Abstract
A study of the in vitro nanoparticle-templated assembly of a mutant of cowpea chlorotic mottle virus lacking most of the N-terminal domain (residues 4–37), NΔ34, is presented. Mutant empty proteins assemble into empty capsids with a much broader distribution of sizes than the wild-type virus. This increased flexibility in the assembly outcomes is known to be detrimental for the assembly process in the presence of molecular polyanions. However, when rigid polyanionic cores are used, such as nanoparticles, the assembly process is restored and virus-like particles form. Moreover, the breadth of the nanoparticle-templated capsid size distribution becomes comparable with the wild-type virus size distribution.
Introduction
Viruses constitute the largest reservoir of genetic material on the planet.1 In its simplest form, a virus is a nanoscopic container made of a protein shell, the capsid, encapsulating one or several molecules of nucleic acid.2 Despite their apparent simplicity on a biological scale, viruses are uniquely apt at delivering their own genome to the cells of virtually any living host. As a consequence, virus-based platforms were the first to be tested for gene therapy and they still account for the main body of gene delivery vectors.3 Other platforms for nucleic acid or non-nucleic acid delivery have also been pursued, such as nanoparticles,4 liposomes,5 and polymeric vesicles with amphiphilic walls.5 Even though these alternatives potentially avoid some of the immunogenic responses associated with viruses, the appeal for virus-based delivery remains strong because of the following two characteristics, which have not yet been fully exploited:
In contrast with nonviral platforms for targeting and delivery, viral materials are amenable to genetic engineering. Thus, the architecture, stability, and surface chemistry of the capsid proteins can be, in principle, application-optimized through recombinant DNA technology. A mechanistic understanding of the interactions between the vector and its environment, difficult to achieve in other cases, is simplified for viruses, which have benefited from studies with defined genetic modifications in conjunction with high-resolution structural determinations.
For a broad class of viruses, the number of capsid subunits is controlled during self-assembly by the properties of each individual protein subunit. In other words, the self-assembly process has mechanisms of quality check and self-termination. As a consequence, many viruses represent biological nanoparticles of well-defined size. Moreover, by virtue of their symmetry and identical composition, capsids have surface properties that are well-defined down to the atomic level as revealed by structural studies. In contrast, most nanoparticles made in the laboratory exhibit intrinsic size and surface heterogeneities with an exception being atomic clusters stabilized by electronic closed-shell effects.
An interesting question that has important implications for the use of viral nanoparticles is: what determines the size of a capsid? For rod-shaped viruses, the particles may depend on the length of the genome, suggesting that the interaction with the nucleic acid is an important parameter.5 In many single-stranded RNA icosahedral viruses the size of the genome and the net charge on the internal surface of the there capsid are correlated.6 There is some degree of flexibility in the formation of virus-like particles (VLPs) however, since particles of an RNA virus with 120 capsid subunits instead of the usual 180-subunit capsids have been observed in cells7 and can be recapitulated in self-assembly reactions in vitro.8 These studies indicate that the mechanisms of virus architecture control during assembly may be encoded in the molecular building blocks used, including the packaged cargo.9–11 Learning how to change the constraints leading to a particular assembly product is important because it would allow an increase in the building versatility using a given molecular building block. For many icosahedral viruses, a determinant for virion structure is the N-terminus of the capsid subunit, a sequence that is rich in basic amino acids, interacts directly with the genomic RNA and has an important role in determining the amount of genome packaged in a virion.9 The N-terminal sequence has been shown to hold a key role in redirecting the cage-constrained nanoparticle synthesis from nucleation of polyoxometalates with the wt sequence10 to Fe oxides with a mutant.11
We hypothesize that the manipulation of the same sequence could result in altered cargo-carrying capacity in self-assembled particles. In this paper, we present a study of the in vitro assembly of a mutant of cowpea chlorotic mottle virus (CCMV) lacking most of the N-terminal domain (residues 4–37), NΔ34.
CCMV was the first icosahedral virus to be assembled in vitro from separated proteins and nucleic acid.12 Wild-type CCMV has a ∼28 nm diameter capsid exhibiting T = 3 quasi-symmetry and containing 180 protein subunits. Each ca. 20 kDa subunit contains 189 amino acids. The subunit has C- and N-terminal arms extending away from a β-barrel core. The C-terminal tail is involved in strong intersubunit interaction at the origin of protein dimer formation in solution.
