Abstract
1, N2-Propanodeoxyguanosine (PdG) is a stable structural analogue for the 3-(2′-deoxy-β-d-erythro-pentofuranosyl)pyrimido[1,2-α]purin-10(3H)-one (M1dG) adduct derived from exposure of DNA to base propenals and to malondialdehyde. The structures of ternary polymerase–DNA–dNTP complexes for three template–primer DNA sequences were determined, with the Y-family Sulfolobus solfataricus DNA polymerase IV (Dpo4), at resolutions between 2.4 and 2.7 Å. Three template 18-mer–primer 13-mer sequences, 5′-d(TCACXAAATCCTTCCCCC)-3′ • 5′-d(GGGGGAAGGATTT)-3′ (template I), 5′-d(TCACXGAATCCT-TCCCCC)-3′ • 5′-d(GGGGGAAGGATTC)-3′ (template II), and 5′-d(TCATXGAATCCTTCCCCC)-3′ • 5′-d(GGGGGAAGGATTC)-3′ (template III), where X is PdG, were analyzed. With templates I and II, diffracting ternary complexes including dGTP were obtained. The dGTP did not pair with PdG, but instead with the 5′-neighboring template dC, utilizing Watson–Crick geometry. Replication bypass experiments with the template–primer 5′TCACXAAATCCTTACGAGCATCGCCCCC-3′ • 5′-GGGGGCGATGCTCGTAAGGATTT-3′, where X is PdG, which includes PdG in the 5′-CXA-3′ template sequence as in template I, showed that the Dpo4 polymerase inserted dGTP and dATP when challenged by the PdG adduct. For template III, in which the template sequence was 5′-TXG-3′, a diffracting ternary complex including dATP was obtained. The dATP did not pair with PdG, but instead with the 5′-neighboring T, utilizing Watson–Crick geometry. Thus, all three ternary complexes were of the “type II” structure described for ternary complexes with native DNA [Ling, H., Boudsocq, F., Woodgate, R., and Yang, W. (2001) Cell 107, 91–102]. The PdG adduct remained in the anti conformation about the glycosyl bond in each of these threee ternary complexes. These results provide insight into how −1 frameshift mutations might be generated for the PdG adduct, a structural model for the exocylic M1dG adduct formed by malondialdehyde.
The guanine adduct M1dG1 [3-(2′-deoxy-β-d-erythro-pentofuranosyl)pyrimido[1,2-α]purin-10(3H)-one] arises in DNA from multiple sources. One source is exposure to malondialdehyde (MDA), a toxic and mutagenic metabolite produced by lipid peroxidation and prostaglandin biosynthesis (for a review, see refs 1 and 2). In aqueous solution, MDA exists as β-hydroxyacrolein. It reacts with DNA as a bis-electrophile to form M1dG (3–7). Alternatively, M1dG arises as a consequence of oxidative damage to DNA, resulting in the formation of base propenals that subsequently transfer their oxopropenyl group to dG (8) (Scheme 1). The base propenals are significantly more potent than MDA in forming M1dG (9).
Scheme 1.
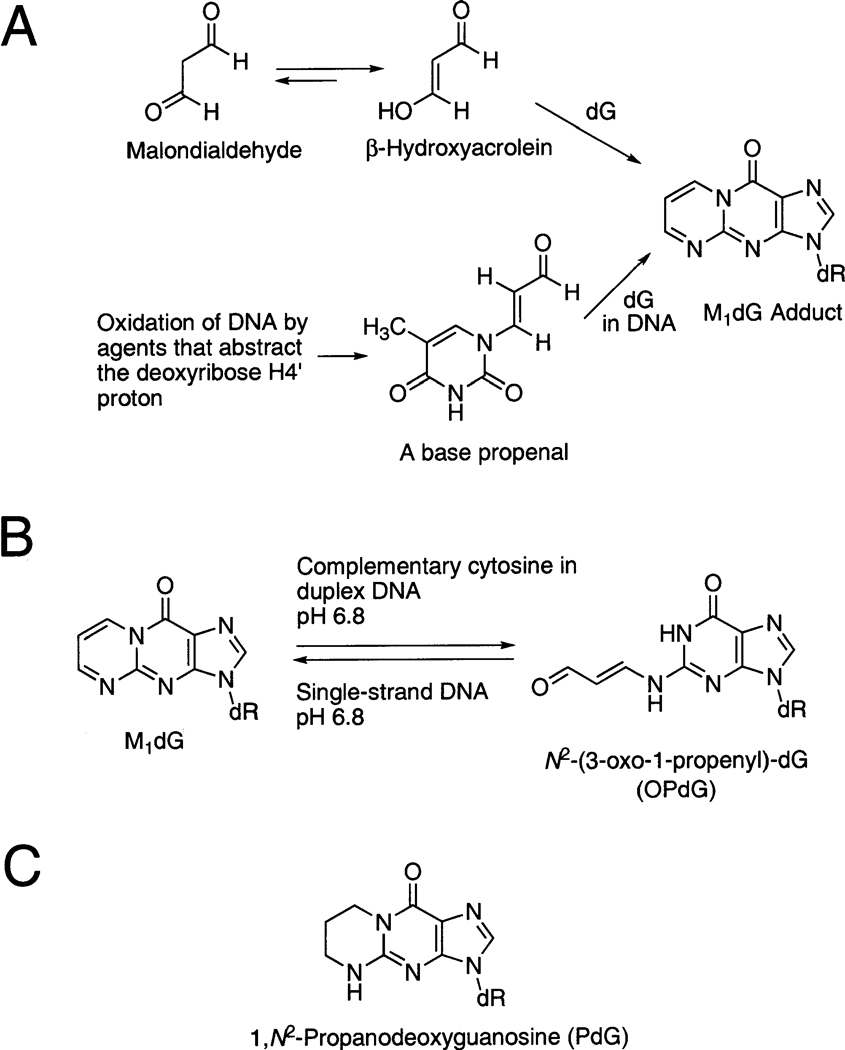
(A) Formation of M1dG from Malondialdehyde or from Base Propenals,a (B) M1dG Which Is Stable in Single-Stranded DNA,b and (C) 1,N2-Propanodeoxyguanosine (PdG), a Structural Model for the M1dG Adduct
a Thymine propenal is shown as a representative propenal.
b In duplex DNA, when placed opposite deoxycytosine, it is spontaneously and quantitatively converted to N2-(3-oxo-1-propenyl)-dG, the OPdG derivative.
M1dG has been identified in DNA from rodent (10) and human (11,12) tissue samples, as have other exocyclic purine lesions (13–16), suggesting their presence in vivo. The levels of M1dG in DNA have been quantified by mass spectrometric (17, 18), postlabeling (19, 20), and immunochemical (21) techniques. This endogenously formed adduct is present in human DNA (18–20, 22). It has also been detected at low levels in human urine (23). The low urinary levels of M1dG likely reflect metabolic conversion to the 6-oxo-M1dG derivative (24). M1dG is an efficient premutagenic lesion in Escherichia coli (25, 26), mammalian (26), and human (27) cells. Thus, M1dG is anticipated to mediate human carcinogenesis.
M1dG is stable in nucleotides and single-stranded DNA at neutral pH. Under basic conditions, it converts to the N2-(3-oxo-1-propenyl)-dG (OPdG) derivative. In contrast, when M1dG is placed at neutral pH into duplex DNA opposite dC, a spontaneous conversion to the OPdG derivative is facilitated. Upon denaturation of the duplex, M1dG is regenerated (Scheme 1). Ring opening does not occur at neutral pH in duplex DNA if thymine is placed opposite M1dG. These observations suggested that dC in duplex DNA catalyzes the transformation of M1dG to its ring-opened OPdG derivative (28). The ring opening of M1dG as a nucleoside or in oligodeoxynucleotides is a reversible second-order reaction with hydroxide ion (29). The reverse ring closure mechanism involves rapid formation of protonated OPdG and 8-hydroxy-6,7-propeno-dG intermediates that slowly convert to M1dG in a general acid-catalyzed reaction (30).
1,N2-Propanodeoxyguanosine (PdG) (Scheme 1) (31) provides a structural model for M1dG. Unlike M1dG, it does not undergo ring opening to OPdG. This chemical difference can be exploited to determine how 1,N2-dG lesions are accommodated by the DNA duplex under conditions in which M1dG spontaneously undergoes ring opening to OPdG when placed opposite dC. When mispaired with dA, PdG exhibited X(syn) • A(anti) pairing at pH 5.8 and simultaneous partial intercalation of the complementary X and A bases at pH 8.9. When mispaired with dG, PdG exhibited X(syn) • G(anti) pairing which was pH-independent (32–35). The exocyclic ring of PdG was inserted into the DNA duplex when positioned opposite an abasic site (36). When placed in 5′-d(CGCXCGGCATG)-3′ at pH 5.8, PdG induced a localized structural perturbation involving the modified base pair and its 3′-neighbor. PdG rotated into the syn conformation about the glycosyl bond, and the 3′-neighbor base pair existed in a mixture of Watson–Crick and Hoogsteen conformations (37). In the 5′-d(CGCGGTXTCCGCG)-3′ • 5′-d(CGCGGACACCGCG)-3′ duplex, PdG introduced a localized perturbation, which was pH-dependent (38). At pH 5.2, PdG rotated into the syn conformation about the glycosyl bond, placing the exocyclic moiety into the major groove. PdG formed a protonated Hoogsteen pair cytosine in the complementary strand. In this sequence, the 3′-neighbor base pair did not equilibrate between Hoogsteen and Watson–Crick base pairing (38). When PdG was placed in 5′-d(ATCGCX-CGGCATG)-3′ opposite a two-base deletion in the complementary strand, PdG was in the anti conformation about the glycosyl bond and inserted into the DNA duplex. The 3′-neighbor dC was extruded toward the major groove (39, 40). PdG reduced the thermal stability, transition enthalpy, and transition free energy of duplex DNA when positioned opposite cytosine or adenine, and the destabilization of the duplex was not sensitive to whether the base opposite the lesion was adenine or cytosine (41). The structure of PdG has also been determined in a three-base hairpin loop formed by d(CGCGGTXTCCGCG). The structure of the PdG-modified hairpin consisted of a 5 bp stem and a three-base loop, with PdG oriented such that the imidazole proton faced the minor groove of the stem and the exocyclic protons projected into the major groove (42).
The Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4) polymerase is a DinB homologue that belongs to the Y-family of DNA polymerases characterized by their low fidelity on undamaged DNA templates and propensity to traverse normally replication-blocking lesions. It possesses a right-hand architecture containing thumb, palm, and finger subdomains in the polymerase domain. The core of the palm appears conserved between Y-family and replicative polymerases, while the thumb and fingers are significantly smaller in the Y-family. Furthermore, Y-family polymerases contain a unique little finger subdomain, which is believed to play an important role in facilitating DNA binding (43–45). The Dpo4 polymerase can insert the correct complementary nucleotide at each of the investigated DNA lesions, but mispairs and frameshifts have also been observed (46). Crystal structures of the Dpo4 polymerase in complex with unmodified DNA and the incoming nucleotide provide excellent models for investigating the structural features that determine lesion bypass efficiency and fidelity (43). The structures determined for the Dpo4 polymerase in the presence of unmodified primer–template DNAs show two modes of entry for dNTPs. In the type I structure (PDB entry 1JX4) (43), the entering dNTP is paired with the normal, 3′-neighbor template base. In the type II structure (PDB entry 1JXL), the entering dNTP skips this base and is paired instead with the 5′-neighboring base. The active sites of Y-family polymerases, in contrast to replicative polymerases, are solvent accessible and more spacious (43–45) and in some cases can accommodate two template bases. Moreover, the nascent base pair is sterically less constrained than in replicative polymerases. A bulky DNA adduct at the active site is bypassed more easily by the Y-family DNA polymerases, and the spacious active site results in a relaxed geometric selection for the incoming 2′-deoxynucleotide 5′-triphosphate (dNTP) (47), thus compromising the efficiency and fidelity of DNA replication. The lesion bypass ability, accuracy, and efficiency of these polymerases vary significantly and depend on the type of DNA lesion (46, 48–58).
At present, structures of the ternary Dpo4–DNA–dNTP complexes for three 18-mer primer • 13-mer sequences modified with PdG (Scheme 2) have been determined, at resolutions between 2.4 and 2.7 Å. These ternary complexes contained PdG in the template 5′-CXA-3′ (template I), 5′-CXG-3′ (template II), and 5′-TXG-3′ (template III) sequence contexts. With templates I and II, diffracting ternary complexes including dGTP were obtained. The dGTP did not pair with PdG, but instead with the 5′-neighboring template dC, utilizing Watson–Crick geometry. Replication bypass experiments utilizing a template–primer sequence which included PdG in the 5′-CXA-3′ template sequence (as in template I) showed that the Dpo4 polymerase inserted dGTP and dATP when challenged by the PdG adduct. For template III, in which the template sequence was 5′-TXG-3′, a diffracting ternary complex including dATP was obtained. The dATP did not pair with PdG, but instead with the 5′-neighboring T, utilizing Watson–Crick geometry. Thus, all three ternary complexes were of the “type II” structure described for ternary complexes with native DNA (43). The PdG adduct remained in the anti conformation about the glycosyl bond in each of these three ternary complexes. The structures analyzed in this work reveal the importance of stacking in the generation of a stable intermediate of the Dpo4 polymerase with primer–template DNA and provide visualization of a possible –1 frameshift mutation mechanism involving the lesion bypass and the template slippage for mutagenic exocyclic DNA adducts.
Scheme 2.

Template (18-mer)—Primer (13-mer) Oligonucleotide Duplexes Used in This Work (X is PdG)
MATERIALS AND METHODS
Materials
The Dpo4 polymerase was expressed in E. coli and purified using heat denaturation, Ni2+-nitriloacetate chromatography, and ion-exchange chromatography as described by Zang et al. (59). All dNTPs were obtained from Amersham Biosciences (Piscataway, NJ). Unmodified oligodeoxynucleotides were synthesized and purified by Midland Certified Reagent Co. (Midland, TX), with analysis by MALDI mass spectrometry. The 13-mer primers used for crystallography were not dideoxy-terminated. PdG was synthesized, purified, and incorporated into oligodeoxynucleotides using an established methodology (31), in house, with analysis by MALDI mass spectrometry. Purity was assessed by capillary gel electrophoresis. Oligodeoxynucleotides were desalted by Sephadex G-25 gel exclusion chromatography (GE Healthcare, Inc., Piscataway, NJ). Oligodeoxynucleotide concentrations were determined by the UV absorption at 260 nm (60).
Replication Bypass Experiment
These experiments utilized either a PdG-adducted or nonadducted 28-mer • 23-mer template • primer 5′-d(TCACXAAATCCTTACGAGCATCGC-CCCC)-3′ • 5′-d(GGGGGCGATGCTCGTAAGGATTT)-3′ sequence, where X is PdG or G. The primers were 5′-end-labeled with [γ-32P]ATP using T4 polynucleotide kinase (T4PNK) (New England BioLabs). The labeled primers were annealed to either unmodified or modified templates in 1:1 molar ratios in 50 mM Tris-HCl buffer (pH 7.8). The unmodified and modified primers were extended in the presence of single dNTPs. Each reaction was initiated by adding 2 µL of dNTP (final concentration of 50 µM) to a preincubated template/primer/polymerase mixture [final concentrations of 50 mM Tris-HCl (pH 7.8), 50 mM NaCl, 5 mM MgCl2, 1 mM DTT, 50 µg/mL BSA, 100 nM DNA duplex, and 150 nM Dpo4] at 37 °C, yielding a total reaction volume of 10 µL. The reaction mixtures were incubated over a time period of 15, 30, or 60 min. Each reaction was quenched with 70 µL of 20 mM Na2EDTA (pH 9.0) in 95% formamide (v/v) and each mixture heated for 10 min at 95 °C. For full-length extension assays, the unmodified and modified primers were extended in the presence of all four dNTPs. Each reaction was initiated by adding 2 µL of all four dNTPs (final concentrations of 50 µM) to a preincubated template/primer/polymerase reaction mixture [final concentrations of 50 mM Tris-HCl (pH 7.8), 50 mM NaCl, 5 mM MgCl2, 1 mM DTT, 50 µg/mL BSA, 100 nM DNA duplex, and 150 nM Dpo4] at 37 °C, yielding a total reaction volume of 10 µL. The reaction mixtures were incubated over a time period of 15, 30, or 60 min. Each reaction was quenched with 70 µL of 20 mM Na2EDTA (pH 9.0) in 95% formamide (v/v) and each mixture heated for 10 min at 95 °C. A denaturing gel containing 8.0 M urea and 16% acrylamide (w/v) (from a 19:1 acrylamide/bisacrylamide solution, Accu-Gel, National Diagnostics, Atlanta, GA) with 80 mM Tris borate buffer (pH 7.8) containing 1 mM Na2EDTA was used for electrophoresis on which aliquots of 6 µL were separated. The gel was exposed to a PhosphorImager screen (Imaging Screen K, Bio-Rad) overnight. The bands were visualized with a PhosphorImaging system (Bio-Rad, Molecular Imager FX) using the manufacturer’s software Quantity One, version 4.3.0.
Crystallization of Dpo4 • DNA Complexes
The 18-mer template containing the PdG adduct was annealed with the 13-mer primer at a 1:1 molar ratio in 0.1 M NaCl, 10 mM NaH2PO4, and 50 µM Na2EDTA (pH 7.2). The duplexes were eluted from DNA grade hydroxylapatite (Bio-Rad Laboratories, Richmond, CA) with a gradient from 10 to 200 mM NaH2PO4 in 10 mM NaH2PO4, 0.1 M NaCl, and 50 µM EDTA (pH 7.0). Duplexes were desalted with Sephadex G-25. The Dpo4 polymerase was concentrated to 300–550 µM (12–22 mg/mL) using a spin concentrator with a 104 Mr Amicon cutoff filter (Millipore, Inc., Billerica, MA) in 50 mM Tris-HCl (pH 7.4 at 25 °C) buffer containing 200 mM NaCl, 5 mM β-mercaptoethanol, and 10% glycerol (v/ v). The Dpo4 polymerase was combined with DNA (1:1.2 molar ratio) and then placed on ice for 1 h prior to incubation with 1 mM d(N)TP and 5 mM CaCl2. Crystals were grown using the sitting drop vapor diffusion method by mixing 1 µL of complex with 1 µL of a solution containing 10% polyethylene glycol 3350 (w/v) and 100 mM Ca(OAc)2 and equilibrated against a well solution containing 25 mM Tris-HCl (pH 7.4 at 25 °C) buffer, 5–10% polyethylene glycol 3350 (w/v), 100 mM Ca(OAc)2, and 2.5% glycerol (v/v). Crystals were soaked in mother liquor containing an additional 25% polyethylene glycol 3350 (w/v) and 15% ethylene glycol (v/v) and then swiped through paratone-N (Hampton Research, Aliso Viejo, CA) and flash-frozen in a stream of liquid nitrogen.
