Summary
In Escherichia coli, translational arrest can elicit cleavage of codons within the ribosomal A site. This A-site mRNA cleavage is independent of RelE, and has been proposed to be an endonucleolytic activity of the ribosome. Here, we show that the 3′→5′ exonuclease RNase II plays an important role in RelE-independent A-site cleavage. Instead of A-site cleavage, translational pausing in ΔRNase II cells produces transcripts that are truncated +12 and +28 nucleotides downstream of the A-site codon. Deletions of the genes encoding polynucleotide phosphorylase (PNPase) and RNase R had little effect on A-site cleavage. However, PNPase overexpression restored A-site cleavage activity to ΔRNase II cells. Purified RNase II and PNPase were both unable to directly catalyze A-site cleavage in vitro. Instead, these exonucleases degraded ribosome-bound mRNA to positions +18 and +24 nucleotides downstream of the ribosomal A site, respectively. Finally, a stable structural barrier to exoribonuclease activity inhibited A-site cleavage when introduced immediately downstream of paused ribosomes. These results demonstrate that 3′→5′ exonuclease activity is an important prerequisite for efficient A-site cleavage. We propose that RNase II degrades mRNA to the downstream border of paused ribosomes, facilitating cleavage of the A-site codon by an unknown RNase.
Keywords: A-site mRNA cleavage, exoribonucleases, ribosome pausing, RNase II, tmRNA
Introduction
A-site mRNA cleavage describes two distinct RNase activities that are specific for codons in the ribosomal A site. A-site cleavage was first identified as an activity of the Escherichia coli RelE protein, which is a sequence-specific endoribonuclease that cleaves A-site codons between the second and third nucleotide (Pedersen et al., 2003). Subsequently, RelE-independent A-site mRNA cleavage was shown to occur during translational pauses in E. coli (Hayes and Sauer, 2003). In contrast to the RelE-mediated activity, A-site cleavage during translational pausing occurs at a wide variety of sense and stop codons (Garza-Sánchez et al., 2008; Hayes and Sauer, 2003). Both A-site cleavage activities result in ribosome stalling on truncated messages (Christensen and Gerdes, 2003; Hayes and Sauer, 2003). In eubacteria, the tmRNA•SmpB quality control system facilitates recycling of such stalled ribosomes in a process termed “ribosome rescue”. tmRNA (or transfer-messenger RNA) is a specialized RNA that acts first as a tRNA to bind the A site of stalled ribosomes, then as an mRNA to direct the co-translational addition of the SsrA peptide degradation tag to the C terminus of the nascent chain (Keiler et al., 1996). SmpB is a tmRNA-binding protein required for ribosome recruitment and translation of the SsrA peptide tag (Karzai et al., 1999; Sundermeier et al., 2005). As a result of tmRNA•SmpB activity, the stalled ribosome undergoes translation termination and recycling, and the SsrA-tagged polypeptide is targeted for proteolysis. Because ribosome pausing can induce both A-site mRNA cleavage and tmRNA•SmpB-mediated peptide tagging, it has been proposed that these two activities occur sequentially as part of a quality control system.
RelE-independent A-site mRNA cleavage is proposed to be the activity of an unknown endonuclease. The initial report characterized the upstream (5′) A-site cleavage fragment, but was unable to detect a downstream (3′) product, presumably because this fragment was not translated and therefore rapidly degraded (Hayes and Sauer, 2003). Since that time, our laboratory has been unable to isolate, stabilize, or detect a downstream fragment from RelE-independent A-site cleavage despite considerable effort. These results led us to consider roles for 3′→5′ exoribonucleases in A-site mRNA cleavage. Here, we report that the 3′→5′ exonuclease RNase II plays an important role in A-site mRNA cleavage in E. coli. Deletion of the genes encoding RNases R, D, PH, T, BN(Z), and PNPase had no significant effect on A-site mRNA cleavage, however the overexpression of PNPase restored A-site cleavage activity to ΔRNase II cells. Although RNase II is critical for A-site mRNA cleavage during ribosome pausing, its structure is incompatible with direct degradation of the A-site codon. RNase II has a relatively long substrate-binding channel, and typically leaves undigested 3′ tails of 6 − 9 nucleotides upon encountering stable secondary structure (Frazao et al., 2006; Spickler and Mackie, 2000; Zuo et al., 2006). Presumably, A-site cleavage occurs immediately adjacent to the codon-anticodon duplex in the P site of paused ribosomes, and thus the role of RNase II is likely indirect. Indeed, we find that RNase II is only able to degrade mRNA to the downstream border of paused ribosomes in vitro. We propose that RNase II-mediated degradation to the ribosome border facilitates the activity of another RNase that cleaves the A-site codon.
Results
The flag-(m)ybeL-PP mini-message undergoes A-site cleavage during translation termination
The consecutive C-terminal Pro residues of the E. coli YbeL-PP protein have been shown to interfere with translation termination and induce A-site cleavage at stop codons (Hayes et al., 2002; Hayes and Sauer, 2003). We sought to identify the downstream (3′) A-site cleavage fragment using a mini-message derived from the ybeL-PP gene (Hayes et al., 2002; Hayes and Sauer, 2003). The flag-(m)ybeL-PP mini-gene encodes the C-terminal 49 residues of YbeL-PP fused to an N-terminal FLAG epitope (Fig. 1A). Like the full-length protein, expression of FLAG-(m)YbeL-PP produced more truncated mRNA in ΔtmRNA cells compared to tmRNA+ cells (Figs. 1B & 1C). A-site cleavage products do not typically accumulate to high levels in tmRNA+ cells, presumably because the tmRNA•SmpB system removes paused ribosomes from truncated mRNA, thereby facilitating the rapid degradation of the cleaved messages (Hayes and Sauer, 2003; Yamamoto et al., 2003). S1 nuclease protection analysis showed that the flag-(m)ybeL-PP transcript from ΔtmRNA cells was truncated primarily within the stop codon (Fig. 2B, RNase+ lane). This pattern of truncated mRNA is characteristic of A-site cleavage during translation termination (Hayes and Sauer, 2003; Sunohara et al., 2004). We note that the truncations extended into the last sense (Pro) codon of the transcript (Fig. 2B), which was not observed previously with the full-length ybeL-PP message (Hayes and Sauer, 2003). These additional cleavages resulted from secondary translational arrest due to the depletion of free tRNA2Pro. A significant proportion of tRNA2Pro was sequestered as peptidyl prolyl-tRNA2Pro on terminating ribosomes (Fig. 1B), reducing the pool of tRNA2Pro available for decoding the final Pro codon (Gong et al., 2006). Overexpression of the cognate tRNA2Pro species prevented cleavage in the Pro codon, but had no effect on stop codon cleavage (data not shown).
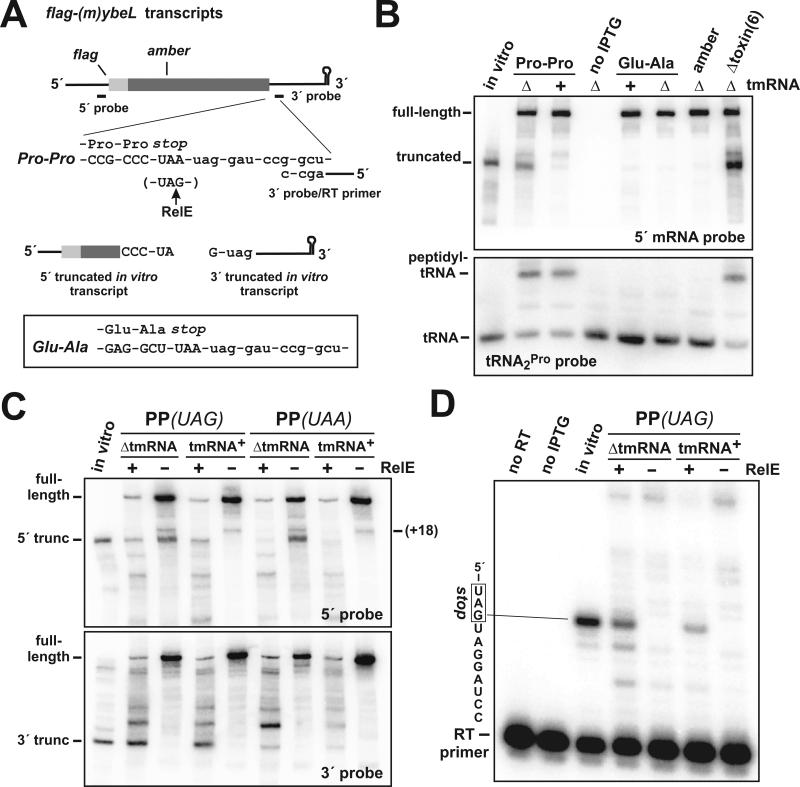
Figure 1. A-site mRNA cleavage induced by ribosome pausing and RelE.
(A) Mini-gene constructs used in this study. The flag-(m)ybeL mini-mRNA variants are depicted with the encoded FLAG epitope and Northern blot probe binding sites. Two versions of flag-(m)ybeL encoding C-terminal Pro-Pro (PP) or Glu-Ala (EA) sequences were examined. An amber mutation was made at position Glu28 in flag-(m)ybeL-PP to generate the flag-(m)ybeL(amber) construct. The 3′ Northern probe also served as the primer for reverse transcriptase (RT) primer extension analysis. The position of RelE cleavage within UAG is indicated by the upward arrow. Two truncated transcripts corresponding to the upstream (5′) and downstream (3′) RelE cleavage fragments were synthesized using bacteriophage T7 RNA polymerase in vitro. (B) Northern analysis of RelE-independent A-site mRNA cleavage. The various flag-(m)ybeL transcripts were expressed in ΔtmRNA or tmRNA+ cells as indicated. The flag-(m)ybeL-PP mRNA was also expressed in ΔtmRNA cells lacking all of the six known toxin endonucleases [Δtoxin(6)]. An in vitro transcript truncated in the flag-(m)ybeL-PP stop codon was mixed with total RNA isolated from uninduced ΔtmRNA cells and run in the lane labeled in vitro. All RNA samples were also hybridized a tRNA2Pro-specific probe to identify peptidyl prolyl-tRNA2Pro. The positions of free tRNA2Pro and peptidyl prolyl-tRNA2Pro are indicated. (C) Northern analysis of flag-(m)ybeL-PP messages containing UAG and UAA stop codons. Total RNA samples from tmRNA+ and ΔtmRNA cells were hybridized with the 5′ and 3′ probes. RelE was expressed from a plasmid-borne arabinose-inducible construct as indicated. The positions of full-length and +18 truncated transcripts are indicated. The positions of stop-codon cleaved (5′ and 3′ truncated) transcripts were determined using control transcripts synthesized in vitro. (D) Primer extension analysis of RelE cleavage. Total RNA from ΔtmRNA and tmRNA+ cells expressing flag-(m)ybeL-PP(UAG) were subjected to primer extension analysis using the 3′ probe depicted in panel A. Control reactions lacking either the flag-(m)ybeL-PP(UAG) transcript (no IPTG), or reverse transcriptase (no RT), showed no primer extension. A positive control reaction containing the 3′ truncated in vitro transcript showed primer extension of nine nucleotides.
