Abstract
Mating in Ustilago maydis requires cross-talk between cAMP and mitogen-activated protein kinase (MAPK) signalling. During this process, pheromone response factor 1 (Prf1) activates transcription of a and b mating type genes by binding to pheromone response elements (PREs) located in regulatory regions of these genes. Here, we show that PREs are also necessary and sufficient to mediate cAMP-induced gene expression. Prf1 interacts with cAMP-dependent protein kinase A (PKA) Adr1 as well as MAPK Kpp2 in vivo, and its central phosphorylation sites that are functionally important are modified by the respective kinases in vitro. PKA sites in Prf1 are essential for induced expression of a and b mating type genes. In contrast, MAPK sites are not required for pheromone-induced expression of a genes but are crucial for pheromone-responsive b gene expression. This illustrates how a single transcription factor can integrate signals from two pathways and how its phosphorylation status can determine different transcriptional responses.
Keywords: cAMP/cross-talk/MAPK/plant pathogen/signalling network
Introduction
Signalling pathways in eukaryotes are not insulated routes but are extensively interwoven to form complex networks that function in a spatiotemporal manner (Hunter, 2000). One of the major challenges in understanding such sophisticated regulatory networks is to identify molecular nodes and mechanisms of interconnection. An intensively studied example is cross-talk between cAMP-dependent kinase and mitogen-activated protein kinase (MAPK) in fungal pathogens (Madhani and Fink, 1998; Lengeler et al., 2000; Lee et al., 2003). Cross-talk is crucial for infection by plant pathogens such as Magnaporthe grisea and Ustilago maydis (Hamer and Talbot, 1998; Kahmann et al., 1999) as well as for morphological changes observed in human pathogens such as Candida albicans and Cryptococcus neoformans (Whiteway, 2000; Sánchez-Martínez and Pérez-Martín, 2001; Hull and Heitman, 2002). The phenomenon of pseudohyphal growth in Saccharomyces cerevisiae serves as a paradigm for these differentiation processes because it shares fundamental molecular principles (D’Souza and Heitman, 2001).
We study the basidiomycete U.maydis that causes smut disease on corn (Banuett, 1992, 1995). A prerequisite for infection is recognition and fusion of two haploid cells and formation of a filamentously growing dikaryon (Kahmann et al., 2000; Bölker, 2001). This process is regulated by a tetrapolar mating system consisting of the a and b mating type loci. The biallelic a locus (a1 and a2) encodes an intercellular recognition system consisting of precursors (mfa1 and mfa2) and receptors (pra1 and pra2) of lipopeptide pheromones (Bölker et al., 1992; Spellig et al., 1994). Pheromone response elicits transcriptional activation of mating type genes as well as formation of conjugation hyphae (Spellig et al., 1994; Snetselaar et al., 1996; Urban et al., 1996). The multiallelic b locus encodes a pair of homeodomain proteins (e.g. bW1 and bE1 in the case of b1) that establish filamentous growth only as heterodimeric transcription factors with subunits derived from different alleles (Gillissen et al., 1992; Kämper et al., 1995). Thus, mating compatibility is regulated by gene products of a and b loci on the pre- and post-fusion level, respectively.
Pheromone-induced gene expression is mediated by pheromone response elements (PREs) present in regulatory regions of a and b mating type genes (Urban et al., 1996). PREs are recognized by transcription factor Prf1, whose activity is regulated transcriptionally as well as post-transcriptionally (Hartmann et al., 1996, 1999). On the transcriptional level, upstream activating sequences in the prf1 promoter determine expression by nutrient signalling, and two PREs are probably involved in autoregulation (Hartmann et al., 1996). Post-transcriptional regulation might occur through phosphorylation since Prf1 contains six putative MAPK phosphorylation sites that fit to the consensus sequence L/PXS/TP (Clark et al., 1995) as well as a presumed MAPK docking site of the FXFP type (Jacobs et al., 1999). Strains expressing a prf1 allele, carrying mutations in these six putative MAPK sites and in the presumed MAPK docking site, were reduced in mating competence (Müller et al., 1999). Recently, it has been shown that a MAPK cascade consisting of MAPKKK Kpp4/Ubc4 (Andrews et al., 2000; Müller et al., 2003), MAPKK Fuz7 (Banuett and Herskowitz, 1994) and MAPK Kpp2/Ubc3 (Mayorga and Gold, 1999; Müller et al., 1999) acts upstream of Prf1 (Müller et al., 2003; Figure 1A).
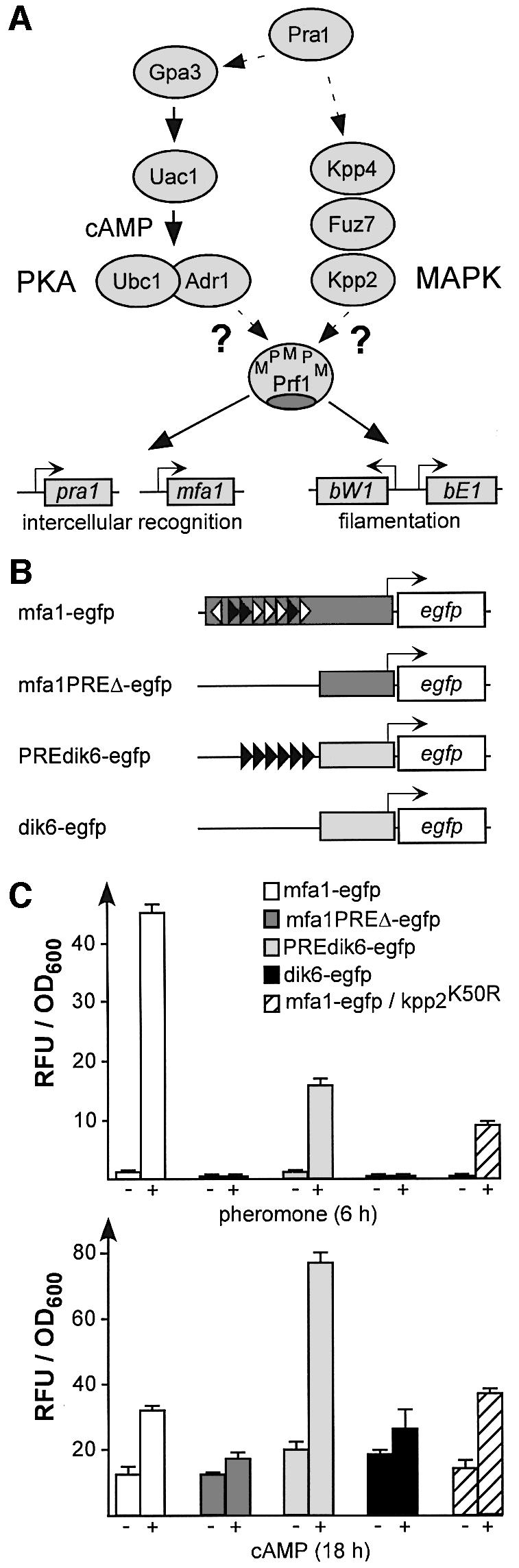
Fig. 1. The pheromone and cAMP response share the same promoter element. (A) Key players in PKA and MAPK signalling are shown schematically. Proteins are drawn as ovals and Prf1 target genes as rectangles with bent arrows. P and M symbolize putative PKA and MAPK sites in Prf1, respectively. (B) Reporter constructs. In mfa1-egfp, 908 bp of the mfa1 promoter (dark grey rectangle), containing eight PREs in the wild type context (filled or open triangles indicate a perfect match or one mismatch to the nonamer PRE ACAAAGGGA, respectively; Urban et al., 1996), were transcriptionally fused to the egfp gene. mfa1PREΔ-egfp contained a 292 bp mfa1 fragment without PREs. PREdik6-egfp harbours six synthetic PREs upstream of a 289 bp basal dik6 promoter (light grey rectangle; Bohlmann, 1996), and dik6-egfp without PREs served as control. (C) Results of fluorimetric measurements using strains harbouring constructs described in (B). Relative fluorescence units (RFU) were measured and normalized to optical density (OD600, see Materials and methods). Results of triplicate experiments are shown.
