Abstract
SIR2-like proteins have been implicated in a wide range of cellular events including chromosome silencing, chromosome segregation, DNA recombination and the determination of life span. We report here the molecular and functional characterization of a SIR2-related protein from the protozoan parasite Trypanosoma brucei, which we termed TbSIR2RP1. This protein is a chromosome-associated NAD-dependent enzyme which, in contrast to other known proteins of this family, catalyses both ADP-ribosylation and deacetylation of histones, particulary H2A and H2B. Under- or overexpression of TbSIR2RP1 decreased or increased, respectively, cellular resistance to DNA damage. Treatment of trypanosomal nuclei with a DNA alkylating agent resulted in a significant increase in the level of histone ADP-ribosylation and a concomitant increase in chromatin sensitivity to micrococcal nuclease. Both of these responses correlated with the level of TbSIR2RP1 expression. We propose that histone modification by TbSIR2RP1 is involved in DNA repair.
Keywords: DNA damage/histone deacetylase/histone ribosyltransferase/SIR2 homologue/Trypanosoma
Introduction
The yeast SIR2 is the paradigmatic member of a large family of proteins present in many organisms from bacteria to metazoan eukaryotes (Brachmann et al., 1995; Frye, 2000). Members of this family share a core domain of ∼250 amino acids that exhibits 25–60% sequence identity between different organisms. SIR2-like proteins have been implicated in a wide range of chromosome-associated phenomena, such as gene silencing, cell cycle progression, chromosome segregation, DNA repair and life span (Brachmann et al., 1995; Tsukamoto et al., 1997; Freeman-Cook et al., 1999; Martin et al., 1999; Xie et al., 1999; Lin et al., 2000; Tissenbaum and Guarente, 2001). The first indication that these proteins might be enzymes came from studies on cobalamine biosynthesis in Salmonella typhimurium, which revealed that CobB, the Salmonella SIR2 homologue protein, was able to compensate in part for the loss of CobT, a protein that functions in the cobalamine biosynthesis pathway as a nicotinate mononucleotide: 5,6-dimethylbenzimidazole phos phoribosyltransferase (Tsang and Escalante-Semerena, 1998). This complementation suggested that SIR2-like proteins might be ribosyltransferases. Initially, biochemical studies on SIR2 from yeast seemed to support this view since the recombinant protein appeared to ADP-ribosylate bovine serum albumin (BSA) and histones in an in vitro assay (Frye, 1999; Tanny et al., 1999). However, it became clear from further studies that the nuclear SIR2 was an NAD-dependent deacetylase and that cleavage of the β-glycosidic linkage between the nicotinamide ring and the ribose sugar of NAD was coupled to the deacetylation reaction liberating free nicotinamide and a novel product, O-acetyl-ADP-ribose (Imai et al., 2000; Landry et al., 2000a,b; Smith et al., 2000; Tanner et al., 2000; Sauve et al., 2001; Tanny and Moazed, 2001; Borra et al., 2002). The consensus view at present is that SIR2-like proteins constitute a new family of NAD-dependent deacetylases, termed class III, but do not possess genuine ADP-ribosyltransferase activity.
The observation that histones at silenced loci in yeast are hypoacetylated (Braunstein et al., 1993, 1996) provided an obvious link between the deacetylase activity of SIR2 and heterochromatin structure. However, the wide phylogenetic distribution of SIR2-like proteins plus their presence in different subcellular compartments within eukaryotes suggests that SIR2-mediated deacetylation might not be restricted to regulation of chromatin structure (Zemzoumi et al., 1998; Afshar and Murnane, 1999; Perrod et al., 2001). For example, it was demonstrated recently that the human SIR2 orthologue hSIRT1 negatively regulates the tumour suppressor p53 by deacetylation of a known control site (Luo et al., 2001; Vaziri et al., 2001; Langley et al., 2002), and that hSIRT2 deacetylates tubulin in vivo (North et al., 2003), while the murine mSIR2a represses Pol I transcription by deacetylation of the transcription factor TAFI68 (Muth et al., 2001). Futhermore, in bacteria, CobB inactivates acetyl-CoA synthetase by deacetylation of an active lysine (Starai et al., 2002) and ssSIR2 from Sulfolobus solfataricus mediates transcriptional repression by deacetylating the major archaeal chromatin protein (Bell et al., 2002). In summary, the evidence emerging from the literature is that the SIR2 proteins regulate the structure/function of multiple targets by deacetylation of specific residues.
African trypanosomes, e.g. Trypanosoma brucei, are protozoan flagellates that grow in the mammalian vasculature and cause sleeping sickness in man and Nagana in cattle. These parasites are transmitted between hosts by the tsetse fly, where they develop as procyclic forms in the insect midgut (Vickerman, 1985). In the bloodstream, they evade the immune response of the host by continually changing their variant surface glycoprotein (VSG) (reviewed in Cross et al., 1998; Pays and Nolan, 1998; Borst and Ulbert, 2001). Individual bloodstream forms express a single VSG at a time, and the expressed VSG gene is located at the end of a telomeric expression site harbouring a polycistronic transcription unit. Variation of the VSG occurs either by gene conversion or by the transcriptional activation of a new expression site. The genome of T.brucei contains 11 diploid pairs of megabase chromosomes (1–6 Mbp), which carry the essential genes (Melville et al., 1998), and ∼100 minichromosomes (50–150 kb) which consist predominantly of a tandemly repeated 177 bp element (Weiden et al., 1991). Both megabase chromomes and minichromosomes have canonical telomeres at both ends containing long TTAGGG repeats. As in yeast, the trypanosome chromosomes never condense and the nuclear envelope remains intact during mitosis (Solari, 1980, 1983). In order to characterize factors that influence chromatin structure and the regulation of gene expression in T.brucei, we initiated a search for SIR2 homologues in this parasite. Here we show that a homologue of SIR2 from T.brucei, termed TbSIR2RP1, is a chromosome-associated protein that catalyses the NAD-dependent ADP ribosylation and deacetylation of histones. TbSIR2RP1 appears to have a role in the mechanism of DNA repair probably by converting target regions of chromatin into a more relaxed or open state. Significantly, there was a marked increase in the level of histone ribosylation and a concomitant increase in chromatin sensitivity to micrococcal nuclease (MNase) in response to the oxidizing DNA damage agent methyl methanosulfonate (MMS). Both of these responses correlated with the level of TbSIR2RP1 expression. We propose that histone modification by TbSIR2RP1 is critical for DNA repair.
Results
Identification of a gene coding for a homologue to SIR2 in T.brucei
We employed degenerate primers based on two highly conserved motifs of the SIR2 sequence (Brachmann et al., 1995) to amplify a 250 bp fragment from T.brucei genomic DNA. This amplification product subsequently was used to isolate a full-length cDNA that we have named TbSIR2RP1, for T.brucei SIR2-related protein 1. During the course of these studies, the sequence of TbSIR2RP1 was deposited in the data bank with the accession No. AF102869 (M.Hoek and G.A.M.Cross, unpublished). In addition, a search of the current T.brucei genome database allowed the identification of two other SIR2-related genes that we termed TbSIR2RP2 and TbSIR2RP3 (accession Nos AC119406 and AF102869). These genes shared 45.7 and 43.2% nucleotide sequence identity, respectively, with TbSI2RP1over the entire open reading frames (ORFs).
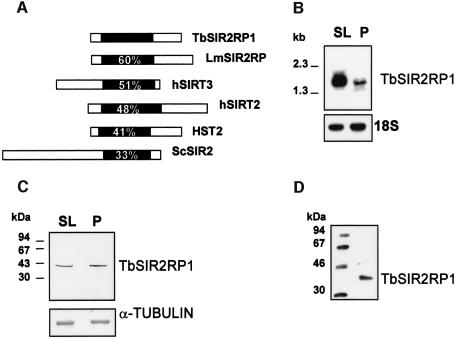
TbSIR2RP1 encodes a protein of 351 amino acids that, like all members of this family, contains a core of 250 amino acids including the GAG and NID motifs and a four-Cys-zinc finger as well as the HG residues adjacent to the zinc finger that have been shown to be essential for its enzymatic activities (Frye, 1999; Tanny et al., 1999; Imai et al., 2000). The primary sequence of TbSIR2RP1 predicted a mol. wt of ∼40 kDa and a pI of 5.7. Comparison of the sequence of TbSIR2RP1 with SIR2-like proteins from other organisms revealed that sequence homology was restricted to the 250 amino acid core domain (Figure 1A). Within this conserved domain, TbSIR2RP1 had the highest sequence homology (between 41 and 60%) with LmSIR2RP (Yahiaoui et al., 1996), human SIRT3 and SIRT2 (Frye, 1999), and the yeast HST2 (Brachmann et al., 1995) (Figure 1A). Together, this group of proteins represents a branch of the SIR2 family tree that has been designated class Ib (Frye, 2000) or HST2-like proteins (Perrod et al., 2001).
