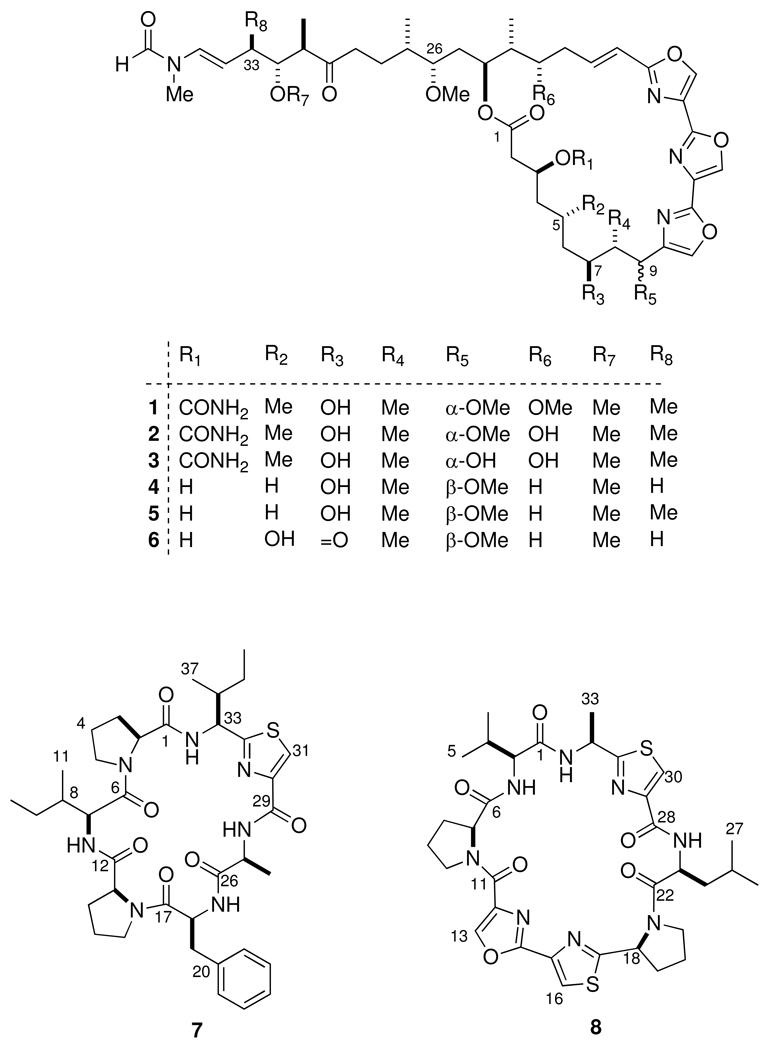
Abstract
A single specimen of Hexabranchus sanguineus, a nudibranch from the Indo-Pacific that is known to sequester kabiramides B, C and other trisoxazole macrolides, yielded new kabiramide analogs – 9-desmethylkabiramide B and 33-methyltetrahydrohalichondramide – and two new unexpected thiazole-containing cyclic peptides in sub-micromole amounts. The structures of these cyclic peptides were determined by analyses of 1D and 2D NMR spectra recorded with a state-of-the-art 1-mm 1H NMR high-temperature superconducting micro-cryoprobe, together with mass spectra. In addition to two proline residues, each peptide contains a thiazole- or oxazole-modified amino acid residue, together with conventional amino acid residues. All of the amino acid residues were L- as determined by Marfey’s analysis of the acid hydrolysates of the peptides. This is the first report of cyclic thiazole peptides from H. sanguineus. Since thiazole-oxazole modified peptides are typically associated with cyanobacteria and tunicates, the finding may imply a dietary component of the H. sanguineus that was previously overlooked.
The Indo-Pacific nudibranch Hexabranchus sanguineus (a shell-less opistobranch mollusc) and its egg masses contain extraordinary bioactive polyketide macrolide natural products known as ‘trisoxazole’ macrolides. The three contiguous 2,4-disubstituted oxazole rings, which constitute the outstanding structural feature of trisoxazole macrolides, are integrated within a larger macrolide ring that is further appended by an aliphatic chain terminated by an N-methyl-N-vinylformamide. The first examples of trisoxazoles, ulapualides A and Bi and kabiramide C (1), were reported in 1986,ii however, a subsequent report described the related analog halichondramide from the sponge Halichondria sp. collected in Palau.iii At about the same time, dihydrohalichondramide and tetrahydrohalichondramide were found in specimens of H. sanguineus, iv collected in Kwajalein atoll, and also in egg-masses laid by live individuals in captivity. Like many dorid nudibranchs, H. sanguineus is a specialist predator with a spongiverous diet; in an aquarium, the nudibranch was found to consume only Halichondria containing trisoxazole macrolides and not sponges of other species or genera.v These observations provided strong evidence that H. sanguineus acquires its suite of trisoxazole compounds from a selective sponge diet, and the first confirmatory evidence of the passage of trisoxazoles macrolides from sponge to nudibranch to progeny.
Trisoxazole macrolides exhibit extremely potent antifungal and cytotoxic activities – properties that appear to correlate with their tight binding to G-actin, depolymerization of F-actin and disruption of actin filament formation and organization.vi Finally, trisoxazoles are concentrated in tissue in Hexabranchus at levels higher than those found in the sponge and provide the animal with a potent chemical defense against crustaceans and fish. For example, the common Pacific wrasse Thalasomma lunare was deterred from consuming krill pellets that were treated with trisoxazoles at concentrations as low as 10 ppm.v
Since 1986, over two dozen analogs, including kabiramide B (2)vii have been described from H. sanguineus,viii and other sponges of the genera Jaspis,ix Mycale,x and Pachastrissa.xi Our own investigations of the extract from a single specimen of H. sanguineus collected in Fiji uncovered 1ii and 2vii as the major antifungal metabolites, however at that time NMR instrumentation was insufficient to allow investigation of the minor constituents and the extract fractions was archived (−20 °C) for almost two decades. In 2007–8, we gained access to two 600 MHz NMR spectrometers highly sensitive probes – a custom built 1-mm high-temperature superconducting (HTS) NMR cryoprobexii (National High Magnetic Field Laboratory) and a commercial 1.7-mm cryoprobe – which allowed full characterization of the very minor components of H. sanguineus. The HTS cryoprobe was constructed with four Helmholtz pairs (1H, 2H, 13C, and 15N) of planar sapphire supporting plates that were coated with a thin film of YBCO (yttrium-barium-copper oxide). The HTS coils and conventional cryoprobe preamplifier components are cooled to 20 K using a commercial cryogenic cooling system.xii The combination of high intrinsic coil sensitivity, low noise, and very small fill volume (~5–10 µL) gives 1H NMR spectra with exceptional mass-sensitivity and opens investigations of new natural products with complex structures from rare sources where only as little as a few nanomoles may be available.xiii In this report, we describe our application of the unmatched sensitivity micro-cryoprobes to elucidate the structures of the very minor natural products from a single specimen of H. sanguineus: two new trisoxazole analogs, 9-O-desmethyl kabiramide B (3) and 33-methyl tetrahydrohalichondramide (5) together with two unexpected thiazole-containing peptides, that we named sanguinamides A (7) and B (8). These complex structures were fully characterized on sub-micromole samples by 1D and 2D 1H and 13C NMR data together with MS and degradative correlation.
Results and Discussion
A single specimen of Hexabranchus sanguineus was collected by hand in Fiji (1987) in the Yasawa Island chain. The deep-red acetone extract of H. sanguineus was partitioned between ethyl acetate and water. The ethyl acetate soluble portion was fractionated by silica chromatography followed by C18 reversed-phase chromatography to give the new compounds 9-O-desmethylkabiramide B (3, 0.04% w/w dry weight), 33-methyltetrahydrohalichondramide (5, 0.07%), and sanguinamides A (7, 0.023% ) and B (8, 0.011%) in addition to the known compounds 1 (2.2%),ii 2vii (0.12%), kabiramide Dvii (0.24%) and 33-methyldihydrohalichondramidevii (0.28%).