In addition to MAPK signalling, a conserved cAMP signalling pathway is necessary for pheromone response. It consists of heterotrimeric G protein α subunit Gpa3, adenylate cyclase Uac1, and cAMP-dependent protein kinase A (PKA) composed of regulatory and catalytic subunits, termed Ubc1 and Adr1, respectively (Gold et al., 1994, 1997; Regenfelder et al., 1997; Dürrenberger et al., 1998; Figure 1A). Under conditions that reflect high intracellular cAMP levels, a drastic increase in mfa1 expression is observed (Krüger et al., 1998). Thus, PKA and MAPK signalling converge on mfa1 expression.
Here, we provide evidence that phosphorylation of Prf1 is used to integrate PKA and MAPK signalling during mating. We demonstrate that PKA phosphorylation sites are essential for induced expression of a and b mating type genes, while MAPK phosphorylation sites are used to differentiate between a and b gene expression.
Results
Pheromone response elements are necessary and sufficient for cAMP-induced mfa1 expression
Activated cAMP signalling leads to elevated mfa1 expression (Krüger et al., 1998; Hartmann et al., 1999). To address whether cAMP-induced mfa1 expression relies on PREs (Urban et al., 1996), we generated a set of reporter constructs (Figure 1B). In order to allow direct comparison of egfp expression, all constructs were targeted to the ip locus (encoding the iron–sulfur protein subunit of succinate dehydrogenase) of wild-type strain FB1 (a1b1) by homologous recombination (Loubradou et al., 2001). Dose–response curves using strain FB1mfa1-egfp revealed that 2.5 µg/ml synthetic a2 pheromone resulted in maximal reporter gene expression. For cAMP, the optimal concentration was 6 mM. Under these conditions, egfp expression was elevated without an increase in the amount of cells with cytokinesis defects (Supplementary figure 1A and B, available at The EMBO Journal Online). When strains containing either construct mfa1-egfp or PREdik6-egfp were stimulated for 6 h with synthetic a2 pheromone, fluorescence intensity increased 44- and 16-fold, respectively (Figure 1C, top). Increased fluorescence could not be detected in the case of strains harbouring control constructs mfa1PREΔ-egfp or dik6-egfp (Figure 1C, top). When the same strains were incubated for 18 h with 6 mM cAMP, strains harbouring PRE-containing promoter constructs showed cAMP-induced egfp expression (2.5- and 3.6-fold for mfa1-egfp and PREdik6-egfp, respectively). In FB1mfa1PREΔ-egfp or FB1dik6-egfp, reporter gene expression in both cases was only increased 1.5-fold (Figure 1C, bottom).
To exclude that cAMP positively regulates expression of the mfa1 promoter through cross-talk with MAPK signalling, we also investigated the cAMP response in a strain harbouring mfa1-egfp in combination with a kinase-dead allele kpp2K50R (Müller et al., 2003). In this strain, the cAMP-induced expression was comparable with FB1mfa1-egfp carrying the wild-type allele of kpp2 (Figure 1C, bottom). Interestingly, the same strain still exhibited 21-fold induced egfp expression upon pheromone stimulation (Figure 1C, top), indicating that pheromone-responsive mfa1 expression takes place in the absence of MAPK signalling.
Putative phosphorylation sites in the central part of Prf1 are functionally important
Prf1 contains six putative MAPK phosphorylation sites that are required for mating (Müller et al., 1999) and five putative phosphorylation sites fitting the PKA consensus sequence R/KR/KXS/T (Taylor et al., 1990). To elucidate which of these sites are functionally important during mating, we constructed epitope-tagged prf1 alleles (N-terminal his6 as well as C-terminal myc3 tags) carrying different combinations of alanine mutations in serine and threonine residues that block protein phosphorylation by the respective kinase (Figure 2A). Modified alleles were introduced into the prf1 locus of strains FB1 (a1b1) and FB2 (a2b2) by replacing the wild-type allele (see Materials and methods). Successful formation of dikaryotic hyphae was scored 24 h after mixing compatible strains by the appearance of white, fuzzy colonies (Figure 2B). To assay pheromone production, we used pheromone tester strain CL13 (a1bW2bE1; Bölker et al., 1995). As published, strains expressing prf1eM1-6d (the allele encoding epitope-tagged Prf1 with alanine mutations in MAPK site 1–6 as well as four alanine mutations in the putative docking site) were drastically reduced in mating (Figure 2A). In contrast to FB2prf1Δ, FB2prf1eM1-6d still produced pheromone that stimulated filamentous growth of CL13 (Müller et al., 1999), indicating that point mutations had not affected overall protein conformation. To address whether the putative MAPK docking site was of functional importance, we compared alleles prf1eM1-6 and prf1eMd. Strains expressing prf1eMd were not impaired in mating, indicating that the docking site was dispensable for Prf1 function (Figure 2B). Conversely, strains carrying prf1eM1-6 showed the same reduced filamentous growth as prf1eM1-6d-expressing strains (Figure 2B). To delineate further, which of the six putative phosphorylation sites were needed for function, we generated alleles prf1eM126d, prf1eM345, prf1eM34, prf1eM3, prf1eM4 and prf1eM5 in which single or multiple sites were mutated (Figure 2A and B). Only strains expressing prf1eM345 were affected in mating. Thus, these three central MAPK phosphorylation sites are crucial for full activity of Prf1 during mating (Figure 2B).
