Abstract
Pax genes encode DNA binding proteins that play pivotal roles in the determination of complex tissues. Members of one subclass, Pax6, function as selector genes and play key roles in the retinal development of all seeing animals. Mutations within the Pax6 homologs including fly eyeless, mouse Small eye and human Pax6 lead to severe retinal defects in their respective systems. In Drosophila eyeless and twin of eyeless, play non-redundant roles in the developing retina. One particularly interesting characteristic of these genes is that, although expression of either gene can induce ectopic eye formation in non-retinal tissues, there are differences in the location and frequencies at which the eyes develop. eyeless induces much larger ectopic eyes, at higher frequencies, and in a broader range of tissues than twin of eyeless. In this report we describe a series of experiments conducted in both yeast and flies that has identified protein modules that are responsible for the differences in tissue transformation. These domains appear to contain transcriptional activator and repressor activity of distinct strengths. We propose a model in which the selective presence of these activities and their relative strengths accounts, in part, for the disparity to which ectopic eyes are induced in response to the forced expression of eyeless and twin of eyeless. The identification of both transcriptional activator and repressor activity within the Pax6 protein furthers our understanding of how this gene family regulates tissue determination.
Keywords: Pax6, Eyeless, Twin of eyeless, Eye development, Retina
Introduction
PAX proteins are transcription factors that play key roles in organogenesis, cell proliferation and disease and are found in organisms as diverse as flies, mice and humans (Noll, 1993; Strachan and Read, 1994; Stuart et al., 1994; Callaerts et al., 1997; Mansouri et al., 1999; Chi and Epstein, 2002; Lang et al., 2007; Wang et al., 2008). All PAX proteins contain a 128 amino acid DNA binding domain called the PAIRED domain, named after the founding member of the PAX family, the Drosophila melanogaster segmentation gene, paired (Bopp et al., 1986; Frigerio et al., 1986). The Paired domain is itself subdivided into two separable DNA binding domains, the PAI and the RED motifs (Treisman et al., 1991; Czerny et al., 1993; Cai et al., 1994; Xu et al., 1995; Jun and Desplan, 1996). Vertebrates have nine Pax genes whose encoded products can be divided into four subclasses. What structurally distinguishes individual PAX proteins is the presence or absence of an octapeptide and a complete or partial second DNA binding domain, the homeobox (Walther et al., 1991, Noll, 1993). Nucleic acid binding specificity is determined, in part, by the selective and/or combinatorial use of the three DNA binding domains (Bertuccioli et al., 1996; Jun and Desplan, 1996; Callaerts et al., 1997; Sheng et al., 1997a; Jun et al., 1998).
Pax proteins also affect development by serving as transcriptional activators and/or repressors (Underhill, 2000). Several Pax proteins, such as Drosophila Paired, are thought to function as dedicated activators and completely lack inhibitory functions (Wilson et al., 1993). Other family members such as Drosophila Eyg and Toe as well as vertebrate Pax4 appear to function purely as repressors (Fujitani et al., 1999; Yao and Sun, 2005; Yao et al., 2008). Interestingly, both activation and inhibitory domains are present within the C-terminal segments of Pax3 and members of the Pax2/5/8 subfamily (Chalepakis et al., 1993, 1994; Fickenscher et al., 1993; Stuart et al., 1995; Dorfler and Busslinger, 1996; Lechner and Dressler, 1996). In at least one case this repression is mediated by interactions with members of the Groucho family of co-repressors (Eberhard et al., 2000). Pax6 appears to be an interesting, if not controversial, case. Several reports have proposed that it functions as a dedicated activator with the transactivation domain being localized to the P/S/T rich region of the C-terminal segment (Plaza et al., 1993; Epstein et al., 1994; Czerny and Busslinger, 1995; Altmann et al., 1997; Singh et al., 2000). However, it has also been reported that Pax6 can repress the transcription of the βB1-crystallin gene and that this repression is mediated by binding of the Paired and homeodomains to the target promoter and does not involve an additional inhibitory domain (Duncan et al., 1998).
Pax6 plays a critical role in the development of the visual system in a wide range of seeing animals (Halder et al., 1995; Gehring, 1996; Gehring and Ikeo, 1999; Pichaud and Desplan, 2002; Kozmik, 2005; Callaerts et al., 2006). Loss of Pax6 in flies, mice and human patients lead to severe eye abnormalities (Hill et al., 1991; Ton et al., 1991; Glaser et al., 1992; Jordan et al., 1992; Quiring et al., 1994). However, despite the central role that Pax6 plays in retinal development and the intense attention that it has received over the last twenty-five years, the list of verified transcriptional targets is relatively short (Altmann et al., 1997; Sheng et al., 1997b; Cvekl et al., 1999; Liu et al., 1999; Niimi et al., 1999; Ostrin et al., 2006; Zhang et al., 2006). In many instances it is not clear if Pax6 activates or represses the verified targets.
The role of Pax6 in fly eye development begins during embryogenesis when the transcription of both ey and toy is activated in small populations of cells within the embryonic head (Quiring et al., 1994; Czerny et al., 1999). As development proceeds these cells proliferate, organize themselves into monolayer epithelia called imaginal discs and eventually give rise to the adult compound eyes (reviewed in Cohen, 1993; Held, 2002). Early in this long transition, ey is expressed uniformly throughout the entire eye-antennal disc and is subsequently restricted to the portion of the epithelium that generates the retina (Kumar and Moses, 2001a, 2001b; Kurata et al., 2000; Kenyon et al., 2003). By the beginning of the late second/early third larval instar, pattern formation has initiated at the posterior margin of the eye field and both ey and toy are further confined to the anterior regions of the disc in a swathe of cells adjacent to the advancing morphogenetic furrow (Quiring et al., 1994; Czerny et al., 1999; Bessa et al., 2002). Within this zone both Pax6 genes activate the transcription of a number of downstream targets, including the retinal determination genes eyes absent (eya), dachshund (dac), sine oculis (so) and optix (Halder et al., 1998; Niimi et al., 1999; Punzo et al., 2002; Ostrin et al., 2006). Expression of these genes, in part, prepares undifferentiated cells for the transition across the furrow and directs them towards adopting retinal cell fates. The expression of ey and toy in the developing eye ceases at the morphogenetic furrow. It is not until much later in pupal development that ey reappears in the retina to activate the transcription of the major fly rhodopsin (Kumar and Ready, 1995; Sheng et al., 1997b; Papatsenko et al., 2001).
When forcibly mis-expressed in non-retinal tissues, ey and toy can, cause a change in tissue identity leading to development of ectopic eyes. Interestingly, the Pax6 genes are unequal in their ability to redirect tissue fate. Expression of ey induces eye formation on the antenna, legs, wings and halteres, while toy has only previously been shown to be sufficient to induce eyes on the legs and wings (Halder et al., 1995; Czerny et al., 1999). Furthermore, neither individual gene can induce ectopic eyes in adult tissues that are derived from the abdominal histoblasts or the labial, clypeolabral, humeral and genital discs. It has been speculated that several molecular and biochemical features such as chromatin structure of the targeted tissues, differences in DNA binding specificity and protein binding partner availability can and are likely to contribute to the disparities in the transformative abilities of ey and toy.
In this report we have focused on the intrinsic properties of these two Drosophila Pax6 proteins and demonstrate that internal differences between ey and toy themselves can account for the phenotypic dissimilarity that is observed in forced expression assays. We have constructed minimal Ey and Toy proteins each containing just the Paired DNA binding domain and the C-terminal region. These proteins appear to contain all the structural and biochemical information that is required to induce ectopic eyes in the requisite tissue types. We show that the transcriptional activation domains, located within the C-termini are of significantly different strengths with Ey acting as a more potent transcriptional activator than Toy. We also demonstrate that Ey, but not Toy, may serve as a putative transcriptional repressor. This repressive activity does not map to the C-terminal region, as is the case for the Pax2/5/8 subfamily. Instead the inhibitory activity lies within the non-conserved segments that flank the Paired DNA binding domain. Removal of one of two segments is sufficient for the induction of ectopic eyes within the genital discs, a feat that is not observed with either full-length Ey or Toy proteins. Together, we propose that (1) the relatively high strength of Ey as a transcriptional activator and (2) the ability for Ey to serve as both transcriptional activator and repressor accounts, in part, for the ability of Ey to transform non-retinal tissues and induce ectopic eye formation in a wider range of non-retinal tissues than Toy.
Materials and methods
Fly stocks, crosses and immunohistochemistry
The following stocks were used in this study: ey-GAL4 (Walter Gehring), dpp-GAL4 (Janice Fischer) and ap-GAL4 (Bloomington Stock Center). We conducted all crosses at 18 °C, 25 °C and 29 °C and multiple UAS insertion lines for each construct (see below) were crossed to both dpp and ap-GAL4 drivers. The following antibodies and reagents were used in this study: rat anti-ELAV (1:100, DSHB), mouse anti-DAC (1:5, DSHB), mouse anti-EYA (1:5, DSHB), goat anti-mouse TRITC (1:100, Jackson Labs), goat anti-rat FITC (1:100, Jackson Labs), donkey anti-mouse TRITC (1:10 [subtracted], Jackson Labs), donkey anti-rat FITC (1:10 [subtracted], Jackson Labs), phalloidin-Cy5 (1:50, Invitrogen), phalloidin-TRITC (1:50, Molecular Probes). Imaginal discs were dissected in PBS, fixed in 4% paraformaldehyde, serially incubated with antibodies and viewed on a Zeiss Axioplan II compound microscope. Adult flies were viewed with a Zeiss Discovery light microscope.
