Abstract
The actin cytoskeleton has been implicated in the intra- and intercellular movement of a growing number of plant and animal viruses. However, the range of viruses influenced by actin for movement and the mechanism of this transport are poorly understood. Here we determine the importance of microfilaments and myosins for the sustained intercellular movement of a group of RNA-based plant viruses. We demonstrate that the intercellular movement of viruses from different genera [tobacco mosaic virus (TMV), potato virus X (PVX), tomato bushy stunt virus (TBSV)], is inhibited by disruption of microfilaments. Surprisingly, turnip vein-clearing virus (TVCV), a virus from the same genus as TMV, did not require intact microfilaments for normal spread. To investigate the molecular basis for this difference we compared the subcellular location of GFP fusions to the 126-kDa protein and the homologous 125-kDa protein from TMV and TVCV, respectively. The 126-kDa protein formed numerous large cytoplasmic inclusions associated with microfilaments, whereas the 125-kDa protein formed few small possible inclusions, none associated with microfilaments. The dependence of TMV, PVX, and TBSV on intact microfilaments for intercellular movement led us to investigate the role of myosin motors in this process. Virus-induced gene silencing of the Nicotiana benthamiana myosin XI-2 gene, but not three other myosins, inhibited only TMV movement. These results indicate that RNA viruses have evolved differently in their requirements for microfilaments and the associated myosin motors, in a manner not correlated with predicted phylogeny.
Keywords: cytoskeleton, microfilaments, tobacco mosaic virus, potato virus X, tomato bushy stunt virus
Viruses are obligate parasites that use various host factors for their replication and movement within and between cells (1–5). The host cytoskeleton, a key component of intracellular transport pathways, is implicated in the movement of both plant and animal viruses. For animal viruses, f-actin (microfilaments) and microtubules have been shown to be important throughout the infection process, from virus entry and intracellular transport to virus egress and budding (4, 5). For plant viruses, the movement of tobacco mosaic virus (TMV) has been particularly well-studied and represents a unique situation where both the microtubule and actin cytoskeleton have been implicated in supporting its movement.
Studies investigating TMV movement initially focused on the association of the TMV movement protein (MP) with microtubules (6–8). The role of this interaction in TMV transport remains unclear, since pharmacological and molecular biological studies indicate that microtubules are not involved in its spread (9, 10). Although the TMV MP associates with microfilaments (7), the importance of this interaction in virus movement is not well characterized. However, the intercellular movement of TMV was recently shown to be associated with microfilaments, since treatment with the microfilament inhibitor, latrunculin B (LatB), severely limited TMV spread in the plant (11). The finding that silencing of host actin transcripts resulted in a similar disruption of TMV-GFP cell-to-cell movement suggested that the effect of LatB on virus movement could indeed be attributed to its disruption of the actin cytoskeleton (12). This inhibition of TMV cell-to-cell spread was linked to the ability of LatB to disrupt the movement of TMV viral replication complexes (VRCs) (11, 12). VRCs are large inclusion bodies that form in the cytoplasm of TMV-infected cells. They contain both viral and host components (6, 13–16) and are proposed to be involved with viral replication (6, 16), degradation (17), and movement (11, 13). The TMV 126-kDa protein is a major constituent of VRCs and is capable of forming cytoplasmic bodies (126-bodies) in the absence of other viral proteins (12, 18). Since both 126-bodies and VRCs traffic along microfilaments, it was suggested that the 126-kDa protein may mediate the interaction between the VRCs and microfilaments (12). However, considering the interaction of the 126-kDa protein and homologs with integral membrane proteins (19), VRC-microfilament interactions may be mediated through host membrane-based factors.
In addition to the TMV 126-kDa protein, there are a growing number of diverse plant viral proteins found to associate with microfilaments. These include the TGBp2 movement protein from potato virus X (PVX), both TGBp2 and TGBp3 from potato mop-top virus, the Hsp70 homolog from beet yellows virus (BYV), and the P6 multifunctional protein from cauliflower mosaic virus (CaMV) (20–23). Although some of these proteins have been implicated in intercellular virus movement, a widespread requirement for microfilaments to enable cell-to-cell plant virus movement has not been demonstrated.
The fact that viral proteins associate with and traffic along microfilaments implies the utilization of an actomyosin-based motility system for virus transport. Myosins are a large superfamily of genes with more than 18 distinct classes (24). Higher plant myosins have been grouped into either class VIII or class XI (25). The Arabidopsis genome, for example, encodes 17 myosins (13 class XI and four class VIII) (25). Although the specific functions of individual myosins have only begun to be elucidated, class XI myosins generally influence organelle trafficking (26–28) and the morphogenesis and elongation of tip-growing cells (28–30). Class VIII myosins are believed involved in trafficking to plasmodesmata (PD) as well as endocytosis in plants (31, 32).
It was recently shown that inhibiting the function of class VIII, but not class XI, myosins disrupted the localization of the BYV Hsp70 movement protein to PD (33). In this same study, it was shown that the localization of TMV MP to PD was not affected by inhibition of class VIII myosins. Interestingly, treatment with the inhibitor 2,3-butanedione monoxime, which targets myosins and other proteins, inhibited TMV cell-to-cell spread by 12% (11). These findings raised the intriguing possibility that different viruses might use distinct classes of myosins for their movement. However, the role of specific myosins in the movement of plant viruses has not been determined.
For animal viruses, specific microtubule motors have been implicated in virus movement (5), but despite the findings that numerous viruses rely upon actin for at least some portion of their intracellular movement (4), the role of specific myosins in this process has only begun to be elucidated (34, 35).
Here, we test the effect of pharmacological disruption of actin filaments on the intercellular spread of a number of different plant RNA viruses: TMV, PVX, tomato bushy stunt virus (TBSV), and turnip vein clearing virus (TVCV). For viruses that showed a difference in their requirement for microfilaments, we investigated the molecular basis for this difference by comparing the subcellular localization of their protein homologs. We further investigated the role of myosin motor proteins in the intercellular movement of these viruses by silencing individual Nicotiana benthamiana myosin genes. We relate our findings to those from other studies of plant and animal virus movement.
Results
The Effect of LatB on the Cell-to-Cell Spread of Diverse RNA Viruses.
