). We compared FL-EBNA1, the EBNA1 DBD, and 4×LR1-DBD for their abilities to stimulate an OriP-containing reporter plasmid in HeLa cells (Fig. 4D). We had previously shown that the 4×LR1-DBD construct is sufficient to rescue the levels of replication to nearly 70% of the levels of FL-EBNA1 (37). OriP-containing plasmid DNA was harvested from cells at 3 days posttransfection, and replication was measured by Southern blotting for resistance to methylation-specific DpnI digestion. We found that addition of 10 μM BRACO-19 inhibited OriP replication by ∼3-fold for FL-EBNA1, as well as for 4×LR1-DBD (Fig. 4E). EBNA1 DBD alone did not stimulate significant levels of OriP replication, and BRACO-19 did not have any effect on this background activity. In all cases, 10 μM BRACO-19 did not alter the protein expression level relative to that of the untreated control (Fig. 4E, lower panel). Furthermore, 10 μM BRACO-19 did not grossly alter the HeLa cell cycle profile as evidenced by propidium iodide staining and analysis after 3 days of treatment (Fig. 4F). These findings suggest that BRACO-19 preferentially inhibits EBNA1-dependent DNA replication in cells and does not inhibit cell division generally in HeLa cells.
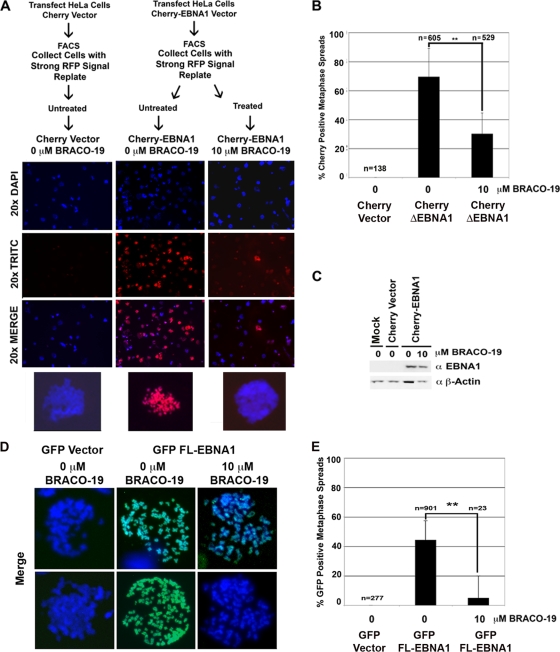
BRACO-19 inhibits EBNA1 metaphase chromosome attachment.
The LR1 and LR2 domains of EBNA1 have also been implicated in metaphase chromosome attachment. To test if structured RNA plays a role in this EBNA1 function, we generated an EBNA1 amino-terminal domain (aa 1 to 440) fusion protein with fluorescent Cherry protein. This Cherry-EBNA1Δ fusion protein (which lacks the DBD but contains LR1 and LR2) was transfected into HeLa cells and compared to the parent Cherry vector in a parallel control. At 24 hours posttransfection, the cells were trypsinized and subjected to fluorescence-activated cell sorting. Cells that expressed high levels of the red fluorescent Cherry fusion protein were collected. These fluorescently positive cells were plated and then treated with either 0 μM (mock treatment) or 10 μM BRACO-19. After 24 h of BRACO-19 treatment, the cells were arrested with colcemid. After 72 h from the initial transfection, cells were examined for epifluorescence of the Cherry proteins and its colocalization to metaphase chromatin. Metaphase spreads were made without fixation to preserve epifluorescence. Representative images are shown in Fig. 5A. As expected, the Cherry peptide alone did not colocalize with metaphase chromatin (Fig. 5A, left column). The Cherry-EBNA1Δ fusion protein containing LR1 and LR2 efficiently attached to metaphase chromatin, and the majority, nearly 70%, of all examined metaphase spreads were positive for EBNA1 colocalization (Fig. 5A middle column, and B). The addition of 10 μM BRACO-19 caused an ∼2-fold reduction in Cherry-EBNA1Δ association with metaphase chromosomes (Fig. 5A, right column, and B). Cherry-EBNA1Δ expression levels were not affected by the addition of BRACO-19 to the culture medium (Fig. 5C).
FIG. 5.