X-ray Diffraction Data Collection and Processing
Diffraction data sets for the ternary Dpo4–DNA–dNTP complexes were collected at 110 K using a synchrotron radiation wavelength of 0.99 Å on the ID-5 beamline (crystals I and III) and a wavelength of 0.92 Å on the ID-22 beamline (crystal II) at the Advanced Photon Source (Argonne, IL). Indexing and scaling were performed using XDS (61) or HKL2000 (62). The data were further processed using CCP4 package programs, and the truncate procedure was performed with TRUNCATE (63).
Structure Determination and Refinement
A refined structure (PDB entry 2BQU) downloaded from Protein Data Bank was used as a starting model by modifying the template and primer and inserting the PdG adduct. In each instance, several rounds of rigid body refinement of the diffraction data, with gradually increasing resolution, optimized the initial positions of the models. The model was refined further using CNS Solve (version 1.1) (64), including simulated annealing, gradient minimization, individual occupancy, and refinement of individual isotropic temperature factors. Manual model building was performed using TURBO-FRODO (65, 66). A total of 5% of the reflections were excluded from the refinement to calculate the cross-validation residual Rfree. Water oxygen atoms were added into positive regions (more than 3.0 standard deviations) of Fo – Fc Fourier difference electron density during the manual model rebuilding steps. The crystallographic figures were prepared using TURBO-FRODO and InsightII (Accelrys, Inc., San Diego, CA).
RESULTS
Replication of a 5′-CXA-3′ PdG-Adducted Template by the Dpo4 Polymerase
Replication bypass experiments were performed at 37 °C using the template • primer 5′TCACXAAATCCTTACGAGCATCGCCCCC-3′ • 5′-GGGGGCGATGCTCGTAAGGATTT-3′ sequence, where X is PdG (Figure 1). This template • primer contained PdG in the 5′-CXA-3′ sequence context. The reactions were monitored at time increments of 0, 15, 30, and 60 min. At this temperature, the Dpo4 polymerase was not highly efficient, but it inserted both dATP and dGTP when challenged by the template PdG. When all four dNTPs were included in the reaction, the Dpo4 polymerase did not extend the primer strand to the full-length product. In the corresponding unmodified template • primer, the Dpo4 polymerase inserted dCTP opposite the template G, and including all four dNTPs in the reaction mixture successfully extended the template • primer to full extension products in 30 min.
FIGURE 1.

(A) Replication bypass of the native primer•template complex with the S. solfataricus P2 DNA polymerase IV (Dpo4) enzyme. (B) Replication bypass of the PdG-adducted primer•template complex with the S. solfataricus P2 DNA polymerase IV (Dpo4) enzyme. The reactions were monitored at time increments of 0, 15, 30, and 60 min. The first four panels in each assay represent insertion of a single dNTP to either the native or the adducted template. The last panel in each assay represents the inclusion of all four dNTPs in the reaction mixture and their addition to either the native or the adducted template. The assays were carried out at 37 °C, using 150 nM Dpo4 and 50 µM dNTPs.
Crystallization, Data Collection, and Data Processing
The three site-specifically PdG-modified 18-mer templates, 5′-TCACXAAATCCTTCCCCC-3′ (template I), 5′-TCACX-GAATCCTTCCCCC-3′ (template II), and 5′-TCATX-GAATCCTTCCCCC-3′ (template III), differed in the nearest-neighbor sequence context at the PdG-modified site, having 5′-CXA-3′, 5′-CXG-3′, and 5′-TXG-3′ sequences, respectively (Scheme 2). These were annealed with the complementary 13-mer primers 5′-GGGGGAAGGATTT-3′, 5′-GGGGGAAGGATTC-3′, and 5′-GGGGGAAGGATTC-3′, respectively, which terminated at the base pair 3′ to PdG in the templates. The resulting template • primer complexes were primed for insertion of dNTP opposite PdG in the template. Crystallization trials were conducted for all three template • primer complexes, in the presence of Ca2+, an inhibitor of DNA polymerases, and Na2EDTA to chelate traces of Mg2+, each in the presence of four possible dNTPs. A concentration range of 3–15 mg/mL of the Dpo4 polymerase was used to find the optimum concentration for growing crystals that diffracted well. For template I, suitable crystals were found at a concentration of 10 mg/mL polymerase. For template II, suitable crystals were found at a concentration of 3 mg/ mL polymerase. For template III, suitable crystals were found at a concentration of 5 mg/mL polymerase. It was possible to crystallize and diffract ternary Dpo4–DNA–dNTP complexes for templates I and II, each with dGTP, whereas for template III, it was possible to crystallize and diffract a ternary complex with dATP. Ternary complexes with TTP and dGTP as the incoming nucleotides with template III also diffracted, but the crystals were twinned and could not be indexed. Thus, the crystallization and diffraction of ternary complexes corresponded to the identities of the 5′-neighbor nucleotides with respect to PdG in the templates.
The statistics of data processing and data quality are summarized in Table 1. Each of the three ternary Dpo4–DNA–dNTP complexes diffracted to medium resolution, within a range of 2.4–2.7 Å. The highest-resolution shell was from 2.7 to 2.9 Å with a completeness of 88.0% for template I, from 2.5 to 2.6 Å with a completeness of 76.1% for template II, and from 2.4 to 2.5 Å with a completeness of 98.2% for template III. The resulting data sets for three ternary complexes were of good quality as indicated by Rmerge values of 8.1% (template I), 7.0% (template II), and 7.0% (template III), as well as by signal-to-noise ratios (I/σI) of 6.9 (template I), 7.0 (template II), and 7.0 (template III), respectively. Autoindexing and systematic absences indicated space group P21212 for all three sequences.
Table 1.
Crystal Data and Refinement Parameters for the Three Ternary Dpo4–DNA–dNTP Complexes
| crystal I | crystal II | crystal III | |
|---|---|---|---|
| template | template I | template II | template III |
| X-ray source | ID-5 (DND-CAT/APS) | ID–22 (SER–CAT/APS) | ID-5 (DND-CAT/APS) |
| wavelength (Å) | 0.92 | 0.99 | 0.92 |
| temperature (K) | 110 | 110 | 110 |
| no. of crystals | 1 | 1 | 1 |
| space group | P21212 | P21212 | P21212 |
| unit cell (a, b, c) (Å) | 93.5, 103.3, 52.8 | 93.4, 103.0, 52.8 | 92.2, 101.9, 52.7 |
| resolution range (Å) | 50.0–2.70 (2.70–2.90)a | 50.0–2.48 (2.48–2.59)a | 50.0–2.38 (2.38–2.48)a |
| no. of measurements | 17770 | 15421 | 25742 |
| no. of unique reflections | 25290 | 23739 | 35739 |
| redundancy | 5.1 | 5.5 | 6.4 |
| completeness (%) | 100.0 (88.0)a | 99.6 (76.1)a | 100.0 (98.2)a |
| Rmergeb(%) | 8.1 (8.6)a | 7.0 (7.4)a | 7.0 (7.2)a |
| signal to noise (I/σI) | 6.9 (7.2)a | 7.0 (7.6)a | 7.0 (7.4)a |
| solvent content (%) | 47.5 | 47.9 | 49.5 |
| model composition (asymmetric unit) | |||
| no. of amino acid residues | 341 | 341 | 341 |
| no. of protein atoms (non-H) | 2744 | 2744 | 2744 |
| no. of water molecules | 105 | 98 | 116 |
| no. of Ca2+ ions | 3 | 3 | 3 |
| no. of template nucleotides | 17 | 17 | 17 |
| no. of primer nucleotides | 13 | 13 | 13 |
| no. of dATPs | 1 | ||
| no. of dGTPs | 1 | 1 | |
| Rfc (%) | 20.8 | 21.5 | 22.0 |
| Rfreed (%) | 25.8 | 27.9 | 26.6 |
| estimated coordinate error (Å) | |||
| from Luzatti plot | 0.38 | 0.43 | 0.35 |
| from Luzatti plot (cross validation) | 0.47 | 0.47 | 0.43 |
| from σ A plot | 0.51 | 0.60 | 0.36 |
| from σ A plot (cross validation) | 0.61 | 0.64 | 0.39 |
| temperature factor from Wilson plot (Å2) | 68.7 | 61.8 | 62.8 |
| mean isotropic temperature factor (Å2) | 58.7 | 56.6 | 49.5 |
| rms deviation in temperature factors (Å2) | |||
| bonded main chain atoms | 1.45 | 1.39 | 1.52 |
| bonded side chain atoms | 2.07 | 2.19 | 2.17 |
| rms deviation from ideal values | |||
| bond lengths (Å) | 0.007 | 0.007 | 0.007 |
| bond angles (deg) | 1.37 | 1.29 | 1.57 |
| dihedral angles (deg) | 21.61 | 22.3 | 21.88 |
| improper angles (deg) | 1.11 | 2.77 | 1.20 |
Values in parentheses correspond to the highest-resolution shells.
Rmerge =Σhkl,j= 1,N |Ihkl — Ihkl,j|/Σhkl,j=1,N|Ihkl,j|, where the outer sum (hkl) is taken over the unique reflections.
Rf = Σhkl ||Fo,hkl — k|Fc,hkl||/Σhkl|Fo,hkl|, where |Fo,hkl| and |Fc,hkl| are the observed and calculated structure factor amplitudes, respectively.
Rfree idem., for the set of reflections (5% of the total) omitted from the refinement process.
