Abstract
GBV-B induces hepatitis in tamarins and marmosets and is a surrogate model for HCV infections. Here, we cloned and characterized the antiviral activity of tamarin and marmoset interferon (IFN)α and IFNγ. Potent antiviral activity was observed for tamarin and marmoset IFNα in primary hepatocyte cultures infected with GBV-B. The antiviral activity was greater in cultures exposed to IFNα prior to GBV-B infection, suggesting that either GBV-B was capable of inhibition of the antiviral activity of exogenous IFNα or that the preexisting endogenous IFN response to the virus reduced efficacy to exogenous IFNα. IFNγ also exhibited antiviral activity in GBV-B infected hepatocytes. The transcriptional response to IFNα in marmoset hepatocytes was characterized using human genome microarrays. Since the GBV-B hepatocyte culture model possesses a functional innate immune response, it will provide opportunities to explore the nature of the antiviral response to a virus closely related to HCV.
Keywords: HCV, hepatitis C virus, primate, microarray, innate immunity, ISG, liver
Introduction
Worldwide, approximately 170 million people are chronically infected with hepatitis C virus (HCV), which frequently progresses to serious liver disease. The current approved therapy involves the combination of pegylated (peg)-IFNα and ribavirin and has response rates for sustained viral clearance of 42% and 82% for genotypes 1 and 2/3, respectively (Manns et al., 2001; Fried et al., 2002). However, a significant proportion of the population still develops serious disease as a consequence of HCV infection. HCV infection is the leading cause for liver transplantation in the US (Thomas and Seeff LB, 2005; Alter HJ and Seeff LB, 2005), and liver cancer due to HCV infection is one of the most rapidly increasing types of cancer in the US (Kim et al., 2005). Little is understood regarding the factors leading to successful or failed viral clearance during IFNα therapy. The only animal model of HCV infection is the chimpanzee. Studies in chimpanzees using pegylated IFNα, traditional IFNα therapy with and without ribavirin, and adenovirus-based IFNα gene therapy, all failed to induce a reduction in viral load despite high levels of circulating IFNα for extended times (Demers et al., 2002; Lanford et al., 2000) (Lanford et al., 2006). Total genome microarray analysis in HCV chronically infected chimpanzees revealed high pretreatment IFN stimulated gene (ISG) expression in the liver and no further induction of ISG transcripts in the liver following IFNα dosing, while the response in PBMC was similar to that in uninfected animals. The IFNα response in HCV chronically infected chimpanzees may be mechanistically similar to the null response in the human population (Lanford et al., 2007). Thus, chimpanzees are not well suited for studies on the mechanism IFNα induced viral clearance in HCV infected individuals and additional animal models are needed.
GBV-B is a hepatotropic virus that causes hepatitis in tamarins and is the virus phylogenetically most closely related to HCV (Muerhoff et al., 1995; Robertson et al., 1998; Ohba et al., 1996), and as such, GBV-B represents an important small primate surrogate model for HCV infections. The history of the GB agent originates with the inoculation of tamarins with serum obtained from a surgeon with hepatitis (for review see Beames et al., 2001); however, there is little doubt that GBV-B is a tamarin virus (Lanford et al., 2003a; Bukh et al., 2001; Weatherford et al., 2009).
The GBV-B model overcomes a number of limitations encountered working with HCV (Beames et al., 2001; Beames et al., 2001; Lanford and Bigger, 2002). In tamarins, GBV-B replicates to levels several logs higher than what is observed in HCV-infected humans and chimpanzees. GBV-B replication in marmosets is typically lower than that observed in tamarins, and reproducible infection profiles in the marmoset requires adaptation to the new host (Weatherford et al., 2009)(Bright et al., 2004). Although robust replication of HCV in vitro is now possible using derivatives of the Huh7 cell line (Blight et al., 2002) and a limited number of viral isolates (Lindenbach et al., 2005; Zhong et al., 2005; Cai et al., 2005; Yi et al., 2006; Wakita et al., 2005), the use of a primary hepatocyte culture system for GBV-B (Beames et al., 2000) may be more suitable for some studies, especially those involving certain aspects of virus-host interactions.
The utility of the GBV-B model as a surrogate for HCV has been demonstrated in a number of studies. HCV and GBV-B polyproteins possess approximately 25-30% homology at the amino acid level (Muerhoff et al., 1995). The GBV-B NS3 protease correctly processes the HCV polyprotein (Scarselli et al., 1997), HCV/GBV-B chimeric NS3 proteins are enzymatically active (Butkiewicz et al., 2000), and HCV NS3 protease inhibitors are active in GBV-B infected marmosets (Bright et al., 2004). Infectious cDNA clones of GBV-B have been produced that induced hepatitis upon intrahepatic inoculation of tamarins and marmosets with in vitro transcribed RNA (Bukh et al., 1999; Sbardellati et al., 2001; Martin et al., 2003). Further modification of the infectious clones was required to render them infectious in marmosets (Weatherford et al., 2009).
In this study, marmoset and tamarin IFNα and IFNγ were cloned, expressed and examined for antiviral activity in the GBV-B primary hepatocyte culture model, as a first step in developing a small primate model for analysis of the mechanism of viral clearance by IFN in a system relevant to HCV. Induction of host gene expression by IFNα was monitored using human total genome microarrays, demonstrating the utility of human long-oligo arrays in a New World primate.
Results
Cloning and expression of tamarin and marmoset IFN genes
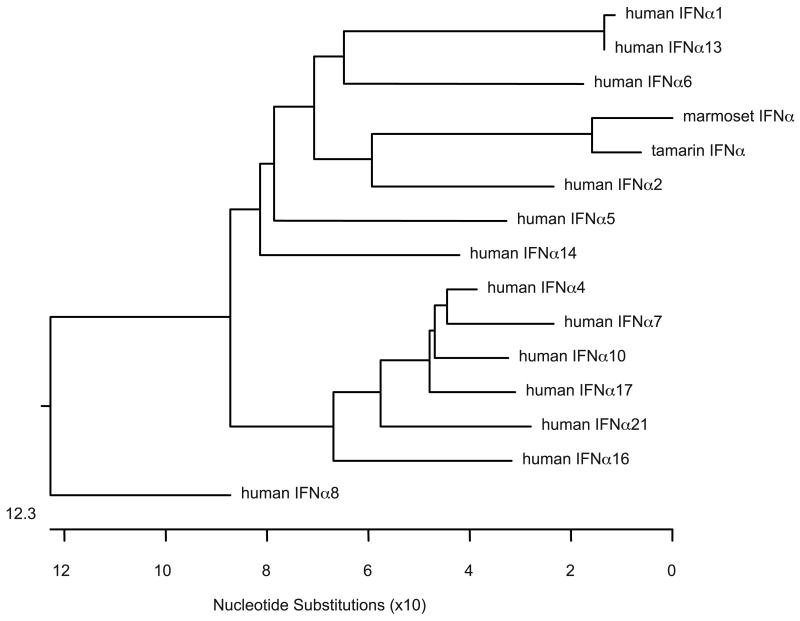
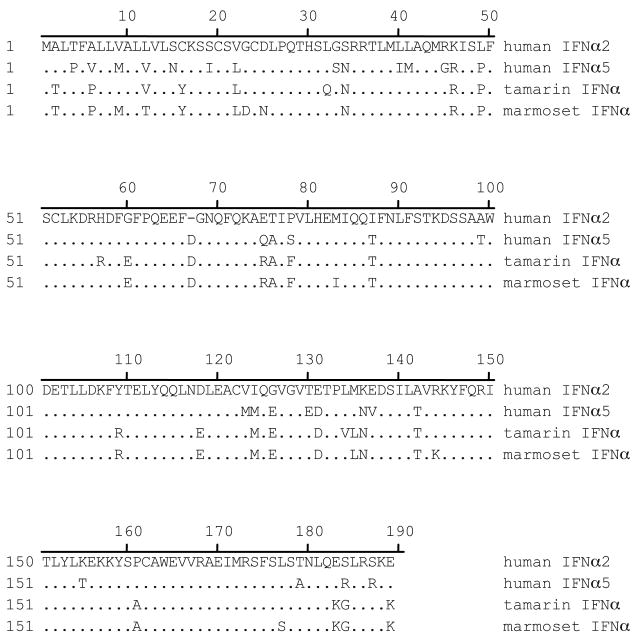
IFNα genes were cloned from tamarin liver, primary tamarin hepatocytes, marmoset liver and marmoset spleen. The same IFNα type was cloned regardless of the tissue source of the RNA. Phylogenetic analysis with all 13 human IFNα genes revealed that the tamarin and marmoset IFNα genes were most closely related to human IFNα2 (Figure 1) and will be referred to as IFNα2 hereafter. Alignment of the tamarin and marmoset IFNα2 amino acid sequences with human IFNα2 and IFNα5 revealed that the marmoset IFNα2 gene differed from the tamarin at 10 residues. Human IFNα2 and IFNα5 differ from each other by 32 residues and differ from marmoset IFNα2 by 32 and 33 residues, respectively. Analysis of sequence pair distances by Clustal using PAM250 residue weights indicated that human IFNα2 and IFNα5 have 84.1 percent identity, while human IFNα2 had 83.1 and 81.5 percent identity to tamarin and marmoset IFNα2.