Fig. 1. Characterization of TbSIR2RP1. (A) Sequence comparison between TbSIR2RP1 and homologous proteins. The position of the conserved core domain is shown in black, and numbers represent the percentage amino acid identity with this region of TbSIR2RP1. The SIR2-like proteins compared are: LmSIR2RP from L.major (Yahiaoui et al., 1996), hSIRT2 and hSIRT3 from man (Frye, 1999) and HST2 and SIR2 from S.cerevisiae (Brachmann et al., 1995). (B) Northern blot analysis of TbSIR2RP1 mRNA. A 10 µg aliquot of poly(A)+ RNA from T.brucei AnTat 1.1 bloodstream long slender (SL) and procyclic forms (P) was separated on a formaldehyde agarose gel, transferred to nitrocellulose and probed with 32P-labelled TbSIR2RP1 cDNA. The filter was subsequently rehybridized with a ribosomal 18S probe, as control for RNA loading. (C) Immunoblot analysis of TbSIR2RP1. Total bloodstream slender (SL) and procyclic (P) cell lysates in SDS–PAGE sample buffer were examined by western blot analysis with affinity- purified anti-TbSIR2RP1. A control analysis was performed on a similar filter with an anti-α-tubulin monoclonal antibody. (D) Immunoprecipitation of in vitro translated TbSIR2RP1 with affinity-purified anti-TbSIR2RP1 antibodies.
Expression of TbSIR2RP1
A Southern blot analysis demonstrated that TbSIR2RP1 is present as a single copy gene (data not shown). A northern blot analysis of poly(A)+ RNA prepared from different stages of the parasite life cycle revealed that the TbSIR2RP1 mRNA was more abundant in bloodstream long slender than in procyclic forms (Figure 1B). However, a western blot analysis using affinity-purified antibodies raised against recombinant TbSIR2RP1 demonstrated that, in contrast to the northern blot data, TbSIR2RP1 was present at the same relative abundance in bloodstream slender and procyclic forms (Figure 1C). In vitro translation followed by immunoprecipitation confirmed the predicted size of TbSIR2RP1 (40 kDa) and demonstrated the specificity of the anti-TbSIR2RP1 antibodies (Figure 1D).
TbSIR2RP1 is a nuclear protein that co-localizes with telomeric sequences and minichromosomes
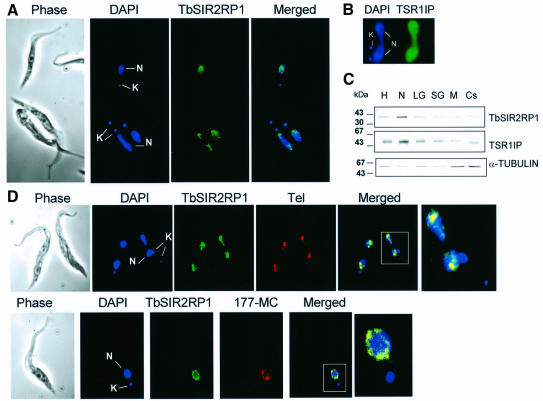
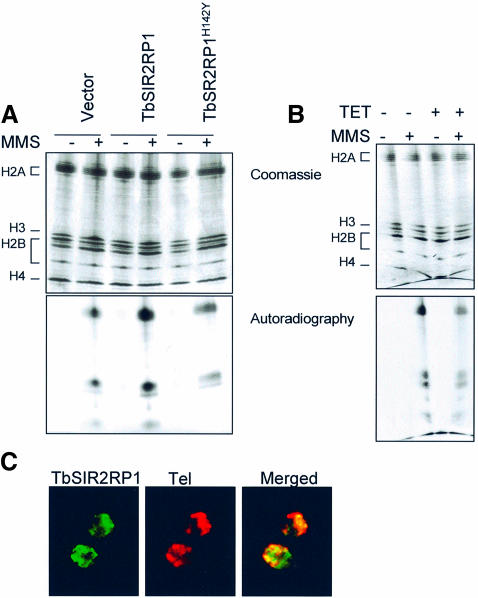
In order to localize TbSIR2RP1, immunofluorescence (IF) microscopy was performed on fixed procyclic cells with anti-TbSIR2RP1 antibodies. TbSIR2RP1 was found in the nucleus where it exhibited a punctate distribution (Figure 2A). In contrast, the transcription factor TbSR1IP used as an internal control showed the expected more diffuse distribution over the entire nucleus (Figure 2B) (Ismaili et al., 2000). Since this was the first example of a member of the HST2 subfamily located in the nucleus, an alternative approach was employed to confirm this location. Western blot analysis of subcellular factions prepared from procyclic forms confirmed that TbSIR2RP1 was recovered mainly in the nuclear fraction (Figure 2C). The distribution of TbSIR2RP1 in these fractions was very similar to that observed for the nuclear TbSR1IP but clearly different from that of α-tubulin. Interestingly, the distribution of TbSIR2RP1 in the nucleus varied depending on the stage of the cell cycle. The cell cycle is easy to follow in trypanosomes by monitoring both nuclear elongation and the segregation of the mitochondrial (kinetoplast) DNA (Sherwin and Gull, 1989). In the G1 phase, trypanosomes have a single kinetoplast and nucleus. After kinetoplast segregation, G2 phase cells contain two kinetoplasts and one nucleus, while after mitosis but before cell division two kinetoplasts and two nuclei are present. In G1 phase cells, the TbSIR2RP1 immunofluorescent signal was always in tight clusters spread throughout the entire nucleus or near the periphery (Figure 2A, top). However, in dividing cells (two kinetoplasts and one or two nuclei), the signal was located at the poles of the dividing nucleus (Figure 2A, bottom, and D, top). This behaviour was very similar to that reported for telomeres (Chung et al., 1990; Ersfeld and Gull, 1997; Perez-Morga et al., 2001) and raised the possibility that TbSIR2RP1 is associated with telomeres. In order to test this possibility, IF was combined with fluorescence in situ hybridization (FISH) using fixed procyclic cells and a fluorescent probe for telomeric repeats (Figure 2D). A punctate staining throughout the nucleus was observed with both TbSIR2RP1 antibodies (green) and the probe for the telomeric repeats (red). The merged image of the two probes showed significant overlap (orange/yellow), suggesting that TbSIR2RP1 is associated with telomeres. Overlapping signals were also obtained with a specific probe for minichromosomes (a 177 bp repeat, 177-MC in Figure 2D). The latter are known to be concentrated in a few foci and to be >10-fold more numerous than large chromosomes (Weiden et al., 1991). Therefore, both telomeric and minichromosome probes essentially recognize spots where chromosomes are concentrated. The minimal conclusion from these data is that TbSIR2RP1 is associated with chromosomes, at least minichromosomes.
Fig. 2. Cellular localization of TbSIR2RP1. (A) Procyclic forms were stained by immunofluorescence with anti-TbSIR2RP1 antibodies and counterstained with DAPI. The DAPI staining reveals the location of the nucleus (N) and kinetoplast (mitochondrial DNA, K). (B) Immunolocalization of TSR1IP nuclear protein. (C) Distribution of TbSIR2RP1 in subcellular fractions from procyclic forms of T.brucei. Subcellular fractions were separated by SDS–PAGE, transferred to nitrocellulose and probed with anti-TbSIR2RP1, anti-α-tubulin and anti-TSR1IP antibodies. Subcellular fraction abbreviations are as follows: H, homogenate; N, nuclear; LG, large granular (mitochondrial); SG, small granular (glycosomal); M, endosomal; Cs, cytoplasmic (soluble). (D) TbSIR2RP1 co-localizes with telomeres and minichromosomes. Procyclic forms were fixed and subjected to IF and FISH using anti-TbSIR2RP1 antibodies and probes for telomeric repeats (Tel) and minichromosome (177-MC). Overlap of the two signals is yellow/orange.
