Abstract
In this study we probe and verify the concept of designing unreactive bioactive metal complexes, in which the metal possesses a purely structural function, by investigating the consequences of replacing ruthenium in a bioactive half-sandwich kinase inhibitor scaffold by its heavier congener osmium. The two isostructural complexes are compared with respect to their anticancer properties in 1205Lu melanoma cells, activation of the wnt signaling pathway, IC50 values against the protein kinases GSK-3β and Pim-1, and binding modes to the protein kinase Pim-1 by protein crystallography. It was found that the two congeners display almost indistinguishable biological activities which can be explained by their nearly identical three-dimensional structures and their identical mode of action as protein kinase inhibitors. This is a unique example in which the replacement of a metal in an anticancer scaffold by its heavier homolog does not alter the biological activity.
Keywords: Bioorganometallic chemistry, kinase inhibitors, anticancer, ruthenium, osmium
Introduction
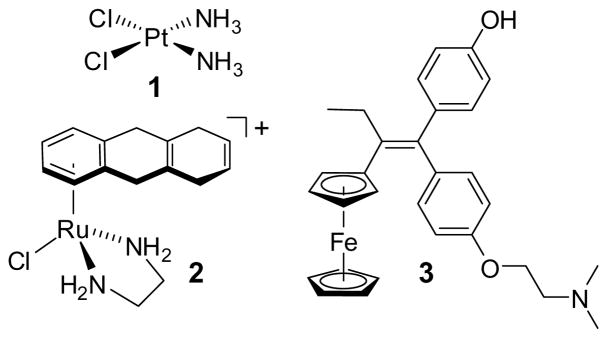
Metal complexes offer vast opportunities for the design of compounds with bioactivity due to the large variety of available metals and the ability to tune the reactivity and structure of the metal complexes by their ligand spheres.[1] Most commonly, the metal is directly involved in the mode of action, either through coordination or redox chemistry. For example, the highly successful anticancer drug cisplatin (1) (Figure 1) crosslinks guanine bases in DNA duplexes.[2] Similarly, the ruthenium arene complex 2 and its derivatives exert their high cytotoxic effects through coordination to DNA,[3] whereas the ferrocene moiety in the breast cancer drug candidate hydroxyferrocifen (3) is believed to serve as a crucial redoxactive moiety.[4] Thus, these reactive anticancer drugs require a finetuned reactivity of the metal center and it is therefore not surprising that in these complexes a substitution of the metal by one of their homologs results in a decline of the anticancer activities: the palladium analog of cisplatin is too hydrolytically unstable to serve as an anticancer drug, whereas the osmium analog of 2 hydrolyses by two orders of magnitude more slowly than the ruthenium congener and therefore should react more slowly with DNA,[5] and the ruthenium analog of hydroxyferrocifen 3, unlike the ferrocifens, does not show antiproliferate effects on estrogen receptor α-negative breast cancer cell lines, most likely due to a modified redox behaviour.[6]
Figure 1.
A selection of metal complexes with anticancer activities.
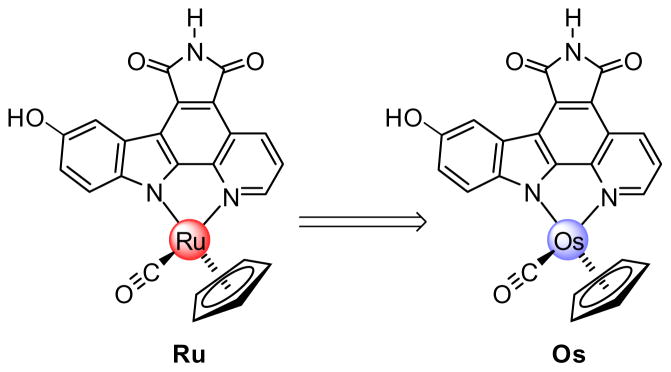
We recently revealed the promising kinase inhibition and anticancer effects of the ruthenium half-sandwich complex Ru and some of its derivatives (Figure 2).[7–12] The organometallic Ru is a highly potent inhibitor for the kinases GSK-3, Pim-1,[8,10,11] and probably some yet unidentified kinases, and induces strong biological responses, such as the activation of the wnt-signaling pathway in mammalian cells,[8] strong pharmacological effects during the development of frog embryos,[8] and the efficient induction of apoptosis in some melanoma cell lines.[12] In contrast to the forementioned examples 1–3, we believe that the ruthenium center in the scaffold Ru has a solely structural role and that it is rather the shape of the organometallic complex which is responsible for all of its bioactivity.[10]
Figure 2.
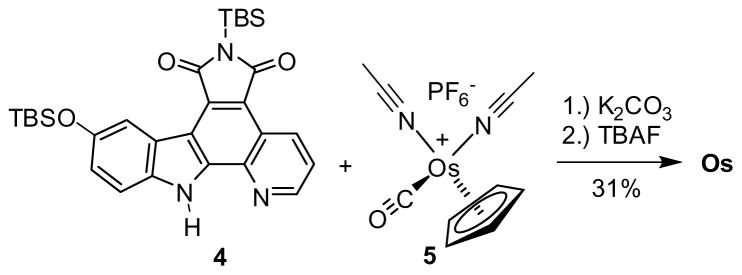
Synthesis of Os. TBAF = tetra-n-butylammonium fluoride, TBS = tert-butyldimethylsilyl.
