Abstract
Geographical separation of host species has shaped the avian influenza A virus gene pool into independently evolving Eurasian and American lineages, although phylogenetic evidence for gene flow and reassortment indicates that these lineages also mix on occasion. While the evolutionary dynamics of the avian influenza gene pool have been described, the consequences of gene flow on virus evolution and population structure in this system have not been investigated. Here we show that viral gene flow from Eurasia has led to the replacement of endemic avian influenza viruses in North America, likely through competition for susceptible hosts. This competition is characterized by changes in rates of nucleotide substitution and selection pressures. However, the discontinuous distribution of susceptible hosts may produce long periods of co-circulation of competing virus strains before lineage extinction occurs. These results also suggest that viral competition for host resources may be an important mechanism in disease emergence.
Keywords: evolution, ecology, population dynamics, influenza A virus, competition
Introduction
Wild waterfowl and shorebirds (Anseriformes and Charadriiformes) have been recognized as the major natural reservoirs of influenza A viruses (Webster et al., 1992). Avian influenza viruses (AIV) in these hosts are maintained asymptomatically, although there is some evidence of clinical effects of these viruses in Anseriformes (van Gils et al., 2007). Phylogenetic analyses have revealed that influenza A viruses found in all other host species, including humans, were ultimately derived from avian viruses (Gorman et al., 1990a, 1990b, 1991; Kawaoka et al., 1989; Okazaki et al., 1989; Schäfer et al., 1993).
Geographical separation of host species has shaped the influenza gene pool into largely independently evolving Eurasian and American lineages (Donis et al., 1989; Obenauer et al., 2006) although gene flow among these regions has been documented (Kishida et al., 2008; Kraus et al., 2007; Liu et al., 2004; Markova et al., 1999; Wallensten et al., 2005). For example, influenza viruses of the H6 subtype derived from Eurasian ancestors were detected in North American wild birds, with subsequent interspecies transmission to local poultry that led to disease outbreaks in California from 2000 to 2002 (Spackman et al., 2001; Webby et al., 2003; Woolcock et al., 2003). These viruses contained the surface hemagglutinin (HA) gene from Eurasian gene pool viruses, while the remaining gene segments were of American origin (Woolcock et al., 2003).
Reassortment between Eurasian and North American lineage viruses have also been documented in wild aquatic bird populations indicating that these two geographically segregated lineages represent mixing populations of viruses, although intercontinental migration of complete viral genomes have not yet been detected (Dugan et al., 2008; Krauss et al. 2007). In Eurasia, the long-term endemicity of H5N1 and H9N2 influenza viruses pose a persistent pandemic threat and has heightened concerns that gene flow may result in natural introductions of H5N1, by means of bird migration, to North America (Olsen et al., 2006; Winker et al., 2007). However, the possible effects of virus gene flow between the Eurasian and American gene pools on influenza virus evolution and population structure have not been explored.
In this study we analyzed 31 years of accumulated influenza virus sequence data to better understand the evolutionary dynamics of influenza viruses, particularly the consequences of gene flow between continents. Our analyses show that viral gene flow from Eurasia has led to the exclusion of endemic AIVs in North America, most likely mediated by competition for susceptible hosts. Furthermore, we show that intercontinental gene flow is frequently mediated through shorebirds, highlighting the need for increased surveillance of influenza viruses in a broader spectrum of potential host species.
Results
Experimental design
All available sequence data from influenza viruses isolated exclusively from North American wild aquatic birds over a 31-year period (1975–2006) were recruited and combined with influenza virus sequence data sets isolated from Eurasia. Extensive preliminary phylogenetic analyses of all influenza subtype viruses detected from wild birds (see Materials and Methods for details) identified two evolutionary scenarios worthy of further investigation: H6-HA phylogenies that showed evidence of two-way gene flow between continents, and H4-HA phylogenies that did not. To explore whether the H6 subtype introductions were associated with specific NA subtype introductions, we also analyzed the most common subtypes of NA in combination with H6-HA found in both gene pools (N1, N2 and N8). Evidence of gene flow was also confirmed for each of the six internal gene segments and is presented below.
It is important to note that analyses of the surface genes presented below, particularly the HA, allows us to assess gene flow and changes in genetic diversity of a specific influenza virus subtype over time. However, the high degree of genetic reassortment observed in the internal genes dictates these results cannot be interpreted in the same manner. Rather the internal gene analyses provide a relative measure of genetic exchange between the Eurasian and North American gene pools, independent of the virus subtype.
Relaxed molecular clock phylogenetic analysis of the H6 hemagglutinin
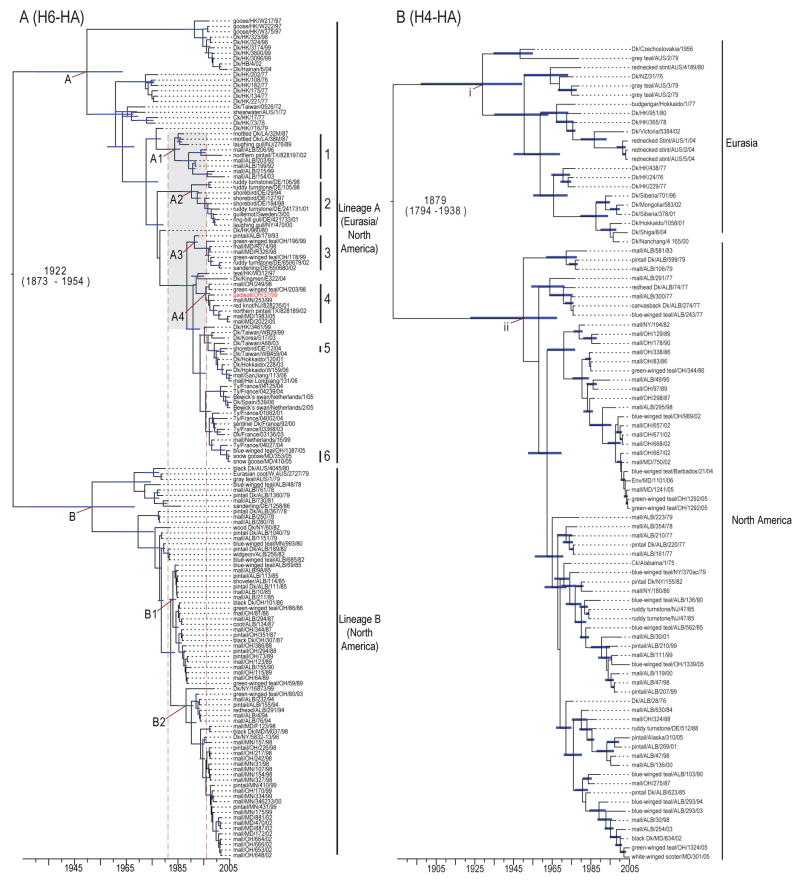
Phylogenetic analysis utilizing Bayesian relaxed molecular clock methods, such that evolutionary rate can vary among branches on the tree, identified two major H6-HA gene lineages (Fig. 1A). Lineage A contained a mixture of H6-HA influenza subtype viruses currently circulating in the Eurasian and North American gene pools, revealing intercontinental gene flow. Interestingly, the H6-HA lineage A phylogeny revealed monophyletic groups containing mixtures of viruses isolated in both continental regions, providing strong evidence for two-way gene flow. In contrast, the H6-HA lineage B was composed exclusively of North American isolates sampled from 1978–2002, with the exception of three closely related viruses from Australian wild ducks (Fig. 1A).