Figure 1.
Phylogenetic analysis of IFNα genes. A phylogenetic tree comparing the thirteen human IFNα genes with the marmoset and tamarin IFNα genes was compiled using software programs in LaserGene. Marmoset and tamarin IFNα cDNAs cloned in this study were most closely related to human IFNα2. Accession numbers for the IFNα genes are: IFNα1 (J00210), IFNα2 (V00549), IFNα4 (X02955), IFNα5 (X02956), IFNα6 (X02958), IFNα7 (M34913), IFNα8 (K01900), IFNα10 (V00551), IFNα13 (X75934), IFNα14 (NM_002172), IFNα16 M28585, IFNα17 (V00532) and IFNα21 (NM_002175). The tamarin IFNα2 (FJ598590) and marmoset IFNα2 (FJ598591) have been deposited with GenBank.
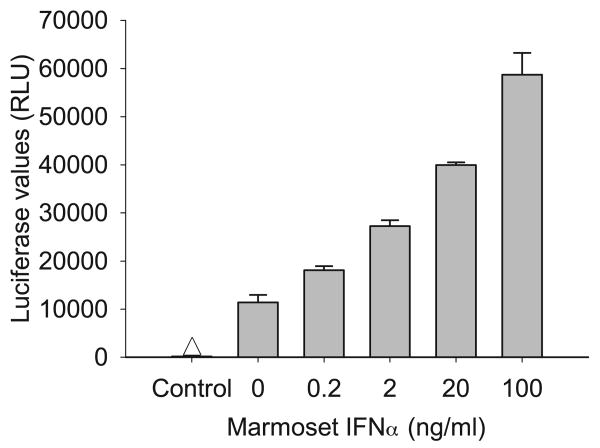
Marmoset and tamarin IFNα2 were expressed in the human liver cell line Huh7, secreted into the serum free medium, and concentrated by ultrafiltration. The level of production was estimated using an ELISA for human IFNα that employed a polyclonal antibody that cross-reacted with both marmoset and tamarin IFNα. Tamarin and marmoset IFNα2 production in Huh7 cells was estimated at 2000 and 700 ng/ml, respectively. Since the ELISA was calibrated with human IFNα2, the values for marmoset and tamarin IFNα2 are approximations. The specific activity was determined in a bioassay using inhibition of EMCV cytopathic effect in human A549 cells. The specific activity of human IFNα2b in the assay was 1.25 units (U)/pg, which is comparable to the 2-5 U/pg normally attributed to purified human IFNα2. Tamarin and marmoset IFNα2 exhibited high activity in human cells with approximately 1.6 U/pg and 0.25 U/pg, respectively (Table 1). In contrast, inhibition of EMCV cytopathic effect on primary hepatocytes required high levels of IFNα2. Human IFNα2 had 30,000-fold decrease in activity on tamarin hepatocytes, while tamarin and marmoset IFNα2 decreased by only 500- and 300-fold, respectively. EMCV cytopathic effect in marmoset hepatocytes could not be effectively inhibited even with marmoset IFNα2. As an alternative to the EMCV assay, the biological activity of marmoset IFNα was tested on marmoset hepatocytes for stimulation of a promoter containing multiple copies of an ISRE (interferon stimulated response element). Primary marmoset hepatocytes were transfected with an ISRE-luciferase reporter construct and treated with various concentrations of marmoset IFNα for 24 hr. Luciferase activity increased by 59% at 0.2 ng/ml IFNα2 (equivalent to 50 u/ml based on activity in A549 cells; Table 1) and by 515% at 100 ng/ml (Figure 3).
Table 1.
Specific activity of IFNα2 on different cell typesa
| IFNα2 | Human A549 |
Tarmarin hepatocyte |
Marmoset hepatocyte |
|---|---|---|---|
| Human | 1.25 | 0.00004 | 0.000001 |
| Tamarin | 1.60 | 0.003 | 0.000004 |
| Marmoset | 0.25 | 0.0008 | 0.000008 |
Values expressed as U/pg based on inhibition of EMCV.
Figure 3.
Transcriptional activation of ISRE promoter by marmoset IFNα2. Primary marmoset hepatocytes were transfected with a pISRE-Luc (Stratagene) which contains five copies of the IFN-stimulated response element (ISRE) derived from the ISG54 gene upstream of a basic promoter element (TATA box) and the firefly luciferase gene. Cells were treated with various concentrations of marmoset IFNα2 and harvested 24 hr later for luciferase assay. Error bars indicate the variation in values from duplicate cultures. The level of variation was not sufficient to yield visible error bars on those were none appear.
Antiviral activity of IFNα in GBV-B infected cultures
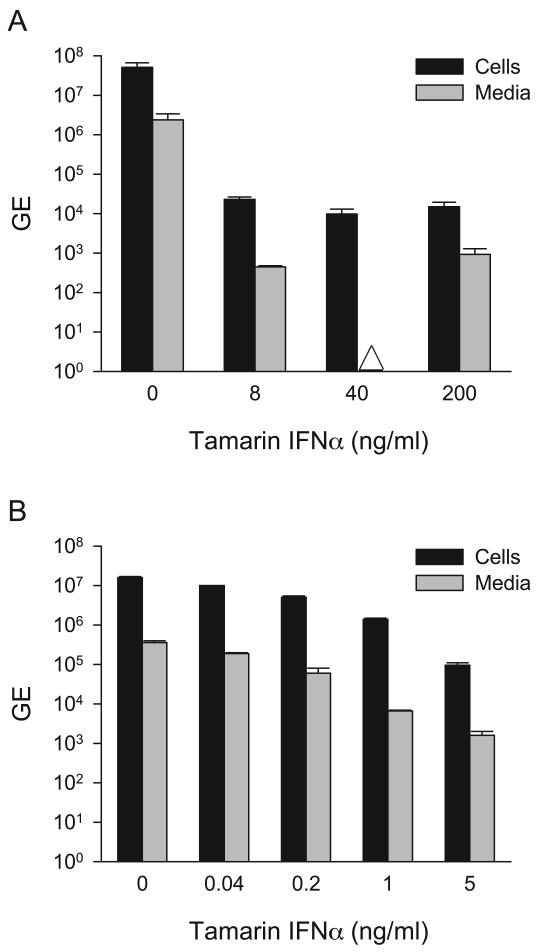
To examine the antiviral activity of tamarin IFNα2 in the GBV-B model, cultures of primary tamarin hepatocytes were treated with five-fold dilutions of IFNα (8, 40 and 200 ng/ml) from 24 hr prior to infection through 72 hr post-infection. At 8 ng/ml of IFNα2, GBV-B cell-associated RNA was suppressed by over 2000-fold, while secreted virus was reduced by 5000-fold (Figure 4A). No further decrease in viral RNA was observed with higher levels of IFNα, suggesting that the residual values for cell-associated RNA probably reflect non-replicating virus adhering to the cultures from the inoculum. Secreted virus was near the limits of detection in all IFNα2 treated cultures and was undetectable at 40 ng/ml of IFNα2 but again detectable at 200 ng/ml of IFNα2. A second experiment was conducted to extend the lower IFNα range using 0.040 to 5.0 ng/ml. No decrease in viral RNA was detected at 0.04 ng/ml of IFNα2, while viral RNA was decreased by 3.1- and 6.0-fold in cells and media at 0.2 ng/ml of IFNα2, and over 2-log reductions of viral RNA were observed in both cells and media at 5.0 ng/ml (Figure 4B). The antiviral activity of marmoset and tamarin IFNα2 were examined in marmoset hepatocytes as well. At 4 ng/ml of marmoset IFNα, cell-associated and secreted GBV-B RNA were suppressed by 4.5-fold and 5.0-fold, respectively, while at 100 ng/ml IFNα2, the cell-associated RNA decreased 357-fold and the secreted virus was undetectable (Figure 5). These data are in agreement with the data from EMCV in which higher levels of IFNα were required to observe inhibition on marmoset hepatocytes. Tamarin IFNα exhibited similar GBV-B antiviral activity as marmoset IFNα on marmoset cells, with 238-fold and 1136-fold suppression of cell and media viral RNA at 200 ng/ml, respectively.
Figure 4.