TbSIR2RP1 is an NAD-dependent ADP-ribosyltransferase
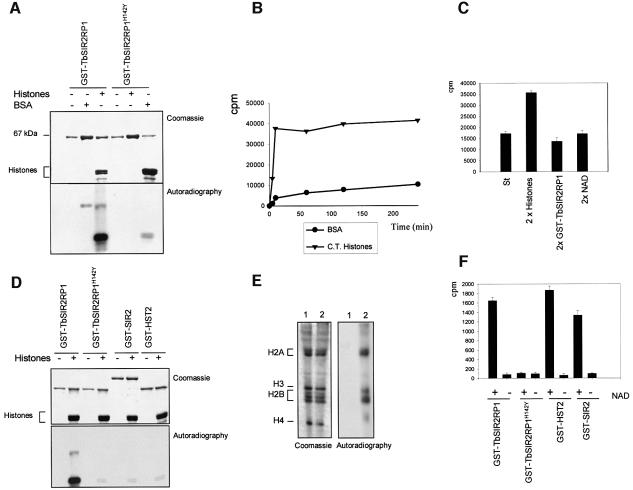
Two reports have suggested that members of the SIR2 family can mediate the transfer of ribose 5′-phosphate from nicotinic acid mononucleotide to amino acid residues of BSA, histones or SIR2 proteins themselves (Frye, 1999; Tanny et al., 1999). To assess whether TbSIR2RP1 also possessed ADP-ribosylase activity, a GST fusion protein containing the entire ORF fused C-terminal to GST was expressed in Escherichia coli and purified to homogeneity by glutathione affinity chromatography. The purified protein was assayed for ADP-ribosyltransferase using [32P]NAD as the donor and BSA or calf thymus histones as acceptor substrates. As a negative control, an essential histidine (H142) required for enzyme activity within the core domain of the protein (Frye, 1999; Tanny et al., 1999; Imai et al., 2000) was mutated by site-directed mutagenesis and converted into tyrosine to generate TbSIR2RP1H142Y. Analysis of the products of these assays by SDS–PAGE/autoradiography revealed the presence of a strongly labelled band corresponding to histones, together with a much weaker band that migrated with the same apparent molecular mass as BSA (67 kDa), but was also present with histones (Figure 3A). Although GST–TbSIR2RP1 migrated with a similar molecular mass to BSA, no signal was detected when the fusion protein was incubated in the absence of substrate. As detailed in the Discussion, at least in the case of histones, this weakly labelled band may reflect transient auto-ADP-ribosylation of TbSIR2RP1 during the course of its activity. Significantly, when the inactive TbSIR2RP1H142Y was employed in the assay, only a very minor incorporation of the 32P label into histones was observed (Figure 3A). This result clearly demonstrated that active TbSIR2RP1 was required to stably and covalently modify the target protein and the modification was not due to some non-specific non-enzymatic reaction. Taken together, these data indicated that TbSIR2RP1 is a genuine ADP-ribosyltransferase with at least two specific substrates, histones and BSA, the affinity for histones being higher than for BSA.
Fig. 3. NAD-dependent ribosylation and deacetylation of proteins by TbSIR2RP1. (A) In vitro ribosylation reactions were performed with 1 µg of GST–TbSIR2RP1 or GST–TbSIR2RP1H142Y, plus 5 µg of BSA and 5 µg of histones using [32P]NAD as donor. The top panel illustrates a Coomassie-stained gel of reaction products resolved by 10% SDS–PAGE, whereas the bottom panel shows the autoradiograph of the gel. (B) Time curves of histone and BSA ribosylation by GST–TbSIR2RP1 at 37°C. Each point represents the ribosylation products of 5 µg of substrate by 0.5 µg of GST–TbSIR2RP1 using 5 µCi of [32P]NAD at the indicated time. Reaction products were precipitated with 20% TCA (w/v), collected and washed on a GF/C glass fibre filter (Whatman), and then counted after addition of liquid scintillation fluid. (C) Ribosylation reactions were performed with 5 µg of histones, 0.5 µg of GST–TbSIR2RP1 and 5 µCi of [32P]NAD (standard reaction, column St) or a double concentration of histone (10 µg, column 2× histones), GST–TbSIR2RP1 (1 µg, column 2× GST–TbSIR2RP1) and [32P]NAD (10 µCi, column 2× NAD). (D) Comparison of TbSIR2RP1 ribosylation activity with that of other members of the SIR2-like family. Ribosylation reactions with 1 µg of GST–TbSIR2RP1, GST–TbSIR2RP1H142Y, GST–SIR2 or GST–HST2 were performed with and without 5 µg of histones using [32P]NAD as donor. The top panel illustrates a Coomassie-stained gel of the reaction products, and the bottom panel shows the autoradiograph of the gel. (E) Triton–acid–urea (TAU) gel of T.brucei histones treated with GST–TbSIR2RP1 or GST–TbHST1H142Y and [32P]NAD, respectively stained with Coomassie blue and autoradiographed. The histone types are indicated on the left. (F) Analysis of the NAD-dependent histone deacetylase activity. Histones were acetylated with [3H]acetyl-CoA by HAT1 and then treated with GST–TbSIR2RP1, GST– TbSIR2RP1H142Y, GST–HST2 and GST–SIR2. The amount of acetate released was measured in the presence or absence of NAD.
In order to compare the rate of ribosylation of histones or BSA by TbSIR2RP1, the assay was stopped at various times and the level of the 32P incorporation was measured after precipitation of the proteins (Figure 3B). The time curves confirmed that TbSIR2RP1 has a far higher activity using histones than BSA (Figure 3B). When histones were the substrate, the reaction progressed much more rapidly and reached a plateau after 10 min. To investigate this plateau further, ribosylation reactions were performed using a double concentration of substrate, enzyme and NAD, respectively. Only a doubling of histone concentration yielded a doubling of 32P incorporation into the substrate (Figure 3C). We conclude that histones are the limiting factor in the ribosylation reaction by TbSIR2RP1.
Comparison of TbSIR2RP1 ribosylation activity with that of other members of the SIR2-like family
It has been proposed that the weak ribosyltransferase activity of other SIR2 proteins is not genuine, but reflects an intermediate step during NAD hydrolysis (Tanner et al., 2000). This suggestion prompted us to compare the ribosyltranferase activity of TbSIR2RP1 with that of other SIR2-like proteins. Two other members of the family, the yeast SIR2 and HST2, were expressed as C-terminal GST fusion proteins exactly as for TbSIR2RP1, and subjected to the identical ADP-ribosylase assay. Consistent with the previous results, a strong ribosylation activity was observed with TbSIR2RP1 (Figure 3D), but not with its H142Y mutant. In contrast, very little ADP-ribosyltransferase activity was observed in the two other cases (Figure 3D), as expected (Landry et al., 2000b).
Histones H2A and H2B are major targets for in vitro TbSIR2RP1 ribosylation
We subsequently analysed whether all of the histones are equally effective as acceptors of ADP-ribosyl groups. Trypanosome histones were incubated with GST–TbSIR2RP1 in the presence of [32P]NAD and the reaction products were separated on a Triton–acid–urea (TAU) gel. These gels allow the separation of the various classes of histones as well as their modified species. Figure 3E demonstrates that two types of histone were ribosylated by TbSIR2RP1 and these appeared to correspond to histones H2A and H2B, respectively (Burri et al., 1994). Protein digestion and mass spectrometry (MALDI-TOF) of the ribosylated bands confirmed the assignment of histones H2A and H2B as the major ribosylation products, whereas histone H4 was ribosylated to a lesser extent. These results indicate that GST–TbSIR2RP1 is an ADP-ribosyltransferase with high affinity for histones H2A and H2B.
TbSIR2RP1 is an NAD-dependent histone deacetylase
It is well established that SIR2-like proteins constitute a new class of protein deacetylases. To analyse whether TbSIR2RP1 also possesses NAD-dependent deacetylase activity, trypanosome histones were first acetylated with [3H]acetyl-CoA using the enzyme HAT1, a yeast histone H4 acetyl transferase (Kleff et al., 1995). Radiolabelled histones were then incubated with GST–TbSIR2RP1, GST–TbSIR2RP1H142Y, GST–SIR2 and GST–HST2 with and without NAD, and the release of [3H]acetyl groups was monitored. As shown in Figure 3F, similar levels of deacetylation were observed with GST–TbSIR2RP1, GST–SIR2 and GST–HST2, and in all cases this activity was absolutely dependent on the presence of NAD. As expected, the H142Y mutant was inactive.