FIG. 1. Phylogeny of H6 and H4 HA genes.
Dated MCC phylogeny of the H6 (A) and H4 (B) HA of viruses. Blue bars at nodes indicate 95% HPD of TMRCAs. Scale bar shows the time-scale in years. In (A) the grey box and dashed red lines correspond to the period when lineage A introductions into the North American gene pool occurred and clades 1–6 indicate isolates with Eurasian ancestry isolated in North American wild birds. Taxa marked indicated in red in (A) is a hemagglutinin lineage introduced with Eurasian ancestry isolated in North American wild birds. This lineage corresponds to the introduction shown in Fig 3.
Date estimates of the ancestral nodes inferred using the same methodology indicated that the Eurasian and North American H6-HA lineages diverged in the 1920’s (time of most recent common ancestor (TMRCA) 1922, 95% highest posterior density (HPD): 1873–1954). Age estimates of Lineage A viruses (TMRCA 1951, 95% HPD: 1930–1965) and lineage B (TMRCA 1956, 95% HPD: 1930–1969) shows that these geographically isolated populations diverged relatively recently. Lineage B consists of two sublineages, B1 and B2. Four novel introductions of Eurasian H6-HA viruses, all within lineage A, resulted in the establishment of viral sublineages in the North American gene pool (Fig. 1A; clades 1–4). Estimates of TMRCAs and 95% HPDs of these clades showed that these introductions most likely occurred between 1981 and 1996 (nodes A1–A4), indicated by the grey box and dashed red lines in Fig. 1A. Lineage B viruses therefore circulated independently in North America until 1981, the earliest possible date that that gene flow of Eurasian lineage A viruses to North America occurred (Fig. 1A; nodes A and B).
Dated phylogenies also revealed corresponding changes in the evolutionary history of H6-HA lineage B viruses with sublineage B1 (TMRCA 1983, 95% HPD: 1982–1984), emerging soon after the introduction of Eurasian lineage A viruses to North America (Fig. 1A). Sublineage B1 was subsequently replaced by sublineage B2 (TMRCA 1987, 95% HPD: 1985–1991), although sublineage B2 also ultimately suffered extinction (Fig. 1A). In these analyses, the time from the lower HPD to the year of the last sampled isolate of a clade provides the maximum time that a lineage may have persisted in a population. In the case of sublineages B1 and B2, maximum persistence time was 1982–1990 (8 years) and 1985–2002 (17 years), respectively (Fig. 1A). The short tree branch lengths further demonstrate the rapid diversification of lineage B viruses. In contrast, longer branch lengths in lineage A, and particularly sublineages A1–A4, show that these viruses generally persisted in the host population for longer periods (A1; 1981–2002, A2; 1986–2001, A3; 1984–2002, A4; 1987–2005) (Fig. 1A). Despite increased surveillance efforts in North America, lineage B has not been detected since 2002 and lineage A viruses predominate in wild aquatic birds. The observed lineage replacement in the North American gene pool following introduction of divergent Eurasian viruses is indicative of a competitive interaction between these lineages.
Genetic diversity of H6 hemagglutinin in the North American gene pool
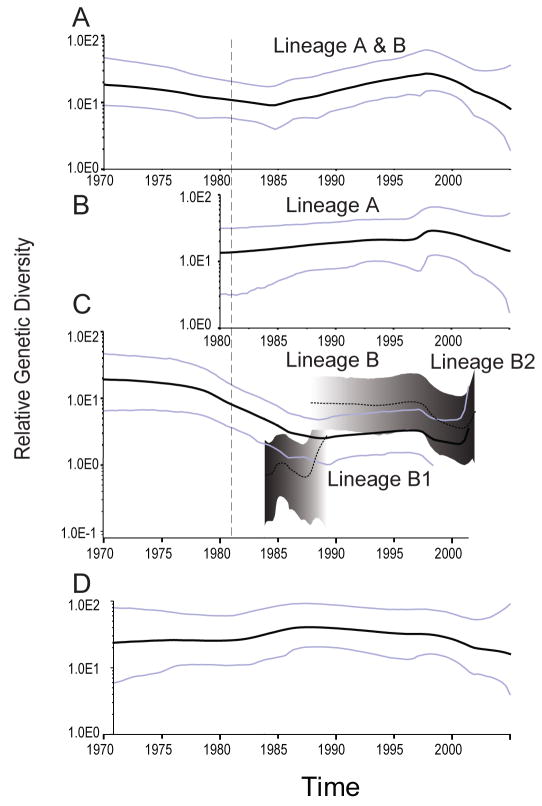
We then used Bayesian skyline plots (BSPs) to visualize temporal changes in genetic diversity (population size) of H6 viruses isolated exclusively from wild birds in North America (Fig. 2). Overall, the BSP showed a slight increase in H6-HA diversity from 1984–1996 that corresponds to the period when Eurasian lineage A viruses were introduced into the North American gene pool (Fig. 2A). When analyzed separately, the BSP of H6 lineage A showed little change in genetic diversity that is consistent with the longer branch lengths observed in the phylogeny (Fig. 2B). In contrast there was a general decrease in diversity of lineage B from 1981 onwards as lineage A replaced lineage B in the North American gene pool (Fig. 2C). The changes in genetic diversity are consistent with the timing of lineage replacement observed in the H6-HA phylogeny.
FIG. 2. Bayesian skyline plots of the H6 and H4 hemagglutinin genes.
(A) Changes in genetic diversity of H6 subtype viruses isolated over the last thirty years from North American wild birds. A measure of genetic diversity is given with the 95% HPD indicated by blue lines. The dashed red lines correspond to the period when lineage A introductions into the North American gene pool occurred. (B) Changes in genetic diversity of H6 subtype lineage A viruses size through time. (C) Changes in genetic diversity of H6 subtype lineage B viruses until time of extinction in 2002. (D) Changes in genetic diversity of H4 subtype viruses. All figures are on the same time-scale in years.