Manipulation of Ey and Toy proteins
All constructs described below were generated using PCR. Individual domains were amplified from plasmids containing the full-length eyeless and twin of eyeless cDNAs. In some cases unique restriction enzyme sites were added to the ends of individual primer sequences in order to facilitate the joining of individual domains while in others primers with overlapping sequences were designed and used to join neighboring gene segments. PCR conditions, cloning strategies, sequence files for each construct and primer sequences are available upon request. Multiple insertion lines for each construct were recovered and tested (Supplemental Tables 1 and 2).
Deletion constructs
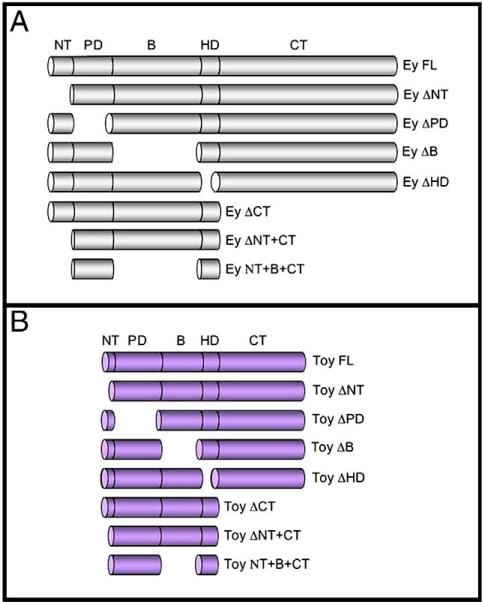
The full-length Ey protein is 838 amino acids in length and can be divided into five segments: the N-terminal (NT), the Paired DNA binding domain (PD), the central linker region (B), the homeobox DNA binding domain (HD) and the C-terminal tail (CT). The NT segment consists of residues 1−36, the PD contains residues 37−164, the B segment contains residues 165−410, the HD contains residues 411−470 and the CT segment contains residues 471−838. The NT deletion (Ey ΔNT) contains amino acids 37−838, the PD deletion (EY ΔPD) contains amino acids 1−36 fused to residues 165−838, the B deletion (Ey ΔB) contains amino acids 1−164 fused to residues 411−838, the HD deletion (Ey ΔHD) contains amino acids 1−410 fused to residues 471−838 and the CT deletion (EY ΔCT) contains amino acids 1−470.
The Ey CT is 370 amino acids in length and consists of residues 471−838. Ey CT-1 contains residues 471−667, Ey CT-2 contains residues 553−779, Ey CT-3 contains residues 596−838 and Ey CT-4 contains residues 668−838. All Ey CT deletion constructs are presented in Fig. 5.
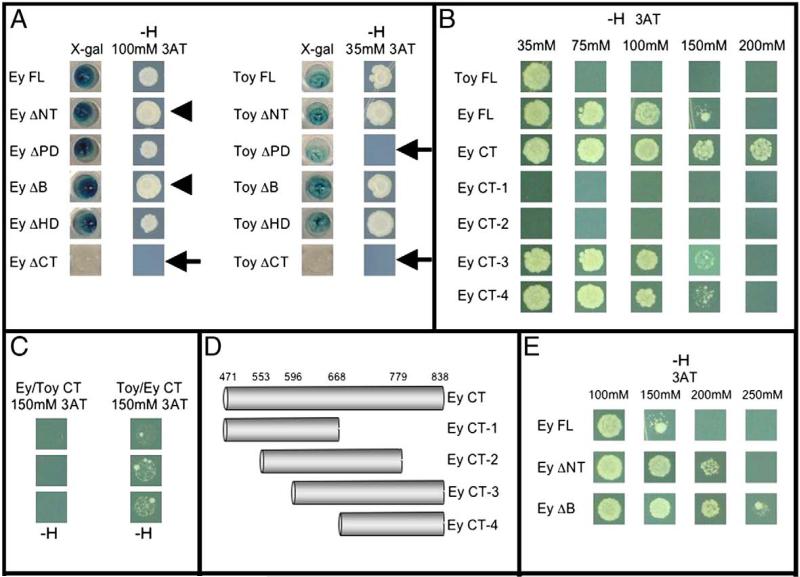
Fig. 5.
Mapping of transactivation potential and repression activity within Ey and Toy. (A–C, E) LacZ and HIS3 reporter assays. (D) Schematic of Ey CT deletion constructs. Arrowheads denote Ey deletion constructs that appear to grow better than full-length Ey. Arrows indicate Ey or Toy deletion constructs that fail to activate either lacZ or HIS3 reporters. –H indicates that cells were grown on histidine deficient plates.
The Toy full-length protein is 543 amino acids in length and, like Ey, can be divided into five segments (see above). The Toy NT segment contains residues 1−28, the PD contains residues 29−156, the B segment contains residues 157−264, the HD contains residues 265−324 and the CT segment contains residues 325−543. The NT deletion (Toy ΔNT) contains amino acids 29−543, the PD deletion (Toy ΔPD) contains amino acids 1−28 fused to residues 157−543, the B deletion (Toy ΔB) contains amino acids 1−156 fused to residues 265−543, the HD deletion (Toy ΔHD) contains amino acids 1−264 fused to residues 325−543 and the CT deletion (Toy ΔCT) contains amino acids 1−324. The deletion constructs are presented in Fig. 1.
Fig. 1.
Schematics of Ey and Toy deletion constructs. (A) Constructs in which protein domains of Ey (grey) have been deleted individually or in combination. (B) Constructs in which protein domains of Toy (purple) have been deleted individually or in combination.
Minimal Pax6 proteins containing just the Paired DNA binding domain and the CT activation domain were generated. In the case of Ey minimal protein (EY PD+CT), amino acids 37−164 were fused to residues 471−838. Similarly for Toy (Toy PD+CT), amino acids 29−156 were fused to residues 325−543. Both minimal molecules are presented in Fig. 4.
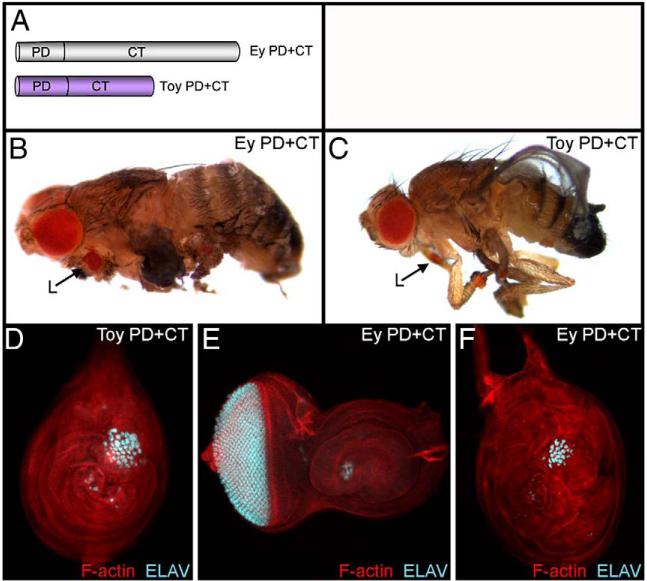
Fig. 4.
Minimal Ey and Toy proteins consisting of just the Paired DNA binding domain and the CT region are sufficient to induce ectopic eye development. (A) Schematic of minimal Ey and Toy proteins. (B–C) Light microscope images of adult flies. (D–F) Confocal images of 3rd instar imaginal discs. Genotypes are listed within each panel. Each construct was expressed along the A/P axis by the dpp-GAL4 driver. Visualized molecules are listed within each panel. L = leg.