Compound 3 was shown to be the homolog of 2 with the formula C46H67N5O14 established by HRESIMS (m/z 936.4574, [M+Na]+) and mostly likely arising from loss of a C-methyl or an O-methyl group. As with all trisoxazoles, the NMR spectra were observed as slowly interconverting rotamers (~2:1) due to restricted rotation about the N-methyl-formamide bond. The 1H NMR spectrum of 3 was similar to that of 2 except only two OMe singlets (δ 3.28, s; 3.29, s) were present instead of three. The COSY, HMBC and HSQC data indicated that 3 had the same carbon framework as 2 including the characteristic 1H NMR singlets for the three oxazole rings (δ 7.72, s, H-11; 8.32, s H-14; 8.27, s, H-17). Since 2 has OMe groups at C-9, C-26 and C-32, one of these must be replaced by an OH group in 3. The HMBC and HSQC data for 3 showed that the position of the new OH group was at C-9 because the two OMe signals (δ 3.28, s; 3.29, s) were located unambiguously at C-26 (δ 56.8, q) and C-32 (δ 60.3, q) through long-range correlations. The remaining 1H NMR signals of 3 were essentially identical to those of 2.
Compound 5 has a molecular formula of C45H66N4O12 as determined by HRESIMS (m/z 855.4738 [M+H]+) with one CH2 unit more than tetrahydrohalichondramide (4).iv The 1H NMR data of 5 revealed five C-methyl signals (δ 1.02, d, J = 6.8 Hz; 0.87, d, J = 7.2 Hz; 0.84, d, J = 7.2 Hz; 0.92, d, J = 6.8 Hz and 1.16, d, J = 6.8 Hz) corresponding to C-8-Me, C-23-Me, C-27-Me, C-31-Me and C-33-Me, respectively. The 1H NMR signal due to H-33 in 5 appeared as a multiplet (δ 2.38, m) and corresponds to replacement by CH for the two diastereotopic CH2 signals in 4 (δ 2.50, m, 1H; 2.17, m, 1H). Careful analysis of COSY and TOCSY spectra revealed correlations of the C-33-Me signal (δ1.16, d, J = 6.8 Hz) to H-32, H-33, H-34 and H-35 and located the additional C-methyl group at C-33. Thus, 5 has the structure 33-methyltetrahydrohalichondramide.
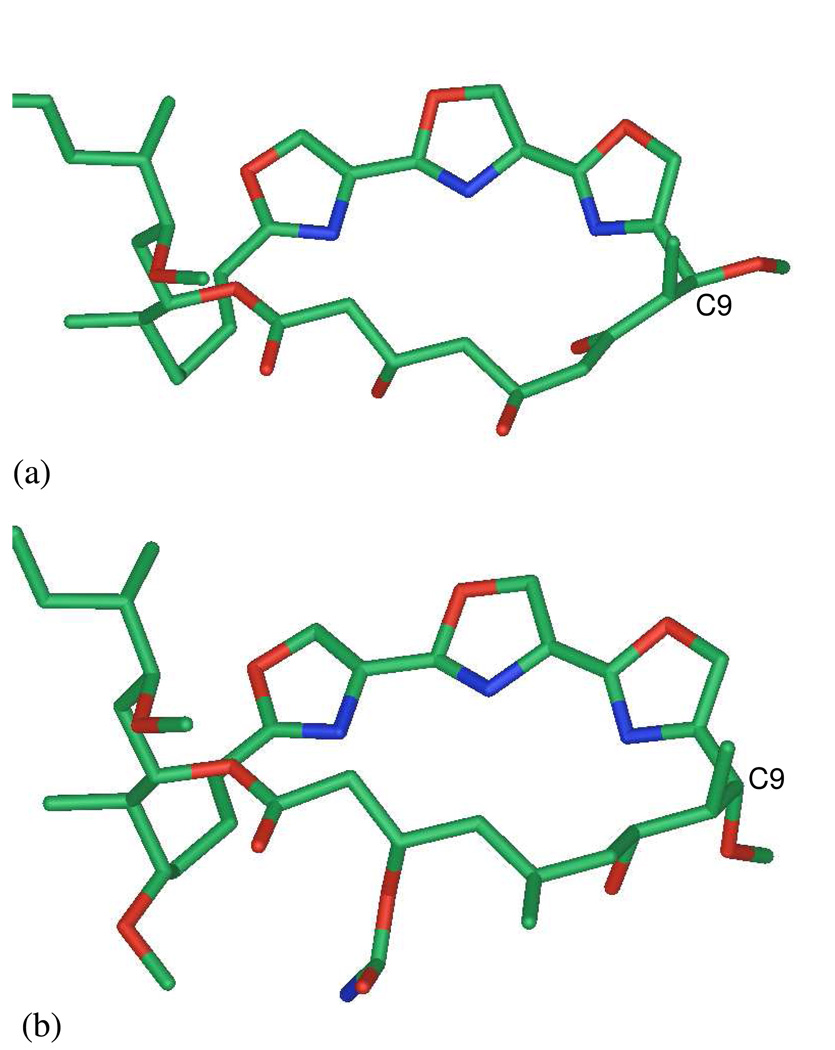
The absolute configurations of 3 and 5 were assigned by comparisons to the trisoxazoles kabiramide C (1) and jaspisamide A (6) and deductions based on conformational analysis and 1H-1H J coupling. The absolute configuration of several of the trisoxazoles have been assigned by a separate groups using different approaches. Panek and coworkers assigned mycalolide A by degradation, synthesis of degradation products and, ultimately, the natural product.xiv None of the trisoxazole macrolides have delivered crystals suitable for X-ray analysis, however the excellent work of Rayment and coworkers provided unambiguous absolute configurations for trisoxazoles by separate single-crystal X-ray analysesvi of G-actin bound with 1, jaspisamide A (6),ix halichondramideiv,v and ulapualide.xvb Not only did this reveal the relationship of the latter natural products to the marine-derived macrolides reidispongiolides and sphinxolides, 14a but this milestone work served to remove stereochemical ambiguities and correctedxvb an anomalous assignmentxvi of at least one trisoxazole macrolide based on the ‘facts and fantasies’xvii of untested metal-chelation hypotheses.xviii
The C-9 configuration changes in different trisoxazolesvi and the C-9-OMe substitutent may be α– or β–, however the configuration of other methyl-branched stereocenters in trisoxazole families appear to be invariant. For reasons of optimum deuterium-lock strength, the 1H NMR spectra of 3 was recorded in CD3CN (600 MHz, 1-mm HTS probe), however NMR data for other trisoxazolesviii have been mostly reported in CDCl3. Comparisons based on chemical shift alone may compromise arguments for the C-9 relative configuration in 3. For example, the polar H-bond acceptor solvent CD3CN may coordinate the free OH groups and influence the local conformation and electronic environment around the 1,3-diol moiety and chemical shifts with respect to 2. In order to resolve this problem, we examined the X-ray structuresvi of jaspisamide A (6) and kabiramide C (1) that share similar C-6–C-9 segments but have epimeric C-9 configurations. Although both X-ray structures were determined for G-actin bound trisoxazoles, the macrolide ring segments show minimal contacts to protein residues and their conformations are likely to be similar to the solution structures. The ring conformations of 1 and 6 (Figure 1) are remarkably similar; the three contiguous oxazole rings form a planar wall, tilted approximately at an angle of ~22° to the averaged macrolide ring plane defined by the ester C=O group and the extended (zig-zag) C-2–C-9 chain. In both molecules, the C-7-O bond of the oxygen substituent (OH in 1 and C=O in 6) points endo to the macrolide ring and the C-8-Me bond is perpendicular to the average macrolide plane. The major difference between the two molecules is the orientation of the C-9 OMe group. In jaspisamide A (6), the β-OMe group lies pseudo-equatorial, almost in-plane with the macrolide ring, but in kabiramide C (1), C-9 has the epimeric configuration with an pseudo-axial α-OMe substitutent almost anti to the C-8 methyl group. The corresponding dihedral angles H-8–C-8–C-9–H-9 for 1 and 6 are −78° and +68°, respectively, and give rise to an H-9 1H NMR signal that is a broad singlet (δ 5.19, bs) for 1 and a doublet for 6 is (δ 4.98, d, J = 4.4 Hz).ix Since the vicinal coupling constant between H-8 and H-9 in 3 is very small (J ~ 0 Hz), this compound must have the same C-9 α-OMe configuration found in 1. Using similar arguments, the J values and chemical shifts 4 and 5 were found to be similar to those of halichondramide,iii,iv and we tentatively assigned the C-9 β-OMe configuration to 5, as depicted here.