EBNA1 attachment to metaphase chromosomes is disrupted by G-quadruplex compounds (A) HeLa cells were transfected with either Cherry-EBNA1 (aa 1\ to 440) (denoted as Cherry-EBNA1Δ) or Cherry vector alone, sorted for red fluorescent protein (RFP) signal, replated, and then treated with 0 or 10 μM BRACO-19. The experimental design is shown in the schematic. Representative images of DNA staining (blue), Cherry protein localization (red), or the merge image are shown. Beneath each column a merge image of one of the metaphase spreads from the field is enlarged for detail. (B) Percentage of nuclei that scored as being positive for Cherry protein colocalization to metaphase spreads. n represents the number of metaphase spreads scored for each condition. The unpaired t test was used to determine statistical significance between treated and untreated Cherry-EBNA1 and is indicated above the bars. Samples with a two-tailed P value of 0.0001 are denoted by **. Error bars indicate standard deviations. (C) To ensure that 10 μM BRACO-19 did not alter Cherry-EBNA1 expression, protein samples were collected and examined with antibody against either EBNA1 or β-actin by Western blotting. (D) GFP-FL-EBNA1 attachment to metaphase chromosomes in the presence of 0 or 10 μM BRACO-19 was also examined. HeLa cells were transfected with either GFP vector or GFP-FL-EBNA1. Metaphase spreads were examined by epifluorescence for GFP association with metaphase chromosomes. Representative metaphase spreads showing the merged image of DNA (blue) and GFP protein colocalization (green) have been enlarged for detail. (E) Quantification of nuclei from the experiment shown in panel D. n represents the number of metaphase spreads scored for each condition. The unpaired t test was used to determine statistical significance between treated and untreated GFP-FL-EBNA1 and is indicated above the bars. Samples with a two-tailed P value of 0.0001 are denoted by **.
To eliminate any concerns that Cherry-EBNA1Δ behaves differently than FL-EBNA1 in metaphase chromosome attachment, we repeated the metaphase attachment assay using GFP-FL-EBNA1 (Fig. 5D and E). We observed that ∼50% of HeLa cells scored positive for GFP-FL-EBNA1 at 24 h posttransfection. Transfected cells were then treated with 0 or 10 μM BRACO-19 and assayed for association with metaphase chromosomes using the same methods as described for Cherry-EBNA1Δ. No GFP localized to metaphase chromosomes in control pEGFP-transfected cells (Fig. 5D, left column). GFP-FL-EBNA1-transfected cells that were mock treated scored ∼50% positive for colocalization with metaphase chromosomes. In contrast, GFP-FL-EBNA1-transfected cells treated with 10 μM BRACO-19 scored ∼10% positive (Fig. 5D and E). These results indicate that BRACO-19 treatment caused a fivefold reduction in GFP-FL-EBNA1 colocalization to metaphase chromosomes. Taken together, these results demonstrate that BRACO-19 treatment can reduce metaphase attachment of EBNA1Δ, as well as FL-EBNA1, suggesting that it disrupts the tethering function of LR1 and LR2.
DISCUSSION
In this work, we investigated the role of EBNA1 RNA binding and found that EBNA1 binds preferentially to RNA capable of forming G-quadruplex structures. The contributions of G-quadruplexes to cellular biology are becoming increasingly appreciated. DNA G-quadruplex structures have been identified as being important in telomere biology as regulators of telomerase activity (39, 61). Recent studies also have shown the emerging importance of G-quadruplexes in promoters as an added layer of transcriptional regulation (reviewed in reference 40). While there are fewer cases of RNA G-quadruplexes, one well-characterized interaction is between the RGG-like protein FMRP and RNA G-quadruplexes (10). FMRP binds to sites within its own transcripts that form G-quadruplexes. It has been demonstrated that FMRP binding to its own transcript acts as an exonic splicing factor rather than as a translational inhibitor (14). However, there are cases of translational inhibition by G-quadruplexes, as seen in the 5′ untranslated region of the N-ras gene (27).