Insight into the nature of protein–protein and protein–RNA interactions is provided by the effects of changes in pH, ionic strength, and temperature on the virus dissociation and assembly. In vitro assembly from purified coat proteins in the presence of viral RNA (vRNA) occurs at neutral pH and high ionic strength.13 Empty capsids do not occur in vivo but have been observed to form at low ionic strength and at pH below 5.5 in vitro.14 These empty capsids are a good starting point to investigate the protein–protein interactions involved in CCMV formation. The dimer interface forms a flexible hinge, which is believed to control the radius of curvature upon the addition of subunits during capsid growth.15 The N-terminal arm extends into the interior of the virus capsid and interacts with the nucleic material by forming β-hexamer structures at the 3-fold axes of the virion to stabilize it.15 Tang et al. found that the truncated NΔ34 assembles into a variety of capsid configurations including, but not predominantly, the T = 3 native structure.16 Thus, the deleted region of the N-terminal domain contributes to the formation of a T = 3 virion. The polymorphism observed for empty capsids of NΔ34 is believed to be the result of a change in the interaction between two dimers, which in turn accounts for increased flexibility in the dihedral angle.
In this work, we examined how the NΔ34 capsid affected the formation of self-assembled VLPs on solid gold nanoparticle (GNP) cores, capped with carboxylate-terminated thiolalkylated tetra(ethylene glycol) (TEG) ligands. These ligands have been shown to induce capsid growth with high efficiency of particle encapsulation for the capsids of the closely related Brome mosaic virus.14 Note that other nonenveloped or enveloped icosahedral virus proteins may require at least part of the nucleic acid to be present to assemble into capsids.17,18 The influence of a foreign core on the result of the assembly of icosahedral capsids has been investigated for polyanions,19 liquid droplets,20 and solid nanoparticles.8 To our knowledge, this is the first report that focuses on what happens to assembly and incorporation efficiency when changes are made in the capsid protein, rather than in the core properties. It was found that, while the NΔ34 capsid subunits do not assemble into capsids in the presence of RNA, they do organize into quasi-spherical capsids if an anionic nanoparticle is used to nucleate VLP formation. Moreover, the size distribution of nanoparticle-templated VLPs is narrower than the size distribution of mutant empty capsids and its median value depends on the diameter of the core particle.
Experimental Section
Twelve nanometers of GNPs was synthesized and stabilized with carboxylate-terminated thiolalkylated tetra(ethylene glycol) (TEG) according to previously published protocol.21 CCMV and NΔ34 VLP assembly was carried out using a slightly modified published protocol.13 Briefly, intact CCMV is dialyzed against disassembly buffer A (0.05 M Tris-HCl, pH 7.5, 0.5 M CaCl2, 0.001 M dithiothreitol (DTT), and 0.0002 M phenylmethylsulfonyl fluoride (PMSF)) overnight at 4 °C and centrifuged at 90000 rpm for 1 h at 4 °C to separate the RNA (Optima TLX Ultracentrifuge, Beckman Instruments, Inc.). The supernatant is dialyzed against buffer B (0.02 M Tris-HCl, pH 7.5, 1.0 M NaCl, 0.001 M DTT and 0.0002 M PMSF) for 12 h at 4 °C. Capsid proteins are further purified by size exclusion chromatography (FPLC Superose 12) and concentrated to approximately 2 mg/mL using Ultracel-10k and Microcon YM-10 centrifugal filter devices (Millipore Corporation). Protein concentration is determined with a ND-1000 spectrometer, (NanoDrop Technologies) using an extinction coefficient of 1.18 cm2/mg for both WT and mutant CCMV.