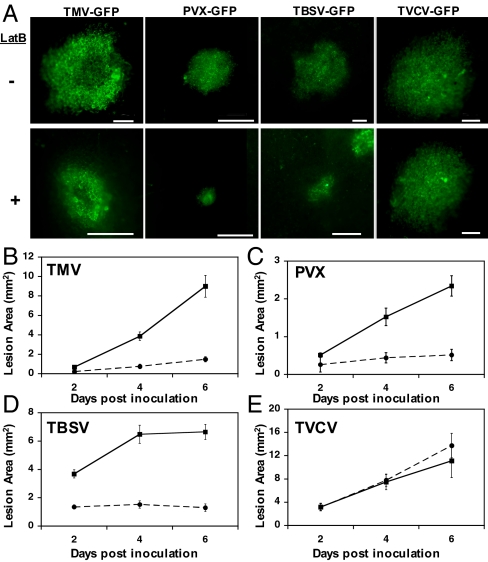
Given the association of a growing number of plant virus proteins with the actin cytoskeleton (12, 20–23), we tested the effect of LatB on the cell-to-cell spread of derivatives of the potexvirus PVX, the tombusvirus TBSV, and the tobamovirus TMV (as a positive control) expressing GFP. To determine variability in movement requirements within a genus, we also constructed a GFP fusion for another tobamovirus, TVCV. Half leaves were infiltrated with either 5 μM LatB or a DMSO buffer control at 3 h before inoculation and again at 3 dpi. The disruption of actin by 5 μM LatB was confirmed by microscopic observation of microfilaments labeled with the actin binding domain 2 of Arabidopsis fimbrin fused to GFP (36) (Fig. S1). Maintenance of cell-to-cell spread of viruses was quantified by imaging GFP lesions between 2 and 6 days post inoculation (dpi) (Fig. 1 A–E). As expected, we observed a significant inhibition of TMV cell-to-cell spread following LatB treatment (Fig. 1B). Inhibited spread was observed for TMV expressing an MP-GFP fusion (Fig. 1B) or a free GFP (Fig. S2). In addition, we observed inhibited spread for wild-type TMV (strain U1; Fig. S3), further validating the use of fluorescent virus derivatives to investigate sustained cell-to-cell movement. We observed a similar inhibited movement for both PVX (Fig. 1C) and TBSV (Fig. 1D) in LatB treated tissue, suggesting that the maintenance of intercellular spread of both of these viruses was also actin-dependent. Surprisingly, the intercellular movement of TVCV, a tobamovirus closely related to TMV was not inhibited by LatB treatment (Fig. 1E).
Fig. 1.
The effect of LatB on virus spread. (A) Representative images showing GFP lesions formed upon infection with the indicated viruses either in the absence (-) or presence (+) of 5 μM LatB. All images were taken at 6 dpi. (Scale bar, 1 mm.) (B–E) Lesion areas were quantified to determine the effect of LatB on the cell-to-cell movement of (B) TMV, (C) PVX, (D) TBSV, and (E) TVCV at 2, 4, and 6 dpi. N. benthamiana leaf tissue was infiltrated with either the actin inhibitor LatB (circles) or a DMSO buffer control (squares). Bars represent standard errors for 15 lesions per treatment.
TVCV 125-kDa and the TMV 126-kDa Protein Homologs Differ in Formation of Cytoplasmic Inclusions and Association with Microfilaments.
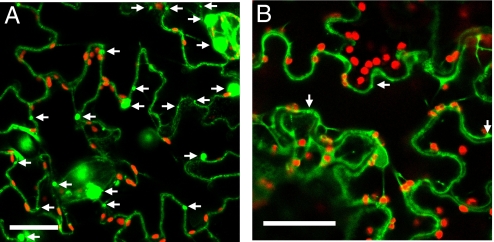
TMV and TVCV cell-to-cell movement require, or are suspected to require, expression of sequences within their 126-kDa and 125-kDa ORFs. The corresponding proteins from the two viruses show sequence and functional homology (37, 38). Given the surprising difference we observed between the dependence of TMV and TVCV upon actin for their intercellular movement, we investigated potential differences in the localization and actin association between the TMV 126-kDa protein and its 125-kDa homolog from TVCV (sequences shown in Fig. S4). We compared the cellular accumulation pattern of a TVCV 125-kDa protein-GFP fusion (TVCV 125-GFP) in N. benthamiana leaf epidermal cells to that of a TMV 126-kDa protein-GFP fusion (TMV 126-GFP) following agro-infiltration (12). Unlike TMV 126-GFP, which formed a large number of cytoplasmic bodies (Fig. 2A, arrows), TVCV 125-GFP fluorescence was generally diffuse throughout the cytoplasm and lacked the appearance of sizeable or numerous visible bodies (Fig. 2B). This difference in accumulation pattern was not due to differences in transcript or protein levels between the two fusion proteins (Fig. S5). We quantified the differences in body formation between the TMV 126-kDa protein and the TVCV 125-kDa protein by counting the number of bodies found in leaf epidermal cell sections and determining their average size (Table 1). The 126-GFP bodies were numerous and large, consistent with previous findings (12), while the 125-GFP fluorescent foci were rare and small (Fig. 2 and Table 1). Since we were unsure of the functional significance of these small 125-GFP foci (possibly representing only areas of enriched cytoplasm) and given the fact that TMV 126-GFP bodies associate with and traffic along actin filaments, we looked for association of TVCV 125-GFP with DSRed-Talin (12) labeled actin microfilaments. Co-localization of the 125-GFP foci with microfilaments was not observed (Table 1).
Fig. 2.
The TVCV 125-kDa protein does not form numerous large inclusions like the TMV 126-kDa protein. Representative images showing TMV 126-kDa protein (A) and TVCV 125-kDa protein (B) GFP fusions expressed in N. benthamiana leaf epidermal cells 3 days following agrobacterium infiltration. The positions of TMV 126-kDa protein bodies (A) and potential TVCV 125-kDa protein bodies (B) are indicated with arrows. Red fluorescent bodies are chloroplasts. (Scale bar, 25 μm.)
Table 1.