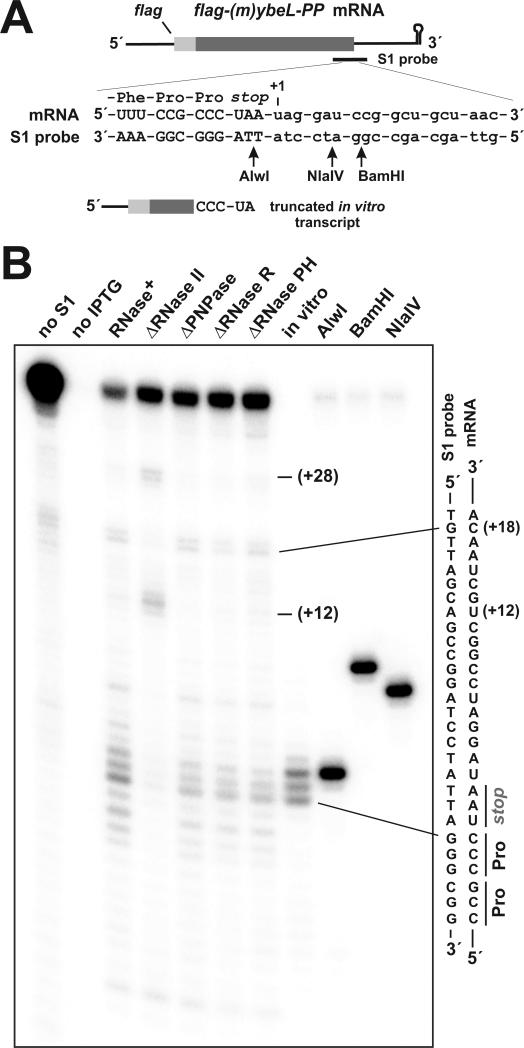
Figure 2. S1 nuclease protection analysis of truncated mRNA.
(A) The model flag-(m)ybeL-PP mRNA is depicted with the S1 nuclease protection probe binding site. Truncation sites are numbered with respect the first nucleotide after the stop codon (+1). Upward arrows indicate the positions of AlwI, NlaIV and BamHI endonuclease cleavages used to generate gel migration standards. The 3′ end of the truncated in vitro transcript used in Northern blot and S1 nuclease protection experiments is shown. (B) S1 nuclease protection analysis of transcripts isolated from ΔtmRNA cells deleted for exonucleases. Total RNA samples were taken from cells induced to express flag-(m)ybeL-PP, and from an uninduced control culture (no IPTG). RNase+ indicates that the cells have the wild-type complement of RNases. Nucleotide positions +12, +18 and +28 (with respect to the stop codon) are indicated. The 5′ truncated in vitro transcript (in vitro) shown in Figs. 1A & 2A was also subjected to S1 protection analysis. The relative positions of AlwI, BamHI and NlaIV digestion sites in the S1 probe with respect to the stop codon are shown in panel A.
To test whether mRNA cleavage occurred during translation termination, we examined two negative control constructs. The flag-(m)ybeL-EA mini-gene encodes C-terminal Glu-Ala residues (Fig. 1A), which facilitate translation termination and do not support A-site cleavage (Hayes et al., 2002; Hayes and Sauer, 2003). In accord with previous studies, truncated transcripts were not observed when FLAG-(m)YbeL-EA was expressed in either ΔtmRNA or tmRNA+ cells (Fig. 1B). The flag-(m)ybeL(amber) construct contains an amber mutation that changes Glu28 to a stop codon (Fig. 1A), and thereby prevents ribosomes from translating the C-terminal Pro residues. No truncated transcripts were detected in ΔtmRNA or tmRNA+ cells expressing the flag-(m)ybeL(amber) transcript (Fig. 1B). Additionally, we expressed FLAG-(m)YbeL-PP in ΔtmRNA cells lacking all six of the known RNase toxins (RelE, ChpBK, YoeB, MazF, YafQ, and YhaV), and observed no decrease in truncated mRNA (Fig. 1B). These results indicate that translation is required for mRNA cleavage, and that this RNase activity is not mediated by any of the known toxins.
We next examined peptidyl-tRNA to demonstrate ribosome pausing during translation termination. We reasoned that ribosomes paused during translation termination should carry peptidyl-tRNA corresponding to the last sense codon of the mRNA. The last sense codon of flag-(m)ybeL-PP is CCC (Fig. 1A), which is decoded exclusively by tRNA2Pro in E. coli (Dong et al., 1996). The remaining five Pro residues of FLAG-(m)YbeL-PP were coded as CCG, which is decoded by both tRNA1Pro and tRNA3Pro (Dong et al., 1996). Therefore, peptidyl prolyl-tRNA2Pro should be a specific marker of ribosomes that are terminating FLAG-(m)YbeL-PP translation. Northern analysis of tRNA2Pro showed significant levels of peptidyl prolyl-tRNA2Pro when FLAG-(m)YbeL-PP was overexpressed (Fig. 1B). In contrast, peptidyl prolyl-tRNA2Pro was not detected in uninduced cells, nor in cells expressing either FLAG-(m)YbeL-EA or the prematurely amber-terminated FLAG-(m)YbeL protein (Fig. 1B). Moreover, there was no evidence of peptidyl alanyl-tRNA1BAla during FLAG-(m)YbeL-EA expression (data not shown), indicating that peptidyl-tRNA does not accumulate during efficient translation termination. Taken together, these data show that ribosomes pause specifically at the flag-(m)ybeL-PP stop codon, strongly suggesting that this codon is cleaved during inefficient translation termination.
Analysis of RelE-mediated and RelE-independent A-site mRNA cleavage
We next sought to identify the downstream A-site cleavage fragment using the 3′ probe in Northern blot hybridizations (Fig. 1A). Although the upstream A-site cleavage fragment was readily detected by Northern blot, we were unable to detect a product corresponding to the predicted downstream fragment (Fig. 1C). To determine whether such a fragment would be stable in vivo, we co-expressed the RelE A-site nuclease with FLAG-(m)YbeL-PP. RelE can act at several codons, but preferentially cleaves UAG and CAG codons (Christensen and Gerdes, 2003; Pedersen et al., 2003). Therefore, we also tested a flag-(m)ybeL-PP construct containing a UAG stop codon (Fig. 1A). We reasoned that pausing during translation termination would facilitate RelE cleavage in the stop codon, and allow us to assess the stability of the resulting downstream fragment. Northern analysis revealed a number of 5′- and 3′-cleavage products upon RelE induction (Fig. 1C). The largest of the RelE 5′-cleavage fragments co-migrated with the upstream A-site cleavage product on polyacrylamide gels (Fig. 1C), suggesting that this species resulted from RelE cleavage in the stop codon. The original construct containing a UAA stop codon was not cleaved as efficiently at this site (Fig. 1C), consistent with the specificity of RelE in vitro (Pedersen et al., 2003). We then looked for the corresponding 3′-cleavage fragment using the 3′ probe. The smallest of the 3′ RelE cleavage fragments co-migrated with a control in vitro transcript that mimics the downstream fragment produced by RelE cleavage in the UAG stop codon (Figs. 1A & 1C). Primer extension analysis identified a RelE-dependent cleavage between the A and G residues of the UAG stop codon (Fig. 1D), which is identical to the published RelE cleavage (Pedersen et al., 2003). Thus, RelE-mediated cleavage in the flag-(m)ybeL-PP stop codon produces a stable downstream fragment in vivo. Although it is possible that RelE-independent cleavage produces a less stable downstream fragment, these results suggest that A-site cleavage in our system involves more than a single endonucleolytic event.
RNase II plays an important role in A-site mRNA cleavage
Four exoribonucleases – RNase II, RNase R, PNPase, and oligoribonuclease – are important for mRNA decay in E. coli (Andrade et al., 2009). We deleted the genes encoding each exonuclease (except for the essential oligoribonuclease gene) in ΔtmRNA cells, and examined A-site mRNA cleavage by Northern blot and S1 nuclease protection. PNPase and RNase R deletions had little effect on A-site mRNA cleavage (Figs. 2B & 3A). In contrast, deletion of the gene encoding RNase II dramatically reduced A-site cleavage, and led to the accumulation of two novel truncated species (Figs. 2B & 3A). S1 nuclease protection analysis showed that these transcripts were truncated at +12 and +27/+28 nucleotides downstream of the stop codon (Fig. 2B). We also examined the role of ATP-dependent RNA helicases in A-site cleavage. RhlB cooperates with PNPase to degrade mRNA containing secondary structure, and HrpA facilitates the ribosome-dependent cleavage of the E. coli daa mRNA (Koo et al., 2004; Liou et al., 2002). However, deletion of either helicase had no effect on A-site mRNA cleavage (Fig. 3A, and data not shown). Poly(A) polymerase I (PAP I) also facilitates the exonucleolytic degradation of messages containing secondary structure (Py et al., 1996). Northern analysis showed that flag-(m)ybeL-PP transcript levels were significantly reduced in cells lacking PAP I (Fig. 3A). The expression defect in ΔPAP I cells was due to the stabilization of plasmid-encoded RNAI, which inhibited the replication of our ColEI-based expression plasmid (He et al., 1993; March et al., 1989; Xu et al., 1993). In an attempt to increase transcript levels, we subcloned the expression construct into pACYC184-derived plasmid containing a different replication origin. However, the resulting pACYC184 plasmid was also maintained at a low copy number in ΔPAP I cells (Fig. 3A, and data not shown). Though total transcript levels were reduced, the ratio of A-site cleaved to full-length message was not affected by PAP I deletion (Fig. 3A, and data not shown).
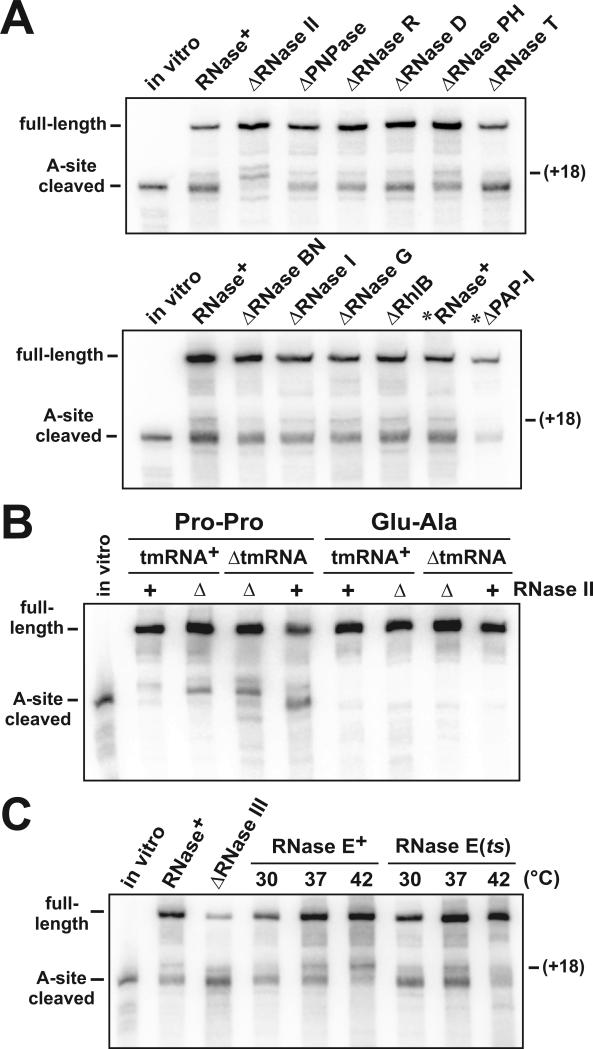
Figure 3. Deletion of RNases and A-site mRNA cleavage.