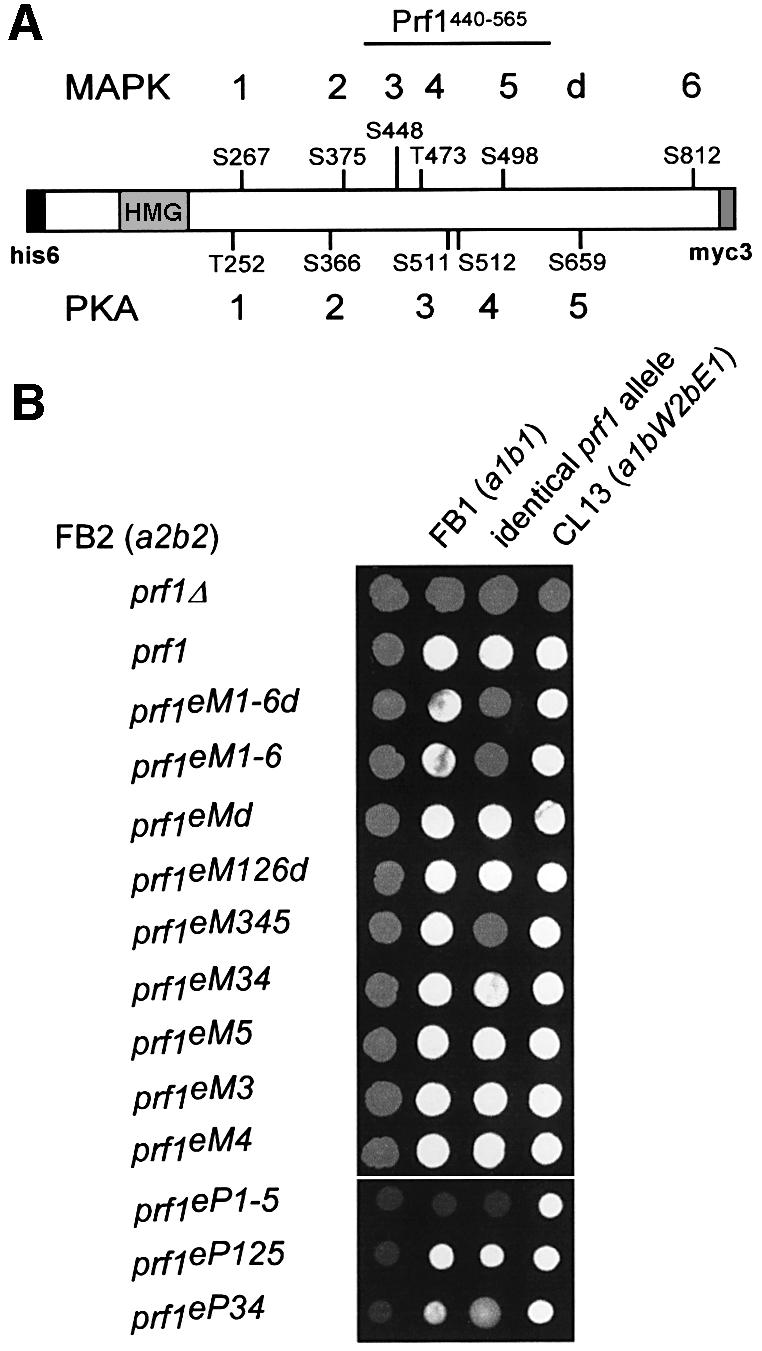
Fig. 2. Mutations in putative phosphorylation sites of MAPK and PKA affect Prf1 activity during mating and tumour formation. (A) Prf1 is depicted schematically. Serine (S) and threonine (T) residues that have been mutated to alanine are shown, with amino acid position and consecutive numbering on the top and bottom for putative MAPK and PKA sites, respectively. A putative MAPK docking site of the FXFP type is marked with a d (586TPNFAFDP592 mutated to 586APNAAAAP592; Müller et al., 1999). The HMG box DNA-binding domain, N-terminal his6 and myc3 epitopes are represented by labelled rectangles. The central 126 amino acids of Prf1 that were expressed as GST fusion protein in E.coli are labelled Prf1440–565. (B) Mating assays on plates containing activated charcoal. White, fuzzy colonies reflect the formation of b-dependent aerial hyphae. Respective FB2 (a2b2) derivatives labelled on the left were either inoculated alone, with FB1 (a1b1), with FB1 derivatives carrying identical prf1 alleles or with pheromone tester strain CL13 (a1bW2bE1) given on the top.
In comparable experiments, we tested the role of putative PKA phosphorylation sites for Prf1 activity. Strains carrying an epitope-tagged prf1 allele with point mutations in putative PKA sites 1–5 (prf1eP1-5; Figure 2A) were severely affected in mating (Figure 2B). However, FB2prf1eP1-5 still elicited filamentous growth of CL13, indicating that basal activity of Prf1 was still present. In crosses with wild-type, strains expressing prf1eP1-5 exhibited attenuated filament formation, suggesting a defect in cell fusion (Figure 2B). Strains expressing prf1eP125 fused efficiently, whereas prf1eP34-harbouring strains were strongly affected in mating. Hence, PKA sites 3 and 4 are important for Prf1 function during mating (Figure 2B).
To eliminate possible complications in this assay that might arise from autoregulation of prf1 (Hartmann et al., 1999), we also performed mating assays with strains expressing prf1 alleles constitutively. As expected, mutations in putative PKA and MAPK sites of prf1 caused reduced mating competence (see Supplementary figure 2). In more sensitive plant infection experiments, we were able to verify these results, showing that post-transcriptional activation of Prf1 is also necessary during tumour formation (see Supplementary table 1).
Prf1 interacts with Adr1 and Kpp2 in vivo and its central part is phosphorylated by both kinases in vitro
In order to demonstrate that Prf1 is a target for Adr1 as well as Kpp2, we performed a yeast two-hybrid analysis. EGY48-derived strains expressing Prf1eNLS [an epitope-tagged version of Prf1 C-terminally fused to the SV40 large T antigen nuclear localization signal (NLS)] in combination with LexA-Adr1 or LexA-Kpp2 (fusion proteins of Adr1 or Kpp2 with the DNA-binding domain LexA at their N-termini) were able to grow on plates in the absence of leucine (Figure 3A). This indicated that the interaction of Prf1 with Adr1 as well as Kpp2 triggered expression of the LEU2 reporter. This was specific, because strains expressing Prf1eNLS in combination with LexA or strains expressing the activation domain B42AD (Gyuris et al., 1993) in combination with LexA, LexA-Adr1 or LexA-Kpp2 were unable to grow in the absence of leucine (Figure 3A).
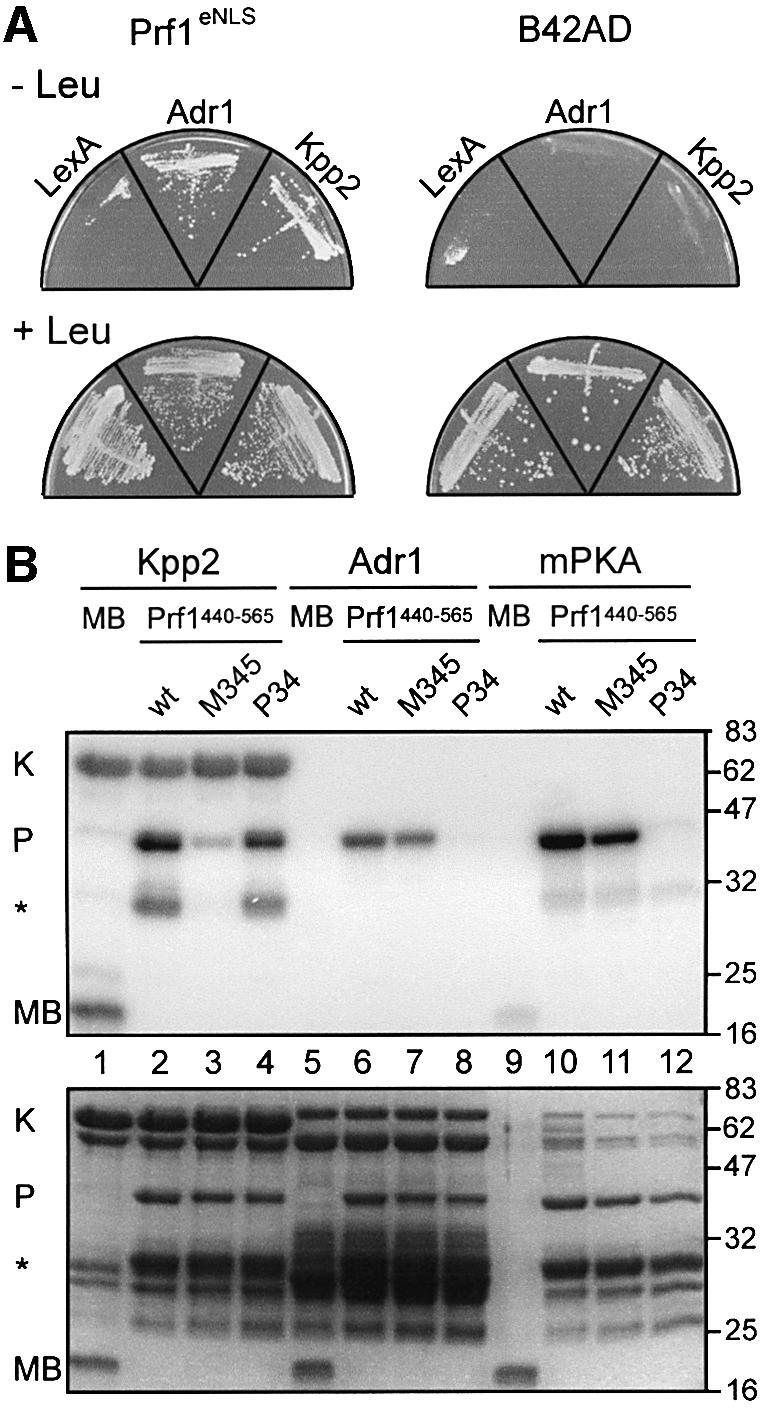
Fig. 3. Prf1 interacts with Kpp2 and Adr1 in vivo and is phosphorylated by the respective kinases at putative phosphorylation sites in vitro. (A) As shown by two-hybrid analysis in yeast strain EGY48, the interaction of Prf1eNLS with LexA-Adr1 and LexA-Kpp2 in vivo triggers expression of the LEU2 reporter. In contrast to plates on the bottom, leucine was omitted in the upper plates. Strains on the left expressed epitope-tagged Prf1 with an additional heterologous N-terminal NLS (Prf1eNLS, see Materials and methods), and strains on the right expressed only the acidic activation domain B42AD (Gyuris et al., 1993). Above each sector, the encoded DNA-binding proteins are indicated. In the case of Adr1 and Kpp2, full-length proteins were C-terminally fused to the LexA DNA-binding domain. (B) Top: SDS–PAGE analysis of proteins phosphorylated in vitro using radioactively labelled γ-ATP. Above each lane, the protein composition of various kinase reactions is indicated. In the case of Kpp2 and Adr1, full-length proteins were C-terminally fused to GST (67 and 73 kDa, respectively). Prf1440–565 (41 kDa; position 440–565) indicates the presence of C-terminal fusion protein GST–Prf1440–565. Either wild-type (wt) or a comparable GST fusion with mutations in the putative MAPK sites 3, 4 and 5 (M345), or PKA sites 3 and 4 (P34) were used. mPKA indicates the use of murine PKA. As unspecific substrate, bovine myelin basic protein, was tested (MB; 18 kDa; Errede et al., 1993). K and P indicate bands of 67 kDa due to MAPK autophosphorylation (Seger et al., 1991) and of 41 kDa corresponding to Prf1440–565, respectively. The asterisk marks a 30 kDa version of Prf1440–565 that, according to its size and co-purification, is presumably truncated at its C-terminus around position 471, still containing MAPK site 3. On the right, molecular weight markers are indicated in kDa. Bottom: the same gel after rehydration and staining with Coomassie brillant blue.
