The step by step assembly process from preribosome in the nucleus to translation-competent 60S ribosome subunit in the cytoplasm is revealed (also see Lo et al. in this issue).
Abstract
Before entering translation, preribosomal particles undergo sequential late maturation steps. In the case of pre-60S particles, these steps involve the release of shuttling maturation factors and transport receptors. In this study, we report a new maturation step in the 60S biogenesis pathway in budding yeast. We show that efficient release of the nucleolar/nuclear ribosomal-like protein Mrt4 (homologous to the acidic ribosomal P-protein Rpp0) from pre-60S particles requires the highly conserved protein Yvh1, which associates only with late pre-60S particles. Cell biological and biochemical analyses reveal that Mrt4 fails to dissociate from late pre-60S particles in yvh1Δ cells, inducing a delay in nuclear pre–ribosomal RNA processing and a pre-60S export defect in yvh1Δ cells. Moreover, we have isolated gain of function alleles of Mrt4 that specifically bypass the requirement for Yvh1 and rescue all yvh1Δ-associated phenotypes. Together, our data suggest that Yvh1-mediated release of Mrt4 precedes cytoplasmic loading of Rpp0 on pre-60S particles and is an obligatory late step toward construction of translation-competent 60S subunits.
Introduction
Eukaryotic cells expend a substantial amount of cellular energy and resources to manufacture ribosomal subunits (Warner, 1999). To produce a ribosome, eukaryotic cells must assemble >70 ribosomal proteins (r-proteins) with four different ribosomal RNA (rRNA) species (Venema and Tollervey, 1999; Tschochner and Hurt, 2003). The concerted actions of all three transcription machineries (RNA polymerases I, II, and III) need to be coordinated to ensure the high efficiency and accuracy of ribosome production: Pol I produces 25S/28S, 5.8S, and 18S rRNA, Pol III synthesizes 5S rRNA, and Pol II transcribes the mRNAs for the r-proteins. Additionally, the process of ribosome assembly requires numerous nonribosomal factors, termed trans-acting factors (Venema and Tollervey, 1999; Tschochner and Hurt, 2003). The final outcome of this spatially and temporally coordinated effort is that every second, a growing yeast cell delivers up to 40 nascent ribosomal subunits from the nucleolus, the primary site of assembly, to the cytoplasm (Tschochner and Hurt, 2003).
Most of our current understanding of this highly dynamic, regulated multistep process comes from studies in budding yeast. The combined use of genetic, cell biological, and proteomic methods has uncovered >150 trans-acting factors that are required for the assembly and maturation of preribosomal particles as they travel from the nucleolus to the cytoplasm (Venema and Tollervey, 1999; Fatica and Tollervey, 2002; Fromont-Racine et al., 2003; Tschochner and Hurt, 2003; Granneman and Baserga, 2004). Moreover, tandem affinity purification (TAP; Rigaut et al., 1999; Puig et al., 2001) and sensitive mass spectrometric methods have allowed refinement of the ribosome assembly scheme, which was originally proposed upon identification of 90S, 66S, and 43S preribosomal particles in the mid-70s (Trapman et al., 1975). These approaches have uncovered changes in protein and rRNA composition of several distinct and successive pre-40S and -60S ribosomal particles (Bassler et al., 2001; Harnpicharnchai et al., 2001; Fatica et al., 2002; Grandi et al., 2002; Nissan et al., 2002; Saveanu et al., 2003; Schäfer et al., 2003). Nevertheless, the functions of the majority of identified trans-acting factors are only starting to be unraveled.
Early pre-60S particles associate with ∼100 trans-acting factors along their biogenesis pathway (Fatica and Tollervey, 2002; Fromont-Racine et al., 2003; Tschochner and Hurt, 2003). As pre-60S particles travel through the nucleoplasm toward the nuclear pore complex, trans-acting factors are released at distinct stages and recycled back to participate in new rounds of biogenesis steps. These events are most likely triggered by transiently associating energy-consuming enzymes, which result in sequential reduction of complexity and acquisition of export competence. Export-competent pre-60S particles associate with transport factors, which promote interactions with the nuclear pore complex. Among these factors is the conserved adaptor Nmd3, which contains a nuclear export sequence that is recognized by the nuclear export receptor Crm1/Xpo1 (Ho et al., 2000; Gadal et al., 2001). Recently, additional shuttling factors such as the HEAT repeat–containing protein Rrp12, the mRNA export factor Mex67-Mtr2, and Arx1 were shown to facilitate the nuclear export of pre-60S particles (Oeffinger et al., 2004; Bradatsch et al., 2007; Yao et al., 2007, 2008; Hung et al., 2008).
The majority of trans-acting factors that associate with pre-60S particles during early biogenesis steps are released in the nucleolus/nucleus. However, few factors (Nmd3, Rlp24, Tif6, Nog1, Arx1, and Alb1) remain associated with pre-60S particles and are escorted to the cytoplasm (Senger et al., 2001; Saveanu et al., 2003; Hedges et al., 2005; Hung and Johnson, 2006; Lebreton et al., 2006; Pertschy et al., 2007). The release and import of these factors to the nucleus constitute late pre-60S maturation steps, which are required for the formation of translation-competent 60S subunits. These steps are crucial for the 60S biogenesis pathway because a recycling failure leads to depletion of these proteins from their nucleolar/nuclear sites of action, resulting in pre-rRNA processing delays, assembly defects, and impaired nuclear export of pre-60S particles. All late pre-60S maturation steps investigated to date have been shown to require cofactors (Kre35, Efl1, Sdo1, Rei1, and Drg1) that transiently associate with late pre-60S particles (Senger et al., 2001; Saveanu et al., 2003; Hedges et al., 2005; Hung and Johnson, 2006; Lebreton et al., 2006; Menne et al., 2007; Pertschy et al., 2007; Zemp and Kutay, 2007). In addition to these late maturation steps, before achieving translation competence, pre-60S particles must associate with the last few remaining r-proteins. Nearly all the r-proteins associate with pre-rRNAs during early nucleolar biogenesis steps (Grandi et al., 2002; Nissan et al., 2002; Schäfer et al., 2003). However, at least three r-proteins, L10, L24, and the acidic r-protein Rpp0, have been suggested to associate at late stages in pre-60S maturation (Zinker and Warner, 1976; Kruiswijk et al., 1978; Saveanu et al., 2003; Kressler et al., 2008).
In this study, we describe a new late maturation step in the 60S biogenesis pathway. We provide multiple lines of evidence that the release of the nucleolar/nuclear pre-60S factor Mrt4, a ribosomal-like protein that is homologous to the acidic r-protein Rpp0, specifically requires the conserved protein Yvh1 that associates transiently with late pre-60S particles. Our data suggest that this late event permits cytoplasmic incorporation of the r-protein Rpp0, a biogenesis step necessary to construct a translation-competent 60S subunit. An accompanying paper (see Lo et al. in this issue) has reached similar conclusions regarding the role of Yvh1.
Results
The evolutionarily conserved dual-specificity phosphatase Yvh1 was found to coenrich with late cytoplasmic pre-60S particles (Nissan et al., 2002). However, its precise role in the biogenesis and nuclear export of pre-60S subunits has remained unclear. In this study, we have used the powerful combination of genetic, cell biological, and biochemical approaches available in budding yeast to gain insights into the requirement of Yvh1 in the 60S pathway.
Yvh1 is required for proper nuclear export of pre-60S particles
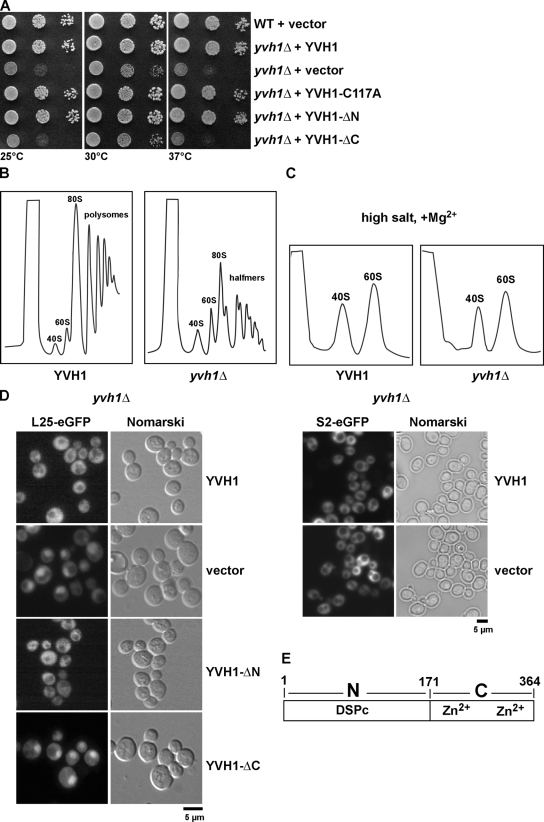
We initiated our analysis by asking whether Yvh1 has a role in promoting the biogenesis and nuclear export of pre-60S subunits. To this end, we disrupted the endogenous YVH1 in wild-type diploid cells. Tetrad analysis yielded two spores with wild-type growth rates and two spores with a slow-growth phenotype that carried the YVH1 deletion (yvh1Δ; unpublished data). As previously reported, yvh1Δ cells exhibited a slow-growth phenotype at all temperatures tested (Fig. 1 A; Beeser and Cooper, 1999; Muda et al., 1999; Aoki et al., 2001; Sakumoto et al., 2001; Liu and Chang, 2009). Next, whole cell lysates derived from YVH1 and yvh1Δ cells were subjected to sucrose gradient centrifugation under polysome-preserving conditions and high salt conditions (800 mM KCl and 10 mM MgCl2 to dissociate the two subunits; Tollervey et al., 1991, 1993). Lysates derived from yvh1Δ cells showed the presence of halfmers, but no significant decrease of free 60S subunits was observed (Fig. 1 B). Dissociative high salt sucrose gradient analyses of lysates derived from yvh1Δ cells also showed no striking deficit in free 60S versus 40S ribosomal subunits (Fig. 1 C). Halfmer polysomes correspond to a 43S complex, consisting of a 40S ribosomal subunit with attached initiation factors awaiting the addition of a 60S ribosomal subunit while stalled at the start codon (Helser et al., 1981). The appearance of halfmers can be attributed either to (1) decreased free 60S pool arising from rapid degradation in the nucleus of defectively assembled pre-60S particles, (2) impaired nuclear export of pre-60S particles, or (3) impaired translation initiation. In the first case, halfmers are accompanied with a net decrease in free 60S (Rotenberg et al., 1988; Deshmukh et al., 1993), whereas in the other cases, there might be no dramatic decrease in free 60S versus 40S subunits observed (Baronas-Lowell and Warner, 1990; Eisinger et al., 1997a,b). In the case of yvh1Δ cells, there is practically no net deficit in free 60S versus 40S ribosomal subunits, which raised the question of whether the halfmer phenotype was derived from impaired subunit joining (a cytoplasmic event) or impaired nuclear export of pre-60S particles. Localizations of the previously described large subunit (60S) reporter constructs L25-GFP and L11-GFP and S2-GFP for the small subunit (40S; Hurt et al., 1999; Stage-Zimmermann et al., 2000; Milkereit et al., 2003) were investigated. As expected, wild-type cells showed cytoplasmic localizations for both 40S and 60S reporter constructs. In contrast, yvh1Δ cells showed nuclear accumulation of both 60S reporters, suggesting an impairment in nuclear export of pre-60S particles (Fig. 1 D, left; and not depicted; Gadal et al., 2001). No nucleolar/nuclear accumulation of the 40S reporter construct was observed in yvh1Δ cells (Fig. 1 D, right).