A comparison of cytoplasmic body number, body size, and actin association between TMV 126-GFP and TVCV 125-GFP proteins in N. benthamiana leaf epidermal cells
| Number of bodies (Average diameter)* | Number of bodies associated with microfilaments† | |
|---|---|---|
| TMV 126-GFP | 134 (6.14) | 82 |
| TVCV 125-GFP | 26 (1.81) | 0 |
*Number of GFP bodies counted in 10 leaf epidermal cell sections. (Average diameter in micrometers).
†Number of GFP bodies found in association with DSRed-Talin labeled microfilaments across 10 random images of cortical actin.
Silencing Individual Myosins in N. benthamiana via Virus-Induced Gene Silencing.
Given the demonstrated dependence of TMV, PVX, and TBSV on microfilaments to maintain intercellular movement, we wanted to investigate the potential role of individual myosin motor proteins in this process. To this end, we used tobacco rattle virus (TRV)-mediated VIGS (TRV-VIGS) (39) to silence individual myosin genes. Since myosins are a large gene family, it was a significant technical challenge to target individual myosin genes for silencing. We used the divergent tail domains of four available N. benthamiana myosin sequences (two class VIII and two class XI) to generate TRV constructs containing portions of these sequences (Fig. S6). In particular, we chose sequences that lacked continuous stretches of identity of 23 bp or more, the length shown to be required for efficient silencing of related transcripts (40).
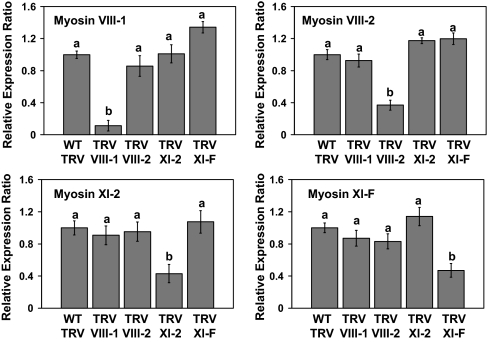
Fig. 3 shows the relative expression ratio of individual myosin genes (myosin VIII-1, VIII-2, XI-2, and XI-F) in plants infected with TRV expressing portions of the myosin genes (TRV-myosin) compared with a TRV control not expressing a myosin fragment. For all four myosin VIGS constructs, we obtained a significant and reproducible silencing of only the intended myosin transcript in N. benthamiana.
Fig. 3.
VIGS of individual myosin genes. Quantitative RT-PCR was used to determine the relative expression ratio of target genes (myosin VIII-1, myosin VIII-2, myosin XI-2, and myosin XI-F) in lines treated with the indicated TRV silencing constructs versus a TRV control not expressing a myosin fragment. Elongation factor 1α served as an internal loading control for each sample. Expression analysis was performed on extracts from systemic leaves at 18 dpi with TRV constructs. Bars represent means ± standard errors for three replicates per treatment. Analysis of variance followed by an lsd calculation was used to determine significant differences between treatments. Different letters above the bars indicate significant differences (P = 0.05). The experiment was repeated at least once for each TRV silencing construct.
TMV Intercellular Movement in N. benthamiana Is Strongly Inhibited By the Silencing of Myosin XI-2.
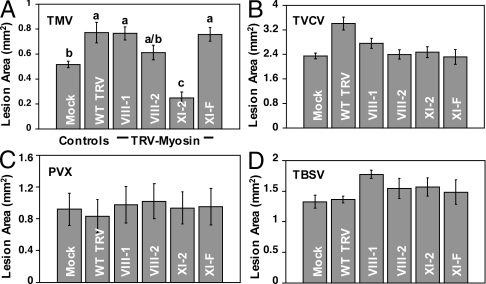
To determine the effect of silencing individual myosin genes on the cell-to-cell movement of diverse viruses, we inoculated the leaves silenced for expression of specific myosins with viruses (TMV, TVCV, PVX, and TBSV) expressing GFP and compared fluorescent lesion sizes at 3 dpi between the plants inoculated with TRV-myosin VIGS constructs and mock-inoculated or TRV controls (Fig. 4). We confirmed that myosin silencing was maintained at 3 dpi with these diverse viruses (Fig. S7). Silencing of myosin XI-2 but not other myosins specifically inhibited the spread of TMV in N. benthamiana (Fig. 4A). Comparison of leaf epidermal cell sizes in control and myosin XI-2 silenced plants confirmed that smaller TMV lesion sizes were not due to a reduction of cell size following myosin silencing (Fig. S8). We did not see an effect of silencing myosins on the cell-to-cell spread of TVCV (Fig. 4B), consistent with our LatB data (Fig. 1E). Importantly, we did not observe reduction in the intercellular movement of PVX (Fig. 4C) or TBSV (Fig. 4D) following myosin silencing, indicating that the effect of myosin XI-2 on cell-to-cell virus movement is not universal among viruses.
Fig. 4.
TMV utilizes a distinct myosin for virus spread in N. benthamiana. GFP lesion areas reflect the cell-to-cell movement of (A) TMV, (B) TVCV, (C) PVX, and (D) TBSV in N. benthamiana leaves silenced for individual myosin genes (VIII-1, VIII-2, XI-2, XI-F) via TRV VIGS. Plants inoculated with wild-type TRV (WT TRV) or buffer (Mock) were controls. The area of GFP fluorescent lesions (mm2) in inoculated leaves was determined at 3 dpi for tissues carrying a systemic infection with the TRV VIGS vector (approximately 18 dpi). Bars represent means ± standard errors for three replicates per treatment. Analysis of variance followed by an lsd calculation was used to determine significant differences between treatments. Different letters above the bars indicate significant differences (P = 0.05). The experiment was repeated at least once for each virus challenge.
Discussion
The Sustained Intercellular Movement of Diverse Plant RNA Viruses Requires Intact Microfilaments.
Although LatB treatment inhibits the cell-to-cell spread of TMV and the DNA plant virus, CaMV (11, 20), the effect of this actin depolymerizing agent on the movement of other plant viruses has not been determined. Here, we show that in addition to TMV, the movement of PVX and TBSV are significantly inhibited by LatB treatment (Fig. 1). These results suggest that utilization of the actin cytoskeleton for sustained transport is a strategy used by a range of RNA plant viruses representing different genera. In the case of PVX, although the intercellular movement of this virus was not known to depend upon actin, the PVX movement protein TGBp2 does associate with actin filaments (22) and TGBp2's ER localization is disrupted by LatB treatment (41). Our finding that PVX movement is disrupted by LatB is consistent with a role for a TGBp2-containing actin-ER network complex in viral cell-to-cell movement.
