Abstract
Human immunodeficiency virus type 1 (HIV-1) group M viruses have achieved a global distribution, while HIV-1 group O viruses are endemic only in particular regions of Africa. Here, we evaluated biological characteristics of group O and group M viruses in ex vivo models of HIV-1 infection. The replicative capacity and ability to induce CD4 T-cell depletion of eight group O and seven group M primary isolates were monitored in cultures of human peripheral blood mononuclear cells and tonsil explants. Comparative and longitudinal infection studies revealed HIV-1 group-specific activity patterns: CCR5-using (R5) viruses from group M varied considerably in their replicative capacity but showed similar levels of cytopathicity. In contrast, R5 isolates from group O were relatively uniform in their replicative fitness but displayed a high and unprecedented variability in their potential to deplete CD4 T cells. Two R5 group O isolates were identified that cause massive depletion of CD4 T cells, to an extent comparable to CXCR4-using viruses and not documented for any R5 isolate from group M. Intergroup comparisons found a five- to eightfold lower replicative fitness of isolates from group O than for isolates from group M yet a similar overall intrinsic pathogenicity in tonsil cultures. This study establishes biological ex vivo characteristics of HIV-1 group O primary isolates. The current findings challenge the belief that a grossly reduced replicative fitness or inherently impaired cytopathicity of viruses from this group underlies their low global prevalence.
Independent cross-species transmission events from simian immunodeficiency virus-infected apes have led to four distinct phylogenetic lineages of human immunodeficiency virus (HIV) in humans (45). The main (M) group of HIV type 1 (HIV-1) is responsible for the HIV pandemic, while HIV-1 group O (outlier) and HIV-2 are endemic only in west and central Africa, and HIV-1 group N (non-M/non-O) infection has been documented only in a small number of Cameroonians (56). These cross-species transmissions are believed to have occurred in western Africa around the same time, but only HIV-1 group M founded the pandemic (33, 37).
The global distribution of HIV-1 group O is remarkably restricted. The relative seroprevalence of group O is reported to be highest in the Republic of Cameroon, Equatorial Guinea, and Gabon (7, 42, 57), implicating this area as the possible starting point of this HIV-1 lineage's epidemic. Rare group O infections have been documented in industrialized countries, the majority comprising patients of Cameroonian descent (8, 25, 30, 40, 46). Notably, the prevalence of group O among HIV-1-positive blood samples in Cameroon showed a marked decline from the period 1986 to 1988 (20.6% of all HIV-1 infections) to the period 1997 to 1998 (1.4%) (7) with evidence of a low, but stabilized, prevalence in the subsequent period up to 2004 (10, 55). Primary isolates from group O and group M display pronounced genetic differences (24, 54), yet the reasons for the decreasing prevalence of HIV-1 group O relative to group M in west Africa and the almost exclusive contribution of group M to the AIDS pandemic are unclear. Many factors could, in principle, have contributed to this variable spread through the human population, including host genetic effects, transmission bottlenecks, behavioral and environmental restrictions, founder effects, and other factors (33, 53).
Clinical observations do not suggest major differences in disease progression in patients infected with HIV-1 groups O and M (23, 24, 35, 39). This notion is based on limited data on the immune status and virological parameters for group O-infected individuals. Few experimental in vitro studies have compared the replicative fitness of HIV types or groups (1, 2, 50, 52, 54). In head-to-head replication competition experiments of pairs of primary isolates from group M and group O in peripheral blood mononuclear cell (PBMC) cultures, Arien et al. reported a greater than 100-fold reduced replicative fitness of group O viruses (2). They suggested that grossly reduced “ex vivo pathogenic fitness” and impaired transmission from dendritic cells to cocultured T cells (“ex vivo transmission fitness”) are intrinsic properties of group O viruses that may contribute to their low prevalence and limited geographical spread (2, 3).
Here, we evaluated characteristics of a panel of primary isolates from HIV-1 group O compared to a panel from group M in three primary cell models of HIV infection. In addition to replication studies in single-donor PBMCs used in a previous fitness study (2), we employed multidonor pools of PBMCs and an ex vivo human tonsil lymphoid aggregate culture (HLAC) model. HIV readily replicates to high titers in tonsil cultures that maintain the cell composition and cytokine milieu of a lymphoid target organ in vivo (17). Previously, studies in this model have shed light on key pathogenic properties of HIV, including cell tropism and cytopathic effects in relation to coreceptor usage, productive infection of resting CD4 T cells, early host responses to infection, and viral coinfections (5, 6, 14, 18-20, 27, 38, 43, 48-50). A unique characteristic of this ex vivo model is that it allows parallel assessment of an isolate's replicative fitness and cytopathicity, the latter determined by its ability to deplete CD4 T cells. The current investigation may enhance our understanding of parameters critical for HIV-1 spread in the human population and could thus potentially also provide clues to prevention and therapy.
MATERIALS AND METHODS
Viral stocks.
The molecular HIV-1 group M clones pNL4-3, p49.5, pYU-2, and pJR-FL were obtained from the NIH AIDS Research and Reference Reagent Program, and virus stocks were generated by calcium-phosphate transfection of 293T cells. HIV-1 Ba-L stocks were from ABI (Columbia, MD). The origin and epidemiological information of HIV-1 group O isolates from Cameroonian patients have been reported previously (41). HIV-1 group infection was confirmed by an HIV-1/HIV-2 plus O enzyme-linked immunosorbent assay (ELISA), competitive HIV-1 ELISA, HIV-1 group O virus immunoblotting (24), and genotyping (Lutz Gürtler, personal communication). The following group O isolates were provided by Gürtler (brackets indicate isolates' abbreviated nomenclature used in figures and text): MVP 2901-94 (2901), MVP 1639-96 (1639), MVP 2549-95 (2549), MVP 6778-94 (6778), MVP 8913-95 (8913), MVP 9435-96 (9435), MVP 13127-96 (13127), and MVP 13740-96 (13740). Group M primary isolates from different subtypes were provided by Valerie Bosch via the NIH AIDS Research and Reference Reagent Program: RW9 (subtype A); 2005, 2044, and D117 (subtype B); IN22 (subtype C); ELI (subtype D); and RU570 (subtype G) (Table 1). All primary isolates were expanded on the same four-donor pool of phytohemagglutinin P/interleukin-2 blasted PBMCs. Close to peak replication, supernatants were harvested, concentrated, and stored in aliquots at −80°C. The p24 concentration and 50% tissue culture infective dose (TCID50) (32) were determined for each stock.
TABLE 1.