Titration of antiviral activity of tamarin IFNα2 on tamarin hepatocytes infected with GBV-B. 4A. Tamarin primary hepatocytes were treated with various concentration of tamarin IFNα2 (0 to 200 ng/ml) for 24 hr prior to infection with GBV-B and for an additional 72 hr after infection. Total RNA was purified from cells and media and analyzed for GBV-B RNA genome equivalents (GE) by quantitative RT-PCR. 4B. Tamarin primary hepatocytes were treated with 0 to 5 ng/ml of tamarin IFNα2 and infected with GBV-B as in 4A. Values are expressed as GE per culture based on 6 well dishes. Black bars = GBV-B GE in cell-associated viral RNA, grey bars = GE of GBV-B secreted into media. The Δ symbol denotes that the sample was below the level of detection. Error bars indicate the variation in values from duplicate cultures. The level of variation was not sufficient to yield visible error bars on those were none appear. Comparison of the values for cell associated viral RNA in the treated and untreated cultures in 4A and 4B using a Student's t-Test yielded p values of p= 0.0002 and 0.002, respectively. Comparison of the values for the media associated viral RNA in the treated and untreated cultures in 4A and 4B using a Student's t-Test yielded p values of p=0.002 and p=0.0007.
Figure 5.
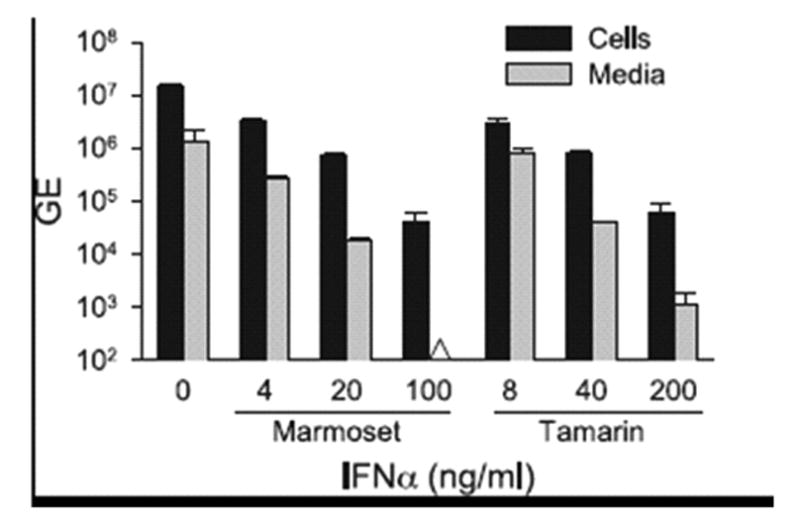
Comparison of antiviral activity of marmoset and tamarin IFNα2 on marmoset hepatocytes infected with GBV-B. Primary marmoset hepatocytes were treated with either marmoset or tamarin IFNα2 at concentrations of 0 to 200 ng/ml for 24 hr prior to infection with GBV-B and for another 72 hr post-infection. Cell and media were analyzed for GBV-B RNA as described for Figure 4. Black bars = GBV-B RNA GE in cell-associated viral RNA, grey bars = GE of GBV-B secreted into media. The Δ symbol denotes that the sample was below the level of detection. Error bars indicate the variation in values from duplicate cultures. The level of variation was not sufficient to yield visible error bars on those were none appear.
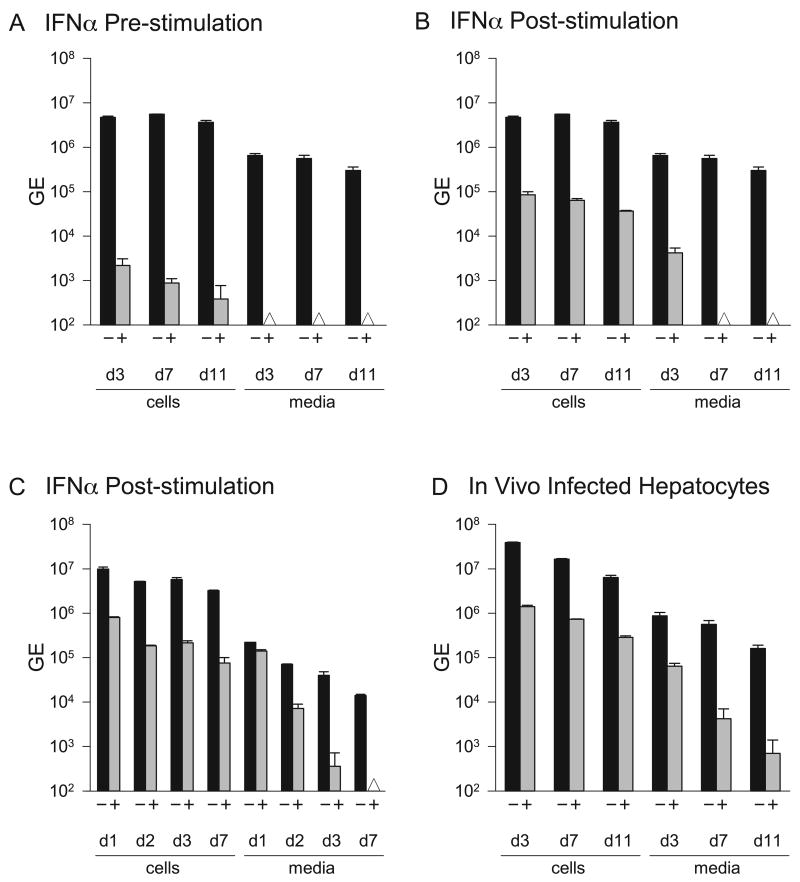
Tamarin hepatocytes were examined in a time course experiment by pre-treating cells at 200 ng/ml for 24 hr prior to infection and then extending treatment for 3, 7 and 11 days post-infection. Cell-associated GBV-B RNA progressively declined over 11 days from 1533-fold at day 3 to 8632-fold at day 11 (Figure 6A). Again, the remaining cell-associated viral RNA probably represents residual inoculum adhering to the cultures, rather than replication, since secreted viral RNA was undetectable at all time points. No apoptosis or cytopathic effect was noted in the cultures exposed to high levels of IFNα for 11 days and for greater times in other studies. Since primary hepatocytes are non-dividing cultures, it was not possible to examine the effects of IFNα on proliferation. To examine the antiviral efficacy of IFNα2 in an established infection, replicate cultures were infected with GBV-B for 3 days and then treated with IFNα2. A marked reduction in the efficacy of the IFNα treatment was noted with only 51- to 97-fold decline in the cell-associated RNA after 3 and 11 days of treatment, respectively (Figure 6B). However, a more pronounced antiviral effect was observed for secretion of viral RNA with a 156-fold decline on day 3 of treatment and no detectable virus secretion on days 7 and 11. Although it was more effective to inhibit GBV-B using pre-infection treatment with greater than 3 logs of inhibition in secreted virus on day 3 (Figure 4A, 5, 6A), treatment of established infections still achieved more than a 2 log decrease in viral secretion by day 3. We repeated the experiment using 5 ng/ml of tamarin IFNα2 to determine if lower levels of IFNα2 could effectively inhibit an established infection. IFNα treatment was initiated 3 days post-infection and cultures were harvested after 1 to 7 days of treatment. Cell-associated viral RNA declined by more than 1 log (12-fold) after 24 hr of treatment with a 42-fold reduction by day 7 (Figure 6C). Secreted virus was reduced to below the levels of detection by day 7. Thus, a clear dose response was observed between experiments conducted with 5 and 200 ng/ml IFNα2 in cultures with an established infection.
Figure 6.
Time course of antiviral activity of tamarin IFNα2 in GBV-B infected tamarin hepatocytes. A. Pre-infection IFNα treatment. Tamarin hepatocytes were either untreated (-) or treated (+) for 24 hr with 200 ng/ml of tamarin IFNα2 prior to infection with GBV-B, and for an additional 3, 7 or 11 days prior to harvest. B. Post-infection IFNα treatment. Tamarin hepatocytes were infected with GBV-B for 3 days to establish the infection and then treated (+) with 200 ng/ml tamarin IFNα2 for 3, 7 or 11 days before harvest. C. Post-infection IFNα treatment. Tamarin hepatocytes were infected with GBV-B for 3 days prior to treatment (+) with 5 ng/ml of tamarin IFNα2 for 1, 2, 3 or 7 days before harvest. D. Post-infection IFNα treatment. Tamarin hepatocyte cultures were prepared from a tamarin during the acute phase of infection with GBV-B to obtain cells with an in vivo established infection. Three days after plating, the hepatocytes were treated with 200 ng/ml of tamarin IFNα2 for 3, 7 or 11 days before harvest. Black bars = GE of cell-associated GBV-B RNA, grey bars = GE of GBV-B secreted into media. The Δ symbol denotes that the sample was below the level of detection. Error bars indicate the variation in values from duplicate cultures. The level of variation was not sufficient to yield visible error bars on those were none appear.
We also examined cultures derived from an infected animal to compare the antiviral activity of IFNα in cultures with infections established in vivo. Surprisingly, the reduction in cell-associated RNA was minimal over the 11 days of treatment at 200 ng/ml IFNα with a 14-fold reduction on day 3 in comparison to untreated cells and no substantial increase in the reduction over 11 days (Figure 6D). Day 11 data were difficult to interpret, since the levels of virus in untreated cultures declined over time in this set of cultures, presumably due to the less robust nature of primary cultures obtained from an animal during acute viral infection. The inhibition of viral secretion was of lesser magnitude than that observed with in vitro infected cultures as well, with a 14-fold reduction on day 3 of treatment (in comparison to 3 logs with pre-infection treatment on day 3).