TbSIR2RP1 confers resistance to DNA damage
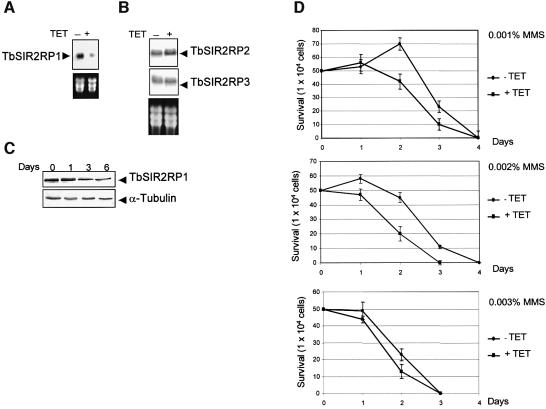
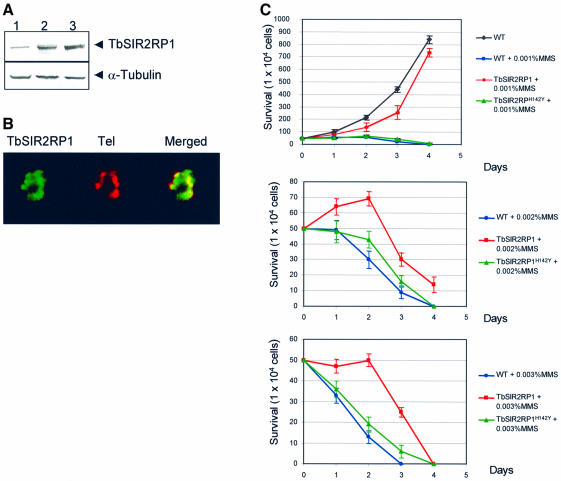
In yeast, SIR proteins have been reported to play a role in DNA repair, although the mechanism remains unclear (Tsukamoto et al., 1997; Lee et al., 1999; Martin et al., 1999; Mills et al., 1999). The human homologue, hSIRT1, is involved in cell survival by inactivating the tumour suppressor p53, the primary mediator of the cellular DNA damage response (Luo et al., 2001; Vaziri et al., 2001; Langley et al., 2002). In mammalian cells, histones are mono- and polyribosylated by a poly(ADP)ribose polymerase in response to DNA damage and oxidative stress. These findings prompted us to consider whether the trypanosomal SIR2 homologue is involved in the response to DNA damage. This question was addressed by specific ablation of the TbSIR2RP1 mRNA using RNA interference (RNAi) (Ngo et al., 1998; Wang et al., 2000). Therefore, we generated a trypanosome cell line that expressed a double-stranded RNA corresponding to the first 600 nucleotides of TbSIR2RP1 under the control of the tetracycline-inducible promoter. The effect of TbSIR2RP1 RNAi was assessed by northern and western blot analysis. TbSIR2RP1 transcripts were reduced to ∼75% after 24 h in the presence of tetracycline (Figure 4A). This RNAi specifically affected TbSIR2RP1 mRNA since no change was observed in the expression of the other two SIR2-related genes found in the T.brucei genome database, which share 47.7 and 46.9% nucleotide sequence identity, respectively, over the region covered in the RNAi construct (Figure 4B). The level of TbSIR2RP1 decreased to approximately one-third of the control level after 3 days of induction (Figure 4C). This decrease did not lead to any detectable effect on trypanosome growth (not shown), but it clearly affected the cell resistance to DNA damage. As shown by the growth curves in Figure 4D, the tetracycline-induced loss of TbSIR2RP1 led to an increased sensitivity to the DNA alkylating agent MMS. This effect was more obvious at lower than at higher concentrations of the drug.
Fig. 4. Effect of TbSIR2RP1 RNAi on cell sensitivity to DNA damage. (A) Northern blot analysis of mRNA levels from cells incubated for 24 h without (–) or with (+) tetracycline. rRNAs stained with ethidium bromide are shown as loading controls. (B) A similar northern blot was hybridized with probes corresponding to the entire ORF of two other trypanosomal homologues of SIR2 (TbSIR2RP2, accession No. AC119406; TbSIR2RP3, accession No. AF102869). (C) Western blot analysis of whole-cell lysate (1 × 106 cell equivalents/lane) from uninduced (day 0) and tetracycline- induced cells (days 1–6) using anti-TbSIR2RP1 and anti-α-tubulin antibodies. (D) Cell survival curves are shown for uninduced (TET–) and tetracycline-induced cells (TET+) treated with different concentrations of MMS.
Since a decrease of TbSIR2RP1 levels correlated with an increase of sensitivity to MMS, we investigated whether overexpression of TbSIR2RP1 could increase resistance to DNA damage. TbSIR2RP1 and TbSIR2RP1H142Y were overexpressed in procyclic forms of T.brucei by targeting both genes into the tubulin locus (Sommer et al., 1992). As shown in Figure 5A, an increase of ∼2- and 3-fold was observed for TbSIR2RP1 and TbSIR2RP1H142Y, respectively. These increased levels of expression did not affect the localization of TbSIR2RP1 (Figure 5B) or cell growth (not shown). However, overexpression of TbSIR2RP1, but not TbSIR2RP1H142Y, conferred resistance to MMS since cells overexpressing TbSIR2RP1 clearly resisted better than the controls in the presence of MMS (Figure 5C). In 0.001% MMS, the cells overexpressing TbSIR2RP1 grew at a rate similar to the parental cell line in the absence of MMS. Altogether, these data support the view that the level of expression of TbSIR2RP1 correlates with cellular tolerance to agents that damage DNA.
Fig. 5. Effect of ectopic expression of TbSIR2RP1 and TbSIR2RP1H142Y on cell sensitivity to MMS. (A) Western blot analysis of control cells (lane 1) or cells overexpressing TbSIR2RP1 (lane 2) and TbSIR2RP1H142Y (lane 3) using anti-TbSIR2RP1 and anti-α-tubulin antibodies. (B) Subcellular localization of TbSIR2RP1 in cells overexpressing the protein. Procyclic forms overexpressing TbSIR2RP1 were subjected to IF and FISH using anti-TbSIR2RP1 antibodies and probes for telomeric repeats (Tel). Overlap of the two signals is yellow/orange. (C) Cell survival in the presence of different concentrations of MMS, comparing wild-type and cells overexpressing TbSIR2RP1 and TbSIR2RP1H142Y.
TbSIR2RP1 enzymatic activities play a role in DNA damage response
Since TbSIR2RP1 possesses an ADP-ribosyltransferase activity, we investigated whether ribosylation of histones was involved in the response to DNA damage caused by MMS. First, we assessed whether DNA damage resulted in ribosylation of endogenous histones in vitro. Nuclei isolated from trypanosomes were incubated for 1 h at 37°C with MMS and radioactive NAD in the presence of 3-aminobenzamide, a specific inhibitor of poly(ADP)ribose polymerase (Farzaneh et al., 1985). TAU gel analysis of histones purified from these nuclei revealed a significant increase in histone ribosylation in response to MMS (Figure 6A). Moreover, the extent of histone ribosylation was higher in nuclei isolated from cells overexpressing TbSIR2RP1 compared with wild-type and mutant overexpressor cells (Figure 6A). In contrast, the bulk of histone ribosylation in TbSIR2RP1 knocked-down cells was lower compared with control cells (Figure 6B). Interestingly, the profile of histone ribosylation under these conditions indicated that histones H2A and H2B were the major products, with histone H4 being modified to a lesser extent. This pattern of modification was similar to that obtained when recombinant TbSIR2RP1 was incubated with a mixture of histones in vitro. Together, these data support the view that histones are ribosylated by TbSIR2RP1 in response to oxidative stress. Finally, we analysed whether like in yeast, TbSIR2RP was mislocalized in response to DNA damage. As illustrated in Figure 6C, most of the signal remained in clusters co-localizing with telomeres.
Fig. 6. Extent of histone ribosylation in response to DNA damage correlates with the cellular concentration of TbSIR2RT1. The results of TAU gel analysis of histone ribosylation in nuclei from trypanosomes overexpressing TbSIR2RP1 and TbSIR2RP1H142Y (A) and from TbSIR2RP1 knocked-down cells (B). Histones were purified from nuclei treated for 1 h with MMS and [32P]NAD in the presence of 5 mM 3-aminobenzamide. The bottom panel shows the autoradiograph of the gel. (C) The localization of TbSIR2RP1 in cells with damaged DNA. Procyclic forms incubated in 0.001% MMS for 12 h were subjected to IF and FISH with anti-TbSIR2RP1 antibodies and telomeric probes, respectively.
Histone modification by TbSIR2RP1 enhances sensitivity to DNases
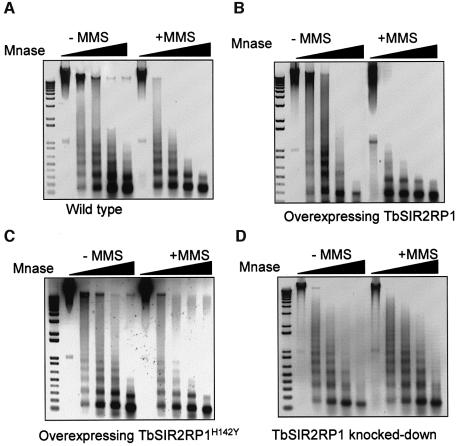
In order to understand how TbSIR2RP1-mediated ribosylation might protect cells from DNA damage, we investigated the effects of this modification on the chromatin structure using MNase. Chromatin was isolated from nuclei treated as described previously but with cold NAD, and then digested with increasing concentrations of MNase. Although the same nucleosome spacing was observed in intact and oxidized (MMS-treated) chromatin, the sensitivity to MNase was significantly greater in the latter case, i.e. damaged DNA was easier to digest (Figure 7A). These results indicated that the majority of damaged chromatin is organized in arrays of regularly spaced nucleosomes but in a more open or relaxed state than in intact chromatin. Similar results were obtained with chromatin extracted from procyclic cells overexpressing TbSIR2RP1 or TbSIR2RP1H142Y but, in the presence of MMS, cells overexpressing TbSIR2RP1 were more sensitive to MNase compared with wild-type and TbSIR2RP1H142Y (Figure 7B and C). In contrast, no significant differences in MNase pattern were observed in cells where TbSIR2RP1 was depleted by RNAi induction (Figure 7D).