One of the goals of this study was to determine if EBNA1 bound to RNA in a sequence- or structure-specific manner. In EMSA, EBNA1 LR1 and LR2 bound most efficiently to RNAs that were predicted to form the most stable G-quadruplex structures. To be sure that EBNA1 binding was driven by G-quadruplex structure and not guanine content, three scrambled versions of the G-quadruplex probe RNA 01 were generated (RNA 05, RNA 06, and RNA 07). These probes are unlikely to form G-quadruplexes and did not bind EBNA1. LR1 and LR2 had no detectable binding affinity for G-quadruplex DNA, indicating that only G-quadruplex RNA is recognized by EBNA1. G-quadruplex RNA is likely to be found among many RNA species in human cells. Interestingly, EBNA1 mRNA sequences that encode LR1 and LR2 are predicted to form G-quadruplex structures, and our EMSAs, shown here and previously, indicate that EBNA1 can bind with moderate affinity to its own transcript (RNA 01) (37). While other G-quadruplex RNAs may mediate interactions with replication factors, like ORC1, and metaphase tethering targets, EBNA1 may also regulate its own mRNA stability, processing, or translational efficiency through LR1 or LR2 binding to the EBNA1 mRNA.
G-quadruplex-interacting molecules were used to further investigate the biochemical and functional properties of EBNA1. We found that the G-quadruplex-specific compounds TMPyP3, TMPyP4, and BRACO-19 interfered with EBNA1 recruitment of ORC from nuclear extracts (Fig. 2). Also, in EMSA studies, the G-quadruplex-interacting compounds interfered with recombinant GST-LR1 (and LR2 [data not shown]) binding to G-quadruplex RNA oligonucleotides (Fig. 1). This appeared to result from the direct binding of these compounds with the RNA, potentially competing with EBNA1 for binding. However, in the absence of exogenous RNA, GST-LR1 was capable of binding to BRACO-19 (Fig. 2C). It is possible that GST-LR1 binds with high avidity to bacterial G-quadruplex RNA during the purification from Escherichia coli and this may account for the interaction with BRACO-19. We also observed that ORC1 (aa 200 to 511) peptide could bind G-quadruplex RNA (Fig. 3). Addition of ORC1 aa 200 to 511 to GST-LR1 prevented the disruptive effects of BRACO-19. The physical basis for these observations is not immediately clear, but one possibility is that ORC1 competes with BRACO-19 for access to G-quadruplex RNA. Combining these results with those of the BRACO-19 retention assay, we hypothesize that the G-quadruplex drugs may stabilize a G-quadruplex RNA conformation that is different from the conformation recognized by EBNA1 or ORC1. Disruption of the interaction between EBNA1 and ORC may be due to this conformation stability or to steric interference that prevents EBNA1 and ORC1 from binding the same G-quadruplex RNA species.
Aside from demonstrating that EBNA1 binds structured RNA, we were interested in the role of G-quadruplex RNA in EBNA1 function during infection. Our first observation was that BRACO-19 treatment moderately reduced EBV genome copy number in Raji cells (Fig. 4A). BRACO-19 treatment also had a modest inhibitory effect on transcription levels of EBNA2 and EBNA3A (Fig. 4B), suggesting that G-quadruplex RNA may be involved in EBNA1 transcription activation functions. Longer-term treatment with BRACO-19 led to a loss of cell viability, with EBV-positive cells showing greater sensitivity to BRACO-19 than EBV-negative cell lines (Fig. 4C). Because G-quadruplexes are involved in various cellular functions, including telomere lengthening by telomerase, it is not surprising that G-quadruplex-interacting compounds inhibit cell viability at some concentration. However, the increased sensitivity of EBV-positive cells to BRACO-19 treatment suggests that there may be a therapeutic window through which they BRACO-19 may preferentially inhibit EBV-specific enhancement of cell viability.
BRACO-19 was also found to inhibit EBNA1-dependent DNA replication (Fig. 4E). The ability of the G-quadruplex-interacting compound BRACO-19 to inhibit ORC recruitment and EBNA1-dependent DNA replication supports the general model that ORC recruitment by EBNA1 is necessary for replication initiation at OriP. Since EBV genomes can initiate replication outside of OriP at some frequency (35, 36), it is not clear whether the loss of ORC recruitment or OriP-dependent DNA replication is sufficient to account for the loss of viability of EBV-positive cells. BRACO-19 may also disrupt EBNA1 interactions with other cellular factors, including transcription factors necessary for EBNA2 activation (Fig. 4B) or factors involved in episome maintenance, such as EBP2. Thus, in addition to ORC recruitment, it is possible that other functional interactions mediated by G-quadruplex RNA are disrupted by BRACO-19 treatment.