Figure 1.
YVH1 is required for proper pre-60S export. (A) The Zn2+-binding domain of YVH1 but not its phosphatase domain is important for cell growth. yvh1Δ cells carrying the indicated plasmids were spotted in serial 10-fold dilutions onto SD-Ura plates and incubated at 25, 30, or 37°C for 3 d. YVH1-C117A (catalytically inactive), YVH1-ΔN (Zn2+ binding), and YVH1-ΔC (phosphatase domain) are shown. (B and C) Analysis of polysome profiles (OD254nm) of the indicated wild-type and yvh1Δ cells by sedimentation centrifugation on sucrose density gradients after cycloheximide treatment and in high salt conditions, respectively. The peaks for 40S and 60S subunits, 80S ribosomes, polysomes, and halfmers are indicated. (D) Nuclear export of pre-60S subunits is impaired in yvh1Δ cells. yvh1Δ cells containing either the 60S subunit reporter L25-GFP (left) or the 40S subunit reporter S2-GFP (right) were transformed with the indicated plasmids. Cells were grown in SD-Leu medium at 30°C, and the subcellular localization of L25-GFP and S2-GFP were visualized by fluorescence microscopy. (E) Schematic of the YVH1 domains. YVH1 contains an N-terminal dual-specificity phosphatase domain (DSPc; catalytic domain) and a C-terminal Zn2+-binding domain.
Yvh1 is a modular two-domain protein that contains an N-terminal dual-specificity phosphatase catalytic domain and a C-terminal Zn2+-binding domain (Fig. 1 E; Beeser and Cooper, 1999; Muda et al., 1999; Aoki et al., 2001; Sakumoto et al., 2001; Liu and Chang, 2009). As previously reported, inactivation of Yvh1 phosphatase activity (Yvh1-C117A) or deletion of the entire N-terminal phosphatase domain (Yvh1-ΔN) did not cause a slow-growth phenotype (Fig. 1 A; Beeser and Cooper, 1999; Muda et al., 1999; Sakumoto et al., 2001; Liu and Chang, 2009). Furthermore, mutations that prevent coordination to Zn2+ or the deletion of the entire C-terminal Zn2+-binding domain (Yvh1-ΔC) were found to be responsible for the yvh1Δ slow-growth phenotype (Fig. 1 A; Beeser and Cooper, 1999; Muda et al., 1999; Aoki et al., 2001; Sakumoto et al., 2001; Liu and Chang, 2009). Thus, we investigated which domain of Yvh1 is required for proper nuclear export of pre-60S particles. Expression of the phosphatase domain alone (Yvh1-ΔC) did not rescue the pre-60S export defect observed in yvh1Δ cells (Fig. 1 D). Interestingly, both Yvh1-C117A and Yvh1-ΔN constructs rescued the 60S export defect (Fig. 1 D and not depicted). Therefore, we conclude that the Zn2+-binding domain, and not the phosphatase domain of Yvh1, is required for proper nuclear export of pre-60S particles. We propose that the yvh1Δ slow-growth phenotype might be a result of impaired nuclear export of pre-60S particles.
Yvh1 coenriches with late pre-60S particles
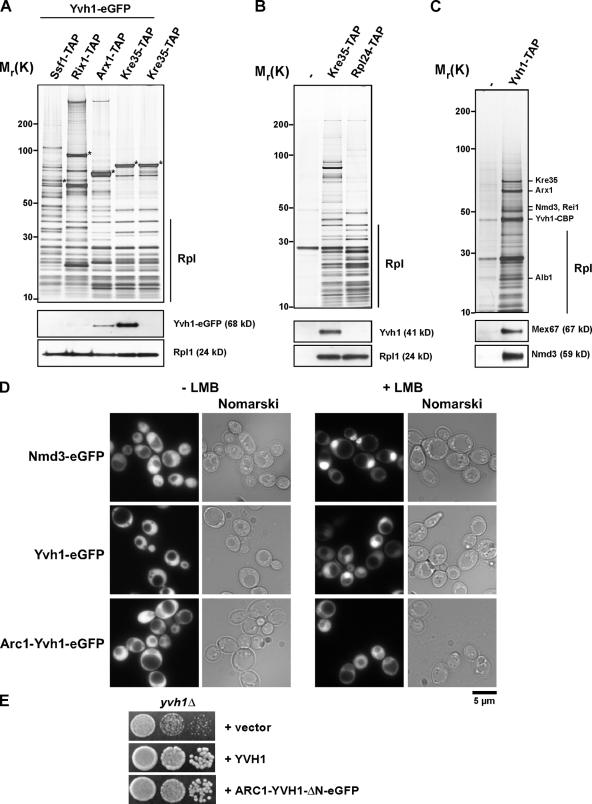
A late pre-60S particle, isolated using the exclusively cytoplasmic GTPase Kre35 as a bait protein for TAP, was reported to coenrich Yvh1 (Nissan et al., 2002). However, the maturation stage on the 60S biogenesis pathway at which Yvh1 joins pre-60S particles was not investigated. To address this, we tagged Yvh1 with GFP at its genomic locus in TAP strains (Ssf1-TAP, Rix1-TAP, Arx1-TAP, and Kre35-TAP) that have been previously used to purify pre-60S particles at different maturation stages, i.e., early to late pre-60S particles (Fig. 2 A; Yao et al., 2007; Kressler et al., 2008). Addition of a C-terminal GFP did not alter the growth rates of the TAP strains, suggesting that the Yvh1-GFP fusion is functional (unpublished data). Pre-60S particles at different stages of maturation were isolated from these strains and analyzed by SDS-PAGE followed by Western blotting for the presence of Yvh1-GFP using an anti-GFP antibody. Yvh1 coenriched only with late preribosomal particles (Arx1-TAP and Kre35-TAP) but not with early preribosomes (Ssf1-TAP and Rix1-TAP; Fig. 2 A). We obtained identical results using antibodies raised against recombinant Yvh1-ΔN (see Fig. 6 D), confirming that the addition of the GFP tag did not alter association with pre-60S particles. Notably, Yvh1 was not found to be associated with mature 60S subunits (Rpl24-TAP; Fig. 2 B). Previously, Yvh1-TAP was detected only in 60S fractions and not in the soluble pool on sucrose density gradient profiles (Liu and Chang, 2009). To further investigate the nature of the pre-60S particle that associates with Yvh1, we isolated Yvh1-TAP from whole cell lysates and analyzed the preparation on SDS-PAGE (Fig. 2 C). Mass spectrometric analysis of the coenriching proteins revealed ribosomal L-proteins and several factors (Arx1, Alb1, Kre35, and Rei1) that are known to associate specifically with late pre-60S particles (Fig. 2 C; Nissan et al., 2002). Additionally, Western analysis showed that Yvh1-TAP coenriched the transport receptors Mex67 and Nmd3 (Fig. 2 C; Yao et al., 2007). Together, these data demonstrate that Yvh1 associates specifically with late pre-60S particles.
Figure 2.
Yvh1 associates with late pre-60S particles in vivo. (A) TAP of early to late pre-60S particles via the indicated TAP-tagged baits from cells expressing Yvh1-GFP. Eluates were separated on 4–12% SDS–polyacrylamide gradient gels and subjected to silver staining (top) or Western blotting (bottom) using GFP antibody to detect Yvh1-GFP. Rpl1 was used as loading control. Asterisks indicate the positions of bait proteins. Rpl, r-proteins of the large (60S) subunit. (B) Cytoplasmic pre-60S particles (Kre35-TAP) and mature 60S subunits (Rpl24-TAP) were isolated via their TAP tag. Wild-type cells (untagged) were used as a negative control. Eluates were separated on 4–12% SDS–polyacrylamide gradient gels and subjected to silver staining (top) or Western blotting (bottom) using antibodies against Yvh1 and Rpl1 (loading control). (C) Protein composition of the Yvh1-TAP purification. Yvh1-TAP was tandem affinity purified and subjected to SDS-PAGE followed by silver staining (top). The indicated protein bands were identified by mass spectrometry. The eluate was further analyzed by Western blotting (bottom) using antibodies against Mex67 and Nmd3. (D) Yvh1 shuttles between the nucleus and cytoplasm in a Crm1-dependent manner. The subcellular localization of Nmd3-GFP, Yvh1-GFP, and Arc1-Yvh1-ΔN-GFP was analyzed by fluorescence microscopy in the LMB-sensitive crm1 mutant (Crm1/Xpo1T539C) in the presence or absence of 100 ng/ml LMB. (E) A cytoplasmic anchored version of Yvh1-ΔN rescues the slow-growth phenotype of yvh1Δ cells. yvh1Δ cells carrying the indicated plasmids were spotted in serial 10-fold dilutions onto SD-Leu plates and incubated at 25, 30, or 37°C for 3 d.
Figure 6.
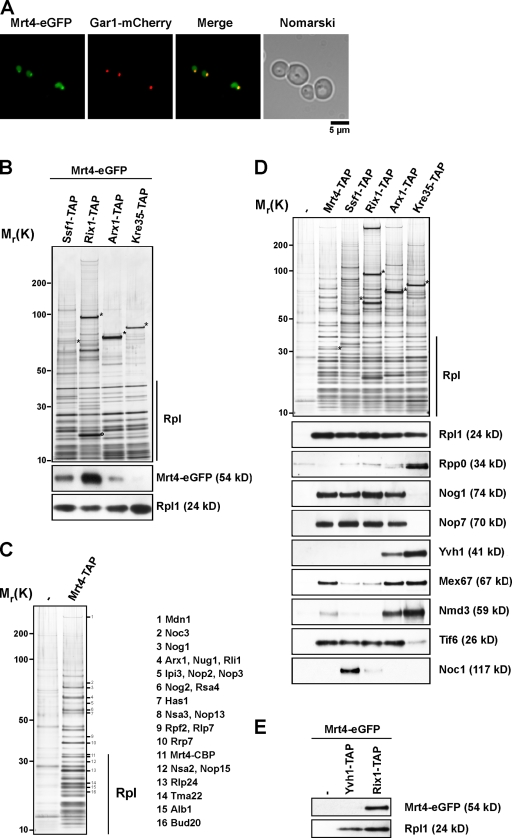
Mrt4 is a nucleolar protein and associates with early pre-60S particles in vivo. (A) The subcellular localization of Mrt4 was determined by fluorescence microscopy from cells expressing Mrt4-GFP and Gar1-mCherry. (B) Mrt4 associates with early pre-60S ribosomal subunits. TAP of early to late pre-60S particles via the indicated TAP-tagged baits from cells expressing Mrt4-GFP. Eluates were separated on 4–12% SDS–polyacrylamide gradient gels and subjected to silver staining (top) or Western blotting (bottom) using antibodies against GFP and Rpl1 (loading control). The circle indicates a Rix1 degradation product as determined by mass spectrometry. Rpl, r-proteins of the large (60S) subunit. (C) Protein composition of the Mrt4-TAP pre-60S particle. Mrt4-TAP was tandem affinity purified and subjected to SDS-PAGE followed by silver staining. The indicated bands (1–16) were excised from the gel, and the proteins were identified by mass spectrometry. (D) TAP of early to late pre-60S particles via the indicated TAP-tagged bait. Eluates were separated on 4–12% SDS–polyacrylamide gradient gels and subjected to silver staining (top) or Western blotting (bottom) using α-Rpl1, α-Rpp0, α-Nog1, α-Nop7, α-Yvh1, α-Mex67, α-Nmd3, α-Tif6, and α-Noc1 antibodies. (E) Yvh1-TAP and Rix1-TAP were tandem affinity purified from strains expressing Mrt4-GFP. Eluates were separated on 4–12% SDS–polyacrylamide gradient gels and subjected to Western blotting using α-GFP and α-Rpl1 antibodies. Wild type (untagged) served as negative control. Asterisks indicate the positions of bait proteins.