(A) Northern analysis of flag-(m)ybeL-PP transcripts isolated from RNase deletion strains. Deletions of genes encoding the various RNases were introduced into ΔtmRNA cells, and transcripts examined using the 5′ probe depicted in Fig. 1A. The migration positions of full-length, +18 truncated, and A-site cleaved transcripts are indicated. Samples marked with an asterisk (*) were taken from cells expressing FLAG-(m)YbeL-PP from a pACYC184-derived plasmid as described in Results. (B) Northern analysis of flag-(m)ybeL transcripts expressed in ΔRNase II cells. FLAG-(m)YbeL-PP (Pro-Pro) and FLAG-(m)YbeL-EA (Glu-Ala) proteins were expressed ΔtmRNA and tmRNA+ cells, with (RNase II+) or without RNase II (ΔRNase II). The positions of full-length and A-site cleaved transcripts are indicated. The assignment of +12, +18, and +28 truncation products in these samples was confirmed by S1 nuclease protection analysis (data not shown). (C) Northern analysis of A-site cleavage in ΔtmRNA cells lacking RNase III and RNase E activity. RNase E activity was assessed by shifting ΔtmRNA RNase E(ts) cells to the non-permissive temperature (42 °C). The migration positions of full-length, +18 truncated, and A-site cleaved transcripts are indicated.
ΔtmRNA ΔRNase II cells exhibited residual A-site mRNA cleavage (Figs. 2B & 3A), so we asked whether the other known 3′→5′ exonucleases contribute to this activity. RNase T, RNase D, and RNase PH are involved in the 3′ processing of stable RNAs (Li and Deutscher, 1995, 1996; Li et al., 1998, 1999), but have no known role in mRNA decay. RNase BN (also known as RNase Z) possesses both 3′→5′ exonuclease and endonuclease activities, and has recently been implicated in mRNA degradation (Dutta and Deutscher, 2009; Ezraty et al., 2005; Perwez and Kushner, 2006). Deletions of the genes encoding RNase T, RNase D, RNase PH and RNase BN(Z) had no significant effect on A-site cleavage (Fig. 3A).
Deletion of RNase II in ΔtmRNA cells not only inhibited A-site cleavage, but also reduced the accumulation of minor products truncated at the +18 and +19 positions (Figs. 2B & 3A). This +18 truncation product accumulates in both ΔtmRNA and tmRNA+ cells (see Fig. 1C), and its formation requires translational pausing (Garza-Sánchez et al., 2008). These observations suggest that RNase II directly produces the +18 truncation product, which was confirmed by in vitro experiments presented below. Therefore, we asked whether the deletion of RNase II provides an opportunity for other RNases to cleave our reporter mRNA in response to translational pausing. We first confirmed ribosome pausing during translation termination in ΔtmRNA ΔRNase II cells using peptidyl prolyl-tRNA2Pro analysis as described above (data not shown). We then expressed FLAG-(m)YbeL-EA in ΔtmRNA ΔRNase II cells, and observed no +12 or +27/+28 truncated transcripts (Fig. 3B), suggesting that ribosome pausing is required to generate these products. Additionally, we examined tmRNA+ ΔRNase II cells and readily detected the +12 and +27/+28 truncation products upon expression of FLAG-(m)YbeL-PP, but not FLAG-(m)YbeL-EA (Fig. 3B). Taken together, these data indicate that the alternative cleavages observed in ΔRNase II cells are dependent on pausing during translation termination.
Role of endonucleases in A-site mRNA cleavage
RNase II cannot initiate degradation on structured RNA, so the intrinsic terminator at the 3′ end of the flag-(m)ybeL-PP transcript must be removed prior to RNase II loading. Indeed, purified RNase II was unable to degrade the full-length flag-(m)ybeL-PP transcript in vitro (data not shown), suggesting that an endonucleolytic cleavage is required for RNase II activity on this message. Therefore, we examined the roles of the RNase I, RNase G, RNase E, and RNase III endonucleases in A-site cleavage. Deletion of the genes encoding RNase I and RNase G had no effect on mRNA cleavage (Fig. 3A). In contrast, Northern analysis of transcripts from ΔtmRNA ΔRNase III cells showed an unusual mRNA cleavage pattern, with an apparent increase in A-site cleavage efficiency, accompanied by the +12 truncation product (Fig. 3C, and data not shown). RNase E is essential, so we used the rne-1 temperature sensitive (ts) allele to inactivate this enzyme in temperature shift experiments (Babitzke and Kushner, 1991). Shifting ΔtmRNA RNase E(ts) cells to the non-permissive temperature (42 °C) resulted in a significant decrease in both A-site cleaved and +18-truncated transcripts (Fig. 3C). However, A-site mRNA cleavage was also inhibited at 42 °C in the control ΔtmRNA RNase E+ cells (Fig. 3C). In contrast to ΔtmRNA RNase E(ts) cells, there was an increase in the +18 truncation product when ΔtmRNA RNase E+ cells were shifted to 42°C (Fig. 3C). These results indicate that RNase E cleavage is required to form the RNase II-dependent +18 truncation product. Additionally, it appears that the A-site nuclease is intrinsically temperature sensitive.
Complementation of A-site mRNA cleavage
To confirm that the lack of 3′→5′ exonuclease activity accounts for the A-site cleavage defect observed in ΔtmRNA ΔRNase II cells, we performed complementation experiments using arabinose-inducible RNase II expression plasmids. Plasmid-borne wild-type RNase II restored A-site mRNA cleavage, whereas the catalytically inactive Asp209Asn (D209N) mutant of RNase II was unable to complement the RNase II deletion (Figs. 4A & 4B) (Amblar and Arraiano, 2005). Arraiano and colleagues have shown that RNase II lacking the first N-terminal cold-shock domain (ΔCSD1) is still active (Amblar et al., 2006), which led to us to examine the role of CSD1 in A-site cleavage. Overexpressed RNase II ΔCSD1 was able to restore A-site cleavage to ΔtmRNA ΔRNase II cells, albeit less efficiently than the wild-type enzyme (Fig. 4A).
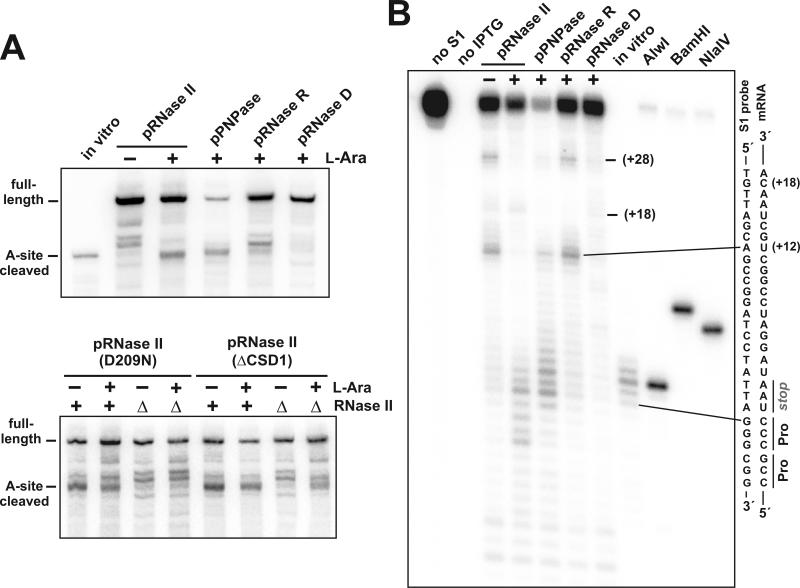
Figure 4. Complementation of A-site mRNA cleavage.
(A) Northern analysis of samples from cells overexpressing 3′→5′ exonucleases. RNase II variants were induced with L-arabinose in ΔtmRNA cells containing (RNase II+) or lacking RNase II (ΔRNase II). RNase II (D209N) is a catalytically inactive enzyme containing the Asp209Asn missense mutation (Amblar and Arraiano, 2005). RNase II (ΔCSD1) contains a deletion of the N-terminal cold-shock domain of RNase II (Amblar et al., 2006). PNPase, RNase R, and RNase D were also overexpressed in ΔRNase II ΔtmRNA cells. The migration positions of full-length and A-site cleaved transcripts are indicated. (B) S1 nuclease protection analysis of mRNA from ΔtmRNA ΔRNase II cells overexpressing exonucleases. Plus (+) and minus (−) symbols indicate induction of exonuclease expression with L-arabinose, or suppression of expression with D-glucose, respectively. Protections corresponding to the +12 and +28 truncations are indicated. No protection was detected from samples in which flag-(m)ybeL-PP expression was not induced (no IPTG). The 5′ truncated in vitro transcript (in vitro) shown in Figs. 1A & 2A was also subjected to S1 protection analysis. Relevant sequences of the flag-(m)ybeL-PP mRNA and the hybridized S1 oligonucleotide probe are depicted. The relative positions of AlwI, BamHI and NlaIV digestion sites in the S1 probe with respect to the stop codon are as shown in Figure 2A.
We also asked whether the overexpression of other E. coli exonucleases could by-pass the apparent RNase II requirement, and restore A-site cleavage to ΔtmRNA ΔRNase II cells. Overexpression of RNase PH, RNase T, RNase BN(Z), and oligoribonuclease had no effect on mRNA cleavage in ΔtmRNA ΔRNase II cells (data not shown). RNase R overexpression appeared to alter mRNA cleavage by Northern blot, but S1 protection analysis showed that the cleavage pattern was essentially the same as that in ΔtmRNA ΔRNase II cells (Figs. 4A & 4B). Somewhat surprisingly, PNPase overexpression partially supported A-site cleavage activity in ΔtmRNA ΔRNase II cells (Figs. 4A & 4B). We wondered whether the association of PNPase with the RNA degradosome complex prevents it from participating in A-site mRNA cleavage under normal circumstances (Carpousis, 2002, 2007). However, deletion of the C-terminus of RNase E, which is required for degradosome assembly (Lopez et al., 1999), failed to restore A-site mRNA cleavage in ΔtmRNA ΔRNase II cells (data not shown). Lastly, we found that RNase D overexpression dramatically reduced the accumulation of all mRNA cleavage products in ΔtmRNA ΔRNase II cells (Figs. 4A & 4B). This result would be expected if high RNase D levels inhibit protein synthesis, or somehow alleviate ribosome pausing in our system. However, control experiments showed that RNase D overproduction had no effect on protein synthesis or translational pausing (data not shown). Instead, it appears that RNase D is able to degrade mRNAs (see below).