Unlike TMV and PVX, TBSV has not been analyzed for its relationship with the cytoskeleton during movement. Our finding that LatB inhibits TBSV intercellular movement suggests that this virus, like TMV, PVX, and CaMV, should have a protein that mediates an association with microfilaments. The movement of TBSV is known to rely upon both the p22 and p19 viral proteins (42, 43). While p19 is a suppressor of silencing found primarily in the cytosol and implicated in long distance transport, p22 is required for cell-to-cell transport and is membrane-associated (42, 44). It will be interesting to determine whether either of these proteins associates with the cytoskeleton, particularly given the close association of microfilaments with membranes. Although the TBSV p33 replication protein has not been associated with movement, the formation of TBSV replication complexes is thought to proceed via a peroxisome-to-endoplasmic reticulum sorting pathway (45). Peroxisome budding in Arabidopsis depends on the actomyosin system (46). Since TMV replication complexes are associated with actin-dependent virus movement (11, 12), it would be interesting to investigate a similar possibility for TBSV.
Surprising Differences between Viruses in the Same Genus for Microfilament-Mediated Movement.
Given the sensitivity of TMV to LatB, it was very surprising to find that the movement of the closely related tobamovirus TVCV was not affected by LatB treatment (Fig. 1). Correlated with this finding was the observation that the TVCV 125-kDa protein, unlike its homolog the TMV 126-kDa protein, did not form cytoplasmic inclusions that associated with microfilaments (Fig. 2 and Table 1). Since the TMV 126-kDa protein, like the TMV VRC, traffics along the microfilament network and it and the TVCV 125-kDa protein are known or suspected to be required for intercellular movement (12, 38), differences between the TMV 126-kDa protein and the TVCV 125-kDa protein may be responsible for the divergent responses of these two viruses to LatB treatment. Although the poorly conserved intervening region (IR) (47) in the center of these protein sequences (Fig. S4) is a candidate as a determinant for inclusion body formation and movement, perhaps a more likely candidate region is the helicase domain, which for TMV is instrumental in 126-kDa protein oligomerization (47). This function may catalyze 126-body and possibly VRC formation and subsequent microfilament association. The TVCV 125-kDa protein may not have the signature sequences necessary for this activity. Indeed, TVCV rarely forms inclusions similar to those formed during TMV infection (48). While the 126- and 125-kDa proteins are primary suspects controlling this differential phenotype between these viruses, their MPs are also potential determinants. It has been shown that the TMV MP requires intact microfilaments for transport to PD (49), and although contrary findings exist (23), differences in MP targeting require investigation.
A second important conclusion to be drawn from the observation that TVCV is unaffected by LatB treatment is that the inhibited movement observed for the other viruses during this treatment was not due to a general inhibition of plant cell processes that would influence virus movement indirectly (20). Here it is pertinent to note that our results contrast with those from recent work showing that intercellular spread of TMV is not inhibited by LatB treatments (50). The reasons for this discrepancy are not known, although there were differences in the experimental systems, notably in the duration of treatment with the inhibitor (24 h versus 6 days) and the time when virus movement was quantified (24 h versus 2–6 days). Antagonists of microfilaments are known to increase the size exclusion limits of PD (51), and the ability of viruses to use this modification for increased short-term movement versus a sustained inhibitory effect of LatB treatment on VRC intracellular and virus intercellular movement is unclear.
Taken together, our results demonstrate that although a dependence upon the actin cytoskeleton for movement appears to be widespread among diverse plant viruses, even closely related viruses may differ in their requirement for actin. Our finding demonstrating PVX and TBSV spread is inhibited by disruption of the actin cytoskeleton does not rule out the possible involvement of microtubules in movement of these viruses. However, a role for microtubules in plant virus cell-to-cell movement remains less supported since TMV (9, 11, 52) and TVCV (Fig. S9) were unaffected in cell-to-cell movement after treatment with the microtubule inhibitor oryzalin. In addition, the targeting of the 48-kDa movement protein of Cowpea mosaic virus to peripheral punctate structures in protoplasts was unaffected after treatment with oryzalin (53) and the P6 protein of CaMV, although associating with microtubules, did not traffic along them (20). These findings suggest that plant viruses use microfilaments to a much greater extent than microtubules for movement compared with animal viruses (4, 5). For TVCV, however, no cytoskeletal element was identified as essential for its movement in our experiments. Clearly, this virus requires further study to determine how it navigates within cells and then to PD for sustained intercellular transport.
An Individual Myosin Motor Influences the Movement of an Individual Plant Virus.
Leaves silenced for individual myosins were inoculated with our collection of RNA viruses (TMV, TVCV, PVX, and TBSV), and only TMV movement was inhibited by silencing individual myosins (Figs. 3 and 4). Based on our current findings and those from earlier work showing that the PD localization of the BYV Hsp70h protein was dependent upon class VIII myosins whereas the PD localization of TMV MP was not (33), it is apparent that plant viruses likely use different myosins to maintain their cell-to-cell movement and that specific viral proteins mediate this response. For the Hsp70h protein, it should be noted that its subcellular localization was affected equally by all class VIII myosins tested. In our studies, it is interesting that although myosin VIII-1 expression was reduced by the greatest amount of the four myosins, silencing this myosin did not have an effect on the movement of the viruses tested, whereas a more modest reduction in myosin XI-2 levels had a significant effect on TMV spread. Although we have tested only two myosins from each class in our current study, our results demonstrate that not all class XI myosins have an identical effect on TMV movement. Whether silencing of N. benthamiana myosin XI-2 resulted in off target silencing of N. benthamiana myosins not studied here cannot be definitively determined, since all of those sequences are not available. By analysis of the A. thaliana myosin gene family homologs, however, off target effects on these other N. benthamiana myosin sequences would be limited to those within classes (i.e., between class XI members) and not between classes (i.e., between class XI and VIII members).