For assembly of empty capsids and VLPs, the final protein concentration is adjusted to within 0.35–0.50 mg/mL. VLPs are assembled using capsid proteins and GNPs in a molar ratio of 180:1 in a total volume of 100 μL. This mixture is dialyzed against reassembly buffer C (0.05 M NaOAc, pH 7.0, 0.05 M NaCl, 0.01 M KCl, 0.005 M MgCl2, 0.001 M DTT, and 0.0002 M PMSF) for 12 h at 4 °C. Empty capsids are assembled by dialyzing capsid proteins against buffer D (0.1 M NaOAc, pH 4.8, 0.1 M NaCl, 0.0002 M PMSF) for 10–12 h at 4 °C. Reassembled particles were not purified by sucrose gradient centrifugation in order to compare the results of assembly. Lastly, the particles were dialyzed against virus buffer (0.1 M NaOAc, pH 4.6, 0.008 M Mg(OAc)2) overnight at 4 °C and stored at 4 °C. The two-step process of first lowering the ionic strength and subsequently the pH has been chosen because it yielded maximum efficiency of incorporation in a series of trials that have tested different buffer exchange pathways across the virus phase diagram. At the first step, there is protein association with the anionic core as seen by transmission electron microscopy (TEM) and dynamic light scattering (DLS), while at the second, there is an apparent redistribution of protein on the surface of nanoparticles (the protein layer becomes more compact as determined by negative-stain TEM).
All dialyses were carried out using Spectra/Por 12–14000 MWCO cellulose membranes (Spectrum Laboratories, Inc.). All reassembled particles are characterized by DLS (Zetasizer Nano, Malvern Instruments) and TEM (80 kV, JEM 1010, JEOL, Inc.). Samples for TEM were stained with 5% uranyl acetate (Mallinckrodt) on Carbon type-B grids (Ted Pella, Inc.).
Three-dimensional reconstruction of NΔ34 VLP was performed with reassembled VLPs that were further purified by centrifugation at 75000g for 1 h over a 10% sucrose gradient. A solution containing an empirically determined particle concentration suitable for electron microscopy was adsorbed to a glow-discharged carbon-coated copper grid, negatively stained with 1% uranyl acetate for 20 s, and visualized at 39000× with a JEOL 1200EX electron microscope at 100 kV. Electron micrographs were digitalized using an Epson (Long Beach, CA) Projection 3200 scanner set at 1200 dpi, corresponding to 5.5 Å/pixel. The EMAN software package was used for single-particle image analysis and three-dimensional (3D) reconstruction.22 The particles were selected from the digitized electron micrographs using EMAN's boxer routine. The images were then filtered to remove high- and low-frequency noise, translationally and rotationally aligned, classified, and averaged without applying symmetry.
An initial model was produced from the centered particles with EMAN's starticos program and subjected to eight cycles of refinement by projection matching. The final 3D reconstruction was scaled to 5.5 Å/pixel with EMAN's proc3d and visualized with the Chimera software.23
Results
The capsid protein from both wild-type CCMV and NΔ34 were used to form VLPs in the absence and presence of TEG-containing cores. Core diameters close to 12 nm have been found to efficiently promote the growth of BMV VLPs that have capsid structures similar to the native T = 3 capsids.8 Figure 1A presents a typical negative-stain transmission electron micrograph of TEG-coated GNPs with a diameter of 11.6 ± 0.9 nm. Results of DLS analysis show that both empty capsids and VLPs have diameters of approximately 30 nm (Figure 1B) demonstrating that the NΔ34 capsid does form both empty capsids and VLPs. However, the actual spread of size distributions is difficult to assess solely from DLS data since the instrumental response is broader or comparable to the particle size spread.
Figure 1.
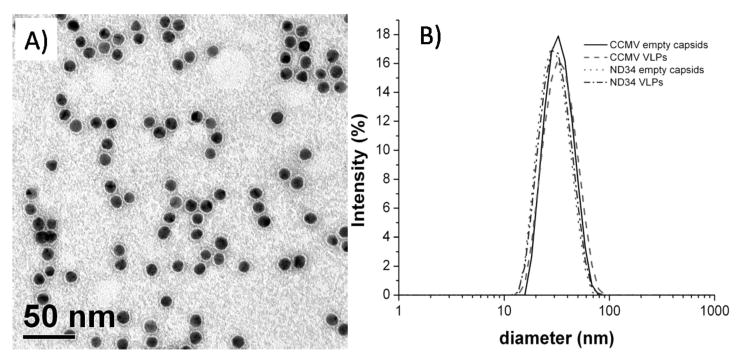
Formation of NΔ34 and CCMV VLPs using TEG-coated gold nanoparticles cores. (A) TEM image of 11.7 nm TEG-coated gold nanoparticles. The particles are stained with 2% uranyl acetate making the PEG layer visible. Scale bar: 50 nm. (B) Dynamic light scattering (DLS) intensity plots of empty and gold-encapsulating wt and mutant capsids taken at 10 °C.