We sought to probe this assumption by replacing ruthenium by its heavier homolog osmium (Ru→Os, Figure 2).[5,13] Ruthenium and osmium, which are located within the same group in the second and third transition metal row, respectively, form isostructural complexes because atomic radii of the two elements are almost identical due to the lanthanide contraction.[14] Despite these structural similarities, third row transition metal ions are generally significantly more substitutionally inert than those of the second row and possess a different redox chemistry.[15] Therefore, if the reactivity of the metal plays at least some role in the mode of action, Ru and Os should differ significantly in their bioactivities. In contrast, if indeed the three-dimensional structures of these organometallic scaffolds determine their bioactivities, the two congeners Ru and Os must display closely related properties. The following study demonstrates that the latter is indeed the case: Ru and Os display almost indistinguishable biological activities, thus verifying the concept of designing unreactive bioactive metal complexes with the metal serving as a key structural center.
Results and Discussion
Synthesis of osmium half-sandwich complexes
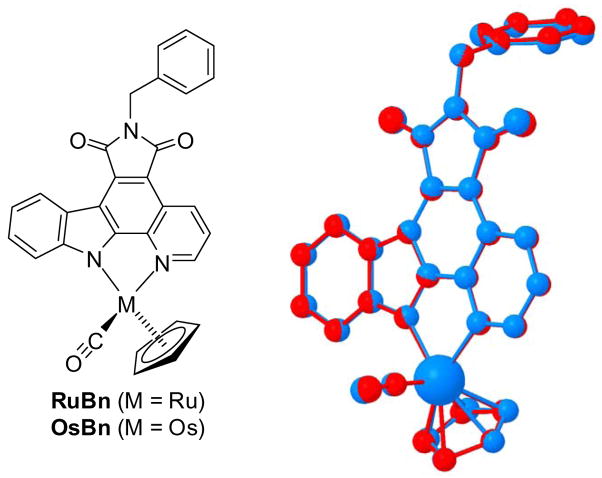
Compound Os was obtained in an analogous fashion to the recently described synthesis of Ru (Scheme 1).[16] Accordingly, the pyridocarbazole ligand 4 was reacted with [Os(η5-C5H5) (CO) (MeCN)2]+PF6− (5)[17] in the presence of one equivalent of K2CO3, followed by a TBAF-induced desilylation, affording Os with a modest yield of 31% over two steps. This synthetic scheme is general and applicable to derivatives of Os, such as OsBn in which a benzyl group at the imide moiety serves as a crystallization handle (see Figure 3).
Scheme 1.
Probing the correlation between shape and function with the isostructural complexes Ru and Os.
Figure 3.
Superimposed crystal structures of RuBn (red) and OsBn (blue).
Structural comparison of ruthenium and osmium complexes
In order to evaluate the structural similarity of Ru and Os, we crystallized the derivative OsBn and compared the obtained crystal structure with the analogous and recently described ruthenium crystal structure RuBn.[7] A superimposition revealed that both complexes are almost indistinguishable in their three-dimensional structures (Figure 3). For example, the differences in the coordinative bond length to the CO (Δ = 0.004 Å), the pyridine nitrogen (Δ = 0.008 Å) and the indole nitrogen (Δ = 0.004 Å) are within three times the estimated standard deviations (3×σ) and therefore identical within experimental errors. The discrepancies to the carbons of the cyclopentadienyl ligand range from 0.001 to 0.032 Å and from less than 1×σ to 7×σ. Altogether, it can therefore be concluded that replacing ruthenium for osmium in this half-sandwich scaffold leads to almost indistinguishable structures.
Configurational stabilities and redox potentials
Despite this desired structural conformity, osmium typically forms coordinative bonds which are more inert than analogous bonds of its lighter homolog ruthenium.[15] In our scaffold this phenomenon is reflected by the significantly higher configurational stability of Os compared to Ru. To evaluate this, a sample of (R)-Os (10 mM in DMSO) was stored in the dark at room temperature for one month and then analyzed with a Daicel Chiralpak 1B HPLC column (45:55→80:20 EtOH:hexane in 20 minutes, flow rate of 0.5 ml/min) after which no trace of the mirror image (S)-Os could be monitored. In contrast, the enantiopure ruthenium complexes racemize at room temperature by about 3% in one week, reflecting the higher inertness of the complex Os compared to Ru. With respect to redox behaviour, Ru and Os differ only slightly: The oxidative peak potential of Os (Epox = 0.312 V vs. Fc/Fc+) is around 50 mV lower compared to Ru (Epox = 0.36 V vs. Fc/Fc+).