We also show that BRACO-19 is a potent inhibitor of EBNA1 metaphase chromosome attachment (Fig. 5). Metaphase attachment has been mapped to the LR1 and LR2 regions of EBNA1, which we have shown can bind to RNA that has the ability to form G-quadruplexes in vitro. The fact that BRACO-19 can disrupt EBNA1 interaction with metaphase chromosomes suggests that G-quadruplex RNA mediates some aspects of this process. Although several subunits of ORC are known to associate with metaphase chromosomes, ORC1 has been shown to be degraded after the completion of S phase (29). Thus, it is unlikely that ORC1 is a metaphase chromosome receptor protein for EBNA1 docking. Other cellular proteins, such as EBP2, have been strongly implicated in EBNA1 metaphase chromosome attachment (23, 24). Interestingly, EBP2 orthologues in yeast have functions in rRNA processing (18), suggesting that RNA may regulate some of EBP2 activities or subcellular localization. It will be interesting to determine if EBP2 interaction with EBNA1 can be regulated by RNA, especially if that RNA is a G-quadruplex species. The metaphase attachment function of LR1 and LR2 may also involve direct interactions with AT-rich DNA. Both LR1 and LR2 can be replaced by the AT hook domain of the histone H1 or HMGA1a (19, 44, 45). Our previous study indicated that the HMGA1a protein could bind G-rich RNA, similar to LR1 and LR2. Thus, it remains possible that metaphase chromosome attachment is mediated by RNA G-quadruplex interactions, in addition to the AT-rich DNA and protein receptors that physically associate with metaphase chromosomes.
In conclusion, we have found that EBNA1 LR1 and LR2 have a strong preference for G-quadruplex RNA and that G-quadruplex RNA-interacting drugs block functions of EBNA1 critical for viral DNA replication and episome maintenance. The common requirement for G-quadruplex RNA in these two seemingly different activities of EBNA1 is not completely understood. The G-quadruplex RNA may regulate EBNA1 function by altering target interaction specificity. The identities of the endogenous RNA molecules that bind EBNA1 LR1 and LR2 in vivo have not yet been determined. Identification of these RNAs may provide additional insights into the mechanisms regulating EBNA1 functions in replication and episome maintenance. They may also provide instruction for the design of more specific interacting compounds that can serve as small-molecule inhibitors of EBNA1 function during latent infection.
Acknowledgments
We acknowledge the essential contribution of the Wistar Microscopy Core and Wistar Flow Cytometry Core. We thank C. Meyers (Fox Chase Cancer Center) for synthesis of BRACO-19.
J.N. is supported by a training grant in virology at the University of Pennsylvania (T32-A107324-17). This work was supported by NIH grant CA093606 to P.M.L.
Footnotes
Published ahead of print on 5 August 2009.
REFERENCES
- 1.Altmann, M., D. Pich, R. Ruiss, J. Wang, B. Sugden, and W. Hammerschmidt. 2006. Transcriptional activation by EBV nuclear antigen 1 is essential for the expression of EBV's transforming genes. Proc. Natl. Acad. Sci. USA 103:14188-14193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atanasiu, C., Z. Deng, A. Wiedmer, J. Norseen, and P. M. Lieberman. 2006. ORC binding to TRF2 stimulates OriP replication. EMBO Rep. 7:716-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avolio-Hunter, T. M., and L. Frappier. 1998. Mechanistic studies on the DNA linking activity of Epstein-Barr nuclear antigen 1. Nucleic Acids Res. 26:4462-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bashaw, J. M., and J. L. Yates. 2001. Replication from oriP of Epstein-Barr virus requires exact spacing of two bound dimers of EBNA1 which bend DNA. J. Virol. 75:10603-10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bochkarev, A., J. A. Barwell, R. A. Pfuetzner, E. Bochkareva, L. Frappier, and A. M. Edwards. 1996. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein, EBNA1, bound to DNA. Cell 84:791-800. [DOI] [PubMed] [Google Scholar]
- 6.Burd, C. G., and G. Dreyfuss. 1994. Conserved structures and diversity of functions of RNA-binding proteins. Science 265:615-621. [DOI] [PubMed] [Google Scholar]