Crystal Structures
(a) Template I
The structure of the ternary Dpo4–DNA–dGTP complex was determined at 2.7 Å resolution (Table 1). The active site of the Dpo4 polymerase (Figure 2A) resembled that in the type II structure with native DNA (Figure 2D; based upon PDB entry 1JXL) (43). Thus, both PdG and the 5′-neighbor template dC were accommodated within the active site. Similar to the native type II structure, the 5′-neighbor template C was directed into the active site. PdG stacked inside the duplex (Figures 3A,B) but did not pair with the incoming dGTP. Instead, the incoming dGTP was inserted opposite the 5′-neighbor template dC to form a Watson–Crick base pair (Figure 3A), leaving a gap of 7.7 Å to the 3′-terminal primer T that was itself paired with the 3′-neighboring template dA. The base moieties of dGTP and the 3′-terminal T in the primer were tilted 3.2° and 18°, respectively, with respect to PdG, reducing the distance between the terminal 3′-hydroxyl group of the primer and the α-phosphate group of dGTP to 6.8 Å (Figure 4A).
FIGURE 2.

Structures of three ternary Dpo4—DNA—dNTP complexes containing PdG-adducted DNA. (A) Template sequence I. (B) Template sequence II. (C) Template sequence III. (D) Type II crystal with native DNA from PDB entry 1JXL (43). The Dpo4 polymerase is colored blue and the PdG adduct red; dNTPs and other DNA bases are colored yellow, and the three Ca2+ ions are shown as red spheres.
FIGURE 3.

Stacking patterns of DNA base pairs at the active site of three ternary Dpo4—DNA—dNTP complexes: (A and B)template sequence I, (C and D) template sequence II, and (E and F) template sequence III. The PdG adduct is colored red. The dGTP•C (template sequences I and II) and dATP•T (template sequence III) base pairs are colored yellow, and the primer 3′-terminal T • A (template sequence I) and C • G (template sequences II and III) base pairs are colored blue.
FIGURE 4.

Electron density and DNA duplex conformations at the active sites in the three ternary Dpo4—DNA—dNTP complexes: (A) template sequence I, (B) template sequence II, and (C) template sequence III. The views are into the minor groove. Fourier 3Fσ — 2Fc sum electron density (light green meshwork) is drawn at the 1σ level. Ca2+ ions and PdG adduct are colored red, and DNA base pairs are colored yellow.
Three calcium ions were coordinated in this structure (Figure 5A). The first two were in the active site for catalysis and dGTP coordination. The first Ca2+ ion was weakly coordinated at a distance of 4.2 Å with respect to the side chain carbonyl oxygen of Glu106. Further contacts by this ion were established at distances of 3.0 and 3.7 Å with respect to the two α-phosphate oxygens of dGTP and at distances of 2.5, 2.7, and 4.4 Å with respect to the three γ-phosphate oxygens of dGTP. The second Ca2+ ion was located 2.9 and 4.5 Å from the two α-phosphate oxygens of dGTP and 3.3 Å from one of the three γ-phosphate oxygens of dGTP. The third Ca2+ ion was located 2.7 Å from the side chain carbonyl oxygen of Ala181 in the thumb domain of the Dpo4 polymerase. The third Ca2+ ion was also located 2.1 and 4.0 Å from two oxygens of the α-phosphate group linking the 12th and 13th primer nucleotides.
FIGURE 5.

Active site structures of three ternary Dpo4—DNA—dNTP complexes containing PdG-adducted DNA, showing the catalytic sites defined by Asp7, Asp105, and Glu106: (A) template sequence I, (B) template sequence II, (C) template sequence III, and (D) type II crystal with native DNA from PDB entry 1JXL (43). The three Ca2+ ions are shown as green spheres.
(b) Template II
The structure of the ternary Dpo4–DNA–dGTP complex was determined at 2.5 Å resolution (Table 1). As for the ternary complex involving template I, the Dpo4 polymerase active site resembled that in the type II structure with native DNA (43) (Figure 2B). Again, PdG stacked inside the duplex (Figures 3C,D) but did not pair with the incoming dGTP. Instead, the incoming dGTP was inserted opposite to the 5′-neighboring template dC to form a Watson–Crick base pair (Figure 3C). This resulted in a gap of 7.4 Å with respect to the 3′-terminal primer dC that was paired with the 3′-neighbor template dG. The base moieties of dGTP and the 3′-terminal dC in the primer were tilted 5.2° and 11°, respectively, with respect to PdG. This reduced the distance between the terminal 3′-hydroxyl group of the primer dC and the α-phosphate group of dGTP to 6.4 Å in the ternary complex (Figure 4B).
Three bound calcium ions were identified (Figure 5B). The first two were in the active site for catalysis and dGTP coordination. The first Ca2+ ion was 3.2 Å from the side chain carbonyl oxygen of Asp7. The first Ca2+ ion was also located 3.0 and 5.0 Å from the two α-phosphate oxygens of dGTP, 3.1 and 4.8 Å from the two β-phosphate oxygens of dGTP, and 3.3, 3.9, and 4.9 Å from the three γ-phosphate oxygens of dGTP. The second Ca2+ ion was 2.9 and 4.5 Å from the two α-phosphate oxygens of dGTP and 4.7 Å from one of the three γ-phosphate oxygens of dGTP. The third Ca2+ ion was 2.5 Å from the side chain carbonyl oxygen of Ala181 in the thumb domain of the Dpo4 polymerase. The third Ca2+ ion was also 4.1 Å from one oxygen of the α-phosphate group linking the 12th and 13th primer nucleotides.
(c) Template III
The structure of the ternary Dpo4–DNA–dATP complex was determined at 2.4 Å resolution (Table 1). Again, the active site of the polymerase resembled that in the type II structure with native DNA (Figure 2C) (43), in which PdG stacked inside the duplex (Figures 3E,F) but did not pair with the incoming dATP. Instead, the incoming dATP was inserted opposite the 5′-neighboring template T to form a Watson–Crick base pair (Figure 3E). This resulted in a 7.5 Å gap with respect to the 3′-terminal primer dC. The latter was Watson–Crick paired with the 3′-neighboring template dG. The base moieties of dATP and the 3′-terminal primer dC were tilted 2.1° and 13°, respectively, with respect to PdG, reducing the distance between the terminal 3′-hydroxyl group of the primer and the α-phosphate group to 6.4 Å (Figure 4C).
Three calcium ions were identified (Figure 5C). The first two were in the active site for catalysis and dATP coordination. The first Ca2+ ion was 2.4 Å from the side chain carbonyl oxygen of Asp7. The first Ca2+ was also 2.4 and 4.6 Å from the two α-phosphate oxygens of dATP, 2.3 and 4.9 Å from the two β-phosphate oxygens of dATP, and 2.5, 3.9, and 5.0 Å from the three γ-phosphate oxygens of dATP. The second Ca2+ ion was 3.0 Å from the side chain carbonyl oxygen of Glu106. The second Ca2+ ion was also 2.7 and 3.7 Å from the two α-phosphate oxygens of dATP, 4.6 Å from one of the two β-phosphate oxygens of dATP, and 4.4 Å from one of the three γ-phosphate oxygens of dATP. The third Ca2+ was 2.7 Å from the side chain carbonyl oxygen of Ala181 in the thumb domain of the Dpo4 polymerase. The third Ca2+ ion was also 4.3 and 4.5 Å from the two oxygens of the α-phosphate group linking the 12th and 13th primer nucleotides.
DISCUSSION
PdG provides a chemically stable structural analogue for structural and biological studies of potential mutagenic exocyclic DNA adducts. In replicative DNA polymerases, the dNTP binding pocket is tightly packed with protein residues on the minor groove side and, to some extent, on the major groove side. These steric restraints, combined with specific hydrogen bonds between amino acid side chains, the primer • template DNA, and the incoming dNTP, strongly favor the incorporation of the correct dNTP. However, because of the smaller thumb and finger subdomains, the Y-family Dpo4 DNA polymerase has been shown to lower the nucleotide selectivity and accommodate two template residues within its active site. The interactions of the Y-family DNA polymerase Dpo4 and the site-specifically modified DNA template • primer containing PdG provide a model of how 1,N2 -dG exocyclic adducts might be processed by related mammalian Y-family polymerases.
Role of the Type II Ternary Complex in Inserting dNTPs Opposite PdG
These three ternary PdG-adducted Dpo4 polymerase complexes are similar to the type II ternary complex between the Dpo4 polymerase and unmodified DNA, in which the incoming dNTP skips the first template nucleotide (in this case, the noninformational PdG nucleotide), to form a Watson–Crick base pair with the 5′-neighboring template nucleotide. Comparisons between the structures of these three ternary PdG-adducted Dpo4 polymerase complexes reveal small differences (Figure 6). The rmsd between superimposed carbon atom positions for the three ternary complexes is 0.4 Å. Figure 6A shows the superimposed structures of the Dpo4 polymerase, in the type II ternary complex with native DNA, and in all three ternary complexes investigated here. From Figure 6A, it is also apparent that the orientations of the DNA primer • template duplexes in all three Dpo4–dNTP complexes differ minimally from the one in the complex with native DNA. The structures of the DNA duplexes at the active sites in the three PdG-modified ternary complexes also differ minimally from the corresponding active site in the type II complex with native DNA (43) (Figure 6B). These structures are unlikely to represent catalytically active conformations since the 3′-hydroxyl of the primer and the α-phosphate of the dGTP remain >6 Å apart. It is unclear what rearrangements would need to occur to bring these atoms close enough so that the reaction could occur.
FIGURE 6.

Superimpositions of the structures of three ternary Dpo4—DNA—dNTP complexes with adducted and native DNA duplexes: (A) overall conformations and (B) DNA conformations at the active site. Template sequence I is colored red. Template sequence II is colored blue. Template sequence III is colored green. The native DNA (PDB entry 1JXL) is colored yellow.