To demonstrate that the putative phosphorylation sites in Prf1 that are important for function in vivo can be modified by Kpp2 and Adr1, we performed in vitro phosphorylation experiments using the respective kinases and the central part of Prf1 from position 440 to 565 as substrate. We affinity purified three Prf1 peptides (41 kDa) expressed as GST fusion proteins in Escherichia coli: Prf1440–565 (wild-type sequence), Prf1440–565M345 (mutated in putative MAPK sites 3, 4 and 5; see Figure 2) and Prf1440–565P34 (mutated in putative PKA sites 3 and 4). In kinase assays with Kpp2 (∼3 µg) and Adr1 (∼1 µg) expressed as GST fusions in E.coli, or with murine PKA, we observed that Prf1440–565 (∼1 µg) served as substrate (Figure 3, lanes 2, 6 and 10). This was specific for the Prf1 part of the fusion protein since GST alone was not phosphorylated (data not shown). In contrast to Prf1440–565 or Prf1440–565P34, phosphorylation of Prf1440–565M345 by Kpp2 was strongly reduced (Figure 3, lanes 2–4). Adr1 was able to phosphorylate wild-type or Prf1440–565M345 efficiently, while phosphorylation of Prf1440–565P34 was barely detectable (Figure 3, lanes 6–8). A comparable pattern of phosphorylation was detected when murine PKA was used as heterologous kinase (Figure 3, lanes 10–12). Thus, Kpp2 as well as Adr1 modified their predicted phosphorylation sites in the central part of Prf1.
MAP kinase sites of Prf1 are necessary for b but not a gene expression
To address how the phosphorylation status of Prf1 determines its function as transcriptional activator, we determined expression of mating type genes in northern analysis with FB1 strains expressing alleles prf1ce, prf1ceM16d or prf1ceP1-5 under control of the constitutively active tef1 promoter (Spellig et al., 1996). Incubation of FB1prf1ce with 6 mM cAMP resulted in elevated mfa1 expression (Figure 4A, left). The induction of mfa1 also occurred in FB1prf1ceM1-6d but was abolished in FB1prf1ceP1-5. Thus, phosphorylation by Adr1 appears necessary to increase mfa1 gene expression. Interestingly, cAMP treatment of strain FB1prf1ce did not result in elevated bW1 and bE1 expression, indicating that under these experimental conditions, Prf1 activation by Adr1 alone was not sufficient to increase b gene expression (Figure 4A, left).
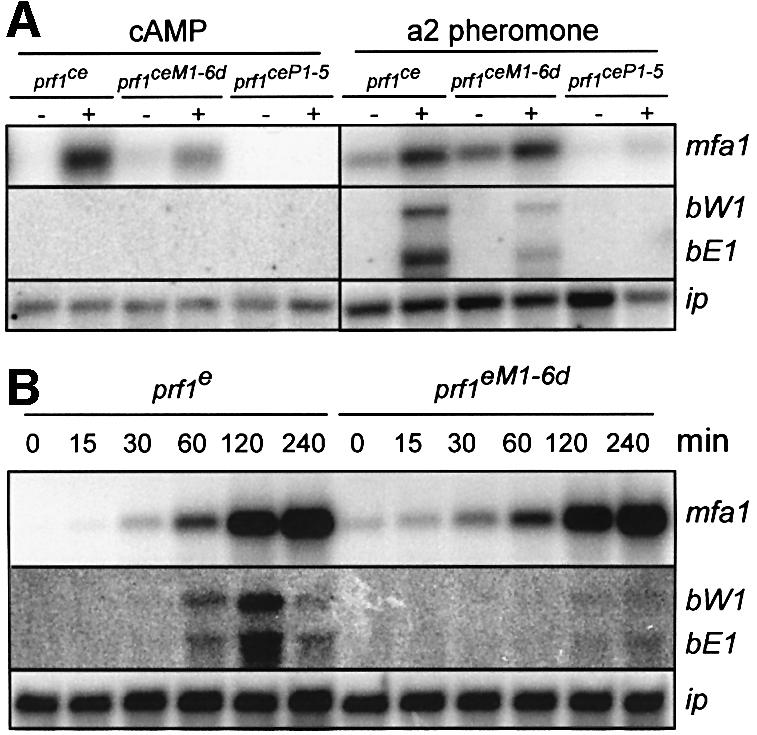
Fig. 4. MAPK phosphorylation sites of Prf1 are important for pheromone-induced b gene expression. In (A), strains indicated at the top were incubated for 18 or 6 h in either the absence or presence of 6 mM cAMP (left) or synthetic a2 pheromone (2.5 µg/ml; right), respectively. Upon harvest, total RNA was extracted and mRNA enriched with Oligotex™ suspension (Qiagen). Approximately 0.5 µg of enriched RNA was loaded per lane and the same filter was hybridized in succession with the probes indicated on the right. Expression of the iron–sulfur protein ip mRNA was used as control for RNA quality and quantity (Keon et al., 1991). In (B), strains indicated at the top were incubated for a time course of 6 h in the presence of synthetic a2 pheromone (2.5 µg/ml). A 15 µg aliquot of total RNA was loaded and the same RNA was analysed with the probes indicated on the right.