Anticancer activities in 1205Lu melanoma cells
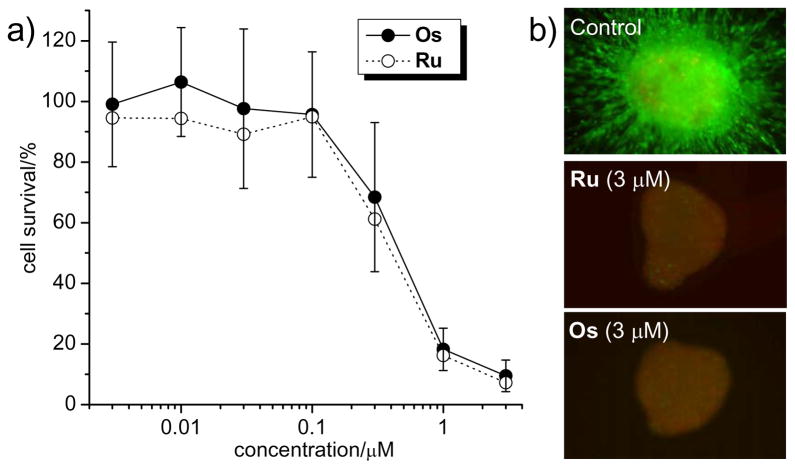
We initiated our comparative bioactivity study by measuring cytotoxicities of Ru and Os in melanoma cells.[12] For this, we incubated 1205Lu cells with different concentrations of organometallic compounds (3 nM to 3 μM) for 72 hours and quantified the reduction of live cells with the MTT method. The results in Figure 4a demonstrate that, within experimental errors, Ru and Os show almost identical concentration dependent cytotoxicity profiles in 1205Lu. For example, at 1 μM the cell survival is at 16 ± 4% and 18 ± 7% for Ru and Os, respectively.
Figure 4.
Anticancer properties of Os and Ru. a) 1205Lu melanoma cells were treated with compounds for 72 h and cell survival was determined using the MTT assay. Shown is the average of 5 independent experiments. b) Collagen-implanted 1205Lu spheroids were overlaid with medium and incubated with compounds for 72 h before treated with calcein-AM and propidium iodide. Green fluorescence indicates viable cells and red fluorescence dead cells.
To investigate this further in a more complex model, Ru and Os were tested in collagen-implanted three-dimensional spheroids of 1205Lu cells.[12] As shown in Figure 4b, at concentrations as low as 3 μM, both compounds decreased cell viability markedly and to a similar extent as visualized by the loss of green fluorescent live cells and the appearance of red fluorescent dead cells.[12] Thus, Ru and Os display highly potent and almost identical antiproliferate properties. Furthermore, cell cycle analyses of 1205Lu cells treated with Ru and Os for 24 hours, revealed a concentration-dependent increase in the number of cells in the sub-G1 population, indicating the induction of apoptosis as the main reason for cell death. Levels of apoptosis were very similar for Ru and Os, reaching 24% and 27% at 1 μM, and 50% and 41% at 3 μM, respectively (see supporting information).
Activation of wnt signaling
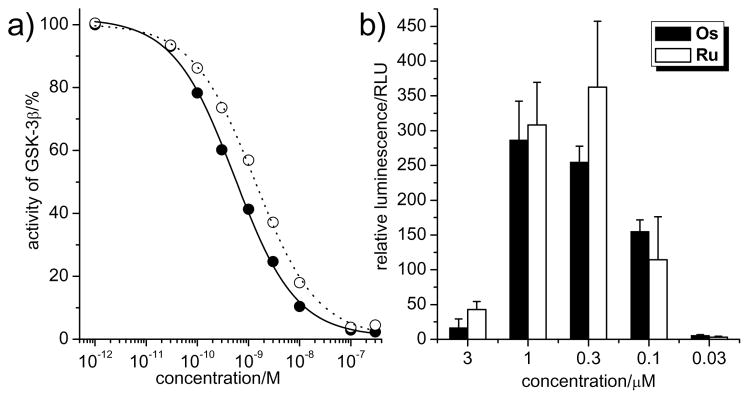
We recently disclosed that the strong apoptotic effect of complex Ru in 1205Lu cells occurs at least in part through p53-induced apoptosis, initiated by the inhibition of glycogen synthase kinase 3β (GSK-3β).[12] We therefore next compared Os and Ru in their ability to inhibit GSK-3β in the test tube and within mammalian cells. We first measured IC50 values against GSK-3β with an enzyme assay and found that both Ru and Os show a very similar binding behaviour, with Os being a slightly more potent inhibitor for GSK-3β with an IC50 of 0.6 ± 0.2 nM versus 1.4 ± 0.4 nM for Ru at 100 μM ATP (Figure 5a).[18] GSK-3β is a negative regulator of the wnt signal transduction pathway and therefore the inhibition of GSK-3β results in the activation of β-catenin dependent transcription.[19] In order to compare the activation of wnt signaling as a response to GSK-3β inhibition inside mammalian cells upon Ru and Os treatment, we used human embryonic kidney cells (HEK293OT) that are stably transfected with a TCF/β-catenin luciferase reporter gene (OT-Luc cells).[20] This system allows the monitoring of β-catenin levels with a luminescent read-out. Accordingly, we incubated OT-Luc cells with different concentrations of Ru and Os for 24 hours and determined luciferase levels from the addition of luciferin to the cell lysate followed by luminescence signal measurement. The results are shown in Figure 5b. Again, Ru and Os show a highly similar bell-shaped concentration-dependent wnt activation profile.
Figure 5.
Inhibition of GSK-3β by Os and Ru in the test tube and within mammalian cells. a) GSK-3β inhibition at 100 μM ATP and 200 pM of GSK-3β. IC50 curves were obtained by phosphorylation of phospho-glycogen synthase peptide-2 with [γ-32P]ATP. Every datapoint was determined from at least two independent measurements with error bars being less than 20%. b) Activation of the wnt signaling pathway by the inhibition of GSK-3β. HEK293 cells, transfected with a β-catenin-responsive luciferase reporter, were treated with different concentrations of compounds for 24 h. Luminescence signals were measured after cell lysis and the addition of luciferin. Shown is the average of 4 individual experiments.