PdG Remains in the anti Conformation about the Glycosyl Bond in the Type II Ternary Complexes with the Dpo4 Polymerase
The exocyclic 1,N2-PdG adduct prevents Watson–Crick hydrogen bonding interactions in DNA. Moreover, the exocyclic ring presents a bulky lesion, which is anticipated to interfere with DNA replication. Consistent with these expectations, PdG disrupted duplex DNA structure (32, 33, 36–38). At neutral pH, it has not been possible to obtain a structure for PdG positioned opposite dC in duplex DNA. The spectrum of PdG opposite dC in DNA sharpened as the pH was lowered (37, 38). Likewise, when PdG was mismatched with dA, an ordered structure was stabilized at pH 5.8 (32, 34). On the other hand, when PdG was mismatched with dG, an ordered structure was formed which was independent of pH (32). The pH-dependent equilibrium observed when PdG was placed opposite dC or dA in DNA arises from its ability to rotate into the syn conformation about the glycosyl bond. In the syn conformation, PdG can form hydrogen bonds with protonated dC and dA. When placed opposite dC, this leads to formation of a Hoogsteen pair (37).
The ability of PdG to rotate into the syn conformation, allowing formation of Hoogsteen-type interactions with protonated cytosine, has led to the question of whether this occurs during lesion bypass in damaged template • primer complexes. In all three type II ternary complexes involving PdG-damaged template • primer complexes and the Dpo4 polymerase studied here, PdG remains interhelical and the glycosyl bond remains in the anti conformation. The data suggest that the Dpo4 polymerase utilizes its large active site to insert an incoming dNTP opposite the 5′-neighboring template base. Thus, dNTP insertion is dependent upon the identity of the 5′-neighboring template base. In the ternary complexes examined here, the 5′-neighboring template base was dC for templates I and II, whereas it was T for template III. Significantly, the replication bypass experiment shown in Figure 1 indicates insertion of dGTP in the 5′-CXA-3′ template sequence similar to the crystallographic template I, which is presumed to have occurred via insertion of dGTP opposite the 5′-neighbor template dC in this sequence, and involving a ternary complex similar to that observed here for template I. Likewise, in the other two ternary complexes examined here, the incoming dNTP was dGTP for template II (5′-CXG-3′ sequence) and it was dATP for template III (5′-TXA-3′ sequence).
The question of whether the Dpo4 polymerase actively stabilizes the anti conformation of the PdG glycosyl bond is of interest. In all three ternary complexes examined here, PdG forms cross-strand stacking interactions with 5′-neighboring incipient base pair dGTP • dC (templates I and II) or dATP • T (template III) and with 3′-neighboring base pair dA • dT (template I) or dG • dC (templates II and III) (Figure 3). These stacking interactions might stabilize the anti orientation of the PdG glycosyl bond. Interestingly, it was reported that in ternary complexes site-specifically modified with 7,8-dihydro-8-oxodeoxyguanosine (8-oxodG), Arg332 of the Dpo4 polymerase assisted in stabilizing the anti conformation of the 8-oxodG glycosyl bond by means of a hydrogen bond or an ion (pole)–dipole pairing interaction, thus increasing the propensity for correct incorporation of dCTP opposite the lesion (59).
Replication Bypass of PdG by the Dpo4 Polymerase
The interactions of M1dG and OPdG with various Y-family polymerases (67, 68) are likely to be crucial in understanding the mechanisms by which these lesions induce both frameshift and base pair substitution mutations in human cells. From the biochemical and crystallographic data presented here, it is inferred that one mechanism by which the Dpo4 polymerase might bypass PdG is by accommodating both PdG and the 5′-neighboring template nucleotide within its enlarged active site, forming a type II dNTP insertion complex, utilizing the 5′-neighboring nucleotide as a templating base. Replication bypass studies (Figure 1) indicated that the Dpo4 polymerase inserted dGTP opposite PdG in the adducted template • primer duplex containing the 5′-CXA-3′ sequence context, in which insertion of dGTP presumably reflected the type II structure using the 5′-neighboring template nucleotide. However, insertion of dATP was also observed in vitro, and the observation that it was possible to crystallize ternary complexes with other dNTPs as incoming nucleotides suggests that the Dpo4 polymerase may utilize additional methods of dNTP insertion when challenged by template PdG. Hence, further in vitro primer extension and kinetic measurements are in progress.
It is of interest to note that when all four dNTPs were included in the reaction mixture, under these conditions (Figure 1), the Dpo4 polymerase did not successfully extend the primer strand to a full-length product. This possibly reflects the fact that the assays in Figure 1 were conducted at 37 °C, conditions under which this polymerase is less efficient. This contrasted with 1,N2-ethenodeoxyguanosine (1,N2-∈dG). 1,N2-∈dG is a mutagenic DNA lesion derived from lipid oxidation products and also from the chemical carcinogen vinyl chloride and is structurally similar to the PdG adduct. With 1,N2-∈dG, analysis of the products of primer extension by the Dpo4 polymerase indicated preferential incorporation of dATP opposite 3′-(1,N2-∈dG)TACT-5′ (59). This was consistent with the Dpo4 polymerase utilizing the 5′-neighboring thymine to template, rather than 1,N2-∈dG. With the template 3′-(1,N2-∈dG)CACT-5′, both dGTP and dATP were incorporated. In the presence of a mixture of all four dNTPs, opposite 3′-(1,N2-∈dG)CACT-5′, the major product was 5′-GTGA-3′ and the minor product was 5′-AGTGA-3′. Again, the major product was consistent with the Dpo4 polymerase skipping the 1,N2-∈dG template and utilizing the 5′-neighboring cytosine as a template (59). With the template 3′-(1,N2-∈dG)TACT-5′, four products were identified: 5′-AATGA-3′, 5′-ATTGA-3′, 5′-ATGA-3′, and 5′-TGA-3′.
Ternary Complexes of 1,N2-∈dG-Adducted Templates with the Dpo4 Polymerase
Structures of the Dpo4 polymerase have been obtained for binary and ternary complexes with primer templates site-specifically modified with 1,N2-∈dG (59, 69). A crystal structure of the Dpo4 polymerase was determined with a primer • template, and A was placed in the primer to be opposite 1,N2-∈dG in the template 3′-(1,N2-∈dG)TACT-5′. The added A in the primer was paired across the template T with Watson–Crick geometry. Similar structures were observed in a ternary Dpo4–DNA–dATP complex and a ternary Dpo4–DNA–ddATP complex, with d(d)ATP opposite the template T. A similar structure was observed with a ddGTP adjacent to the primer and opposite the C next to 1,N2-∈dG in 3′-(1,N2-∈dG)CACT-5′. In concert with the crystallography studies, the replication bypass studies led to the conclusion that the Dpo4 polymerase uses several mechanisms, including incorporation of dATP opposite 1,N2-∈dG and also a variation of dNTP-stabilized misalignment, to generate both base pair and frameshift mutations (59).
Generation of Frameshift Mutations by PdG
The observation that malondialdehyde is a frameshift mutagen in the hisD3052 tester strain of Salmonella typhimurium (70) originally piqued interest in PdG, a structural analogue of the malondialdehyde-induced M1dG adduct. Site-specific mutagenesis studies utilizing PdG (71–74) and M1dG (25, 26) showed that PdG and M1dG induced both base pair substitution and frameshift mutations, in a sequence-dependent manner. For example, M1dG induced −1 and −2 frameshift mutations in vivo when positioned in a reiterated (CpG)4 sequence but not when positioned in a nonreiterated sequence in E. coli and in COS-7 cells (26).
However, relationships between adduct structure and dynamics, and the formation and accommodation of transient dislocation complexes during error-prone DNA replication, are anticipated to be polymerase-specific (75). Indeed, the replication bypass of PdG in vitro depended upon the identity of the polymerase attempting to replicate past the lesion and was also sequence-dependent (76, 77). In the 5′-TXT-3′ sequence, the Klenow fragment of E. coli DNA polymerase I induced −1 base deletions in vitro, which were proposed to occur by misinsertion of dA followed by slippage of the newly inserted adenine to form the dA • dT pair with the 5′-neighbor dT, and subsequent extension (76). In contrast, when PdG was in a 5′-CXC-3′ sequence, −1 and −2 deletions were observed during in vitro lesion bypass (76). A similar result was reported by Shibutani and Grollman (72). With DNA polymerase β, PdG bypass in vitro yielded a mixture of base pair substitutions and deletions (77). Significantly, the incorporated dNTP was complementary to the base 5′ to PdG in the template strand. The replication by pol β of template • primers containing abasic sites showed similar behavior (78). The type II structures reported here for all three PdG-adducted ternary Dpo4–DNA–dNTP complexes suggest a possible mechanism for the generation of −1 bp frameshift mutations during bypass of PdG by the Dpo4 polymerase, in which the polymerase selectively utilizes the 5′-neighboring template base and skips the damage site.
Base Pair Substitution Mutagenesis by PdG and M1dG
The relationship of the structural data presented here to the formation of base pair substitutions by PdG and M1dG is less clear. Site-specific mutagenesis of PdG in E. coli showed approximately equal numbers of PdG → A transitions and PdG → T transversions when 5′-d(CGCGGTXTCCGCG)-3′ was inserted into a viral replication vector and replicated (74, 79). The mutation frequency was 2-fold greater than with M1dG, which is attributed to the fact that M1dG can rearrange to the acyclic N2-OPdG adduct, which supports Watson–Crick base pairing (80). Thus, N2-OPdG is anticipated to be less blocking than M1dG. With the Klenow fragment of DNA polymerase I in a (CpG)4 repeat sequence (80), greater bypass to full-length replication products was observed with N2-OPdG than with M1dG. With Kf exo+, the extended primers contained dC opposite both adducts, whereas with Kf exo−, primers extended past M1dG contained T opposite the adduct but primers extended past N2-OPdG contained dC opposite the adduct. Single-nucleotide incorporation experiments indicated that Kf exo− incorporated all four nucleotides opposite M1dG or N2-OPdG. Kf exo+ removed dA, dG, and T opposite M1dG and N2-OPdG but was less active when dC was opposite M1dG.