The observation that Yvh1-TAP purified shuttling pre-60S factors (Nmd3, Arx1, and Mex67) prompted us to investigate whether Yvh1 is a shuttling protein. To test this, we tagged Yvh1 with GFP in a yeast strain (Crm1/Xpo1T539C) that is sensitive to the inhibitor leptomycin B (LMB; Neville and Rosbash, 1999). Yvh1-GFP, like the pre-60S export factor Nmd3, is excluded from the vacuole and appears to be localized both in the nucleus and cytoplasm (Fig. 2 D). Upon LMB treatment of this strain, nuclear export of pre-60S particles is blocked and thus very rapidly induces nuclear accumulation of the L25-GFP reporter construct and the pre-60S export adaptor Nmd3-GFP (Fig. 2 D and not depicted; Ho and Johnson, 1999; Gadal et al., 2001). In contrast, nuclear accumulation of Yvh1-GFP was observed in ∼70% of the cells after 1 h of LMB treatment (Fig. 2 D). Similar nuclear accumulation was observed upon the overexpression of the dominant-negative nmd3-Δ100 allele (Hedges et al., 2005; Lebreton et al., 2006; unpublished data). These data suggest that Yvh1 can shuttle between the nucleus and cytoplasm.
Next, we asked whether Yvh1 could perform its function in the 60S biogenesis pathway exclusively in the cytoplasm. Previously, a dominant-negative phenotype of a catalytically active domain of the desumoylating enzyme, Ulp1 was rescued by excluding it from the nucleus upon fusion with the exclusively cytoplasmic protein Arc1 (Panse et al., 2003). Thus, to tether Yvh1-ΔN to the cytoplasm, we fused the Zn2+-binding domain of Yvh1 (Yvh1-ΔN) to Arc1 (Arc1-Yvh1-ΔN-GFP). First, we assessed whether the Arc1-Yvh1-ΔN-GFP fusion protein, like Yvh1-GFP, was trapped in the nucleus in the Crm1/Xpo1T539C yeast strain upon LMB treatment. We did not observe any nuclear accumulation of this fusion protein in the nucleus even after LMB treatment for >2 h, strongly suggesting that Arc1 was able to very efficiently anchor Yvh1-ΔN in the cytoplasm. Next, we asked whether the Arc1-Yvh1-ΔN-GFP fusion construct could rescue the yvh1Δ slow-growth phenotype and pre-60S export defect. This was indeed the case; Arc1-Yvh1-ΔN-GFP rescued the slow-growth phenotype of yvh1Δ cells to the same extent as the wild-type Yvh1 at all temperatures tested (Fig. 2 E and not depicted) and rescued the pre-60S export defect (not depicted). These data suggest that Yvh1 can perform its function in the 60S pathway when tethered to the cytoplasm.
Gain of function alleles of Mrt4 rescue slow growth and 60S export defects of yvh1Δ cells
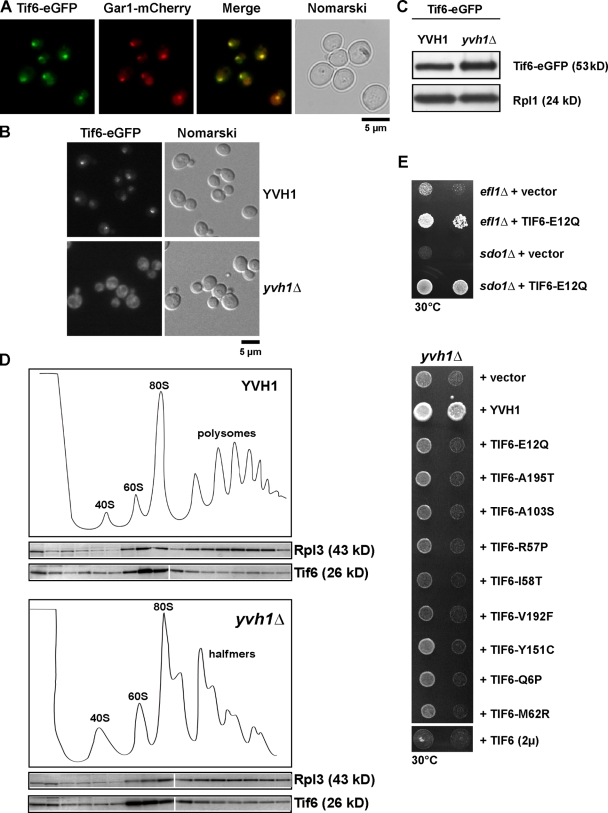
Previous studies demonstrated a specific requirement for the cytoplasmic factors Efl1 and Sdo1 in the release of the nucleolar maturation factor Tif6 from pre-60S particles (Senger et al., 2001; Menne et al., 2007). A consequence of this is that Tif6-GFP mislocalized to the cytoplasm in efl1Δ and sdo1Δ cells (Senger et al., 2001; Menne et al., 2007). We asked whether Yvh1 functions in the recycling of Tif6 from late cytoplasmic pre-60S particles. Tif6-GFP localization was investigated in YVH1 and yvh1Δ cells by fluorescence microscopy. In YVH1 cells, Tif6-GFP at steady state exhibits nucleolar/nucleoplasmic staining as judged by colocalization of Tif6-GFP and the nucleolar marker Gar1-mCherry (Fig. 3 A). However, in yvh1Δ cells, Tif6-GFP mislocalizes to the cytoplasm (Fig. 3 B) as previously observed in sdo1Δ and efl1Δ cells (Senger et al., 2001; Menne et al., 2007). Western analysis showed that Tif6-GFP levels in Yvh1 and yvh1Δ remain unaltered (Fig. 3 C). To determine whether Tif6 remained bound to pre-60S subunits in the cytoplasm in yvh1Δ cells, whole cell lysates derived from yvh1Δ cells were subject to sucrose density gradient analyses. As shown in Fig. 3 D, Tif6 copeaked with 60S fractions in both YVH1 and yvh1Δ, suggesting that Tif6 remains bound to pre-60S particles. These findings raised the possibility that the slow-growth phenotype and 60S export defect observed in yvh1Δ cells might be caused by impaired recycling of Tif6. Like the yvh1Δ strain, both efl1Δ and sdo1Δ strains exhibit a slow-growth phenotype (Fig. 3 E). Previously, it was reported that the slow-growth phenotype of efl1Δ and sdo1Δ strains could be suppressed by either the overexpression of Tif6 (expression of Tif6 on a 2µ plasmid) or by expressing dominant gain of function alleles of Tif6 (Senger et al., 2001; Menne et al., 2007). If Yvh1 was directly involved in the recycling of Tif6, then increased expression of Tif6 or dominant gain of function alleles of Tif6 should rescue the yvh1Δ slow-growth phenotype. Although increased expression of Tif6 or expression of the dominant gain of function alleles of Tif6 rescued the slow-growth phenotype of efl1Δ and sdo1Δ strains (Fig. 3 E and not depicted), it did not rescue the slow-growth phenotype of the yvh1Δ strain. These data suggest that Yvh1 might not be directly involved in the recycling of Tif6 back to the nucleolus.
Figure 3.
Yvh1 is not directly involved in the recycling of Tif6. (A) Tif6 is a predominantly nucleolar protein. Cells expressing Tif6-GFP and the nucleolar marker Gar1-mCherry were analyzed by fluorescence microscopy. (B) Tif6 is mislocalized to the cytoplasm in yvh1Δ cells. Subcellular localization of Tif6-GFP was analyzed by fluorescence microscopy in wild-type and yvh1Δ cells. (C) Tif6-GFP protein levels were analyzed by Western blotting (α-GFP) in whole cell extracts prepared from wild-type and yvh1Δ cells. Rpl1 serves as loading control. (D) Sucrose density gradient localization of Tif6 in YVH1 and yvh1Δ cells. Whole cell lysates derived from YVH1 and yvh1Δ cells were applied to a sucrose density gradient and fractionated. The collected fractions were analyzed on a 4–12% gradient SDS-PAGE and subjected to Western analyses using the indicated antibodies. White lines indicate that intervening lanes have been spliced out. (E) efl1Δ and sdo1Δ cells were transformed with empty vector or a plasmid expressing the gain of function allele Tif6-E12Q. The resulting strains were spotted in serial 10-fold dilutions onto SD-Leu plates and incubated at 30°C for 3 d (top). yvh1Δ cells were transformed with the indicated plasmids. The resulting strains were spotted in serial 10-fold dilutions onto SD-Leu plates and incubated at 30°C for 3 d (bottom).
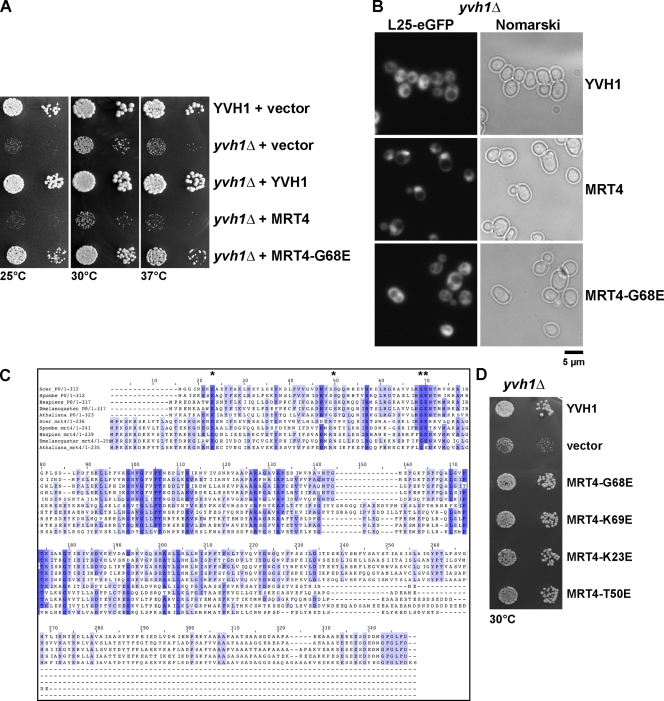
Thus, to get further insights into the function of Yvh1 in 60S biogenesis, we resorted to a previously described genetic approach (Senger et al., 2001). We observed that, like the efl1Δ and sdo1Δ cells, the yvh1Δ strain gave rise to spontaneous suppressors (fast-growing colonies), albeit with a lower frequency. Four fast-growing colonies were obtained after incubating ∼109 yvh1Δ cells at 25°C. Standard yeast genetic analyses showed that one was a dominant suppressor. Cloning of this dominant suppressor was pursued because the recessive suppressors did not exhibit any observable phenotype on their own. A genomic library was constructed from the dominant suppressor strain and transformed into yvh1Δ cells. Plasmids from fast-growing colonies were rescued. Sequence analyses revealed that only a mutated version of the pre-60S factor Mrt4 (Mrt4-G68E), not the wild-type Mrt4, rescued both the yvh1Δ slow-growth and pre-60S export defect (Fig. 4, A and B).