Protein kinase binding
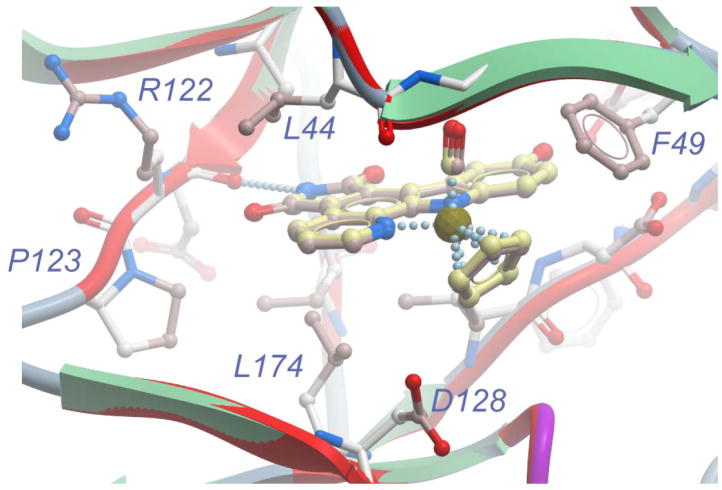
Altogether, the presented data imply that the almost identical anticancer properties of Ru and Os are the result of their highly similar protein kinase inhibition profiles. To support this conclusion, we attempted to compare the binding modes of the two congeners to a protein kinase, using the protein kinase Pim-1 as a model system. Accordingly, we cocrystallized (S)-Os with full-length human Pim-1, solved the structure to a resolution of 2.35 Å (Table 1), and compared it with the recently obtained structure of (S)-Ru and Pim-1.[10] To our knowledge, this crystal structure represents the first disclosed structure of an osmium complex bound to an enzyme. Upon superimposing the main chain atoms of both cocrystal structures (r.m.s. deviation of 0.24 Å), (S)-Ru and (S)-Os occupy almost indistinguishable binding positions within the ATP binding site. The same van der Waals interactions that (S)-Ru establishes with Pim-1 are preserved in the structure with (S)-Os. The pyridocarbazole moiety of (S)-Os is nicely placed in the hydrophobic pocket formed by the residues from the N-terminal and C-terminal domain, the cyclopentadienyl ring stacks against Phe49, while the CO group is positioned in close proximity to Gly45 (Figure 6). In the same way as (S)-Ru, (S)-Os forms a characteristic hydrogen bond between the maleimide NH-group and the carbonyl oxygen of Glu121 and the indole hydroxyl group is involved in two additional hydrogen bonds (to Lys67 and water-mediated to Glu89). This is in slight variation to the ruthenium structure in which both of these contacts are water mediated. Importantly, the metal centers are not involved in any direct interaction with the kinase active site and the two crystal structures are consistent with an experimentally verified identical binding affinity of Ru and Os to Pim-1 (IC50 values of 200 pM at 100 μM ATP).
Table 1.
Crystallographic data and refinement statistics of (S)-Os with Pim-1.a
| Parameters | |
|---|---|
| space group | P65 |
| cell dimensions [Å] | a,b = 98.45, c =80.36 |
| resolution [Å] | 2.35 |
| total observation (unique, redundancy) | 212458 (18397, 11.5) |
| completeness (outer shell) [%] | 97.7 (100.0) |
| Rmerge (outer shell) [%] | 14.9 (78.9) |
| I/σ (outer shell) | 18.1 (3.5) |
| Rwork (Rfree) [%] | 16.8 (22.5) |
| hetero groups | (S)-Os |
| rmsd bond length [Å] | 0.016 |
| rmsd bond angle [°] | 1.447 |
| Ramachandran [%](allowed/generally allowed/disallowed) | 92.8/7.2/0 |
rmsd = root−mean−square deviation.
Figure 6.
Superimposition of the cocrystal structures of Pim-1 with (S)-Os (PDB code 3BWF) and (S)-Ru (PDB code 2BZI). Amino acid side chains are only displayed for the structure with (S)-Os since the positions are virtually identical. Color coding: Ribbon in red for the ruthenium structure and for the osmium structure green (sheets) and blue (loops). Carbons of (S)-Ru and (S)-Os in pink and yellow, respectively. (S)-Os is displayed with slightly bigger stick and ball radii in order to distinguish it from (S)-Ru.
Conclusion
In summary, swapping ruthenium for the isostructural but chemically distinguished osmium in the organometallic protein kinase inhibitor scaffold (Ru→Os) enabled us to probe and verify our concept of designing unreactive bioactive metal complexes. To our knowledge, this is a unique example in which the replacement of a metal in an anticancer scaffold by its heavier homolog does not significantly alter the biological activity. This phenomenon can be explained by the almost identical three-dimensional structures of the two complexes and their identical mode of action as protein kinase inhibitors.[21] In fact, an osmium complex with such high antiproliferative effect in two- and three-dimensional cell culture is without any precedence. Ru and Os may thus be members of a new class of bioactive organometallic agents.
Experimental Section
Materials and general methods
NMR spectra were recorded on a Bruker AM-500 (500 MHz) or DMX-360 (360 MHz) spectrometer. Infrared spectra were recorded on a Perkin Elmer 1600 series FTIR spectrometer and high-resolution mass spectra were obtained with a Waters LCT Premier instrument using ESI ionization and TOF analyzer. Solvents and reagents were used as supplied from Fisher, Sigma-Aldrich, Acros, or Strem. Protein kinases (human) and substrates were purchased from Upstate Biotechnology, USA.