Figure 1.
X-ray structures of trisoxazoles bound to G-actin (data adapted from Rayment et al. Ref vi. (a) jaspisamide A (6): torsional angles C8(Me)-C8-C9-C10 = −57.4°, H8-C8-C9-H9 =+67.9° (b) kabiramide C (1): torsional angles C8(Me)-C8-C9-C10 = −67.3°, H8-C8-C9-H9 =+75.8° Side chains of each compound (upper left) and protein residues have been removed for clarity
Further examination of the minor components of H. sanguineus gave, unexpectedly, two new peptides 7 and 8. Sanguinamide A (7) was isolated as a minor component from a fraction containing the trisoxazole compounds. HRESIMS gave the formula C37H52N7O6S (m/z 722.3683 [M+H]+) with 16 degrees of unsaturation. The 1H NMR data of 7 revealed signals typical of a peptide; four amide NH proton signals (Table 1, δ 9.19, d, J = 7.2 Hz; 8.78, d, J = 7.2 Hz; 8.11, d, J = 7.8 Hz; 6.11, bs) and six α-CH signals (δ 3.38 d, J = 7.6 Hz, H-13; 4.22, ddd, J = 10.1, 5.8, 1.7 Hz, H-18; 4.26, dd, J = 10.0, 7.2 Hz, H-7; 4.67, d, J = 7.9 Hz, H-2; 4.69, qd, J = 7.8, 6.4 Hz, H-27; 5.09, t, J = 7.2 Hz, H-33). HSQC showed a downfield broad 1H singlet (δ 8.01, s, H-31) coupled to a 13C signal (δ 123.1, d, C-31) characteristic of C-4 in a 2-substituted thiazole-4-carboxamide. This was verified by HMBC data that showed correlations from H-31 to other thiazole ring carbon signals at δ 148.5, 160.3 and 168.1.
Table 1.
NMR data of sanguinamide A, 7 (600 MHz, CDCl3)
| # | δCa, mult. | δH, mult. (J in Hz) | HMBC b | |
|---|---|---|---|---|
| L-Pro-1 | ||||
| 1 | 170.6, qC | |||
| 2 | 60.8, CH | 4.67, d (7.9) | 1, 6 | |
| 3 | 30.5, CH2 | 1.81, m | 1, 4, 5 | |
| 2.68, dd (12.0, 5.2) | ||||
| 4 | 25.0, CH2 | 1.90, 2.02, m | 2 | |
| 5 | 48.1, CH2 | 3.67, 4.00, m | 4 | |
| L-Ile-1 | ||||
| 6 | 174.5, qC | |||
| 7 | 56.3, CH | 4.26, dd (10.0, 7.2) | 6, 8, 9, 12 | |
| 8 | 35.5, CH | 2.30, m | ||
| 9 | 25.3, CH2 | 1.24, 1.70, m | 8, 10 | |
| 10 | 15.1, CH3 | 1.02, d (6.8) | 7, 8, 9 | |
| 11 | 10.3, CH3 | 0.91, t (7.6) | 8, 9 | |
| NH | 9.19, d (7.2) | 12 | ||
| L-Pro-2 | ||||
| 12 | 171.50, qC | |||
| 13 | 60.9, CH | 3.38, d (7.6) | 12, 17 | |
| 14 | 30.4, CH2 | 0.93, | 12, 16 | |
| m 2.10, dd (12.2, 6.1) | ||||
| 15 | 22.0, CH2 | 1.70, 1.50, m | 13 | |
| 16 | 46.3, CH2 | 3.50, 3.53, m | 15 | |
| L-Phe | ||||
| 17 | 169.2, qC | |||
| 18 | 54.5, CH | 4.22, ddd (10.1, 5.8, 1.7) | 19, 26 | |
| 19 | 37.9, CH2 | 2.98, dd (11.9, 5.8) | 17 | |
| 3.08, (11.9, 11.9) | ||||
| 20 | 134.3, qC | |||
| 21 | 129.3, CH | 7.16, d (7.10) | 20, 22, 23 | |
| 22 | 127.8, CH | 7.24, m | ||
| 23 | 129.1, CH | 7.28, m | ||
| NH | 6.11, br s | |||
| L-Ala | ||||
| 26 | 171.0, qC | |||
| 27 | 49.1, CH | 4.69, qd (7.8, 6.4) | 26, 28, 29 | |
| 28 | 17.9, CH3 | 1.48, d (6.4) | 26, 27 | |
| NH | 8.11, d (7.8) | 26, 29 | ||
| Thz | ||||
| 29 | 160.3, qC | |||
| 30 | 148.5, qC | - | ||
| 31 | 123.1, CH | 8.01, s | 29, 30, 32 | |
| 32 | 168.1, qC | |||
| L-Ile-2 | ||||
| 33 | 56.3, CH | 5.09, dd (7.2, 7.2) | 1, 32, 34, 35 | |
| 34 | 40.9, CH | 1.80, m | 35 | |
| 35 | 25.9, CH2 | 1.17, 1.50, m | 34, 36 | |
| 36 | 15.0, CH3 | 0.77, d (6.7) | 33, 34, 35 | |
| 37 | 10.3, CH3 | 0.91, t (7.6) | 34, 35 | |
| NH | 8.78, d (7.2) | 1, 32 |
δ’s measured by indirect detection, HSQC and HMBC (1JCH= 6 Hz)
1H->13C.
The identities of six remaining amino acid residues – two Pro, two Ile, Ala and Phe – were revealed by analysis of COSY, TOCSY, HSQC and HMBC data (Table 1). Therefore, sanguinamide A (7) is a modified heptapeptide. The amino acid sequence of 7 was defined from the HMBC correlations between the α-CH and amide-NH proton signals with the adjacent C=O group. An HMBC correlation from the α-CH of Pro-1 (H-2) and the C=O of Ile-1 (δ 174.5, s, C-6). The amide NH proton of Ile-1 (δ 9.19) showed a correlation to -CO of Pro-2 (δ 171.5, s, C-12). In turn, the α-proton of Pro-2 (H-13) showed a cross peak to the C=O of Phe (δ 169.2, s, C-17), and the α-CH signal of Phe (H-18) showed connectivity to the C=O group of Ala (δ 171.0, s, C-26).