Figure 4.
MRT4 gain of function alleles can rescue the slow growth and 60S export defects of yvh1Δ cells. (A) YVH1 and yvh1Δ cells carrying the indicated plasmids were spotted in serial 10-fold dilutions onto SD-Leu plates and incubated at 25, 30, or 37°C for 3 d. (B) yvh1Δ cells containing the 60S subunit reporter L25-GFP were transformed with the indicated plasmids. The subcellular localization of L25-GFP was visualized by fluorescence microscopy. (C) Sequence alignments of RPP0 and MRT4 from the indicated organisms were performed using the program ClustalX. Asterisks indicate the conserved amino acids that make contact with 25S rRNA that have been subjected to site-directed mutagenesis. (D) Rescue of the slow-growth phenotype by mutations in 25S rRNA–interacting amino acid residues. yvh1Δ cells carrying the indicated plasmids were spotted in serial 10-fold dilutions onto SD-Leu plates and incubated at 30°C for 3 d.
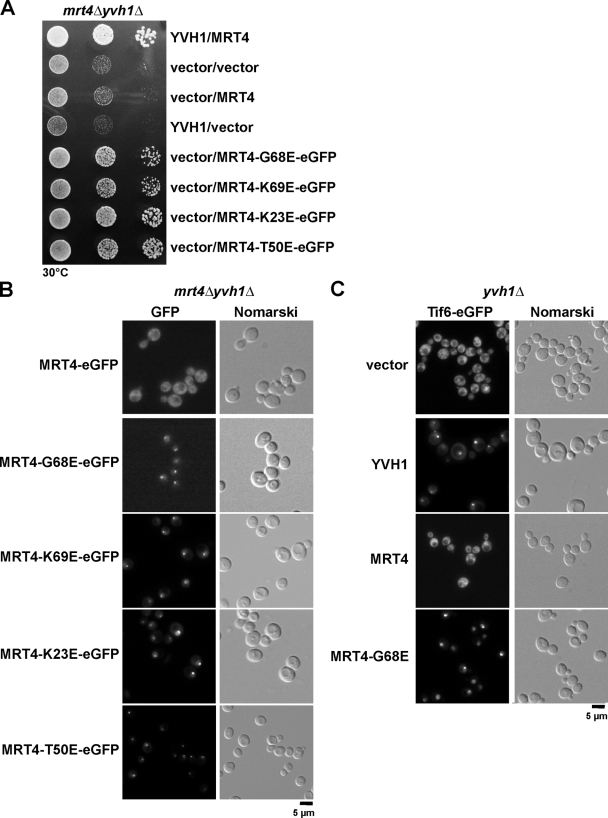
Mrt4 encodes a ribosomal-like protein, which was previously shown to be associated with early pre-60S particles and is highly homologous to the acidic ribosomal P-protein Rpp0 (Zuk et al., 1999; Rodriguez-Mateos et al., 2009). Mrt4 and its ribosomal counterpart Rpp0 are highly conserved proteins and have homologues in all eukaryotes (Fig. 4 C; Wahl and Möller, 2002; Diaconu et al., 2005). The prokaryotic equivalent for Rpp0 is L10e. Crystallographic experiments on the 50S subunit of the Escherichia coli ribosome have shed light on the electrostatic interactions between specific amino acid residues on L10e and the phosphate backbone of 23S rRNA (Diaconu et al., 2005). Interestingly, residues on L10e that make contact with the phosphate backbone of 23S rRNA are also highly conserved in the eukaryotic Rpp0 and its ribosomal-like protein homologue, Mrt4 (Zuk et al., 1999; Diaconu et al., 2005; Rodriguez-Mateos et al., 2009). Notably, we found that the dominant G68E mutation in Mrt4, which rescued the yvh1Δ-associated phenotypes, maps to one of its conserved 25S rRNA–binding residues (Fig. 4 C). We asked whether similar point mutations in other 25S rRNA–binding patches of Mrt4 could also rescue the slow growth and 60S export defect of yvh1Δ cells (Zuk et al., 1999; Diaconu et al., 2005). To this end, we created three other point mutations in the conserved patches K69E, K23E, and T50E (Fig. 4 C, asterisks). Intriguingly, like the MRT4-G68E allele, all three point mutations rescued the slow-growth phenotype and 60S export defect observed in yvh1Δ cells (Fig. 4 D and not depicted). We conclude that the gain of function alleles of Mrt4, which harbor mutations in the 25S rRNA–binding patches, rescue both the slow-growth phenotype and the 60S export defect seen in yvh1Δ cells.
Mrt4 is required for proper nuclear export of pre-60S particles
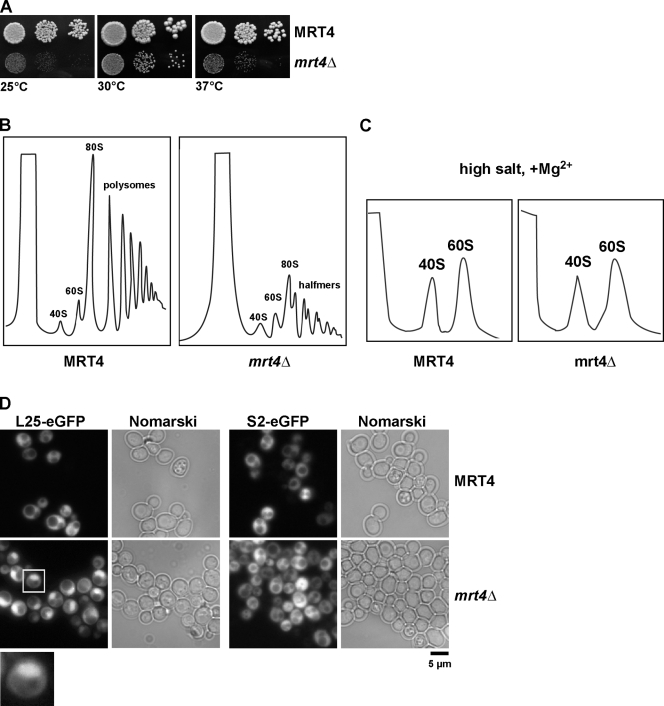
Proteomic approaches have revealed that several early pre-60S particles coenrich the ribosomal-like protein Mrt4 (Bassler et al., 2001; Harnpicharnchai et al., 2001; Nissan et al., 2002). We asked whether Mrt4, like Yvh1, was required for proper nuclear export of pre-60S particles. To address this, we first disrupted MRT4 in diploid wild-type cells. As previously reported, tetrad analysis revealed that mrt4Δ is not an essential gene and induces a slow-growth phenotype at all temperatures tested (Fig. 5 A; Zuk et al., 1999). Sucrose density gradient analysis of yeast lysates derived from mrt4Δ cells under polysome-preserving conditions showed the presence of halfmers and no significant decrease in 60S subunits (Fig. 5 B; Harnpicharnchai et al., 2001). Furthermore, sucrose gradient analyses under dissociative high salt gradients showed a relatively subtle decrease in free 60S subunits in mrt4Δ-derived lysates (Fig. 5, B and C). Next, we investigated the localization of the 60S (L25-GFP and L11-GFP) and 40S (S2-GFP) reporter constructs in wild-type and mrt4Δ cells. As expected, mrt4Δ cells accumulated only the 60S subunit reporter constructs in the nucleoplasm but not the 40S reporter S2-GFP (Fig. 5 D and not depicted). We conclude that like Yvh1, Mrt4 is required for proper nuclear export of pre-60S particles.
Figure 5.
MRT4 is required for proper pre-60S export. (A) Wild-type and mrt4Δ cells were spotted in serial 10-fold dilutions onto YPD plates and incubated at 25, 30, and 37°C for 3 d. (B and C) Analysis of polysome profiles (OD254nm) of the indicated wild-type and mrt4Δ cells by sedimentation centrifugation on sucrose density gradients after cycloheximide treatment and under high salt conditions, respectively. The peaks for 40S and 60S subunits, 80S ribosomes, polysomes, and halfmers are indicated. (D) Nuclear export of 60S subunits is impaired in mrt4Δ cells. MRT4 and mrt4Δ cells containing either the 60S subunit reporter L25-GFP (left) or the 40S subunit reporter S2-GFP (right) were grown in SD medium at 30°C, and the subcellular localization of L25-GFP and S2-GFP was visualized by fluorescence microscopy. The inset shows a magnified image of the nuclear accumulation of the L25-GFP reporter (boxed region).