Synthesis of compound Os
A round bottom flask was charged with the pyridocarbazole ligand 4[16a] (50 mg, 0.094 mmol), [Os(η5-C5H5) (CO) (MeCN)2]+PF6− (5)[17] (48 mg, 0.094 mmol), K2CO3 (13 mg, 0.094 mmol), and a magnetic stir bar. The flask was purged with argon and solvent was added (4 ml MeCN and 1 ml MeOH). The reaction was stirred at room temperature for 2 h during which time the reaction became purple. The solvent was removed and the crude material was purified via silica gel chromatography eluting with hexanes:EtOAc 5:1, later 3:1. This yielded the product as a mixture with unreacted ligand. This mixture (36 mg) was carried forward without further purification and dissolved in methylene chloride (3 ml) and purged with argon. To this solution was added TBAF (97 μl of a 1 M solution in THF). The solution immediately became black and turbid. The reaction was stirred for 30 min at room temperature. Acetic acid was added to quench the reaction and the solution became purple. The solvent was removed and the crude material was purified via silica gel chromatography eluting with benzene:acetone 10:1, later 5:1. Compound Os was isolated as a purple solid (17 mg, 31% over 2 steps).
1H-NMR (500 MHz, [D8] DMSO): δ = 11.04 (s, 1H), 9.41 (d, J = 4.6 Hz, 1H), 9.23 (s, 1H), 9.00 (d, J = 8.2 Hz, 1H), 8.09 (d, J = 2.4 Hz, 1H), 7.65 (dd, J = 8.3, 5.2 Hz, 1H), 7.51 (d, J = 8.7 Hz, 1H), 7.12 (dd, J = 8.7, 2.5 Hz, 1H), 5.72 (s, 5H); 13C-NMR (125 MHz, [D6] DMSO): δ = 182.2, 170.6, 170.4, 157.8, 156.8, 151.8, 146.30, 146.27, 133.2, 131.3, 123.23, 123.20, 121.1, 116.5, 116.4, 114.5, 112.2, 108.2, 78.2; IR (thin film): v(cm−1) = 3392 (br), 2921, 2850, 1916, 1747, 1689, 1643, 1504, 1470, 1339, 1212, 668; HRMS calculated for C30H20N3O3Os (MH)+ 588.0599, found 588.0602.
Cyclic voltammetry of Os and Ru
Voltammetric experiments were conducted with a computer controlled Eco Chemie μ Autolab III potentiostat with 1 mm diameter planar Pt and glassy carbon (GC) working electrodes, a Pt wire auxiliary electrode and with an Ag wire reference electrode (isolated by a salt bridge containing 0.5 M Bu4NPF6 in CH3CN). Potentials were referenced to the ferrocene/ferrocenium redox couple. The electrochemical cell was thermostated at 293 K using an Eyela PSL-1000 variable temperature cooling bath. Data were obtained at a scan rate of 1 V s−1 in CH2Cl2 with 0.25 M Bu4NPF6 as the supporting electrolyte. Oxidative peak potentials (Epox): Ru = +0.36 V, Os = +0.31 V.
Pim-1 expression, purification, and cocrystallization with Os
Pim-1 was expressed and purified with some modifications as described previously.[22] Briefly, expression of the protein in BL21(DE3) pLysS cells was induced with 2 mM IPTG for 5 h at 18 °C. Cells were collected by centrifugation, resuspended in 50 mM HEPES pH 7.5, 500 mM NaCl, 5% glycerol, and lysed applying high pressure (French-press). The lysate was purified with a DEAE cellulose column (DE52 Whatmann) and Ni-NTA chromatography (Qiagen). The protein was treated overnight with lambda phosphatase and TEV protease to remove phosphate and His-tag, respectively. Further purification was achieved with a Mono-Q column (Amersham Biosiences), which separated dephosphorylated and phosphorylated fractions, and an additional Ni-NTA affinity column to ensure separation of Pim-1 from His-tag. Separated dephosphorylated and phosphorylated fractions were concentrated to 5 mg/ml in crystallography buffer (50 mM HEPES pH 7.5, 250 mM NaCl, 5% glycerol, 10 mM DTT). The osmium complex (S)-Os was added to the protein from a 10 mM DMSO stock solution to a final concentration of 1 mM. Crystals of nonphosphorylated Pim-1 with (S)-Os were grown at 4 °C in 4 μL sitting drops where 2 μL of protein solution was mixed with 2 μL of the precipitate stock containing 0.2 M lithium sulfate, 100 mM BisTrisPropane (pH 7.0), 20% PEG3350, 10% ethylene glycol and 0.3% DMSO. Crystals were cryoprotected and flesh cooled in liquid nitrogen.
Data collection and structure determination
Cryoprotected crystals yielded x-ray diffraction to 2.35 Å on a X12C beam line at National Synchrotron Light Source (Upton, NY). Data were indexed and merged using HKL2000.[23] The structure was solved by molecular replacement using a crystal structure of Pim-1 (PDB code 1YWV) as a search model for rotation and translation functions calculated with the program AmoRe.[24] Iterative cycles of refinement and manual rebuilding of the model were performed using the program REFMAC5 and O, respectively.[25,26] The structure has been deposited at the RCSB Protein Data Bank under the PDB code 3BWF.