- 7.Burger, A. M., F. Dai, C. M. Schultes, A. P. Reszka, M. J. Moore, J. A. Double, and S. Neidle. 2005. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 65:1489-1496. [DOI] [PubMed] [Google Scholar]
- 8.Chaudhuri, B., H. Xu, I. Todorov, A. Dutta, and J. L. Yates. 2001. Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 98:10085-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins, C. M., and P. G. Medveczky. 2002. Genetic requirements for the episomal maintenance of oncogenic herpesvirus genomes. Adv. Cancer Res. 84:155-174. [DOI] [PubMed] [Google Scholar]
- 10.Darnell, J. C., K. B. Jensen, P. Jin, V. Brown, S. T. Warren, and R. B. Darnell. 2001. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell 107:489-499. [DOI] [PubMed] [Google Scholar]
- 11.Deng, Z., C. Atanasiu, J. S. Burg, D. Broccoli, and P. M. Lieberman. 2003. Telomere repeat binding factors TRF1, TRF2, and hRAP1 modulate replication of Epstein-Barr virus OriP. J. Virol. 77:11992-12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng, Z., L. Lezina, C. J. Chen, S. Shtivelband, W. So, and P. M. Lieberman. 2002. Telomeric proteins regulate episomal maintenance of Epstein-Barr virus origin of plasmid replication. Mol. Cell 9:493-503. [DOI] [PubMed] [Google Scholar]
- 13.Dhar, S. K., K. Yoshida, Y. Machida, P. Khaira, B. Chaudhuri, J. A. Wohlschlegel, M. Leffak, J. Yates, and A. Dutta. 2001. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell 106:287-296. [DOI] [PubMed] [Google Scholar]
- 14.Didiot, M. C., Z. Tian, C. Schaeffer, M. Subramanian, J. L. Mandel, and H. Moine. 2008. The G-quartet containing FMRP binding site in FMR1 mRNA is a potent exonic splicing enhancer. Nucleic Acids Res. 36:4902-4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldsmith, K., L. Bendell, and L. Frappier. 1993. Identification of EBNA1 amino acid sequences required for the interaction of the functional elements of the Epstein-Barr virus latent origin of DNA replication. J. Virol. 67:3418-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grasser, F. A., P. G. Murray, E. Kremmer, K. Klein, K. Remberger, W. Feiden, G. Reynolds, G. Niedobitek, L. S. Young, and N. Mueller-Lantzsch. 1994. Monoclonal antibodies directed against the Epstein-Barr virus-encoded nuclear antigen 1 (EBNA1): immunohistologic detection of EBNA1 in the malignant cells of Hodgkin's disease. Blood 84:3792-3798. [PubMed] [Google Scholar]
- 17.Hanakahi, L. A., H. Sun, and N. Maizels. 1999. High affinity interactions of nucleolin with G-G-paired rDNA. J. Biol. Chem. 274:15908-15912. [DOI] [PubMed] [Google Scholar]
- 18.Huber, M. D., J. H. Dworet, K. Shire, L. Frappier, and M. A. McAlear. 2000. The budding yeast homolog of the human EBNA1-binding protein 2 (Ebp2p) is an essential nucleolar protein required for pre-rRNA processing. J. Biol. Chem. 275:28764-28773. [DOI] [PubMed] [Google Scholar]
- 19.Hung, S. C., M. S. Kang, and E. Kieff. 2001. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc. Natl. Acad. Sci. USA 98:1865-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huppert, J. L. 2008. Four-stranded nucleic acids: structure, function and targeting of G-quadruplexes. Chem. Soc. Rev. 37:1375-1384. [DOI] [PubMed] [Google Scholar]
- 21.Izbicka, E., R. T. Wheelhouse, E. Raymond, K. K. Davidson, R. A. Lawrence, D. Sun, B. E. Windle, L. H. Hurley, and D. D. Von Hoff. 1999. Effects of cationic porphyrins as G-quadruplex interactive agents in human tumor cells. Cancer Res. 59:639-644. [PubMed] [Google Scholar]
- 22.Kanda, T., M. Kamiya, S. Maruo, D. Iwakiri, and K. Takada. 2007. Symmetrical localization of extrachromosomally replicating viral genomes on sister chromatids. J. Cell Sci. 120:1529-1539. [DOI] [PubMed] [Google Scholar]