Chimeric proteins
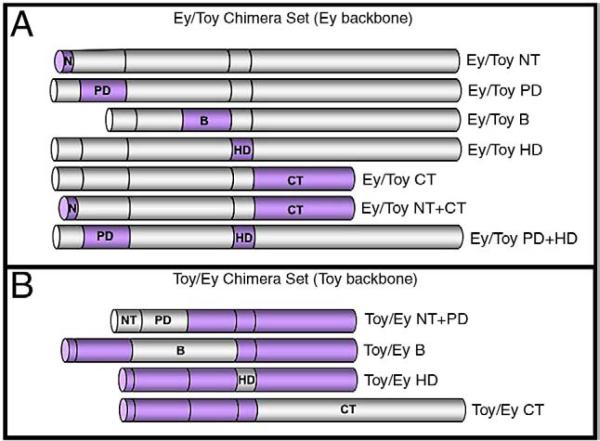
We generated a set of chimeric molecules in which individual segments of Ey were deleted and replaced with the corresponding region of Toy. The Ey/Toy NT chimera was created by replacing the NT segment of Ey with amino acids 1−28 of Toy. The Ey/Toy PD chimera was created by replacing the PD of Ey with amino acids 29−156 of Toy. The Ey/Toy B chimera was created by replacing the B segment of Ey with amino acids 157−264 of Toy. The Ey/Toy HD chimera was created by replacing the HD of Ey with amino acids 265−324 of Toy. The Ey/Toy CT chimera was generated by replacing the CT segment of Ey with amino acids 325−543 of Toy. Similarly, we generated a set of chimeric molecules in which individual segments of Toy were deleted and replaced with the corresponding region of Ey. The Toy/Ey NT+PD chimera was created by replacing the NT and PD segments of Toy with amino acids 1−165 of Ey. The Toy/Ey B chimera was created by replacing the B segment of Toy with amino acids 165−410 of Ey. The Toy/Ey HD chimera was created by replacing the HD of Toy with amino acids 411−470 of Ey. The Toy/Ey CT chimera was created by replacing the CT segment of Toy with amino acids 471−838 of Ey. The chimeric proteins are presented in Fig. 8.
Fig. 8.
Schematic of Ey and Toy chimeric proteins. (A) Chimeric proteins in which segments of Ey (grey) have been removed and replaced with the corresponding segments from Toy (purple). (B) Chimeric proteins in which segments of Toy (purple) have been removed and replaced with the corresponding segments from Ey (grey).
Identification of activation and repression activity in yeast
Each of the deletion constructs and selected chimeric protein constructs (see above) were assayed in yeast for regions with either transcriptional activation or repression activity. Each construct was cloned into the pDEST32 bait vector, which contains the GAL4 DNA binding domain and the ADH1 promoter allowing for constitutive expression of the cDNA of interest to ensure consistent expression of each construct. These ARS/CEN based vectors are low copy number expression vectors, which result in the bait proteins being expressed at relatively low levels. This bait plasmid was co-transformed with the pEXP AD502 vector into the MaV203 yeast strain (Invitrogen, ProQuest Two Hybrid System). All transformations were plated on media deficient for the amino acids tryptophan and leucine to ensure incorporation of both plasmids into the yeast cells. UAS sites were incorporated into the regulatory regions of the lacZ and HIS3 reporters. These reporters were then stably integrated into the yeast genome. The strength of the activation was measured by the ability of transformed cells to grow on increasing levels of 3-amino-1,2,4 triazol (3AT). Each assay was replicated at least five times. Each well and yeast colony in Fig. 5 is a representative example of the five replicates. A detailed procedure describing yeast transformations and X-gal assays are available upon request.
Results
The non-conserved segments of Ey and Toy are crucial for function
Several reports have demonstrated that ey and toy are limited in their ability to induce eye development in non-retinal tissues with ey being a more potent and prolific inducer of ectopic eyes than toy (Halder et al., 1995; Czerny et al., 1999). Furthermore, developing tissues such as the abdominal histoblasts and the labial, clypeolabral, humeral and genital imaginal discs appear to be resistant to ectopic eye formation by either gene (Weasner, B.M., Salzer, C.L. and Kumar, J.P., unpublished results). We set out to determine if these limitations are intrinsic properties of the Pax6 proteins themselves. Individual conserved domains and non-conserved segments were deleted from each Pax6 protein (Figs. 1A, B) and expressed within the dorsal sector of the wing, haltere and leg discs via the ap-GAL4 driver and along the A/P axis of the eye-antenna, leg, wing, haltere and genital discs via the dpp-GAL4 driver (Supplemental Figs. 1A–H). Our a priori expectation was that deletion one of more protein regions would be sufficient to either (1) narrow the range of tissues that can be transformed by Ey; (2) expand the array of tissues that can be converted by Toy; or (3) broaden the collection of tissues that can be forced into supporting ectopic eye formation by both Ey and Toy proteins. Based solely on the results of our deletion analysis, which are summarized in Table 1, it appears that, broadly speaking, the tissue specificity of ectopic eye formation cannot be mapped to a single region of either Ey or Toy. Deletion of individual domains had, in most cases, either no effect on tissue specificity or adversely affected the ability of the protein to induce ectopic eyes in selected tissues in a non-specific manner (Table 1). We observed only two instances (expression of Toy ΔNT and Toy ΔB) in which ectopic eyes were recovered in a location that is unique compared to the full-length wild type molecule (Fig. 2D; Table 1, blue boxes).
Table 1.
Minima Ey and Toy proteins consisting of just the Paired DNA binding domains and the CT segments are sufficient to induce ectopic eyes.
| dpp-GAL4 |
ap-GAL4 |
||||||
|---|---|---|---|---|---|---|---|
| Eye-antenna | Leg | Wing | Haltere | Genital | Wing | Haltere | |
| Ey FL | + | + | + | + | − | + | + |
| Ey ΔNT | + | + | + | − | − | + | + |
| Ey ΔPD | − | − | − | − | − | − | − |
| Ey ΔB | + | + | + | − | − | + | − |
| Ey ΔHD | + | + | + | − | − | + | − |
| Ey ΔCT | − | − | − | − | − | − | − |
| Ey ΔNY+CT | − | − | − | − | − | + | − |
| Ey ΔNT+B+CT | − | − | − | − | − | − | − |
| Ey PD+CT | + | + | + | − | − | + | + |
| Toy FL | − | + | + | − | − | + | − |
| Toy ΔNT | + | + | − | − | − | + | − |
| Toy ΔPD | − | − | − | − | − | − | − |
| Toy ΔB | − | + | + | − | − | + | + |
| Toy ΔHD | − | + | − | − | − | + | − |
| Toy ΔCT | − | − | − | − | − | + | − |
| Toy ΔNT+CT | − | − | − | − | − | + | − |
| Toy ΔNT+B+CT | − | − | − | − | − | − | − |
| Toy PD+CT | − | + | − | − | − | + | − |
Fig. 2.
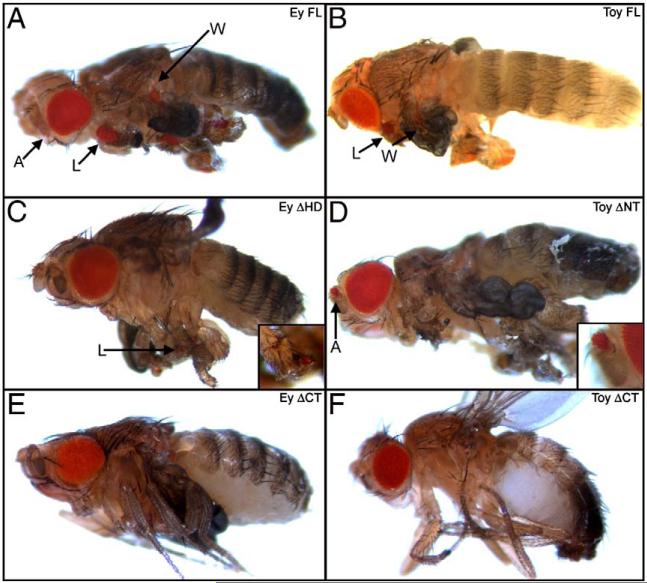
The non-conserved CT segments are required for ectopic eye induction. (A–F) Light microscope images of adult flies. A = antenna, W = wing, L = leg. Genotypes are listed at the top of each panel. Each construct was expressed along the A/P axis by the dpp-GAL4 driver. Anterior is to the left.
These results have provided further insights into which regions of Ey and Toy are required and/or necessary for ectopic eye formation. A previous report indicated that the Paired domain (PD) is essential for normal and ectopic eye formation, which our results confirm (Punzo et al., 2001; 2004). These reports also demonstrate that the HD is completely dispensable for the induction of ectopic eyes. In contrast to these reports, we have observed that the HD is required for inducting ectopic eyes in some tissues. For instance, expression of EY ΔHD is sufficient to support ectopic eye formation in the antenna and the legs but not within the wings and halteres (Table 1). Additionally, the ectopic eyes that are generated by expression of EY ΔHD are also smaller than those seen with the full-length protein (Figs. 2A, C). Interestingly, our deletion analysis also indicates that the non-conserved C-terminal (CT) is required for Pax6 proteins to induce eye formation. Removal of these domains (Ey ΔCT, Toy ΔCT) eliminates the ability of either Ey or Toy to induce ectopic eyes within the dpp expression domain (Table 1, Figs. 2E, F).