It seems reasonable that different individual myosins may be targeted for use by different plant viruses since no individual myosin has been found to be essential for plant growth (28, 29). During our experiments, we also did not observe any visible phenotype associated with the silencing of these individual myosins. Myosins XI-2 and XI-K are among the most abundant and widely expressed myosins in Arabidopsis (28). In Arabidopsis, myosin XI-2 influences organelle trafficking and polar tip growth (28). However, in N. benthamiana plants, where myosins XI-2 and XI-F were subjected to dominant negative inhibition or RNAi, only modest effects on organelle transport were observed compared with those observed after myosin XI-K inhibition (27). It may be that the targeting by TMV of this apparently least essential class XI plant myosin, which is expressed in most plant tissues, occurred through plant-virus co-evolution to allow maximum TMV movement with minimum impact on the host. Although further studies will be required to determine the effect of silencing myosin XI-2 and XI-K on the intracellular movement of VRCs, a myosin capable of transporting organelles would be a good candidate to transport large TMV VRCs that contain ER (16, 18).
In addition to expanding on the findings with TMV, it will be important to determine which myosin influences movement of PVX and TBSV. Although myosin XI-K has a larger influence than myosin XI-2 or XI-F on organelle trafficking in N. benthamiana, it still is not essential for plant growth and is expressed in leaf cells, a necessity for viruses that infect this tissue. This reasoning also would suggest that myosins expressed only in flowers or other reproductive tissue [e.g., XI-A, -B, -C, -D, and -J in Arabidopsis; (28)] would not be targeted by plant viruses for movement. It also remains a possibility that the percentage knockdown of the individual myosins was not sufficient to modify PVX and TBSV movement. Such a finding, however, would still indicate a significant difference in threshold myosin levels necessary to support intercellular movement between virus species.
It is interesting to note that although actin has been implicated in many aspects of animal virus infections such as virus entry, replication complex intracellular movement, and virus release (4, 54), the role of specific myosins has only been identified in processes such as filopodial “surfing” or cellular budding (35, 55), events which have no equivalent during the infection cycle in plants. Although the role of specific myosins in the intracellular transport of animal viruses remains to be determined, there is evidence that myosin-dependent movement is important for the trafficking of animal viruses within cells. Myosin inhibitor studies with 2,3-butanedione monoxime have revealed that myosin-based movement is required for the transport of herpes simplex virus-1 capsids within the nucleus (34). Our findings should have the greatest influence on those studying the mechanism by which animal viruses modify the cortical microfilament network to reach or exit the long-distance microtubule transport system within the cell (4), a phenomenon that, like plant virus movement, occurs in the cytoplasm.
Our findings have practical significance for those in agriculture investigating methods to produce virus-resistant plants. Decreased expression of nonessential host myosins should lead to increased resistance to different plant viruses. Although a knockdown of a single myosin may not inhibit the spread of many viruses, knockdowns of a small variety of myosins could lead to resistance against particular viruses for specific plants of economic interest.
Materials and Methods
Viruses Used in This Study.
For construction of TVCV-GFP, pTVCV50 (38) was modified to include a duplication of the CP subgenomic promoter between the MP and CP ORFs driving enhanced GFP (EGFP) expression. Construction of TMV-GFP (previously TMV-MP-GFP-CP) is described elsewhere (56). TMV expressing free GFP (p4GD-GFP; Fig. S2) was a gift of Curtis Holt and is described elsewhere (57). TBSV-GFP was a gift from H. Scholthof (Texas A&M University), and PVX-GFP (originally pPC2S) (58) was obtained from J. Verchot-Lubicz (Oklahoma State University). For virus inoculations, infectious transcripts were generated from 1 μg linearized plasmid DNA using the mMessage mMachine in vitro transcription kit (Ambion). Half of each transcript reaction was used to rub-inoculate individual N. benthamiana leaves. For analysis of TMV local lesions, N. tabacum cv. Xanthi-NN were rub-inoculated with TMV (strain U1) virions. Greenhouse conditions were 24 ± 2 °C with 60% humidity and 16 h supplemental lighting (400 μmol m−2s−1).
Cloning and Ectopic Expression of 126-kDa and 125-kDa Protein-GFP Fusions.
For construction of TVCV 125-GFP, the TVCV 125-kDa protein ORF (minus a stop codon) from pTVCV50 (38) was cloned into the pRTL2 vector (59) containing an enhanced 35S promoter. EGFP was added in-frame at the 3′ end of the TVCV 125-kDa protein, and the promoter-gene-terminator fragment was removed from pRTL2 and cloned into binary vector pGA482 (60). The resulting construct was sequenced to confirm that no modification occurred. TMV 126-GFP construction is described elsewhere (12, 61). Following transformation into Agrobacterium spp. (strain LBA4404), constructs were grown under selection and infiltrated into N. benthamiana leaves at a final optical density (600 nm) of 0.5 as described (62).
Inhibitor Treatments.
LatB infiltrations were carried out as described (20). Disruption of actin by LatB was confirmed by microscopic observation of microfilaments labeled with the actin binding domain 2 of Arabidopsis fimbrin fused to GFP (36). This construct was expressed transiently following Agrobacterium spp. infiltration as described above.
Microscopy.
Fluorescent viral lesions were imaged on an SZX-12 fluorescent stereo microscope (Olympus) equipped with a Dxm 1200c digital camera (Nikon Instruments). Lesion sizes were calculated using ImageJ software (version 1.38e; http://rsbweb.nih.gov/ij/). For visualization of TMV 126-GFP and TVCV 125-GFP, images were acquired on a Bio-Rad 1024ES confocal imaging system (Bio-Rad Laboratories) or a Leica TCS SP2 confocal imaging system (Leica Microsystems). GFP was excited at 488 nm and captured at 522 nm. Images were processed using Adobe Photoshop (Adobe). GFP body diameters were determined using Image J software.
VIGS and Quantitative RT-PCR.