Measurements of protein kinase inhibition
GSK-3β, Pim-1, and substrates were purchased from Upstate Biotechnology USA. For measuring IC50 values of Os and Ru against Pim-1, various concentration of inhibitors were incubated at room temperature with 80 pM kinase in 20 mM MOPS pH 7, 1 mM EDTA, 0.8 μg/μL bovine serum albumin, 5% DMSO (resulting from the inhibitor stock solution), in the presence of 50 μM S6 kinase/Rsk2 substrate peptide and 100 μM ATP including [γ-32P]ATP (0.2 μCi/μL). Reactions were initiated after 20 minutes by adding MgCl2 to a final concentration of 30 mM in the total reaction volume of 25 μL. The reactions were terminated by spotting 17.5 μL onto a circular P81 phosphocellulose paper (diameter 2.1 cm, Whatman) followed by washing with 0.75% phosphoric acid (three times) and acetone (once). The dried P81 papers were transferred to scintillation vials and 5ml of scintillation cocktail was added. The counts per minute (CPM) were measured with a Beckmann 6000 scintillation counter and IC50 values were defined as the concentration of inhibitor at which the CPM was 50% of the control sample corrected by the background.
For determining IC50 values of Os and Ru against GSK-3β, various concentration of compounds were incubated with 200 pM kinase in 20 mM MOPS pH 7, 30 mM MgCl2 1 mM EDTA, 0.8 μg/μL bovine serum albumin, 5% DMSO, in the presence of 20 μM phosphoglycogen synthase kinase-2 substrate for 20 minutes and reactions were initiated by adding ATP to a final concentration of 100 μM including 0.2 μCi/μL [γ-32P]ATP to a final volume of 25 μL. Reaction termination, measurements and IC50 determinations were performed in the same way as for Pim-1.
Cell cycle analysis
Cell cycle analysis was performed after treatment with kinase inhibitors (Ru, 1 and 3 μM, for 24 hrs; or Os, 1 and 3 μM). 1–2 × 106 cells grown adherently on a culture dish were harvested, washed in cold PBS and resuspended in 200 μl cold PBS. Cells were fixed by adding 200 μl of above-mentioned cell solution to 4 ml of 70% ethanol and incubated on ice for at least 1 hour. Intracellular DNA was labeled with 200 μl propidium iodide solution, containing 40 μg/ml propidium iodide, 100 μg/ml RNase in PBS and incubated at 37°C for 30 min in the darkness. Samples were analyzed using an EPICS XL (Beckman-Coulter, Inc., Miami, FL). The cell cycle profile was obtained by analyzing 15000 cells. Data was analyzed using WinMDI software and apoptosis was defined by the percentage of cells in the sub-G1 fraction of the cell cycle.
Cytotoxicity measurements in 1205Lu cell culture
1205Lu melanoma cells were maintained in DMEM medium plus 2% fetal bovine serum (FBS) at 37 °C under an atmosphere of 5% CO2 and constant humidity. For each experiment, cells were plated into a 96-well plate with 2500–3000 cells/well and left to grow for 24 h. Thereafter, cells were treated with increasing concentrations of Os and Ru and 1% DMSO (resulting from inhibitor stock solution) for 72 h. As a control, the same number of cells was treated with 1% DMSO. After the treatment, 20 μL of 5 mg/ml of 3-(4,5- dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) was added to each well. Cells were incubated with MTT for 3 h and after that time the media was removed followed by the addition of 100 μL of DMSO to solubilize the resulting purple crystals. Absorbance of each well was measured at 540 nm in a plate reader, and the cell survival in the presence of inhibitors was calculated as a percentage of control absorbance. Experiments were repeated 5 times and the average was taken.
Cytotoxicity measurements in 1205Lu spheroids
Melanoma spheroids were prepared using the liquid overlay method. Briefly, 200 μl of melanoma cells (25000 cells per ml) were added to a 96-well plate coated with 1.5% agar (Difco, Sparks MD). Plates were left to incubate for 72 h, by which time cells had organized into 3D spheroids. Spheroids were then harvested using a P1000 pipette. The media was removed and the spheroids were implanted into a gel of bovine collagen I containing EMEM, L-glutamine, and 2% FBS. Normal 2% melanoma media was overlayed on top of the solidified collagen. Spheroids were treated with either Ru (1–3 μM) or Os (1–3 μM), before being left to grow for 72 hours. Spheroids were then washed twice in PBS before being treated with calcein-AM, ethidium bromide (Molecular Probes, Eugene, OR) for 1 hour at 37 °C, according to the manufacturers instruction. After this time, pictures of the invading spheroids were taken using a Nikon-300 inverted fluorescence microscope.
Wnt activation in cell culture
OT Luc cells, kindly provided by Dr. Peter Klein (University of Pennsylvania, USA), were maintained in DMEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin at 37 °C under atmosphere containing 5% CO2 at constant humidity. Cells were plated on six-well plates (250000 cells/well in 2 mL medium) and let attach for 24 hours. Thereafter, the medium was exchanged with fresh medium (2 mL) and inhibitors were added (10 μL, 200 times concentrated in 100% DMSO). After incubation with different concentrations of inhibitor for 24 hours, cells were washed with 1 mL cold PBS. The luciferase assay system (Promega) was used for cells lysis and the luciferase assay. Accordingly, cells were harvested and lysed with 200 μL of lysis buffer supplemented with protease inhibitor cocktail and phosphatase inhibitor cocktails I and II from Sigma. The lysates were transferred into 1.5 mL tubes vortexed for 10 sec and left on ice for 30 min to ensure complete lysis. Cells were spun down at 10000 rpm at 4 °C for 20 minutes. Supernatants were stored at −80 °C until luminescence measurements. For luminescence measurements, luciferase substrate was dissolved in assay buffer according to the manufacturers protocol (5 μL of lysate was added into 100 μL of substrate) and luminescence signals measured immediately with a Monolight 3010 Luminometer from BD Biosciences.