- 23.Kapoor, P., B. D. Lavoie, and L. Frappier. 2005. EBP2 plays a key role in Epstein-Barr virus mitotic segregation and is regulated by aurora family kinases. Mol. Cell. Biol. 25:4934-4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kapoor, P., K. Shire, and L. Frappier. 2001. Reconstitution of Epstein-Barr virus-based plasmid partitioning in budding yeast. EMBO J. 20:222-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kennedy, G., and B. Sugden. 2003. EBNA-1, a bifunctional transcriptional activator. Mol. Cell. Biol. 23:6901-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiledjian, M., and G. Dreyfus. 1992. Primary structure and binding activity of hnRNP U protein: binding RNA through RGG box. EMBO J. 11:2655-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumari, S., A. Bugaut, J. L. Huppert, and S. Balasubramanian. 2007. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 3:218-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, M. A., M. E. Diamond, and J. L. Yates. 1999. Genetic evidence that EBNA-1 is needed for efficient, stable latent infection by Epstein-Barr virus. J. Virol. 73:2974-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, C. J., and M. L. DePamphilis. 2002. Mammalian Orc1 protein is selectively released from chromatin and ubiquitinated during the S-to-M transition in the cell division cycle. Mol. Cell. Biol. 22:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu, C. C., C. W. Wu, S. C. Chang, T. Y. Chen, C. R. Hu, M. Y. Yeh, J. Y. Chen, and M. R. Chen. 2004. Epstein-Barr virus nuclear antigen 1 is a DNA-binding protein with strong RNA-binding activity. J. Gen. Virol. 85:2755-2765. [DOI] [PubMed] [Google Scholar]
- 31.Mackey, D., T. Middleton, and B. Sugden. 1995. Multiple regions within EBNA1 can link DNAs. J. Virol. 69:6199-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackey, D., and B. Sugden. 1999. The linking regions of EBNA1 are essential for its support of replication and transcription. Mol. Cell. Biol. 19:3349-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marechal, V., A. Dehee, R. Chikhi-Brachet, T. Piolot, M. Coppey-Moisan, and J. C. Nicolas. 1999. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J. Virol. 73:4385-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nanbo, A., A. Sugden, and B. Sugden. 2007. The coupling of synthesis and partitioning of EBV's plasmid replicon is revealed in live cells. EMBO J. 26:4252-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norio, P., and C. L. Schildkraut. 2004. Plasticity of DNA replication initiation in Epstein-Barr virus episomes. PLoS Biol. 2:e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Norio, P., and C. L. Schildkraut. 2001. Visualization of DNA replication on individual Epstein-Barr virus episomes. Science 294:2361-2364. [DOI] [PubMed] [Google Scholar]
- 37.Norseen, J., A. Thomae, V. Sridharan, A. Aiyar, A. Schepers, and P. M. Lieberman. 2008. RNA-dependent recruitment of the origin recognition complex. EMBO J. 27:3024-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parkinson, G. N., M. P. Lee, and S. Neidle. 2002. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 417:876-880. [DOI] [PubMed] [Google Scholar]
- 39.Phillips, K., Z. Dauter, A. I. Murchie, D. M. Lilley, and B. Luisi. 1997. The crystal structure of a parallel-stranded guanine tetraplex at 0.95 A resolution. J. Mol. Biol. 273:171-182. [DOI] [PubMed] [Google Scholar]
- 40.Qin, Y., and L. H. Hurley. 2008. Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie 90:1149-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Read, M., R. J. Harrison, B. Romagnoli, F. A. Tanious, S. H. Gowan, A. P. Reszka, W. D. Wilson, L. R. Kelland, and S. Neidle. 2001. Structure-based design of selective and potent G quadruplex-mediated telomerase inhibitors. Proc. Natl. Acad. Sci. USA 98:4844-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ritzi, M., K. Tillack, J. Gerhardt, E. Ott, S. Humme, E. Kremmer, W. Hammerschmidt, and A. Schepers. 2003. Complex protein-DNA dynamics at the latent origin of DNA replication of Epstein-Barr virus. J. Cell Sci. 116:3971-3984. [DOI] [PubMed] [Google Scholar]