Exonuclease-mediated degradation of mRNA in vitro
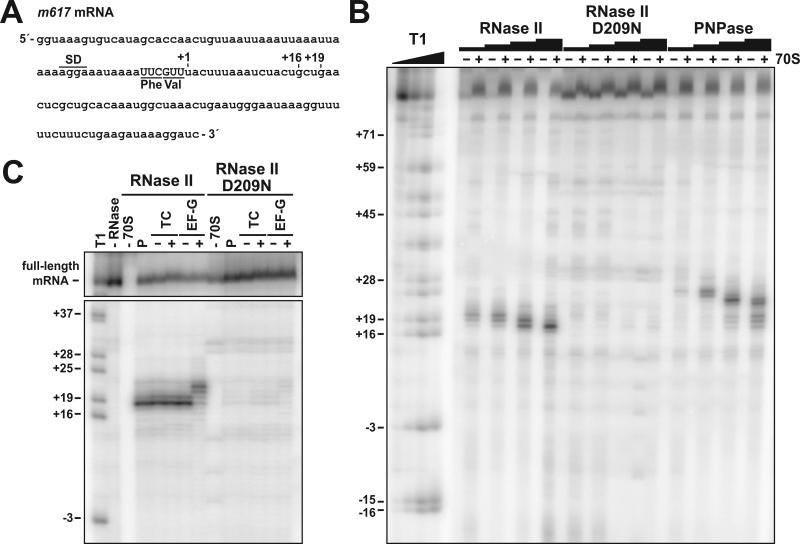
Although efficient A-site cleavage depends on RNase II, its biochemical properties seem incompatible with direct degradation of the A-site codon (see Introduction). To examine the role of exonucleases in A-site cleavage, we digested ribosome-mRNA complexes with purified RNase II and PNPase in vitro. Ribosomes were positioned on a model mRNA derived from the phage T4 gene 32 by binding N-acetyl-phenylalanyl-tRNAPhe to the P site (Fig. 5A) (Shoji et al., 2006). Ribosome-mRNA complexes were then treated with increasing concentrations of purified exonucleases. RNase II digestion produced transcripts that were truncated primarily at positions +18 and +19 nucleotides downstream of the A-site codon (Figs. 5A & 5B). These degradation products were dependent upon the addition of 70S ribosomes, and were not observed when the catalytically inactive RNase II (D209N) enzyme was used (Figs. 5B & 5C). In contrast, PNPase degraded mRNA to positions +24 to +26 (Fig. 5B). At higher concentrations (2.5 μM), PNPase was able to degrade up to the +18 and +19 positions (Fig. 5B). These results suggest that RNase II and PNPase are only able to degrade mRNA to the 3′ leading edge of the ribosome. To test this hypothesis, ternary complexes and elongation factor-G (EF-G) were added to translocate the ribosome forward by one codon. Addition of valyl-tRNAVal•EF-Tu•GTP ternary complexes to the ribosome had no effect on the RNase II-mediated truncations (Fig. 5C). Addition of both ternary complex and EF-G shifted the truncation pattern by three nucleotides toward the 3′ end of the mRNA (Fig. 5C). However, there was no evidence of A-site cleavage under any of the examined conditions in vitro (Figs. 5B & 5C). These data strongly suggest that the downstream border of the ribosome limits the progress of RNase II and PNPase.
Figure 5. Exonuclease digestion of mRNA in vitro.
(A) Sequence of the bacteriophage T4 gene 32-derived m617 mRNA. The positions of the Shine-Dalgarno sequence (SD), P-site Phe codon, and A-site Val codon are indicated. Nucleotide positions are numbered with respect to the first nucleotide (+1) after the A-site codon. (B) In vitro exonuclease digestion of ribosome-bound mRNA. 5′-labeled mRNA was incubated with N-acetyl-phenylalanyl-tRNAPhe (AcPhe-tRNAPhe) with (+) or without (−) 70S ribosomes. Reactions were split and incubated with purified RNase II (4, 20, 100, and 500 nM, final concentrations throughout), RNase II (D209N) (4, 20, 100, and 500 nM), or PNPase (20, 100, 500, 2500 nM) as described in Experimental Procedures. Molecular standards were generated by digesting m617 mRNA with RNase T1 (Ambion), which cuts after G residues. (C) RNase II cleaves mRNA to the 3′ border of the ribosome. Ribosomal complexes with AcPhe-tRNAPhe in the P site (P) were incubated with valyl-tRNAVal•EF-Tu•GTP ternary complexes (+TC) to form the pretranslocation complex. In control reactions lacking ternary complexes (-TC), valyl-tRNAVal was omitted. Pre-translocation complexes were then incubated with GTP in the absence or presence of EF-G to promote translocation. Aliquots were removed after each step and incubated with 500 nM of purified RNase II or RNase II (D209N), as indicated.
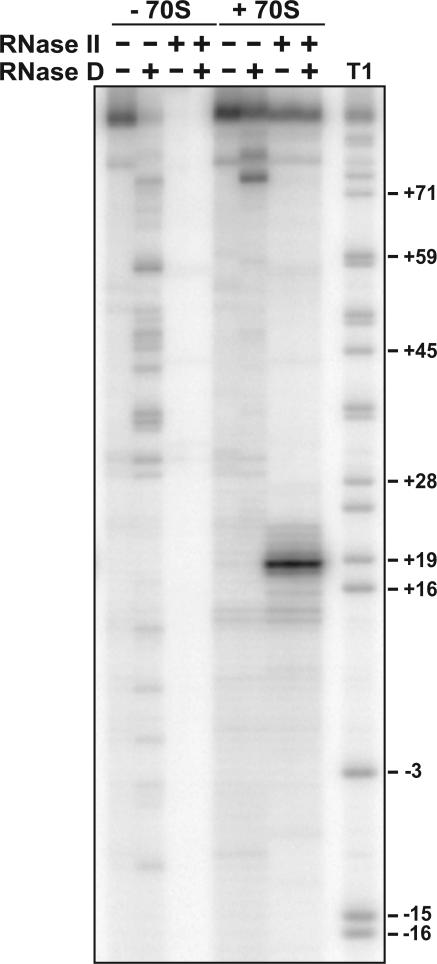
Because RNase D overexpression had a significant effect on mRNA cleavage in vivo (Figs. 5A & 5B), we also examined RNase D activity on ribosome-mRNA complexes in vitro. In the absence of added ribosomes, RNase D was able to extensively degrade the m617 transcript (Fig. 6). In contrast, RNase D-mediated degradation was limited to position +74 in the presence of 70S ribosomes (Fig. 6). When ribosome-mRNA complexes were digested sequential with RNase II and RNase D, degradation was indistinguishable from digestion with RNase II alone (Fig. 6). These data demonstrate that RNase D is capable of degrading mRNA, but do not support a direct role for this RNase in A-site mRNA cleavage.
Figure 6. Digestion of ribosome-mRNA complexes with RNase II and RNase D.
In vitro exonuclease digestion of mRNA within ribosome complexes. 5′-labeled m617 mRNA and AcPhe-tRNAPhe were incubated with (+ 70S), or without (− 70S) ribosomes. Reactions were then treated with 500 nM RNase II and/or 500 nM RNase D as indicated. Double digests were performed sequentially with RNase II treatment preceding the addition of RNase D. Molecular standards were generated by digesting m617 mRNA with RNase T1 (Ambion), which cuts after G residues.
Downstream mRNA secondary structure inhibits A-site mRNA cleavage
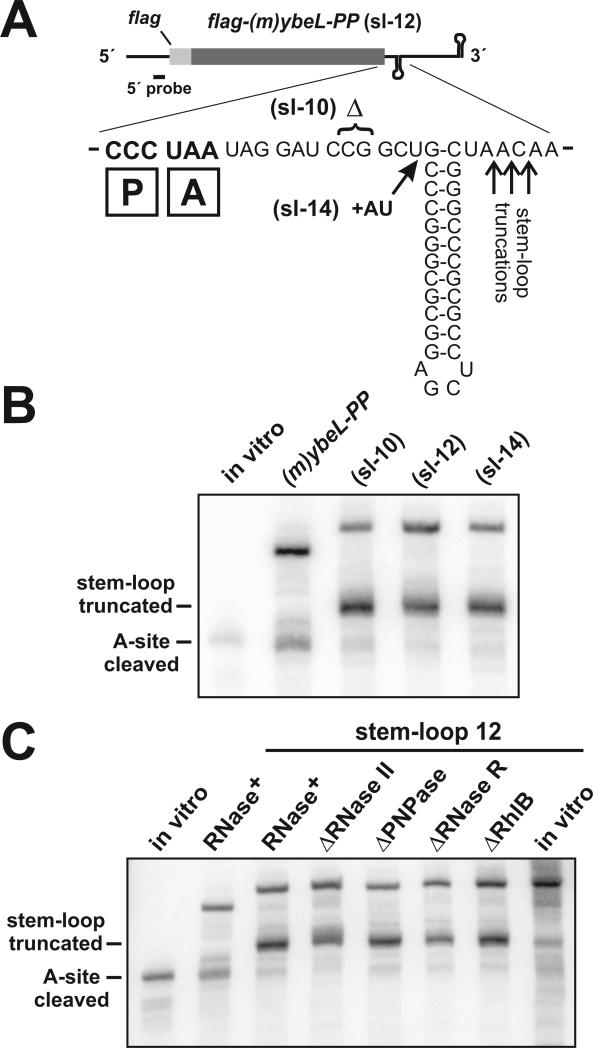
Because the in vitro degradation assays showed that RNase II degrades mRNA until it encounters the ribosome, we asked whether this ribosome-border degradation is a prerequisite for A-site mRNA cleavage. If so, then A-site cleavage should be reduced if RNase II is prevented from approaching the paused ribosome. Because RNase II is unable to degrade through extensive secondary structure, we reasoned that a stable stem-loop positioned immediately downstream of paused ribosomes would act as a physical barrier to RNase II, and suppress A-site cleavage. We generated three messages containing a stable stem-loop inserted into the 3′ UTR at positions 10, 12, and 14 nucleotides downstream of the stop codon (Fig. 7A). The ribosome binds approximately 9 − 10 nucleotides downstream of the A-site codon (Yusupova et al., 2001), so each stem-loop should be adjacent to, but not disrupted by the paused ribosome (Fig. 7A). Expression of each stem-loop message led to peptidyl-tRNA2Pro accumulation (data not shown), indicating that ribosome pausing during translation termination was unaffected by the stem-loops. However, there was significantly less A-site cleavage with the stem-loop messages compared to the original flag-(m)ybeL-PP transcript (Fig. 7B). Instead of undergoing A-site cleavage, the stem-loop transcripts were truncated at a site downstream of the stop codon (Fig. 7B). S1 nuclease protection analysis revealed that the stem-loop transcripts were truncated primarily 2 − 4 nucleotides downstream of the stem helix (Fig. 7A, and data not shown). Some of the stem-loop truncated messages probably arose from premature transcription termination, because these truncation products were also observed when the flag-(m)ybeL-PP(sl-12) message was synthesized in vitro using T7 RNA polymerase (Fig. 7C, last lane). However, there was much less stem-loop truncated message (with respect to full-length transcript) in the in vitro transcription reaction compared to mRNA synthesized in vivo (Fig. 7C). These data argue that most of the stem-loop truncated product was generated by RNase activity in vivo. Additionally, we note that premature transcription termination would also inhibit RNase II (and PNPase) activity, because the 3′ stem-loop structure prevents the binding of these RNases (Spickler and Mackie, 2000).