Mrt4-TAP coenriches early- to late-associating factors on the 60S pathway
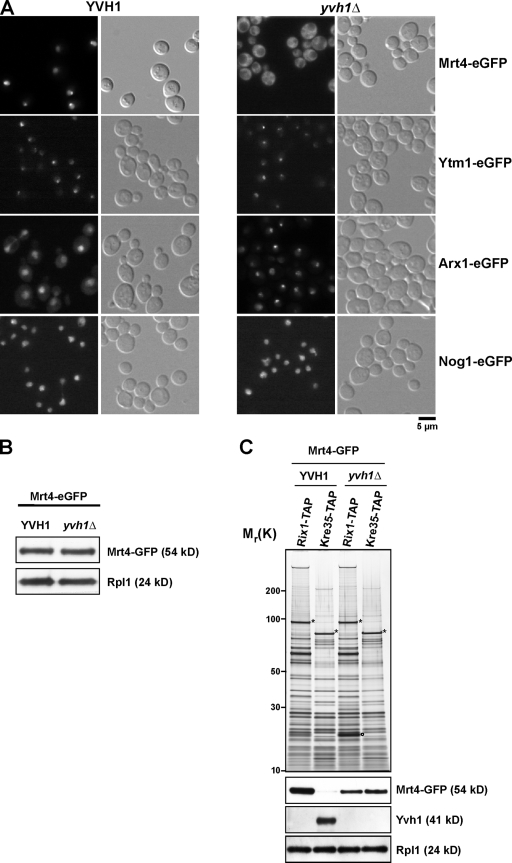
Mrt4-GFP is enriched in the nucleolus with nucleoplasmic staining at steady state (Fig. 6 A). Consistent with this, previous studies have shown that Mrt4 coenriches with several early pre-60S particles (Bassler et al., 2001; Harnpicharnchai et al., 2001; Nissan et al., 2002). However, the question of when Mrt4 joins and leaves the pre-60S particles has not been investigated. To address this, we tagged Mrt4 with GFP in TAP strains (Ssf1-TAP, Rix1-TAP, Arx1-TAP, and Kre35-TAP) that purify early to late pre-60S particles. Addition of a C-terminal GFP did not alter the growth characteristics of the TAP strains, suggesting that Mrt4-GFP fusion is functional (unpublished data). We isolated pre-60S particles from these strains and analyzed the preparations on an SDS-PAGE and by Western blotting for the presence of Mrt4-GFP using anti-GFP antibody. Mrt4 coenriched only with early and nuclear preribosomal particles on Ssf1-TAP, Rix1-TAP, and is found in low amounts on the pre-60S isolated from Arx1-TAP. Mrt4-GFP is completely absent on an exclusively cytoplasmic pre-60S particle (Kre35-TAP; Fig. 6 B). Next, we isolated Mrt4-TAP and analyzed the preparation by SDS-PAGE. The identity of coenriching proteins was determined by mass spectrometry (Fig. 6 C). Mrt4-TAP coenriched ribosomal L-proteins and myriad early to late pre-60S factors (Fig. 6 C). To better characterize the Mrt4-TAP purification in relation to other TAP-tagged bait proteins (Ssf1, Rix1, Arx1, and Kre35), which are well-described markers for different (early to late) pre-60S particles, we performed Western analysis using antibodies against several pre-60S marker proteins to further distinguish between nucleolar, late nucleolar/early nucleoplasmic, and late nucleoplasmic pre-60S particles (Fig. 6 D; Kressler et al., 2008). Mrt4-TAP did not purify Rpp0, which is consistent with the notion that Rpp0 and Mrt4 might mutually exclude each other on the pre-60S (Fig. 6 D; Rodriguez-Mateos et al., 2009). Importantly, as previously observed, Rpp0 coenriches with cytoplasmic pre-60S particles and not with early pre-60S particles (Fig. 6 D; Kressler et al., 2008). Mrt4-TAP coenriches several nucleolar/nucleoplasmic pre-60S factors such as Nog1, Nop7, and Tif6 but not the early nucleolar factor Noc1 (Milkereit et al., 2001). Remarkably, Mrt4-TAP reproducibly coenriched low amounts of pre-60S export factors Mex67 and Nmd3 and late-associating shuttling pre-60S factors Arx1, Alb1, Rli1, and Rlp24 (Fig. 6, C and D). Interestingly, Mrt4-TAP did not coenrich Yvh1 (Fig. 6 D). Moreover, in a reciprocal experiment, Mrt4-GFP also did not coenrich in the Yvh1-TAP purification (Fig. 6 E). We conclude that Mrt4-TAP coenriches several early- and few late-associating factors on the 60S biogenesis pathway.
Yvh1 is required for the release of Mrt4 from cytoplasmic pre-60S particles
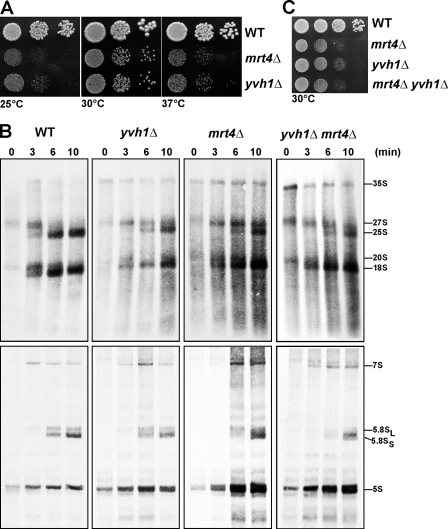
Intriguingly, we found that the slow-growth phenotype of mrt4Δ cells was very similar to that of yvh1Δ cells (Fig. 7 A). Moreover, pulse-chase analysis revealed that both yvh1Δ and mrt4Δ cells showed a very similar delay in the processing of 27S rRNA to 25S rRNA (Fig. 7 B, top). No delay in the processing of the small subunit 20S rRNAs and small rRNAs (5.8S and 5S) was observed (Fig. 7 B, bottom). Notably, the mrt4Δyvh1Δ double-deletion strain was viable and showed similar growth characteristics and a very similar delay in processing of 27S rRNA to 25S rRNA as the individual deletion yvh1Δ and mrt4Δ strains (Fig. 7, B and C), suggesting that Yvh1 and Mrt4 might not act on parallel pathways. These data raised the question of how the nucleolar pre-60S factor Mrt4 is functionally linked to the pre-60S factor Yvh1. Two observations prompted us to investigate whether Yvh1 was required for the nucleolar recycling of Mrt4: (1) dominant gain of function alleles of Mrt4 that rescue the yvh1Δ-associated phenotypes map into the conserved 25S rRNA–binding sites, and (2) Mrt4-TAP coenriches low amounts of late pre-60S factors. We asked whether the steady-state nucleolar localization of Mrt4-GFP was perturbed in yvh1Δ cells. This was indeed the case, as Mrt4-GFP mislocalizes to the cytoplasm in yvh1Δ cells (Fig. 8 A). Importantly, protein levels of Mrt4-GFP in Yvh1 and yvh1Δ remain unaltered (Fig. 8 B). Next, we investigated whether the cytoplasmic Mrt4-GFP still remained bound to late pre-60S particles in yvh1Δ cells. We purified Rix1-TAP and Kre35-TAP from yvh1Δ cells that expressed Mrt4-GFP. In wild-type cells, Mrt4-GFP was found maximally enriched in the Rix1-TAP and absent in the Kre35-TAP purification (Fig. 6 B and Fig. 8 C). However, in yvh1Δ cells, a marked decrease in the amount of Mrt4-GFP in Rix1-TAP accompanied with an increase in the Kre35-TAP particle was seen (Fig. 8 C). Is the mislocalization of Mrt4-GFP observed in yvh1Δ cells specific? The localization of several nucleolar pre-60S factors (Nsa3, Nop7, and Ytm1) was not affected in yvh1Δ cells (Fig. 8 A and not depicted). Previous studies have shown that the pre-60S factors Nog1 and Arx1 travel to the cytoplasm and that the exclusively cytoplasmic localized proteins AAA-ATPase Drg1 and Rei1 were required for their release from pre-60S (Hung and Johnson, 2006; Lebreton et al., 2006; Pertschy et al., 2007). In yvh1Δ cells, Nog1-GFP localization remained completely unaltered. However, Arx1-GFP showed a nucleolar staining as opposed to the nucleoplasmic/cytoplasmic staining seen in wild-type cells (Fig. 8 A). Thus, Yvh1 is not required for the recycling of Nog1 and Arx1 back to the nucleus. All of these data strongly suggest that Yvh1 is required for the release of Mrt4 from late pre-60S particles.
Figure 7.
mrt4Δ and yvh1Δ cells show similar phenotypes. (A) Wild-type (WT), mrt4Δ, and yvh1Δ cells were spotted in serial 10-fold dilutions onto YPD plates and incubated at 25, 30, or 37°C for 3 d. (B) Logarithmically growing cultures of wild-type, yvh1Δ, mrt4Δ, and yvh1Δmrt4Δ cells were pulse labeled with 100 µCi [3H]uridine for 1 min and chased with an excess of nonradioactive uridine for 0, 3, 6, and 10 min. RNA was extracted by the hot phenol method and was separated on a 1.2% agarose formaldehyde gel to follow the processing of high molecular weight rRNA precursors (top) or on an 8% acrylamide 7 M urea gel to detect the formation of low molecular weight RNAs (bottom). The gels were blotted, and the radioactivity was detected by autoradiography. (C) Wild-type, mrt4Δ, yvh1Δ, and mrt4Δyvh1Δ cells were spotted in serial 10-fold dilutions onto YPD and incubated at 30°C for 3 d.
Figure 8.
Yvh1 is required for nucleolar recycling of Mrt4. (A) The distribution of Mrt4-GFP, Ytm1-GFP, Arx1-GFP, and Nog1-GFP in wild-type and yvh1Δ cells was visualized by fluorescence microscopy. (B) Whole cell extracts were prepared from wild-type and yvh1Δ strains expressing Mrt4-GFP. The extracts were analyzed by SDS-PAGE and subjected to Western analysis using α-GFP and α-Rpl1 antibodies. (C) Nuclear (Rix1-TAP) and cytoplasmic (Kre35-TAP) pre-60S particles were purified from wild-type and yvh1Δ strains expressing Mrt4-GFP. Eluates were separated on 4–12% SDS–polyacrylamide gradient gels and subjected to silver staining (top) or Western blotting (bottom) using α-GFP, α-Yvh1, and α-Rpl1 antibodies. Asterisks indicate the positions of bait proteins. The circle indicates a Rix1 degradation product as determined by mass spectrometry.
Mrt4 gain of function alleles bypass the requirement for Yvh1 recycling function
Finally, we investigated the basis underlying the rescue of the slow-growth phenotype and 60S export defects observed in yvh1Δ cells by the dominant gain of function alleles of Mrt4. A plausible explanation could be that these gain of function alleles of Mrt4 were efficiently released from pre-60S particles independent of Yvh1 and recycled back to the nucleolus to carry out an important yet unknown nucleolar/nucleoplasmic function. To test this, we created C-terminal GFP fusions of wild type and all dominant alleles of Mrt4 and analyzed their growth characteristics after transformation into yvh1Δ and yvh1Δmrt4Δ strains. Wild-type Mrt4-GFP did not rescue the yvh1Δ and yvh1Δmrt4Δ slow-growth phenotypes. However, all of the GFP-tagged dominant alleles of Mrt4 rescued the slow-growth phenotype of both yvh1Δ and yvh1Δmrt4Δ cells (Fig. 9 A and not depicted). Next, we assessed their localization by fluorescence microscopy. As expected, the plasmid-borne Mrt4-GFP mislocalizes to the cytoplasm in the yvh1Δmrt4Δ cells (Fig. 9 B). Interestingly, all of the dominant alleles localized to the nucleolus at steady state in the yvh1Δmrt4Δ strain. Thus, the dominant gain of function alleles of Mrt4 completely bypassed the requirement of Yvh1 for their cytoplasmic release. Moreover, we observed that the untagged dominant alleles of Mrt4, not the wild type, rescued the cytoplasmic mislocalization of Tif6-GFP seen in yvh1Δ cells (Fig. 9 C). We conclude that the dominant gain of function alleles of Mrt4 bypass the requirement of Yvh1 and therefore rescue all the yvh1Δ-associated phenotypes.
Figure 9.