Classification and characterization of virus stocks of HIV-1 group M and group O isolates used in this study
| Virus isolate | Group | Subtype | Coreceptor usagea | p24 CA level (ng/ml)b | TCID50 (IU/ml)c | Infectivity (IU/ng of p24 CA)d |
|---|---|---|---|---|---|---|
| NL4-3 | M | B | X4 | 35,144 | 68,080 | 1.94 |
| 2005 | M | B | X4 | 3,827 | 1,540 | 0.40 |
| 2044 | M | B | X4 | 1,743 | 1,778 | 1.02 |
| ELI | M | D | X4 | 4,570 | 178 | 0.04 |
| JR-FL | M | B | R5 | 4,315 | 41,248 | 9.56 |
| Ba-L | M | B | R5 | 9,400 | 316,228 | 33.64 |
| IN22 | M | C | R5 | 1,448 | 2,610 | 1.80 |
| RU570 | M | G | R5 | 442 | 562 | 1.27 |
| RW9 | M | A | R5 | 440 | 1,000 | 2.27 |
| D117 | M | B | R5 | 2,870 | 1,778 | 0.62 |
| 2901 | O | X4 | 55 | 1,540 | 28.00 | |
| 1639 | O | R5 | 56 | 562 | 10.04 | |
| 2549 | O | R5 | 117 | 562 | 4.80 | |
| 6778 | O | R5 | 405 | 178 | 0.44 | |
| 8913 | O | R5 | 58 | 178 | 3.07 | |
| 13127 | O | R5 | 193 | 3,162 | 16.38 | |
| 13740 | O | R5 | 124 | 562 | 4.53 | |
| 9435 | O | R5 | 500 | ND | ND |
Coreceptor usage was determined as described in Materials and Methods, and representative data are shown in Fig. 1.
CA, capsid protein.
The infectious titer was determined on a pool of phytohemagglutinin/IL-2-activated PBMCs from six donors by end-point limiting dilution 5 days after inoculation as reported previously (32). ND, not determined; IU, infectious units.
The arithmetic mean ± standard error of the mean of relative infectivity (IU/ng of p24 CA) was 5.26 ± 3.27 for group M isolates and 9.6 ± 3.65 for group O isolates. ND, not determined; IU, infectious units.
PBMCs.
PBMCs from healthy blood donors were purified by Ficoll-Hypaque gradient centrifugation and typically stimulated with phytohemagglutinin P/interleukin-2 (IL-2) (32).
Ex vivo HLACs from tonsil.
Tonsil tissue was removed during therapeutic tonsillectomy from HIV-, hepatitis B virus-, and hepatitis C virus-negative patients with informed consent. Use of anonymous surgical waste for experimental HIV infections was approved by the ethics committee of Heidelberg University (077/2005). To prepare HLACs, tonsil tissue was mechanically dispersed and passed through 40-μm-pore-size cell strainers (BD Falcon). Cells were washed in phosphate-buffered saline and plated in culture medium in 96-well V-bottom plates (Corning, Inc.) (14, 28). One day after tonsil preparation, HLACs were inoculated with HIV-1 (5 ng of p24 per 2 × 106 cells per well in 200 μl). After overnight infection, cells were washed, and the medium was subsequently changed on days 4, 7, and 11 postinfection (p.i.) without dispersing the pellet. At the same intervals, supernatant samples were harvested and stored at −20°C for subsequent analysis by p24 ELISA (31). Of note, the coating anti-p24 monoclonal antibody 183 recognizes the 9-amino-acid sequence RAEQASQEV, which is highly conserved among HIV-1 group M and group O isolates.
Assessment of HIV-1 replication.
As a measure of overall virus production over the course of infection, values for the integral area under the curve (AUC) of p24 concentrations in culture supernatants were determined as reported previously (6, 26).
Assessment of CD4 T-cell depletion.
CD4 T-cell depletion was determined as reported previously (29, 50). Briefly, on day 11 p.i., HLACs were fixed and stained with anti-human CD3-phycoerythrin (clone HIT3a; BD Pharmingen) and anti-human CD8-allophycocyanin (clone RPA-T8; BD Pharmingen). Flow cytometry was performed with a FACSCalibur with BD CellQuest Pro, version 4.0.2, software (BD Pharmingen).
Determination of coreceptor usage.
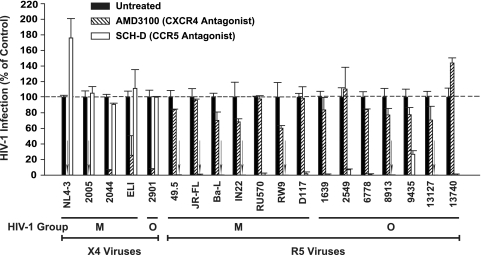
Expanded virus stocks were analyzed for utilization of the major coreceptors on TZM-bl reporter cells (16). Briefly, cells were treated for 30 min with either the CXCR4-antagonist AMD3100 or the CCR5-antagonist SCH-D (both, 5 μM) (gifts from Matthias Dittmar, London, United Kingdom) or left untreated. After overnight challenge with HIV-1 (15 ng of p24/well), cells were washed with medium, and 36 to 48 h p.i. a luciferase reporter assay system (Promega) was used to monitor the reporter activity in infected cells. In addition, for group O viruses 2901, 2549, and 13740, the ability to replicate was monitored on CCR5-negative A3.01 T cells (see Fig. 4A).
FIG. 4.
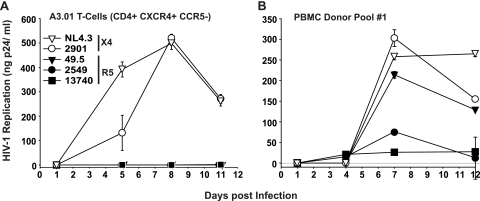
HIV-1 group O isolates 2549 and 13740 do not replicate on CCR5-negative A3.01 T cells. (A) Human A3.01 T cells (CD4+ CXCR4+ CCR5−) or (B) a blasted PBMC donor pool were challenged with either the HIV-1 group O isolates 2901, 2549, or 13740 or with the group M reference viruses NL4-3 (X4) or 49.5 (R5) (5 ng of p24 per well). After overnight infection, washed cells were cultivated for 11 to 12 more days, and replication was monitored at the indicated time points by quantification of the p24 concentration in culture supernatants. Shown are the arithmetic means ± SDs (n = 3).
Statistical analysis.
Statistical analysis of depletion and replication data was done in the statistical environment R, version 2.8.0 (44). Triplicate measurements of viral replication for each time point were summarized using the arithmetic mean, cumulative replication (AUC) computed by linear interpolation between time points, and integration of replication over time. Triplicate measurements for CD4 T-cell depletion were summarized using the arithmetic mean, normalized to the CD4/CD8 T-cell ratio of uninfected cells, and expressed as percentages. In ensuing analyses over subgroups (viral strain, group, coreceptor usage, and sample type), biological replicates for replication and depletion were summarized using the median. Box plots were produced in R to visualize results. Since some of the distributions of replication and depletion were highly non-Gaussian, we used a Wilcoxon rank sum test to assess the significance of differences in replication and depletion between different subgroups. Relative changes in replication or depletion were calculated as ratios on the medians.