Figure 7. Stable secondary structure inhibits A-site mRNA cleavage.
(A) Expanded sequence view of the stem-loop 12 (sl-12) mRNA. The (sl-10) and (sl-14) transcripts were derived from (sl-12) by a two base deletion and insertion, respectively. Because the stem-loop was introduced into the 3′ UTR, A-site cleavage products are the same size for each transcript. The position of the paused ribosomal P and A sites are shown as boxed P and A, respectively. S1 nuclease protection showed that stem-loop truncated transcripts were cleaved primarily at positions indicated by the upward arrows (data not shown). (B) Northern analysis of stem-loop mRNA. Stem-loop messages were expressed in ΔtmRNA cells, and compared to the original flag-(m)ybeL-PP transcript. Stem-loop truncated transcripts correspond to the species indicated in panel A. The position of A-site cleaved mRNA is indicated by the migration of the control in vitro transcript. (C) Northern analysis of stem-loop 12 mRNA expressed in ΔtmRNA cells lacking RNase II, RNase R, PNPase, and RhlB. A control in vitro transcript marks the migration position of A-site cleaved mRNA (lane 1). RNase+ indicates samples were taken from cells containing the wild-type complement of exonucleases. In vitro transcription of the flag-(m)ybeL-PP(sl-12) message showed that the stem-loop induced some transcription termination (final lane).
Although the stem-loop structure significantly inhibited A-site cleavage, there appeared to be residual activity with each message (Fig. 7B). Purified RNase II and PNPase are unable to degrade this structure in vitro (Spickler and Mackie, 2000), but PNPase can degrade structured mRNAs with the assistance of the RhlB helicase in vivo (Liou et al., 2002; Py et al., 1996). Moreover, RNase R can efficiently degrade mRNA containing extensive secondary structure (Cheng and Deutscher, 2002; Vincent and Deutscher, 2009). We examined the flag-(m)ybeL-PP(sl-12) message expressed in ΔtmRNA cells deleted for PNPase, RNase R, and RhlB, and observed no significant changes in the relative proportions of A-site cleavage and stem-loop truncation (Fig. 7C). In contrast, residual A-site cleavage was further reduced upon deletion of RNase II (Fig. 7C). Additionally, we noted that the stem-loop truncated message appeared to be somewhat larger in ΔtmRNA ΔRNase II cells (Fig. 7C). We suspect this represents polyadenylation at the 3′ end of the stem-loop. Such poly(A) tails have been previously shown to accumulate in ΔRNase II cells (Marujo et al., 2000).
Discussion
The results presented here demonstrate that the 3′→5′ exonuclease activity of RNase II is required for RelE-independent A-site mRNA cleavage. RNase II is unable to directly degrade the A-site codon, thus we propose that RNase II-catalyzed degradation of mRNA to the ribosome border is a prerequisite for A-site cleavage. Deletion of RNase II also destabilizes a large number of transcripts in E. coli, (Mohanty and Kushner, 2003), and therefore it is formally possible that RNase II influences A-site cleavage through changes in global gene expression. However, two observations argue that RNase II is integral to the mRNA degradation pathway leading to A-site cleavage. Firstly, in addition to the inhibition of A-site cleavage, ΔRNase II cells do not accumulate the +18-truncated transcript. Formation of the +18 truncation product depends on translational pausing (Garza-Sánchez et al., 2008), and RNase II digestion of ribosome-mRNA complexes in vitro produces a similar +18 truncation product. These data strongly suggest that RNase II directly produces the +18 degradation product upon encountering paused ribosomes in vivo. Secondly, the introduction of a stable secondary structure element that prevents RNase II from approaching close to paused ribosomes also strongly inhibits A-site mRNA cleavage. This specific inhibition of A-site cleavage activity in RNase II+ cells argues against an indirect effect via altered gene expression. Therefore, RelE-independent A-site cleavage appears to be a complex process requiring at least two RNase activities. We propose calling this process the “A-site degradation pathway” to differentiate it from the RelE-mediated endonuclease activity.
Exonuclease activity is clearly an important component of the A-site degradation pathway, but it is unclear why RNase II-mediated degradation would be specifically required for subsequent A-site cleavage. In ΔRNase II cells, an unknown RNase truncates mRNA at the +12 position, which is very close to the leading edge of the paused ribosome. Therefore, mRNA degradation adjacent to paused ribosomes is not, per se, sufficient to induce A-site cleavage. Perhaps a specific contact between RNase II and the ribosome facilitates A-site mRNA cleavage. Butland et al. observed a direct interaction between RNase II and ribosomal protein S5 (Butland et al., 2005). Ribosomal proteins S3, S4, and S5 form the tunnel through which downstream mRNA enters the ribosome (Yusupova et al., 2001). Therefore, S5 is ideally positioned to interact with RNase II as it degrades mRNA to the border of paused ribosomes. It is possible that, upon encountering the ribosome, RNase II induces a conformational change in the complex that allows a second RNase to gain access to the A-site codon. Alternatively, the length of the exonucleolytic degradation product may be critical, because degradation to the +18 position appears to be correlated with A-site cleavage. For example, PNPase only plays a role in the A-site degradation pathway when overproduced, and is able to degrade mRNA to the +18 position only at high concentrations in vitro. In this model, the length of mRNA protruding from the ribosome would presumably influence recruitment of the A-site nuclease.
Which RNase catalyzes RelE-independent A-site mRNA cleavage? The A-site degradation pathway produces heterogeneous truncation products (Garza-Sánchez et al., 2008; Hayes and Sauer, 2003; Sunohara et al., 2004). This heterogeneity is suggestive of exonuclease activity, although it is possible for a non-specific endonuclease to produce a similar pattern. We have ruled out all of the known toxin endonucleases, but of course other uncharacterized toxins could be responsible for this activity. Additionally, none of the other non-essential 3′→5′ exonucleases – RNases D, PH, T, and BN(Z) – appears to play a direct role in A-site cleavage. The only known E. coli 3′→5′ exonuclease that we have not examined in vivo is the essential oligoribonuclease. However, oligoribonuclease has very low activity on polynucleotides greater than five residues (Datta and Niyogi, 1975; Ghosh and Deutscher, 1999), and therefore is unlikely to function as an A-site nuclease. Indeed, we have purified oligoribonuclease and find that is inactive on mRNA in vitro (S.S., F.G.-S., K.F., and C.S.H., unpublished results). Taken together, it appears that aside from RelE, none of the known endonucleases or exonucleases has A-site nuclease activity. These findings suggest that RelE-independent A-site cleavage is catalyzed by an uncharacterized RNase. Of course, it is still possible that the ribosome itself catalyzes A-site mRNA cleavage as we and others have suggested (Hayes and Sauer, 2003; Sunohara et al., 2004). If the ribosome does indeed mediate this activity, then there must be additional requirements because we have seen no evidence of A-site cleavage in our in vitro reactions.
The A-site degradation pathway appears to be a relatively rare consequence of ribosome pausing in bacteria. Several mRNA processing events are known to result from translational pausing, but most involve cleavage at, or near, the upstream border of the paused ribosome (Bjornsson and Isaksson, 1996; Loomis et al., 2001; Yao et al., 2008). Cleavage of the ermC leader transcript during erythromycin-induced ribosome pausing in Bacillus subtilis is perhaps the best understood of these processing events. Bechhofer and colleagues have recently shown that RNase J1 catalyzes an endonucleolytic cleavage 15 nucleotides upstream of the A-site codon (Yao et al., 2008). This processing is strikingly similar to a ribosome-dependent cleavage that occurs during translation of the DaaP peptide in uropathogenic E. coli (Loomis et al., 2001). Although, the precise position of the DaaP-paused ribosome is not known, the importance of nascent peptide residues Gly49-Pro50-Pro51 suggest that the cleavage occurs 12 − 15 nucleotides upstream of the A-site codon (Loomis et al., 2001). Bjornsson and Isaksson have also reported a cleavage 13 nucleotides upstream of the A-site UGA stop codon during inefficient translation termination (Bjornsson and Isaksson, 1996). Although the RNase(s) responsible for these latter cleavages is unknown, it is presumably an endonuclease because 5′→3′ exoribonucleases have not been identified in E. coli. In each of these instances, the paused ribosome stabilizes the downstream cleavage fragment. In contrast, the SecM-mediated translational arrest in E. coli allows 3′→5′ exonucleolytic degradation of mRNA to the downstream border of paused ribosomes, leaving a paused ribosome on the upstream degradation product (Garza-Sánchez et al., 2006). However, in this instance, degradation does not extend into the A site (Garza-Sánchez et al., 2006), indicating that A-site cleavage is not an obligatory result of border degradation. Presumably, A-site cleavage is inhibited in this system by prolyl-tRNAPro bound in the A site of SecM-arrested ribosomes (Garza-Sánchez et al., 2006; Muto et al., 2006). However, it is unclear why endonucleolytic processing at the upstream ribosome border appears to be favored over the A-site degradation pathway during the other translational arrests. Perhaps RNase II is unable to load onto the transcript in these instances. Given the complexity of prokaryotic mRNA turnover pathways, more work will be required to understand the molecular determinants controlling these processing events.
Experimental Procedures
Bacterial strains and plasmids
All bacterial strains were derivatives of E. coli strain X90 (DE3) and listed in Table I. Deletions of the rnb and pnp genes have been previously described (Garza-Sánchez et al., 2006). The Δrnc-38 RNase III deletion strain (SK7621), and the temperature sensitive rne-1 allele of RNase E (SK5704) were generously provided by Sidney Kushner. Deletions of all other genes were obtained from the Keio Collection of single-gene knockout mutants (Baba et al., 2006). Chromosomal deletions were introduced into strain CH113 by bacteriophage P1-mediated generalized transduction. The rne-1 allele was P1-transduced using a kanamycin resistance marker in the linked yceF gene. All of the resulting transductants were subjected to whole-cell PCR to confirm chromosomal deletion.