N. benthamiana myosin tail sequences outlined in Fig. S6 were amplified by PCR from cDNA using exTaq polymerase (Takara). The resulting fragments were sequenced and cloned into pTRV2 (63) using Gateway cloning technology (Invitrogen). TRV infections were established in N. benthamiana by co-agroinfiltration of pTRV1 and pTRV2. To confirm silencing of specific myosin transcripts, we prepared RNA from TRV-infected systemic leaves (three plants/construct) using the RNeasy plant mini kit (Qiagen). For lesion analysis (Fig. S7B), fluorescent lesions were visualized with a UV lamp and excised with a scalpel. DNase-treated RNA (3 μg) was used to generate cDNA with M-MLV reverse transcriptase (Promega). Fifteen- to eighteen-base Oligo (dT) (Invitrogen) was used for first strand cDNA synthesis. Following a 20-fold dilution of the cDNA, 2 μL were used for quantitative RT-PCR with an ABI Prism 7900 HT sequence detection system (Applied Biosystems). Primers used to detect N. benthamiana myosins are given (Fig. S6). GFP primers 38016: 5′-GTCCGCCCTGAGCAAAGA-3′ and 38017: 5′-TCCAGCAGGACCATGTGATC-3′ were used to detect 126-GFP and 125-GFP. Transcript levels were adjusted for loading differences after comparison with EF1α transcript internal control values and were calculated using a relative quantification method (64).
Amino Acid Alignments.
Amino acid sequences were aligned using clustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html) with default settings.
Supplementary Material
Acknowledgments.
We thank Ulrich Melcher (Oklahoma State University), Jeanmarie Verchot-Lubicz, and Herman Scholthof for providing pTVCV50, PVX-GFP, and TBSV-GFP constructs, respectively; Rujin Chen, Jim Schoelz, and Julia Dyachok for critical reading of the manuscript; Sumana Bhat for molecular cloning work; Stacy Allen for assistance with real-time PCR; Frank Coker and Vicki Barrett for plant care; and Stephen Mannas for technical support. This work was supported by the Samuel Roberts Noble Foundation, Inc.; National Science Foundation Multiuser Instrumentation Program Grant DBI-0400580 (to R.S.N.); and the United Kingdom Biotechnology and Biological Sciences Research Council through a grant-in-aid to the John Innes Centre (to A.J.M.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909239106/DCSupplemental.
References
- 1.Verchot-Lubicz J, Ye CM, Bamunusinghe D. Molecular biology of potexviruses: Recent advances. J Gen Virol. 2007;88:1643–1655. doi: 10.1099/vir.0.82667-0. [DOI] [PubMed] [Google Scholar]
- 2.Hofmann C, Sambade A, Heinlein M. Plasmodesmata and intercellular transport of viral RNA. Biochem Soc Trans. 2007;35:142–145. doi: 10.1042/BST0350142. [DOI] [PubMed] [Google Scholar]
- 3.Lucas WJ. Plant viral movement proteins: Agents for cell-to-cell trafficking of viral genomes. Virology. 2006;344:169–184. doi: 10.1016/j.virol.2005.09.026. [DOI] [PubMed] [Google Scholar]
- 4.Radtke K, Dohner K, Sodeik B. Viral interactions with the cytoskeleton: A hitchhiker's guide to the cell. Cell Microbiol. 2006;8:387–400. doi: 10.1111/j.1462-5822.2005.00679.x. [DOI] [PubMed] [Google Scholar]
- 5.Greber UF, Way M. A superhighway to virus infection. Cell. 2006;124:741–754. doi: 10.1016/j.cell.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 6.Heinlein M, et al. Changing patterns of localization of the tobacco mosaic virus movement protein and replicase to the endoplasmic reticulum and microtubules during infection. Plant Cell. 1998;10:1107–1120. doi: 10.1105/tpc.10.7.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McLean BG, Zupan J, Zambryski PC. Tobacco mosaic virus movement protein associates with the cytoskeleton in tobacco cells. Plant Cell. 1995;7:2101–2114. doi: 10.1105/tpc.7.12.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyko V, Ferralli J, Ashby J, Schellenbaum P, Heinlein M. Function of microtubules in intercellular transport of plant virus RNA. Nat Cell Biol. 2000;2:826–832. doi: 10.1038/35041072. [DOI] [PubMed] [Google Scholar]
- 9.Gillespie T, et al. Functional analysis of a DNA-shuffled movement protein reveals that microtubules are dispensable for the cell-to-cell movement of tobacco mosaic virus. Plant Cell. 2002;14:1207–1222. doi: 10.1105/tpc.002303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curin M, et al. MPB2C, a microtubule-associated plant factor, is required for microtubular accumulation of tobacco mosaic virus movement protein in plants. Plant Physiol. 2007;143:801–811. doi: 10.1104/pp.106.091488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawakami S, Watanabe Y, Beachy RN. Tobacco mosaic virus infection spreads cell to cell as intact replication complexes. Proc Natl Acad Sci USA. 2004;101:6291–6296. doi: 10.1073/pnas.0401221101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu JZ, Blancaflor EB, Nelson RS. The tobacco mosaic virus 126-kilodalton protein, a constituent of the virus replication complex, alone or within the complex aligns with and traffics along microfilaments. Plant Physiol. 2005;138:1853–1865. doi: 10.1104/pp.105.065722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szecsi J, et al. Development of tobacco mosaic virus infection sites in Nicotiana benthamiana. Mol Plant Microbe Interact. 1999;12:143–152. [Google Scholar]
- 14.Hills GJ, et al. Immunogold localization of the intracellular sites of structural and nonstructural tobacco mosaic virus proteins. Virology. 1987;161:488–496. doi: 10.1016/0042-6822(87)90143-7. [DOI] [PubMed] [Google Scholar]
- 15.Saito T, Hosokawa D, Meshi T, Okada Y. Immunocytochemical localization of the 130K and 180K proteins (putative replicase components) of tobacco mosaic virus. Virology. 1987;160:477–481. doi: 10.1016/0042-6822(87)90020-1. [DOI] [PubMed] [Google Scholar]