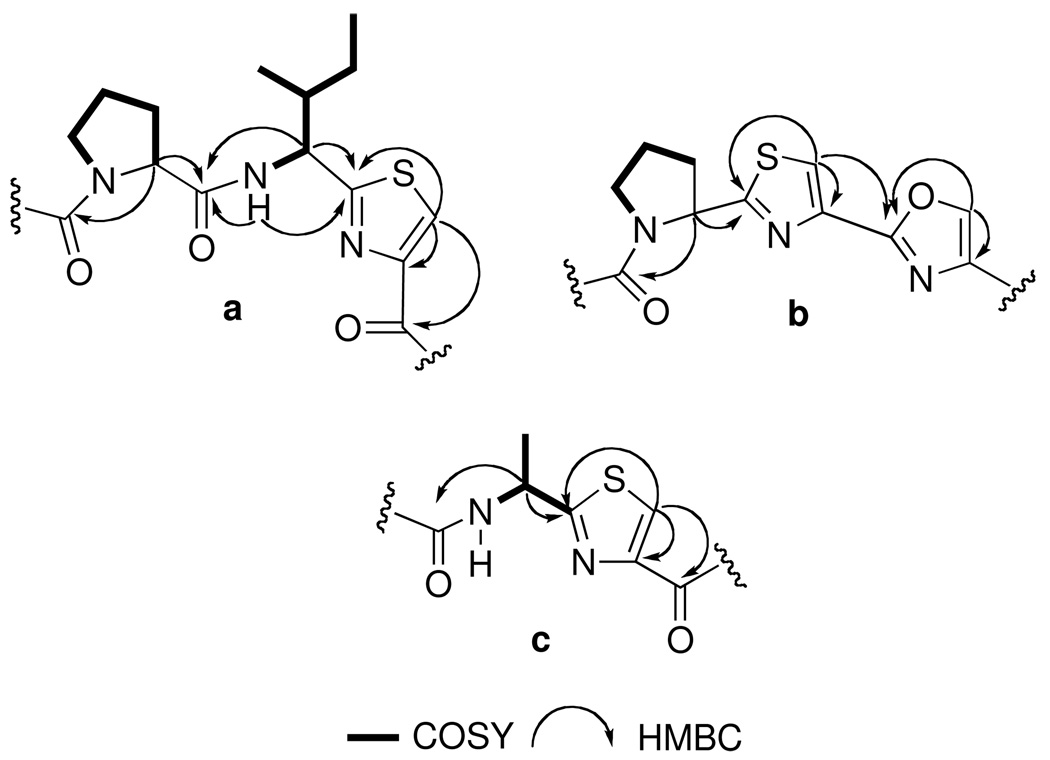
The above HMBC correlations provided the partial structure Pro1-Ile1-Pro2-Phe-Ala that was connected to the thiazole-modified amino acid as follows. The H-31 singlet showed an HMBC correlation to the Thz C=O (δ 160.3, s, C-29) that in turn was correlated to 1H NMR signals of the Ala α-CH (H-27) and NH signals (δ 8.11). The last residue, Ile-2 identified through COSY correlations, constitute the thiazole-modified amino acid residue. Mutual correlations (Fig 2a) were observed from the Ile-2 NH (δ 8.78), α-CH (H-33) and H-31 signals to the thiazole carbon C-32 (δ 168.1, s). Finally, an HMBC correlation between the amide proton at (δ 8.78) and α-CH signal (H-33) of Ile2-thiazole fragment and C=O of Pro-1 (δ 170.6, s, C-1) returns to the first residue, closes the ring and completes the structure of cyclic peptide 7.
Figure 2.
Subunits of sanguinamide A (7, a) and sanguinamide B (8, b–c), showing 1H-1H COSY and HMBC correlations.
Sanguinamide B (8) C33H43N8O6S2, (HRESIMS m/z 711.2733 [M+H]+) is a larger peptide than 7 with 17 degrees of unsaturation. The 1H NMR features of 8 are indicative of differences in amino acid composition from 7, in particular the presence of three azole-modified amino acids instead of one. The backbone 1H NMR signals included three amide-NH protons, confirmed by MS deuterium exchange (δ 9.60, d, J = 7.2 Hz; 9.26, d, J = 6.6 Hz; 8.42, d, J = 6.6 Hz), and five α-CH proton resonances (δ 4.39, d, J = 6.6 Hz, H-7; 4.61, d, J = 6.0 Hz, H-2; 5.29, dd, J = 16.8, 7.8 Hz, H-23; 5.56, dq, J = 7.2, 6.6, Hz, H-32). Evidence for the presence of three five-membered ring heterocycles include three downfield 1H NMR singlets (δ 7.95, s, H-16; 8.12, s, H-30; 8.27, s, H-13) that were coupled (HSQC) to their respective 13C nuclei (δ 121.8, d, C-16; 124.1 d, C-30; 141.5 d, C-13), accounting for two thiazoles and one 2-substituted oxazole 4-carboxamide. Long-range H-H couplings from the two broad singlets H-16 and H-30 to those 13C signals in the two thiazole rings (Table 2) were identified as before with 7, however the third singlet H-13 showed long-range couplings to carbon signals which were significantly shifted downfield (δ 136.7, C-12; 157.7, C-14) indicative of oxazole. Spin systems for five additional amino acid residues – two Pro, Val, Leu and Ala – were identified by 1H NMR, COSY and TOCSY experiments and the sequence was solved as before using HMBC correlations of the α-CH and -NH resonances to the adjacent C=O 13C signals. The -NH of Val (δ 9.26) was correlated to the C=O of Pro-1 (δ 170.4, s, C-6). The -NH of Val (δ 9.26) was correlated to the C=O of Pro-1 (δ 170.4, s, C-6), the α-CH of Pro-1 (H-7) was correlated to the C=O of the oxazole-4-carboxamide (δ 169.2, s, C-11). The H16 signal showed an HMBC correlation to heterocyclic 13C nuclei (δ 157.7, C-14; 142.3, C-15; 172.9, C-17) indicating that the oxazole and thiazole rings in 8 were connected to each other as shown (Fig 2b). Extending the sequence, the α-CH of Pro-2 (H-18) was correlated with C-17 (δ 172.9, s), showing that four of the five cyclic amino acid residues were contiguous.
Table 2.