Upon treatment with synthetic a2 pheromone (2.5 µg/ml), FB1prf1ce strains exhibited pheromone-induced gene expression of mating type genes mfa1, bW1 and bE1 (Figure 4A, right). However, in pheromone-stimulated FB1prf1ceP1-5 strains, neither mfa1 nor b genes were upregulated. This indicates that phosphorylation of PKA sites in Prf1 was necessary to observe pheromone-responsive expression. FB1prf1ceM1-6d responded with increased mfa1 expression upon pheromone stimulation, while induced expression of bW1 and bE1 genes was attenuated (Figure 4A, right). Hence, MAPK signalling is important for pheromone-induced b gene expression, but not for mfa1 expression.
To investigate the pheromone response in more detail, we compared the pheromone-induced gene expression in FB1prf1e and FB1prf1eM1-6d at different time points after pheromone addition. Pheromone-induced mfa1 expression was comparable in both strains over the course of the experiment. However, pheromone-induced expression of b genes was severely reduced in FB1prf1eM1-6d, while it was unaffected in FB1prf1e (Figure 4B). In summary, PKA sites in Prf1 are needed for mfa1 as well as b gene expression, whereas MAPK sites are important for b gene expression only.
MAP kinase signalling regulates prf1 on the transcriptional level
To verify our observation that MAPK phosphorylation sites are specifically needed for pheromone-induced b gene expression, we used a previously characterized constitutively active allele of the MAPK kinase Fuz7 (termed fuz7DD), whose expression is controlled by the arabinose-regulated crg1 promoter (Müller et al., 2003). crg1-fuz7DD was targeted to the ip locus of strains expressing alleles prf1e, prf1eM16d or prf1eP1-5. Upon 4 h shift to arabinose, fuz7DD strains harbouring prf1e showed increased mRNA accumulation of mfa1, prf1, bW1 and bE1 (Figure 5A). In the respective prf1eP1-5 background, fuz7DD-mediated expression of mfa1, bW1 and bE1 was almost abolished. In prf1eM1-6d-expressing strains, fuz7DD-induced expression of mfa1 still occurred, while induction of bW1 and bE1 was no longer detectable (Figure 5A). This confirmed that MAPK sites are important for b gene expression during MAPK signalling. In addition, mfa1 levels in prf1eM1-6d-expressing strains under non-induced conditions were elevated in comparison with control strains (Figure 5A).
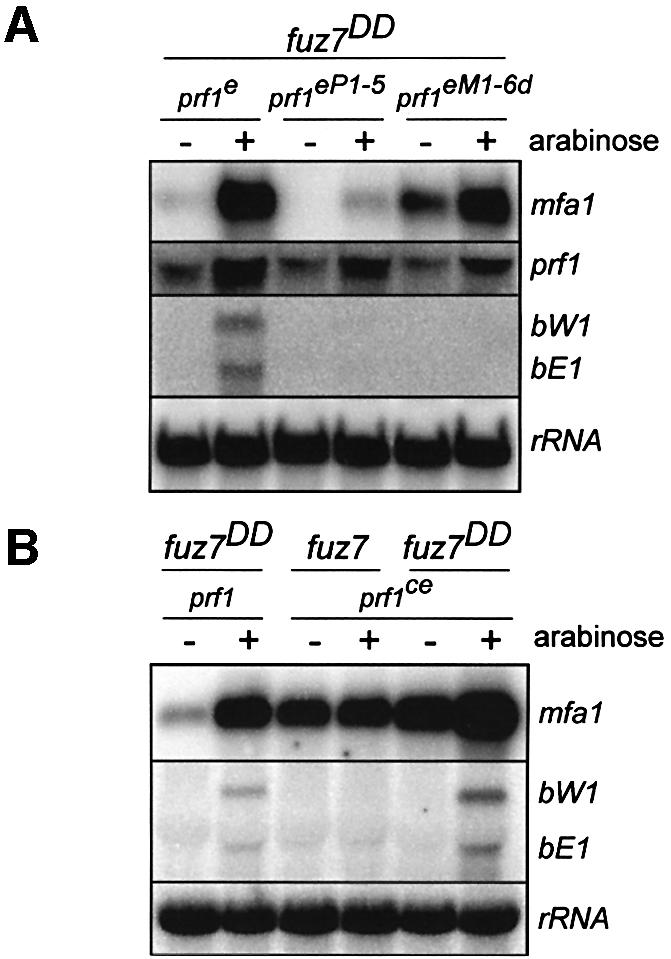
Fig. 5. A constitutively active MAPK kinase induces prf1 expression on the transcriptional level. In (A) and (B), strains indicated at the top were incubated in either the absence or presence of arabinose for 4 h. The activated crg1 promoter drives expression of fuz7DD encoding a constitutively active MAPK kinase (Müller et al., 2003). A 10 µg aliquot of total RNA was loaded and the same filter was hybridized in succession with the probes indicated on the right. Since the construct crg1-fuz7DD was integrated in single copy at the ip locus by homologous recombination (Müller et al., 2003), rRNA was probed as control for RNA quality and quantity.
The finding that prf1 and mfa1 expression was fuz7DD-induced in prf1eP1-5-and in prf1eM1-6d-expressing strains, respectively, can most easily be explained by assuming transcriptional activation of prf1 by MAPK signalling. To test this, we investigated fuz7DD-mediated expression of mating type genes in a strain expressing prf1 constitutively (Figure 5B). In control strain FB1crg1-fuz7DD, mfa1 and b genes were strongly expressed after induction of fuz7DD. In FB1prf1ce, mfa1 levels were elevated, but neither mfa1 nor b gene expression was inducible by arabinose (Figure 5B). In prf1ce-expressing strains carrying crg1-fuz7DD, expression of bW1 and bE1, on the other hand, was strongly induced, but fold induction of mfa1 expression was diminished (13-fold in FB1crg1-fuz7DD compared with 3-fold in FB1crg1-fuz7DDprf1ce according to quantification of phosphorimager data). Hence, induction of mfa1 gene expression by MAPK signalling is mediated mainly by transcriptional activation of prf1.
MAP kinase sites in Prf1 are important for pheromone-induced expression of the b heterodimer
We have shown above that mutations in MAPK sites of Prf1 affect pheromone-induced b but not a gene expression. In order to assess the biological relevance of this finding, we assayed pheromone-induced increases of b activity in strain CL13 (a1bW2bE1). CL13 carries a hybrid b locus with encoded bW2 and bE1 genes controlled by wild-type promoters. Pheromone-induced expression of the b heterodimer can be scored as filament formation independently of cell fusion. To monitor mfa1 expression, we targeted construct mfa1-egfp (Figure 1B) to the ip locus of CL13. In this strain CL13mfa1-egfp, we replaced prf1 with prf1e, prf1eM1-6d or prf1eP1-5. The resulting strains were grown in the presence of synthetic a2 pheromone or FB2 (a2b2) on plates containing activated charcoal (Figure 6A). As expected, CL13mfa1-egfp/prf1e formed white, fuzzy colonies, indicative of an active b heterodimer. The deletion of prf1 abolished this reaction. In CL13mfa1-egfp/prf1eM1-6d or CL13mfa1-egfp/prf1eP1-5, the formation of fuzzy colonies was drastically reduced (Figure 6A), demonstrating that MAPK as well as PKA sites were essential for b gene expression, respectively. The same set of strains was tested for green fluorescence after pheromone addition (Figure 6B). Fluorescence intensity increased ∼3-fold in prf1e- and prf1eM1-6d-expressing strains, whereas no induction was observed in strains harbouring prf1Δ and prf1eP1-5 alleles. Thus, MAPK phosphorylation sites were required for full pheromone-induced expression of the b heterodimer but not for pheromone-induced mfa1 expression.

Fig. 6. MAPK sites in Prf1 are important for pheromone-induced activity of the b heterodimer but not for mfa1 expression. In (A), colonies of CL13-derived strains (a1bW2bE1) harbouring construct mfa1-egfp are documented that were incubated on plates containing activated charcoal in the absence or presence of a2 pheromone (2.5 µg/ml) or FB2 (a2b2). Formation of white, fuzzy colonies is indicative of an active b heterodimer. On the left, the respective prf1 alleles tested are indicated. In (B), the strains indicated above were either untreated (white bars) or incubated for 6 h with synthetic a2 pheromone (2.5 µg/ml; grey bars). Relative fluorescence units (RFU) were measured and normalized to the optical density (OD600, see Materials and methods). Results of triplicate experiments are shown.