To visualize the VLPs formed with NΔ34 and wild-type capsid proteins, TEM images of the unpurified assembly reactions were obtained (Figure 2A–C). The empty capsids assembled from the NΔ34 capsid confirm the previously reported formation of a variety of capsid sizes (Figure 2A). In the NΔ34 containing the GNP, however, we observed a homogeneous population of VLP particles formed without further purification (Figure 2B). Encapsulation was also less complete in the unpurified assembly reaction with NΔ34 when compared to the wild-type CCMV (Figure 2C). Additionally, upon purification of the assembly reaction over a 10% sucrose density gradient, the NΔ34 VLPs can be enriched and show a fairly homogeneous preparation (Figure 2D).
Figure 2.
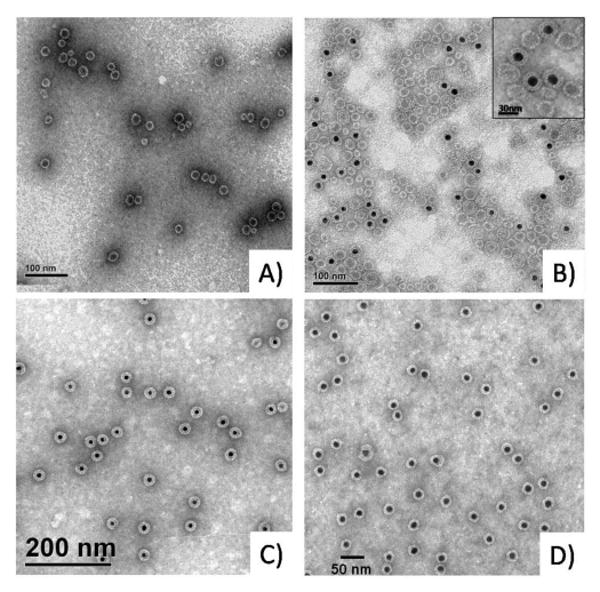
TEM pictures of assembly products: (A) Empty NΔ34 capsids showing significant size variability. (B) NΔ34 VLPs from an unpurified assembly reaction showing incorporation of gold cores although at a lower efficiency. Inset: higher magnification micrograph for a more detailed view of the complexes. (C) VLPs from an unpurified assembly reaction with wild-type CCMV capsid. (D) NΔ34 VLPs purified over a sucrose density gradient.
A quantitative comparison of the size distributions of assembly products for empty capsids and capsids encapsulating the 11.7 nm gold cores of NΔ34 is provided in Figure 3A. Similar experimental size distribution probabilities for wild-type CCMV empty capsids and capsids with 11.6 nm cores are given in Figure 3B. To confirm the effects of the cores further, assembly of the VLPs were performed with GNPs of size 6 nm and the unpurified VLPs formed were measured and plotted in Figure 3C in direct comparison with VLPs having 11.7 nm cores. Table 1 contains numerical values for the mean and sample variance for each size distribution. The largest variance is observed for the empty NΔ34 capsids.
Figure 3.
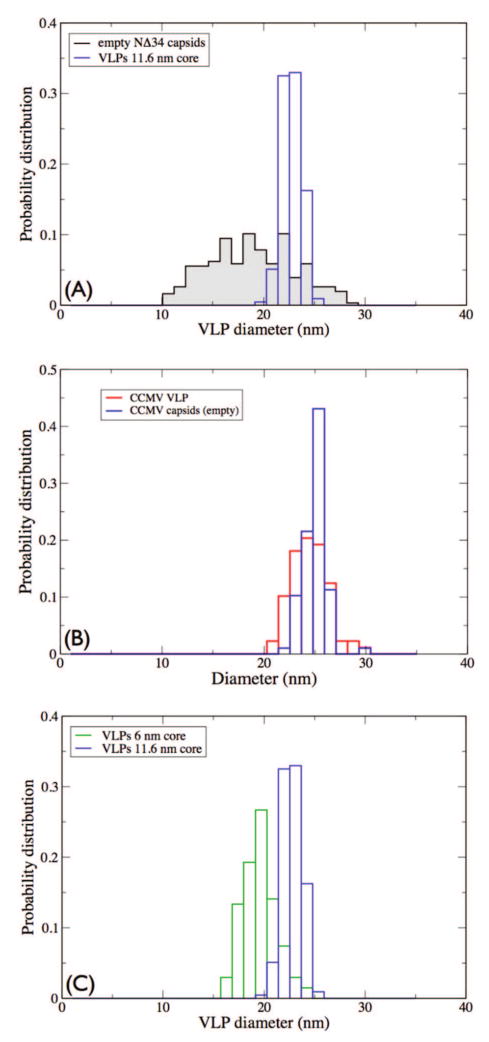
Probability distributions of diameters for empty and nanoparticle-encapsulating capsids: (A) mutant complexes showing a broader variation for empty capsids (solid) compared to VLPs; (B) wild-type empty capsids (solid) and VLPs (11.6 nm core) with similar size distributions; (C) mutant VLP complexes with 11.6 and 6 nm cores indicating the effect on the mean diameter of the complexes of varying the core size.