Antiviral Activity of IFNγ in GBV-B infected cultures
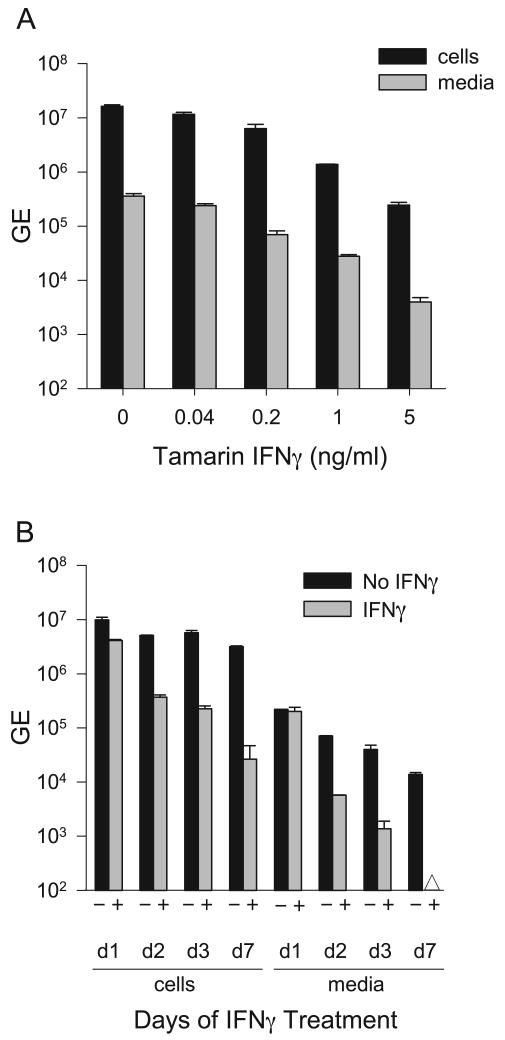
The tamarin and marmoset IFNγ genes were amplified and cloned from PBMC stimulated with phorbol myristate acetate (PMA) and ionamycin. Marmoset and tamarin IFNγ differed by only 3 amino acids (Figure 7). Human IFNγ differed from marmoset and tamarin IFNγ by 18 and 16 residues, respectively, and thus the New World monkey IFNγ gene had diverged less than IFNα2 in comparison to human. IFNγ was produced in Huh7 cells using the same procedure as described for IFNα. ELISA values indicated that marmoset and tamarin IFNγ were secreted in transfected Huh7 cells at 200 and 80 ng/ml, respectively. In the EMCV inhibition assay using A549 cells, tamarin and marmoset IFNγ exhibited specific activities of 1 U/pg and 0.5 U/pg, respectively. Tamarin IFNγ was compared with commercially obtained human IFNγ (R&D Systems; produced in bacteria) in GBV-B infected hepatocytes. Cultures were pretreated with 0.04 to 5.0 ng/ml and harvested 3 days post infection. Inhibition of GBV-B was first detected for tamarin IFNγ at 0.2 ng/ml with 2.6-and 5.1-fold reduction in the cell and media viral RNA, respectively. These values are remarkably close to the inhibition with 0.2 ng/ml of tamarin IFNα2 (Fig. 4B). A dose response was observed with higher concentrations of IFNγ with 5.0 ng/ml reducing GBV-B RNA by 67-fold and 90-fold in the cells and media, respectively (Figure 8A). Human IFNγ exhibited no antiviral effect in tamarin hepatocytes at the concentrations tested (data not shown). The specific activity of the human IFNγ (0.002 U/pg) was much lower that the tamarin IFNγ when calibrated in the same ELISA and bioassay, and this alone could explain the lack of activity in the GBV-B assay even if no species difference in activity were present.
Figure 7.
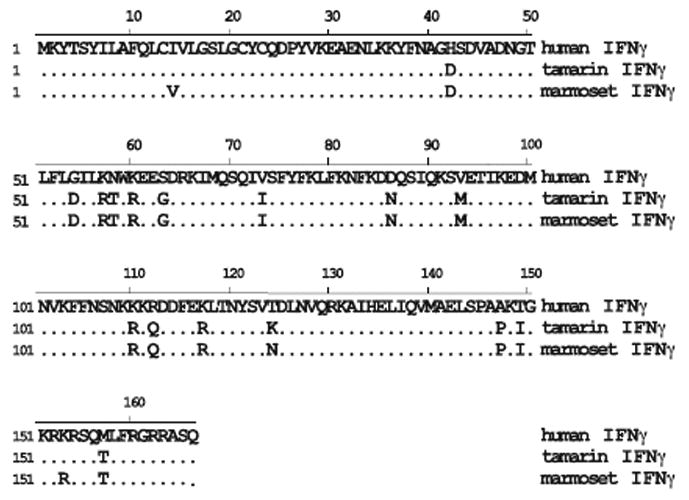
Alignment of amino acid sequence of marmoset, tamarin and human IFNγ genes. The human IFNγ sequence is used as the consensus sequence with identity to this sequence indicated by a dot. Tamarin and marmoset IFNγ genes differ by 3 residues, while they differ from the human sequence at 16 and 18 residues, respectively. The full length proteins are 166 amino acids with the first 23 residues representing the cleaved signal sequence.
Figure 8.
Antiviral activity of tamarin IFNγ in tamarin hepatocytes infected with GBV-B. A. Pre-infection IFNγ treatment. Tamarin primary hepatocytes were treated with various concentration of tamarin IFNγ (0 to 5.0 ng/ml) for 24 hr prior to infection with GBV-B and for an additional 72 hr after infection. Total RNA was purified from cells and media and analyzed for GBV-B RNA genome equivalents (GE) by quantitative RT-PCR. B. Post-infection IFNγ treatment. Tamarin hepatocytes were infected with GBV-B for 3 days to establish the infection and then treated with tamarin IFNγ (+) at 5.0 ng/ml for 1, 2, 3 or 7 days before harvest. The Δ symbol denotes that the sample was below the level of detection. Error bars indicate the variation in values from duplicate cultures. The level of variation was not sufficient to yield visible error bars on those were none appear.
A time course experiment was performed in cultures with established infections. Cultures were treated at 5.0 ng/ml for 1 to 7 days beginning 3 days after infection. At day 3 of treatment, the fold reduction in cells and medium was only 3-fold lower than that observed in pre-treated cultures. A maximum reduction in viral RNA occurred at 7 days of treatment, with 120-fold inhibition of cell-associated RNA and undetectable secreted virus. The data suggest that GBV-B may have a more limited ability to block the antiviral activity of exogenous IFNγ in cultures with established infections. The antiviral activity of marmoset IFNγ in marmoset hepatocytes was examined in a titration experiment with pre-treatment for 24 hr prior to infection. The values were similar to that observed with tamarin IFNγ with a 221-fold reduction in cell-associated RNA on day 3 at 5.0 ng/ml (data not shown).
Antiviral activity of marmoset and tamarin IFNα and IFNγ in HCV replicon cells
The inhibition of EMCV cytopathic effect in human A549 cells suggested that both tamarin and marmoset IFNs should have antiviral activity against the HCV replicon in Huh7 cells. We examined tamarin and marmoset IFNα in the replicon model by treating a cell line containing a genotype 1b replicon for 72 hrs. Tamarin IFNα2 (200 ng/ml) and marmoset IFNα2 (100 ng/ml) reduced replicon RNA levels by 114- and 96-fold, respectively. Human IFNα2 was used as a positive control at 1000 U/ml (0.8 ng/ml), a level we had previously observed to have potent antiviral activity in the replicon model (Lanford et al., 2003b). Human IFNα2 reduced replicon levels by 37-fold and was probably less active due to the concentration difference. The values for marmoset and tamarin IFNα and IFNγ were comparable to the inhibition of GBV-B in cultures with established infections treated at the same level of IFN. IFNα2 induction of transcripts for ISG12 and MX1 were monitored in the replicon cell line as well. Tamarin IFNα2 yielded the highest induction of ISG12 (596-fold) and MX1 (1012-fold), while human IFNα2 induced the transcripts by 276- and 112-fold.