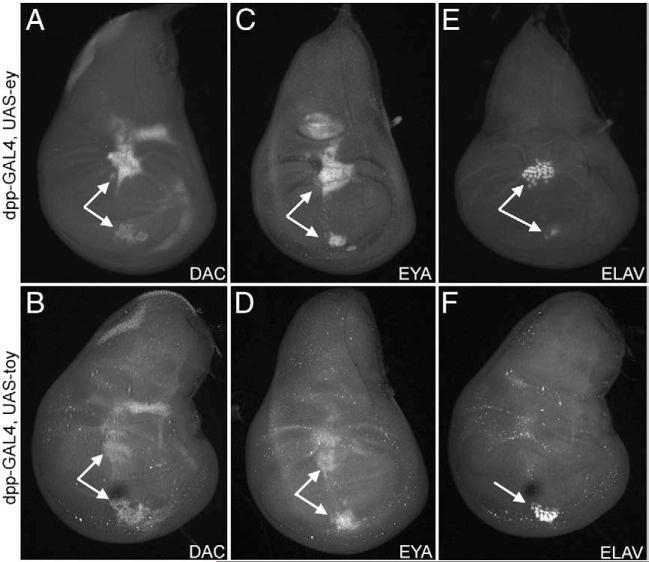
We set out to determine if Ey and Toy differentially activate the expression of downstream retinal determination genes accounting for the differences in the spectrums of ectopic eye generation. We expressed each gene individually along the A/P axis of a variety of tissues via the dpp-GAL4 driver (Supplemental Figs. 1D–H) and assayed for distribution of the Dac and Eya proteins. When ey and toy are expressed within the entire dpp expression domain, both genes are expressed in a smaller subpopulation of cells along the A/P axis (Figs. 3A–D). There appears to be only slight differences in the ability of either gene to activate dac and eya expression. However, within the wing disc, for example, Ey induces relatively large ectopic eyes (as assayed for the expression of the panneuronal marker, Elav) in two regions of the disc while Toy only weakly stimulates retinal development in one subset of dpp expressing cells (Figs. 3E, F). We find it interesting that all dac and eya positive cells are not also Elav positive suggesting that only a smaller subset of cells is converted into photoreceptors (Figs. 3E, F). We reported a similar phenomenon in an earlier publication that described the ability of So to induce ectopic eye development (Weasner et al., 2007). Consistent with the previous result that expression of either Ey ΔCT or Toy ΔCT failed to induce ectopic eyes in the adult, these two proteins also fail to activate downstream RD genes in the disc (data not shown).
Fig. 3.
Induction of RD genes by Ey and Toy in the developing wing. (A–F) Confocal images of 3rd instar wing imaginal discs. Genotypes are listed to left of each row. Each construct was expressed along the A/P axis by the dpp-GAL4 driver. Visualized molecules are listed at the bottom of each panel. Arrows indicate regions of ectopic Dac, Eya or Elav expression. Anterior to the right.
Identification of the minimal Ey and Toy proteins required to induce retinal development
The only constructs that eliminated the ability of either gene to promote ectopic eye development within the dpp expression zone are ones in which either the Paired DNA binding domain or the CT segment is deleted (Table 1, yellow boxes). Based on these results we reasoned that minimal Ey and Toy proteins consisting of just these two domains (Fig. 4A) should be sufficient to induce retinal development. Minimal Ey (Ey PD+CT) and Toy (Toy PD+CT) proteins were expressed in developing imaginal discs and, indeed ectopic eyes were generated (Figs. 4B–F). While both minimal proteins can induce ectopic eyes they are not capable of fully recapitulating the effects of expressing the full-length proteins. The minimal Ey protein is only sufficient to induce ectopic eyes in the developing wing, leg and antenna while the minimal Toy protein can induce ectopic eyes only on the legs (Supplemental Table 1). Furthermore, the ectopic eyes often are smaller in size and are present in less than 100% of the examined animals (Ey PD+CT: 100% in legs, 30% in antennae, 15% in wings; Toy PD+CT: 100% in legs and 0% in wings). Like the deletion analysis, these results further suggest that other portions of the Ey and Toy proteins are required to induce ectopic eyes in a full spectrum of tissues.
Ey and Toy contain transactivation domains
We set out to identify potential role(s) for the non-conserved CT regions in modulating the formation of ectopic eyes. Two putative roles could be in transcriptional activation and/or repression of target genes. Vertebrate Pax6 and Drosophila Ey both have been demonstrated to harbor a transactivation domain within the CT segment (Plaza et al., 1993; Glaser et al., 1994; Czerny and Busslinger, 1995; Altmann et al., 1997; Singh et al., 2000; Punzo et al., 2001). Pax6 also has the ability to repress the expression of the βB1 crystallin gene (Duncan et al., 1998). While this repressive activity has yet to be mapped to a specific domain within Pax6, there is evidence from Pax3 and the Pax2/5/8 subfamily that both activation and repressive domains map to the same segment of the protein, the non-conserved CT segment (Chalepakis et al., 1993, 1994; Fickenscher et al., 1993; Stuart et al., 1995; Dorfler and Busslinger, 1996; Lechner and Dressler, 1996). Using a yeast transcriptional activity assay we set out to determine (1) if Ey and Toy contain single or multiple activation domains; (2) whether the transactivation potentials of the two proteins are equivalent to each other; (3) does either protein harbor regions with repressive activity; and (4) do such activities plot to the non-conserved CT segments.
Both full-length genes and all deletion constructs (Fig. 1A) were fused to the GAL4 DNA binding domain (GAL4 BD) and then expressed in MaV203 yeast cells containing GAL4 binding sites combined to either lacZ or HIS3 reporters. The full-length proteins are capable of activating expression of both β-galactosidase, which can break down exogenously added X-gal, and HIS3, which allows yeast cells to grow on media deficient for the amino acid histidine. Additionally, increasing amounts of 3AT, an inhibitor of histidine biosynthesis and growth, were added to the media plates lacking histidine as an indication of strength of activation potential. Cells expressing proteins exhibiting strong transactivation potential will be able to grow on increasing amounts of 3AT whereas proteins with weak or no transactivation potential will be inhibited by relatively low amounts of 3AT. We demonstrate here that both Ey and Toy harbor transactivation domains within the CT segment, as cells expressing the Ey ΔCT and Toy ΔCT proteins failed to either break down X-gal or grow on histidine deficient plates (Fig. 5A, arrows). Additionally, expression of just the CT of EY is capable of activating expression of both lacZ and HIS3 (Fig. 5B). However, the CT of Toy alone failed to activate expression of either reporter (data not shown). These results are consistent with data identifying a transactivation domain in the CT region of vertebrate Pax6. From our results the transactivating activity of Ey requires only the CT, whereas the activity of Toy requires additional domains. We have identified the PD as a potential domain that contributes to the transactivation potential of Toy as yeast cells expressing Toy ΔPD also fail to grow on 35 mM 3AT and only weakly activate lacZ expression (Fig. 5A, arrow).
Punzo et al. suggested that the CT of Ey and Toy functionally distinguish the two proteins from each other. They hypothesized that the CT regions may bind to different sets of co-factors or transcription factors (Punzo et al., 2004). We demonstrate here that differences in the strength of the transactivating potential of the CT also contribute to the differences in Ey and Toy function. Cells expressing either full-length Ey-GAL4 (BD) or Toy-GAL4 (BD) were grown on histidine deficient plates in increasing amounts of 3AT. Cells expressing Toy were only able to grow on media containing 35 mM 3AT or less. In contrast, cells expressing Ey grew on media containing up to 150 mM 3AT suggesting that the Ey is a stronger transcriptional activator (Fig. 5B). This was further confirmed by the activation of the HIS3 reporter by chimeric Ey and Toy proteins, in which the CT regions of the two proteins were switched (Figs. 5C, 7A, B). A chimeric protein containing the Ey backbone fused to the CT of Toy (Ey/Toy CT) activated transcription of HIS3 at lower levels of 3AT than the opposite chimera in which the Toy protein backbone is fused to the CT of Ey (Toy/Ey CT; Fig. 5C).
Fig. 7.
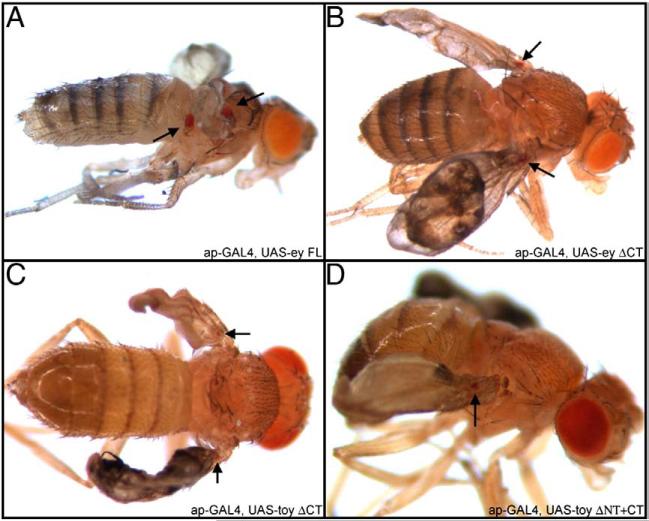
Pax6 proteins lacking the CT transactivation potential induces ectopic eyes via the ap-GAL4 driver. (A–D) Light microscope images of adult flies. Genotypes are listed within the bottom right of each image. Anterior is to the right.