Table 1.
Statistics of the Assembly Products Sizes from Negative Staining TEM
| NΔ34emptycapsids | NΔ346 nmVLPs | NΔ3411.7 nmVLPs | CCMVemptycapsids | CCMV11.7 nmVLPs | |
|---|---|---|---|---|---|
| mean diameter (nm) | 18.9 | 19.6 | 22.7 | 24.5 | 25.0 |
| sample variance (nm2) | 17.1 | 2.5 | 0.9 | 3.2 | 1.0 |
VLPs encapsulating gold cores have variances an order of magnitude smaller. These results suggest that the anionic GNPs are organizing the formation of the VLPs and resulting in a more homogeneous preparation.
While these results demonstrate that the anionic nanogold particles can impose homogeneity in the results of capsid assembly even to a defective capsid subunit, we addressed whether any of the VLPs recapitulate the normal virion structure. Over 3800 particles of the VLPs with the 11.6 nm cores were individually picked and subjected to image reconstruction. The class averages are shown in Figure 4A and they show a good representation of the available views (Figure 4B). The model for the final reconstruction is shown in Figure 4C and shows a pseudo T = 2 particle, consistent with the ∼20 nm diameter particles formed. A comparison with the wild-type CCMV model (T = 3) generated by Wikoff et al.24 can be readily observed by the arrangement of the capsomeres, (Figure 4D). On the basis of the size distribution in Figure 3, NΔ34 may also form T = 1 and T = 3, but at this point, we do not have reconstruction data supporting this hypothesis. Moreover, the prevalence of the pseudo-T = 2 structure may be a protein concentration effect. Further studies are underway to test this hypothesis.
Figure 4.
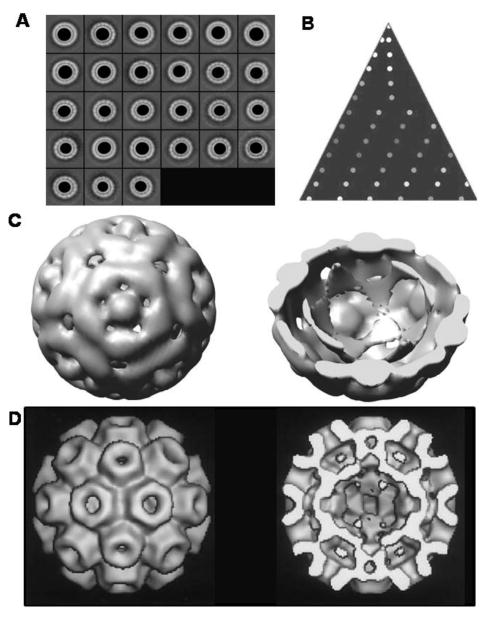
Negative-stain TEM reconstruction of the VLPs formed by NΔ34. (A) Class averages generated from ∼3800 VLPs formed with the 11.7 nm cores.(B) An asymmetric triangle depicting anisotropy with regards to the sampling of the 3D space. (C) 3D reconstruction of the VLPs formed with 11.7 nm showing the pentamer. The surface rendering threshold was set to correspond to a molecular mass of 240 kDa. The model to the right shows a cut-away structure of the VLP. (D) The structure of the CCMV particle generated by Wikoff et al.24
Discussion
The importance of the N-terminal of the capsid protein for icosahedral plant virion assembly has been well established.2,15,25 Residues 1–27 of the CCMV capsid are disordered in the crystal structure but account for the bulk positive charge carried by the floppy segment of the N-terminal. Deletion of the entire N-terminal arm prevents the formation of both empty capsids and virions.22,23 Mutant NΔ34 has residues 4–37 deleted and does not form the β-hexamer structure required to stabilize the wild-type virion or encapsidate RNA in vitro. The latter fact is not surprising, since in a capsid with 180 subunits, the NΔ34 will cause a cumulative decrease of ∼1500 elementary charges. However, we showed with two different anionic nanoparticle sizes that the defect in ordered assembly by the NΔ34 capsid could be restored. This is not a purely electrostatic effect: while the nanoparticle surface contains at saturation a few hundred elementary charges, the native RNA carries more than a thousand. While these results show that core–capsid shell interactions drive at least the initial stages of assembly, there might be mutual adjustment of portions of the capsid and the genomic RNA that are not solely based on charge.