Analysis of Interferon Stimulated Gene Expression in Marmoset Hepatocytes
One of the obstacles of working with GBV-B in tamarins and marmosets is the lack of reagents for New World primates. The marmoset genome has recently been sequenced, but at this time, it has not been annotated. The genetic divergence between humans and marmosets presents difficulties in the use of quantitative RT-PCR and microarrays for analysis of gene expression during GBV-B infections. We designed an experiment to test whether array platforms using the long-oligo approach may offer an opportunity to examine GBV-B infections with human arrays. We have previously utilized human microarrays to examine changes in liver gene expression in HCV acute and chronic infections in chimpanzees and during IFNα therapy (Bigger et al., 2001; Bigger et al., 2004; Lanford et al., 2006; Lanford et al., 2007). In one of these studies, primary human and chimpanzee hepatocytes were induced with IFNα2 and changes in gene expression were determined at 4 and 8 hr post treatment (Lanford et al., 2006). Thus, a database of human genes verified to be induced by IFNα2 in our primary hepatocyte culture system was available for comparison. The identical experiment was conducted in marmoset hepatocytes using marmoset IFNα2. Cultures were treated with or without 20 ng/ml of marmoset IFNα2 and were harvested at 4 hr post treatment. Total cell RNA was derived from 18 cultures with pools of 3 cultures each used to probe 6 arrays, 3 untreated and 3 IFNα treated. The ABI long-oligo arrays representative of the entire human genome were utilized. Our previous studies in human and chimpanzee hepatocytes were performed with Affymetrix microarrays which utilize shorter oligos and were found to be unsuitable for analysis of marmoset hepatocytes under standard hybridization conditions. In contrast, the ABI arrays scored a large number of transcripts as present and significantly induced by IFNα2.
Our previous studies in chimpanzee and human hepatocytes detected 384 ISGs (positively induced genes), while our studies in the chimpanzee detected 500 ISGs in the liver and over 1000 total ISGs detected in combined data from PBMC and liver (Lanford et al., 2006). Of the ISGs detected in chimpanzee hepatocytes, only 227 were detected at 4 hr post-treatment, the only valid time point for comparison to the marmoset study, and of these 83 (37%) were detected and induced in the marmoset hepatocytes. ISGs detected in both human/chimpanzee hepatocytes and marmoset hepatocytes represented many of the commonly recognized ISGs (Supplement Table 1), and 30 of the genes with the largest fold induction in the chimpanzee/human data set are shown in Table 2. An additional 21 ISGs detected in the marmoset had been detected in chimpanzees under conditions other than the 4 hr time point (either in hepatocytes at 8 hr post-IFN or in liver of IFNα treated chimpanzees) confirming their ISG designation. Thus, 104 ISGs were detected by the marmoset array. Reproducibly down-regulated transcripts were detected as well that were common to the marmoset and chimpanzee/human data sets (Supplement Table 1). There were 366 ISGs in the chimpanzee/human data set that were not detected in the marmoset hepatocytes (Supplement Table 2), but only 144 of these ISGs were detected at 4 hr in the in vitro chimpanzee/human hepatocytes experiments. The lack of detection of many ISGs with the ABI arrays is likely due to sequence divergence of the marmoset. Thirty of the most highly induced genes in chimpanzee and human hepatocytes not detected in marmosets are shown in Table 3.
Table 2.
Interferon Stimulated Genes Detected in Marmoset Hepatocytesa
| ABI Microarray | Affymetrix U133 Plus 2.0 Array | ||||
|---|---|---|---|---|---|
| Marmoset hepatocytes | Chimpanzee and human hepatocytes | Uninfected chimpanzee liver | |||
| Gene symbolb | 4 hr | 4 hr | 8 hr | 4 hr | 8 hr |
| CXCL11 (I-TAC) | 11 | 161 | 172 | 21 | 25 |
| RSAD2 (Viperin) | 117 | 130 | 116 | 13 | 15 |
| IFIT2 (ISG54) | 34 | 55 | 16 | 38 | 7 |
| CXCL10 (IP-10) | 18 | 50 | 46 | 24 | 6 |
| IFIT1 (ISG56) | 55 | 44 | 44 | 18 | 13 |
| GBP5 | 11 | 37 | 37 | 6 | |
| IFIT3 (RIG-G) | 112 | 30 | 21 | 26 | 9 |
| PML | 3 | 26 | 10 | ||
| SP110 | 4 | 23 | 10 | 4 | |
| HK2 | 2 | 22 | 15 | ||
| LOC129607 | 11 | 20 | 18 | 28 | 17 |
| DDX58 (RIG-I) | 28 | 19 | 13 | 14 | 10 |
| IFIH1 (MDA5) | 2 | 14 | 13 | 13 | 8 |
| HERC6 | 10 | 13 | 16 | 30 | 15 |
| IL28RA | 2 | 13 | 3 | ||
| OASL | 132 | 13 | 14 | 9 | 4 |
| ETV7 | 5 | 13 | 12 | ||
| ZC3HAV1 | 2 | 12 | 5 | 4 | |
| MX1 | 26 | 12 | 14 | 19 | 18 |
| ISG15 | 19 | 12 | 14 | 20 | 23 |
| HERC5 | 6 | 11 | 15 | 12 | 9 |
| TNFSF10 (TRAIL) | 3 | 10 | 5 | 6 | |
| SECTM1 | 3 | 10 | 5 | 5 | 4 |
| SAMD9L | 11 | 8 | 10 | 5 | 5 |
| DHX58 (LGP2) | 11 | 8 | 8 | 6 | 7 |
| EPSTI1 | 68 | 8 | 12 | 7 | 7 |
| IL1RN | 5 | 7 | 4 | 6 | 3 |
| NT5C3 | 4 | 7 | 5 | 7 | 3 |
| OAS1 | 30 | 7 | 10 | 12 | 8 |
| OAS2 | 17 | 7 | 9 | 11 | 11 |
Comparison of microarray data from marmoset hepatocytes, chimpanzee and human hepatocytes, and chimpanzee liver following treatment with IFNα2. Data shown are fold-induction of genes in comparison to untreated cultures or chimpanzees.
Common gene names are shown in parenthesis.
Table 3.
Interferon Stimulated Genes Not Detected in Marmoset Hepatocytesa
| ABI Microarray | Affymetrix U133 Plus 2.0 Array | ||||
|---|---|---|---|---|---|
| Marmoset hepatocytes | Chimpanzee and human hepatocytes | Uninfected chimpanzee liver | |||
| Gene symbol | 4 hr | 4 hr | 8 hr | 4 hr | 8 hr |
| IFI44L | NDb | 112 | 175 | 24 | 31 |
| SOCS1 | ND | 28 | |||
| ANGPTL1 | ND | 21 | 34 | ||
| CCL8 | ND | 20 | 15 | 16 | 7 |
| LAMP3 | ND | 19 | 33 | ||
| IFIT3 | ND | 18 | 14 | 26 | 13 |
| TREX1 | ND | 17 | 15 | 7 | 7 |
| LINCR | ND | 17 | |||
| CX3CL1 | ND | 16 | |||
| --- | ND | 16 | 36 | ||
| MAB21L2 | ND | 15 | 5 | ||
| IFI16 | ND | 15 | 18 | 8 | 5 |
| SLFN5 | ND | 14 | 16 | ||
| USP18 | ND | 13 | 14 | 10 | 4 |
| ISG20 | ND | 13 | 21 | 28 | 45 |
| RNF24 | ND | 13 | |||
| BATF2 | ND | 12 | 6 | 13 | |
| HES4 | ND | 12 | 11 | ||
| FAM26F | ND | 11 | 5 | 3 | |
| SAMD9 | ND | 10 | 10 | 10 | 5 |
| NOD2 | ND | 9 | |||
| USP42 | ND | 9 | |||
| SDS | ND | 9 | 6 | ||
| CYP1B1 | ND | 9 | |||
| PRIC285 | ND | 9 | 8 | 9 | 5 |
| CXCL9 | ND | 8 | 6 | ||
| SLFN5 | ND | 8 | 6 | 4 | |
| GBP4 | ND | 8 | 5 | 5 | 3 |
| IFI44 | ND | 8 | 12 | 8 | 9 |
| GBP1 | ND | 8 | 7 | 8 | 6 |
Comparison of microarray data from marmoset hepatocytes, chimpanzee and human hepatocytes, and chimpanzee liver following treatment with IFNα2. Data shown are fold induction of genes in comparison to untreated cultures or chimpanzees.
ND, No change greater than two-fold detected.
There were 1660 genes detected using the ABI arrays that were not detected in our previous studies with Affymetrix arrays in chimpanzees (Supplement Table 3), 675 of these have no gene symbol and are denoted as nulls. Genes that were only detected using the ABI array with marmoset hepatocytes were for the most part not known ISGs, and their presence may reflect a less stringent selection criteria and an under-powering of the experiment using 3 treated and 3 untreated arrays. Since many of these transcripts are not represented on the Affymetrix array, the reason for their detection with the ABI array using marmoset hepatocytes is uncertain. Resolution of this issue would require repetition of the chimpanzee and human in vitro experiments using the ABI arrays. The detection of a large set of ISGs in marmoset cells under conditions known to induce this set of genes suggests that human long-oligo arrays are suitable for pathway analysis in GBV-B infections, even though many genes will be missed.