Summary
The Dpo4 polymerase may utilize a type II ternary complex to insert dNTPs when challenged by template PdG. In this scenario, the Dpo4 polymerase skips the PdG base and utilizes the 5′-neighbor template base to form a Watson–Crick hydrogen bonding interaction with the incoming dNTP. PdG remains in the anti conformation about the glycosyl bond. These data also reveal the importance of stacking for generating stable ternary Dpo4–DNA–dNTP complexes. They provide insight into how −1 frameshift mutations might be generated for PdG, a structural model for the M1dG adduct formed by malondialdehyde.
ACKNOWLEDGMENT
Dr. Ivan Kozekov synthesized the PdG-modified oligodeoxynucleotide template. Ms. Wen Xu assisted with the purification of the Dpo4 polymerase. Dr. Adriana Irimia and Dr. Joel Harp assisted with the crystal screening. Dr. Zhongmin Jin assisted with the collection of X-ray data at Argonne National Laboratory (www.aps.anl.gov).
Footnotes
This work was supported by NIH Grants ES-05355 (M.E. and M.P.S.), CA-55678 (M.P.S.), and CA-87819 (L.J.M.), the Vanderbilt Center in Molecular Toxicology (Grant ES-00267), and the Vanderbilt-Ingram Cancer Center (Grant CA-68485). Vanderbilt University and the Vanderbilt Center for Structural Biology assisted with the purchase of in-house crystallographic instrumentation. Crystallographic data were collected on the 5-ID (DND-CAT) and 22-ID beamlines of the Southeast Regional Collaborative Access Team (SER-CAT) at the Advanced Photon Source (Argonne National Laboratory, Argonne, IL). Supporting institutions may be found at www.ser-cat.org/members.html. The DND-CAT Synchrotron Research Center is supported by E. I. DuPont de Nemours & Co., the Dow Chemical Co., the National Science Foundation, and the State of Illinois. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract W-31-109-Eng-38.
Atomic coordinates and measured structure factor amplitudes for all three complexes have been deposited in the Protein Data Bank as entries 2R8G, 2R8H, and 2R8I.
Abbreviations: PdG, 1,N2-propanodeoxyguanosine; M1dG, 3-(β-d-ribofuranosyl)pyrimido[1,2-a]purin-10(3H)-one; OPdG, N2-(3-oxo-1-propenyl)deoxyguanosine; EDTA, ethylenediaminetetraacetic acid; HPLC, high-performance liquid chromatography; dNTP, nucleoside triphosphate; Dpo4, Sulfolobus solfataricus P2 DNA polymerase IV; rmsd, root-mean-square deviation.
NOTE ADDED IN PROOF
Recently, the structure of a PdG-modified template in complex with the yeast Rev1 DNA polymerase has also been reported by Aggarwal and coworkers (81). Of interest is the observation that in the Rev1 complex, template PdG was evicted from the polymerase active site and incoming dCTP was paired with a “surrogate” arginine residue. This provides an interesting precedent as to how the same damaged DNA template could be differentially bypassed by different bypass polymerases.
SUPPORTING INFORMATION AVAILABLE
Details on quality assurance (Figure S1), reference materials, sediment core data (Table S1), and OctaBDE congener patterns (Table S2 and Figure S2). This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Marnett LJ. Lipid peroxidation-DNA damage by malondialdehyde. Mutat. Res. 1999;424:83–95. doi: 10.1016/s0027-5107(99)00010-x. [DOI] [PubMed] [Google Scholar]
- 2.Marnett LJ. Chemistry and biology of DNA damage by malondialdehyde. IARC Sci. Publ. 1999;150:17–27. [PubMed] [Google Scholar]
- 3.Basu AK, Essigmann JM. Site-specifically modified oligodeoxynucleotides as probes for the structural and biological effects of DNA-damaging agents. Chem. Res. Toxicol. 1988;1:1–18. doi: 10.1021/tx00001a001. [DOI] [PubMed] [Google Scholar]
- 4.Marnett LJ, Basu AK, O’Hara SM, Weller PE, Rahman AFMM, Oliver JP. Reaction of malondialdehyde with guanine nucleosides: Formation of adducts containing oxadiazabicyclononene residues in the base-pairing region. J. Am. Chem. Soc. 1986;108:1348–1350. [Google Scholar]
- 5.Seto H, Okuda T, Takesue T, Ikemura T. Reaction of malonaldehyde with nucleic acid. I. Formation of fluorescent pyrimido [1,2-α ]purin-10(3H)-one nucleosides. Bull. Chem. Soc. Jpn. 1983;56:1799–1802. [Google Scholar]
- 6.Seto H, Seto T, Takesue T, Ikemura T. Reaction of malonaldehyde with nucleic acid. III. Studies of the fluorescent substances released by enzymatic digestion of nucleic acids modified with malonaldehyde. Chem. Pharm. Bull. 1986;34:5079–5085. [PubMed] [Google Scholar]
- 7.Reddy GR, Marnett LJ. The mechanism of reaction of β-aryloxyacroleins with nucleosides. Chem. Res. Toxicol. 1996;9:12–15. doi: 10.1021/tx950105t. [DOI] [PubMed] [Google Scholar]
- 8.Dedon PC, Plastaras JP, Rouzer CA, Marnett LJ. Indirect mutagenesis by oxidative DNA damage: Formation of the pyrimidopurinone adduct of deoxyguanosine by base propenal. Proc. Natl. Acad. Sci. U.S.A. 1998;95:11113–11116. doi: 10.1073/pnas.95.19.11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plastaras JP, Riggins JN, Otteneder M, Marnett LJ. Reactivity and mutagenicity of endogenous DNA oxopropenylating agents: Base propenals, malondialdehyde, and N∈-oxopropenyllysine. Chem. Res. Toxicol. 2000;13:1235–1242. doi: 10.1021/tx0001631. [DOI] [PubMed] [Google Scholar]
- 10.Wang MY, Liehr JG. Lipid hydroperoxide-induced endogenous DNA adducts in hamsters: Possible mechanism of lipid hydroperoxide-mediated carcinogenesis. Arch. Biochem. Biophys. 1995;316:38–46. doi: 10.1006/abbi.1995.1007. [DOI] [PubMed] [Google Scholar]
- 11.Chaudhary AK, Nokubo M, Reddy GR, Yeola SN, Morrow JD, Blair IA, Marnett LJ. Detection of endogenous malondialdehyde-deoxyguanosine adducts in human liver. Science. 1994;265:1580–1582. doi: 10.1126/science.8079172. [DOI] [PubMed] [Google Scholar]
- 12.Wang M, Dhingra K, Hittleman WN, Liehr JG, de Andrade M, Li D. Lipid peroxidation-induced putative malondialdehyde-DNA adducts in human breast tissues. Cancer Epidemiol. 1996;5:705–710. [PubMed] [Google Scholar]
- 13.Nath RG, Chung FL. Detection of 1, N2-propanodeoxyguanosine adducts in rodent and human liver DNA by 32P-postlabeling. Proc. Am. Assoc. Cancer Res. 1993;34:137. [Google Scholar]
- 14.Nath RG, Chung F-L. Detection of exocyclic 1, N2-propanodeoxyguanosine adducts as common DNA lesions in rodents and humans. Proc. Natl. Acad. Sci. U.S.A. 1994;91:7491–7495. doi: 10.1073/pnas.91.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Nair J, Barbin A, Guichard Y, Bartsch H. 1,N6-Ethenodeoxyadenosine and 3,N4-ethenodeoxycytidine in liver DNA from humans and untreated rodents detected by immunoaffinity/ 32P-postlabeling. Carcinogenesis. 1995;16:613–617. doi: 10.1093/carcin/16.3.613. [DOI] [PubMed] [Google Scholar]
- 16.Zhang S, Villalta PW, Wang M, Hecht SS. Analysis of crotonaldehyde-and acetaldehyde-derived 1,N2-propanodeoxyguanosine adducts in DNA from human tissues using liquid chromatography electrospray ionization tandem mass spectrometry. Chem. Res. Toxicol. 2006;19:1386–1392. doi: 10.1021/tx060154d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaudhary AK, Nokubo M, Marnett LJ, Blair IA. Analysis of the malondialdehyde-2′-deoxyguanosine adduct in rat liver DNA by gas chromatography/electron capture negative chemical ionization mass spectrometry. Biol. Mass Spectrom. 1994;23:457–464. doi: 10.1002/bms.1200230802. [DOI] [PubMed] [Google Scholar]
- 18.Rouzer CA, Chaudhary AK, Nokubo M, Ferguson DM, Reddy GR, Blair IA, Marnett LJ. Analysis of the malondialdehyde-2′-deoxyguanosine adduct pyrimidopurinone in human leukocyte DNA by gas chromatography/electron capture-negetiave chemical ionization/mass spectrometry. Chem. Res. Toxicol. 1997;10:181–188. doi: 10.1021/tx9601216. [DOI] [PubMed] [Google Scholar]
- 19.Vaca CE, Fang JL, Mutanen M, Valsta L. 32P-postlabelling determination of DNA adducts of malonaldehyde in humans: Total white blood cells and breast tissue. Carcinogenesis. 1995;16:1847–1851. doi: 10.1093/carcin/16.8.1847. [DOI] [PubMed] [Google Scholar]