Supplementary Material
Acknowledgments
E.M. gratefully acknowledges support for this work from the US National Institutes of Health (GM071695), the Camille and Henry Dreyfus Foundation, and the Alfred P. Sloan Foundation (BR-4634). We thank Dr. Judit É. Debreczeni and Dr. Luigi Di Costanzo for initial assistance with refining the Pim-1 structure.
Footnotes
Supporting Information Available: Crystallographic data for OsBn and cell cycle analysis data.
References
- 1.Metal-based drugs: Orvig C, Abrams MJ, editors. Chem Rev. Vol. 99. 1999. pp. 2201–2842.Guo Z, Sadler PJ. Angew Chem Int Ed. 1999;38:1512–1531. doi: 10.1002/(SICI)1521-3773(19990601)38:11<1512::AID-ANIE1512>3.0.CO;2-Y.Farrell N, editor. Coord Chem Rev. Vol. 232. 2002. pp. 1–230.Schlawe D, Majdalani A, Velcicky J, Heßler E, Wieder T, Prokop A, Schmalz HG. Angew Chem Int Ed. 2004;43:1731–1734. doi: 10.1002/anie.200353132.Dyson PJ, Sava G. Dalton Trans. 2006:1929–1933. doi: 10.1039/b601840h.Schatzschneider U, Metzler-Nolte N. Angew Chem Int Ed. 2006;45:1504–1507. doi: 10.1002/anie.200504604.Ang WH, Dyson PJ. Eur J Inorg Chem. 2006:4003–4018.
- 2.a) Wang D, Lippard SJ. Nature Rev Drug Discov. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]; b) Kelland L. Nature Rev Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 3.Yan YK, Melchart M, Habtemariam A, Sadler PJ. Chem Commun. 2005;38:4764–4776. doi: 10.1039/b508531b. [DOI] [PubMed] [Google Scholar]
- 4.Hillard E, Vessières A, Thouin L, Jaouen G, Amatore C. Angew Chem Int Ed. 2006;45:285–290. doi: 10.1002/anie.200502925. [DOI] [PubMed] [Google Scholar]
- 5.a) Peacock AFA, Habtemariam A, Moggach SA, Prescimone A, Parsons S, Sadler PJ. Inorg Chem. 2007;46:4049–4059. doi: 10.1021/ic062350d. [DOI] [PubMed] [Google Scholar]; b) Peacock AFA, Habtemariam A, Fernández R, Walland V, Fabbiani FPA, Parsons S, Aird RE, Jodrell DI, Sadler PJ. J Am Chem Soc. 2006;128:1739–1748. doi: 10.1021/ja055886r. [DOI] [PubMed] [Google Scholar]
- 6.Pigeon P, Top S, Vessières A, Huché M, Hillard EA, Salomon E, Jaouen G. J Med Chem. 2005;48:2814–2821. doi: 10.1021/jm049268h. [DOI] [PubMed] [Google Scholar]
- 7.Bregman H, Williams DS, Atilla GE, Carroll PJ, Meggers E. J Am Chem Soc. 2004;126:13594–13595. doi: 10.1021/ja046049c. [DOI] [PubMed] [Google Scholar]
- 8.Williams DS, Atilla GE, Bregman H, Arzoumanian A, Klein PS, Meggers E. Angew Chem Int Ed. 2005;44:1984–1987. doi: 10.1002/anie.200462501. [DOI] [PubMed] [Google Scholar]
- 9.Atilla-Gockumen GE, Williams DS, Bregman H, Pagano N, Meggers E. ChemBioChem. 2006;7:1443–1450. doi: 10.1002/cbic.200600117. [DOI] [PubMed] [Google Scholar]
- 10.Debreczeni JÉ, Bullock AN, Atilla GE, Williams DS, Bregman H, Knapp S, Meggers E. Angew Chem Int Ed. 2006;45:1580–1585. doi: 10.1002/anie.200503468. [DOI] [PubMed] [Google Scholar]
- 11.Meggers E, Ekin Atilla-Gokcumen G, Bregman H, Maksimoska J, Mulcahy SP, Pagano N, Williams DS. Synlett. 2007;8:1177–1189. [Google Scholar]
- 12.Smalley KSM, Contractor R, Haass NK, Kulp AN, Atilla-Gokcumen GE, Williams DS, Bregman H, Flaherty KT, Soengas MS, Meggers E, Herlyn M. Cancer Res. 2007;67:209–217. doi: 10.1158/0008-5472.CAN-06-1538. [DOI] [PubMed] [Google Scholar]
- 13.a) Peacock AFA, Parsons S, Sadler PJ. J Am Chem Soc. 2007;129:3348–3357. doi: 10.1021/ja068335p. [DOI] [PubMed] [Google Scholar]; b) Dorcier A, Ang WH, Bolano S, Gonsalvi L, Juillerat-Jeannerat L, Laurenczy G, Peruzzini M, Phillips AD, Zanobini F, Dyson PJ. Organometallics. 2006;25:4090–4096. [Google Scholar]; c) Peacock AFA, Melchart M, Deeth RJ, Habtemariam A, Parsons S, Sadler PJ. Chem Eur J. 2007;13:2601–2613. doi: 10.1002/chem.200601152. [DOI] [PubMed] [Google Scholar]; d) Cebrián-Losantos B, Krokhin AA, Stepanenko IN, Eichinger R, Jakupec MA, Arion VB, Keppler BK. Inorg Chem. 2007;46:5023–5033. doi: 10.1021/ic700405y. [DOI] [PubMed] [Google Scholar]