We further investigated the significance of these observed fluctuations by testing the fit of three generalized parametric coalescent models (constant size, exponential growth, expansion growth) to the different H6-HA lineages (Table S1). For the complete North American H6-HA the constant population was strongly supported. For lineage A there was no statistical difference between the constant and expansion coalescent models although the exponential growth model was rejected. There was no significant difference between generalized models for lineage B (Table S1).
Changing evolutionary rates and selection pressures in the North American H6 hemagglutinin
Analysis of the North American H6-HA showed high rates of nucleotide substitution (4.2 × 10−3 substitutions per site per year (subs/site/year), 95% HPD 3.3–5.0 × 10−3 subs/site/year), typical for influenza virus (Table 1). However, comparison of the major H6-HA sublineages (A, B1, and B2) revealed that substitution rates for lineage B2 were significantly lower (2.0 × 10−3 subs/site/year, 95% HPD 1.6–2.4 × 10−3 subs/site/year) compared to A and B1 (Table 1).
TABLE 1.
Rates of nucleotide substitution and selection pressures in the North American influenza gene pool
| Gene | Substitution rate (10−3 subs/site/year) | Mean dN/dS [95% CI] | |
|---|---|---|---|
| Mean | 95% HPD | ||
| H4 | 2.5 | 2.0–3.1 | 0.092 [0.077, 0.108] |
| H6 | 4.2 | 3.3–5.0 | 0.146 [0.132, 0.161] |
| H6 lineage A | 4.4 | 3.5–5.5 | 0.125 [0.132, 0.146] |
| H6 lineage B | 4.1 | 2.9–5.6 | 0.211 [0.183, 0.240] |
| H6 lineage B1 | 4.6 | 2.8–6.6 | 0.284 [0.179, 0.423] |
| H6 lineage B2 | 2.0 | 1.6–2.4 | 0.262 [0.205, 0.329] |
To determine how these differences in evolutionary rate relate to selection pressures we measured the mean numbers of non-synonymous (dN) and synonymous (dS) substitutions per site (ω = dN/dS) in each lineage. Overall, the HA of North American H6 viruses (ω = 0.146, 95% CI: 0.132, 0.161) was under relatively strong purifying selection. However, one amino acid residue (position 277; ω = 4.28, P<0.05) was under positive selection (Table 1). No positively selected sites were detected in lineage B, or sublineages B1 (ω = 0.284, 95% CI: 0.179, 0.423) and B2 (ω = 0.262, 95% CI: 0.205, 0.329). A likelihood ratio test (P<0.01) revealed significant differences in the relative selection pressures between North American isolates of lineage A (ω = 0.125, 95% CI: 0.106, 0.146) and lineage B (ω = 0.210, 95% CI: 0.183, 0.240) (Table 1). The lower dN/dS in H6 lineage A relative to lineage B suggests that the former is subject to weaker positive selection pressure.
Evolutionary dynamics of the H4 hemagglutinin
Phylogenetic analysis of the H4-HA genes showed two major lineages that correspond to the segregation of the Eurasian gene pools (TMRCA 1930, 95% HPD: 1898–1951) and North American (TMRCA 1953, 95% HPD: 1928–1969) with no evidence of gene flow (Fig. 1B). These lineages diverged in 1879 (95% HPD: 1794–1938) and have therefore been geographically isolated and evolving independently for at least 68 years (Fig. 1B).
Analysis of North American H4-HA viruses using a Bayesian skyline plot revealed minor fluctuations in relative genetic diversity (Fig. 2D). Comparison of the three generalized parametric coalescent models indicated a constant population size best described the data set (Table S1). Analysis of the North American H4-HA again revealed high rates of nucleotide substitution (2.5 × 10−3 subs/site/year, 95% HPD: 2.0–3.1 × 10−3 subs/site/year), although, notably, these rates were significantly lower than those observed in H6 (Table 1). The H4-HA in wild aquatic birds in North America was also under stronger purifying selection than the H6 viruses with no positively selected sites detected (ω = 0.092, 95% CI: 0.077, 0.108) (Table 1). These results demonstrate the relative stability of H4 viruses in natural populations where no gene flow, and hence no inter-lineage competition, has been observed.
Evolutionary dynamics and selection pressures in the N1, N2 and N8 neuraminidase
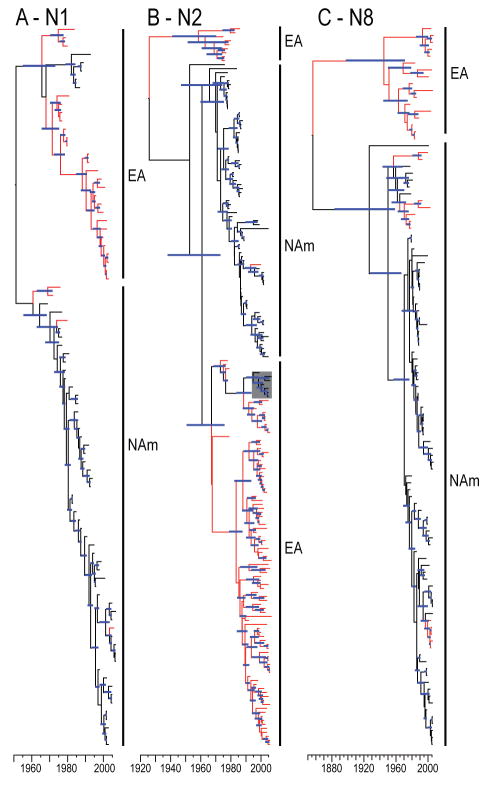
N1, N2 and N8 subtype neuraminidases are most frequently associated with H6 viruses from wild aquatic birds. We therefore investigated, and subsequently observed, evidence of gene flow between continents for each of these NA genes (Fig. 3 and S1). Despite the identification of gene flow, only one North American H6N2 virus (gadwall/Ohio/37/1999) contained both surface genes derived from a Eurasian ancestor (Fig. 3B). Analysis of these NA genes also showed high rates of nucleotide substitution (2.1–4.7 × 10−3 subs/site/year) that are consistent with the HA (Table 2). Selection analyses revealed that each gene was under strong purifying selection.
FIG. 3. Phylogeny of N1, N2, and N8 neuraminidase genes.
Dated phylogenies of N1 (A), N2 (B), N3 (C) NA gene of viruses. Red branches indicate sequence data with geographical origins other than North America (i.e. Eurasian or Oceanic isolates). Blue bars at nodes indicate 95% HPD of TMRCAs. Scale bar shows the time-scale in years. Full phylogenetic trees with sequence identifiers are provided in the supplementary material (Fig. S2). The lineage indicated in the grey box in (B) is a neuraminidase lineage introduced with Eurasian ancestry isolated in North American wild birds. This lineage corresponds to the introduction number 4 (represented by gadwall/OH/37/99) shown in Fig. 1.