RESULTS
Replication of HIV-1 group O and group M isolates in primary cell culture systems of infection.
All primary HIV-1 isolates were expanded on multidonor PBMCs to preserve their biological characteristics, and virus stocks were subsequently analyzed for p24 concentration, TCID50, and coreceptor usage (Table 1 and Fig. 1). We first tested the ability of a panel of HIV-1 group O primary isolates (one CXCR4-using [X4] and seven CCR5-using [R5] viruses) and a panel of HIV-1 group M primary isolates (three X4 and four R5 viruses) from subtypes A, B, C, D, or G, to replicate in various primary cell culture models of infection. Prototypic HIV-1 M strains served as additional references. We analyzed in parallel infections of single-donor PBMCs, three- to four-donor pools of PBMCs, and HLACs. In contrast to PBMCs, HLACs are permissive for HIV-1 infection independent of exogenous stimuli such as mitogens or IL-2 (14, 28).
FIG. 1.
Coreceptor usage of HIV-1 group M and group O isolates. The ability of the expanded virus stocks to utilize the major coreceptors CXCR4 and/or CCR5 for infection was examined on TZM-bl reporter cells in the presence or absence of the indicated coreceptor antagonists (see Materials and Methods for details). For each isolate, luciferase counts determined for lysates from untreated, infected cells were set to 100%. Shown are the arithmetic means ± SDs (n = 4).
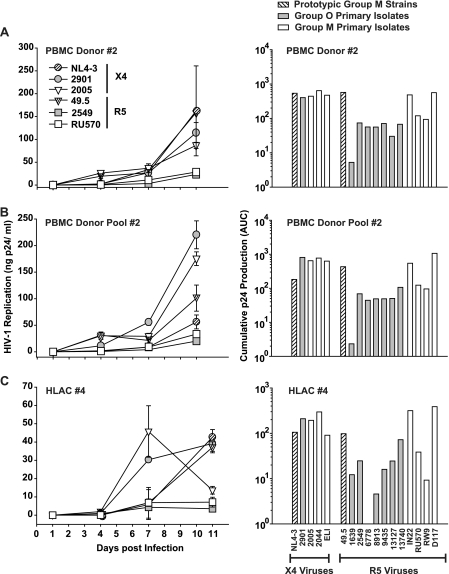
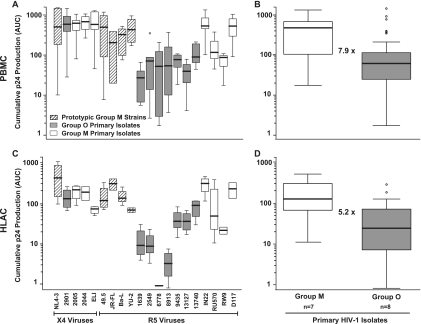
HLACs and activated PBMCs were challenged with HIV-1 stocks, and 24 h later, cells were washed and cultivated for 10 more days. As a means of standardizing the inocula of the different isolates, we decided to use identical amounts of p24 (5 ng per well) rather than identical numbers of infectious units. This avoids an artificial masking of the isolates' potential infectivity and replication phenotypes by normalization based on end-point limiting dilution analyses performed in the very same culture model. Of note, the relative infectivity of the panel of HIV-1 group O isolates compared to the panel of group M isolates used in the current study differed by less than twofold (Table 1). At the indicated days p.i. (Fig. 2), supernatants were harvested from the triplicate parallel infections and analyzed to quantify HIV-1 production. Quantification of p24 antigen over the time course of infection showed considerable differences between the different HIV-1 strains and primary isolates, both in terms of replication kinetics and peak concentrations of p24. As an example, replication curves for six viruses in these three primary cell models are depicted in Fig. 2 (left panels). As a measure of overall virus production, AUC values were calculated based on the plots of these p24 replication kinetics, in principle as reported previously (6, 26). This longitudinal and cumulative p24 evaluation provides a marker for an isolate's replicative ex vivo fitness. Interestingly, the pattern of AUC values found for the panel of viruses in one culture model largely matched that seen in the other two models (Fig. 2, right panels). Group O isolates 1639 and 6778 were notably different, showing higher or lower relative replicative fitness than in HLACs and PBMC cultures, respectively. The variability in AUC values was considerably more pronounced among R5 viruses than X4 viruses, and individual, but not all, group M R5 viruses replicated to higher levels than group O R5 viruses in these culture models.
FIG. 2.
Replication of HIV-1 group M and group O viruses in single-donor PBMCs, a PBMC donor pool, and a culture of human tonsil tissue ex vivo. PBMCs from a single donor (A), a pool of PBMCs from four different donors (B), and an HLAC from a routine therapeutic tonsillectomy (C) were infected in triplicates with identical amounts of prototypic HIV-1 M strains (X4: NL4-3; R5: 49.5), HIV-1 group O primary isolates (X4: 2901; R5: 1639, 2549, 6778, 8913, 9435, 13127, and 13740), or HIV-1 group M primary isolates (X4: 2005, 2044, 2044, and ELI; R5: IN22, RU570, RW9, and D117) (each with 5 ng of p24 per well). Viral replication was monitored by assessing the concentration of p24 in the culture medium between successive medium changes on days 1, 4, 7, and 11 p.i. and is depicted for six selected viruses (left panels). Given are the arithmetic means ± SDs from one of three to four different donors analyzed for each primary cell system. The corresponding cumulative p24 production over the culture period was determined for each virus as the AUC of the mean replication kinetic (left panels), in principle as reported previously (6, 26), and is shown in the panels on the right.
Cytopathic phenotype of HIV-1 group O and group M primary isolates in lymphoid tissue ex vivo.
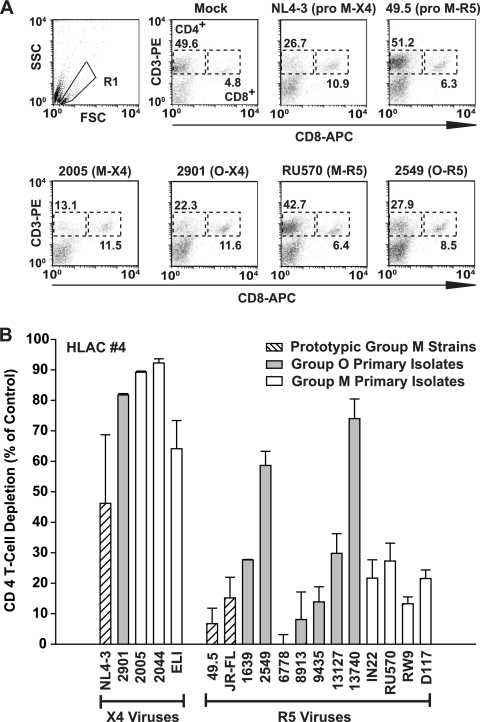
To assess virus-induced cytopathicity and CD4 T-cell depletion for these HIV-1 isolates and prototypic reference strains, HLACs from tonsil were harvested on day 11 after infection and immunostained with anti-CD3 and anti-CD8 antibodies. CD4 T cells in infected cultures were identified as CD3+ CD8− (17) since CD4 levels on productively infected cells can be downregulated by the viral Nef, Vpu, and Env proteins. Fluorescence-activated cell sorting (FACS) analysis was used to measure the ratio of CD4 T cells to CD8 T cells in infected and uninfected cultures, and CD4 T-cell depletion was detected as a decrease in the CD4/CD8 ratio in infected HLACs, as reported previously (14, 26, 28).