Discussion
In this study, we demonstrate that Prf1 integrates PKA and MAPK signalling during mating. The central domain of Prf1 is phosphorylated by both kinases at distinct sites and, depending on its phosphorylation status, Prf1 is able to activate either a or b mating type genes.
PKA and MAPK signalling converge at Prf1
Deletion of PREs in the natural context of the mfa1 promoter resulted in loss of pheromone- as well as cAMP-induced reporter gene expression, while grafting six PREs on the heterologous dik6 basal promoter enabled pheromone- and cAMP-induced expression. cAMP induction is independent of MAPK signalling since it occurs in strain expressing an inactive MAPK. Therefore, PREs that have been identified initially as pheromone response elements (Urban et al., 1996) also function as cAMP response elements. Strains expressing prf1 alleles mutated in either putative PKA or MAPK phosphorylation sites were impaired in mating, indicating that post-translational modification by phosphorylation through both kinases is important for Prf1 function. Mapping the crucial PKA as well as MAPK sites revealed that both are located in the central part of Prf1 and are phosphorylated by the respective kinases in vitro. In addition, PKA Adr1 and MAPK Kpp2 both interact with full-length Prf1 in yeast two-hybrid assays. Thus, in U.maydis, PKA and MAPK phosphorylation are of functional importance for the key transcription factor regulating mating. MAPK phosphorylation had been postulated for Ste12p in S.cerevisiae and Ste11 in Schizosaccharomyces pombe (Kurjan, 1992; Herskowitz, 1995; Davey, 1998; Elion, 2000), although according to current views MAPKs Fus3p and Kss1p regulate Ste12p activity indirectly by phosphorylation of two inhibitors, Dig1p and Dig2p (Pi et al., 1997; Tedford et al., 1997; Bardwell et al., 1998; Olson et al., 2000).
The phosphorylation status of Prf1 is used to differentiate between a and b gene expression
Pheromone stimulation induces a and b gene expression (Urban et al., 1996). In this study, we demonstrate that induction of a genes requires PKA sites in Prf1 while induction of b genes is dependent on the integrity of both PKA and MAPK sites. This differential response was unexpected and led us to investigate the contributions of each signalling pathway separately.
Elevated cAMP levels induce expression of pheromone genes, as has been observed before (Krüger et al., 1998; Hartmann et al., 1999), but b gene expression did not increase. cAMP-induced expression of the pheromone gene mfa1 was dependent on the integrity of PKA sites in Prf1, suggesting that cAMP-activated PKA phosphorylates Prf1, resulting in transcriptional activation of the a genes (Figure 7A). Surprisingly, pheromone-induced mfa1 expression was also dependent on intact PKA sites, while requiring neither MAPK sites in Prf1 nor an active MAPK Kpp2. This suggests that the pheromone signal activates cAMP signalling by an unknown mechanism which induces a gene expression independently of MAPK signalling (Figure 7A).

Fig. 7. The activity of Prf1 is regulated by the concerted action of PKA and MAPK signalling. A model proposing how nutritional or mate sensing is transduced to differential expression of pre- and post-fusion genes. Signalling events during PKA signalling (A) are compared with those during MAPK signalling (B). The prf1-labelled rectangle with a bent arrow symbolizes the prf1 gene. The letters P and M represent PKA and MAPK sites, respectively, whose phosphorylation state is indicated by circled Ps. See text for discussion.
Under conditions where prf1 expression is made constitutively, mfa1 expression is increased and this depends on intact PKA sites, indicating that basal activity of Prf1 is dependent on Adr1 phosphorylation. On the other hand, we observe that a constitutively active form of MAPKK, Fuz7, induces transcriptional induction of prf1 also when PKA sites are mutated. This hints at the existence of another route of pheromone signalling where an activated MAPK Kpp2 activates prf1 transcription through an as yet unknown regulator and this is sufficient to cause increased expression of a genes (Figure 7B). This would also explain why pheromone-responsive mfa1 promoter activity is reduced in strains expressing an inactive MAPK (Figure 1C).
Contrary to this mode of regulation, b genes can be induced neither by an activated cAMP pathway nor by increasing transcription of prf1. Instead, PKA sites and MAPK sites both are required to observe induction. This is most easily explained by assuming that Prf1 has to be phosphorylated by both kinases to become an activator of b genes (Figure 7B). The fact that PKA sites are needed for b gene expression although cAMP signalling is unable to induce b gene expression indicates that cAMP signalling is prerequisite for Prf1 activation. This is supported by the observation that induction of b gene expression by a genetically activated MAPKK also needs intact PKA sites in Prf1.
The scenario in which cAMP signalling is prerequisite for the pheromone response is reminiscent of mating in S.pombe (Davey, 1998). In this ascomycete, nutritional signalling relies on cAMP to regulate the activity of the HMG box protein Ste11, the key transcription factor for mating. However, in contrast to cAMP signalling in U.maydis, high cAMP levels inhibit mating in S.pombe by regulating Ste11 activity mainly on the transcriptional level (Sugimoto et al., 1991; Mochizuki and Yamamoto, 1992).
In U.maydis, promoter discrimination by Prf1 might be aided through the different location of PREs in the regulatory regions of target genes. In the mfa1 promoter, eight PREs are clustered ∼500 bp upstream of the transcriptional start site. In contrast, in the divergently transcribed b genes, PREs are not found in the intergenic promoter region but reside in the open reading frame and in an intron of bW as well as downstream of bE (Hartmann et al., 1996). For mfa1 gene induction, the amount of PKA-activated Prf1 alone might suffice, while for activation of the b promoter Prf1 phosphorylated at MAPK as well as PKA sites might have to interact with a second transcription factor, as has been described for Ste12p and Ste12α in S.cerevisiae and Cryptococcus neoformans, respectively (Madhani et al., 1997; Davidson et al., 2003). In S.cerevisiae, clustering of PREs occurs in promoters of pheromone-responsive genes that are activated by Ste12p alone, whereas PREs adjacent to Tec1p-binding sites form composite response elements in regulatory regions of genes involved in filamentation (Madhani et al., 1997; Zeitlinger et al., 2003).
Why should a network consisting of PKA and MAPK signalling be needed to regulate mating in U.maydis? Since mating in this fungus usually occurs on the plant surface, it would be advantageous to sense the presence of the plant, which could involve cAMP signalling. Elevated cAMP levels would result in PKA-activated Prf1, thereby increasing expression of pheromone and pheromone receptor genes. This would increase the chances of being recognized by a mating partner. Once a compatible mate has perceived the pheromone, one branch would feed into cAMP signalling to increase Prf1 activity and the other branch would stimulate the pheromone-responsive MAPK module leading to increased prf1 transcription and formation of conjugation hyphae (Müller et al., 2003). In addition, fully phosphorylated Prf1 would trigger b gene expression in preparation for post-fusion events (Figure 7).