1H and 13C NMR data of sanguinamide B, 8 (600 MHz, CDCl3)
| # | δCa, mult. | δH, mult. (J in Hz) | HMBC b | |
|---|---|---|---|---|
| L-Val | ||||
| 1 | 159.1, qC | |||
| 2 | 58.7, CH | 4.61, d (6.0) | 1 | |
| 3 | 30.6, CH | 2.93, m | ||
| 4 | 20.4, CH3 | 0.87, d (6.6) | ||
| 5 | 20.2, CH3 | 1.15, d (6.6) | ||
| - NH | 9.26, d (6.6) | 2, 3, 6 | ||
| L-Pro-1 | ||||
| 6 | 170.4, qC | |||
| 7 | 60.9, CH | 4.39, d (6.6) | 6, 11 | |
| 8 | 29.4, CH2 | 1.74, 2.80, m | 6 | |
| 9 | 25.2, CH2 | 2.22, 1.94, m | ||
| 10 | 45.4, CH2 | 3.75, 3.34, m | ||
| Oxz | ||||
| 11 | 169.2, qC | |||
| 12 | 136.7, qC | |||
| 13 | 141.5, CH | 8.27, s | 12, 14 | |
| 14 | 157.7, qC | |||
| Thz-1 | ||||
| 15 | 142.3, qC | |||
| 16 | 121.8, CH | 7.95, s | 14, 15, 17 | |
| 17 | 172.9, qC | |||
| L-Pro-2 | ||||
| 18 | 58.7, CH | 4.57, d (6.0) | 17, 22 | |
| 19 | 31.1, CH2 | 2.94, 2.07, m | 17 | |
| 20 | 28.7, CH2 | 2.10, 1.81, m | ||
| 21 | 48.0, CH2 | 3.81, 3.97, m | ||
| L- Leu | ||||
| 22 | 170.8, qC | |||
| 23 | 48.4, CH | 5.29, dd (16.8, 7.8) | 22, 28 | |
| 24 | 41.4, CH | 2.02, m | ||
| 25 | 25.6, CH2 | 1.63, m | ||
| 26 | 23.2, CH3 | 0.99, d (6.6) | ||
| 27 | 22.0, CH3 | 1.03, d (6.7) | ||
| -NH | 9.60, d (7.2) | |||
| Thz-2 | ||||
| 28 | 160.1, qC | |||
| 29 | 149.2, qC | |||
| 30 | 124.1, CH | 8.12, s | 28, 29, 31 | |
| 31 | 170.6, qC | |||
| L-Ala | ||||
| 32 | 47.8, CH | 5.56, qd (7.2 6.6) | 1, 31 | |
| 33 | 24.8, CH3 | 1.68, d (6.6) | 31 | |
| -NH | 8.42, d (7.2) | 1, 31 |
δ’s measured by indirect detection, HSQC and HMBC (1JCH= 6 Hz)
1H->13C
Completion of the sequence was made by correlating the α-CH of Pro-2 (H-18) to the C=O of Leu and the α-CH of Leu to the C=O of the Thz-2 ring. The thiazole-modified amino acid Ala-Thz (Thz-1) was established by identifying correlations from α-CH (H-32), NH and β-Me (δ 1.68, d, J = 6.4 Hz) to 13C resonances within the heterocyclic ring and C-31 (δ 170.6, Fig. 2c), as with 7. Finally, the macrolactam ring of the octapeptide 8 was completed by connecting the Ala α-CH (H-32) and the amide proton (δ 8.42, d) to the C=O signal of Val (δ 159.1, s, C-1).
Rotation about tertiary proline amide bonds is restricted and, in peptides, the Pro residue may adopt either s-trans or s-cis conformations. The syn β-CH2 group is shielded by the C=O group in s-cis Pro residues with respect to the γ-CH2 group, therefore the difference in 13C chemical shifts between the Cβ and Cγ signals in Pro amino acid residues (Δδβγ = δβ − δγ) in peptides is diagnostic of s-cis and s-trans conformations of the Pro amide bond; s-cis Pro residues have a larger difference than s-trans Pro residues.xix The Δδβγ values measured for Pro-1 and Pro-2 in the heptapeptide 7, (5.5 ppm, and 8.4 ppm, respectively) indicate the s-cis conformation for both residues. In contrast, the more flexible octapeptide 8 has smaller Δδβγ's for Pro-1 (4.2 ppm) and Pro-2 (2.4 ppm) which is consequently assigned the s-trans configuration at each residue.
The absolute configuration of both 7 and 8 was determined by total hydrolysis of the peptides followed Marfey’s analysis.xx Given the known propensity for racemization at α-CH centers of thiazole-modified peptides, samples of 7 and 8 were split into two pools, one of which was first subjected to ozonolysis prior to total hydrolysis (6M HCl, 110 °C, 16 h) to degrade the heterocyclic rings. The limited supply of 7 and 8 precluded the standard the Marfey’s methodxx and required optimized derivatization followed by sub-nanomole detection of the each amino acid derivative using ultra-high pressure liquid chromatography-mass spectrometry (UPLC-MS) and single-ion monitoring (SIM). These criteria were fulfilled and complete configurational analysis was achieved on ~30–50 µg of each peptide; all the amino acid residues in 7 and 8 were found to have the L- configuration.
Some comment is in order with respect to the absolute mass sensitivity of a 600 MHz spectrometer equipped with a 1-mm HTS cryoprobe (measurement of compounds 3 and 5) or a commercial 1.7-mm cryoprobe (compounds 7 and 8). Both probes have the significant advantage of cryocooled (~20 K) preamplifier and probe components and fill volumes far smaller than conventional 5 mm 1H inverse-detect cryoprobes (~600 µL). Mass sensitivity is inversely proportional to the diameter of the coil, but this increased mass sensitivity comes at a cost of reduced volume. The smaller diameter of the HTS 1-mm probe, with a fill volume of ~5–7 µL, provides high mass sensitivity but also requires a relatively high sample concentration. The mass sensitivity of the two probes can be compared from S/N values of 0.1% ethylbenzene, CDCl3: 292:1 and 1120:1 for the 1-mm and 1.7-mm probes, respectively. When normalizing for the total volume of the sample, the 1-mm HTS probe has a slightly higher mass sensitivity than the 1.7-mm cryoprobe: 292/7 µL ≈ 42 µL−1 versus 1120/30 µL ≈ 37 µL−1. Thus, there is a slight mass sensitivity advantage for the 1-mm probe, but the larger volume of the 1.7-mm probe provides greater overall sensitivity and can be used to study less concentrated samples. In practice both probes gave exceptionally high mass sensitivity for compounds of relatively high MW and a large number of resonances. Perhaps more significant is the fact that complete NMR data sets of sub-micromolar amounts of molecules 3, 5 and 7, 8 (C37–C45) – greater in complexity than the majority of sub-micromolar natural products measured with capillary or microprobesxxi,xiii – were easily obtained in time frames similar to those encountered in conventional NMR. Clearly, this portends success for natural products discovery that previously were unattainable, particularly applications in characterization of novel drug leads from rare marine organisms.xxii
The antifungal mechanism of action of trisoxazole macrolides is likely related to tight binding to G-actin, depolymerization of F-actin, and disruption of actin filament formation and organization.vi Compounds 1–3, 5, and the clinical antifungal agent amphotericin B (AMB) were assayed for antifungal activity against Candida sp. and Cryptococcus neoformans (Table 3). Compound 1 showed the most potency against all fungal strains tested with an MIC nearly equal to the AMB. Compound 3 was least inhibitory against the strains of Candida and Cryptococcus.
Table 3.
Minimum inhibitory activity, MIC (µg/mL) of trisoxazole macrolides against pathogenic Candida and Cryptococcus neoformans strains (positive control, amphotericin B, AMB).
|
Candida albicans ATCC 14503 |
C. glabrata |
C. neoformans var. grubii |
|
|---|---|---|---|
| 1 | 0.016 | 0.25 | 0.125 |
| 2 | 0.50 | 0.50 | 1.00 |
| 3 | 2.00 | 1.00 | 2.00 |
| 5 | 0.250 | 0.125 | 0.50 |
| AMB | 0.016 | 0.25 | 0.016 |
Conclusions
Two new antifungal trisoxazole macrolides 3 and 5 were isolated from Hexabranchus sanguineus in addition to two unexpected cyclic peptides, 7 and 8. The structures of all four compounds were solved by MS and NMR, aided by data collected on extremely mass-sensitive NMR cryo-microprobes with limited materials at the nanomole scale. Cyclic peptides 7 and 8 have not been found in sponges consumed by H. sanguineus suggesting either a different as-yet unidentified dietary source or de novo biosynthesis.