Figure 3A depicts representative FACS plots of these analyses. While mock-infected HLACs harbored higher relative levels of CD4 T cells (49.6%) than CD8 T cells (4.8%) at the time of harvest, infection with all X4 viruses, including primary isolates 2005 (group M; X4) and 2901 (group O; X4), severely depressed the CD4/CD8 ratio (Fig. 3). In contrast, the majority of R5 viruses (Fig. 3B), including 49.5 (NL4-3 recombinant with V3 loop sequence from Ba-L env; prototypic group M, R5 [11]) and isolate RU570 (group M; R5) (Fig. 3) exerted markedly less effect or virtually none on the relative levels of CD4 T cells. Of particular interest, in this cross-sectional analysis, two R5 primary isolates of group O, isolates 2549 and 13740, displayed a high degree of CD4 T-cell depletion that was indistinguishable from X4 viruses in HLACs from donor 4 (Fig. 3). Notably, the infection of TZM-bl reporter cells by these two O-type viruses was completely blocked by the CCR5-antagonist SCH-D and unaffected by the CXCR4-antagonist AMD3100 (Fig. 1), and they did not spread on CCR5-negative A3.01 T-cells (Fig. 4A), confirming the absence of a relevant X4 or dual-tropic quasi-species (data not shown). Collectively, group O viruses replicate in lymphoid tissue ex vivo, and certain isolates massively deplete CD4 T cells. Thus, we found no systematic difference in cytopathicity for viruses from groups O and M.
FIG. 3.
Depletion of CD4 T cells in HLACs from tonsil by HIV-1 group M and O viruses. CD4 T-cell depletion in infected HLACs (HLAC donor 4) was assessed on day 11 p.i. by measuring the relative percentage of CD4 T cells and CD8 T cells by flow cytometry. (A) FACS dot plots of dispersed HLACs costained for CD3-phycoerythrin and CD8-allophycocyanin. Viable lymphocytes were identified by forward scatter (FSC) and side scatter (SSC) characteristics (gate R1) and then classified as CD4 T cells (CD3+ CD8−) or CD8 T cells (CD3+ CD8+). The relative percentage of these T-cell subsets of all lymphocytes is indicated next to the FACS gates. CD3− CD8− cells to large extent represent B cells and NK cells (data not shown). (B) The CD4/CD8 ratios of HLACs infected with the different HIV-1 strains and primary isolates were calculated and plotted as percent CD4 T-cell depletion relative to the ratios observed in mock-infected cultures. Shown are the arithmetic means ± SDs from one of four different donors analyzed for each virus, performed in triplicate.
Cross-donor and HIV-1 group stratification of HIV-1 replication in PBMCs and HLACs.
To corroborate the results for replicative ex vivo fitness (Fig. 2) and account for donor variability, the replicative capacities of all 20 viruses were analyzed in three single-donor PBMC cultures, three multidonor pools of PBMCs, and tonsil-derived HLACs from four patients by measuring the cumulative p24 production (AUC values).
The relative replicative fitness of individual viruses was quite similar in blasted PBMCs (combined analysis of single- and multidonor pools) and in HLACs (Fig. 5A and C). The group M primary X4 isolate ELI and the group O R5 isolates 6778 and 8913 were notable exceptions, displaying a lower relative fitness in HLACs than PBMCs. Stratification of these data by HIV-1 groups demonstrated a 7.9-fold (PBMCs; P = 7.8 × 10−8) and 5.2-fold (HLACs; P = 2.9 × 10−5) lower replicative ex vivo fitness of primary isolates from group O than from group M (Fig. 5B and D). No significant difference was seen between the only X4 HIV-1 O isolate available and the three HIV-1 M isolates tested, while the cumulative p24 production of the R5 virus-only panels was threefold (PBMCs; P = 5.5 × 10−6) and ninefold (HLACs; P = 0.00055) lower for group O than group M.
FIG. 5.
Cross-donor and HIV-1 group-stratified analyses of HIV-1 replication. The analysis of HIV-1 replication kinetics based on p24 concentrations in culture supernatants and corresponding AUC determinations were performed on primary cells from multiple donors as described in the legend to Fig. 2. Box plots were used as a descriptive statistical representation of the AUC values obtained for each virus (A) in a combined analysis of results for single-donor PBMCs (n = 3) and multidonor pools of PBMCs (n = 3) and in a combined analysis of results for HLACs established from four different donors (C). A stratification of results in panels A and C was performed according to the viruses' HIV-1 group classification and is represented as box plots for PBMCs (B) and HLACs (D). Box plots depict the median (horizontal line) and upper and lower quartiles. Whiskers define the border for the 1.5× interquartile distance. Circles represent outliers. Values given within panels B and D represent the relative differences between all group M and group O viruses analyzed: M > O, 7.9-fold (PBMCs; P = 7.8 × 10−8) (B) and 5.2-fold (HLACs; P = 2.9 × 10−5) (D).
CD4 T-cell depletion in lymphoid tissue depends on coreceptor usage but not group classification.
Cross-donor evaluation of virus-induced CD4 T-cell depletion in HLACs showed that, overall, R5 viruses only mildly affected their levels while X4 viruses strongly depleted CD4 T cells (Fig. 6C). In particular, all prototypic and primary R5 viruses from group M had little (up to 20% depletion) or no effect (Fig. 6A), whereas X4 viruses consistently led to a reduction in CD4 T-cell counts by ≥60% (Fig. 6B). This strong coreceptor dependence of cytopathicity in ex vivo tonsil cultures confirms previous observations for isolates from this predominant HIV-1 group (19, 20, 34, 43, 50, 51). Surprisingly, a quite heterogeneous picture emerged for the R5 viruses from group O, with depletion rates ranging from 5% (isolate 8913) to 60% (isolate 2549) (Fig. 6A). Importantly, HIV-1 group stratification independent of coreceptor usage demonstrated that cumulative CD4 T-cell depletion was similar for groups M and O (Fig. 6D), indicating that a lower cytopathicity is not an integral feature of group O viruses.
FIG. 6.