Mechanisms of cross-talk between PKA and MAPK signalling in eukaryotes
Cross-talk between PKA and MAPK signalling is a widespread phenomenon in eukaryotic cells (Stork and Schmitt, 2002). In higher eukaryotes, it is used to regulate cell proliferation and, depending on the cell type, hormonal stimulation of cAMP/PKA signalling has either stimulatory or inhibitory effects, e.g. PKA phosphorylation of Ser259 of MAPKKK Raf1 inhibits its activity (Dhillon et al., 2002) or PKA phosphorylation of Ser23 of haematopoietic protein tyrosine phosphatase releases MAPK resulting in transcriptional activation of target genes (Saxena et al., 1999). In S.cerevisiae, at least two levels of cross-talk are realized, the MAPKs Fus3p and Kss1p act upstream of the Ras/cAMP signalling pathway regulating survival (Cherkasova et al., 2003), and PKA as well as MAPK signalling converge on distinct promoter elements of the FLO11 gene during filamentous growth (Rupp et al., 1999). In this developmental programme, MAPK Kss1p activates a transcription factor complex consisting of Ste12p and Tec1p that recognizes filament response elements (Lo and Dranginis, 1998; Pan and Heitman, 1999, 2002; Rupp et al., 1999) and PKA Tpk2p phosphorylates transcription factors Sfl1p and Flo8p that bind to a different region in the FLO11 promoter (Pan and Heitman, 2002). In U.maydis, a novel concept of cross-talk is realized in which PKA and MAPK phosphorylation of a single transcription factor is used to discriminate promoters. Future research should reveal whether such a mechanism is widespread in eukaryotes.
Materials and methods
Strains and growth conditions
The E.coli K-12 derivatives DH5α (Bethesda Research Laboratories) and Top10 (Invitrogen) were used for cloning purposes. BL21(DE3)pLysS (Novagen) was used for protein expression, and S.cerevisiae strain EGY48[p8op-lacZ] (Clontech) for the two-hybrid analysis. Ustilage maydis strains were constructed by transformation of progenitor strains with linearized plasmids (see Table I). Homologous integration events at the ip, prf1 or kpp2 locus were verified by Southern analysis (Müller et al., 1999, 2003; Loubradou et al., 2001). Growth conditions for U.maydis strains and source of antibiotics are described in Brachmann et al. (2003).
Table I. Ustilago maydis strains used in this study.
| Strains |
Relevant genotype |
UMa |
Reference |
Plasmid transformed |
Locus |
Progenitor strain |
| FB1 | a1 b1 | 51 | Banuett and Herskowitz (1989) | |||
| FB2 | a2 b2 | 52 | Banuett and Herskowitz (1989) | |||
| CL13 | a1 bE1bW2 | 66 | Bölker et al. (1995) | |||
| FB1prf1Δ | a1 b1 prf1Δ | 36 | Müller et al. 1999 | |||
| FB2prf1Δ | a2 b2 prf1Δ | 37 | Müller et al. 1999 | |||
| FB1prf1e | a1 b1 prf1e | 38 | Müller et al. 1999 | |||
| FB2prf1e | a2 b2 prf1e | 39 | Müller et al. 1999 | |||
| FB1prf1eM1-6d | a1 b1 prf1eM1-6d | 48 | Müller et al. 1999 | |||
| FB2 prf1eM1-6d | a2 b2 prf1eM1-6d | 49 | Müller et al. 1999 | |||
| FB1Pcrg1:fuz7DD | a1 b1 Pcrg1:fuz7DD | 265 | Müller et al. (2003) | |||
| FB1mfa1-egfp | a1 b1 Pmfa1:egfp | 9 | This study | pmfa1-egfp-cbx | ip | FB1 |
| FB1mfa1PREΔ-egfp | a1 b1 PmfaPREΔ:egfp | 116 | This study | pmfa1PREΔ-egfp-cbx | ip | FB1 |
| FB1PREdik6-egfp | a1 b1 PPREdik6:egfp | 118 | This study | pPREdik6-egfp-cbx | ip | FB1 |
| FB1dik6-egfp | a1 b1 Pdik6:egfp | 148 | This study | pdik6-egfp-cbx | ip | FB1 |
| FB1mfa1-egfp/kpp2K50R | a1 b1 Pmfa1:egfp kpp2K50R | 280 | This study | pkpp2K50R-nat | kpp2 | FB1mfa1-egfp |
| FB1prf1eM1-6 | a1 b1 prf1eM1-6 | 91 | This study | pprf1eM1-6-hyg | prf1 | FB1prf1Δ |
| FB2prf1eM1-6 | a2 b2 prf1eM1-6 | 95 | This study | pprf1eM1-6-hyg | prf1 | FB2prf1Δ |
| FB1prf1eMd | a1 b1 prf1eMd | 92 | This study | pprf1eMd-hyg | prf1 | FB1prf1Δ |
| FB2prf1eMd | a2 b2 prf1eMd | 96 | This study | pprf1eMd-hyg | prf1 | FB2prf1Δ |
| FB1prf1eM126d | a1 b1 prf1eM126d | 93 | This study | pprf1eM126d-hyg | prf1 | FB1prf1Δ |
| FB2prf1eM126d | a2 b2 prf1eM126d | 97 | This study | pprf1eM126d-hyg | prf1 | FB2prf1Δ |
| FB1prf1eM345 | a1 b1 prf1eM345 | 94 | This study | pprf1eM345-hyg | prf1 | FB1prf1Δ |
| FB2prf1eM345 | a2 b2 prf1eM345 | 98 | This study | pprf1eM345-hyg | prf1 | FB2prf1Δ |
| FB1prf1eM34 | a1 b1 prf1eM34 | 103 | This study | pprf1eM34-hyg | prf1 | FB1prf1Δ |
| FB2prf1eM34 | a2 b2 prf1eM34 | 104 | This study | pprf1eM34-hyg | prf1 | FB2prf1Δ |
| FB1prf1eM5 | a1 b1 prf1eM5 | 105 | This study | pprf1eM5-hyg | prf1 | FB1prf1Δ |
| FB2prf1eM5 | a2 b2 prf1eM5 | 106 | This study | pprf1eM5-hyg | prf1 | FB2prf1Δ |
| FB1prf1eM3 | a1 b1 prf1eM3 | 107 | This study | pprf1eM3-hyg | prf1 | FB1prf1Δ |
| FB2prf1eM3 | a2 b2 prf1eM3 | 108 | This study | pprf1eM3-hyg | prf1 | FB2prf1Δ |
| FB1prf1eM4 | a1 b1 prf1eM4 | 109 | This study | pprf1eM4-hyg | prf1 | FB1prf1Δ |
| FB2prf1eM4 | a2 b2 prf1eM4 | 110 | This study | pprf1eM4-hyg | prf1 | FB2prf1Δ |
| FB1prf1eP1-5 | a1 b1 prf1eP1-5 | 45 | This study | pprf1eP1-5-hyg | prf1 | FB1prf1Δ |
| FB2prf1eP1-5 | a2 b2 prf1eP1-5 | 46 | This study | pprf1eP1-5-hyg | prf1 | FB2prf1Δ |
| FB1prf1eP125 | a1 b1 prf1eP125 | 111 | This study | pprf1eP125-hyg | prf1 | FB1prf1Δ |
| FB2prf1eP125 | a2 b2 prf1eP125 | 112 | This study | pprf1eP125-hyg | prf1 | FB2prf1Δ |
| FB1prf1eP34 | a1 b1 prf1eP34 | 113 | This study | pprf1eP34-hyg | prf1 | FB1prf1Δ |
| FB2prf1eP34 | a2 b2 prf1eP34 | 114 | This study | pprf1eP34-hyg | prf1 | FB2prf1Δ |
| FB1prf1ce | a1 b1 prf1ce | 44 | This study | pprf1ce-hyg | prf1 | FB1prf1Δ |
| FB2prf1ce | a2 b2 prf1ce | 42 | This study | pprf1ce-hyg | prf1 | FB2prf1Δ |
| FB1prf1ceM1-6d | a1 b1 prf1ceM1-6d | 86 | This study | pprf1ceM1-6d-hyg | prf1 | FB1prf1Δ |
| FB2prf1ceM1-6d | a2 b2 prf1ceM1-6d | 88 | This study | pprf1ceM1-6d-hyg | prf1 | FB2prf1Δ |
| FB1prf1ceP1-5 | a1 b1 prf1ceP1-5 | 85 | This study | pprf1ceP1-5-hyg | prf1 | FB1prf1Δ |
| FB2prf1ceP1-5 | a2 b2 prf1ceP1-5 | 87 | This study | pprf1ceP1-5-hyg | prf1 | FB2prf1Δ |
| FB1Pcrg1:fuz7DD/prf1e | a1 b1 Pcrg1:fuz7DD prf1e | 266 | This study | Pcrg1:fuz7DD-cbx | ip | FB1prf1-E |