Table I.
Bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Reference |
|---|---|---|
| Strains | ||
| CH12 | X90 (DE3) | (Hayes et al., 2002) |
| CH113 | X90 (DE3) ssrA::cat, Cmr | (Hayes et al., 2002) |
| CH1207 | CH113 Δrnb::kan, Cmr, Kanr | (Garza-Sánchez et al., 2006) |
| CH1208 | CH113 Δpnp::kan, Cmr, Kanr | (Garza-Sánchez et al., 2006) |
| CH1916 | CH113 Δrna::kan, Cmr, Kanr | (Garza-Sánchez et al., 2006) |
| CH2790 | CH12 Δrnb::kan, Kanr | This study |
| CH3295 | CH113 Δrnr::kan, Cmr, Kanr | This study & (Baba et al., 2006) |
| CH3550 | CH113 ΔrelBE ΔchpBIK ΔmazEF ΔyefM-yoeB ΔmazEF ΔdinJ-yafQ ΔyhaV, Cmr | (Garza-Sánchez et al., 2008) |
| CH3574 | CH113 Δrnc-38, Cmr, Kanr | This study & (Babitzke et al., 1993) |
| CH3575 | CH113 Δrng::kan, Cmr, Kanr | This study & (Baba et al., 2006) |
| CH3583 | CH113 ΔyceF::kan rne-1, Cmr, Kanr | This study & (Babitzke and Kushner, 1991) |
| CH4312 | CH113 Δrnd::kan, Cmr, Kanr | This study & (Baba et al., 2006) |
| CH4318 | CH113 ΔpcnB::kan, Cmr, Kanr | This study & (Baba et al., 2006) |
| CH4456 | CH113 ΔrhlB::kan, Cmr, Kanr | This study & (Baba et al., 2006) |
| CH4463 | CH113 Δrnt::kan, Cmr, Kanr | This study & (Baba et al., 2006) |
| CH4465 | CH113 Δrph::kan, Cmr, Kanr | This study & (Baba et al., 2006) |
| CH4704 | CH113 ΔelaC::kan, Cmr, Kanr | This study & Baba et al., 2006) |
| CH5363 | CH113 Δrne515::kan, Cmr, Kanr | This study & (Garza-Sánchez et al., 2006) |
| CH5866 | CH113 ΔhrpA::kan, Cmr, Kanr | This study & (Baba et al., 2006) |
| Plasmids | ||
| pFLAG-(m)YbeL-PP | T7 promoter construct expressing C-terminal 49 residues of YbeL(PP) with N-terminal FLAG epitope, Ampr | This study |
| pFLAG-(m)YbeL-EA | T7 promoter construct expressing C-terminal 49 residues of YbeL(EA) with N-terminal FLAG epitope, Ampr | This study |
| pFLAG-(m)YbeL(amber) | pFLAG-(m)YbeL-PP with Glu28 amber point mutation, Ampr | This study |
| pFLAG-(m)YbeL-PP(UAG) | FLAG-mYbeL-PP construct with UAG stop codon, Ampr | This study |
| pCH405Δ-(m)YbeL-PP | pACYC184 derivative expressing FLAG-(m)YbeL-PP, Tetr | This study |
| pFLAG-(m)YbeL-PP(sl-10) | Contains GC-rich stem-loop 10 nucleotides downstream of stop codon, Ampr | This study |
| pFLAG-(m)YbeL-PP(sl-12) | Contains GC-rich stem-loop 12 nucleotides downstream of stop codon, Ampr | This study |
| pFLAG-(m)YbeL-PP(sl-14) | Contains GC-rich stem-loop 14 nucleotides downstream of stop codon, Ampr | This study |
| pRelE | pACYC184 derivative expressing RelE under araBAD promoter, Tetr | This study |
| pRNase II | pACYC184 derivative expressing RNase II under araBAD promoter, Tetr | This study |
| pRNase II (D209N) | pACYC184 derivative expressing inactive RNase II under araBAD promoter, Tetr | This study |
| pRNase II (ΔCSD1) | pACYC184 derivative expressing RNase II lacking cold shock domain-1 under araBAD promoter, Tetr | This study |
| pPNPase | pACYC184 derivative expressing PNPase under araBAD promoter, Tetr | This study |
| pRNase R | pACYC184 derivative expressing RNase R under araBAD promoter, Tetr | This study |
| pRNase D | pACYC184 derivative expressing RNase D under araBAD promoter, Tetr | This study |
| pRNase PH | pACYC184 derivative expressing RNase PH under araBAD promoter, Tetr | This study |
| pRNase T | pACYC184 derivative expressing RNase T under araBAD promoter, Tetr | This study |
| pRNase BN | pACYC184 derivative expressing RNase BN(Z) under araBAD promoter, Tetr | This study |
| pORNase | pACYC184 derivative expressing oligoribonuclease under araBAD promoter, Tetr | This study |
| pET-RNase II | pET21d with the wild-type rnb gene ligated via NcoI and XhoI, Ampr | This study |
| pET-RNase II (D209N) | pET21d with the rnb(D209N) gene ligated via NcoI and XhoI, Ampr | This study |
| pET-PNPase | pET21b with the pnp gene ligated via NdeI and XhoI, Ampr | This study |
| pET-RNase D | pET21b with the rnd gene ligated via NdeI and XhoI, Ampr | This study |
Abbreviations: Ampr, ampicillin resistant; Cmr, chloramphenicol resistant; Kanr, kanamycin resistant; Tetr, tetracycline resistant
Plasmids expressing A-site mRNA cleavage substrates were derivatives of pYbeL(PP), a pET11d-derived T7 RNA polymerase expression construct (Hayes et al., 2002; Hayes and Sauer, 2003). The (m)ybeL-PP and (m)ybeL-EA mini-genes were generated by PCR amplification of plasmids pYbeL(PP) and pYbeL(EA), respectively, using oligonucleotides, ybeL(49)-NdeI, (5′ - GGA CCT CAA TCA TAT GGG GGT TTA TCA CAG) and pET-Eco, (5′ - CGT CTT CAA GAA TTC TCA TGT TTG ACA GC), digested with NdeI and EcoRI, and ligated to plasmid pFG11 (Garza-Sánchez et al., 2008). The resulting plasmids, pFLAG-mYbeL-PP and pFLAG-mYbeL-EA express the C-terminal 49 residues of YbeL-PP and YbeL-EA fused to an N-terminal FLAG epitope. The Glu28 amber mutation was made within plasmid pFLAG-mYbeL-PP by the Stratagene QuickChange protocol using oligonucleotides, Glu28Amb, (5′ - GGC TGG GAA ATC TGG TCT GCT AGA AAT GTC ACT TCC ATC TCC); and Glu28Amb-GC, (5′ - GGA GAT GGA AGT GAC ATT TCT AGC AGA CCA GAT TTC CCA GCC).
The sl-12 stem-loop was introduced into the 3′-UTR of pFLAG-mYbeL-PP using two PCR amplification and ligation steps. The 5′-PCR fragment was produced using oligonucleotides, pET-Pst, (5′ - CAA GGA ATG GTG CAT GCC TGC AGA TGG CGC CC), and SL12-rev, (5′ - AAA GAG CTC CGC GCC CGG GCA GCC GGA TCC TAT TAG G); and the 3′-fragment amplified using oligonucleotides, SL12-for, (5′ - TTT GAG CTC CGC GCC CGG GCT AAC AAA GCC CGA AAG G), and pET-Eco. The two PCR products were sequentially ligated to pFG21P: the 5′-fragment with PstI/SacI and the 3′-fragment with SacI/EcoRI. The PstI/EcoRI fragment from the resulting plasmid was subcloned into pET11P to generate plasmid pFLAG-mYbeL-PP(sl-12). Plasmids pFLAG-mYbeL-PP(sl-10) and pFLAG-mYbeL(sl-14) were constructed from PCR fragments amplified with oligonucleotide pET-Eco, in conjunction with either SL10-Bam, (5′ - TTT CCG CCC TAA TAG GAT CCC TGC CCG G); or SL14-Bam, (5′ - ATA GGA TCC GGC TAT GCC CGG GCG C).
Genes encoding 3′→5′ exoribonucleases were PCR amplified using the following oligonucleotide primers: RNase II - rnb-NcoI, (5′ - CAA CCA TGG CAT TTC AGG ACA ACC CGC TGC TAG CGC) and rnb-XhoI, (5′ - TGA CTC GAG ATT ACG CGA CCG GGC GC); PNPase – pnp-EcoRI, (5′ - AGG GAA TTC ATA TGC TTA ATC CGA TCG TTC GTA AAT TCC) and pnp-XhoI, (5′ - AAT CTC GAG CTT ACT CGC CCT GTT CAG CAG CCG); RNase R – rnr-EcoRI, (5'- GAG GAA TTC ATA TGT CAC AAG ATC CTT TCC AGG AAC G) and rnr-XhoI, (5′ - AGG CTC GAG ATC ACT CTG CCA CTT TTT TCT TCG C); RNase D – rnd-EcoRI, (5′ - AGA GAA TTC CAT ATG AAT TAC CAA ATG ATT ACC ACG) and rnd-XhoI, (5′ - TTC TCG AGA TTT TAC TGC GGA TAT TCC TGC); RNase PH – rph-EcoRI, (5′ - GGA GAA TTC CAT ATG CGT CCA GCA GGC CG) and rph-XhoI, (5′ - TTT CTC GAG ACA AAA AAA AGG CGA CTC); RNase T – rnt-EcoRI, (5′ - TAA GAA TTC CAT ATG TCC GAT AAC GCT CAA CTT ACC) and rnt-XhoI, (5′ - GGC CTC GAG TCG ATT ACA CCT CTT CGG C); RNase BN – elaC-EcoRI, (5′ - CGT GAA TTC CAT ATG GAA TTA ATT TTT TTA GGT AC) and elaC-XhoI, (5′ - TAA TAA CTC GAG TTA AAC GTT AAA CAC GG); and oligoribonuclease – orn-EcoRI, (5′ -GTG GAA TTC CAT ATG AGT GCC AAT GAA AAC AAC C) and orn-XhoI, (5′ - CGC CGC TCG AGG ATT CCG GCG CTT ACG). The E. coli relE gene was PCR amplified using primers relE-NdeI, (5′ - GGA TCA ACT CAT ATG GCG TAT TTT CTG GAT TTT G), and relE-SacI, (5′ - ACA GAG CTC TCA TGC TTT GGT TCA GAG AAT GC). RNase II lacking the N-terminal cold shock domain was amplified with oligonucleotide, rnb-ΔCSD-NcoI, (5′ - CGT CCA TGG CTC GTT TCG TGG GTA AGG), and rnb-XhoI. The catalytically inactive Asp209Asn variant of RNase II was made by megaprimer PCR using oligonucleotide, rnb(D209N), (5′ - CGA AAA GGG CGT CAT TCA TAT CTT CTG TGC TG), in conjunction with rnb-NcoI and rnb-XhoI. All PCR products were ligated to plasmid pCH450, which is a derivative of pACYC184 containing the L-arabinose-inducible araBAD promoter (Table I). The pET21b and pET21d-derived overexpression constructs were generated by subcloning from the pCH450-based plasmids. All plasmid constructions were confirmed by DNA sequencing.