TABLE 2.
Rates of nucleotide substitution and selection pressures of the neuraminidase and internal genes
| Gene | Substitution rate (10−3 subs/site/year) | Mean dN/dS [95% CI] | |
|---|---|---|---|
| Mean | 95% HPD | ||
| N1 | 4.7 | 3.4–6.0 | 0.115 [0.105, 0.126] |
| N2 | 3.4 | 2.8–4.0 | 0.168 [0.159, 0.178] |
| N8 | 2.1 | 1.4–2.7 | 0.105 [0.097, 0.113] |
| PB2 | 2.7 | 2.4–3.1 | 0.030 [0.028, 0.035] |
| PB1 | 3.0 | 2.5–3.4 | 0.029 [0.027, 0.031] |
| PA | 2.1 | 1.8–2.4 | 0.042 [0.039, 0.045] |
| NP | 2.4 | 2.1–2.7 | 0.037 [0.034, 0.040] |
| M | 1.3 | 1.2–1.5 | 0.022 [0.018, 0.026 |
| NS1 allele A | 1.3 | 0.9–1.6 | 0.122 [0.104, 0.141] |
| NS1 allele B | 2.0 | 1.7–2.3 | 0.147 [0.134, 0.161] |
Gene flow and selection pressures in the internal genes
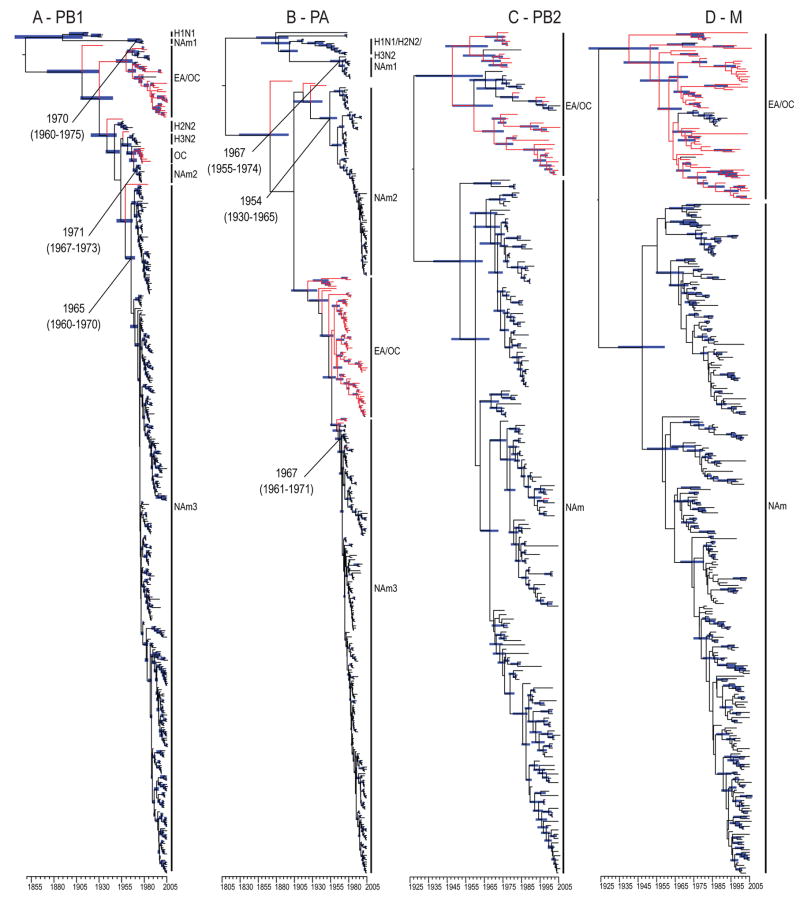
Analyses of the internal gene segments demonstrated that one-way and two-way gene flow was regularly detected throughout the thirty years of systematic surveillance of avian influenza in the wild aquatic birds (Fig. 4 and S2–8). Gene flow was detected in each gene segment analyzed but no entire genome with a recent Eurasian common ancestor was detected. This finding is consistent with the high frequency of reassortment observed in viruses from these host populations (Dugan 2008 and Krauss 2007).
FIG. 4. Dated internal gene phylogenies of viruses isolated from North American and Eurasian gene pools.
PB1 (A), PA (B), PB2 (C) and M (D) genes. Red branches indicate sequence data with geographical origins other than North America (i.e. Eurasian or Oceanic isolates). Blue bars at nodes indicate 95% HPD of TMRCAs. Scale bar shows the time-scale in years.
Interestingly, phylogenies revealed that all PB1 and PA genes currently circulating in North American wild aquatic birds were derived from Eurasian introductions that occurred sometime in the 1960’s (clade NAm3 in Fig. 4A and B). The previous North American PB1 and PA genes were replaced by the introduced lineage in all virus subtypes, suggesting a fitness advantage to North American viruses that incorporated these genes through reassortment.
It is also noteworthy that gene flow was detected in both waterfowl and shorebird hosts and occurred on both the eastern and western seaboards of North America. For example, in the PB2 phylogeny influenza viruses isolated from shorebirds sampled from the North American Atlantic coast contained gene segments with recent Eurasian ancestors (Fig. 4C). In contrast, only a single bird sampled from the Pacific coast contained this Eurasian derived gene segment. Similar patterns were observed in the PA, NP and M genes (Fig. 4B–D and Fig. S2, 4–6). These results suggest that avenues of virus introduction to North America are not restricted to ducks or the Pacific migratory corridor.
Analyses of internal gene sequences revealed relatively little variation in substitution rates across genes (1.3–3.0 × 10−3 subs/site/year) (Table 2). The mean dN/dS was low for all ribonucleoprotein and M genes (ω = 0.022–0.040) (Table 2). The mean dN/dS was higher in both NS alleles (allele A ω = 0.144; allele B ω = 0.122) compared to other genes. Hence, even though there is some variation in selection pressures of the internal gene segments, they are largely still under strong purifying selection indicating the genes encoded are functionally conserved.
Discussion
The present study demonstrates that occasional gene flow between geographically segregated gene pools of AIV has resulted in competition for limited resources – most likely susceptible avian hosts – between introduced and endemic virus lineages. This process has long lasting effects on AIV population structure, most notably the extinction of lineage B of the H6-HA. However, this selection is only likely to occur in cases where the viruses in question are sufficiently antigenically similar (i.e. those of the same subtype) to induce a cross-protective immune response. In contrast antigenically dissimilar viruses (i.e. viruses of different subtypes) would be able to infect the same bird and not experience immune selection. Our results show these lineages co-circulated in the host population for as long as 20 years before lineage B went extinct.