- 20.Fang JL, Vaca CE, Valsta LM, Mutanen M. Determination of DNA adducts of malonaldehyde in humans: Effects of dietary fatty acid composition. Carcinogenesis. 1996;17:1035–1040. doi: 10.1093/carcin/17.5.1035. [DOI] [PubMed] [Google Scholar]
- 21.Sevilla CL, Mahle NH, Eliezer N, Uzieblo A, O’Hara SM, Nokubo M, Miller R, Rouzer CA, Marnett LJ. Development of monoclonal antibodies to the malondialdehyde-deoxyguanosine adduct, pyrimidopurinone. Chem. Res. Toxicol. 1997;10:172–180. doi: 10.1021/tx960120d. [DOI] [PubMed] [Google Scholar]
- 22.Chaudhary AK, Reddy RG, Blair IA, Marnett LJ. Characterization of an N6-oxopropenyl-2′-deoxyadenosine adduct in malondialdehyde-modified DNA using liquid chromatography/electrospray ionization tandem mass spectrometry. Carcinogenesis. 1996;17:1167–1170. doi: 10.1093/carcin/17.5.1167. [DOI] [PubMed] [Google Scholar]
- 23.Hoberg AM, Otteneder M, Marnett LJ, Poulsen HE. Measurement of the malondialdehyde-2′-deoxyguanosine adduct in human urine by immuno-extraction and liquid chromatography/atmospheric pressure chemical ionization tandem mass spectrometry. J. Mass Spectrom. 2004;39:38–42. doi: 10.1002/jms.547. [DOI] [PubMed] [Google Scholar]
- 24.Otteneder MB, Knutson CG, Daniels JS, Hashim M, Crews BC, Remmel RP, Wang H, Rizzo C, Marnett LJ. In vivo oxidative metabolism of a major peroxidation-derived DNA adduct, M1dG. Proc. Natl. Acad. Sci. U.S.A. 2006;103:6665–6669. doi: 10.1073/pnas.0602017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fink SP, Reddy GR, Marnett LJ. Mutagenicity in Escherichia coli of the major DNA adduct derived from the endogenous mutagen malondialdehyde. Proc. Natl. Acad. Sci. U.S.A. 1997;94:8652–8657. doi: 10.1073/pnas.94.16.8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.VanderVeen LA, Hashim MF, Shyr Y, Marnett LJ. Induction of frameshift and base pair substitution mutations by the major DNA adduct of the endogenous carcinogen malondialdehyde. Proc. Natl. Acad. Sci. U.S.A. 2003;100:14247–14252. doi: 10.1073/pnas.2332176100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niedernhofer LJ, Daniels JS, Rouzer CA, Greene RE, Marnett LJ. Malondialdehyde, a product of lipid peroxidation, is mutagenic in human cells. J. Biol. Chem. 2003;278:31426–31433. doi: 10.1074/jbc.M212549200. [DOI] [PubMed] [Google Scholar]
- 28.Mao H, Schnetz-Boutaud NC, Weisenseel JP, Marnett LJ, Stone MP. Duplex DNA catalyzes the chemical rearrangement of a malondialdehyde deoxyguanosine adduct. Proc. Natl. Acad. Sci. U.S.A. 1999;96:6615–6620. doi: 10.1073/pnas.96.12.6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riggins JN, Daniels JS, Rouzer CA, Marnett LJ. Kinetic and thermodynamic analysis of the hydrolytic ring-opening of the malondialdehyde-deoxyguanosine adduct, 3-(2′-deoxy-β-D-erythro-pentofuranosyl)-pyrimido[1,2-α ]purin-10(3H)-one. J. Am. Chem. Soc. 2004;126:8237–8243. doi: 10.1021/ja040009r. [DOI] [PubMed] [Google Scholar]
- 30.Riggins JN, Pratt DA, Voehler M, Daniels JS, Marnett LJ. Kinetics and mechanism of the general-acid-catalyzed ring-closure of the malondialdehyde-DNA adduct N2-(3-oxo-1-propenyl)deoxyguanosine (N 2OPdG-), to 3-(2′-deoxy-β-D-erythro-pentofuranosyl)pyrimido[1,2-α]purin-10(3H)-one (M1dG) J. Am. Chem. Soc. 2004;126:10571–10581. doi: 10.1021/ja040010q. [DOI] [PubMed] [Google Scholar]
- 31.Marinelli ER, Johnson F, Iden CR, Yu PL. Synthesis of 1,N2-(1,3-propano)-2′-deoxyguanosine and incorporation into oligodoexynucleotides: A model for exocyclic acrolein-DNA adducts. Chem. Res. Toxicol. 1990;3:49–58. doi: 10.1021/tx00013a009. [DOI] [PubMed] [Google Scholar]
- 32.Kouchakdjian M, Marinelli E, Gao X, Johnson F, Grollman A, Patel D. NMR studies of exocyclic 1, N 2-propan-odeoxyguanosine adducts (X) opposite purines in DNA duplexes: Protonated X(syn):A(anti) pairing (acidic pH) and X(syn):G(anti) pairing (neutral pH) at the lesion site. Biochemistry. 1989;28:5647–5657. doi: 10.1021/bi00439a047. [DOI] [PubMed] [Google Scholar]
- 33.Kouchakdjian M, Eisenberg M, Live D, Marinelli E, Grollman AP, Patel DJ. NMR studies of an exocyclic 1,N2-propanodeoxyguanosine adduct (X) located opposite deoxyadenosine (A) in DNA duplexes at basic pH: Simultaneous partial intercalation of X and A between stacked bases. Biochemistry. 1990;29:4456–4465. doi: 10.1021/bi00470a028. [DOI] [PubMed] [Google Scholar]
- 34.Huang P, Eisenberg M. The three-dimensional structure in solution (pH 5.8) of a DNA 9-mer duplex containing 1, N2-propanodeoxyguanosine opposite deoxyadenosine. Restrained molecular dynamics and NOE-based refinement calculations. Biochemistry. 1992;31:6518–6532. doi: 10.1021/bi00143a023. [DOI] [PubMed] [Google Scholar]
- 35.Huang P, Patel DJ, Eisenberg M. Solution structure of the exocyclic 1, N 2-propanodeoxyguanosine adduct opposite deoxyadenosine in a DNA nonamer duplex at pH 8.9. Model of pH-dependent conformational transition. Biochemistry. 1993;32:3852–3866. doi: 10.1021/bi00066a004. [DOI] [PubMed] [Google Scholar]
- 36.Kouchakdjian M, Eisenberg M, Johnson F, Grollman AP, Patel DJ. Structural features of an exocyclic adduct positioned opposite an abasic site in a DNA duplex. Biochemistry. 1991;30:3262–3270. doi: 10.1021/bi00227a014. [DOI] [PubMed] [Google Scholar]
- 37.Singh US, Moe JG, Reddy GR, Weisenseel JP, Marnett LJ, Stone MP. 1H NMR of an oligodeoxynucleotide containing a propanodeoxyguanosine adduct positioned in a (CG)3 frameshift hotspot of Salmonella typhimurium hisD3052: Hoogsteen base-pairing at pH 5.8. Chem. Res. Toxicol. 1993;6:825–836. doi: 10.1021/tx00036a012. [DOI] [PubMed] [Google Scholar]
- 38.Weisenseel JP, Reddy GR, Marnett LJ, Stone MP. Structure of an oligodeoxynucleotide containing a 1, N2-propanodeoxyguanosine adduct positioned in a palindrome derived from the Salmonella typhimurium hisD3052 gene: Hoogsteen pairing at pH 5.2. Chem. Res. Toxicol. 2002;15:127–139. doi: 10.1021/tx0101090. [DOI] [PubMed] [Google Scholar]
- 39.Moe JG, Reddy GR, Marnett LJ, Stone MP. 1H NMR characterization of a duplex oligodeoxynucleotide containing propanodeoxyguanosine opposite a two-base deletion in the (CpG)3 frameshift hotspot of Salmonella typhimurium hisD3052. Chem. Res. Toxicol. 1994;7:319–328. doi: 10.1021/tx00039a008. [DOI] [PubMed] [Google Scholar]
- 40.Weisenseel JP, Moe JG, Reddy GR, Marnett LJ, Stone MP. Structure of a duplex oligodeoxynucleotide containing propanodeoxyguanosine opposite a two-base deletion in the (CpG)3 frameshift hotspot of Salmonella typhimurium hisD3052 determined by 1H NMR and restrained molecular dynamics. Biochemistry. 1995;34:50–64. doi: 10.1021/bi00001a007. [DOI] [PubMed] [Google Scholar]
- 41.Plum GE, Grollman AP, Johnson F, Breslauer KJ. Influence of an exocyclic guanine adduct on the thermal stability, conformation, and melting thermodynamics of a DNA duplex. Biochemistry. 1992;31:12096–12102. doi: 10.1021/bi00163a019. [DOI] [PubMed] [Google Scholar]
- 42.Weisenseel JP, Reddy GR, Marnett LJ, Stone MP. Structure of the 1, N 2-propanodeoxyguanosine adduct in a three-base DNA hairpin loop derived from a palindrome in the Salmonella typhimurium hisD3052 gene. Chem. Res. Toxicol. 2002;15:140–152. doi: 10.1021/tx010107f. [DOI] [PubMed] [Google Scholar]
- 43.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: A mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 44.Silvian LF, Toth EA, Pham P, Goodman MF, Ellenberger T. Crystal structure of a DinB family error-prone DNA polymerase from Sulfolobus solfataricus. Nat. Struct. Biol. 2001;8:984–989. doi: 10.1038/nsb1101-984. [DOI] [PubMed] [Google Scholar]