Cell growth, isolation and analysis of RNA and protein
Cultures were seeded at OD600 = 0.05 in LB medium supplemented with the appropriate antibiotics (ampicillin, 150 μg/mL; tetracycline, 12.5 μg/mL; kanamycin, 50 μg/mL; chloramphenicol, 66 μg/mL) and incubated at 37 °C with aeration. Upon reaching OD600 of ∼ 0.2 − 0.25, IPTG was added to a final concentration of 2 mM to induce synthesis of the FLAG-mYbeL variants. For exonuclease overproduction, cells were grown in LB medium containing 0.4% L-arabinose prior to addition of IPTG.
RNA isolations, T7 RNA polymerase in vitro transcriptions, and Northern blot analyses were conducted as described previously (Hayes and Sauer, 2003). The 5′ truncated in vitro transcription template was generated by PCR using oligonucleotides, (m)ybeL-PP trunc, (5′ - TAG GGC GGA AAC GGG CTT TTC TGG) and pET-Pst. The 3′ truncated in vitro transcription template was generated by PCR using oligonucleotides T7−3′-UTR, (5′ - GAA TTC TAA TAC GAC TCA CTA TAG TAG GAT CCG GCT GCT AAC AAA GC), and pET-Eco. Oligonucleotide pET-rbs, (5′ - GTA TAT CTC CTT CTT AAA GTT AAA C), and oligonucleotide pET-3′ probe, (5′ - TCA GCT TCC TTT CGG GCT TTG TTA GCA GCC) were used as 5′ and 3′ probes (respectively) for Northern blot hybridization with mRNA. Oligonucleotide proL probe (5′ - CAC CCC ATG ACG GTG CG) was used to detect tRNA2Pro by Northern blot. The S1 nuclease protection analysis was conducted essentially as described (Hayes and Sauer, 2003) using the oligonucleotide, ybeL-PP S1, (5′ - CTC AGC TTC CTT TCG GGC TTT GTT AGC AGC CGG ATC CTA TTA GGG CGG AAA CGG GCT TTT CTG GAA CTG GTC ATG ACC AC) as a probe to map the 3′-ends of flag-(m)ybeL-PP cleavage products. Primer extension analysis was performed with Superscript III (Invitrogen, Carlsbad, CA) reverse transcriptase and oligonucleotide pET-3′ probe as described (Diner and Hayes, 2009). The flag-(m)ybeL-PP(sl-12) cleavage product was mapped by S1 nuclease protection using oligonucleotide, SL-12 S1, (5′ - GCA GCC AAC TCA GCT TCC TTT CGG GCT TTG TTA GCC CGG GCG CGG AGC TCC GCG CCC GGG CAG CCG GAT CCT ATT).
Purification of exoribonucleases
RNase II, RNase II (D209N), and PNPase were overexpressed from pET plasmid constructs in CH12 Δrna cells. Cultures were harvested over ice, collected by centrifugation, and frozen at − 80 °C. Frozen cell pellets were resuspended in the appropriate purification buffer supplemented with 2 mM phenylmethylsulfonyl fluoride (PMSF). Buffer A [50mM Tris-HCl (pH 8.0) − 5 mM MgCl2 − 1 mM DTT − 5% glycerol] was used for PNPase purification, and buffer B [20mM Tris-HCl (pH 8.0) − 100 mM KCl] for RNase II purifications. Cells were broken by two French press passages at 20,000 psi. Lysates were clarified by centrifugation at 15,000 rpm for 15 min at 4 °C in an SS-34 rotor. PNPase-containing lysates were dialyzed against buffer A supplemented with 0.1 mM PMSF and 10 mM potassium phosphate (pH 8.0) at 37 °C for 1 hr as described (Jones et al., 2003). Native RNase II and PNPase were both purified by sequential QAE-cellulose ion exchange, and butyl-sepharose hydrophobic interaction chromatography. PNPase was loaded onto a QAE-cellulose column in buffer A and the column was developed with a linear gradient of 100 − 500 mM NaCl. RNase II was loaded onto the QAE-cellulose column in buffer B and eluted with a linear gradient of 100 − 500 mM KCl. Fractions were pooled and dialyzed against buffer C [25 mM bis-Tris (pH 6.5) − 1.2 M (NH2) 2SO4 − 0.5 M NaCl − 1 mM DTT] overnight at 4 °C. Dialyzed RNase preps were loaded onto HiTrap™ butyl-sepharose columns (GE Healthcare) pre-equilibrated in buffer C. RNases were eluted by step gradients with progressively lower salt concentrations. Fractions were screened by SDS-PAGE, pooled, and dialyzed against buffer A (for PNPase) or buffer B supplemented with 1 mM DTT (for RNase II). RNase II and RNase II (D209N) were quantified by absorbance at 280 nm using a molar extinction coefficient (ε) of 65,890 M-1 cm−1, and PNPase was quantified using ε = 30,370 M−1 cm−1.
Native RNase D was overexpressed from plasmid pET21b in CH12 Δrna Δrnb cells. Cell lysates were prepared in buffer D [20 mM Tris-HCl (pH 7.5) − 5 mM MgCl2 − 0.1 mM DTT − 0.1 mM EDTA] supplemented with 150 mM KCl and 0.1 mM PMSF as described above. The clarified lysate was diluted to 60 mM KCl with 1 × buffer D and loaded onto a QAE-cellulose column pre-equilibrated in the same buffer. The column was washed extensively with buffer D + 60 mM KCl, followed by washing with buffer D + 100 mM KCl. Protein was eluted with a linear gradient of 100 − 500 mM KCl in buffer D. Fractions containing RNase D were pooled and dialyzed against buffer C as described above. The dialyzed RNase D prep was loaded onto a butyl-sepharose column (GE Healthcare) pre-equilibrated in buffer C. Protein was eluted with step gradients of progressively lower salt concentration. Purified RNase D was dialyzed against buffer D supplemented with 100 mM KCl. RNase D was quantified by absorbance at 280 nm using a molar extinction coefficient (ε) of 79,419 M−1 cm−1.
Preparation of translation components
Ribosomes were prepared from E. coli strain CSH142 [F− ara Δ(gpt-lac)5] as described (Fredrick and Noller, 2002), dialyzed against polymix buffer [5 mM potassium phosphate (pH 7.3) − 95 mM KCl − 5 mM Mg(OAc)2 − 0.5 mM CaCl2 − 5 mM (NH4)Cl − 8 mM putrescine − 1 mM spermidine − 1 mM DTT] (Ehrenberg et al., 1990), and stored at −70 °C. tRNAPhe and tRNAVal were purchased from Chemical Block (Moscow, Russia). tRNAs were aminoacylated and acetylated as described (Walker and Fredrick, 2008). Aminoacylation and acetylation approached 100% efficiency, as determined by acid gel electrophoresis. Phage T4 gene 32-derived mRNA (m617) was made by in vitro transcription as described (Shoji et al., 2006). Transcripts were dephosphorylated using calf intestinal alkaline phosphatase, and 5′ end-labeled using polynucleotide kinase and γ-[32P]-ATP.
In vitro RNase digestions
Ribosomes (0.7 μM) were incubated in polymix buffer with radiolabeled m617 transcript and 1 μM N-acetyl-Phe-tRNAPhe (AcPhe-tRNAPhe) at 37°C for 20 min to bind the P site. For experiments shown in Fig. 5B, aliquots of P-site binding complexes were removed, and RNases added at the various concentrations. Reactions were incubated at 37 °C for 10 min, then quenched with formamide-containing loading dye (95% formamide − 20 mM EDTA − 0.05% xylene cyanol FF − 0.05% bromophenol blue) on ice. The degradation products were heated at 95 °C for 3 min, resolved by 6% denaturing gel electrophoresis, and visualized by phosphorimaging analysis (GE Healthcare). Radiolabeled m617 transcript was digested by RNase T1 (Ambion) to generate molecular size markers. To monitor translocation of ribosomal complexes as shown in Fig. 5C, 4 μM EF-Tu•GDP was first activated with 3 μM phosphoenolpyruvate and 0.33 mg/ml pyruvate kinase in polymix buffer to convert GDP to GTP. Ternary complexes were then formed by addition of Val-tRNAVal to a final concentration of 2 μM. As a negative control, Val-tRNAVal was omitted from the reaction. Equal volumes of ternary complexes were then added to P-site binding complexes and incubated at 37 °C for 10 min to form pre-translocation complexes. Pretranslocation complexes were incubated with 1 μM GTP in the absence or presence of 1 mM EF-G at 37 °C for 10 min. Aliquots were removed after each step and RNases added to a final concentration of 0.5 μM.
Acknowledgments
This work is supported by the National Institutes of Health through grants GM072528 (to K.F.), and GM078634 (to C.S.H.). We thank Sidney Kushner for generously providing bacterial strains; and Elie Diner, Laura Holberger, Brian Janssen, and Zach Ruhe for helpful discussions. We extend additional thanks to Zach Ruhe for strain construction, Brian Janssen for providing plasmid constructs, and Michael Hayes for RNase II and PNPase purification.