Population genetics offers a number of concepts and principles to explain the evolutionary behavior of RNA viruses. For example, the competitive exclusion principle states that when two species compete for limited resources one species will eventually outcompete the other and become dominant in the population (Hardin, 1960). In the case of RNA viruses, a combination of high replication numbers and high nucleotide substitution rates may render the prolonged co-existence of two or more genetically distinct viral populations unlikely (Clarke et al., 1994; Moya et al., 2000). In our study we observed that lineage A out-competed lineage B for susceptible hosts with corresponding changes in the evolutionary dynamics of the H6 influenza gene pool.
Most notably our study shows that the introduction and establishment of Eurasian H6 subtype viruses (lineage A) in the North American gene pool dramatically changed the evolutionary dynamics of influenza virus in wild birds. Specifically, following its introduction, the genetic diversity (which can also be interpreted as a measure of population size) of the introduced H6 lineage A was stable or slowly expanded, while the endemic North American H6 lineage B viruses showed variation in genetic diversity until extinction as they unsuccessfully competed with the introduced viruses. During this extended period of competition both H6 lineages A and B exhibited combined high nucleotide substitution rates and relatively high values of dN/dS, particularly in comparison to those seen in H4. Hence, the introduction of divergent H6-HA genes into the North American gene pool appears to have exerted a direct selection pressure on endemic H6 viruses.
In contrast, the evolutionary dynamics of H4 subtype viruses, where no gene flow between the Eurasian and North American gene pools has been detected in the last 30 years, likely reflects a population having reached an optimal local fitness peak in its environment. Indeed, compared to H6 evolutionary parameters, nucleotide substitution rates and positive selection pressures were substantially lower for H4 viruses. However, our data imply that competition resulting from gene migration would disrupt this equilibrium.
In theory, the competitive pressure exerted by an invading influenza virus will select for viruses with increased reproduction and transmissibility (Bremermann and Pickering, 1983; Bremermann and Thieme, 1989; Clarke et al., 1994). Interestingly, the introduced H6 lineage A virus caused low pathogenic poultry disease in California (Webby et al., 2003; Woolcock et al., 2003). We therefore hypothesize that the adaptive advantage conferred through competition may contribute to influenza disease emergence in poultry populations.
The effective habitat of AIV, such as host species, is similar in both the Eurasian and North American gene pools (Ito et al., 1995; Krauss et al., 2007; Obenauer et al., 2006; Okazaki et al., 2000; Olsen et al., 2006; Webster et al., 1992; Widjaja et al., 2004). However, the outcome of interactions of the two gene pools revealed in this study indicate that prolonged geographic separation has resulted in fundamental differences in the fitness of each viral lineage. The high frequency of reassortment of Eurasian derived internal gene segments with North American influenza viruses indicates that the fitness landscape of these genes between the two natural gene pools is relatively flat, consistent with previous studies (Dugan et al. 2008). Therefore, this competition is most probably due to antigenic differences between H6 lineages.
Our results show intercontinental transmission of genes was frequently mediated through shorebird (Charadriiformes) hosts and was detected on both eastern and western North America. However, the epidemiology of gene migration is difficult to assess with the sampling bias for western and central continental North American regions. While surveillance in wild bird populations in North America has intensified due to concern of natural introductions of H5N1 into the North American gene pool, these efforts have focused on waterfowl, particularly mallard and pintail ducks in Alaska (Dugan et al., 2008; Krauss et al., 2007; Obenauer et al., 2006; Winker et al., 2007, 2008). Therefore, increased full genome surveillance of influenza viruses in other bird populations is critical for understanding the effects of gene flow between populations.
The extent of viral competition in avian populations infected with influenza remains unknown. In Asia, the long-term endemicity of H5N1 appears to have replaced, most probably through competitive selection, low pathogenic H5 subtype viruses that have been only rarely isolated from poultry in Asia since 2000 when compared to previous surveillance in the 1970’s (Duan et al., 2007). While these birds are often vaccinated and the selection pressures on avian influenza viruses in poultry are higher (Vijaykrishna et al., 2008), this study provides a possible mechanism for disease emergence and transmission from natural reservoir hosts.
Materials and Methods
Data sets
Influenza viruses isolated exclusively from North American wild aquatic birds over a 31-year period (1975–2006) were recruited and combined with influenza virus sequence data sets isolated from Eurasia. To determine the degree of gene flow between Eurasia and North America, initial analyses included all available influenza A sequence data from GenBank.
Preliminary phylogenetic analysis and data preparation
To identify specific influenza virus populations we analyzed all full-length publicly available sequence data from all hosts and all geographic regions (>18 000 gene segments). For each gene segment (including polymerase basic 2 (PB2), polymerase basic 1 (PB1), polymerase acidic (PA), nucleoprotein (NP), membrane (M), non–structural (NS), hemagglutinin (HA) and neuraminidase (NA)) full-length gene sequence data was downloaded and aligned using the NCBI Influenza Virus Resource (Bao et al., 2007). Final data sets were restricted to coding regions read in the first reading frame for each gene. Sequences with insertions or deletions resulting in frame shifts or amino acid insertions were excluded from the analyses.
Provisional phylogenetic inference was carried out using neighbor-joining methods assuming the Hasegawa–Kishino–Yano 85 (HKY85) substitution model available in PAUP* 4b10 (Swofford, 2001). The purpose of these large-scale phylogenetic analyses was to identify individual ‘gene pools’ – that is, phylogenetically distinct clusters of sequences – and determine the degree of interaction, if any, between these populations. Gene pools were identified in each tree as monophyletic groups with bootstrap support ≥80%. The phylogenies were also used to identify virus genes that have transmitted from Eurasia to North America or vice versa.
Data sets were limited to only wild aquatic hosts from North America, while sequence data originating from Eurasia was limited to aquatic birds (if possible) as limited surveillance data of influenza in the wild population is available. Duplicate sequences and those with 100% similarity isolated from the same region and same year were excluded from the analyses while ensuring representative virus sequences of each monophyletic clade, determined from the phylogenetic analysis described above, were included in all analyses.
To understand the effects of the gene flow between the Eurasian and American gene pools on influenza virus evolution and population structure, two scenarios, in which sufficient data was available, were identified for further investigation; H6 viruses that showed evidence of two-way gene flow between continents, and H4 viruses that did not. To explore whether the H6 subtype introductions were associated with specific NA subtype introductions, we also analyzed the most common subtypes of NA in combination with H6-HA found in both gene pools (N1, N2 and N8). For the analyses of the H4-HA and H6-HA, all full-length North American sequence data were recruited along with additional Eurasian isolates: H4, 92 and H6, 156 sequences. Data sets consisting of full-length N1, N2 and N8 sequences (n = 87, 182 and 112 respectively) were analyzed to determine if any H6 introduction was associated with a particular NA introduction. Gene flow was also confirmed for each of the six internal gene segments. Final data sets of the following size were then compiled: PB2, 196; PB1, 400; PA, 383; NP, 219; M, 324; NS allele A, 132; and NS allele B, 268 sequences.