Discussion
The GBV-B model provides a powerful surrogate system for HCV. GBV-B is the most closely related virus to HCV phylogenetically, and it displays a liver tropism similar to HCV. The use of small non-human primates as the animal model for GBV-B, in contrast to chimpanzees for HCV, has distinct advantages. However, the fact that tamarins and marmosets are New World monkeys presents challenges as well, since they have not been as extensively characterized as models for infectious disease, and immunological and molecular reagents are limited. This deficiency is being overcome rapidly due to an increased interest in the marmoset as a model for numerous biomedical studies, and the recent sequencing of the marmoset genome. In this report, we have cloned and expressed IFNα and IFNγ from the tamarin and marmoset to advance studies on the mechanism of antiviral response to GBV-B as a surrogate for HCV, and to compare and contrast the effects of type I and II IFNs in these closely related models.
The IFNα genes cloned from several tissues from both marmosets and tamarins were most closely related to human IFNα2 showing 17 to 18% divergence from the human sequence. Surprisingly, the marmoset and tamarin clones differed by 5% at the amino acid level. Both marmoset and tamarin IFNα2 displayed a high level of cross-species functionality in human cells when tested in the EMCV inhibition assay and for the inhibition of the HCV replicon in Huh7 cells. High levels of IFNα2 were required to inhibit EMCV in primary hepatocytes from marmosets and tamarins, suggesting that hepatocytes may have a reduced sensitivity to IFNα. Inhibition of EMCV in marmoset hepatocytes required 1 million-fold more human IFNα2 in comparison to human cells. This was not simply a lack of cross-reactivity of human IFNα to marmoset cells, since over 30,000-fold more marmoset IFNα2 was required in marmoset cells in comparison to human cells. The A549 cells have been selected for their sensitivity to IFN in this assay, and this may account for much of the differences observed. The different sensitivities of the cell lines to IFNα may be a reflection of differences in the levels of various components of the Jak-STAT pathway. The enhancement of the IFNα response following priming with IFNγ demonstrates the impact of levels of Jak-STAT signaling proteins on the intensity of the IFNα response (Levy et al., 1990).
Actual estimation of the levels of IFNα and the specific activity were complicated by the lack of an immunological assay specific for marmoset and tamarin IFNα and appropriate standards.Although the dilution of each IFN possessing 1 unit of activity (inhibition of EMCV cytopathic effect on human A549 cells) is easily measured, the concentration of IFNα represented by 1 unit could only be estimated using an ELISA for human IFNα and human IFNα standards. The fact that tamarin IFNα had higher activity than human IFNα on human A549 cells could be due to an underestimation of the tamarin IFNα levels in the ELISA, or an increase in the specific activity of IFN produced and secreted in mammalian cell culture in comparison to protein synthesized in bacterial culture (human IFNα). Nonetheless, pretreatment of primary hepatocytes with tamarin and marmoset IFNα2 revealed potent antiviral activity with complete inhibition of GBV-B secretion and reduction of cell-associated viral RNA to levels approaching the background from viral particles from the inoculum adhering to the cultures. In contrast, the antiviral activity in cultures with established GBV-B infections was reduced.
The innate immune response of the host recognizes a variety of pathogen-associated molecular patterns. Viral RNA can interact with a number of cellular proteins to trigger the innate response through the toll-like receptors TLR3 and TLR7/8, and the intracellular sensing molecules RIGI and MDA5 (Yoneyama et al., 2004; Yoneyama et al., 2005; Foy et al., 2005) which lead to downstream activation of IRF3 and NF-κB and induction of Type I IFN synthesis. Most viruses have evolved mechanisms to block activation of the innate responses. The HCV NS3/4a protease blocks this pathway (Foy et al., 2005; Foy et al., 2003; Li et al., 2005a) via cleavage of the mitochondrial adapter protein (CARDIF, IPS-1, MAVS, VISA) (Xu et al., 2005; Seth et al., 2005; Kawai et al., 2005; Meylan et al., 2005). The GBV-B NS3 protein has been shown to similarly cleave the tamarin IPS protein (Chen et al., 2007). HCV inhibits the innate response by other mechanisms that are likely shared by GBV-B as well. However, further studies will be required to determine the mechanism by which GBV-B interferes with the antiviral activity of exogenously added IFNα.
Studies with GBV-B not only offer the advantage of a small primate model, but the GBV-B model also offers the advantage of an in vitro culture system using primary hepatocytes. Although the use of primary cultures is a disadvantage in routine virological studies, continuous cell lines are subject to many alterations that affect viral-host interactions. Recent studies with in vitro cultivation of HCV in Huh7.5 cells have provided new opportunities to examine aspects of HCV replication and viral-host interactions (Lindenbach et al., 2005; Zhong et al., 2005; Cai et al., 2005; Yi et al., 2006; Wakita et al., 2005), but this model is limited in its potential for use in some aspects of the interaction of the virus with the innate immunity. Our early studies in the replicon demonstrated that Huh7 cells were defective in the response to dsRNA (poly-IC) in comparison to primary hepatocytes and other hepatoma cell lines (Lanford et al., 2003b). Others have found Huh7 to be defective in innate responses, as well (Li et al., 2005b; Preiss, 2008) and more recent studies have suggested that Huh7.5 cells are even further defective in these pathways, thus perhaps partially explaining their uniquely high permissiveness to JFH1 infection and replication (Saito et al., 2007). Thus, improved in vitro models are needed for analysis of the myriad of viral-host interactions that appear to be regulating the innate response to HCV infection. Primary hepatocytes may also reveal viral-host interactions involved in replication and pathogenesis beyond the innate immunity that are not represented in the current established cell lines.
Although, the GBV-B model offers distinct advantages for analysis of some viral-host interactions, as mentioned above, the analysis of viral-host interactions in GBV-B is limited by the availability of reagents. In this study, we examined human microarrays for use in the marmoset model. Using long-oligo arrays, we were able to detect increases in many of the same genes detected in human and chimpanzees hepatocytes in response to IFNα. This will provide an opportunity to initiate studies on the host response to GBV-B both in vivo and in vitro, until the marmoset genome has been annotated and a marmoset specific array has been generated. It has only recently become possible to examine pre- and post-infection changes in gene expression with HCV using the JFH1-Huh7.5 culture system. However, for reasons still not appreciated, the JFH-1 virus replicates well in vitro but poorly in vivo in chimpanzees (Lindenbach et al., 2006), while GBV-B exhibits robust replication both in vivo and in vitro, allowing direct comparison to changes in gene expression under both conditions. Again, despite the current limitations of working in New World primates, the GBV-B model offers distinct advantages over HCV for certain types of studies.
The mechanism of the antiviral response to IFNα has been highly characterized in a number of model systems, yet the reasons for failed IFNα therapy in humans infected with HCV are still poorly understood. Recently, a correlation was observed between failed response to IFNα and increased pretreatment levels for some hepatic ISG transcripts (Chen et al., 2005). Studies in HCV chronically infected chimpanzees demonstrated that high pretreatment levels of liver transcripts for ISGs correlated with a lack of antiviral response to IFNα therapy and a lack of transcriptional response of the liver to IFNα, while PBMC in the same chronically infected animals responded to IFNα in a manner similar to uninfected animals (Lanford et al., 2006; Lanford et al., 2007). The recent use of paired biopsies in humans undergoing IFNα therapy demonstrated that individuals responding to IFNα therapy have low pretreatment levels of ISGs and undergo further activation of hepatic ISGs upon dosing with IFNα, while individuals that failed therapy were similar to chimpanzees and had high pretreatment levels of hepatic ISG transcripts and no further induction upon dosing with IFNα (Sarasin-Filipowicz et al., 2008). A number of factors have been shown to correlate with response to IFNα therapy including the HCV genotype, NS5A ISDR sequence, viral load, age, race, weight and fibrosis. Presently, the interaction of each of these factors with the antiviral response initiated by exogenous IFNα is incompletely understood. Although the marmoset small size lends itself well to studies on the antiviral response to IFNα, currently it is not possible to examine chronic infections in the GBV-B model. Persistent infections have been rarely observed and eventually resulted in viral clearance (Martin et al., 2003; Nam et al., 2004). Ongoing efforts to generate chronic infections in this model may overcome this limitation in the near future, whereas the lack of chronic infection in the GBV-B model may offer insight into the roles of the innate and adaptive immune response required for viral clearance and persistence.
Materials and Methods
Animals and Hepatocytes
Tamarins (Saguinus mystax) and marmosets (Callithrix jacchus) were housed at the Southwest National Primate Research Center at the Southwest Foundation for Biomedical Research. The animals were cared for in accordance with the Guide for the Care and Use of Laboratory Animals, and all protocols were approved by the Institutional Animal Care and Use Committee. Liver and spleen samples were obtained by either open biopsy or at the time of sacrifice. Total cell RNA was purified from tissues using RNAzol B (Leedo, Houston, TX, USA) following the manufacturers protocol and quantified by optical density. Primary hepatocytes were isolated by collagenase perfusion and were maintained in a hormonally defined, serum free medium (Lanford et al., 2003a) (Beames et al., 2000; Lanford and Estlack, 1998). Cells were grown in 6 well dishes for 3 days prior to infection. Inoculations were performed with 5 μl of GBV-B containing serum (approximately 2×105 to 4×106 GE) in 1 ml of serum free medium for 6 hr at 37 °C followed by two washes to remove residual inoculum. Culture medium was changed three times per week.