Yeast cells expressing only the CT segment of Ey (Ey CT) were also capable of surviving on media containing 200 mM 3AT confirming that the CT is both necessary and sufficient for the transactivation potential of Ey. In order to more precisely map the source of the transactivation activity, we made a series of finer deletions of the CT tail (Fig. 5D) and assayed the ability of each construct to activate the HIS3 reporter in the presence of increasing concentrations of 3AT. Our analysis indicates that the last 59 amino acids of the CT segment are necessary (Figs. 5B, D) but not sufficient for activating transcription of our reports in yeast (data not shown). We therefore report that the transactivation domain is located within the last 170 amino acids of the protein (Figs. 5B, D). We were able to confirm these results by assaying the ability of each construct to activate the lacZ reporter (data not shown).
Given these observations regarding the differing transactivating potentials, we sought to broaden the spectrum of tissues that can be converted into ectopic eyes by Toy by expressing a chimeric protein in which the Toy CT has been replaced with that of the Ey (Toy/Ey CT, Fig. 8B). We reasoned that since this chimera has a higher transactivation potential than wild type Toy (Figs. 5A, C) it should be sufficient to induce ectopic eyes in a greater number of tissues than wild type Toy. Expression of Toy/Ey CT did, in fact, induce ectopic eyes within the antenna, leg and wings at a frequency and size similar to full-length Ey (Figs. 6A–D). We also observe ectopic eyes in a broad range of tissues when the Ey CT was replaced with the weaker transactivation domain of Toy (Ey/Toy CT; Table 2). However, consistent with the Toy CT having weaker transactivation potential these eyes are significantly smaller in size and individual flies with ectopic eyes in multiple adult tissues were found in far lower frequencies than wild type Ey (Ey/Toy CT: 5% in antennae, 100% in legs and 100% in wings).
Fig. 6.
The transactivation potential of Ey and Toy mediates tissue specificity of ectopic eye formation in Drosophila. (A) Light microscope image of an adult fly. (B–D) Confocal images of 3rd instar imaginal discs. Genotypes are listed within each panel. Each construct was expressed along the A/P axis by the dpp-GAL4 driver. Visualized molecules are listed within each panel. A = antenna, W = wing, L = leg.
Table 2.
A novel Pax6 protein can induce ectopic eyes within the developing genital discs of Drosophila.
| dpp-GAL4 |
ap-GAL4 |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Eye-antenna | Leg | Wing | Haltere | Genital | Head | Wing | Haltere | Leg | |
| Ey FL | + | + | + | + | − | − | + | + | − |
| Ey/Toy NT | + | + | + | + | − | − | + | + | − |
| Ey/Toy PD | + | + | + | + | − | − | + | + | − |
| Ey/Toy B | + | + | + | + | + | + | + | + | − |
| Ey/Toy HD | + | + | + | + | − | − | + | + | + |
| Ey/Toy CT | + | + | + | − | − | − | + | + | − |
| Ey/Toy NT+CT | + | + | + | − | − | − | + | + | − |
| Ey/Toy PD+HD | + | + | + | + | − | − | + | + | − |
| Toy FL | − | + | + | − | − | − | + | − | − |
| Toy/Ey NT+PD | − | + | − | − | − | − | + | − | − |
| Toy/Ey B | − | + | + | − | − | − | + | + | − |
| Toy/Ey HD | − | + | + | − | − | − | + | − | − |
| Toy/Ey CT | + | + | + | − | − | − | + | − | + |
It has been previously reported that differences in function between Ey and Toy are, in part, due to dissimilarities in binding partner interactions (Punzo et al., 2004). If protein–protein interactions are a necessity for Ey and Toy function then expressing constructs lacking the transactivation domains may still be able to induce ectopic eyes (due to interactions with endogenous transcriptional co-activators). We were concerned that the dpp expression domain is too narrow and might exclude cells that express potential co-activators. To address this concern we expressed our Ey and Toy deletion proteins within the apterous (ap) expression domain, which is significantly broader than dpp (Supplemental Figs. 1A–H). Expression of Ey ΔNT+CT, Toy ΔCT and Toy ΔNT+CT (each lacking the CT segments and thus transactivation potential) were capable of inducing retinal development within the ap domain. It should be noted that the eyes are significantly smaller than the full-length proteins (Fig. 7). Intriguingly, although the ap promoter is activated in the third larval instar wing disc (Supplemental Fig. 1) we have been unable to observe ectopic eyes at this stage of development (data not shown). It is likely that the ectopic eyes are formed during the pupal stage of development. Since the CT transactivation domain is missing from these constructs, we have to conclude that both Ey and Toy can interact with other, unidentified, endogenously expressed transcriptional co-activators and that this interaction is sufficient to induce ectopic eyes. It is likely that, in addition to DNA binding specificity, both mechanisms (transactivation potential and interactions with co-factors) contribute to ectopic eye formation by Ey and Toy proteins. These results also indicate that since these deletion proteins are capable of inducing ectopic eye development they are likely to be stable, fold correctly and are functional.
Ey, but not Toy, contains repressive activity
In the course of identifying the transactivation domain in the C-terminus of Ey we uncovered two regions within the Ey protein that putatively harbor transcriptional repressive activity. We observed that while cells expressing just the Ey CT grew in 200 mM 3AT, cells expressing the full-length protein failed to grow when the concentration of 3AT rose above 150 mM. In the presence of 100 mM 3AT, yeast cells expressing Ey proteins lacking either the NT or B non-conserved segments (Ey ΔNT, Ey ΔB) grew better than cells expressing wild type Ey protein (Fig. 5A, arrowhead). Cells expressing Ey ΔNT and Ey ΔB continued to grow in the presence of 200 mM and 250 mM 3AT respectively, whereas cells expressing wild type Ey failed to survive at this concentration (Fig. 5E). The repressive activity of the B segment appears to be stronger than that of the NT since yeast cells expressing constructs lacking the B segment survive on media containing a higher concentration of the 3AT inhibitor than cells expressing a construct lacking the NT segment (Fig. 5E). Together, these results suggest that the non-conserved NT and B segments of Ey harbor transcriptional repressive activity. It should be noted that unlike Pax3 and the Pax2/5/8 subfamily, the putative repressor activity in Ey does not reside in the same segment of the molecule as the transactivation domain. Interestingly, Toy does not seem to have any repressive activity since yeast cells expressing the various deletion constructs either grew as well or worse than wild type Toy (Fig. 5A).
A chimeric Ey/Toy protein induces eyes in the genitals
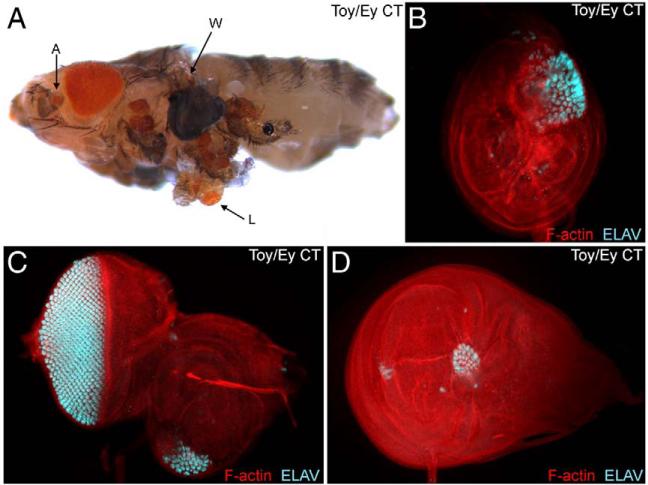
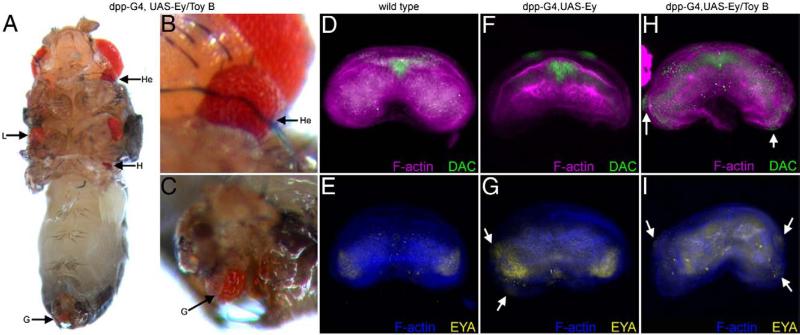
The genital imaginal disc is one of the tissues in which ectopic eye formation is not initiated in response to the forced expression of either Ey or Toy (Halder et al., 1995; Czerny et al., 1999). We hypothesized that in the case of Ey this may be due to the presence of strong repressor domains within the non-conserved NT and B segments and in the instance of Toy, the relatively weak transactivation potential may be insufficient to compensate for the lack of repressor activity. Simple removal of the putative repressor activity within Ey (Ey ΔNT, EY ΔB) or addition of a strong transactivation domain to Toy (Toy/Ey CT chimera) is insufficient to induce ectopic eyes within the genitals (Tables 1 and 2). It is likely that these deletions and chimeras do not have the exact biological information necessary for overcoming a molecular block in the genitals. We therefore reasoned that a chimeric protein that had the (1) correct DNA binding domains; (2) strong transactivation potential; and (3) reduced repressor activity would be more likely to promote eye formation. Indeed, a chimeric molecule in which one of the segments of the Ey protein that carries repressive activity has been replaced with the homologous region of Toy (Ey/Toy B chimera) can now induce ectopic formation in the genitals (Table 2, green box; Figs. 8A, 9A,C). Since the Ey/Toy B chimera contains the two DNA binding domains and the strong CT transactivation domain from Ey we conclude that induction of ectopic eyes by this chimeric protein is not due to differences in DNA binding specificity. Instead, we conclude that the removal of the repressor domain and/or differences in protein–protein interactions via the B domain may account for the induction of ectopic eyes. In order to assess the difference between the effect that wild type Ey and the Ey/Toy B chimera had on the genital disc we looked at the expression of eya and dac. During normal eye development eya is a direct transcriptional target of Ey and dac lies genetically downstream of Ey (Shen and Mardon, 1997; Bonini et al., 1997; Halder et al., 1998; Pappu et al., 2005; Ostrin et al., 2006). In wild type (female discs shown here) both dac and eya are expressed in non-overlapping patterns within the genital discs (Figs. 9D, E). Expression of wild type Ey within the dpp expression pattern (Supplemental Fig. 1) is sufficient to induce eya, but not dac, expression (Figs. 9F, G). The lack of co-expression is a potential explanation for why ectopic eyes are not induced in response to ey expression. In contrast, expression of the Ey/Toy B chimeric protein was sufficient to induce both downstream genes (Figs. 9H, I). Although the mechanism is unclear, it appears that the repressive activity located within the B region works, in part, to inhibit the expression of dac within the genital disc. Curiously, we also observed the formation of ectopic eyes on the ventral side of the head. This is another tissue that is normally not converted by the expression of full-length Ey (Figs. 9A, B).