References
- Amblar M, Arraiano CM. A single mutation in Escherichia coli ribonuclease II inactivates the enzyme without affecting RNA binding. FEBS J. 2005;272:363–374. doi: 10.1111/j.1742-4658.2004.04477.x. [DOI] [PubMed] [Google Scholar]
- Amblar M, Barbas A, Fialho AM, Arraiano CM. Characterization of the functional domains of Escherichia coli RNase II. J. Mol. Biol. 2006;360:921–933. doi: 10.1016/j.jmb.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Andrade JM, Pobre V, Silva IJ, Domingues S, Arraiano CM. The role of 3′-5′ exoribonucleases in RNA degradation. Prog. Mol. Biol. Transl. Sci. 2009;85:187–229. doi: 10.1016/S0079-6603(08)00805-2. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:2006–0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babitzke P, Kushner SR. The Ams (altered mRNA stability) protein and ribonuclease E are encoded by the same structural gene of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1991;88:1–5. doi: 10.1073/pnas.88.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsson A, Isaksson LA. Accumulation of a mRNA decay intermediate by ribosomal pausing at a stop codon. Nucleic Acids Res. 1996;24:1753–1757. doi: 10.1093/nar/24.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butland G, Peregrin-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, Davey M, Parkinson J, Greenblatt J, Emili A. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- Carpousis AJ. The Escherichia coli RNA degradosome: structure, function and relationship in other ribonucleolytic multienzyme complexes. Biochem. Soc. Trans. 2002;30:150–155. [PubMed] [Google Scholar]
- Carpousis AJ. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 2007;61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- Cheng ZF, Deutscher MP. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J. Biol. Chem. 2002;277:21624–21629. doi: 10.1074/jbc.M202942200. [DOI] [PubMed] [Google Scholar]
- Christensen SK, Gerdes K. RelE toxins from bacteria and Archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol. Microbiol. 2003;48:1389–1400. doi: 10.1046/j.1365-2958.2003.03512.x. [DOI] [PubMed] [Google Scholar]
- Datta AK, Niyogi K. A novel oligoribonuclease of Escherichia coli. II. Mechanism of action. J. Biol. Chem. 1975;250:7313–7319. [PubMed] [Google Scholar]
- Diner EJ, Hayes CS. Recombineering reveals a diverse collection of ribosomal proteins L4 and L22 that confer resistance to macrolide antibiotics. J. Mol. Biol. 2009;386:300–315. doi: 10.1016/j.jmb.2008.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Nilsson L, Kurland CG. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- Dutta T, Deutscher MP. Catalytic properties of RNase BN/RNase Z from Escherichia coli: RNase BN is both an exo- and endoribonuclease. J. Biol. Chem. 2009;284:15425–15431. doi: 10.1074/jbc.M109.005462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenberg M, Rojas AM, Weiser J, Kurland CG. How many EF-Tu molecules participate in aminoacyl-tRNA binding and peptide bond formation in Escherichia coli translation? J. Mol. Biol. 1990;211:739–749. doi: 10.1016/0022-2836(90)90074-V. [DOI] [PubMed] [Google Scholar]
- Ezraty B, Dahlgren B, Deutscher MP. The RNase Z homologue encoded by Escherichia coli elaC gene is RNase BN. J. Biol. Chem. 2005;280:16542–16545. doi: 10.1074/jbc.C500098200. [DOI] [PubMed] [Google Scholar]
- Frazao C, McVey CE, Amblar M, Barbas A, Vonrhein C, Arraiano CM, Carrondo MA. Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature. 2006;443:110–114. doi: 10.1038/nature05080. [DOI] [PubMed] [Google Scholar]
- Fredrick K, Noller HF. Accurate translocation of mRNA by the ribosome requires a peptidyl group or its analog on the tRNA moving into the 30S P site. Mol. Cell. 2002;9:1125–1131. doi: 10.1016/s1097-2765(02)00523-3. [DOI] [PubMed] [Google Scholar]
- Garza-Sánchez F, Janssen BD, Hayes CS. Prolyl-tRNAPro in the A-site of SecM-arrested ribosomes inhibits the recruitment of transfer-messenger RNA. J. Biol. Chem. 2006;281:34258–34268. doi: 10.1074/jbc.M608052200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza-Sánchez F, Gin JG, Hayes CS. Amino acid starvation and colicin D treatment induce A-site mRNA cleavage in Escherichia coli. J. Mol. Biol. 2008;378:505–519. doi: 10.1016/j.jmb.2008.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Deutscher MP. Oligoribonuclease is an essential component of the mRNA decay pathway. Proc. Natl. Acad. Sci. U. S. A. 1999;96:4372–4377. doi: 10.1073/pnas.96.8.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong M, Gong F, Yanofsky C. Overexpression of tnaC of Escherichia coli inhibits growth by depleting tRNA2Pro availability. J. Bacteriol. 2006;188:1892–1898. doi: 10.1128/JB.188.5.1892-1898.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes CS, Bose B, Sauer RT. Proline residues at the C terminus of nascent chains induce SsrA tagging during translation termination. J. Biol. Chem. 2002;277:33825–33832. doi: 10.1074/jbc.M205405200. [DOI] [PubMed] [Google Scholar]
- Hayes CS, Sauer RT. Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Mol. Cell. 2003;12:903–911. doi: 10.1016/s1097-2765(03)00385-x. [DOI] [PubMed] [Google Scholar]
- He L, Soderbom F, Wagner EG, Binnie U, Binns N, Masters M. PcnB is required for the rapid degradation of RNAI, the antisense RNA that controls the copy number of ColE1-related plasmids. Mol. Microbiol. 1993;9:1131–1142. doi: 10.1111/j.1365-2958.1993.tb01243.x. [DOI] [PubMed] [Google Scholar]
- Jones GH, Symmons MF, Hankins JS, Mackie GA. Overexpression and purification of untagged polynucleotide phosphorylases. Protein Expr. Purif. 2003;32:202–209. doi: 10.1016/j.pep.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Karzai AW, Susskind MM, Sauer RT. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA). EMBO J. 1999;18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- Koo JT, Choe J, Moseley SL. HrpA, a DEAH-box RNA helicase, is involved in mRNA processing of a fimbrial operon in Escherichia coli. Mol. Microbiol. 2004;52:1813–1826. doi: 10.1111/j.1365-2958.2004.04099.x. [DOI] [PubMed] [Google Scholar]
- Li Z, Deutscher MP. The tRNA processing enzyme RNase T is essential for maturation of 5S RNA. Proc. Natl. Acad. Sci. U. S. A. 1995;92:6883–6886. doi: 10.1073/pnas.92.15.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Deutscher MP. Maturation pathways for E. coli tRNA precursors: a random multienzyme process in vivo. Cell. 1996;86:503–512. doi: 10.1016/s0092-8674(00)80123-3. [DOI] [PubMed] [Google Scholar]
- Li Z, Pandit S, Deutscher MP. 3′ exoribonucleolytic trimming is a common feature of the maturation of small, stable RNAs in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1998;95:2856–2861. doi: 10.1073/pnas.95.6.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Pandit S, Deutscher MP. Maturation of 23S ribosomal RNA requires the exoribonuclease RNase T. RNA. 1999;5:139–146. doi: 10.1017/s1355838299981669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou GG, Chang HY, Lin CS, Lin-Chao S. DEAD box RhlB RNA helicase physically associates with exoribonuclease PNPase to degrade double-stranded RNA independent of the degradosome-assembling region of RNase E. J. Biol. Chem. 2002;277:41157–41162. doi: 10.1074/jbc.M206618200. [DOI] [PubMed] [Google Scholar]
- Loomis WP, Koo JT, Cheung TP, Moseley SL. A tripeptide sequence within the nascent DaaP protein is required for mRNA processing of a fimbrial operon in Escherichia coli. Mol. Microbiol. 2001;39:693–707. doi: 10.1046/j.1365-2958.2001.02241.x. [DOI] [PubMed] [Google Scholar]
- Lopez PJ, Marchand I, Joyce SA, Dreyfus M. The C-terminal half of RNase E, which organizes the Escherichia coli degradosome, participates in mRNA degradation but not rRNA processing in vivo. Mol. Microbiol. 1999;33:188–199. doi: 10.1046/j.1365-2958.1999.01465.x. [DOI] [PubMed] [Google Scholar]
- March JB, Colloms MD, Hart-Davis D, Oliver IR, Masters M. Cloning and characterization of an Escherichia coli gene, pcnB, affecting plasmid copy number. Mol. Microbiol. 1989;3:903–910. doi: 10.1111/j.1365-2958.1989.tb00239.x. [DOI] [PubMed] [Google Scholar]
- Marujo PE, Hajnsdorf E, Le Derout J, Andrade R, Arraiano CM, Regnier P. RNase II removes the oligo(A) tails that destabilize the rpsO mRNA of Escherichia coli. RNA. 2000;6:1185–1193. doi: 10.1017/s135583820000073x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty BK, Kushner SR. Genomic analysis in Escherichia coli demonstrates differential roles for polynucleotide phosphorylase and RNase II in mRNA abundance and decay. Mol. Microbiol. 2003;50:645–658. doi: 10.1046/j.1365-2958.2003.03724.x. [DOI] [PubMed] [Google Scholar]
- Muto H, Nakatogawa H, Ito K. Genetically encoded but nonpolypeptide prolyl-tRNA functions in the A site for SecM-mediated ribosomal stall. Mol. Cell. 2006;22:545–552. doi: 10.1016/j.molcel.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Pedersen K, Zavialov AV, Pavlov MY, Elf J, Gerdes K, Ehrenberg M. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell. 2003;112:131–140. doi: 10.1016/s0092-8674(02)01248-5. [DOI] [PubMed] [Google Scholar]
- Perwez T, Kushner SR. RNase Z in Escherichia coli plays a significant role in mRNA decay. Mol. Microbiol. 2006;60:723–737. doi: 10.1111/j.1365-2958.2006.05124.x. [DOI] [PubMed] [Google Scholar]
- Py B, Higgins CF, Krisch HM, Carpousis AJ. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- Shoji S, Walker SE, Fredrick K. Reverse translocation of tRNA in the ribosome. Mol. Cell. 2006;24:931–942. doi: 10.1016/j.molcel.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spickler C, Mackie GA. Action of RNase II and polynucleotide phosphorylase against RNAs containing stem-loops of defined structure. J. Bacteriol. 2000;182:2422–2427. doi: 10.1128/jb.182.9.2422-2427.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundermeier TR, Dulebohn DP, Cho HJ, Karzai AW. A previously uncharacterized role for small protein B (SmpB) in transfer messenger RNA-mediated trans-translation. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2316–2321. doi: 10.1073/pnas.0409694102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunohara T, Jojima K, Yamamoto Y, Inada T, Aiba H. Nascent-peptide-mediated ribosome stalling at a stop codon induces mRNA cleavage resulting in nonstop mRNA that is recognized by tmRNA. RNA. 2004;10:378–386. doi: 10.1261/rna.5169404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent HA, Deutscher MP. Insights into how RNase R degrades structured RNA: analysis of the nuclease domain. J. Mol. Biol. 2009;387:570–583. doi: 10.1016/j.jmb.2009.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SE, Fredrick K. Preparation and evaluation of acylated tRNAs. Methods. 2008;44:81–86. doi: 10.1016/j.ymeth.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Lin-Chao S, Cohen SN. The Escherichia coli pcnB gene promotes adenylylation of antisense RNAI of ColE1-type plasmids in vivo and degradation of RNAI decay intermediates. Proc. Natl. Acad. Sci. U. S. A. 1993;90:6756–6760. doi: 10.1073/pnas.90.14.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Sunohara T, Jojima K, Inada T, Aiba H. SsrA-mediated trans-translation plays a role in mRNA quality control by facilitating degradation of truncated mRNAs. RNA. 2003;9:408–418. doi: 10.1261/rna.2174803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, Blaustein JB, Bechhofer DH. Erythromycin-induced ribosome stalling and RNase J1-mediated mRNA processing in Bacillus subtilis. Mol. Microbiol. 2008;69:1439–1449. doi: 10.1111/j.1365-2958.2008.06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- Zuo Y, Vincent HA, Zhang J, Wang Y, Deutscher MP, Malhotra A. Structural basis for processivity and single-strand specificity of RNase II. Mol. Cell. 2006;24:149–156. doi: 10.1016/j.molcel.2006.09.004. [DOI] [PubMed] [Google Scholar]