- 16.Mas P, Beachy RN. Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein in intracellular distribution of viral RNA. J Cell Biol. 1999;147:945–958. doi: 10.1083/jcb.147.5.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Padgett HS, et al. Distribution of tobamovirus movement protein in infected cells and implications for cell-to-cell spread of infection. Plant J. 1996;10:1079–1088. doi: 10.1046/j.1365-313x.1996.10061079.x. [DOI] [PubMed] [Google Scholar]
- 18.dos Reis Figueira A, Golem S, Goregaoker SP, Culver JN. A nuclear localization signal and a membrane association domain contribute to the cellular localization of the Tobacco mosaic virus 126-kDa replicase protein. Virology. 2002;301:81–89. doi: 10.1006/viro.2002.1560. [DOI] [PubMed] [Google Scholar]
- 19.Yamanaka T, et al. TOM1, an Arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein. Proc Natl Acad Sci USA. 2000;97:10107–10112. doi: 10.1073/pnas.170295097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harries PA, Karuppaiah P, Yu W, Schoelz JE, Nelson RS. The cauliflower mosaic virus protein P6 forms motile inclusions that traffic along actin microfilaments and stabilize microtubules. Plant Physiol. 2009;149:1005–1016. doi: 10.1104/pp.108.131755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haupt S, et al. Two plant-viral movement proteins traffic in the endocytic recycling pathway. Plant Cell. 2005;17:164–181. doi: 10.1105/tpc.104.027821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ju HJ, et al. The potato virus X TGBp2 movement protein associates with endoplasmic reticulum-derived vesicles during virus infection. Plant Physiol. 2005;138:1877–1895. doi: 10.1104/pp.105.066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prokhnevsky AI, Peremyslov VV, Dolja VV. Actin cytoskeleton is involved in targeting of a viral Hsp70 homolog to the cell periphery. J Virol. 2005;79:14421–14428. doi: 10.1128/JVI.79.22.14421-14428.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foth BJ, Goedecke MC, Soldati D. New insights into myosin evolution and classification. Proc Natl Acad Sci USA. 2006;103:3681–3686. doi: 10.1073/pnas.0506307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reddy AS, Day IS. Analysis of the myosins encoded in the recently completed Arabidopsis thaliana genome sequence. Genome Biol. 2001;2:research0024.0021–research0024.0017. doi: 10.1186/gb-2001-2-7-research0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sparkes IA, Teanby NA, Hawes C. Truncated myosin XI tail fusions inhibit peroxisome, Golgi, and mitochondrial movement in tobacco leaf epidermal cells: A genetic tool for the next generation. J Exp Bot. 2008;59:2499–2512. doi: 10.1093/jxb/ern114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Avisar D, Prokhnevsky AI, Makarova KS, Koonin EV, Dolja VV. Myosin XI-K Is required for rapid trafficking of Golgi stacks, peroxisomes, and mitochondria in leaf cells of Nicotiana benthamiana. Plant Physiol. 2008;146:1098–1108. doi: 10.1104/pp.107.113647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peremyslov VV, Prokhnevsky AI, Avisar D, Dolja VV. Two class XI myosins function in organelle trafficking and root hair development in Arabidopsis. Plant Physiol. 2008;146:1109–1116. doi: 10.1104/pp.107.113654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prokhnevsky AI, Peremyslov VV, Dolja VV. Overlapping functions of the four class XI myosins in Arabidopsis growth, root hair elongation, and organelle motility. Proc Natl Acad Sci USA. 2008;105:19744–19749. doi: 10.1073/pnas.0810730105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ojangu EL, Jarve K, Paves H, Truve E. Arabidopsis thaliana myosin XIK is involved in root hair as well as trichome morphogenesis on stems and leaves. Protoplasma. 2007;230:193–202. doi: 10.1007/s00709-006-0233-8. [DOI] [PubMed] [Google Scholar]
- 31.Sattarzadeh A, Franzen R, Schmelzer E. The Arabidopsis class VIII myosin ATM2 is involved in endocytosis. Cell Motil Cytoskeleton. 2008;65:457–468. doi: 10.1002/cm.20271. [DOI] [PubMed] [Google Scholar]
- 32.Golomb L, Abu-Abied M, Belausov E, Sadot E. Different subcellular localizations and functions of Arabidopsis myosin VIII. BMC Plant Biol. 2008;8:3. doi: 10.1186/1471-2229-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Avisar D, Prokhnevsky AI, Dolja VV. Class VIII myosins are required for plasmodesmatal localization of a closterovirus Hsp70 homolog. J Virol. 2008;82:2836–2843. doi: 10.1128/JVI.02246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forest T, Barnard S, Baines JD. Active intranuclear movement of herpesvirus capsids. Nat Cell Biol. 2005;7:429–431. doi: 10.1038/ncb1243. [DOI] [PubMed] [Google Scholar]
- 35.Sasaki H, et al. Myosin-actin interaction plays an important role in human immunodeficiency virus type 1 release from host cells. Proc Natl Acad Sci USA. 1995;92:2026–2030. doi: 10.1073/pnas.92.6.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang YS, Yoo CM, Blancaflor EB. Improved imaging of actin filaments in transgenic Arabidopsis plants expressing a green fluorescent protein fusion to the C- and N-termini of the fimbrin actin-binding domain 2. New Phytol. 2008;177:525–536. doi: 10.1111/j.1469-8137.2007.02261.x. [DOI] [PubMed] [Google Scholar]
- 37.Hirashima K, Watanabe Y. RNA helicase domain of tobamovirus replicase executes cell-to-cell movement possibly through collaboration with its nonconserved region. J Virol. 2003;77:12357–12362. doi: 10.1128/JVI.77.22.12357-12362.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Lartey RT, Hartson SD, Voss TC, Melcher U. Limitations to tobacco mosaic virus infection of turnip. Arch Virol. 1999;144:957–971. doi: 10.1007/s007050050558. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 2002;30:415–429. doi: 10.1046/j.1365-313x.2002.01297.x. [DOI] [PubMed] [Google Scholar]