Relaxed molecular clock phylogenetic tree inference, estimation of nucleotide substitution rates and population dynamics
To estimate the rates of nucleotide substitution in avian influenza A virus and to compare the population dynamics of virus genes, we used a Bayesian Markov chain Monte Carlo (MCMC) coalescent approach as implemented in the program BEAST version 1.4.8 (Drummond et al., 2002; Drummond and Rambaut, 2007). As data sets were restricted to coding regions the codon based Shapiro-Rambaut-Drummond-2006 (SRD06) nucleotide substitution model assuming the HKY85 substitution model for first/second and third codon positions with unlinked base frequencies was used to analyze the datasets (Shapiro et al., 2006).
The performance of three molecular clock models: the strict clock that assumes a single evolutionary rate along all branches, and the uncorrelated lognormal relaxed clock and uncorrelated exponential relaxed clock that allow evolutionary rates to vary along branches within lognormal and exponential distributions, respectively, was compared for each data (Drummond et al., 2002, 2005, 2006, 2007; Shapiro et al., 2008). Comparison of model fit to the data in a Bayesian framework was accomplished with a Bayes factor (BF) test, the ratio of marginal likelihoods of two models being compared. We calculated the approximate marginal likelihoods for each coalescent model (Suchard et al., 2001). Strength of model selection was assessed (with respect to the model with the lower likelihood) in the following way: 2≤ 2× ln BF ≤6 indicates positive evidence for the null model; 6≤ 2× ln BF ≤0 indicates strong evidence for the null model; 2× ln BF >10 indicates very strong evidence for the null model (Salemi et al., 2008). For each analysis a Bayesian skyline coalescent tree prior was used as it makes the fewest a priori assumptions about the data (Drummond et al., 2005). The results of the BF test showed that both relaxed clocks fit the data significantly better than the strict clock. However, the uncorrelated exponential relaxed clock model was most appropriate for each data set. Finally, the degree of statistical uncertainty in each parameter estimate is reflected in values of the 95% Highest Probability Density (HPD).
Three independent analyses were carried out for 20–60 million generations sampled to produce at least 10,000 trees for each data set to ensure adequate sample size of the posterior, prior, nucleotide substitution rates and likelihoods (effective sample size >200). The mean substitution rates, TMRCAs and Maximum Clade Credibility (MCC) phylogenetic trees were then calculated after the removal of an appropriate burnin (10–15% of the samples in most cases, with one exception where ~20% was removed for analyses of the PB1 gene) following visual inspection in TRACER version 1.4 (Rambaut and Drummond, 2007).
Introductions of H6 subtype viruses from Eurasia have been reported previously (Dugan et al., 2008; Spackman et al., 2005; Webby et al., 2003; Woolcock et al., 2003). Analyses of the mixed Eurasian and North American H6 gene segments were used to estimate the TMRCAs for introductions into North America. The distribution of TMRCA estimates were then enforced as TMRCA priors for analyzing H6-HA data isolated purely from North America, including rate comparisons of each lineage and demographic models (monophyletic nodes and TMRCAs shown in Fig. 1 and Fig. S1).
After our initial analysis of population dynamics using the non-parametric Bayesian skyline plot (BSP) model, which depicts changing levels of genetic diversity through time (and which can be considered as a measure of effective population size assuming neutral evolution) we undertook an additional analysis of lineage-specific epidemiological behavior. Approximate marginal likelihoods of three parametric coalescent models (constant population size, exponential population growth and expansion population growth) enforcing the best-fit clock model for each data set was assessed using a BF as described above.
Comparison of nucleotide substitution rates
Finally, the BF test was used to evaluate whether the mean nucleotide substitution rates (r) for specific monophyletic groups were significantly different. In general, given two monophyletic groups, the significant differences in r is estimated by the ratio of posterior probabilities (r1 > r2|Data)/(r2 > r1|Data) by prior probabilities (r1 > r2)/(r2 > r1) (Jeffreys, 1935; Kass and Raftery, 1995), i.e. analyses was conducted independently with and without data to sample from the posterior and the prior, respectively.
Analysis of selection pressures
An analysis of selection pressures was undertaken using a codon-based approach as implemented in HYPHY (Kosakovsky et al., 2005a). The single likelihood ancestor counting (SLAC) method was employed (Kosakovsky et al., 2005b), with a critical p-value of 0.05. Based on the most appropriate model of nucleotide substitution and a given tree topology, the SLAC method calculates a global dN/dS rate ratio (with 95% confidence intervals) and tests selection of variable codon sites based on the inference of the ancestral sequence. To determine if selection was acting differentially on lineages introduced to the North American gene pool dN/dS rate ratio estimates (with confidence intervals) were calculated for introduced lineages and endemic lineages separately. The dN/dS rate ratio estimates from the endemic lineage was applied to the introduced lineage and vice versa. A likelihood ratio test was conducted to test for significance in dN/dS rate ratio estimates of different lineages of a gene data set, with critical p-value of 0.01. This test was repeated using the upper and lower limits of the confidence intervals.
Supplementary Material
Figures S1–S8 show complete dated phylogenetic trees including sequence identifiers. Table S1 shows Bayes factor comparisons of generalized coalescent models for H4 and H6 viruses from the North American gene pool.