Experimental Section
General Procedures
All solvents for HPLC purification were HPLC grade. CD spectra were recorded on a Jasco J810 spectropolarimeter in 0.2 cm quartz cells at 23 °C unless otherwise stated. UV-Vis spectra were recorded in a dual beam Jasco V630 spectrometer in 1 cm quartz cells. 1H and 13C NMR spectra were recorded in CDCl3 using either a Varian Mercury-400 (400 MHz), Varian Unity-500 (500 MHz), Bruker DMX-600 (600 MHz) equipped with a 1.7-mm {13C}1H CPTCI probe, or a custom built high-temperature superconducting probe (1-mm {13C,15N}1H HTS, 600 MHz; design and performance features of this probe are described elsewhere).xii NMR spectra were measured in CDCl3 and referenced to residual solvent signals (1H, δ 7.26 ppm; 13C, δ 77.16 ppm). HRMS measurements were measured at The Scripps Research Institute (TOF-MS) or University of California, San Diego (EI-MS) mass spectrometry facilities. IR spectra were recorded on on thin films using a Jasco 4100 FTIR and attenuated total reflectance (ATR, 3 mm ZnSe plate). LCMS was carried out on a ThermoFisher Accela UPLC coupled to an MSQ single quadrupole mass spectrometer operating in positive ion mode, unless otherwise stated. Semi-preparative HPLC was carried out on a Varian SD200 system equipped with a dual-pump and UV-1 UV detector under specified conditions.
Extraction and Isolation
The nudibranch Hexabranchus sanguineus was collected in the Yasawa Islands, Fiji in 1987. The sample (N= 1) was immediately frozen and stored at −20 °C until extraction (~12 months). Frozen tissue was thawed and extracted with acetone (2x times) and each acetone extract was filtered and concentrated before partitioning between EtOAc and water. The EtOAc layers were concentrated under reduced pressure to give a dark orange oil. The second batch of acetone extract was separated by silica chromatography using a shallow gradient (1% MeOH/CH2Cl2, 2% MeOH/ CH2Cl2, 4% MeOH/ CH2Cl2 and 50% MeOH/ CH2Cl2) giving six fractions. Fraction 90.001.6 was purified twice by reversed-phase HPLC (Dynamax, 5µm, C18 column, 10 × 250 mm, 80:20 MeOH:H2O, 3.5 mL/min) and (Dynamax, 5µm, C18 column, 4.6 × 250 mm, 80:20 MeOH:H2O, 1.5 mL/min) yielded 620 µg of pure 2 (tR = 12.10 min). Fraction 90.001.5 was separated under reversed-phase HPLC (Dynamax semiprep, 25 × 1 cm; 83:17 MeOH:H2O, 3.5 mL/min) to obtain 3 fractions. Fraction 90.001.5A was purified by reversed-phase HPLC (Zorbax Agilent, 5µm, C18 column, 4.6 × 250 mm, 50:50 CH3CN:H2O) yielded two major fractions of 1 (16.64 mg) and crude 4. Final purification by reversed-phase HPLC (Phenylhexyl Luna, 4.6 × 250 mm, 50:50 CH3CN:H2O, 1 mL/min, detection at 230 nm) yielded 3 (620 µg, tR = 15.2 min), additional 1 (5.64 mg, tR = 16.40 min) and pure 4, (1.30 mg, tR = 21.04 min). Fraction 90.001.5B was further purified by reversed-phase HPLC (Phenylhexyl Luna, 4.6 × 250 mm, 50:50 CH3CN:H2O, 1.5 mL/min) to provide 8 (190 µg, tR = 11.0 min), 7 (390 µg, tR = 12.30 min) and 5 (1.3 mg, tR = 18.0 min).
9-O-desmethyl kabiramide B (3)
Colorless film; UV (MeOH) λmax (log ε) 243 nm (4.38); IR (ATR): ν 3200–3600 br, 1618 cm−1; 1H NMR (600 MHz, CD3CN, 1mm-HTS probe) δ8.36 (0.7H, s, H-42), 8.32 (1H, s, H-14), 8.27 (1H, s, H-17), 8.04 (0.3H, s, H-42), 7.72 (1H, s, H-11), 7.25 (1H, ddd, J = 14.0, 8.5, 5.8 Hz, H-20), 7.07 (0.3H, d, J = 14.0 Hz, H-35), 6.39 (1H, d, J = 14.0 Hz, H-19), 6.62 (0.7H, d, J = 14.0 Hz, H-35), 5.19 (1H, br s, H-9), 5.15 (1H, m, H-3), 5.16 (0.3H, d, J = 14.0 Hz, H-34), 5.08 (0.7H, d, J = 14.0 Hz, H-34), 5.07 (1H, dd, J = 10.7, 7.0 Hz, H-24), 3.99 (1H, m, H-22), 3.92 (1H, br s, H7), 3.84 (1H, br s, C9-OH), 3.50 (1H, br s, OH7), 3.36 (1H, d, J = 6.0 Hz, OH-22), 3.29 (3H, s, C32-MeO), 3.28 (3H, s, C26-OMe), 3.27 (1H, m, H-32), 3.09 (3H, m, C35-NMe), 3.06 (1H, m, H-26), 2.74 (1H, dd, J = 7.2, 6.5 Hz, H-31), 2.58 (0.3H, m, H-33), 2.52 (1H, m, H-29’), 2.51 (1H, m, H-21’), 2.50 (1H, m, H-29), 2.50 (1H, m, H-2’), 2.45 (0.7H, m, H-33), 2.31 (1H, m, H-21), 2.30 (1H, m, H-2), 2.15 (1H, dd J = 8.0, 7.0 Hz, H-8), 1.81 (1H, m, H-5), 1.81 (1H, m, H-6), 1.81 (1H, m, H-6’), 1.72 (1H, m, H-27), 1.71 (1H, m, H-28), 1.65 (1H, m, H-23), 1.64 (1H, m, H-25’), 1.58 (1H, m, H-4’), 1.50 (1H, m, H-25), 1.31 (1H, m, H-4), 1.22 (1H, m, H-28’), 1.36 (3H, d, J = 7.0 Hz, H-41), 0.95 (3H, d, J = 7.0 Hz, H-36), 0.91 (3H, d, J = 7.0 Hz, H-40), 0.90 (3H, d, J = 7.0 Hz, H-38), 0.87 (3H, d, J = 8.0 Hz, H-37), 0.82 (3H, d, J = 7.0 Hz, H-39); 13C NMR, (indirect detection, HSQC and HMBC (JCH = 6 Hz), 600 MHz). δ 214.1 (C, C-30), 175.8 (C, C-1), 163.5 (NHCHO), 145.4 (CH, C-20), 138.3 (CH, C-17), 137.9 (CH, C-14), 136.9 (C, C-10), 135.9 (CH, C-11), 131.4 (C, C-13), 130.7 (C, C-16), 111.4 (113.7*) (CH, C-34), 88.4 (89.6*) (CH, C-32), 82.02 (CH, C-26), 74.5 (CH, C-24), 74.0 (CH, C-9), 69.5 (CH, C-22), 68.7 (CH, C-7), 67.6 (CH, C-3), 60.3 (OMe32), 56.8 (OMe26), 49.7 (CH, C-31), 45.1 (CH2, C-4), 43.1 (CH2, C-6), 42.9 (CH, C-23), 41.3 (CH2, C-29), 41.2 (CH2, C-2), 38.3 (CH, C-8), 38.1 (38.0*) (CH, C33), 36.1 (CH2, C-21), 35.0 (CH, C-27), 33.7 (CH2, C-25), 27.7 (NMe), 26.5 (CH, C-5), 25.6 (CH2, C-28), 18.3 (19.3*) (CH3, C-41), 18.1 (CH3, C-36), 15.7 (CH3, C-40), 15.5 (CH3, C-39), 9.5 (CH3, C-38) (*minor isomer); HREIMS m/z 936.4574 [M+Na]+ (calcd for C46H67N5O14, 936.4582).