- 14.Structural comparison of homologous complexes of osmium and ruthenium: Gregory UA, Ibekwe SD, Kilbourn BT, Russell DR. J Chem Soc A. 1971:1118–25.Roesky HW, Pandey KK, Clegg W, Noltemeyer M, Sheldrick GM. Dalton Trans. 1984;4:719–21.Albers MO, Liles DC, Robinson DJ, Shaver A, Singleton E, Wiege MB, Boeyens JCA, Levendis DC. Organometallics. 1986;5:2321–7.Cotton FA, Diebold MP, Matusz M. Polyhedron. 1987;6:1131–4.Cheng CC, Goll JG, Neyhart GA, Welch TW, Singh P, Thorp HH. J Am Chem Soc. 1995;117:2970–80. doi: 10.1021/ja00110a001.Sellmann D, Wemple MW, Donaubauer W, Heinemann FW. Inorg Chem. 1997;36:1397–1402. doi: 10.1021/ic961240b.Adams RD, Bunz U, Captain B, Fu W, Steffen W. J Organomet Chem. 2000;614–615:75–82.Sugimoto H, Matsunami C, Koshi C, Yamasaki M, Umakoshi K, Sasaki Y. Bull Chem Soc Jpn. 2001;74:2091–2099.Baitalik S, Florke U, Nag K. Inorg Chimica Acta. 2002;337:439–449.Cabeza JA, del Rio I, Riera V, Suarez M, Alvarez-Rua C, Garcia-Granda S, Chuang SH, Hwu JR. Eur J Inorg Chem. 2003:4159–4165.Finze M, Bernhardt E, Willner H, Lehmann CW, Aubke F. Inorg Chem. 2005;44:4206–4214. doi: 10.1021/ic0482483.
- 15.Comparison of the kinetic inertness of ruthenium and osmium complexes: Richens DT. The Chemistry of Aqua Ions. Wiley; Chichester: 1997. pp. 421–429.Griffith WP. In: Comprehensive Coordination Chemistry. Wilkinson G, editor. Vol. 4. Pergamon; Oxford: 1987. pp. 519–633. ch. 46.Lay PA, Harman WD. Adv Inorg Chem. 1991;37:219–379.Halpern J, Cai L, Desrosiers PJ, Lin Z. Dalton Trans. 1991:717–723.George R, Andersen JM, Moss JR. J Organomet Chem. 1995;505:131–133.
- 16.a) Bregman H, Williams DS, Meggers E. Synthesis. 2005:1521–1527. [Google Scholar]; b) Pagano N, Maksimoska J, Bregman H, Williams DS, Webster RD, Xue F, Meggers E. Org Biomol Chem. 2007;5:1218–1227. doi: 10.1039/b700433h. [DOI] [PubMed] [Google Scholar]
- 17.a) Freedman DA, Gill TP, Blough AM, Koefod RS, Mann KR. Inorg Chem. 1997;36:95–102. [Google Scholar]; b) Dwyer FP, Hogarth JW, Rhoda RN. Inorg Syn. 1957;5:206–207. [Google Scholar]
- 18.A slight difference in polarity of Ru and Os may account for these differences in the binding affinity to GSK-3. To compare the molecular polarities of Os and Ru, we coinjected a mixture of Ru and Os on a Varian Microsorb MV 100–5 Si HPLC column (250 × 4.6 mm) eluting with EtOH:Hexanes 1:3 at a flow rate of 1 ml/min. Retention time for Os was 11.1 min compared to 11.8 min for Ru, thus, indicating a slightly higher hydrophobicity of Os over Ru.
- 19.a) Cohen P, Frame S. Nat Rev Mol Cell Biol. 2001;2:769–776. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]; b) Jope RS, Johnson GVW. Trends Biochem Sci. 2004;29:95–102. doi: 10.1016/j.tibs.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 20.Zhang F, Phiel CJ, Spece L, Gurvich N, Klein PS. J Biol Chem. 2003;278:33067–33077. doi: 10.1074/jbc.M212635200. [DOI] [PubMed] [Google Scholar]
- 21.It is noteworthy that altering the structure of Ru by replacing the imide nitrogen for a methyl group abolishes the bioactivity almost completely. For example, this methylated derivative does not affect cell growth and survival at all even at concentrations as high as 3 μM. This again illustrates that it is not the metal itself that is responsible for the bioactivity but clearly the overall structure of the organometallic complex.
- 22.Bullock AN, Debreczeni JÉ, Fedorov OY, Nelson A, Marsden BD, Knapp S. J Med Chem. 2005;48:7604–7614. doi: 10.1021/jm0504858. [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski Z, Minor W. Methods in Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 25.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 26.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.