Cross-donor, HIV-1 group-, and coreceptor-stratified analyses of HIV-1-mediated CD4 T-cell depletion. CD4 T-cell depletion in HIV-infected tonsil HLACs from four patients was quantified as described in the legend to Fig. 3. Combined depletion analysis of results for individual R5 viruses (n = 14) (A) or X4 viruses (n = 5) (B) from prototypic group M strains, group O primary isolates, or group M primary isolates are represented as box plots. (C) Stratification of results in panels A and B according to the viruses' coreceptor usage (C) or HIV-1 group classification (D). ***, P < 0.0001; n.s., not significant.
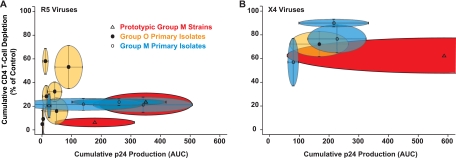
Correlation of ex vivo replicative fitness and ex vivo cytopathicity for R5 HIV-1 primary isolates reveals a group-specific signature.
Finally, we examined the relationship of replicative capacity and pathogenic potential of isolates from the two HIV-1 groups in HLACs. In Fig. 7, each elliptical symbol depicts the cumulative information derived from four independent HLACs for individual HIV-1 isolates, with standard deviations (SD) for both parameters indicated by the horizontal and vertical lines. The intragroup comparison of R5 primary isolates from group M showed marked differences in their replicative capacity but little variability in their ability to deplete CD4 T cells (Fig. 7A). In stark contrast, intragroup comparison of R5 primary isolates from group O highlighted strong differences in their ex vivo cytopathicity yet a relatively low variance in their replicative ability (Fig. 7A). Based on all the elliptical symbols for each HIV-1 group, an almost horizontal vector for the relationship of replicative ex vivo fitness and cytopathicity for R5 viruses of group M was contrasted by a nearly vertical vector for group O viruses. This evaluation of primary R5 HIV-1 isolates revealed that replicative fitness and the ability to deplete CD4 T cells in lymphoid cultures ex vivo do not show a linear correlation. On the contrary, markedly divergent patterns of this relationship appear to be specific for groups M and O and may thus constitute biological signature characteristics.
FIG. 7.
R5 primary isolates from HIV-1 groups O and M differ strikingly in their relationship of CD4 T-cell depletion and viral replication in ex vivo cultures from human tonsil. Mean values for the cumulative p24 production (AUC) (Fig. 5C) and for the cumulative CD4 T-cell depletion (Fig. 6A and B) of individual viruses in HIV-infected tonsil HLACs from four patients were plotted against each other and are shown for R5 viruses (A) or X4 viruses (B). Each elliptical symbol depicts one isolate and is numbered according to increasing values for cumulative CD4 T-cell depletion (group O) or increasing values for cumulative p24 production (group M). Lines within the elliptical symbols reflect the SD of values calculated for both parameters.
DISCUSSION
Few studies have investigated the biological properties of the HIV-1 group O viruses (1, 2, 54). Here, we establish virological characteristics of group O primary isolates in ex vivo models of HIV-1 infection. Compared to pandemic group M viruses, group O viruses show a similar ability to deplete CD4 T cells and display a modestly reduced replicative fitness. This challenges the belief that a severe inherent impairment of replicative fitness or pathogenicity in this HIV-1 group underlies its low prevalence in the human population.
Our HLAC studies demonstrate that the relationship of these two properties, replication and cytopathicity, is highly variable and that they are poor predictors of each other. This finding does not support the conclusion from earlier studies that an HIV-1 isolates's ex vivo pathogenic fitness can be deduced from its replicative fitness in PBMCs (1-3). Our results suggest that a more refined and discriminatory assessment of these characteristics is feasible and warranted. Interestingly, their relationship revealed a group-specific signature pattern: R5 viruses from group M varied considerably in their replicative capacity but showed a low and relatively stable cytopathicity (0 to 20% CD4 T-cell depletion), consistent with previous reports (19, 20, 28, 29, 43, 50). In stark contrast, R5 isolates from group O displayed a high variability in their potential to deplete CD4 T cells, with lower variance in their replicative fitness in human lymphoid tissue ex vivo.
The relationship between the replicative fitness and cytopathicity of groups O and M and their global distribution appears to be more complex than a simple correlation between these ex vivo characteristics and disease progression and/or transmission efficiency in humans (4). Several mechanistic scenarios can be envisioned that may underlie the limited contribution of HIV-1 group O viruses to the pandemic. Important for a first scenario, clinical studies indicate that a patient's viral load is a good predictor of vertical and horizontal HIV-1 transmission (36, 47). An earlier study reported that the replicative fitness of isolates from group O is over 100-fold less than that of group M (2) and suggested that such a severe replicative defect in tissue culture could translate into lower viral loads in infected individuals and, consequently, lower transmission efficiencies. However, we found only a five- to eightfold reduced ex vivo replicative fitness of group O isolates in PBMC cultures or HLACs. Notably, for most primary isolates, replicative fitness turned out to be an intrinsic, largely donor-independent characteristic in all three experimental infection models. The reason for these striking quantitative differences in the intergroup comparison in our and the previous study (2) is not known but may relate to different panels of primary isolates or to specific features of the assays. It is not apparent why head-to-head replication competitions of viruses (1, 2) should provide a more faithful predictor for their replicative in vivo fitness than the side-by-side replication studies in our study. The former model emphasizes the first few rounds of replication with possible target-cell competition and superinfection interference. However, the vast majority of infections in humans are the result of the transmission of individual HIV-1 viruses. This may be better reflected in a parallel, yet separate, infection setup for two different viruses in a lymphoid organ culture model. It is difficult to predict whether the modest replicative differences seen for group O isolates in our study could translate into a relevant effect in vivo and thus account for the drastic prevalence phenotype. It should also be noted that the expansion and ex vivo experiments for all isolates were conducted on primary cells from Caucasian donors; this could be a confounding problem since group O viruses may be adapted to individuals of African descent.
Relevant for a second scenario, the diagnosis of AIDS and thus overt HIV pathogenesis goes along with a higher risk for the sexual partner of acquiring infection (9, 13, 47). Our tonsil explant infections show that low cytopathicity is not an intrinsic property of group O viruses. In fact, two R5 isolates from group O induced massive CD4 T-cell depletion, comparable to X4 viruses and not seen in the current study or previously with R5 viruses from group M (19, 20, 28, 29, 43, 50). Previous reports have estimated that approximately 15 to 20% of CD4 T cells express CCR5 (14, 22) and suggested that R5 viruses almost exclusively kill this subset of productively infected cells. The unprecedented CD4 T-cell depletion (up to 70%) induced by the two R5 O types 2549 and 13740 could be explained by either a marked bystander killing, specific to infections with these isolates, or by their efficient utilization of surface CCR5 for entry, levels of which on CD4 T cells are below the limit of detection. The comparable and, for some isolates, even higher cytopathicity of group O isolates argues against this characteristic as responsible for their limited contribution to the HIV pandemic. However, host immune responses, which are not apparent in short-term ex vivo tonsil cultures and may target HIV-1 group-specific epitopes, could operate to control group O replication in patients. Detailed clinical analyses of the immune status and virus load in plasma and genital fluids of a larger number of group O-infected individuals are needed to probe this hypothesis. The required technology is available including an in-house HIV-1 RNA load protocol (23) or the recently approved Cobas TaqScreen MPX test, both of which have been validated to detect genetically divergent group O isolates.