MRT4 gain of function alleles relocalize back to the nucleolus in the absence of YVH1. (A) yvh1Δmrt4Δ cells carrying the indicated plasmids were spotted in serial 10-fold dilutions onto SD-Leu-Ura plates and incubated at 30°C for 3 d. (B) yvh1Δmrt4Δ cells were transformed with the indicated plasmids. The subcellular localization of MRT4 and dominant MRT4 alleles was visualized by fluorescence microscopy. (C) yvh1Δ cells expressing Tif6-GFP were transformed with the indicated plasmids. The subcellular localization of Tif6-GFP was visualized by fluorescence microscopy.
Discussion
In contrast to prokaryotes, eukaryotic ribosome biogenesis requires a large number of trans-acting factors that aid proper assembly and maturation of preribosomal subunits (Tschochner and Hurt, 2003). These factors associate with preribosomal particles at distinct stages and participate in specific yet unknown maturation events. Upon accomplishing their task, these maturation factors are released and recycled to participate in the next round of maturation. Little is known regarding the mechanisms or the cofactors that are required for the release of trans-acting factors from pre-60S particles at distinct maturation steps. In this study, we provide several lines of evidence that Yvh1 is specifically required for the release of the nucleolar trans-acting factor Mrt4 from pre-60S particles. First, we show that Mrt4-GFP, which at steady state localizes to the nucleolus, is mislocalized to the cytoplasm in yvh1Δ cells (Fig. 8 A). Notably, nucleolar/nuclear trans-acting factors such as Nog1 and Arx1 that travel to the cytoplasm were not mislocalized to the cytoplasm in yvh1Δ cells. Additionally, localization of several other tested nucleolar factors involved in pre-60S biogenesis remained unaltered. Thus, the mislocalization of Mrt4 to the cytoplasm in yvh1Δ cells is specific. The reason why Arx1 shows a more nucleolar localization in yvh1Δ cells remains unclear and needs further investigation. Second, biochemical analyses revealed that Mrt4-GFP failed to be released from late pre-60S particles in yvh1Δ cells (Fig. 8 C). This release step appears to be an extremely efficient process because Kre35-TAP does not coenrich Mrt4-GFP in wild-type cells (Fig. 6 B and Fig. 8 C). Third, via an unbiased genetic screen, we identified a dominant gain of function allele of Mrt4 that rescued the slow-growth phenotype and pre-60S export defect of yvh1Δ cells. Interestingly, a very similar mutation in Mrt4 (MRT4-G68D) was reported to rescue the slow-growth phenotype of yvh1Δ cells (Nugroho, S., N. Sakumoto, and S. Harashima. 2003. International Conference on Yeast Genetics and Molecular Biology. Abstr. 10-42). However, the nature of this genetic interaction was not further examined. Curiously, the dominant mutation in Mrt4 (MRT4-G68E) maps into a conserved 25S rRNA–binding patch. These mutations are predicted to lower the affinity of Mrt4 for pre-60S particles. Consistent with this, we observed that other mutations introduced into conserved residues, which are predicted to make contact with 25S rRNA, also rescued yvh1Δ-associated phenotypes (Fig. 4). Finally, these dominant alleles were able to recycle without the requirement of Yvh1 and relocalize to their site of action, the nucleolus (Fig. 9 B). Altogether, these data strongly suggest that the late-associating pre-60S factor Yvh1 is specifically required for release of Mrt4 from pre-60S particles.
In which cellular compartment does the release of Mrt4 occur? Yvh1 was previously shown to coenrich with late cytoplasmic pre-60S particles. Yvh1 is present in low amounts in Arx1-TAP and maximally enriched in Kre35-TAP (Fig. 2 A and Fig. 6 D). Interestingly, Yvh1 does not coenrich with mature 60S subunits (Rpl24-TAP), suggesting that Yvh1 visits late pre-60S particles during a short time window to perform its role in Mrt4 release (Fig. 2 B). Our cell biological data suggest that Yvh1 is a shuttling protein because Yvh1-GFP accumulated in the nucleus after nuclear export of pre-60S was impaired either by LMB treatment or overexpression of the dominant-negative nmd3-Δ100 allele (Fig. 2 D and not depicted). We found that a nonshuttling, exclusively cytoplasmically tethered Yvh1-ΔN can rescue the yvh1Δ slow-growth phenotype, suggesting that Yvh1 can also perform its role in the cytoplasm. In the accompanying paper, Lo et al. (2009) show that a nuclear enriched Yvh1 can rescue yvh1Δ-associated phenotypes. These data suggest that the release of the nucleolar pre-60S factor Mrt4 might occur in both nuclear and cytoplasm compartments.
When is the acidic r-protein Rpp0 incorporated into pre-60S particles? Nearly all 60S r-proteins are incorporated during early nucleolar pre-60S biogenesis steps (Nissan et al., 2002). However, few r-proteins are incorporated at late stages of maturation of the pre-60S particles (L10, L24, and the acidic ribosomal P-protein Rpp0; Zinker and Warner, 1976; Kruiswijk et al., 1978; Saveanu et al., 2003). As previously observed, we find that Rpp0 coenriches only with late cytoplasmic pre-60S particles (Fig. 6 D; Kressler et al., 2008). Moreover, Mrt4 and Rpp0 mutually exclude each other on pre-60S particles because Mrt4-TAP did not coenrich Rpp0 (Fig. 6 D). Thus, the incorporation of Rpp0 requires the prior removal of Mrt4 from pre-60S particles. Because the release of Mrt4 from pre-60S particles requires a late-associating pre-60S factor, Yvh1, the incorporation of Rpp0 into pre-60S particles is likely to be a late cytoplasmic event on the 60S maturation pathway. This is in line with the observation made by Lo et al. (2009) that Rpp0-GFP does not accumulate in the nucleus upon impairment of 60S export.
How is Mrt4 released from late pre-60S particles? We asked whether the release of Mrt4 from pre-60S in yvh1Δ might be promoted by providing increased amounts of Rpp0. However, overexpression of Rpp0 did not rescue yvh1Δ-associated phenotypes in the 60S biogenesis pathway (unpublished data). Yvh1 is a dual-specificity phosphatase. Unexpectedly, the C-terminal Zn2+-binding domain, not the phosphatase activity, was found to be necessary for proper pre-60S export (Fig. 1 D). Next, we speculated that upon binding the pre-60S, Yvh1might induce a conformational change in the pre-60S particle that reduces its affinity for Mrt4. However, Mrt4-GFP was not released from pre-60S particles upon the addition of yeast lysates supplemented with energy (GTP and ATP) and/or recombinant Zn2+-binding domain of Yvh1 (unpublished data). It is possible that this release might involve additional components that might not efficiently purify with late pre-60S particles. Determining the precise mechanism by which the Zn2+-binding domain of Yvh1 stimulates the release of Mrt4 from late pre-60S particles remains a challenge for the future.
In conclusion, this study has uncovered a late maturation step in the 60S biogenesis pathway. Notably, DUSP12, the human homologue of Yvh1, can rescue the yvh1Δ slow-growth phenotype (Muda et al., 1999). Thus, the role of Yvh1 in late maturation of pre-60S particles might be conserved from yeast to humans.
Materials and methods
Yeast strains and yeast genetic methods
The S. cerevisiae strains used in this study are listed in Table S1. Genomic disruptions and C-terminal tagging at the genomic loci were performed as described previously (Longtine et al., 1998; Puig et al., 2001; Janke et al., 2004). Preparation of media, yeast transformations, and genetic manipulations were performed according to established procedures.
Genetic screening and analysis
Four fast-growing colonies (revertants) were obtained after plating ∼109 yvh1Δ cells at 25°C for 3 d on YPD. Using standard yeast genetic analysis, we tested whether the revertant loci were dominant or recessive; for this, we mated the fast-growing strains with yvh1Δ of the opposite mating type and asked whether the slow-growth phenotype of the resulting diploid yvh1Δ cells were rescued or not. These analyses showed that the slow-growth phenotype of only one revertant colony was rescued. Next, we dissected tetrads from this diploid strain and showed that a single locus was able to suppress the yvh1Δ slow-growth phenotype. Thus, one of the four revertants rescued the yvh1Δ slow-growth phenotype in a dominant manner. A genomic library was constructed from the original revertant strain and transformed into the yvh1Δ strain to clone the dominant suppressor. Plasmids from transformants that rescued the yvh1Δ slow-growth phenotype at 25°C were extracted and sequenced to determine the suppressor mutation. Finally, a side-directed point mutation in Mrt4 was independently made to confirm that it is indeed a dominant suppressor mutation.
Plasmid constructs and site-directed mutagenesis
Plasmids and oligonucleotides used in this study are listed in Tables S2 and S3, respectively. All recombinant DNA techniques were performed according to established procedures using E. coli XL1 blue cells for cloning and plasmid propagation. Cloned DNA fragments generated by PCR amplification were verified by sequencing. Site-directed mutations were created using QuickChange (Agilent Technologies) according to the manufacturer′s instructions. Mutagenized plasmids were verified by sequencing.
Fluorescence microscopy and L25-GFP reporter assay
The L25-GFP and S2-GFP reporter assays to analyze ribosomal subunit export were performed as previously described (Panse et al., 2006; Yao et al., 2007, 2008). In brief, wild-type, mrt4Δ, and yvh1Δ cells transformed with GFP reporter constructs were grown to OD600 >8, subsequently diluted to fresh medium, and grown for 2 h before the cells were examined by fluorescence microscopy at room temperature. A microscope (DM6000B; Leica) fitted with a 63× 1.25 NA or a 100× 1.30–0.60 NA oil immersion lens (HCX PL Fluotar; Leica) was used to visualize the cells. Pictures were acquired with a digital camera (ORCA-ER; Hamamatsu Photonics) and Openlab software (PerkinElmer).
Sucrose gradient analysis and fractionation
Before isolation of ribosomes either in polysome-preserving conditions or high salt conditions, wild-type, mrt4Δ, and yvh1Δ cells were grown to OD600 >8, subsequently diluted to fresh medium, and grown for 2 h before treatment with cycloheximide or before harvesting for high salt gradients. For polysome-preserving conditions, 100 µg/ml cycloheximide was added to the culture, and it was cooled on ice for 5 min. Cells were harvested by centrifugation, washed once in lysis buffer (10 mM Tris-Cl, pH 7.4, 100 mM NaCl, 30 mM MgCl2, and 100 µg/ml cycloheximide), and disrupted by glass-bead lysis in 1 ml of lysis buffer. After clarification of the lysate (14,000 rpm for 10 min at 4°C), the supernatant corresponding to an OD260 of 4 was loaded onto a 7–50% sucrose density gradient containing 50 mM Tris-acetate, pH 7.4, 50 mM NH4Cl, and 12 mM MgCl2. The gradient was centrifuged at 39,000 rpm for 165 min in a rotor (SW41; Beckman Coulter) and analyzed at 254 nm using a density gradient fractionator. For polysome runoff experiments under high salt conditions (10 mM Tris-Cl, pH 7.4, 800 mM KCl, and 10 mM MgCl2), extracts were prepared in the absence of cycloheximide. For Western analysis, 500 µl of fractions was collected and TCA was precipitated. The acetone-washed pellets were resuspended in 100 µl of sample loading buffer and subsequently separated on NuPAGE 4–12% SDS– polyacrylamide gradient gels (Invitrogen) and analyzed by Western blotting.