Fig. 9.
Expression of a novel Pax6 protein induces ectopic eyes in the genital disc. (A–C) Light microscope images of adult flies. (D–I) Confocal images of 3rd instar female genital discs. Genotypes are listed above each panel. Each construct was expressed along the A/P axis by the dpp-GAL4 driver. Visualized molecules are listed within each panel. Arrows in panel G–I indicate ectopic Dac or Eya expression. W = wing, L = leg, H = haltere, He = head, G = genital.
Punzo et al. have raised the issue that the difference in the expression levels of randomly inserted UAS-ey and UAS-toy lines can influence the penetrance and tissue range of ectopic eye induction (Punzo et al., 2004). Potential differences in expression can be minimized by using the phiC31 site-specific integration system. In order to mitigate the potential differences associated with distinct integration sites we have generated multiple insertion lines for the constructs reported herein (Supplemental Tables 1 and 2). Additionally, the results from the in vivo ectopic eye assays are completely congruent (for each construct) with the data from our yeast transcriptional activation assays where the expression level of each construct is consistent with the others. The plasmids used in the yeast transactivation assays contain the ADH1 promoter, which allows for constitutive expression of the cDNA of interest to ensure consistent expression of each construct. As the data sets from these two very different systems are in agreement with one another we conclude that the differences that we observe amongst individual Ey and Toy proteins are likely to be due, in large part, to the activation and repression activities that we have described here. In particular, both sets of data point to transactivation potentials within the C-terminal segments and the presence of repressive activity within the B segment.
Discussion
Members of the retinal determination cascade in Drosophila are known to induce ectopic eye development with varying proficiencies. It also has been observed that only a limited number of tissues can be redirected to adopt an eye fate. Attempts to explain these two observations have often relied upon discussions of (1) the relative position of each gene within the regulatory network hierarchy; (2) putative interactions with differing sets of protein interaction partners; (3) differences in the target genes of individual transcription factors within the network; and (4) potential modulations of chromatin structure. Here we report on two potential mechanisms that are distinct from these factors. We propose that the differences between the ability of the Drosophila Ey and Toy to induce ectopic eyes are due, in significant part to, the differences in transactivation potential and the presence of repressive activity within Ey.
In particular, we are able to show that the expression of mutant Ey and Toy proteins lacking the non-conserved CT segments (Ey ΔCT, Toy ΔCT) fails both to induce ectopic eyes in flies and to activate the transcription of several reporter constructs in yeast. The activation domain plots to the CT segments of both proteins. Toy appears to also have activation activity within the Paired DNA binding domain. The transactivation potential of Ey is significantly higher. Minimal Ey and Toy proteins consisting of just the Paired DNA binding domain and the CT transactivation domains are sufficient to induce ectopic eye formation. And finally, a chimeric protein in which the CT of Toy was replaced with that of Ey (Toy/Ey CT) can now induce eyes in a wider than normal range of tissues in flies and can, in the presence of high concentrations of inhibitors, activate the transcription of reporters in yeast. Taken together these results suggest a mechanism in which the strength of an activation domain plays a major, but not exclusive, role in modulating the activity of individual transcription factors.
In addition, our results revealed that the presence of repressive activity within Ey contributes to differences in ectopic eye formation. Yeast cells expressing Ey proteins that lack either the non-conserved NT and B segments (Ey ΔNT, EY ΔB) grow significantly better overall and in the presence of higher concentrations of inhibitor than the wild type full-length Ey. Additionally, a chimeric protein in which one putative repressor domain of Ey has been removed and replaced with the corresponding region of Toy (Ey/Toy B) can induce ectopic eyes in the developing genital disc, a tissue that normally cannot be converted into retinal tissue by either wild type gene alone. The mechanism by which transcriptional repression is achieved by Ey is unclear.
The ability of Ey to function as both transcriptional activator and repressor raises interesting issues about its role in normal development. A number of putative target genes have been identified based on (1) direct in vitro/in vivo binding to defined enhancer elements (2) interrogations of the genome for Pax6 binding sites; and (3) transcriptional profiling using DNA microarrays (Sheng et al., 1997a; Niimi et al., 1999; Papatsenko et al., 2001; Michaut et al., 2003; Pauli et al., 2005; Ostrin et al., 2006). The extent to which Ey represses transcription of these putative targets and under what developmental circumstances is not clear. Our results suggest that this mechanism may be important for preventing Ey, which is normally expressed in a wide range of non-retinal tissues, from inappropriately inducing eye development during normal development. Despite these limitations, it appears that Ey is still a stronger transcriptional activator than Toy. The repressive activity within the B domain, while potentially moderating the activation potential of Ey, is not sufficient to completely counteract the activation domain, which our yeast assay shows is intrinsically stronger than that of Toy.
Vertebrate Pax6 may also have similar bivalent transcriptional activities. Most reports have suggested that vertebrate Pax6 is a dedicated transcriptional activator (Plaza et al., 1993; Glaser et al., 1994; Czerny and Busslinger, 1995; Altmann et al., 1997; Singh et al., 2000). A lone report suggests that the transcription of at least one target gene (βB1-crystallin) can be inhibited (Duncan et al., 1998). One implication of the results presented here is that vertebrate Pax6 may indeed also repress sets of developmental genes. It should be noted that while some members of the Pax superfamily appear to function as either dedicated activators (Paired) and repressors (Eyg, Toe) there are several other members (Pax3, Pax2/5/8) that contain both types of activities (Chalepakis et al., 1993, 1994; Fickenscher et al., 1993; Stuart et al., 1995; Dorfler and Busslinger, 1996; Lechner and Dressler, 1996). The ability of a single Pax protein to activate and/or repress transcription in a context dependent manner would provide a mechanism for increasing its functional diversity.
Supplementary Material
Acknowledgments
We would like to thank Claire Salzer for comments and suggestions on the manuscript and for providing the images (Figs. 3B, D, F) of ectopic eye generation in the developing wing imaginal disc by expression of Toy. This research was supported in part by the Indiana METACyt Initiative of Indiana University, funded in part through a major grant from the Lilly Endowment, Inc. This work was supported by grants to Scott D. Michaels from the NSF (IOB-0447583) and the NIH (R01GM075060) and to Justin P. Kumar from NIH (R01EY014863).
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ydbio.2009.04.027.