Fig. 7. Alkylated chromatin is more sensitive to MNase. Untreated and MMS-treated chromatin from wild-type procyclic cells (A) and from cell overexpressing TbSIR2RP1 (B) or TbSIR2RP1H142Y (C), or from TbSIR2RP1 knocked-down cells (D) were incubated with MNase. All digestion reactions contained 4 µg of DNA. Chromatin was digested with 0.00025, 0.0005, 0.0015 and 0.005 U of MNase for 10 min at 30°C. A negative print of the ethidium-stained DNA is shown. A 1 kb PLUS DNA ladder (Invitrogen) is on the left of each panel.
Thus, chromatin appears to be more open in response to DNA damage, and when cells overexpress TbSIR2RP1.
Discussion
Two NAD-dependent enzymatic activities in TbSIR2RP1
TbSIR2RP1 appears to be an unusual member of the SIR2 family since this homologue possesses NAD-dependent histone ADP-ribosyltransferase as well as deacetylase activities. Repeated in vitro assays using [α-32P]NAD demonstrated that TbSIR2RP1 catalyses the stable covalent incorporation of 32P label, consistent with ADP-ribosyl transfer, into histones and to a lesser extent BSA. However, this activity was not detected with GST-tagged yeast SIR2 or HST2 homologues under the same assay conditions. This latter result was in agreement with those of other workers who employed histidine-tagged versions of the proteins and concluded that these proteins are not ADP-ribosyltransferases (Landry et al., 2000b; Tanner et al., 2000). ADP-ribosylation in our assay was not due to non-specific chemical modification since it was not observed when a mutant form of the protein (TbSIR2RP1H142Y) was employed. The activity was also detected using low amounts of TbSIR2RP1 (∼100 ng). Moreover, the reaction was relatively specific since histones H2A and H2B were the major ribosylation products and the labelling of histones was ∼10-fold higher than that of BSA. The final level of histone ADP-ribosylation was reached rapidly and appeared to depend solely on the amount of histones present in the assay, which suggested that only a limited number of specific sites in the substrate are available for modification by TbSIR2RP1. These assays also demonstrated an apparent auto-ADP-ribosylation of TbSIR2RP1 during the course of the assay. This observation is consistent with the generation of an ADP-ribosyl-TbSIR2RP1 intermediate during the course of the reaction. These intermediates are predicted to form part of the catalytic mechanism of ADP-ribosyltransferases due to nucleophilic attack on the C-1 of ribose. The nucleophilic group from the ADP-ribosyltransferase carries the ADP-ribosyl group that can then be transferred by a second displacement onto a suitable nucleophilic acceptor group (Koch-Nolte et al., 1996). The observation that the TbSIR2RP1 ADP-ribosyl intermediate was relatively stable, i.e. resistant to boiling in SDS–PAGE sample buffer, suggests that direct transfer to water to give free ADP-ribose does not occur but that transfer of the ADP-ribose group requires a specific nucleophilic acceptor group in the histone substrate. Presumably the build up of this intermediate during the reaction occurred because of the limiting amount of specific nucleophilic acceptor group in the histone substrate.
We also demonstrated that TbSIR2RP1 possesses the expected deacetylase activity associated with the SIR2 family and readily catalysed the release of 3H-labelled acetyl groups from histones. In common with other members of the family, the activity required NAD and was not observed using the TbSIR2RP1H142Y mutant. This finding agrees with the current view that the deacetylation step is coupled to the cleavage of the glycosidic bond and suggets that histone deacetylation and ADP-ribosylation activities are also coupled in the case of TbSIR2RP1. At present, it is unclear whether the same residues or even the same histones are undergoing coupled deacetylation/ADP-ribosylation reactions. For example, it was notable that histones H2A and H2B were the major ribosylation products, whereas histone H4 was the substrate for deacetylation since the enzyme used to acetylate histones, HAT1, mainly acetylates histone H4 (Kleff et al., 1995). However, other workers have noted that the deacetylase activity of SIR2 proteins may be relatively relaxed in these assays (Landry et al., 2000a).
The identity between TbSIR2RP1 and eukaryotic homologues within the core catalytic domain is between 30 and 50% with no significant homology outside this region. This degree of homology is typical of the conservation observed within the catalytic core of other enzymes, which raises the question of why is the ADP-ribosylation activity apparently unique to TbSIR2RP1. The simple answer might be that the ADP-ribosyltransferase activity is contained within the non-homologous region rather than the conserved core domain. It is difficult to assess this possibility given the lack of a well established consensus motif for ADP-ribosyltransferases (Takada et al., 1993), but it is clear that a mutation within the conserved core region (TbSIR2RP1H142Y) also abolished ADP-ribosyl transfer activity.
A role for TbSIR2RP1 in DNA repair
There are indications in the literature that SIR2-like proteins are involved in cellular responses to DNA damage and oxidative stress, but clear evidence as to their precise role has remained elusive (Tsukamoto et al., 1997; Lee et al., 1999; Martin et al., 1999; Mills et al., 1999; Luo et al., 2001; Langley et al., 2002). Our data suggest a correlation between the activity of TbSIR2RP1 and extent of chromatin ADP-ribosylation and the sensitivity of trypanosomes to agents that damage DNA, i.e. the ability to mount an effective repair response. For example, a decrease in the level of TbSIR2RP1 resulted in an increased sensitivity to the DNA alkylating agent MMS, while overexpression of TbSIR2RP1 resulted in an increased resistance to the agent. Moreover, both effects correlated with the extent of ADP-ribosylation of histones. In contrast, overexpression of an inactive mutant form of the protein did not increase resistance to MMS or the ribosylation status of histones. Incubation with MMS resulted in changes in the organization of trypanosomal chromatin, which appeared to adopt a more relaxed or less condensed state as it became more sensitive to MNase. Although the repair of damaged DNA is obviously a complex process, a key requirement appears to be a relaxation or reduction in the condensation state of chromatin in the region of the damage. Indeed, the efficiency of nucleotide excision repair (NER) mechanisms of UV-induced DNA lesions is reduced in chromatin substrates (Smerdon and Conconi, 1999; Green and Almouzni, 2002), nucleosome core particles (Hara et al., 2000; Liu and Smerdon, 2000; Kosmoski et al., 2001) and within SV40 minichromosomes compared with naked DNA (Wang et al., 1991; Sugasawa et al., 1993). In view of these considerations, we propose that TbSIR2RP1 is a component of the chromatin remodelling machinery that disassembles nucleosomes, by affecting the acetylation and ribosylation status of specific residues, to create enough space for subsequent binding of other NER factors.
There is evidence for a functional link between deacetylation and ADP-ribosylation in other cells. Repair of DNA damage is associated with increased mono-ADP-ribosylation of core histones as well other nuclear proteins (Bredehorst et al., 1981; Wielckens et al., 1982; Kreimeyer et al., 1984; Boulikas, 1988, 1989). Interest ingly, histone H2B served as the major acceptor of ADP-ribosyl groups in mammalian nuclei in vivo (Burzio et al., 1979; Ogata et al., 1980; Adamietz and Rudolph, 1984). There is also evidence for a link between deacetylation and ribosylation since deacetylated and ribosylated histones appear to be associated with silent regions of the mammalian genome (Tikoo and Ali, 1997). In Physarum polycephalum, inhibition of histone deacetylases by n-butyrate was accompanied by an increase in ADP-ribose incorporation into highly acetylated histone H4 subspecies (Golderer and Grobner, 1991). In addition, n-butyrate also increased the utilization of NAD and ADP-ribosylation of proteins in HeLa, V79, mouse B16, mouse Fib/T and human T1 kidney cells in culture (Bohm et al., 1997). All of these findings support the view that connection of ADP-ribosylation and deacetylation of proteins is a feature of all cells. It is tempting to speculate that one member of the SIR2 family, among the seven identified so far in human, may possess the same activities as TbSIR2RP1.
Finally, the histone code hypothesis predicts that modification marks on the histone tails provide binding sites for effector proteins (Jenuwein and Allis, 2001). Hence, histone mono- and polyribosylation may act as markers for damaged chromatin.