Preparation of total yeast protein extracts and Western analysis
Whole cell lysates from yeast cells were prepared by the alkaline lysis method. In brief, cultures were grown to an OD600 of 0.5–0.8. An equivalent of two OD600 of cells was harvested by centrifugation, and the pellet was lysed by the addition of 150 µl 1.85 M NaOH/8% β-mercaptoethanol. After a 10-min incubation on ice, 150 µl 50% TCA was added (10 min at 4°C). The lysate was centrifuged (14,000 rpm at 4°C for 10 min), and the acetone-washed pellet was resuspended in 140 µl of 4× SDS sample loading buffer, boiled for 5 min at 95°C, and pelleted again. Typically, 5–10 µl of the supernatant was loaded per lane for a standard minigel. Western blot analysis was performed according to standard protocols. The following primary antibodies were used in this study: mouse monoclonal α-GFP (1:3,000; Roche), chicken polyclonal α-Tif6 (1:1,000; Genway), rabbit polyclonal α-Yvh1 (1:1,000; this study, raised against the Zn2+-binding domain of Yvh1), α-Nmd3 (1:5,000; provided by A.W. Johnson, University of Texas, Austin, TX), α-Noc1 (1:500; provided by H. Tschochner, Universität Regensburg, Regensburg, Germany), α-Nog1 (1:10,000; provided by M. Fromont-Racine, Institut Pasteur, Paris, France), α-Nop7 (1:10,000; provided by B. Stillman, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY), α-Rpp0 (1:2,000; provided by J.P.G. Ballesta, Universidad Autonoma, Madrid, Spain), α-Mex67 (1:5,000; provided by C. Dargemont, Institut Jacques Monod, Paris, France), and α-Rpl1 (1:10,000; provided by F. Lacroute, Centre de Génétique Moléculaire, Gif-sur-Yvette, France). The secondary HRP-conjugated α-rabbit, α-mouse, and α-chicken antibodies (Sigma-Aldrich) were used at a 1:5,000 dilution. Proteins were visualized using enhanced chemiluminescence detection kits (GE Healthcare).
TAP and mass spectrometry
Pre-60S particles were purified in lysis buffer containing 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1.5 mM MgCl2, and 0.15% NP-40 (plus complete protease inhibitor cocktail [Roche]) by affinity purification of TAP-tagged proteins as previously described (Rigaut et al., 1999; Puig et al., 2001). In brief, for each TAP purification, 2–4 liters of yeast culture was harvested for 30 min at 5,000 rpm and 4°C. The cell pellet was resuspended in 25 ml lysis buffer plus 1 mM DTT (including protease inhibitors) and lysed with glass beads for 20 min at 4°C and 500 rpm (Pulverissette 6; Fritsch). The lysate was clarified at 20,000 rpm and 4°C for 30 min, and 300 µl of preequilibrated rabbit IgG Sepharose beads was added. After a 1-h incubation at 4°C, the beads were collected in a chromatography column (Bio-Rad Laboratories) and washed with 15 ml of lysis buffer and 0.5 mM DTT. The bait complex was subsequently eluted from the IgG beads by incubation with tobacco etch virus protease at 16°C overnight. The tobacco etch virus eluate was incubated with preequilibrated calmodulin Sepharose for 1 h at 4°C in 6 ml lysis buffer plus 2 mM CaCl2 and 1 mM DTT. The beads were washed with lysis buffer plus 2 mM CaCl2 and 1 mM DTT, and the bait complex was finally eluted twice with elution buffer (10 mM Tris-Cl, pH 8.0, 5 mM EGTA, and 100 mM NaCl) for 10 min at 30°C. The final EGTA eluate was TCA precipitated, and the acetone-washed pellet was dissolved in SDS sample buffer for further analysis. Proteins were visualized by NuPAGE 4–12% gradient SDS-PAGE (Invitrogen) followed by silver staining or Western blotting. For mass spectrometry analyses, tryptic peptides from Coomassie/silver-stained proteins were prepared as described previously in Shevchenko et al. (1996) with the exception that pipette tips (C18 ZipTip; Millipore) were used for the final purification according to the manufacturer's instructions. Analysis was performed on a matrix-assisted laser desorption/ionization time of flight instrument (Reflex III; Brücker), and proteins were identified using Mascot and the Mass Spec Database (Matrix Science).
rRNA labeling and pulse-chase analysis
For the pulse-chase analyses, strains were grown at 30°C to an OD600 of 0.5–0.8 in SD-Ura medium. Cells were harvested by centrifugation, resuspended in 1 ml SD medium, and pulse labeled for 1 min by the addition of 100 µCi [5,6 3H]uracil (Hartmann). The chase was initiated by diluting 200 µl aliquots of the pulse-labeled cells in 1.8 ml of SD medium containing 1 mg/ml uracil. The cells were harvested after 0, 3, 6, and 10 min of chase, washed with ice-cold water, and frozen in liquid nitrogen. Total RNA was extracted by the hot phenol method (Schmitt et al., 1990) and loaded on a 1.2% agarose formaldehyde gel or on an 8% acrylamide 7 M urea gel. RNA was transfered to a nylon membrane (Hybond-N+; GE Healthcare) by capillary blotting and subsequently UV cross-linked to the membrane (Stratalinker 1800; Agilent Technologies). Membranes were sprayed with EN3HANCE (PerkinElmer) and exposed to x-ray films for 3–5 d at −80°C with an intensifying screen.
Online supplemental material
Table S1 lists all yeast strains, Table S2 lists all plasmids, and Table S3 lists all oligonucleotides used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200904111/DC1.
Acknowledgments
We are grateful to E. Hurt, M. Peter, A. Johnson, J. Warner, P. Milkereit, J. Woolford, A. Warren, M. Fromont-Racine, J. Ballesta, F. Lacroute, B. Stillman, and C. Dargemont for generously sharing strains, plasmids, and antibodies. We thank C. Azzalin and D. Kressler for help with pulse-chase experiments, C. Weirich for sequence alignments, and J. Pfannstiel for mass spectrometry analysis. We thank A. Johnson, U. Kutay, and all members of the Panse laboratory for enthusiastic discussions.
V.G. Panse is supported by grants from the Swiss National Science Foundation and the Swiss Federal Institute of Technology.
Footnotes
Abbreviations used in this paper:
- LMB
- leptomycin B
- r-protein
- ribosomal protein
- rRNA
- ribosomal RNA
- TAP
- tandem affinity purification
References
- Aoki N., Aoyama K., Nagata M., Matsuda T. 2001. A growing family of dual specificity phosphatases with low molecular masses.J. Biochem. 130:133–140 [DOI] [PubMed] [Google Scholar]
- Baronas-Lowell D.M., Warner J.R. 1990. Ribosomal protein L30 is dispensable in the yeast Saccharomyces cerevisiae.Mol. Cell. Biol. 10:5235–5243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler J., Grandi P., Gadal O., Lessmann T., Petfalski E., Tollervey D., Lechner J., Hurt E. 2001. Identification of a 60S preribosomal particle that is closely linked to nuclear export.Mol. Cell. 8:517–529 doi:10.1016/S1097-2765(01)00342-2 [DOI] [PubMed] [Google Scholar]
- Beeser A.E., Cooper T.G. 1999. The dual-specificity protein phosphatase Yvh1p acts upstream of the protein kinase mck1p in promoting spore development in Saccharomyces cerevisiae.J. Bacteriol. 181:5219–5224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradatsch B., Katahira J., Kowalinski E., Bange G., Yao W., Sekimoto T., Baumgärtel V., Boese G., Bassler J., Wild K., et al. 2007. Arx1 functions as an unorthodox nuclear export receptor for the 60S preribosomal subunit.Mol. Cell. 27:767–779 doi:10.1016/j.molcel.2007.06.034 [DOI] [PubMed] [Google Scholar]
- Deshmukh M., Tsay Y.F., Paulovich A.G., Woolford J.L., Jr 1993. Yeast ribosomal protein L1 is required for the stability of newly synthesized 5S rRNA and the assembly of 60S ribosomal subunits.Mol. Cell. Biol. 13:2835–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaconu M., Kothe U., Schlünzen F., Fischer N., Harms J.M., Tonevitsky A.G., Stark H., Rodnina M.V., Wahl M.C. 2005. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation.Cell. 121:991–1004 doi:10.1016/j.cell.2005.04.015 [DOI] [PubMed] [Google Scholar]
- Eisinger D.P., Dick F.A., Denke E., Trumpower B.L. 1997a. SQT1, which encodes an essential WD domain protein of Saccharomyces cerevisiae, suppresses dominant-negative mutations of the ribosomal protein gene QSR1.Mol. Cell. Biol. 17:5146–5155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisinger D.P., Dick F.A., Trumpower B.L. 1997b. Qsr1p, a 60S ribosomal subunit protein, is required for joining of 40S and 60S subunits.Mol. Cell. Biol. 17:5136–5145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A., Tollervey D. 2002. Making ribosomes.Curr. Opin. Cell Biol. 14:313–318 doi:10.1016/S0955-0674(02)00336-8 [DOI] [PubMed] [Google Scholar]
- Fatica A., Cronshaw A.D., Dlakić M., Tollervey D. 2002. Ssf1p prevents premature processing of an early pre-60S ribosomal particle.Mol. Cell. 9:341–351 doi:10.1016/S1097-2765(02)00458-6 [DOI] [PubMed] [Google Scholar]
- Fromont-Racine M., Senger B., Saveanu C., Fasiolo F. 2003. Ribosome assembly in eukaryotes.Gene. 313:17–42 doi:10.1016/S0378-1119(03)00629-2 [DOI] [PubMed] [Google Scholar]
- Gadal O., Strauss D., Kessl J., Trumpower B., Tollervey D., Hurt E. 2001. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p.Mol. Cell. Biol. 21:3405–3415 doi:10.1128/MCB.21.10.3405-3415.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P., Rybin V., Bassler J., Petfalski E., Strauss D., Marzioch M., Schäfer T., Kuster B., Tschochner H., Tollervey D., et al. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors.Mol. Cell. 10:105–115 doi:10.1016/S1097-2765(02)00579-8 [DOI] [PubMed] [Google Scholar]
- Granneman S., Baserga S.J. 2004. Ribosome biogenesis: of knobs and RNA processing.Exp. Cell Res. 296:43–50 doi:10.1016/j.yexcr.2004.03.016 [DOI] [PubMed] [Google Scholar]
- Harnpicharnchai P., Jakovljevic J., Horsey E., Miles T., Roman J., Rout M., Meagher D., Imai B., Guo Y., Brame C.J., et al. 2001. Composition and functional characterization of yeast 66S ribosome assembly intermediates.Mol. Cell. 8:505–515 doi:10.1016/S1097-2765(01)00344-6 [DOI] [PubMed] [Google Scholar]