Acknowledgments
This work was supported by the Area of Excellence Scheme of the University Grants Committee (Grant AoE/M-12/06) of the Hong Kong SAR Government, the National Institutes of Health (NIAID contract HHSN266200700005C), and the Li Ka Shing Foundation. GJDS is supported by a career development award under NIAID contract HHSN266200700005C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Azemi A, Bahl J, Al-Zenki S, Al-Shayji Y, Al-Ahmad S, Chen H, Guan Y, Peiris JSM, Smith GJD. Avian influenza A virus (H5N1) outbreaks, Kuwait, 2007. Emerg Infect Dis. 2008;14:958–961. doi: 10.3201/eid1406.080056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Bolotov P, Dernovoy D, Kiryutin, Zaslavsky L, Tatusova T, Ostell J, Lipman D. The Influenza Virus Resource at the National Center for Biotechnology Information. J Virol. 2007;82:596 –601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremermann HJ, Pickering J. A game-theoretical model of parasite virulence. J Theor Biol. 1983;100:411–426. doi: 10.1016/0022-5193(83)90438-1. [DOI] [PubMed] [Google Scholar]
- Bremermann HJ, Thieme HR. A competitive exclusion principle for pathogen virulence. J Math Biol. 1989;27:179–190. doi: 10.1007/BF00276102. [DOI] [PubMed] [Google Scholar]
- Brown JD, Stallknecht DE, Beck JR, Suarez DL, Swayne DE. Susceptibility of North American ducks and gulls to H5N1 highly pathogenic avian influenza viruses. Emerg Infec Dis. 2006;12:1663–70. doi: 10.3201/eid1211.060652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JD, Stallknecht DE, Swayne DE. Experimental infections of herring gulls (Larus argentatus) with H5N1 highly pathogenic avian influenza viruses by intranasal inoculation of virus and ingestion of virus-infected chicken meat. Avian Pathology. 2007;37:393–397. doi: 10.1080/03079450802216595. [DOI] [PubMed] [Google Scholar]
- Brown JD, Stallknecht DE, Swayne DE. Experimental infection of swans and geese with highly pathogenic avian influenza virus (H5N1) of Asian lineage. Emerg Infec Dis. 2008;14:136–142. doi: 10.3201/eid1401.070740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Smith GJD, Zhang SY, Qin K, Wang J, Li KS, Webster RG, Peiris JSM, Guan Y. H5N1 virus outbreak in migratory waterfowl. Nature. 2005;436:191–192. doi: 10.1038/nature03974. [DOI] [PubMed] [Google Scholar]
- Chen R, Holmes EC. Avian influenza virus exhibits rapid evolutionary dynamics. Mol Biol Evol. 2006;23:2336–2341. doi: 10.1093/molbev/msl102. [DOI] [PubMed] [Google Scholar]
- Clarke DK, Duarte EA, Elena SF, Moya A, Domingo E, Holland JJ. The red queen reigns in the kingdom of RNA viruses. Proc Natl Acad Sci USA. 1994;91:4821–4824. doi: 10.1073/pnas.91.11.4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis RO, Bean WJ, Kawaoka Y, Webster RG. Distinct lineages of influenza virus H4 hemagglutinin genes in different regions of the world. Virology. 1989;169:408–417. doi: 10.1016/0042-6822(89)90166-9. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics. 2002;161:1307–1320. doi: 10.1093/genetics/161.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 2005;22:1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- Duan L, Campitelli L, Fan XH, Leung YHC, Vijaykrishna D, Zhang JX, Donatelli I, Delogu M, Li KS, Foni E, Chiapponi C, Wu WL, Kai H, Webster RG, Shortridge KF, Peiris JSM, Smith GJD, Chen H, Guan Y. Characterization of low pathogenic H5 subtype influenza viruses from Eurasia: Implications for the origin of highly pathogenic H5N1 viruses. J Virol. 2007;81:7529–7539. doi: 10.1128/JVI.00327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugan VG, Chen R, Spiro DJ, Sengamalay N, Zaborsky J, Ghedin E, Nolting J, Swayne DE, Runstadler JA, Happ GM, Senne DA, Wang R, Slemons RD, Holmes EC, Taubenberger JK. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Pathog. 2008;4:e1000076. doi: 10.1371/journal.ppat.1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman OT, Bean WJ, Kawaoka Y, Donatelli I, Gou Y, Webster RG. Evolution of influenza A virus nucleoprotein genes: implications for the origin of H1N1 human and classical swine viruses. J Virol. 1991;65:3704–3714. doi: 10.1128/jvi.65.7.3704-3714.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman OT, Bean WJ, Kawaoka Y, Webster RG. Evolution of the nucleoprotein gene of influenza A virus. J Virol. 1990a;64:1487–1497. doi: 10.1128/jvi.64.4.1487-1497.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman OT, Donis RO, Kawaoka Y, Webster RG. Evolution of influenza A virus PB2 genes: implications for evolution of the ribonucleoprotein complex and origin of human influenza A virus. J Virol. 1990b;64:4893–4902. doi: 10.1128/jvi.64.10.4893-4902.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin G. The Competitive Exclusion Principle. Science. 1960;131:1292–1297. doi: 10.1126/science.131.3409.1292. [DOI] [PubMed] [Google Scholar]
- Ito T, Okazaki K, Kawaoka Y, Takada A, Webster RG, Kida H. Perpetuation of influenza A viruses in Alaskan waterfowl reservoirs. Arch Virol. 1995;140:1163–1172. doi: 10.1007/BF01322743. [DOI] [PubMed] [Google Scholar]
- Jenkins GM, Rambaut A, Pybus OG, Holmes EC. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J Mol Evol. 2002;54:156–165. doi: 10.1007/s00239-001-0064-3. [DOI] [PubMed] [Google Scholar]
- Jeffreys H. Some tests of significance, treated by the theory of probability. Math Proc Cambridge Philos Soc. 1935;31:203–222. [Google Scholar]
- Kass RE, Raftery AE. Bayes Factors. J Amer, Statistical Assoc. 1995;90:773–795. [Google Scholar]
- Kawaoka Y, Krauss S, Webster RG. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol. 1989;63:4603–4608. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keawcharoen J, van Riel D, van Amerongen G, Bestebroer T, Beyes WE, van Lavieren R, Osterhaus ADME, Fouchier RAM, Kuiken T. Wild ducks as long-distance vectors of highly pathogenic avian influenza virus (H5N1) Emerg Infec Dis. 2008;14:600–607. doi: 10.3201/eid1404.071016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick AM, Chmura AA, Gibbons DW, Fleischer RC, Marra PP, Daszak P. Predicting the global spread of H5N1 avian influenza. Proc Natl Acad Sci USA. 2006;103:19368–19373. doi: 10.1073/pnas.0609227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishida N, Sakoda Y, Shiromoto M, Bai GR, Isoda N, Takada A, Laver G, Kida H. H2N5 influenza virus isolates from terns in Australia: genetic reassortants between those of the Eurasian and American lineages. Virus Genes. 2008;37:16–21. doi: 10.1007/s11262-008-0235-z. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW, Muse SV. HyPhy: Hypothesis testing using phylogenies. Bioinformatics. 2005a;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW. Not so different after all: A comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005b;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- Krauss S, Obert CA, Franks J, Walker D, Jones K, Seiler P, Niles L, Pryor SP, Obenauer JC, Naeve CW, Widjaja L, Webby RJ, Webster RG. Influenza in migratory birds and evidence of limited intercontinental virus exchange. PLoS Pathog. 2007;3:e167. doi: 10.1371/journal.ppat.0030167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JH, Okazaki K, Bai GR, Shi WM, Mweene A, Kida H. Interregional transmission of the internal protein genes of H2 influenza virus in migratory ducks from North America to Eurasia. Virus Genes. 2004;29:81–86. doi: 10.1023/B:VIRU.0000032791.26573.f1. [DOI] [PubMed] [Google Scholar]