- 45.Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Replication by human DNA polymerase-𝜄 occurs by Hoogsteen base-pairing. Nature. 2004;430:377–380. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- 46.Boudsocq F, Iwai S, Hanaoka F, Woodgate R. Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): An archaeal DinB-like DNA polymerase with lesion-bypass properties akin to eukaryotic pol eta. Nucleic Acids Res. 2001;29:4607–4616. doi: 10.1093/nar/29.22.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 48.Johnson RE, Prakash S, Prakash L. The human DINB1 gene encodes the DNA polymerase pol θ. Proc. Natl. Acad. Sci. U.S.A. 2000;97:3838–3843. doi: 10.1073/pnas.97.8.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gerlach VL, Feaver WJ, Fischhaber PL, Friedberg EC. Purification and characterization of pol κ, a DNA polymerase encoded by the human DINB1 gene. J. Biol. Chem. 2001;276:92–98. doi: 10.1074/jbc.M004413200. [DOI] [PubMed] [Google Scholar]
- 50.Frank EG, Sayer JM, Kroth H, Ohashi E, Ohmori H, Jerina DM, Woodgate R. Translesion replication of benzo[a]pyrene and benzo[c]phenanthrene diol epoxide adducts of deoxyadenosine and deoxyguanosine by human DNA polymerase 𝜄. Nucleic Acids Res. 2002;30:5284–5292. doi: 10.1093/nar/gkf643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Yuan F, Wu X, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z. Error-prone lesion bypass by human DNA polymerase η. Nucleic Acids Res. 2000;28:4717–4724. doi: 10.1093/nar/28.23.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ohashi E, Ogi T, Kusumoto R, Iwai S, Masutani C, Hanaoka F, Ohmori H. Error-prone bypass of certain DNA lesions by the human DNA polymerase κ. Genes Dev. 2000;14:1589–1594. [PMC free article] [PubMed] [Google Scholar]
- 53.Washington MT, Minko IG, Johnson RE, Wolfle WT, Harris TM, Lloyd RS, Prakash S, Prakash L. Efficient and error-free replication past a minor-groove DNA adduct by the sequential action of human DNA polymerases ι and κ. Mol. Cell. Biol. 2004;24:5687–5693. doi: 10.1128/MCB.24.13.5687-5693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Washington MT, Minko IG, Johnson RE, Haracska L, Harris TM, Lloyd RS, Prakash S, Prakash L. Efficient and error-free replication past a minor-groove N2-guanine adduct by the sequential action of yeast Rev1 and DNA polymerase ζ. Mol. Cell. Biol. 2004;24:6900–6906. doi: 10.1128/MCB.24.16.6900-6906.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang X, Kolbanovskiy A, Wu X, Zhang Y, Wang Z, Zhuang P, Amin S, Geacintov NE. Effects of base sequence context on translesion synthesis past a bulky (+)-trans-anti-B[a]P-N2-dG lesion catalyzed by the Y-family polymerase pol κ. Biochemistry. 2003;42:2456–2466. doi: 10.1021/bi026912q. [DOI] [PubMed] [Google Scholar]
- 56.Rechkoblit O, Zhang Y, Guo D, Wang Z, Amin S, Krzeminsky J, Louneva N, Geacintov NE. trans-Lesion synthesis past bulky benzo[a]pyrene diol epoxide N2-dG and N6-dA lesions catalyzed by DNA bypass polymerases. J. Biol. Chem. 2002;277:30488–30494. doi: 10.1074/jbc.M201167200. [DOI] [PubMed] [Google Scholar]
- 57.Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase η. Nat Genet. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 58.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 59.Zang H, Goodenough AK, Choi JY, Irimia A, Loukachevitch LV, Kozekov ID, Angel KC, Rizzo CJ, Egli M, Guengerich FP. DNA adduct bypass polymerization by Sulfolobus solfataricus DNA polymerase Dpo4: Analysis and crystal structures of multiple base pair substitution and frameshift products with the adduct 1, N2-ethenoguanine. J. Biol. Chem. 2005;280:29750–29764. doi: 10.1074/jbc.M504756200. [DOI] [PubMed] [Google Scholar]
- 60.Cavaluzzi MJ, Borer PN. Revised UV extinction coefficients for nucleoside-5′-monophosphates and unpaired DNA and RNA. Nucleic Acids Res. 2004;32:13. doi: 10.1093/nar/gnh015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kabsch W. Evaluation of single-crystal X-ray-diffraction data from a position-sensitive detector. J. Appl. Crystallogr. 1988;21:916–924. [Google Scholar]
- 62.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 63.French S, Wilson K. Treatment of negative intensity observations. Acta Crystallogr. 1978;A34:517–525. [Google Scholar]
- 64.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 65.Vellieux FMD, Dijkstra BW. Computation of Bhat’s OMIT maps with different coefficients. J. Appl. Crystallogr. 1997;30:396–399. [Google Scholar]
- 66.Cambillau C, Roussel A. TURBO-FRODO, version OpenGL.1. Marseille, France: Université Aix-Marseille II; 1997. [Google Scholar]
- 67.Tippin B, Pham P, Goodman MF. Error-prone replication for better or worse. Trends Microbiol. 2004;12:288–295. doi: 10.1016/j.tim.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 68.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 69.Irimia A, Zang H, Loukachevitch LV, Eoff RL, Guengerich FP, Egli M. Calcium is a cofactor of polymerization but inhibits pyrophosphorolysis by the Sulfolobus solfataricus DNA polymerase Dpo4. Biochemistry. 2006;45:5949–5956. doi: 10.1021/bi052511+. [DOI] [PubMed] [Google Scholar]
- 70.O’Hara SM, Marnett LJ. DNA sequence analysis of spontaneous and β-methoxy-acrolein-induced mutations in Salmonella typhimurium hisD3052. Mutat. Res. 1991;247:45–56. doi: 10.1016/0027-5107(91)90032-j. [DOI] [PubMed] [Google Scholar]
- 71.Benamira M, Singh U, Marnett LJ. Site-specific frameshift mutagenesis by a propanodeoxyguanosine adduct positioned in the (CpG)4 hot-spot of Salmonella typhimurium hisD3052 carried on an M13 vector. J. Biol. Chem. 1992;267:22392–22400. [PubMed] [Google Scholar]
- 72.Shibutani S, Grollman AP. On the mechanism of frameshift (deletion) mutagenesis in vitro. J. Biol. Chem. 1993;268:11703–11710. [PubMed] [Google Scholar]
- 73.Moriya M, Zhang W, Johnson F, Grollman AP. Mutagenic potency of exocyclic DNA adducts: Marked differences between Escherichia coli and simian kidney cells. Proc. Natl. Acad. Sci. U.S.A. 1994;91:11899–11903. doi: 10.1073/pnas.91.25.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Burcham PC, Marnett LJ. Site-specific mutagenesis by a propanodeoxyguanosine adduct carried on an M13 genome. J. Biol. Chem. 1994;269:28844–28850. [PubMed] [Google Scholar]
- 75.Tippin B, Kobayashi S, Bertram JG, Goodman MF. To slip or skip, visualizing frameshift mutation dynamics for error-prone DNA polymerases. J. Biol. Chem. 2004;279:45360–45368. doi: 10.1074/jbc.M408600200. [DOI] [PubMed] [Google Scholar]
- 76.Hashim MF, Marnett LJ. Sequence-dependent induction of base pair substitutions and frameshifts by propanodeoxyguanosine during in vitro DNA replication. J. Biol. Chem. 1996;271:9160–9165. doi: 10.1074/jbc.271.15.9160. [DOI] [PubMed] [Google Scholar]
- 77.Hashim MF, Schnetz-Boutaud N, Marnett LJ. Replication of template-primers containing propanodeoxyguanosine by DNA polymerase β. Induction of base pair substitution and frameshift mutations by template slippage and deoxynucleoside triphosphate stabilization. J. Biol. Chem. 1997;272:20205–20212. doi: 10.1074/jbc.272.32.20205. [DOI] [PubMed] [Google Scholar]
- 78.Efrati E, Tocco G, Eritja R, Wilson SH, Goodman MF. Abasic translesion synthesis by DNA polymerase β violates the ’A-rule′. Novel types of nucleotide incorporation by human DNA polymerase β at an abasic lesion in different sequence contexts. J. Biol. Chem. 1997;272:2559–2569. doi: 10.1074/jbc.272.4.2559. [DOI] [PubMed] [Google Scholar]
- 79.Fink SP, Reddy GR, Marnett LJ. Relative contribution of cytosine deamination and error-prone replication to the induction of propanodeoxyguanosine to deoxyadenosine mutations in Escherichia coli. Chem. Res. Toxicol. 1996;9:277–283. doi: 10.1021/tx950060w. [DOI] [PubMed] [Google Scholar]
- 80.Hashim MF, Riggins JN, Schnetz-Boutaud N, Voehler M, Stone MP, Marnett LJ. In vitro bypass of malondialdehyde-deoxyguanosine adducts: Differential base selection during extension by the Klenow fragment of DNA polymerase I is the critical determinant of replication outcome. Biochemistry. 2004;43:11828–11835. doi: 10.1021/bi049360f. [DOI] [PubMed] [Google Scholar]
- 81.Nair DK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structure. 2008;16:239–245. doi: 10.1016/j.str.2007.12.009. [DOI] [PubMed] [Google Scholar]