Materials and methods
Trypanosomes
Procyclic T.brucei from strains EATRO 1125 and 29-13 were grown in SDM-79 medium (Brun and Schonenberger, 1979) supplemented with 15% fetal bovine serum. Strain 29-13, which harbours integrated genes for T7 polymerase and tetracycline repressor (Wirtz et al., 1999), was used for RNAi. Transfection of procyclic cells was as described previously (García-Salcedo et al., 2002). The transfectants were selected with 2.5 µg/ml phleomycin for RNAi analysis (Wang et al., 2000) or 50 µg/ml hygromycin for overexpression studies.
Cloning a SIR2 homologue from T.brucei
A partial TbSIR2RP1 sequence was isolated by PCR amplification of T.brucei genomic DNA using degenerate oligonucleotide primers based on two highly conserved regions within the catalytic core domain of the SIR2-like proteins family (Brachmann et al., 1995). The PCR amplification was performed with 100 ng of genomic DNA as template and 30 pmol of primers 5′-GGNATYCCNGAYTTYMG-3′ and 5′-CKR AARTCNGGRATNCC-3′ with an initial 2 min of denaturation at 94°C followed by 35 cycles of 1 min at 90°C, 1 min at 5°C, 1 min at 72°C and a final elongation step of 10 min at 72°C. The amplification products were subcloned into pCR2.1 vector (Invitrogen). A 250 bp fragment amplified using this approach was used to screen a λgt10 cDNA library from T.brucei procyclic forms, allowing the isolation of a full-length cDNA clone.
Plasmid constructions and expression of recombinant proteins
Plasmids pGEX-TbSIR2RP1 and pMAL-TbSIR2RP1 were generated by cloning the entire TbSIR2RP1 coding sequence in-frame with the C-terminus of GST into the BamHI–XhoI site of pGEX5. Saccharomyces cerivisieae SIR2 and HST2 genes were PCR amplified from yeast genomic DNA as template and cloned into the EcoRI–XhoI sites of pGEX5 to yield pGEX-HST2 and pGEX-SIR2, respectively. Site-directed mutations were generated in the plasmid pGEX-TbSIR2RP1 using the Gene Editor system (Promega) according to the manufacturer’s instructions and verified by sequencing. TbSIR2RP1 and the mutated gene were cloned in pTSA-HYGRO for overexpression in trypanosomes (Sommer et al., 1992). The HAT1 E.coli expression plasmid, pSTT21, was a gift from Dr R.Sternglanz (Kleff et al., 1995).
Expression of the recombinant proteins was induced in mid-logarithmic phase DH5α cells, by incubation with 0.1 mM isopropyl-β-d-thiogalactopyranoside for 5 h at 37°C. The bacteria were lysed by sonication and the fusion proteins purified on glutathione–agarose (Pharmacia) as described by the manufacturer.
Immunofluorescence and in situ hybridization
Trypanosoma brucei cells were harvested by centrifugation. The pellets were resuspended in phosphate-buffered saline (PBS; 1.7 mM NaH2PO4, 9.1 mM Na2HPO4, 150 mM NaCl pH 7.5) and settled onto poly-l-lysine-coated slides. Trypanosomes were fixed in 100% methanol for 4 min at –20°C, rehydrated for 20 min in PBS and processed for IF using a 1:500 dilution of anti-TbSIR2RP1 antibodies. Rabbit anti-TbSIR2RP1 was raised against recombinant maltose-binding peptide fused to TbSIR2RP1 expressed in E.coli. FISH was performed according to instructions provided by the manufacturer (Dako, Glodstrup Demmark) using a PNA (peptide nucleic acid) fluorescein isothiocyanate (FITC)-labelled probe against the human telomeric repeats, which are identical to those in T.brucei. When IF and FISH were combined, samples were first processed as described for IF and then for FISH. Images were taken on a Zeiss Axioskop 2 microscope coupled to a CCD camera and processed by ISIS 3.
Enzyme activity assays
Protein ADP-ribosylation assays were performed in a volume of 20 µl containing 0.1–1 µg of each GST fusion protein, 5 µCi of [32P]NAD and 5 µg of calf thymus (Sigma) or T.brucei histones and BSA as indicated. The reaction buffer contained 150 mM NaCl, 10 mM dithiothreitol (DTT), 50 mM Tris–HCl pH 8.8 (Tanny et al., 1999). Samples were incubated for the indicated period of time at 37°C and reactions were stopped by the additon of Laemmli gel loading buffer. After electrophoresis, gels were stained with Coomassie brilliant blue, destained, dried on Whatman paper and exposed to film for 4–12 h at –80°C.
For deacetylation assays, calf thymus and T.brucei histones were first acetylated in vitro with [3H]acetyl-CoA and the yeast histone acetyltransferase HAT 1 (Kleff et al., 1995) as described elsewhere (Landry et al., 2000b). Reactions were incubated at 37°C for 30 min and then heated at 55°C for 30 min to inactivate the histone acetyltransferase. Labelled histones were precipitated by adding trichloroacetic acid (TCA) to a final concentration of 20% (w/v) and incubating on ice for 1 h. Precipitates were collected by centrifugation, washed with 10% TCA (w/v) and dissolved in 100 mM Tris pH 8.0. Approximately 10 µg of labelled histones (∼30 000 c.p.m.) were assayed for deacetylase activity in 50 µl reactions including 0.5 mM NAD, 0.5 mM DTT, 50 mM sodium phosphate pH 7.2 and 0.5 µg of enzyme to be tested. Reactions were stoped by addition of 18 µl of 0.1 M HCl/0.4 M acetic acid. Released acetyl groups were extracted by adding 400 µl of ethyl acetate. Afer 5 min centrifugation in a microfuge, 300 µl was counted in 4 ml of scintillation fluid (Insta-gel II, Packard).
Nuclear extraction and histone and chromatin purification
Trypanosome nuclei were prepared as described previously (Murphy et al., 1987) and then lysed in 100 mM NaCl, 0.1% Triton X-100, 1% CHAPS, 50 mM Tris pH 7.8 at 4°C for 1 h. Histones were extracted with 0.4 M H2SO4 for 2 h at 4°C. After centrifugation at 10 000 g for 10 min at 4°C, histones were recovered from the supernatant by precipitation in 20% (w/v) TCA. Precipitated histones were washed successively with acidic acetone (1% HCl in acetone) and acetone, desiccated, and dialysed against 10 mM Tris pH 8. A cocktail of aminoethylbenzenesulfonyl fluoride (AEBSF), E64 and pepstatin protease inhibitors was used during the purification process.
For ribosylation reactions, nuclei from 1 × 108 cells were incubated with 0.3% MMS and 5 µCi of [32P]NAD in 20 µl of ribosylation buffer for 1 h at 37°C. Nuclei were then treated with 10 U of DNase I for 1 h at 37°C. Proteins were precipitated with 20% TCA (w/v), washed with 10% TCA (w/v), 10% ethanol (v/v) and resuspended in TAU loading buffer. Afer electrophoresis, TAU gels were stained with Coomassie brilliant blue, dried on Whatman paper and exposed to Hyperfilm film (Amersham Pharmacia) for 1–3 weeks at –80°C.
Microccal muclease digestion of chromatin
A 4 µg aliquot of chromatin was digested in 20 µl of 250 mM sucrose, 5 mM MgCl2, 1 mM CaCl2, 50 mM Tris pH 7.5 with 0.00025, 0.0005, 0.0015 and 0.005 U of MNase at 30°C for 10 min. The reaction was stopped by addition of 5 mM EDTA. DNA was extracted, precipitated, washed, dried and resolved on a 1% agarose–TBE gel. The gel was stained with ethidium bromide and digitally scanned.
Acknowledgments
Acknowledgements
We thank Dr Paul Englund (Johns Hopkins Medical School, Baltimore) for providing us with the constructs and cell lines to perform the RNAi silencing, Dr Rolf Sternglanz (University of California, San Diego) for HAT plasmid, Dr David Pérez-Morga, Dr Luc Vanhamme and Annette Pays for help and advice, and Dr Ali Ouaissi (IRD, Montpellier) for sharing unpublished data. This work was supported by the Interuniversity Poles of Attraction Programme of the Belgian State Prime Minister’s Office–the Federal Office for Scientific, Technical and Cultural Affairs.