We postulate that the radius of curvature of the mutant capsid seems to be determined by the internal scaffold as demonstrated in Figure 3 and Table 1. This supports the idea of structural support role and geometrical constraints restored by the hard surface core. In the case of the wild-type virus, this structural support could come from the spherical polyelectrolyte layer formed through the association between the N-terminal and RNA. Atomic force microscopy studies have found that RNA-filled capsids of CCMV are 30% stiffer to nanoindentation than empty capsids at acidic pH.26 The reason for this might be different from the case of viruses such as the bacteriophage lambda, for which the resistance to nanoindentation could be entirely explained by an osmotic pressure effect.27 However, the CCMV capsid is permeable28 and dynamic with respect to the compact, resilient capsid of the lambda phage that actively packs its genome at pressures of ∼20 atm.27
Another feature that differentiates NΔ34 from the wild-type CCMV is the greater presence of empty capsids under conditions of VLP assembly in vitro (Figure 2B). At stoichiometric gold to protein assembly ratios, no free TEG-capped nanoparticles were observed for the wild-type correlating to 100% incorporation efficiency whereas NΔ34 only assembles 72% of the total gold nanoparticles into VLPs. Also in wild-type VLP assembly, hardly any empty capsids are observed (<0.5% per 100 total assembled products) in contrast to mutant VLP assemblies in which 30% of all assembly products are empty capsids at stoichiometric ratios. Therefore, in the absence of the N-terminal and under the conditions for virus assembly (not empty capsid assembly), mutant capsid protein interactions allow for the formation of empty shells whereas the wild-type proteins simply aggregate. Interestingly, even with ∼80% of the basic residues removed, the truncated N-terminal arm is able to bias the assembly toward the formation of intact viruses or VLPs under in vitro assembly conditions and away from empty capsid formation. These observations suggest that there is a smaller free energy difference between VLPs and empty capsids in the case of the mutant. This difference could come from changes in both the protein–nanoparticle interactions or protein–protein interactions. Removing charges should result in a weaker electrostatic interaction with core surfaces, thus reducing the driving force for the capsid protein to assemble on the nanoparticle surface. At the same time, however, deleting 80% of the positive charges on the N-terminal arm would reduce intersubunit electrostatic repulsions and thus increase the driving force for empty shell formation. What weight may carry each of these mechanisms remains to be determined by further experiments on the influence of pH and ionic strength on mutant VLP and empty capsid assembly.
The observed reduced encapsidation efficiency of NΔ34 is consistent with the model proposed by Willits et al. to explain the increased symptom expression by mutant-infected hosts resulting from β-hexamer deletions.25 According to these authors, mutations may reduce the rate of assembly and subsequently increase the cellular concentrations of unassembled viral components (empty capsids are not stable in vivo), which in turn would elicit an increased host response to infection.