In a third scenario, the efficiency of sexual transmission, independent of the virus load, could be the bottleneck and underlie group O's low prevalence. However, it is still controversial which experimental in vitro and ex vivo models incorporate the relevant primary target cells and recapitulate the key events of horizontal transmission of HIV-1 (12, 15, 21). Quantifying HIV-1 transfer from dendritic cells to CD4 T cells, Arien and colleagues reported a 10- to 100-fold impaired transmission of group O isolates compared to that observed with group M isolates (2). Further studies in different experimental transmission models should follow up on this interesting observation.
Collectively, this report establishes virological ex vivo characteristics of HIV-1 group O primary isolates which challenge the assumption that an inherently reduced replicative fitness or pathogenicity of viruses from this HIV-1 group underlies their limited spread in the human population.
Acknowledgments
We thank Hans-Georg Kräusslich for support. We are grateful to Valerie Bosch, Matthias Dittmar, Beatrice Hahn, and Malcom Martin for the gift of reagents. We are indebted to Lutz Gürtler for providing HIV-1 group O primary isolates. We thank Oliver T. Fackler and Jason Kreisberg for comments on the manuscript and Gary Howard for editorial assistance.
This study was supported by the Deutsche Forschungsgemeinschaft, SFB544, project B17 to O.T.K., and BMBF, grant 0313923 Forsys/ViroQuant, to L.K.
Footnotes
Published ahead of print on 12 August 2009.
REFERENCES
- 1.Abraha, A., I. L. Nankya, R. Gibson, K. Demers, D. M. Tebit, E. Johnston, D. Katzenstein, A. Siddiqui, C. Herrera, L. Fischetti, R. J. Shattock, and E. J. Arts. 2009. CCR5- and CXCR4-tropic subtype C human immunodeficiency virus type 1 isolates have a lower level of pathogenic fitness than other dominant group M subtypes: implications for the epidemic. J. Virol. 83:5592-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arien, K. K., A. Abraha, M. E. Quinones-Mateu, L. Kestens, G. Vanham, and E. J. Arts. 2005. The replicative fitness of primary human immunodeficiency virus type 1 (HIV-1) group M, HIV-1 group O, and HIV-2 isolates. J. Virol. 79:8979-8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arien, K. K., G. Vanham, and E. J. Arts. 2007. Is HIV-1 evolving to a less virulent form in humans? Nat. Rev. Microbiol. 5:141-151.17203103 [Google Scholar]
- 4.Arts, E. J., and M. E. Quinones-Mateu. 2003. Sorting out the complexities of HIV-1 fitness. AIDS 17:780-781. [DOI] [PubMed] [Google Scholar]
- 5.Audige, A., E. Schlaepfer, A. Bonanomi, H. Joller, M. C. Knuchel, M. Weber, D. Nadal, and R. F. Speck. 2004. HIV-1 does not provoke alteration of cytokine gene expression in lymphoid tissue after acute infection ex vivo. J. Immunol. 172:2687-2696. [DOI] [PubMed] [Google Scholar]
- 6.Audige, A., M. Urosevic, E. Schlaepfer, R. Walker, D. Powell, S. Hallenberger, H. Joller, H. U. Simon, R. Dummer, and R. F. Speck. 2006. Anti-HIV state but not apoptosis depends on IFN signature in CD4+ T cells. J. Immunol. 177:6227-6237. [DOI] [PubMed] [Google Scholar]
- 7.Ayouba, A., P. Mauclere, P. M. Martin, P. Cunin, J. Mfoupouendoun, B. Njinku, S. Souquieres, and F. Simon. 2001. HIV-1 group O infection in Cameroon, 1986 to 1998. Emerg. Infect. Dis. 7:466-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barin, F., F. Cazein, F. Lot, J. Pillonel, S. Brunet, D. Thierry, F. Damond, F. Brun-Vezinet, J. C. Desenclos, and C. Semaille. 2007. Prevalence of HIV-2 and HIV-1 group O infections among new HIV diagnoses in France: 2003-2006. AIDS 21:2351-2353. [DOI] [PubMed] [Google Scholar]
- 9.Boily, M. C., R. F. Baggaley, L. Wang, B. Masse, R. G. White, R. J. Hayes, and M. Alary. 2009. Heterosexual risk of HIV-1 infection per sexual act: systematic review and meta-analysis of observational studies. Lancet Infect. Dis. 9:118-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brennan, C. A., P. Bodelle, R. Coffey, S. G. Devare, A. Golden, J. Hackett, Jr., B. Harris, V. Holzmayer, K. C. Luk, G. Schochetman, P. Swanson, J. Yamaguchi, A. Vallari, N. Ndembi, C. Ngansop, F. Makamche, D. Mbanya, L. G. Gurtler, L. Zekeng, and L. Kaptue. 2008. The prevalence of diverse HIV-1 strains was stable in Cameroonian blood donors from 1996 to 2004. J. Acquir. Immune Defic. Syndr. 49:432-439. [DOI] [PubMed] [Google Scholar]
- 11.Chesebro, B., J. Nishio, S. Perryman, A. Cann, W. O'Brien, I. S. Chen, and K. Wehrly. 1991. Identification of human immunodeficiency virus envelope gene sequences influencing viral entry into CD4-positive HeLa cells, T-leukemia cells, and macrophages. J. Virol. 65:5782-5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Jong, M. A., L. de Witte, M. J. Oudhoff, S. I. Gringhuis, P. Gallay, and T. B. Geijtenbeek. 2008. TNF-alpha and TLR agonists increase susceptibility to HIV-1 transmission by human Langerhans cells ex vivo. J. Clin. Investig. 118:3440-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Vincenzi, I. 1994. A longitudinal study of human immunodeficiency virus transmission by heterosexual partners. European Study Group on Heterosexual Transmission of HIV. N. Engl. J. Med. 331:341-346. [DOI] [PubMed] [Google Scholar]
- 14.Eckstein, D. A., M. L. Penn, Y. D. Korin, D. D. Scripture-Adams, J. A. Zack, J. F. Kreisberg, M. Roederer, M. P. Sherman, P. S. Chin, and M. A. Goldsmith. 2001. HIV-1 actively replicates in naive CD4+ T cells residing within human lymphoid tissues. Immunity 15:671-682. [DOI] [PubMed] [Google Scholar]