References
- Altmann CR, Chow RL, Lang RA, Hemmati-Brivanlou A. Lens induction by Pax-6 in Xenopus laevis. Dev. Biol. 1997;185:119–123. doi: 10.1006/dbio.1997.8573. [DOI] [PubMed] [Google Scholar]
- Bertuccioli C, Fasano L, Jun S, Wang S, Sheng G, Desplan C. In vivo requirement for the paired domain and homeodomain of the paired segmentation gene product. Development. 1996;122:2673–2685. doi: 10.1242/dev.122.9.2673. [DOI] [PubMed] [Google Scholar]
- Bessa J, Gebelein B, Pichaud F, Casares F, Mann RS. Combinatorial control of Drosophila eye development by eyeless, homothorax, and teashirt. Genes Dev. 2002;16:2415–2427. doi: 10.1101/gad.1009002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonini NM, Bui QT, Gray-Board GL, Warrick JM. The Drosophila eyes absent gene directs ectopic eye formation in a pathway conserved between flies and vertebrates. Development. 1997;124:4819–4826. doi: 10.1242/dev.124.23.4819. [DOI] [PubMed] [Google Scholar]
- Bopp D, Burri M, Baumgartner S, Frigerio G, Noll M. Conservation of a large protein domain in the segmentation gene paired and in functionally related genes of Drosophila. Cell. 1986;47:1033–1040. doi: 10.1016/0092-8674(86)90818-4. [DOI] [PubMed] [Google Scholar]
- Cai J, Lan Y, Appel LF, Weir M. Dissection of the Drosophila paired protein: functional requirements for conserved motifs. Mech. Dev. 1994;47:139–150. doi: 10.1016/0925-4773(94)90086-8. [DOI] [PubMed] [Google Scholar]
- Callaerts P, Halder G, Gehring WJ. PAX-6 in development and evolution. Annu. Rev. Neurosci. 1997;20:483–532. doi: 10.1146/annurev.neuro.20.1.483. [DOI] [PubMed] [Google Scholar]
- Callaerts P, Clements J, Francis C, Hens K. Pax6 and eye development in Arthropoda. Arthropod Struct. Dev. 2006;35:379–391. doi: 10.1016/j.asd.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Chalepakis G, Stoykova A, Wijnholds J, Tremblay P, Gruss P. Pax: gene regulators in the developing nervous system. J. Neurobiol. 1993;24:1367–1384. doi: 10.1002/neu.480241009. [DOI] [PubMed] [Google Scholar]
- Chalepakis G, Goulding M, Read A, Strachan T, Gruss P. Molecular basis of splotch and Waardenburg Pax-3 mutations. Proc. Natl. Acad. Sci. U. S. A. 1994;91:3685–3689. doi: 10.1073/pnas.91.9.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi N, Epstein JA. Getting your Pax straight: Pax proteins in development and disease. Trends Genet. 2002;18:41–47. doi: 10.1016/s0168-9525(01)02594-x. [DOI] [PubMed] [Google Scholar]
- Cohen SM. In: Imaginal Disc Development. Bate M, Martinez Arias A, editors. InCold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1993. pp. 747–841. [Google Scholar]
- Cvekl A, Kashanchi F, Brady JN, Piatigorsky J. Pax-6 interactions with TATA-box-binding protein and retinoblastoma protein. Invest. Ophthalmol. Vis. Sci. 1999;40:1343–1350. [PubMed] [Google Scholar]
- Czerny T, Busslinger M. DNA-binding and transactivation properties of Pax-6: three amino acids in the paired domain are responsible for the different sequence recognition of Pax-6 and BSAP (Pax-5). Mol. Cell. Biol. 1995;15:2858–2871. doi: 10.1128/mcb.15.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerny T, Schaffner G, Busslinger M. DNA sequence recognition by Pax proteins: bipartite structure of the paired domain and its binding site. Genes Dev. 1993;7:2048–2061. doi: 10.1101/gad.7.10.2048. [DOI] [PubMed] [Google Scholar]
- Czerny T, Halder G, Kloter U, Souabni A, Gehring WJ, Busslinger M. Twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol. Cell. 1999;3:297–307. doi: 10.1016/s1097-2765(00)80457-8. [DOI] [PubMed] [Google Scholar]
- Dorfler P, Busslinger M. C-terminal activating and inhibitory domains determine the transactivation potential of BSAP (Pax-5), Pax-2 and Pax-8. EMBO J. 1996;15:1971–1982. [PMC free article] [PubMed] [Google Scholar]
- Duncan MK, Haynes JI, II, Cvekl A, Piatigorsky J. Dual roles for Pax-6: a transcriptional repressor of lens fiber cell-specific beta-crystallin genes. Mol. Cell. Biol. 1998;18:5579–5586. doi: 10.1128/mcb.18.9.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard D, Jimenez G, Heavey B, Busslinger M. Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family. EMBO J. 2000;19:2292–2303. doi: 10.1093/emboj/19.10.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein JA, Glaser T, Cai J, Jepeal L, Walton DS, Maas RL. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes Dev. 1994;8:2022–2034. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- Fickenscher HR, Chalepakis G, Gruss P. Murine Pax-2 protein is a sequence-specific trans-activator with expression in the genital system. DNA Cell Biol. 1993;12:381–391. doi: 10.1089/dna.1993.12.381. [DOI] [PubMed] [Google Scholar]
- Frigerio G, Burri M, Bopp D, Baumgartner S, Noll M. Structure of the segmentation gene paired and the Drosophila PRD gene set as part of a gene network. Cell. 1986;47:735–746. doi: 10.1016/0092-8674(86)90516-7. [DOI] [PubMed] [Google Scholar]
- Fujitani Y, Kajimoto Y, Yasuda T, Matsuoka TA, Kaneto H, Umayahara Y, Fujita N, Watada H, Miyazaki JI, Yamasaki Y, et al. Identification of a portable repression domain and an E1A-responsive activation domain in Pax4: a possible role of Pax4 as a transcriptional repressor in the pancreas. Mol. Cell. Biol. 1999;19:8281–8291. doi: 10.1128/mcb.19.12.8281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring WJ. The master control gene for morphogenesis and evolution of the eye. Genes Cells. 1996;1:11–15. doi: 10.1046/j.1365-2443.1996.11011.x. [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Ikeo K. Pax 6: mastering eye morphogenesis and eye evolution. Trends Genet. 1999;15:371–377. doi: 10.1016/s0168-9525(99)01776-x. [DOI] [PubMed] [Google Scholar]
- Glaser T, Walton DS, Maas RL. Genomic structure, evolutionary conservation and aniridia mutations in the human PAX6 gene. Nat. Genet. 1992;2:232–239. doi: 10.1038/ng1192-232. [DOI] [PubMed] [Google Scholar]
- Glaser T, Jepeal L, Edwards JG, Young SR, Favor J, Maas RL. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat. Genet. 1994;7:463–471. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Flister S, Walldorf U, Kloter U, Gegring WJ. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development. 1998;125:2181–2191. doi: 10.1242/dev.125.12.2181. [DOI] [PubMed] [Google Scholar]
- Held LI. Dev. Cell Biol. Ser. Vol. 39. Cambridge Press; 2002. Imaginal discs: the genetic and cellular logic of pattern formation. p. 460. [Google Scholar]
- Hill RE, Favor J, Hogan BL, Ton CC, Saunders GF, Hanson IM, Prosser J, Jordan T, Hastie ND, van Heyningen V. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- Jordan T, Hanson I, Zaletayev D, Hodgson S, Prosser J, Seawright A, Hastie N, van Heyningen V. The human PAX6 gene is mutated in two patients with aniridia. Nat. Genet. 1992;1:328–332. doi: 10.1038/ng0892-328. [DOI] [PubMed] [Google Scholar]
- Jun S, Desplan C. Cooperative interactions between paired domain and homeodomain. Development. 1996;122:2639–2650. doi: 10.1242/dev.122.9.2639. [DOI] [PubMed] [Google Scholar]
- Jun S, Wallen RV, Goriely A, Kalionis B, Desplan C. Lune/eye gone, a Pax-like protein, uses a partial paired domain and a homeodomain for DNA recognition. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13720–13725. doi: 10.1073/pnas.95.23.13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon KL, Ranade SS, Curtiss J, Mlodzik M, Pignoni F. Coordinating proliferation and tissue specification to promote regional identity in the Drosophila head. Dev. Cell. 2003;5:403–414. doi: 10.1016/s1534-5807(03)00243-0. [DOI] [PubMed] [Google Scholar]
- Kozmik Z. Pax genes in eye development and evolution. Curr. Opin. Genet. Dev. 2005;15:430–438. doi: 10.1016/j.gde.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Kumar JP, Moses K. EGF receptor and Notch signaling act upstream of Eyeless/Pax6 to control eye specification. Cell. 2001a;104:687–697. doi: 10.1016/s0092-8674(01)00265-3. [DOI] [PubMed] [Google Scholar]
- Kumar JP, Moses K. Expression of evolutionarily conserved eye specification genes during Drosophila embryogenesis. Dev. Genes Evol. 2001b;211:406–414. doi: 10.1007/s004270100177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP, Ready DF. Rhodopsin plays an essential structural role in Drosophila photoreceptor development. Development. 1995;121:4359–4370. doi: 10.1242/dev.121.12.4359. [DOI] [PubMed] [Google Scholar]