- 43.Schepers, A., M. Ritzi, K. Bousset, E. Kremmer, J. L. Yates, J. Harwood, J. F. Diffley, and W. Hammerschmidt. 2001. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 20:4588-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sears, J., J. Kolman, G. M. Wahl, and A. Aiyar. 2003. Metaphase chromosome tethering is necessary for the DNA synthesis and maintenance of oriP plasmids but is insufficient for transcription activation by EBNA1. J. Virol. 77:11767-11780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sears, J., M. Ujihara, S. Wong, C. Ott, J. Middeldorp, and A. Aiyar. 2004. The amino terminus of Epstein-Barr virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J. Virol. 78:11487-11505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shammas, M. A., R. J. Shmookler Reis, M. Akiyama, H. Koley, D. Chauhan, T. Hideshima, R. K. Goyal, L. H. Hurley, K. C. Anderson, and N. C. Munshi. 2003. Telomerase inhibition and cell growth arrest by G-quadruplex interactive agent in multiple myeloma. Mol. Cancer Ther. 2:825-833. [PubMed] [Google Scholar]
- 47.Shi, D. F., R. T. Wheelhouse, D. Sun, and L. H. Hurley. 2001. Quadruplex-interactive agents as telomerase inhibitors: synthesis of porphyrins and structure-activity relationship for the inhibition of telomerase. J. Med. Chem. 44:4509-4523. [DOI] [PubMed] [Google Scholar]
- 48.Shire, K., D. F. Ceccarelli, T. M. Avolio-Hunter, and L. Frappier. 1999. EBP2, a human protein that interacts with sequences of the Epstein-Barr virus nuclear antigen 1 important for plasmid maintenance. J. Virol. 73:2587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siddiqui-Jain, A., C. L. Grand, D. J. Bearss, and L. H. Hurley. 2002. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA 99:11593-11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simonsson, T., and M. Henriksson. 2002. c-Myc suppression in Burkitt's lymphoma cells. Biochem. Biophys. Res. Commun. 290:11-15. [DOI] [PubMed] [Google Scholar]
- 51.Snudden, D. K., J. Hearing, P. R. Smith, F. A. Grasser, and B. E. Griffin. 1994. EBNA-1, the major nuclear antigen of Epstein-Barr virus, resembles ‘RGG’ RNA binding proteins. EMBO J. 13:4840-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sugden, B., K. Marsh, and J. Yates. 1985. A vector that replicates as a plasmid and can be efficiently selected in B lymphocytes transformed by Epstein-Barr virus. Mol. Cell. Biol. 5:410-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomae, A. W., D. Pich, J. Brocher, M. P. Spindler, C. Berens, R. Hock, W. Hammerschmidt, and A. Schepers. 2008. Interaction between HMGA1a and the origin recognition complex creates site-specific replication origins. Proc. Natl. Acad. Sci. USA 105:1692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Scoy, S., I. Watakabe, A. R. Krainer, and J. Hearing. 2000. Human p32: a coactivator for Epstein-Barr virus nuclear antigen-1-mediated transcriptional activation and possible role in viral latent cycle DNA replication. Virology 275:145-157. [DOI] [PubMed] [Google Scholar]
- 55.Wang, Y., J. E. Finan, J. M. Middledorp, and S. D. Hayward. 1997. P32/TAP, a cellular protein that interacts with EBNA-1 of Epstein-Barr virus. Virology 236:18-29. [DOI] [PubMed] [Google Scholar]
- 56.Wu, H., P. Kapoor, and L. Frappier. 2002. Separation of the DNA replication, segregation, and transcriptional activation functions of Epstein-Barr nuclear antigen 1. J. Virol. 76:2480-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wysokenski, D. A., and J. L. Yates. 1989. Multiple EBNA1-binding sites are required to form an EBNA1-dependent enhancer and to activate a minimal replicative origin within oriP of Epstein-Barr virus. J. Virol. 63:2657-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yates, J. L., N. Warren, P. Reisman, and B. Sugden. 1984. A cis-acting element from Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc. Natl. Acad. Sci. USA 81:3806-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yates, J. L., N. Warren, and B. Sugden. 1985. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 313:812-815. [DOI] [PubMed] [Google Scholar]
- 60.Young, L. S., and A. B. Rickinson. 2004. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4:757-768. [DOI] [PubMed] [Google Scholar]
- 61.Zahler, A. M., J. R. Williamson, T. R. Cech, and D. M. Prescott. 1991. Inhibition of telomerase by G-quartet DNA structures. Nature 350:718-720. [DOI] [PubMed] [Google Scholar]
- 62.Zhou, J., A. Snyder, and P. M. Lieberman. 2008. Epstein-Barr virus episome stability is coupled to a delay in replication timing. J. Virol. 83:2154-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]