References
- Adamietz P. and Rudolph,A. (1984) ADP-ribosylation of nuclear proteins in vivo. Identification of histone H2B as a major acceptor for mono- and poly(ADP-ribose) in dimethyl sulfate-treated hepatoma AH 7974 cells. J. Biol. Chem., 259, 6841–6846. [PubMed] [Google Scholar]
- Afshar G. and Murnane,J.P. (1999) Characterization of a human gene with sequence homology to Saccharomyces cerevisiae SIR2. Gene, 234, 161–168. [DOI] [PubMed] [Google Scholar]
- Bell S.D., Botting,C.H., Wardleworth,B.N., Jackson,S.P. and White,M.F. (2002) The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science, 296, 148–151. [DOI] [PubMed] [Google Scholar]
- Bohm L., Schneeweiss,F.A., Sharan,R.N. and Feinendegen,L.E. (1997) Influence of histone acetylation on the modification of cytoplasmic and nuclear proteins by ADP-ribosylation in response to free radicals. Biochim. Biophys Acta, 1334, 149–154. [DOI] [PubMed] [Google Scholar]
- Borra M.T., O’Neill,F.J., Jackson,M.D., Marshall,B., Verdin,E., Foltz,K.R. and Denu,J.M. (2002) Conserved enzymatic production and biological effect of O-acetyl-ADP-ribose by silent information regulator 2-like NAD+-dependent deacetylases. J. Biol. Chem., 277, 12632–12641. [DOI] [PubMed] [Google Scholar]
- Borst P. and Ulbert,S. (2001) Control of VSG gene expression sites. Mol. Biochem. Parasitol., 114, 17–27. [DOI] [PubMed] [Google Scholar]
- Boulikas T. (1988) At least 60 ADP-ribosylated variant histones are present in nuclei from dimethylsulfate-treated and untreated cells. EMBO J., 7, 57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulikas T. (1989) DNA strand breaks alter histone ADP-ribosylation. Proc. Natl Acad. Sci. USA, 86, 3499–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann C.B., Sherman,J.M., Devine,S.E., Cameron,E.E., Pillus,L. and Boeke,J.D. (1995) The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression and chromosome stability. Genes Dev., 9, 2888–2902. [DOI] [PubMed] [Google Scholar]
- Braunstein M., Rose,A.B., Holmes,S.G., Allis,C.D. and Broach,J.R. (1993) Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev., 7, 592–604. [DOI] [PubMed] [Google Scholar]
- Braunstein M., Sobel,R.E., Allis,C.D., Turner,B.M. and Broach,J.R. (1996) Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol. Cell. Biol., 16, 4349–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredehorst R., Wielckens,K., Adamietz,P., Steinhagen-Thiessen,E. and Hilz,H. (1981) Mono(ADP-ribosyl)ation and poly(ADP-ribosyl)ation of proteins in developing liver and in hepatomas: relation of conjugate subfractions to metabolic competence and proliferation rates. Eur. J. Biochem., 120, 267–274. [DOI] [PubMed] [Google Scholar]
- Brun R. and Schonenberger. (1979) Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication. Acta Trop., 36, 289–292. [PubMed] [Google Scholar]
- Burri M., Schlimme,W., Betschart,B. and Hecker,H. (1994) Characterization of the histones of Trypanosoma brucei brucei bloodstream forms. Acta Trop., 58, 291–305. [DOI] [PubMed] [Google Scholar]
- Burzio L.O., Riquelme,P.T. and Koide,S.S. (1979) ADP ribosylation of rat liver nucleosomal core histones. J. Biol. Chem., 254, 3029–3037. [PubMed] [Google Scholar]
- Chung H.M., Shea,C., Fields,S., Taub,R.N., Van der Ploeg,L.H. and Tse,D.B. (1990) Architectural organization in the interphase nucleus of the protozoan Trypanosoma brucei: location of telomeres and mini-chromosomes. EMBO J., 9, 2611–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross G.A., Wirtz,L.E. and Navarro,M. (1998) Regulation of vsg expression site transcription and switching in Trypanosoma brucei. Mol. Biochem. Parasitol., 91, 77–91. [DOI] [PubMed] [Google Scholar]
- Ersfeld K. and Gull,K. (1997) Partitioning of large and minichromosomes in Trypanosoma brucei. Science, 276, 611–614. [DOI] [PubMed] [Google Scholar]
- Farzaneh F., Shall,S., Michels,P. and Borst,P. (1985) ADP-ribosyl transferase activity in Trypanosoma brucei. Mol. Biochem. Parasitol., 14, 251–259. [DOI] [PubMed] [Google Scholar]
- Freeman-Cook L.L., Sherman,J.M., Brachmann,C.B., Allshire,R.C., Boeke,J.D. and Pillus,L. (1999) The Schizosaccharomyces pombe hst4(+) gene is a SIR2 homologue with silencing and centromeric functions. Mol. Biol. Cell, 10, 3171–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye R.A. (1999) Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun., 260, 273–279. [DOI] [PubMed] [Google Scholar]
- Frye R.A. (2000) Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun., 273, 793–798. [DOI] [PubMed] [Google Scholar]
- García-Salcedo J.A., Nolan,D.P., Gijón,P., Gómez-Rodríguez,J. and Pays,E. (2002) A protein kinase specifically associated with proliferative forms of Trypanosoma brucei is functionally related to a yeast kinase involved in the co-ordination of cell shape and division. Mol. Microbiol., 45, 307–319. [DOI] [PubMed] [Google Scholar]
- Golderer G. and Grobner,P. (1991) ADP-ribosylation of core histones and their acetylated subspecies. Biochem. J., 277, 607–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C.M. and Almouzni,G. (2002) When repair meets chromatin. First in series on chromatin dynamics. EMBO Rep., 3, 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara R., Mo,J. and Sancar,A. (2000) DNA damage in the nucleosome core is refractory to repair by human excision nuclease. Mol. Cell. Biol., 20, 9173–9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S., Armstrong,C.M., Kaeberlein,M. and Guarente,L. (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature, 403, 795–800. [DOI] [PubMed] [Google Scholar]
- Ismaili N., Perez-Morga,D., Walsh,P., Cadogan,M., Pays,A., Tebabi,P. and Pays,E. (2000) Characterization of a Trypanosoma brucei SR domain-containing protein bearing homology to cis-spliceosomal U1 70 kDa proteins. Mol. Biochem. Parasitol., 106, 109–120. [DOI] [PubMed] [Google Scholar]
- Jenuwein T. and Allis,C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Kleff S. andrulis,E.D. anderson,C.W. and Sternglanz,R. (1995) Identification of a gene encoding a yeast histone H4 acetyltransferase. J. Biol. Chem., 270, 24674–24677. [DOI] [PubMed] [Google Scholar]
- Koch-Nolte F., Petersen,D., Balasubramanian,S., Haag,F., Kahlke,D., Willer,T., Kastelein,R., Bazan,F. and Thiele,H.G. (1996) Mouse T cell membrane proteins Rt6-1 and Rt6-2 are arginine/protein mono(ADPribosyl)transferases and share secondary structure motifs with ADP-ribosylating bacterial toxins. J. Biol. Chem., 271, 7686–7693. [DOI] [PubMed] [Google Scholar]
- Kosmoski J.V., Ackerman,E.J. and Smerdon,M.J. (2001) DNA repair of a single UV photoproduct in a designed nucleosome. Proc. Natl Acad. Sci. USA, 98, 10113–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreimeyer A., Wielckens,K., Adamietz,P. and Hilz,H. (1984) DNA repair-associated ADP-ribosylation in vivo. Modification of histone H1 differs from that of the principal acceptor proteins. J. Biol. Chem., 259, 890–896. [PubMed] [Google Scholar]
- Landry J., Slama,J.T. and Sternglanz,R. (2000a) Role of NAD(+) in the deacetylase activity of the SIR2-like proteins. Biochem. Biophys. Res. Commun., 278, 685–690. [DOI] [PubMed] [Google Scholar]
- Landry J., Sutton,A., Tafrov,S.T., Heller,R.C., Stebbins,J., Pillus,L. and Sternglanz,R. (2000b) The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl Acad. Sci. USA, 97, 5807–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley E., Pearson,M., Faretta,M., Bauer,U.M., Frye,R.A., Minucci,S., Pelicci,P.G. and Kouzarides,T. (2002) Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J., 21, 2383–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.E., Paques,F., Sylvan,J. and Haber,J.E. (1999) Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr. Biol., 9, 767–770. [DOI] [PubMed] [Google Scholar]
- Lin S.J., Defossez,P.A. and Guarente,L. (2000) Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science, 289, 2126–2128. [DOI] [PubMed] [Google Scholar]
- Liu X. and Smerdon,M.J. (2000) Nucleotide excision repair of the 5S ribosomal RNA gene assembled into a nucleosome. J. Biol. Chem., 275, 23729–23735. [DOI] [PubMed] [Google Scholar]