33-Methyl tetrahydrohalichondramide (5)
Colorless film; UV (MeOH) λmax (log ε) 233 nm (4.53); IR (ATR): ν3200–3600 br, 1584 cm−1 ; 1H NMR 600 MHz (CDCl3) δ 8.28 (0.7H, s, H-41), 8.10 (1H, s, H-17), 8.07 (0.3H, s, H-41), 8.05 (1H, m, H-14), 7.63 (1H, s, H-11), 7.13 (0.3H, d, J = 14.0 Hz, H-35), 6.99 (1H, dt, J = 16.0, 8.0 Hz, H-20), 6.46 (1H, d, J = 0.7 Hz, H-35), 6.43 (1H, d, J = 16.0 Hz, H-19), 5.20 (1H, m, H-24), 5.09 (1H, m, H-34), 4.39 (1H, d, J = 3.0 Hz, H-9), 4.22 (1H, m, H-3), 3.93 (1H, m, H-7), 3.30 (1H, m, H-32), 3.50 (3H, s, C9-OMe), 3.35 (3H, s, C26-OMe), 3.34 (3H, s, C32-OMe), 3.04 (3H, s, NMe), 2.97 (1H, br d, J = 10.0 Hz, H-26), 2.64 (1H, dq, J = 9.0, 7.0 Hz, H-31), 2.69 (1H, m, H-6), 2.67 (1H, m, H-6'), 2.53 (1H, m, H-2), 2.48 (1H, m, H-21), 2.48 (2H, m, H-29), 2.43 (1H, br qd, J = 7.0, 4.5 Hz, H-8), 2.38 (1H, m, H-33), 2.26 (1H, m, H-21’), 1.85 (2H, m, H-5), 1.79 (1H, m, H-28’), 1.77 (2H, m, H-23), 1.71 (2H, m, H-27), 1.71 (1H, m, H-4), 1.71 (m, 1H, H-22’), 1.61 (m, 1H, H-4’), 1.56 (m, 1H, H-25), 1.34 (m, 1H, H-22’), 1.16 (3H, d, J = 7.0 Hz, H-40), 1.02 (3H, d, J = 7.0 Hz, H-36), 0.92 (3H, d, J = 7.0 Hz, H-39), 0.87 (3H, d, J = 7.0 Hz, H-37), 0.84 (3H, d, J = 7.0 Hz, H-38); 13C NMR, (indirect detection, HSQC and HMBC (JCH = 8 Hz), 600 MHz). δ 213.6 (C, C-30), 173.2 (C, C-1), 162.1(160.8*) (NHCHO), 162.0 (C, C-18), 156.3 (C, C-15), 155.0 (C, C-12), 143.2 (CH, C-20), 141.3 (C, C-10), 137.7 (CH, C-17), 137.4 (CH, C-14), 136.7 (CH, C-11), 130.9 (C, C-13), 129.9 (124.6*) (CH, C-35), 129.7 (C, C-16), 116.3 (CH, C-19), 111.0 (113.0*) (CH, C-34), 82.4 (CH, C-32), 82.4 (CH, C-9), 81.9 (CH, C-26), 73.9 (CH, C-24), 71.0 (CH, C-7), 68.7 (CH, C-3), 61.2 (C9-OMe), 58.9 (C26-OMe), 57.9 (C32-OMe), 48.9 (CH, C-31), 42.5 (CH2, C-2), 42.1 (CH2, C-29), 38.8 (CH, C-8), 37.6 (CH, C-33), 36.7 (CH2, C-4), 35.7 (CH, C-23), 34.7 (CH, C-27), 33.3 (CH2, C-6), 32.1 (CH2, C-25), 31.5 (CH2, C-22), 28.8 (CH2, C-21), 27.6 (33.4*) (NMe), 25.3 (CH2, C-28), 22.4 (CH2, C-5), 19.4 (CH3, C-40), 15.6 (CH3, C-38), 13.7 (CH3, C-37), 13.3 (CH3, C-39), 9.1 (CH3, C-36) (*minor isomer); HREIMS m/z 855.4738 [M+H]+ (calcd. for C45H67N4O12, 855.4750)
Sanguinamide A (7)
colorless film; IR (ATR): ν2922, 2852, 1733, 1646, 1541 cm−1 ; CD (MeOH) ) λmax (Δε) 224 nm (−14.5); 1H (600 MHz, 1-mm probe) and 13C NMR, see Table 1; HREIMS m/z 722.3694 [M+H]+ (calcd. for C34H54N6O9S, 722.3683)
Sanguinamide B (8)
colorless film; UV (MeOH) λmax (log ε) 250 nm (4.7) ; IR (ATR): ν2980, 2357, 1589 cm−1 ; CD (MeOH) ) λmax (Δε) 235 (−4.30); 1H (600 MHz, 1-mm probe) and 13C NMR, see Table 2); HREIMS m/z 711.2741 [M+H]+ (calcd for C33H43N8O6S2,711.2733)
Determination of absolute configuration of 7 and 8
(a) Ozonolysis and Hydrolysis of Peptides
Samples of peptide were split into two pools (A) and (B). The first pool (A) was ozonolyzed prior to hydrolysis as follows: 7 (50 µg) and 8 (30 µg), were individually suspended in ozone saturated MeOH (150 µL) at −78 °C for 5 min before concentration of the solvent under a N2 stream at −78 °C to remove ozone then to dryness at 0 °C. Pool (B) samples, 7 (50 µg), 8 (30 µg), and ozonolyzed samples from pool (A) were each individually resuspended in degassed, freshly distilled constant-boiling 6N HCl (200 µL) and heated in a flame-sealed glass tube at 110 °C for 15 h. The mixtures were concentrated to dryness under a stream of N2 stream and redissolved in 100 µL of H2O.
(b) Marfey’s derivatization with L-FDLA.xx
Each of the above hydrolysate solutions were treated with a solution of 2,4-dinitrophenyl-5-fluoro-L-leucinamide (100µL, 1% w/v in acetone), followed by 1.0 M NaHCO3 (25 µL), then heated in a sealed tube at 80 °C for 10 min. The mixture was cooled and quenched with 1.0 M HCl (25 µL). Authentic l- and d-amino acid standards were treated under identical conditions.
(c) LCMS Analysis
The Marfey’s derivatives (see above) were analysed by LC-MS using a ThermoElectron Accela series ultra-high pressure liquid chromatograph (UPLC) with a Thermo Hypersil Gold C-18 column (50 mm × 2.1-mm, 1.9 µm) or Agilent Zorbax SB-Aq C18 column (250 mm × 4.6 mm, 5 µm) connected to a PDA and ThermoFinnigan MSQ quadrapole mass spectrometer. LC parameters were as follows; Hypersil: Flow rate 0.5 mL/min, initial 80% solvent A (H2O + 0.1% formic acid) 20% solvent B (acetonitrile), hold 0.3 min, at 9 min 40% A, at 10 min 100% B hold for 3 min, at 14 min 80% A hold for 2 min; Zorbax SB-Aq: Flow rate 1.0 mL/min, initial 95% solvent A (H2O + 0.1% formic acid) 5% solvent B (acetonitrile), hold 2 min, at 65 min 40% A, at 67 min 100% B hold for 5 min, at 73 min 95% A hold for 5 min Injection volume was 3 µL. PDA parameters were as follows; Channel A 340 nm, Channel B 254 nm. MSQ parameters were as follows; ESI-MS, selected ion monitoring at m/z 384.15, 410.17, 412.18, 426.20, 460.18 [M+H]+, span 1.5 amu, dwell 0.6 sec, cone 75 V, probe temperature 450 °C. Retention times for the amino acids on the Hypersil in min tR= (l/d) were as follows; alanine (4.28/5.16), proline (4.32/4.99), valine (5.07/6.61), phenylalanine (5.85/7.12), allo-isoleucine (5.64/7.26), isoleucine (5.67/7.36), leucine (5.75/7.39). Because of overlap of peaks of the allo-isoleucine and isoleucine FDLA derivatives on the C18 Hypersil column, these substituted amino acids were analyzed on the Zorbax SB-Aq column which gave the following retention times; allo-isoleucine (55.79/62.38), isoleucine (56.17/62.42). Co-injections of l-allo-isoleucine and l-isoleucine FDLA derivatives with the corresponding derivatives from 7 and 8 were used to verify the assignments.