- 15.Fletcher, P., Y. Kiselyeva, G. Wallace, J. Romano, G. Griffin, L. Margolis, and R. Shattock. 2005. The nonnucleoside reverse transcriptase inhibitor UC-781 inhibits human immunodeficiency virus type 1 infection of human cervical tissue and dissemination by migratory cells. J. Virol. 79:11179-11186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geuenich, S., C. Goffinet, S. Venzke, S. Nolkemper, I. Baumann, P. Plinkert, J. Reichling, and O. T. Keppler. 2008. Aqueous extracts from peppermint, sage and lemon balm leaves display potent anti-HIV-1 activity by increasing the virion density. Retrovirology 5:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glushakova, S., B. Baibakov, L. B. Margolis, and J. Zimmerberg. 1995. Infection of human tonsil histocultures: a model for HIV pathogenesis. Nat. Med. 1:1320-1322. [DOI] [PubMed] [Google Scholar]
- 18.Glushakova, S., B. Baibakov, J. Zimmerberg, and L. B. Margolis. 1997. Experimental HIV infection of human lymphoid tissue: correlation of CD4+ T cell depletion and virus syncytium-inducing/non-syncytium-inducing phenotype in histocultures inoculated with laboratory strains and patient isolates of HIV type 1. AIDS Res. Hum. Retrovir. 13:461-471. [DOI] [PubMed] [Google Scholar]
- 19.Glushakova, S., J. C. Grivel, W. Fitzgerald, A. Sylwester, J. Zimmerberg, and L. B. Margolis. 1998. Evidence for the HIV-1 phenotype switch as a causal factor in acquired immunodeficiency. Nat. Med. 4:346-349. [DOI] [PubMed] [Google Scholar]
- 20.Glushakova, S., Y. Yi, J. C. Grivel, A. Singh, D. Schols, E. De Clercq, R. G. Collman, and L. Margolis. 1999. Preferential coreceptor utilization and cytopathicity by dual-tropic HIV-1 in human lymphoid tissue ex vivo. J. Clin. Investig. 104:R7-R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grivel, J. C., and L. Margolis. 2009. Use of human tissue explants to study human infectious agents. Nat. Protoc. 4:256-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grivel, J. C., and L. B. Margolis. 1999. CCR5- and CXCR4-tropic HIV-1 are equally cytopathic for their T-cell targets in human lymphoid tissue. Nat. Med. 5:344-346. [DOI] [PubMed] [Google Scholar]
- 23.Gueudin, M., J. C. Plantier, F. Damond, P. Roques, P. Mauclere, and F. Simon. 2003. Plasma viral RNA assay in HIV-1 group O infection by real-time PCR. J. Virol. Methods 113:43-49. [DOI] [PubMed] [Google Scholar]
- 24.Gurtler, L. G., P. H. Hauser, J. Eberle, A. von Brunn, S. Knapp, L. Zekeng, J. M. Tsague, and L. Kaptue. 1994. A new subtype of human immunodeficiency virus type 1 (MVP-5180) from Cameroon. J. Virol. 68:1581-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hampl, H., D. Sawitzky, M. Stoffler-Meilicke, A. Groh, M. Schmitt, J. Eberle, and L. Gurtler. 1995. First case of HIV-1 subtype 0 infection in Germany. Infection 23:369-370. [DOI] [PubMed] [Google Scholar]
- 26.Homann, S., N. Tibroni, I. Baumann, S. Sertel, O. T. Keppler, and O. T. Fackler. 2009. Determinants in HIV-1 Nef for enhancement of virus replication and depletion of CD4+ T lymphocytes in human lymphoid tissue ex vivo. Retrovirology 6:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jayakumar, P., I. Berger, F. Autschbach, M. Weinstein, B. Funke, E. Verdin, M. A. Goldsmith, and O. T. Keppler. 2005. Tissue-resident macrophages are productively infected ex vivo by primary X4 isolates of human immunodeficiency virus type 1. J. Virol. 79:5220-5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jekle, A., O. T. Keppler, E. De Clercq, D. Schols, M. Weinstein, and M. A. Goldsmith. 2003. In vivo evolution of human immunodeficiency virus type 1 toward increased pathogenicity through CXCR4-mediated killing of uninfected CD4 T cells. J. Virol. 77:5846-5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jekle, A., B. Schramm, P. Jayakumar, V. Trautner, D. Schols, E. De Clercq, J. Mills, S. M. Crowe, and M. A. Goldsmith. 2002. Coreceptor phenotype of natural human immunodeficiency virus with Nef deleted evolves in vivo, leading to increased virulence. J. Virol. 76:6966-6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jonassen, T. O., K. Stene-Johansen, E. S. Berg, O. Hungnes, C. F. Lindboe, S. S. Froland, and B. Grinde. 1997. Sequence analysis of HIV-1 group O from Norwegian patients infected in the 1960s. Virology 231:43-47. [DOI] [PubMed] [Google Scholar]
- 31.Keppler, O. T., I. Allespach, L. Schuller, D. Fenard, W. C. Greene, and O. T. Fackler. 2005. Rodent cells support key functions of the human immunodeficiency virus type 1 pathogenicity factor Nef. J. Virol. 79:1655-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keppler, O. T., W. Yonemoto, F. J. Welte, K. S. Patton, D. Iacovides, R. E. Atchison, T. Ngo, D. L. Hirschberg, R. F. Speck, and M. A. Goldsmith. 2001. Susceptibility of rat-derived cells to replication by human immunodeficiency virus type 1. J. Virol. 75:8063-8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korber, B., M. Muldoon, J. Theiler, F. Gao, R. Gupta, A. Lapedes, B. H. Hahn, S. Wolinsky, and T. Bhattacharya. 2000. Timing the ancestor of the HIV-1 pandemic strains. Science 288:1789-1796. [DOI] [PubMed] [Google Scholar]
- 34.Kreisberg, J. F., D. Kwa, B. Schramm, V. Trautner, R. Connor, H. Schuitemaker, J. I. Mullins, A. B. van't Wout, and M. A. Goldsmith. 2001. Cytopathicity of human immunodeficiency virus type 1 primary isolates depends on coreceptor usage and not patient disease status. J. Virol. 75:8842-8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laurent, C., A. Bourgeois, M. A. Faye, R. Mougnutou, M. Seydi, M. Gueye, F. Liegeois, C. T. Kane, C. Butel, J. Mbuagbaw, L. Zekeng, S. Mboup, E. Mpoudi-Ngole, M. Peeters, and E. Delaporte. 2002. No difference in clinical progression between patients infected with the predominant human immunodeficiency virus type 1 circulating recombinant form (CRF) 02_AG strain and patients not infected with CRF02_AG, in Western and West-Central Africa: a four-year prospective multicenter study. J. Infect. Dis. 186:486-492. [DOI] [PubMed] [Google Scholar]