- Luo J., Nikolaev,A.Y., Imai,S., Chen,D., Su,F., Shiloh,A., Guarente,L. and Gu,W. (2001) Negative control of p53 by Sir2α promotes cell survival under stress. Cell, 107, 137–148. [DOI] [PubMed] [Google Scholar]
- Martin S.G., Laroche,T., Suka,N., Grunstein,M. and Gasser,S.M. (1999) Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell, 97, 621–633. [DOI] [PubMed] [Google Scholar]
- Melville S.E., Leech,V., Gerrard,C.S., Tait,A. and Blackwell,J.M. (1998) The molecular karyotype of the megabase chromosomes of Trypanosoma brucei and the assignment of chromosome markers. Mol. Biochem. Parasitol., 94, 155–173. [DOI] [PubMed] [Google Scholar]
- Mills K.D., Sinclair,D.A. and Guarente,L. (1999) MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell, 97, 609–620. [DOI] [PubMed] [Google Scholar]
- Murphy N.B., Pays,A., Tebabi,P., Coquelet,H., Guyaux,M., Steinert,M. and Pays,E. (1987) Trypanosoma brucei repeated element with unusual structural and transcriptional properties. J. Mol. Biol., 195, 855–871. [DOI] [PubMed] [Google Scholar]
- Muth V., Nadaud,S., Grummt,I. and Voit,R. (2001) Acetylation of TAF(I)68, a subunit of TIF-IB/SL1, activates RNA polymerase I transcription. EMBO J., 20, 1353–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo H., Tschudi,C., Gull,K. and Ullu,E. (1998) Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl Acad. Sci. USA, 95, 14687–14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North B.J., Marshall,B.L., Borra,M.T., Denu,J.M. and Verdin,E. (2003) The human Sir2 ortholog, SIRT2, is an NAD(+)-dependent tubulin deacetylase. Mol. Cell, 11, 437–444. [DOI] [PubMed] [Google Scholar]
- Ogata N., Ueda,K. and Hayaishi,O. (1980) ADP-ribosylation of histone H2B. Identification of glutamic acid residue 2 as the modification site. J. Biol. Chem., 255, 7610–7615. [PubMed] [Google Scholar]
- Pays E. and Nolan,D.P. (1998) Expression and function of surface proteins in Trypanosoma brucei. Mol. Biochem. Parasitol., 91, 3–36. [DOI] [PubMed] [Google Scholar]
- Perez-Morga D., Amiguet-Vercher,A., Vermijlen,D. and Pays,E. (2001) Organization of telomeres during the cell and life cycles of Trypanosoma brucei.J. Eukaryot. Microbiol., 48, 221–226. [DOI] [PubMed] [Google Scholar]
- Perrod S., Cockell,M.M., Laroche,T., Renauld,H., Ducrest,A.L., Bonnard,C. and Gasser,S.M. (2001) A cytosolic NAD-dependent deacetylase, Hst2p, can modulate nucleolar and telomeric silencing in yeast. EMBO J., 20, 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauve A.A., Celic,I., Avalos,J., Deng,H., Boeke,J.D. and Schramm,V.L. (2001) Chemistry of gene silencing: the mechanism of NAD+-dependent deacetylation reactions. Biochemistry, 40, 15456–15463. [DOI] [PubMed] [Google Scholar]
- Sherwin T. and Gull,K. (1989) The cell division cycle of Trypanosoma brucei brucei: timing of event markers and cytoskeletal modulations. Philos. Trans. R. Soc. Lond. B Biol. Sci., 323, 573–588. [DOI] [PubMed] [Google Scholar]
- Smerdon M.J. and Conconi,A. (1999) Modulation of DNA damage and DNA repair in chromatin. Prog. Nucleic Acid Res. Mol. Biol., 62, 227–255. [DOI] [PubMed] [Google Scholar]
- Smith J.S. et al. (2000) A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl Acad. Sci. USA, 97, 6658–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solari A.J. (1980) Function of the dense plaques during mitosis in Trypanosoma cruzi. Exp. Cell Res., 127, 457–460. [DOI] [PubMed] [Google Scholar]
- Solari A.J. (1983) The ultrastructure of mitotic nuclei of Blastocrithidia triatomae. Z. Parasitenkd., 69, 3–15. [DOI] [PubMed] [Google Scholar]
- Sommer J.M., Cheng,Q.L., Keller,G.A. and Wang,C.C. (1992) In vivo import of firefly luciferase into the glycosomes of Trypanosoma brucei and mutational analysis of the C-terminal targeting signal. Mol. Biol. Cell, 3, 749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starai V.J., Celic,I., Cole,R.N., Boeke,J.D. and Escalante-Semerena,J.C. (2002) Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science, 298, 2390–2392. [DOI] [PubMed] [Google Scholar]
- Sugasawa K., Masutani,C. and Hanaoka,F. (1993) Cell-free repair of UV-damaged simian virus 40 chromosomes in human cell extracts. I. Development of a cell-free system detecting excision repair of UV-irradiated SV40 chromosomes. J. Biol. Chem., 268, 9098–9104. [PubMed] [Google Scholar]
- Takada T., Iida,K. and Moss,J. (1993) Cloning and site-directed mutagenesis of human ADP-ribosylarginine hydrolase. J. Biol. Chem., 268, 17837–17843. [PubMed] [Google Scholar]
- Tanner K.G., Landry,J., Sternglanz,R. and Denu,J.M. (2000) Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl Acad. Sci. USA, 97, 14178–14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny J.C. and Moazed,D. (2001) Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: evidence for acetyl transfer from substrate to an NAD breakdown product. Proc. Natl Acad. Sci. USA, 98, 415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny J.C., Dowd,G.J., Huang,J., Hilz,H. and Moazed,D. (1999) An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell, 99, 735–745. [DOI] [PubMed] [Google Scholar]
- Tikoo K. and Ali,Z. (1997) Structure of active chromatin: covalent modifications of histones in active and inactive genes of control and hypothyroid rat liver. Biochem. J., 322, 281–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum H.A. and Guarente,L. (2001) Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature, 410, 227–230. [DOI] [PubMed] [Google Scholar]
- Tsang A.W. and Escalante-Semerena,J.C. (1998) CobB, a new member of the SIR2 family of eucaryotic regulatory proteins, is required to compensate for the lack of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase activity in cobT mutants during cobalamin biosynthesis in Salmonella typhimurium LT2. J. Biol. Chem., 273, 31788–31794. [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y., Kato,J. and Ikeda,H. (1997) Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature, 388, 900–903. [DOI] [PubMed] [Google Scholar]
- Vaziri H., Dessain,S.K., Ng Eaton,E., Imai,S.I., Frye,R.A., Pandita,T.K., Guarente,L. and Weinberg,R.A. (2001) hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell, 107, 149–159. [DOI] [PubMed] [Google Scholar]
- Vickerman K. (1985) Developmental cycles and biology of pathogenic trypanosomes. Br. Med. Bull., 41, 105–114. [DOI] [PubMed] [Google Scholar]
- Wang Z., Morris,J.C., Drew,M.E. and Englund,P.T. (2000) Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem., 275, 40174–40179. [DOI] [PubMed] [Google Scholar]
- Wang Z.G., Wu,X.H. and Friedberg,E.C. (1991) Nucleotide excision repair of DNA by human cell extracts is suppressed in reconstituted nucleosomes. J. Biol. Chem., 266, 22472–22478. [PubMed] [Google Scholar]
- Weiden M., Osheim,Y.N., Beyer,A.L. and Van der Ploeg,L.H. (1991) Chromosome structure: DNA nucleotide sequence elements of a subset of the minichromosomes of the protozoan Trypanosoma brucei. Mol. Cell. Biol., 11, 3823–3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielckens K., Bredehorst,R., Adamietz,P. and Hilz,H. (1982) Mono ADP-ribosylation and poly ADP-ribosylation of proteins in normal and malignant tissues. Adv. Enzyme Regul., 20, 23–37. [DOI] [PubMed] [Google Scholar]
- Wirtz E., Leal,S., Ochatt,C. and Cross,G.A. (1999) A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol., 99, 89–101. [DOI] [PubMed] [Google Scholar]
- Xie J., Pierce,M., Gailus-Durner,V., Wagner,M., Winter,E. and Vershon,A.K. (1999) Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J., 18, 6448–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahiaoui B., Taibi,A. and Ouaissi,A. (1996) A Leishmania major protein with extensive homology to silent information regulator 2 of Saccharomyces cerevisiae. Gene, 169, 115–118. [DOI] [PubMed] [Google Scholar]
- Zemzoumi K., Sereno,D., Francois,C., Guilvard,E., Lemesre,J.L. and Ouaissi,A. (1998) Leishmania major: cell type dependent distribution of a 43 kDa antigen related to silent information regulatory-2 protein family. Biol. Cell, 90, 239–245. [DOI] [PubMed] [Google Scholar]