In vitro antifungal testing
The fungal isolates used in this study were strains of Candida albicans (2 clinical isolates which are fluconazole-resistant, strain UCDFR1 and 96–489 and a reference strain, ATCC 14503), clinical isolates of Candida glabrata, Candida krusei, Cryptococcus neoformans var. grubii and C. neoformans var. gattii. The fungi were grown and maintained in Sabouraud dextrose agar, SDA plates (BBL, 211584) and incubated at 30 °C for 24 h (Candida sp.) or 48 h (C. neoformans). The in vitro susceptibility of each compound was determined by the broth micro dilution method according to the guidelines National Committee for Clinical Laboratory Standards (NCCLS).xxiii Briefly, 2-fold serial dilutions of compounds were prepared in 96-well microtiter plates (Corning Incorporated, 3595) from stock solutions in an RPMI-1640 broth medium (Sigma) buffered to a final pH of 7.0 with 0.165 M morpholinepropane-sulfonic acid (MOPS; Sigma) to a final volume of 100 µL. A stock solution was prepared in dimethyl sulfoxide (DMSO, Sigma) for the various compounds and for amphotericin B, AMB (Sigma) that was used as positive control. The final drug concentrations tested were from 0.062 to 64 µg/mL and from 0.0078 to 8 µg/mL for amphotericin B. All fungal strains were tested in replicate in each run of the experiments. Cell growth was determined by the OD at 600 nm using a Spectramax Plus 384 microplate reader (Molecular Devices, CA). The MIC endpoint was defined as the lowest concentration with complete (≥ 90%) growth inhibition.
Supplementary Material
Acknowledgments
We are grateful to T. M. Zabriskie (Oregon State University) for collection of H. sanguineus and C. M. Ireland (University of Utah) for expedition logistics, T. Kühn (Bruker, Switzerland) for some of the 1.7-mm cryoprobe NMR data and Brandon I. Morinaka for helpful discussions. We thank J. Pawlik (University of North Carolina, Wilmington) for the image of H. sanguineus (graphic abstract). Mass spectra were provided by Scripps Research Institute MS Facility and Y. Su (UCSD MS Facility). This research was supported grants from the NIH (RO1 AI 039987, CA122256 to TFM) and National High Magnetic Field Laboratory support to ASE. The 1-mm HTS instrument time was provided by the NSF-funded NHMFL external user program.
Footnotes
Supporting Information. 1H NMR, 2D NMR and CD spectra of 3, 5, 7 and 8. This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- i.Roesener JA, Scheuer PJ. J. Am. Chem. Soc. 1986;108:846–847. [Google Scholar]
- ii.Matsunaga S, Fusetani N, Hashimoto K, Koseki K, Noma M. J. Am. Chem. Soc. 1986;108:847–849. [Google Scholar]
- iii.a Kernan MR, Faulkner DJ. Tetrahedron Lett. 1987;28:2809–2812. [Google Scholar]; b Kernan MR. Ph.D. thesis. San Diego: University of California; 1988. [Google Scholar]
- iv.Kernan MR, Molinski TF, Faulkner DJ. J. Org. Chem. 1988;53:5014–5020. [Google Scholar]
- v.Pawlik JR, Kernan MK, Molinski TF, Harper MK, Faulkner DJ. J. Exp. Mar. Biol. Ecol. 1988;119:99–109. [Google Scholar]
- vi.Klenchin VA, Allingham JS, King R, Tanaka J, Marriott G, Rayment I. Nat. Struct. Biol. 2003;10:1058–1063. doi: 10.1038/nsb1006. [DOI] [PubMed] [Google Scholar]
- vii.Matsunaga S, Fusetani N, Hashimoto K, Koseki K, Noma M, Noguchi H, Sankawa U. J. Org Chem. 1989;54:1360–1363. [Google Scholar]
- viii.Matsunaga S. Prog. Mol. Subcell. Biol. 2006;43:241–260. doi: 10.1007/978-3-540-30880-5_11. [DOI] [PubMed] [Google Scholar]
- ix.Kobayashi J, Murata O, Shigemori H, Sasaki T. J. Nat. Prod. 1993;56:787–791. doi: 10.1021/np50095a021. [DOI] [PubMed] [Google Scholar]
- x.Fusetani N, Yasumuro K, Matsunaga S, Hashimoto K. Tetrahedron Lett. 1989;30:2809–2812. [Google Scholar]
- xi.Petchprayoon C, Asato Y, Higa T, Garcia-Fernandez LF, Pedpradab S, Marriott G, Suwanborirux K, Tanaka J. Heterocycles. 2006;69:447. [Google Scholar]
- xii.Brey WW, Edison AS, Nast RE, Rocca JR, Saha S, Withers RS. J. Magn. Reson. 2006;179:290–293. doi: 10.1016/j.jmr.2005.12.008. [DOI] [PubMed] [Google Scholar]
- xiii.Molinski TF. Curr. Opin. Drug Disc. Dev. 2009 in press. [PubMed] [Google Scholar]
- xiv.Matsunaga S, Liu P, Celatka CA, Panek JS, Fusetani N. J. Am. Chem. Soc. 1999;121:5605–5606. [ibid. J. Am. Chem. Soc. 1999, 121, 8969–8969]. [Google Scholar]
- xv.a Allingham JS, Zampella A, D'Auria MV, Rayment I. Proc. Nat. Acad. Sci. USA. 2005;102:14527–14532. doi: 10.1073/pnas.0502089102. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Allingham JS, Tanaka J, Marriott G, Rayment I. Org. Lett. 2004;6:597–599. doi: 10.1021/ol036458y. [DOI] [PubMed] [Google Scholar]
- xvi.Chattopadhyay SK, Pattenden G. Tetrahedron Lett. 1998:6095–6098. [Google Scholar]
- xvii.Michael JP, Pattenden G. Angew. Chem. Intl. Ed. 1993;32:1–130. [Google Scholar]
- xviii. Maddock J, Pattenden G, Wight PG. Computer-Aided Molecular Design. 1993;7:573–585. doi: 10.1007/BF00124363. In fact, experimental evidence based on fluorescence measurements suggest that trisoxazole macrolides do not chelate divalent metal ions and, with the exception of Hg2+, ligate poorly with common row 2 transition metal ions. James, D. M.; Wintner, E.; Faulkner, D. J.; Siegel, J. S. Heterocycles 1993, 35, 675–678.
- xix.a Dorman DE, Bovey FA. J. Org. Chem. 1973;38:2379–2383. and references cited within. [Google Scholar]; b Siemion IZ, Wieland T, Pook K. Angew. Chem., Int. Ed. 1975;14:702–703. doi: 10.1002/anie.197507021. [DOI] [PubMed] [Google Scholar]
- xx.Marfey P. Carlsberg. Res. Commun. 1984;49:591–596. [Google Scholar]
- xxi.Schroeder FC, Gronquist M. Angew. Chem. Intl. Ed. 2006;45:7122–7131. doi: 10.1002/anie.200601789. [DOI] [PubMed] [Google Scholar]
- xxii.Molinski TF, Dalisay DS, Lievens SL, Saludes JP. Nat. Rev. Drug. Discov. 2009;8:69–85. doi: 10.1038/nrd2487. [DOI] [PubMed] [Google Scholar]
- xxiii.National Committee for Clinical Laboratory Standards. Wayne, Pa: National Committee for Clinical Laboratory Standards; Reference method for broth dilution antifungal susceptibility testing of yeast, 2nd ed. Approved standard M27-A2. 2002
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.