Interferon cloning and sequence analysis
Primers for cDNA cloning of IFNα were designed by aligning all human IFNα genes and the tamarin IFNα sequence (12). The forward primer included a KpnI site and the first 22 nucleotides of the IFNα open reading frame The reverse primer included an EcoRI site and the last 27 nucleotides of the anti-sense strand cDNAs from primary tamarin hepatocytes, tamarin liver, marmoset liver, and marmoset spleen were amplified using the Superscript One-Step RT-PCR for Long Templates kit (Invitrogen, Carlsbad, CA, USA) and cloned into the mammalian expression vector pcDNA4/TO (Invitrogen, Carlsbad, CA, USA). The IFNγ sequence was cloned from tamarin and marmoset PBMC using a similar design. PBMC from marmoset and tamarins were cultivated 24 hr in RPMI with 10% FBS and then treated for 24 hr with 10 ng/ml phorbol myristate acetate and 1 μg/ml ionamycin to induce IFNγ transcription. The primers were based on the sequence of the human IFNγ gene. The forward primer was 5′-GGGGTACCACAATGAAATATACAAGTTATATCTTGGC-3′ and the reverse primer was 5′-GGAATTCTTATTGGGACGCTCTTCGACCTCG-3′. GenBank accession numbers for tamarin and marmoset consensus sequences are: Tamarin IFNα2 = FJ598590, Marmoset IFNα2 = FJ598591, Tamarin IFNγ = FJ598592, Marmoset IFNγ = FJ598593.
Production and Assay of recombinant interferon
A single clone was selected for expression of marmoset and tamarin IFNα and IFNγ. Huh7 cells were transfected using Mirus II Trans-It transfection kit (Mirus Corp., Madison, WI, USA) following the manufacturer's protocol. Huh7 cells were tranfected in the same manner using the vector without an insert as a negative control. At 24 hours post transfection, the medium was replaced with the serum-free medium formulated for primary hepatocytes (Lanford et al., 2003a), and the culture medium was harvested at 48 hr post transfection. Secreted IFNα was concentrated by ultrafiltration (10,000 molecular weight cut-off; Millipore, Bedford, MA), and the final concentration was determined by ELISA (Human Interferon Alpha ELISA Kit; R&D Systems, Minneapolis, MN, USA). The specific activity of IFNα2 was determined using a bioassay for inhibition of EMCV induced cytopathic effect in the A549 human cell line. A549 cells growing in 96 well plates were pretreated for 24 hr with 3-fold dilutions of IFNα2, cells were infected with 104 TCID50 of EMCV per well, and monolayers were stained with crystal violet at 24 hr post-infection. One unit (U) was the amount required to reduce cytopathic effect by approximately 50%. The absence of non-IFNα contaminants with ISG inducing activity was demonstrated by immuno-depletion of ISG-inducing activity with rabbit anti-IFNα (data not shown) and production of media lacking ISG-inducing activity following transfection of the same cells with the vector without an insert. IFNγ was also quantified using an ELISA based on a polyclonal antiserum that cross-reacted with tamarin and marmoset IFNγ (Quantikine; Human IFNγ Immunoassay; R&D Systems, Minneapolis MN).
RT-PCR for GBV-B
GBV-B RNA was isolated from cells or medium by extraction with RNazol (Biotecx Laboratories, Leedo, TX), and total cell RNA was quantified by optical density. GBV-B RNA was quantified by a real time, 5′ exonuclease RT-PCR (TaqMan) assay using a primer/probe combination that recognized a portion of the GBV-B capsid gene as previously described (Beames et al., 2000).
Microarray analysis
Primary marmoset hepatocytes were cultivated in serum free medium in 6 well dishes. Nine wells each were untreated or treated with 1000 U/ml of marmoset IFNα for 4 hr prior to harvest for total cellular RNA. Each sample for array analysis was comprised of a pool of three wells for a total of 6 arrays, 3 treated and 3 untreated. Raw data was processed using the AB 1700_Algorithm. The data was normalized using ABI's quantile-based normalization, and the normalized signal intensities were used to generate fold changes between the treated and untreated samples. Each of the 3 treated arrays was compared to each of the control arrays, and genes that differed by 2-fold or more in 6 of 9 comparisons were included in the comparison with chimpanzee and human hepatocytes. Analysis of primary chimpanzee and human hepatocytes using Affymetrix arrays has been previously published (Lanford et al., 2006). The chimpanzee hepatocytes were cultivated under identical conditions as used for marmoset hepatocytes.
Supplementary Material
Figure 2.
Alignment of amino acid sequence of IFNα genes. Human IFNα2 and IFNα5 amino acid sequences were aligned with the marmoset and tamarin IFNα2 sequences. The human IFNα2 was used as the consensus sequence with sequence identity to this sequence indicated by a dot. The full length intact proteins are 189 amino acids (188 amino acids for human IFNα2, note internal deletion) with the first 23 amino acids being the cleaved signal sequence. The marmoset and tamarin IFNα2 genes differ at 10 residues.
Acknowledgments
This work was supported by grant RO1 AI49574 and P51 RR13986 from the National Institutes of Health. This investigation was conducted in facilities constructed with support from Research Facilities Improvement Program Grant Number C06 RR 12087 from the National Center for Research Resources of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Alter HJ, Seeff LB. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin Liver Dis. 2005;20:17–35. doi: 10.1055/s-2000-9505. [DOI] [PubMed] [Google Scholar]
- Beames B, Chavez D, Guerra B, Notvall L, Brasky KM, Lanford RE. Development of a primary tamarin hepatocyte culture system for GB virus-B: a surrogate model for hepatitis C virus. Journal of Virology. 2000;74:11764–11772. doi: 10.1128/jvi.74.24.11764-11772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beames B, Chavez D, Lanford RE. GB virus B as a model for hepatitis C virus. ILAR J. 2001;42:152–160. doi: 10.1093/ilar.42.2.152. [DOI] [PubMed] [Google Scholar]
- Bigger CB, Brasky KM, Lanford RE. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. Journal of Virology. 2001;75:7059–7066. doi: 10.1128/JVI.75.15.7059-7066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigger CB, Guerra B, Brasky KM, Hubbard G, Beard MR, Luxon BA, Lemon SM, Lanford RE. Intrahepatic gene expression during chronic hepatitis C virus infection in chimpanzees. Journal of Virology. 2004;78:13779–13792. doi: 10.1128/JVI.78.24.13779-13792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. Journal of Virology. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright H, Carroll AR, Watts PA, Fenton RJ. Development of a GB virus B marmoset model and its validation with a novel series of hepatitis C virus NS3 protease inhibitors. Journal of Virology. 2004;78:2062–2071. doi: 10.1128/JVI.78.4.2062-2071.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukh J, Apgar CL, Govindarajan S, Purcell RH. Host range studies of GB virus-B hepatitis agent, the closest relative of hepatitis C virus, in New World monkeys and chimpanzees. J Med Virol. 2001;65:694–697. doi: 10.1002/jmv.2092. [DOI] [PubMed] [Google Scholar]
- Bukh J, Apgar CL, Yanagi M. Toward a surrogate model for hepatitis C virus: An infectious molecular clone of the GB virus-B hepatitis agent. Virology. 1999;262:470–478. doi: 10.1006/viro.1999.9941. [DOI] [PubMed] [Google Scholar]
- Butkiewicz N, Yao N, Zhong W, Wright-Minogue J, Ingravallo P, Zhang R, Durkin J, Strandring DN, Baroudy BM, Sangar DV, Lemon SM, Lau JYN, Hong Z. Virus-specific cofactor requirement and chimeric hepatitis C virus/GB virus B nonstructural protein 3. Journal of Virology. 2000;74:4291–4301. doi: 10.1128/jvi.74.9.4291-4301.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, Zhang C, Chang KS, Jiang J, Ahn BC, Wakita T, Liang TJ, Luo G. Robust Production of Infectious Hepatitis C Virus (HCV) from Stably HCV cDNA-Transfected Human Hepatoma Cells. The Journal of Virology. 2005;79:13963–13973. doi: 10.1128/JVI.79.22.13963-13973.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LM, Borozan I, Feld J, Sun J, Tannis LL, Coltescu C, Heathcote J, Edwards AM, McGilvray ID. Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology. 2005;128:1437–1444. doi: 10.1053/j.gastro.2005.01.059. [DOI] [PubMed] [Google Scholar]
- Chen Z, Benureau Y, Rijnbrand R, Yi J, Wang T, Warter L, Lanford RE, Weinman SA, Lemon SM, Martin A, Li K. GB Virus B Disrupts RIG-I Signaling by NS3/4A-Mediated Cleavage of the Adaptor Protein MAVS. Journal of Virology. 2007;81:964–976. doi: 10.1128/JVI.02076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demers GW, Sugarman BJ, Beltran JC, Westreich LN, Iqbal Ahmed CM, Lau JY, Hong Z, Lanford RE, Maneval DC. Interferon-a2b secretion by adenovirus-mediated gene delivery in rat, rabbit, and chimpanzee results in similar pharmacokinetic profiles. Toxicology and Applied Pharmacology. 2002;180:36–42. doi: 10.1006/taap.2002.9372. [DOI] [PubMed] [Google Scholar]