- Makarova NV, Kaverin NV, Krauss S, Senne D, Webster RG. Transmission of Eurasian avian H2 influenza virus to shorebirds in North America. J Gen Virol. 1999;80:3167–3171. doi: 10.1099/0022-1317-80-12-3167. [DOI] [PubMed] [Google Scholar]
- Moya A, Elena SF, Bracho A, Miralles R, Barrio E. The evolution of RNA viruses: A population genetics view. Proc Natl Acad Sci USA. 2000;97:6967–6973. doi: 10.1073/pnas.97.13.6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obenauer JC, Denson J, Mehta PK, Su X, Mukatira S, Finkelstein DB, Xu X, Wang J, Ma J, Fan Y, Rakestraw KM, Webster RG, Hoffman E, Krauss S, Zheng J, Zhang Z, Naeve CW. Large-scale sequence analysis of avian influenza isolates. Science. 2006;311:1576–1580. doi: 10.1126/science.1121586. [DOI] [PubMed] [Google Scholar]
- Okazaki K, Kawaoka Y, Webster RG. Evolutionary pathways of the PA genes of influenza a viruses. Virology. 1989;172:601–608. doi: 10.1016/0042-6822(89)90202-x. [DOI] [PubMed] [Google Scholar]
- Okazaki K, Takada A, Ito T, Imai M, Takakuwa H, Hatta M, Ozaki H, Tanizaki T, Nagano T, Ninomiya A, Demenev VA, Tyaptirganpv MM, Karatayeva TD, Yamnikova SS, Lvov DK, Kida H. Precursor genes of future pandemic influenza viruses are perpetuated in ducks nesting in Siberia. Arch Virol. 2000;145:885–893. doi: 10.1007/s007050050681. [DOI] [PubMed] [Google Scholar]
- Olsen B, Munster VJ, Wallensten A, Waldenström J, Osterhaus ADME, Fouchier RA. Global patterns of influenza A virus in wild birds. Science. 2006;312:384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond A. [Accessed 20 June 2008];Tracer v1.4: MCMC trace analyses tool. 2007 Available: http://beast.bio.ed.ac.uk/Tracer.
- Rambaut A, Pybus OG, Nelson MI, Viboud C, Taubenberger JA, Holmes ECH. The genomic and epidemiological dynamics of human influenza A virus. Nature. 2008;439:615–619. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salemi M, de Oliveira T, Ciccozzi M, Rezza G, Goodenow MM. High-resolution molecular epidemiology and evolutionary history of HIV-1 subtypes in Albania. PLoS ONE. 2008;3(1):e1390. doi: 10.1371/journal.pone.0001390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer JR, Kawaoka Y, Bean WJ, Süss J, Senne D, Webster RG. Origin of the pandemic 1957 H2 influenza A virus and the persistence of its possible progenitors in the avian reservoir. Virology. 1993;194:781–788. doi: 10.1006/viro.1993.1319. [DOI] [PubMed] [Google Scholar]
- Shapiro B, Rambaut A, Drummond AJ. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol Biol Evol. 2006;23:7–9. doi: 10.1093/molbev/msj021. [DOI] [PubMed] [Google Scholar]
- Spackman E, Stallknecht DE, Slemons RD, Winker K, Suarez DL, Scott M, Swayne DE. Phylogenetic analyses of type A influenza genes in natural reservoir species in North America reveals genetic variation. Virus Res. 2005;114:89–100. doi: 10.1016/j.virusres.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Suchard MA, Weiss RE, Sinsheimer JS. Bayesian selection of continuous-time Markov chain evolutionary models. Mol Biol Evol. 2001;18:1001–1013. doi: 10.1093/oxfordjournals.molbev.a003872. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (and other methods) 4.0 Beta. Sunderland, MA: Sinauer Associates; 2001. [Google Scholar]
- van Gils JA, Munster VJ, Radersma R, Liefhebber D, Fouchier RA, Klaassen M. Hampered foraging and migratory performance in swans infected with low-pathogenic avian influenza A virus. PLoS ONE. 2007;2:e184. doi: 10.1371/journal.pone.0000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijaykrishna D, Bahl J, Riley S, Duan L, Zhang JX, Chen H, Peiris JSM, Smith GJD, Guan Y. Evolutionary dynamics and emergence of panzootic H5N1 influenza viruses. PLoS Pathog. 2008;4(9):e1000161. doi: 10.1371/journal.ppat.1000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallensten A, Munster VJ, Elmberg J, Osterhaus AD, Fouchier RA, Olsen B. Multiple reassortment between Eurasian and American lineages of influenza A virus (H6N2) in Guillemot (Uria aalge) Arch Virol. 2005;150:1685–1692. doi: 10.1007/s00705-005-0543-8. [DOI] [PubMed] [Google Scholar]
- Webby RJ, Woolcock PR, Krauss SL, Walker DB, Chin PS, Shortridge KF, Webster RG. Multiple genotypes of nonpathogenic H6N2 influenza viruses isolated from chickens in California. Avian Dis. 2003;47:905–910. doi: 10.1637/0005-2086-47.s3.905. [DOI] [PubMed] [Google Scholar]
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microb Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widjaja L, Krauss S, Webby RJ, Xie T, Webster RG. Matrix gene of influenza A viruses isolated from wild aquatic birds: Ecology and emergence of influenza A viruses. J Virol. 2004;78:8771–8779. doi: 10.1128/JVI.78.16.8771-8779.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winker K, McCracken KG, Gibson DD, Pruett CL, Meier R, Huettmann F, Wege M, Kulikova IV, Zhuravlev YN, Perdue M, Spackman E, Suarez DL, Swayne DE. Movements of birds and avian influenza from Asia into Alaska. Emerg Infect Dis. 2007;13:547–552. doi: 10.3201/eid1304.061072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winker K, Spackman E, Swayne DE. Rarity of influenza A virus in spring shorebirds, southern Alaska. Emerg Infect Dis. 2008;14:1314–1316. doi: 10.3201/eid1408.080083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolcock PR, Suarez DL, Kuney D. Low-pathogenicity avian influenza virus (H6N2) in chickens in California, 2000–02. Avian Dis. 2003;47:872–881. doi: 10.1637/0005-2086-47.s3.872. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1–S8 show complete dated phylogenetic trees including sequence identifiers. Table S1 shows Bayes factor comparisons of generalized coalescent models for H4 and H6 viruses from the North American gene pool.