Conclusion
A CCMV mutant capsid protein lacking most of the basic N-terminal residues has been shown to unexpectedly self-assemble around solid nongenomic cores, resulting in structure sizes that can be tuned by varying the core size. In the context of size-dependent properties, a recent study in nanoparticle-assisted transfection indicated a strong dependence of membrane translocation efficiency on size.29 In light of this result, one could hypothesize that virus shapes and sizes have also evolved for optimal specific interactions with the host, and an ability to control this size as indicated in this work would be advantageous for virus applications in biomedicine. These results also have relevance in the engineering of VLPs as delivery vectors. The fact that a capsid with a significant N-terminal deletion can retain the ability to form homogeneous VLPs should allow the deleted sequence to be replaced with some molecule that has desired biotechnological applications. These manipulations should enable tuning of the size of VLP by manipulating capsid sequence and possibly directing evolution of the capsid to desired cargos. For instance, insight into the design of virus-based drug delivery systems30 and hierarchical materials8,26,27 could be gained from having a single molecular building block (capsid protein) from which a variety of capsule sizes could be built. Lastly, we note that viral RNA, despite carrying more charge than the nanoparticle, cannot rescue viral encapsulation with the NΔ34 capsid protein. It is interesting to observe that the association between RNA and the internal N-terminal may result in a quasi-rigid polyelectrolyte scaffold that induces the particle formation of a given size. The rigid nanoparticle core can similarly guide the efficient interlocking of the structural parts of wild-type capsid protein subunits.
Acknowledgments
The authors thank Ayaluru Murali of Texas A&M for his help with the 3D reconstructions. This research was supported in part by the Indiana METACyt Initiative of Indiana University, funded through a major grant from the Lilly Endowment, Inc. We are gratefully acknowledging partial support for this work from the Center for Hierarchical Manufacturing, DMI-0531171, the National Science Foundation (Grant 0631982), and the National Institutes of Health (Grant GM081029-01).
References
- 1.Suttle CA. Viruses in the sea. Nature. 2005;437:356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- 2.Casjens S, editor. Virus Structure and Assembly. Jones and Bartlett; Boston: 1985. [Google Scholar]
- 3.Edelstein ML, Abedi MR, Wixon J, Edelstein RM. Gene therapy clinical trials worldwide 1989–2004–an overview. J Gene Med. 2004;6:597–602. doi: 10.1002/jgm.619. [DOI] [PubMed] [Google Scholar]
- 4.Kumar M, Hellermann G, Lockey RF, Mohapatra SS. Nanoparticle-mediated gene delivery: state of the art. Expert Opin Biol Ther. 2004;4:1213–1224. doi: 10.1517/14712598.4.8.1213. [DOI] [PubMed] [Google Scholar]
- 5.Hwang DJ, Roberts IM, Wilson TM. Expression of tobacco mosaic virus coat protein and assembly of pseudovirus particles in Escherichia coli. Proc Natl Acad Sci USA. 1994;91:9067–9071. doi: 10.1073/pnas.91.19.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belyi VA, Muthukumar M. Electrostatic origin of the genome packing in viruses. Proc Natl Acad Sci USA. 2006;103:17174–17178. doi: 10.1073/pnas.0608311103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krol MA, Olson NH, Tate J, Johnson JE, Baker TS, Ahlquist P. RNA-Controlled Polymorphism in the in vivo Assembly of 180-Subunit and 120-Subunit Virions from a Single Capsid Protein. Proc Natl Acad Sci USA. 1999;96:13650–13655. doi: 10.1073/pnas.96.24.13650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun J, DuFort C, Daniel MC, Murali A, Chen C, Gopinath K, Stein B, De M, Rotello VM, Holzenburg A, Kao CC, Dragnea B. Core-controlled polymorphism in virus-like particles. Proc Natl Acad Sci USA. 2007;104:1354–1359. doi: 10.1073/pnas.0610542104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belyi VA, Muthukumar M. Electrostatic origin of the genome packing in viruses. Proc Natl Acad Sci USA. 2006;103:17174–17178. doi: 10.1073/pnas.0608311103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douglas T, Young M. Host-guest encapsulation of materials by assembled virus protein cages. Nature (London) 1998;393:152–155. [Google Scholar]