- Foy E, Li K, Sumpter R, Loo YM, Johnson CL, Wang CF, Fish PM, Yoneyama M, Fujita T, Lemon SM, Gale M. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:2986–2991. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foy E, Li K, Wang CF, Sumpter R, Ikeda M, Lemon SM, Gale M. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science. 2003;300:1145–1148. doi: 10.1126/science.1082604. [DOI] [PubMed] [Google Scholar]
- Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Goncales FL, Haussinger D, Diago M, Carosi G, Dhumeaux D, Craxi A, Lin A, Hoffman J, Yu J. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. New England Journal of Medicine. 2002;347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Kim WR, Gores GJ, Benson JT, Therneau TM, Melton LJ., III Mortality and Hospital Utilization for Hepatocellular Carcinoma in the United States. Gastroenterology. 2005;129:486–493. doi: 10.1016/j.gastro.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Lanford RE, Bigger C. Advances in model systems for hepatitis C virus research. Virology. 2002;293:1–9. doi: 10.1006/viro.2001.1316. [DOI] [PubMed] [Google Scholar]
- Lanford RE, Chavez D, Notvall L, Brasky KM. Comparison of tamarins and marmosets as hosts for GBV-B infections and the effect of immunosuppression on duration of viremia. Virology. 2003a;311:72–80. doi: 10.1016/s0042-6822(03)00193-4. [DOI] [PubMed] [Google Scholar]
- Lanford RE, Estlack LE. A cultivation method for highly differentiated primary chimpanzee hepatocytes permissive for hepatitis C virus replication. Methods Mol Med. 1998;19:501–516. doi: 10.1385/0-89603-521-2:501. [DOI] [PubMed] [Google Scholar]
- Lanford RE, Guerra B, Bigger CB, Lee H, Chavez D, Brasky KM. Lack of response to exogenous interferon-alpha in the liver of HCV chronically infected chimpanzees. Hepatology. 2007;46:999–1008. doi: 10.1002/hep.21776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford RE, Guerra B, Demers GW, Chang S, Sugarman BJ, Maneval DC, Hong Z, Lau JYN. Gene therapy for chronic hepatitis C based on adenoviral-mediated transfer of interferon-2b gene study based on the chimpanzee model. Hepatology Abtracts to AASLD Meeting. 2000;32:1141. Part 2. [Google Scholar]
- Lanford RE, Guerra B, Lee H, Averett DR, Pfeiffer B, Chavez D, Notvall L, Bigger C. Antiviral effect and virus-host interactions in response to alpha interferon, gamma interferon, poly(I)-poly(C), tumor necrosis factor alpha, and ribavirin in hepatitis C virus subgenomic replicons. Journal of Virology. 2003b;77:1092–1104. doi: 10.1128/JVI.77.2.1092-1104.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford RE, Guerra B, Lee H, Chavez D, Brasky K, Bigger CB. Genomic response to interferon-alpha in chimpanzees: Implications of rapid downregulation for hepatitis C kinetics. Hepatology. 2006;43:961–972. doi: 10.1002/hep.21167. [DOI] [PubMed] [Google Scholar]
- Levy DE, Lew DJ, Decker T, Kessler DS, Darnell JE., Jr Synergistic interaction between interferon-alpha and interferon-gamma through induced synthesis of one subunit of the transcription factor ISGF3. Embo Journal. 1990;9:1105–1111. doi: 10.1002/j.1460-2075.1990.tb08216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Foy E, Ferreon JC, Nakamura M, Ferreon ACM, Ikeda M, Ray SC, Gale M, Lemon SM. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci USA. 2005a;102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Chen Z, Kato N, Gale M, Jr, Lemon SM. Distinct Poly(I-C) and Virus-activated Signaling Pathways Leading to Interferon-{beta} Production in Hepatocytes. Journal of Biological Chemistry. 2005b;280:16739–16747. doi: 10.1074/jbc.M414139200. [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. Complete Replication of Hepatitis C Virus in Cell Culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Meuleman P, Ploss A, Vanwolleghem T, Syder AJ, McKeating JA, Lanford RE, Feinstone SM, Major ME, Leroux-Roels G, Rice CM. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proceedings of the National Academy of Sciences. 2006;103:3805–3809. doi: 10.1073/pnas.0511218103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, Goodman ZD, Ling MH, Albrecht JK. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: A randomised trial. The Lancet. 2001;358:958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- Martin A, Bodola F, Sangar DV, Goettge K, Popov V, Rijnbrand R, Lanford RE, Lemon SM. Chronic hepatitis associated with GB virus B persistence in a tamarin after intrahepatic inoculation of synthetic viral RNA. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9962–9967. doi: 10.1073/pnas.1731505100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Muerhoff AS, Leary TP, Simons JN, Pilot-Matias TJ, Dawson GJ, Erker JC, Chalmers ML, Schlauder GG, Desai SM, Mushahwar IK. Genomic organization of GB viruses A and B: Two new members of the Flaviviridae associated with GB agent hepatitis. Journal of Virology. 1995;69:5621–5630. doi: 10.1128/jvi.69.9.5621-5630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam JH, Faulk K, Engle RE, Govindarajan S, St Claire M, Bukh J. In vivo analysis of the 3 ′ untranslated region of GB virus B after in vitro mutagenesis of an infectious cDNA clone: Persistent infection in a transfected tamarin. Journal of Virology. 2004;78:9389–9399. doi: 10.1128/JVI.78.17.9389-9399.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba K, Mizokami M, Lau JYN, Orito E, Ikeo K, Gojobori T. Evolutionary relationship of hepatitis C, pesti-, flavi-, plantviruses, and newly discovered GB hepatitis agents. FEBS Letters. 1996;378:232–234. doi: 10.1016/0014-5793(95)01441-1. [DOI] [PubMed] [Google Scholar]
- Preiss AT. Characterization of the innate immune signalling pathways in hepatocyte cell lines. J Viral Hepat. 2008;15:888–900. doi: 10.1111/j.1365-2893.2008.01001.x. [DOI] [PubMed] [Google Scholar]
- Robertson B, Myers G, Howard C, Brettin T, Bukh J, Gaschen B, Gojobori T, Maertens G, Mizokami M, Nainan O, Netesov S, Nishioka K, Shin, Simmonds P, Smith D, Stuyver L, Weiner A. Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: proposals for standardization. International Committee on Virus Taxonomy [news] Archives of Virology. 1998;143:2493–2503. doi: 10.1007/s007050050479. [DOI] [PubMed] [Google Scholar]
- Saito T, Hiral R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M. Regulation of innate antiviral defense through a shared repressor domain in RIG-1 and LGP-2. Proceedings of National Academic Sciences USA. 2007;104:582–857. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarasin-Filipowicz M, Oakeley EJ, Duong FHT, Christen V, Terracciano L, Filipowicz W, Heim MH. Interferon signaling and treatment outcome in chronic hepatitis C. Proceedings of the National Academy of Sciences. 2008;105:7034–7039. doi: 10.1073/pnas.0707882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbardellati A, Scarselli E, Verschoor E, De Tomassi A, Lazzaro D, Traboni C. Generation of infectious and transmissible virions from a GB virus B full-length consensus clone in tamarins. Journal of General Virology. 2001;82:2437–2448. doi: 10.1099/0022-1317-82-10-2437. [DOI] [PubMed] [Google Scholar]
- Scarselli E, Urbani A, Sbardellati A, Tomei L, De Francesco R, Traboni C. GB virus B and hepatitis C virus NS3 serine proteases share substrate specificity. Journal of Virology. 1997;71:4985–4989. doi: 10.1128/jvi.71.7.4985-4989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and Characterization of MAVS, a Mitochondrial Antiviral Signaling Protein that Activates NF-kB and IRF3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Thomas D, Seeff LB. Natural history of hepatitis C. Clin Liver Dis. 2005;9:383–398. doi: 10.1016/j.cld.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherford T, Chavez D, Brasky KM, Lanford RE. The Marmoset Model of GBV-B Infections: Adaptation to Host Phenotypic Variation. Journal of Virology. 2009 doi: 10.1128/JVI.00033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA Is an Adapter Protein Required for Virus-Triggered IFN-beta Signaling. Molecular Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Yi M, Villanueva RA, Thomas DL, Wakita T, Lemon SM. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2310–2315. doi: 10.1073/pnas.0510727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Jr, Akira S, Yonehara S, Kato A, Fujita T. Shared and Unique Functions of the DExD/H-Box Helicases RIG-I, MDA5, and LGP2 in Antiviral Innate Immunity1. The Journal of Immunology. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:699–701. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.