- 40.Thomas CL, Jones L, Baulcombe DC, Maule AJ. Size constraints for targeting post-transcriptional gene silencing and for RNA-directed methylation in Nicotiana benthamiana using a potato virus X vector. Plant J. 2001;25:417–425. doi: 10.1046/j.1365-313x.2001.00976.x. [DOI] [PubMed] [Google Scholar]
- 41.Mitra R, et al. The potato virus X TGBp2 protein association with the endoplasmic reticulum plays a role in but is not sufficient for viral cell-to-cell movement. Virology. 2003;312:35–48. doi: 10.1016/s0042-6822(03)00180-6. [DOI] [PubMed] [Google Scholar]
- 42.Scholthof HB, Scholthof KB, Kikkert M, Jackson AO. Tomato bushy stunt virus spread is regulated by two nested genes that function in cell-to-cell movement and host-dependent systemic invasion. Virology. 1995;213:425–438. doi: 10.1006/viro.1995.0015. [DOI] [PubMed] [Google Scholar]
- 43.Turina M, et al. A newly identified role for Tomato bushy stunt virus P19 in short distance spread. Mol Plant Path. 2003;4:67–72. doi: 10.1046/j.1364-3703.2003.00151.x. [DOI] [PubMed] [Google Scholar]
- 44.Canto T, Uhrig JF, Swanson M, Wright KM, MacFarlane SA. Translocation of Tomato bushy stunt virus P19 protein into the nucleus by ALY proteins compromises its silencing suppressor activity. J Virol. 2006;80:9064–9072. doi: 10.1128/JVI.00953-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCartney AW, Greenwood JS, Fabian MR, White KA, Mullen RT. Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell. 2005;17:3513–3531. doi: 10.1105/tpc.105.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jedd G, Chua NH. Visualization of peroxisomes in living plant cells reveals acto-myosin-dependent cytoplasmic streaming and peroxisome budding. Plant Cell Physiol. 2002;43:384–392. doi: 10.1093/pcp/pcf045. [DOI] [PubMed] [Google Scholar]
- 47.Goregaoker SP, Culver JN. Oligomerization and activity of the helicase domain of the tobacco mosaic virus 126- and 183-kilodalton replicase proteins. J Virol. 2003;77:3549–3556. doi: 10.1128/JVI.77.6.3549-3556.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Resconich EC. Interpretation of the forms of inclusions in bean systemically infected with tobacco mosaic virus. Virology. 1961;15:16–21. doi: 10.1016/0042-6822(61)90071-x. [DOI] [PubMed] [Google Scholar]
- 49.Wright KM, et al. Targeting of TMV movement protein to plasmodesmata requires the actin/ER network: Evidence from FRAP. Traffic. 2007;8:21–31. doi: 10.1111/j.1600-0854.2006.00510.x. [DOI] [PubMed] [Google Scholar]
- 50.Hofmann C, Niehl A, Sambade A, Steinmetz A, Heinlein M. Inhibition of tobacco mosaic virus movement by expression of an actin-binding protein. Plant Physiol. 2009;149:1810–1823. doi: 10.1104/pp.108.133827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ding B, Kwon MO, Warnberg L. Evidence that actin filaments are involved in controlling the permeability of plasmodesmata in tobacco mesophyll. Plant J. 1996;10:157–164. [Google Scholar]
- 52.Liu JZ, Blancaflor E, Nelson RS. In: Biology of Molecular Plant-Microbe Interaction. Sanchez F, Quinto C, Lopez-Lara IM, Geiger O, editors. St. Paul, MN: IS-MPMI; 2006. pp. 410–415. [Google Scholar]
- 53.Pouwels J, Van Der Krogt GN, Van Lent J, Bisseling T, Wellink J. The cytoskeleton and the secretory pathway are not involved in targeting the cowpea mosaic virus movement protein to the cell periphery. Virology. 2002;297:48–56. doi: 10.1006/viro.2002.1424. [DOI] [PubMed] [Google Scholar]
- 54.Lai CK, Jeng KS, Machida K, Lai MM. Association of hepatitis C virus replication complexes with microtubules and actin filaments is dependent on the interaction of NS3 and NS5A. J Virol. 2008;82:8838–8848. doi: 10.1128/JVI.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lehmann MJ, Sherer NM, Marks CB, Pypaert M, Mothes W. Actin- and myosin-driven movement of viruses along filopodia precedes their entry into cells. J Cell Biol. 2005;170:317–325. doi: 10.1083/jcb.200503059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oparka KJ, Prior DA, Santa Cruz S, Padgett HS, Beachy RN. Gating of epidermal plasmodesmata is restricted to the leading edge of expanding infection sites of tobacco mosaic virus (TMV) Plant J. 1997;12:781–789. doi: 10.1046/j.1365-313x.1997.12040781.x. [DOI] [PubMed] [Google Scholar]
- 57.Casper SJ, Holt CA. Expression of the green fluorescent protein-encoding gene from a tobacco mosaic virus-based vector. Gene. 1996;173:69–73. doi: 10.1016/0378-1119(95)00782-2. [DOI] [PubMed] [Google Scholar]
- 58.Baulcombe DC, Chapman S, Santa Cruz S. Jellyfish green fluorescent protein as a reporter for virus infections. Plant J. 1995;7:1045–1053. doi: 10.1046/j.1365-313x.1995.07061045.x. [DOI] [PubMed] [Google Scholar]
- 59.Restrepo MA, Freed DD, Carrington JC. Nuclear transport of plant potyviral proteins. Plant Cell. 1990;2:987–998. doi: 10.1105/tpc.2.10.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.An GP, Ebert R, Mitra A, Ha SB. In: Plant Molecular Biology Manual. Gelvin SB, Schilperoort RA, Verma DPS, editors. A3. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1988. pp. 1–19. [Google Scholar]
- 61.Ding XS, et al. The Tobacco mosaic virus 126-kDa protein associated with virus replication and movement suppresses RNA silencing. Mol Plant Microbe Interact. 2004;17:583–592. doi: 10.1094/MPMI.2004.17.6.583. [DOI] [PubMed] [Google Scholar]
- 62.Harries PA, Palanichelvam K, Bhat S, Nelson RS. Tobacco mosaic virus 126-kDa protein increases the susceptibility of Nicotiana tabacum to other viruses and its dosage affects virus-induced gene silencing. Mol Plant Microbe Interact. 2008;21:1539–1548. doi: 10.1094/MPMI-21-12-1539. [DOI] [PubMed] [Google Scholar]
- 63.Liu Y, Schiff M, Dinesh-Kumar SP. Virus-induced gene silencing in tomato. Plant J. 2002;31:777–786. doi: 10.1046/j.1365-313x.2002.01394.x. [DOI] [PubMed] [Google Scholar]
- 64.Pfaffl MW. A new mathematical model for relative quantification in real- time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.