- 11.Allen M, Willits D, Mosolf J, Young M, Douglas T. Protein cage constrained synthesis of ferrimagnetic iron oxide nanoparticles. Adv Mater. 2002;14:1562. [Google Scholar]
- 12.Bancroft JB, Hills GJ, Markham R. The self-assembly process in a small spherical virus. Formation of organized structures from protein subunits in vitro. Virology. 1967;31:354–79. doi: 10.1016/0042-6822(67)90180-8. [DOI] [PubMed] [Google Scholar]
- 13.Zhao X, Fox JM, Olson NH, Baker TS, Young MJ. In Vitro Assembly of Cowpea Chlorotic Mottle Virus from Coat Protein Expressed in Escherichia coli and in Vitro-Transcribed Viral cDNA. Virology. 1995;207:486–494. doi: 10.1006/viro.1995.1108. [DOI] [PubMed] [Google Scholar]
- 14.Bancroft JB, Hiebert E. Formation of an infectious nucleoprotein from protein and nucleic acid isolated from a small spherical virus. Virology. 1967;32:354. doi: 10.1016/0042-6822(67)90284-x. [DOI] [PubMed] [Google Scholar]
- 15.Speir JA, Munshi S, Wang G, Baker TS, Johnson JE. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure. 1995;3:63–78. doi: 10.1016/s0969-2126(01)00135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang J, et al. The role of subunit hinges and molecular “switches” in the control of viral capsid polymorphism. J Struct Biol. 2006;154:59–67. doi: 10.1016/j.jsb.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 17.Loo L, Guenther RH, Basnayake VR, Lommel SA, Franzen S. Controlled encapsidation of gold nanoparticles by a viral protein shell. J Am Chem Soc. 2006;128:4502–4503. doi: 10.1021/ja057332u. [DOI] [PubMed] [Google Scholar]
- 18.Goicochea NL, De M, Rotello VM, Mukhopadhyay S, Dragnea B. Core-like particles of an enveloped animal virus can self-assemble efficiently on artificial templates. Nano Lett. 2007;7:2281–2290. doi: 10.1021/nl070860e. [DOI] [PubMed] [Google Scholar]
- 19.Hu YF, Zandi R, Anavitarte A, Knobler CM, Gelbart WM. Packaging of a polymer by a viral capsid: The interplay between polymer length and capsid size. Biophys J. 2008;94:1428–1436. doi: 10.1529/biophysj.107.117473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang CB, Knobler CM, Gelbart WM, Mason TG. Curvature dependence of viral protein structures on encapsidated nanoemulsion droplets. ACS Nano. 2008;2:281–286. doi: 10.1021/nn700385z. [DOI] [PubMed] [Google Scholar]
- 21.Chen C, Daniel MC, Quinkert ZT, De M, Stein B, Bowman VD, Chipman PR, Rotello VM, Kao CC, Dragnea B. Nanoparticle-templated assembly of viral protein cages. Nano Lett. 2006;6:611–615. doi: 10.1021/nl0600878. [DOI] [PubMed] [Google Scholar]
- 22.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 23.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera - A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 24.Wikoff WR, Tsai CJ, Wang G, Baker TS, Johnson JE. The structure of cucumber mosaic virus: cryoelectron microscopy, X-ray crystallography, and sequence analysis. Virology. 1997;232:91–97. doi: 10.1006/viro.1997.8543. [DOI] [PubMed] [Google Scholar]
- 25.Willits D, et al. Effects of the Cowpea chlorotic mottle bromovirus [beta]-hexamer structure on virion assembly. Virology. 2003;306:280–288. doi: 10.1016/s0042-6822(02)00054-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michel JP, Ivanovska IL, Gibbons MM, Klug WS, Knobler CM, Wuite GJL, Schmidt CF. Nanoindentation studies of full and empty viral capsids and the effects of capsid protein mutations on elasticity and strength. Proc Natl Acad Sci USA. 2006;103:6184–6189. doi: 10.1073/pnas.0601744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evilevitch A, Lavelle L, Knobler CM, Raspaud E, Gelbart WM. Osmotic pressure inhibition of DNA ejection from phage. Proc Natl Acad Sci USA. 2003;100:9292–9295. doi: 10.1073/pnas.1233721100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Durham ACH, Witz J, Bancroft JB. The semipermeability of simple spherical virus capsids. Virology. 1984;133:1–8. doi: 10.1016/0042-6822(84)90419-7. [DOI] [PubMed] [Google Scholar]
- 29.Jiang W, KimBetty YS, Rutka JT, ChanWarren CW. Nanoparticle-mediated cellular response is size-dependent. Nat Nanotechnol. 2008;3:145–150. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- 30.Georgens C, Weyermann J, Zimmer A. Recombinant virus like particles as drug delivery system. Curr Pharm Biotechnol. 2005;6:49–55. doi: 10.2174/1389201053167202. [DOI] [PubMed] [Google Scholar]