- Hedges J., West M., Johnson A.W. 2005. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p.EMBO J. 24:567–579 doi:10.1038/sj.emboj.7600547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helser T.L., Baan R.A., Dahlberg A.E. 1981. Characterization of a 40S ribosomal subunit complex in polyribosomes of Saccharomyces cerevisiae treated with cycloheximide.Mol. Cell. Biol. 1:51–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J.H., Johnson A.W. 1999. NMD3 encodes an essential cytoplasmic protein required for stable 60S ribosomal subunits in Saccharomyces cerevisiae.Mol. Cell. Biol. 19:2389–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J.H., Kallstrom G., Johnson A.W. 2000. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit.J. Cell Biol. 151:1057–1066 doi:10.1083/jcb.151.5.1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung N.J., Johnson A.W. 2006. Nuclear recycling of the pre-60S ribosomal subunit-associated factor Arx1 depends on Rei1 in Saccharomyces cerevisiae.Mol. Cell. Biol. 26:3718–3727 doi:10.1128/MCB.26.10.3718-3727.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung N.J., Lo K.Y., Patel S.S., Helmke K., Johnson A.W. 2008. Arx1 is a nuclear export receptor for the 60S ribosomal subunit in yeast.Mol. Biol. Cell. 19:735–744 doi:10.1091/mbc.E07-09-0968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt E., Hannus S., Schmelzl B., Lau D., Tollervey D., Simos G. 1999. A novel in vivo assay reveals inhibition of ribosomal nuclear export in ran-cycle and nucleoporin mutants.J. Cell Biol. 144:389–401 doi:10.1083/jcb.144.3.389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes.Yeast. 21:947–962 doi:10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Kressler D., Roser D., Pertschy B., Hurt E. 2008. The AAA ATPase Rix7 powers progression of ribosome biogenesis by stripping Nsa1 from pre-60S particles.J. Cell Biol. 181:935–944 doi:10.1083/jcb.200801181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruiswijk T., Planta R.J., Krop J.M. 1978. The course of the assembly of ribosomal subunits in yeast.Biochim. Biophys. Acta. 517:378–389 [DOI] [PubMed] [Google Scholar]
- Lebreton A., Saveanu C., Decourty L., Rain J.C., Jacquier A., Fromont-Racine M. 2006. A functional network involved in the recycling of nucleocytoplasmic pre-60S factors.J. Cell Biol. 173:349–360 doi:10.1083/jcb.200510080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Chang A. 2009. A mutant plasma membrane protein is stabilized upon loss of Yvh1, a novel ribosome assembly factor.Genetics. 181:907–915 doi:10.1534/genetics.108.100099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo K.-Y., Li Z., Wang F., Marcotte E.M., Johnson A.W. 2009. Ribosome stalk assembly requires the dual-specificity phosphatase Yvh1 for the exchange of Mrt4 with P0.J. Cell Biol. 186:849–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae.Yeast. 14:953–961 doi:10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U [DOI] [PubMed] [Google Scholar]
- Menne T.F., Goyenechea B., Sánchez-Puig N., Wong C.C., Tonkin L.M., Ancliff P.J., Brost R.L., Costanzo M., Boone C., Warren A.J. 2007. The Shwachman-Bodian-Diamond syndrome protein mediates translational activation of ribosomes in yeast.Nat. Genet. 39:486–495 doi:10.1038/ng1994 [DOI] [PubMed] [Google Scholar]
- Milkereit P., Gadal O., Podtelejnikov A., Trumtel S., Gas N., Petfalski E., Tollervey D., Mann M., Hurt E., Tschochner H. 2001. Maturation and intranuclear transport of pre-ribosomes requires Noc proteins.Cell. 105:499–509 doi:10.1016/S0092-8674(01)00358-0 [DOI] [PubMed] [Google Scholar]
- Milkereit P., Strauss D., Bassler J., Gadal O., Kühn H., Schütz S., Gas N., Lechner J., Hurt E., Tschochner H. 2003. A Noc complex specifically involved in the formation and nuclear export of ribosomal 40 S subunits.J. Biol. Chem. 278:4072–4081 doi:10.1074/jbc.M208898200 [DOI] [PubMed] [Google Scholar]
- Muda M., Manning E.R., Orth K., Dixon J.E. 1999. Identification of the human YVH1 protein-tyrosine phosphatase orthologue reveals a novel zinc binding domain essential for in vivo function.J. Biol. Chem. 274:23991–23995 doi:10.1074/jbc.274.34.23991 [DOI] [PubMed] [Google Scholar]
- Neville M., Rosbash M. 1999. The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae.EMBO J. 18:3746–3756 doi:10.1093/emboj/18.13.3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan T.A., Bassler J., Petfalski E., Tollervey D., Hurt E. 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm.EMBO J. 21:5539–5547 doi:10.1093/emboj/cdf547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M., Dlakic M., Tollervey D. 2004. A pre-ribosome-associated HEAT-repeat protein is required for export of both ribosomal subunits.Genes Dev. 18:196–209 doi:10.1101/gad.285604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse V.G., Küster B., Gerstberger T., Hurt E. 2003. Unconventional tethering of Ulp1 to the transport channel of the nuclear pore complex by karyopherins.Nat. Cell Biol. 5:21–27 doi:10.1038/ncb893 [DOI] [PubMed] [Google Scholar]
- Panse V.G., Kressler D., Pauli A., Petfalski E., Gnädig M., Tollervey D., Hurt E. 2006. Formation and nuclear export of preribosomes are functionally linked to the small-ubiquitin-related modifier pathway.Traffic. 7:1311–1321 doi:10.1111/j.1600-0854.2006.00471.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertschy B., Saveanu C., Zisser G., Lebreton A., Tengg M., Jacquier A., Liebminger E., Nobis B., Kappel L., van der Klei I., et al. 2007. Cytoplasmic recycling of 60S preribosomal factors depends on the AAA protein Drg1.Mol. Cell. Biol. 27:6581–6592 doi:10.1128/MCB.00668-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Séraphin B. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification.Methods. 24:218–229 doi:10.1006/meth.2001.1183 [DOI] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. 1999. A generic protein purification method for protein complex characterization and proteome exploration.Nat. Biotechnol. 17:1030–1032 doi:10.1038/13732 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Mateos M., Abia D., Garcia-Gomez J.J., Morreale A., de la Cruz J., Santos C., Remacha M., Ballesta J.P. 2009. The amino terminal domain from Mrt4 protein can functionally replace the RNA binding domain of the ribosomal P0 protein.Nucleic Acids Res. 37:3514–3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotenberg M.O., Moritz M., Woolford J.L., Jr 1988. Depletion of Saccharomyces cerevisiae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of half-mer polyribosomes.Genes Dev. 2:160–172 doi:10.1101/gad.2.2.160 [DOI] [PubMed] [Google Scholar]
- Sakumoto N., Yamashita H., Mukai Y., Kaneko Y., Harashima S. 2001. Dual-specificity protein phosphatase Yvh1p, which is required for vegetative growth and sporulation, interacts with yeast pescadillo homolog in Saccharomyces cerevisiae.Biochem. Biophys. Res. Commun. 289:608–615 doi:10.1006/bbrc.2001.6021 [DOI] [PubMed] [Google Scholar]
- Saveanu C., Namane A., Gleizes P.E., Lebreton A., Rousselle J.C., Noaillac-Depeyre J., Gas N., Jacquier A., Fromont-Racine M. 2003. Sequential protein association with nascent 60S ribosomal particles.Mol. Cell. Biol. 23:4449–4460 doi:10.1128/MCB.23.13.4449-4460.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer T., Strauss D., Petfalski E., Tollervey D., Hurt E. 2003. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes.EMBO J. 22:1370–1380 doi:10.1093/emboj/cdg121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M.E., Brown T.A., Trumpower B.L. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae.Nucleic Acids Res. 18:3091–3092 doi:10.1093/nar/18.10.3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger B., Lafontaine D.L., Graindorge J.S., Gadal O., Camasses A., Sanni A., Garnier J.M., Breitenbach M., Hurt E., Fasiolo F. 2001. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis.Mol. Cell. 8:1363–1373 doi:10.1016/S1097-2765(01)00403-8 [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels.Anal. Chem. 68:850–858 doi:10.1021/ac950914h [DOI] [PubMed] [Google Scholar]
- Stage-Zimmermann T., Schmidt U., Silver P.A. 2000. Factors affecting nuclear export of the 60S ribosomal subunit in vivo.Mol. Biol. Cell. 11:3777–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D., Lehtonen H., Carmo-Fonseca M., Hurt E.C. 1991. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast.EMBO J. 10:573–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D., Lehtonen H., Jansen R., Kern H., Hurt E.C. 1993. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly.Cell. 72:443–457 doi:10.1016/0092-8674(93)90120-F [DOI] [PubMed] [Google Scholar]
- Trapman J., Retèl J., Planta R.J. 1975. Ribosomal precursor particles from yeast.Exp. Cell Res. 90:95–104 doi:10.1016/0014-4827(75)90361-4 [DOI] [PubMed] [Google Scholar]
- Tschochner H., Hurt E. 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm.Trends Cell Biol. 13:255–263 doi:10.1016/S0962-8924(03)00054-0 [DOI] [PubMed] [Google Scholar]
- Venema J., Tollervey D. 1999. Ribosome synthesis in Saccharomyces cerevisiae.Annu. Rev. Genet. 33:261–311 doi:10.1146/annurev.genet.33.1.261 [DOI] [PubMed] [Google Scholar]
- Wahl M.C., Möller W. 2002. Structure and function of the acidic ribosomal stalk proteins.Curr. Protein Pept. Sci. 3:93–106 doi:10.2174/1389203023380756 [DOI] [PubMed] [Google Scholar]
- Warner J.R. 1999. The economics of ribosome biosynthesis in yeast.Trends Biochem. Sci. 24:437–440 doi:10.1016/S0968-0004(99)01460-7 [DOI] [PubMed] [Google Scholar]
- Yao W., Roser D., Köhler A., Bradatsch B., Bassler J., Hurt E. 2007. Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67-Mtr2.Mol. Cell. 26:51–62 doi:10.1016/j.molcel.2007.02.018 [DOI] [PubMed] [Google Scholar]
- Yao W., Lutzmann M., Hurt E. 2008. A versatile interaction platform on the Mex67-Mtr2 receptor creates an overlap between mRNA and ribosome export.EMBO J. 27:6–16 doi:10.1038/sj.emboj.7601947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemp I., Kutay U. 2007. Nuclear export and cytoplasmic maturation of ribosomal subunits.FEBS Lett. 581:2783–2793 doi:10.1016/j.febslet.2007.05.013 [DOI] [PubMed] [Google Scholar]
- Zinker S., Warner J.R. 1976. The ribosomal proteins of Saccharomyces cerevisiae. Phosphorylated and exchangeable proteins.J. Biol. Chem. 251:1799–1807 [PubMed] [Google Scholar]
- Zuk D., Belk J.P., Jacobson A. 1999. Temperature-sensitive mutations in the Saccharomyces cerevisiae MRT4, GRC5, SLA2 and THS1 genes result in defects in mRNA turnover.Genetics. 153:35–47 [DOI] [PMC free article] [PubMed] [Google Scholar]