- Kurata S, Go MJ, Artavanis-Tsakonas S, Gehring WJ. Notch signaling and the determination of appendage identity. Proc. Natl. Acad. Sci. U. S. A. 2000;97:2117–2122. doi: 10.1073/pnas.040556497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D, Powell SK, Plummer RS, Young KP, Ruggeri BA. PAX genes: roles in development, pathophysiology, and cancer. Biochem. Pharmacol. 2007;73:1–14. doi: 10.1016/j.bcp.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Lechner MS, Dressler GR. Mapping of Pax-2 transcription activation domains. J. Biol. Chem. 1996;271:21088–21093. doi: 10.1074/jbc.271.35.21088. [DOI] [PubMed] [Google Scholar]
- Liu JJ, Kao WW, Wilson SE. Corneal epithelium-specific mouse keratin K12 promoter. Exp. Eye Res. 1999;68:295–301. doi: 10.1006/exer.1998.0593. [DOI] [PubMed] [Google Scholar]
- Mansouri A, Goudreau G, Gruss P. Pax genes and their role in organogenesis. Cancer Res. 1999;59:1707s–1709s. discussion 1709s–1710s. [PubMed] [Google Scholar]
- Michaut L, Flister S, Neeb M, White KP, Certa U, Gehring WJ. Analysis of the eye developmental pathway in Drosophila using DNA microarrays. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4024–4029. doi: 10.1073/pnas.0630561100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimi T, Seimiya M, Kloter U, Flister S, Gehring WJ. Direct regulatory interaction of the eyeless protein with an eye-specific enhancer in the sine oculis gene during eye induction in Drosophila. Development. 1999;126:2253–2260. doi: 10.1242/dev.126.10.2253. [DOI] [PubMed] [Google Scholar]
- Noll M. Evolution and role of Pax genes. Curr. Opin. Genet. Dev. 1993;3:595–605. doi: 10.1016/0959-437x(93)90095-7. [DOI] [PubMed] [Google Scholar]
- Ostrin EJ, Li Y, Hoffman K, Liu J, Wang K, Zhang L, Mardon G, Chen R. Genome-wide identification of direct targets of the Drosophila retinal determination protein Eyeless. Genome Res. 2006;16:466–476. doi: 10.1101/gr.4673006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli T, Seimiya M, Blanco J, Gehring WJ. Identification of functional sine oculis motifs in the autoregulatory element of its own gene, in the eyeless enhancer and in the signalling gene hedgehog. Development. 2005;132:2771–2782. doi: 10.1242/dev.01841. [DOI] [PubMed] [Google Scholar]
- Papatsenko D, Nazina A, Desplan C. A conserved regulatory element present in all Drosophila rhodopsin genes mediates Pax6 functions and participates in the fine-tuning of cell-specific expression. Mech. Dev. 2001;101:143–153. doi: 10.1016/s0925-4773(00)00581-5. [DOI] [PubMed] [Google Scholar]
- Pappu KS, Ostrin EJ, Middlebrooks BW, Sili BT, Chen R, Atkins MR, Gibbs R, Mardon G. Dual regulation and redundant function of two eye-specific enhancers of the Drosophila retinal determination gene dachshund. Development. 2005;132:2895–2905. doi: 10.1242/dev.01869. [DOI] [PubMed] [Google Scholar]
- Pichaud F, Desplan C. Pax genes and eye organogenesis. Curr. Opin. Genet. Dev. 2002;12:430–434. doi: 10.1016/s0959-437x(02)00321-0. [DOI] [PubMed] [Google Scholar]
- Plaza S, Dozier C, Saule S. Quail Pax-6 (Pax-QNR) encodes a transcription factor able to bind and trans-activate its own promoter. Cell Growth Differ. 1993;4:1041–1050. [PubMed] [Google Scholar]
- Punzo C, Kurata S, Gehring WJ. The eyeless homeodomain is dispensable for eye development in Drosophila. Genes Dev. 2001;15:1716–1723. doi: 10.1101/gad.196401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punzo C, Plaza S, Seimiya M, Schnupf P, Kurata S, Jaeger J, Gehring WJ. Functional divergence between eyeless and twin of eyeless in Drosophila melanogaster. Development. 2004;131:3943–3953. doi: 10.1242/dev.01278. [DOI] [PubMed] [Google Scholar]
- Punzo C, Seimiya M, Flister S, Gehring WJ, Plaza S. Differential interactions of eyeless and twin of eyeless with the sine oculis enhancer. Development. 2002;129:625–634. doi: 10.1242/dev.129.3.625. [DOI] [PubMed] [Google Scholar]
- Quiring R, Walldorf U, Kloter U, Gehring WJ. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [see comments] [DOI] [PubMed] [Google Scholar]
- Shen W, Mardon G. Ectopic eye development in Drosophila induced by directed dachshund expression. Development. 1997;124:45–52. doi: 10.1242/dev.124.1.45. [DOI] [PubMed] [Google Scholar]
- Sheng G, Harris E, Bertuccioli C, Desplan C. Modular organization of Pax/homeodomain proteins in transcriptional regulation. Biol. Chem. 1997a;378:863–872. doi: 10.1515/bchm.1997.378.8.863. [DOI] [PubMed] [Google Scholar]
- Sheng G, Thouvenot E, Schmucker D, Wilson DS, Desplan C. Direct regulation of rhodopsin 1 by Pax-6/eyeless in Drosophila: evidence for a conserved function in photoreceptors. Genes Dev. 1997b;11:1122–1131. doi: 10.1101/gad.11.9.1122. [DOI] [PubMed] [Google Scholar]
- Singh S, Stellrecht CM, Tang HK, Saunders GF. Modulation of PAX6 homeodomain function by the paired domain. J. Biol. Chem. 2000;275:17306–17313. doi: 10.1074/jbc.M000359200. [DOI] [PubMed] [Google Scholar]
- Strachan T, Read AP. PAX genes. Curr. Opin. Genet. Dev. 1994;4:427–438. doi: 10.1016/0959-437x(94)90032-9. [DOI] [PubMed] [Google Scholar]
- Stuart ET, Kioussi C, Gruss P. Mammalian Pax genes. Annu. Rev. Genet. 1994;28:219–236. doi: 10.1146/annurev.ge.28.120194.001251. [DOI] [PubMed] [Google Scholar]
- Stuart ET, Haffner R, Oren M, Gruss P. Loss of p53 function through PAX-mediated transcriptional repression. EMBO J. 1995;14:5638–5645. doi: 10.1002/j.1460-2075.1995.tb00251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton CC, Hirvonen H, Miwa H, Weil MM, Monaghan P, Jordan T, van Heyningen V, Hastie ND, Meijers-Heijboer H, Drechsler M, et al. Positional cloning and characterization of a paired box- and homeobox-containing gene from the aniridia region. Cell. 1991;67:1059–1074. doi: 10.1016/0092-8674(91)90284-6. [DOI] [PubMed] [Google Scholar]
- Treisman J, Harris E, Desplan C. The paired box encodes a second DNA-binding domain in the paired homeo domain protein. Genes Dev. 1991;5:594–604. doi: 10.1101/gad.5.4.594. [DOI] [PubMed] [Google Scholar]
- Underhill DA. Genetic and biochemical diversity in the Pax gene family. Biochem. Cell Biol. 2000;78:629–638. [PubMed] [Google Scholar]
- Walther C, Guenet JL, Simon D, Deutsch U, Jostes B, Goulding MD, Plachov D, Balling R, Gruss P. Pax: a murine multigene family of paired box-containing genes. Genomics. 1991;11:424–434. doi: 10.1016/0888-7543(91)90151-4. [DOI] [PubMed] [Google Scholar]
- Wang Q, Fang WH, Krupinski J, Kumar S, Slevin M, Kumar P. Pax genes in embryogenesis and oncogenesis. J. Cell. Mol. Med. 2008 doi: 10.1111/j.1582-4934.2008.00427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weasner B, Salzer C, Kumar JP. Sine oculis, a member of the SIX family of transcription factors, directs eye formation. Dev. Biol. 2007;303:756–771. doi: 10.1016/j.ydbio.2006.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D, Sheng G, Lecuit T, Dostatni N, Desplan C. Cooperative dimerization of paired class homeo domains on DNA. Genes Dev. 1993;7:2120–2134. doi: 10.1101/gad.7.11.2120. [DOI] [PubMed] [Google Scholar]
- Xu W, Rould MA, Jun S, Desplan C, Pabo CO. Crystal structure of a paired domain-DNA complex at 2.5 A resolution reveals structural basis for Pax developmental mutations. Cell. 1995;80:639–650. doi: 10.1016/0092-8674(95)90518-9. [DOI] [PubMed] [Google Scholar]
- Yao JG, Sun YH. Eyg and Ey Pax proteins act by distinct transcriptional mechanisms in Drosophila development. EMBO J. 2005;24:2602–2612. doi: 10.1038/sj.emboj.7600725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao JG, Weasner BM, Wang LS, Jang CC, Weasner B, Tang CY, Salzer CL, Chen CH, Hay BA, Sun YH, et al. Differential requirements for the Pax6(5a) genes eyegone and twin of eyegone during eye development in Drosophila. Dev. Biol. 2008;315:535–551. doi: 10.1016/j.ydbio.2007.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Ranade S, Cai CQ, Clouser C, Pignoni F. Direct control of neurogenesis by selector factors in the fly eye: regulation of atonal by Ey and So. Development. 2006;133:4881–4889. doi: 10.1242/dev.02669. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.