- 36.Lee, T. H., N. Sakahara, E. Fiebig, M. P. Busch, T. R. O'Brien, and S. A. Herman. 1996. Correlation of HIV-1 RNA levels in plasma and heterosexual transmission of HIV-1 from infected transfusion recipients. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 12:427-428. [DOI] [PubMed] [Google Scholar]
- 37.Leitner, T., and J. Albert. 1999. The molecular clock of HIV-1 unveiled through analysis of a known transmission history. Proc. Natl. Acad. Sci. USA 96:10752-10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Llano, A., J. Barretina, A. Gutierrez, B. Clotet, and J. A. Este. 2003. Interleukin-7-dependent production of RANTES that correlates with human immunodeficiency virus disease progression. J. Virol. 77:4389-4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loussert-Ajaka, I., M. L. Chaix, B. Korber, F. Letourneur, E. Gomas, E. Allen, T. D. Ly, F. Brun-Vezinet, F. Simon, and S. Saragosti. 1995. Variability of human immunodeficiency virus type 1 group O strains isolated from Cameroonian patients living in France. J. Virol. 69:5640-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mas, A., E. Quinones-Mateu, V. Soriano, and E. Domingo. 1996. Env gene characterization of the first HIV type 1 group O Spanish isolate. AIDS Res. Hum. Retrovir. 12:1647-1649. [DOI] [PubMed] [Google Scholar]
- 41.Neumann, T., I. Hagmann, S. Lohrengel, M. L. Heil, C. A. Derdeyn, H. G. Krausslich, and M. T. Dittmar. 2005. T20-insensitive HIV-1 from naive patients exhibits high viral fitness in a novel dual-color competition assay on primary cells. Virology 333:251-262. [DOI] [PubMed] [Google Scholar]
- 42.Nkengasong, J. N., M. Peeters, M. vanden Haesevelde, S. S. Musi, B. Willems, P. M. Ndumbe, E. Delaporte, J. L. Perret, P. Piot, and G. van den Groen. 1993. Antigenic evidence of the presence of the aberrant HIV-1ANT70 virus in Cameroon and Gabon. AIDS 7:1536-1538. [PubMed] [Google Scholar]
- 43.Penn, M. L., J. C. Grivel, B. Schramm, M. A. Goldsmith, and L. Margolis. 1999. CXCR4 utilization is sufficient to trigger CD4+ T cell depletion in HIV-1-infected human lymphoid tissue. Proc. Natl. Acad. Sci. USA 96:663-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.R Development Core Team. 2008. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- 45.Robertson, D. L., J. P. Anderson, J. A. Bradac, J. K. Carr, B. Foley, R. K. Funkhouser, F. Gao, B. H. Hahn, M. L. Kalish, C. Kuiken, G. H. Learn, T. Leitner, F. McCutchan, S. Osmanov, M. Peeters, D. Pieniazek, M. Salminen, P. M. Sharp, S. Wolinsky, and B. Korber. 2000. HIV-1 nomenclature proposal. Science 288:55-56. [DOI] [PubMed] [Google Scholar]
- 46.Rowe, P. M. 1996. HIV-1 group O infection identified in USA. Lancet 348:116. [DOI] [PubMed] [Google Scholar]
- 47.Royce, R. A., A. Sena, W. Cates, Jr., and M. S. Cohen. 1997. Sexual transmission of HIV. N. Engl. J. Med. 336:1072-1078. [DOI] [PubMed] [Google Scholar]
- 48.Rucker, E., J. C. Grivel, J. Munch, F. Kirchhoff, and L. Margolis. 2004. Vpr and Vpu are important for efficient human immunodeficiency virus type 1 replication and CD4+ T-cell depletion in human lymphoid tissue ex vivo. J. Virol. 78:12689-12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schlaepfer, E., A. Audige, H. Joller, and R. F. Speck. 2006. TLR7/8 triggering exerts opposing effects in acute versus latent HIV infection. J. Immunol. 176:2888-2895. [DOI] [PubMed] [Google Scholar]
- 50.Schramm, B., M. L. Penn, E. H. Palacios, R. M. Grant, F. Kirchhoff, and M. A. Goldsmith. 2000. Cytopathicity of human immunodeficiency virus type 2 (HIV-2) in human lymphoid tissue is coreceptor dependent and comparable to that of HIV-1. J. Virol. 74:9594-9600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schramm, B., M. L. Penn, R. F. Speck, S. Y. Chan, E. De Clercq, D. Schols, R. I. Connor, and M. A. Goldsmith. 2000. Viral entry through CXCR4 is a pathogenic factor and therapeutic target in human immunodeficiency virus type 1 disease. J. Virol. 74:184-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Talbott, R., G. Kraus, D. Looney, and F. Wong-Staal. 1993. Mapping the determinants of human immunodeficiency virus 2 for infectivity, replication efficiency, and cytopathicity. Proc. Natl. Acad. Sci. USA 90:4226-4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tebit, D. M., I. Nankya, E. J. Arts, and Y. Gao. 2007. HIV diversity, recombination and disease progression: how does fitness “fit” into the puzzle? AIDS Rev. 9:75-87. [PubMed] [Google Scholar]
- 54.Tebit, D. M., L. Zekeng, L. Kaptue, L. Gurtler, O. T. Fackler, O. T. Keppler, O. Herchenroder, and H. G. Krausslich. 2004. Construction and characterization of an HIV-1 group O infectious molecular clone and analysis of Vpr- and Nef-negative derivatives. Virology 326:329-339. [DOI] [PubMed] [Google Scholar]
- 55.Vergne, L., A. Bourgeois, E. Mpoudi-Ngole, R. Mougnutou, J. Mbuagbaw, F. Liegeois, C. Laurent, C. Butel, L. Zekeng, E. Delaporte, and M. Peeters. 2003. Biological and genetic characteristics of HIV infections in Cameroon reveals dual group M and O infections and a correlation between SI-inducing phenotype of the predominant CRF02_AG variant and disease stage. Virology 310:254-266. [DOI] [PubMed] [Google Scholar]
- 56.Yamaguchi, J., R. Coffey, A. Vallari, C. Ngansop, D. Mbanya, N. Ndembi, L. Kaptue, L. G. Gurtler, P. Bodelle, G. Schochetman, S. G. Devare, and C. A. Brennan. 2006. Identification of HIV type 1 group N infections in a husband and wife in Cameroon: viral genome sequences provide evidence for horizontal transmission. AIDS Res. Hum. Retrovir. 22:83-92. [DOI] [PubMed] [Google Scholar]
- 57.Zekeng, L., J. Obiang Sima, H. Hampl, J. M. Ndemesogo, J. Ntutumu, V. Sima, S. Devare, L. Kaptue, and L. Gurtler. 1997. Update on HIV-1 group O infection in Equatorial Guinea, Central Africa. AIDS 11:1410-1412. [PubMed] [Google Scholar]