| FB1Pcrg1:fuz7DD/prf1eM1-6d | a1 b1 Pcrg1:fuz7DD prf1eM1-6d | 267 | This study | Pcrg1:fuz7DD-cbx | ip | FB1prf1-M1 |
| FB1Pcrg1:fuz7DD/prf1eP1-5 | a1 b1 Pcrg1:fuz7DD prf1eP1-5 | 268 | This study | Pcrg1:fuz7DD-cbx | ip | FB1prf1-P1 |
| FB1Pcrg1:fuz7DD/prf1ce | a1 b1 Pcrg1:fuz7DD prf1ce | 35 | This study | Pcrg1:fuz7DD-cbx | ip | FB1prf1-Ec |
| CL13prf1Δ | a1 bE1bW2 prf1Δ | 16 | This study | pprf1Δ-nat | prf1 | CL13 |
| CL13prf1Δ/mfa1-egfp | a1 bE1bW2 prf1Δ Pmfa1:egfp | 235 | This study | pmfa1-egfp-cbx | ip | CL13prf1Δ |
| CL13prf1e/mfa1-egfp | a1 bE1bW2 Pmfa1:egfp prf1e | 237 | This study | pprf1e-hyg | prf1 | CL13prf1Δ/mfa1-eGFP |
| CL13prf1eM1-6d/mfa1-egfp | a1 bE1bW2 Pmfa1:egfp prf1eM1-6d | 236 | This study | pprf1eM1-6d-hyg | prf1 | CL13prf1Δ/mfa1-eGFP |
| CL13prf1eP1-5/mfa1-egfp | a1 bE1bW2 Pmfa1:egfp prf1eP1-5 | 238 | This study | pprf1eP1-5-hyg | prf1 | CL13prf1Δ/mfa1-eGFP |
Nucleic acid procedures
Plasmids and plasmid constructions are described in detail in the Supplementary data. Transformation, and DNA and RNA isolation from U.maydis were performed as described in Brachmann et al. (2003). mRNA was enriched using Oligotex™ suspension (Qiagen) according to the manufacturer’s instructions. Total RNA (15 µg/lane) or enriched mRNA (∼0.5 µg/lane) was separated on MOPS-buffered 1% agarose gels and transferred to Hybond-N+ membranes (Amersham Biosciences). Double-stranded probes were used for northern analysis: mfa1 (Bölker et al., 1992); bE1 and bW1 (Kämper et al., 1995); ip (Müller et al., 1999); and rRNA (Bottin et al., 1996).
In vitro kinase assay
Protein expression and affinity purification using glutathione–Sepharose beads (Amersham Biosciences) was performed according to the manufacturer’s instructions (see Supplementary data). Equal amounts of substrate (Prf1440–565, Prf1440–565M345 or Prf1440–565P34; ∼1 µg) were incubated with GST–Kpp2 (∼3 µg), GST–Adr1 (∼1 µg) coupled to glutathione–Sepharose beads or recombinant murine PKA (250 U; NEB Biolabs) in kinase buffer containing 20 mM HEPES, 15 mM MgCl2, 5 mM EGTA, 1 mM dithiothreitol (DTT), 50 mM Na-β-glycerolphosphate, 5 mM NaVO3, 50 µM ATP and 4 µCi of [γ-32P]ATP (6000 Ci/mmol). As control, bovine myelin basic protein (2 µg; Sigma) was used. After 20 min at 28°C kinase reactions were terminated by adding 10 µl of SDS–PAGE loading buffer and samples were frozen. After SDS–PAGE, gels were dried and analysed using a Storm phosphoimager (Molecular Dynamics). Equal loading was verified by staining with Coomassie brilliant blue after rehydration.
Yeast two-hybrid analysis
The yeast two-hybrid analysis was carried out using the MATCHMAKER LexA two-hybrid system (Clontech) according to the manufacturer’s instructions. Plasmids pPrf1eNLS or pJG4-5 (Gyuris et al., 1993) were transformed in strain EGY48[p8op-lacZ] in combination with pEG202 (pLexA; Gyuris et al., 1993), pLexA-Kpp2 or pLexA-Adr1. Transformants were transferred on synthetic dropout medium plates either containing or not containing leucine and incubated for 48 or 72 h at 28°C, respectively.
Mating, filamentation, pathogenicity assay, pheromone and cAMP treatment
Mating and filamentation assays were performed by co-spotting respective strains on charcoal-containing PD plates that were sealed with parafilm and incubated at 22°C for 24–48 h. Plant infections of corn variety Early Golden Bantam (Olds Seeds, Madison, WI) were performed as described (Brachmann et al., 2003). Tumour formation was scored after 14 days. For pheromone or cAMP stimulation, strains were grown in CM medium to an OD600 of 0.5. Synthetic a2 pheromone dissolved in dimethylsulfoxide (Szabo et al., 2002) was added to a final concentration of 2.5 µg/ml and cells were incubated for 6 h at 28°C in a 15 ml plastic tube rotating at 20 r.p.m.. For cAMP stimulation, cells were harvested by centrifugation and resuspended in CM medium containing 6 mM cAMP, and incubated by shaking (200 r.p.m.) for 18 h at 28°C.
Fluorimetric measurements
After respective incubation, 200 µl of cell suspension were transferred in a microtitre plate and fluorescence was measured in a TECAN Saphire fluorescence reader. GFP fluorescence was measured at a wavelength of 485 nm for excitation and 520 nm for emission, with a bandwidth of 7.5 nm in both cases. Optical density was measured as absorbance at 600 nm. Fluorescence was normalized to OD600. Three pheromone and cAMP treatments were performed in parallel and measured in triplicate.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Drs A.Brachmann, M.Bölker and lab members for valuable discussion and critical reading of the manuscript, S.Hester, J.Schwarz and T.Schlunck for excellent technical assistance, and Drs C.Aichinger for generating construct mfa1-egfp, N.Loubradou for strain CL13prf1Δ and G.Weinzierl for the dik6 basal promoter. This work was supported through SFB369 of the DFG and by Bayer CropScience.
References
- Andrews D.L., Egan,J.D., Mayorga,M.E. and Gold,S.E. (2000) The Ustilago maydis ubc4 and ubc5 genes encode members of a MAP kinase cascade required for filamentous growth. Mol. Plant-Microbe Interact., 13, 781–786. [DOI] [PubMed] [Google Scholar]
- Banuett F. (1992) Ustilago maydis, the delightful blight. Trends Genet., 8, 174–180. [DOI] [PubMed] [Google Scholar]
- Banuett F. (1995) Genetics of Ustilago maydis, a fungal pathogen that induces tumors in maize. Annu. Rev. Genet., 29, 179–208. [DOI] [PubMed] [Google Scholar]
- Banuett F. and Herskowitz,I. (1989) Different a alleles are necessary for maintenance of filamentous growth but not for meiosis. Proc. Natl Acad. Sci. USA, 86, 5878–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuett F. and Herskowitz,I. (1994) Identification of fuz7, a Ustilago maydis MEK/MAPKK homolog required for a-locus-dependent and -independent steps in the fungal life cycle. Genes Dev., 8, 1367–1378. [DOI] [PubMed] [Google Scholar]
- Bardwell L., Cook,J.G., Voora,D., Baggott,D.M., Martinez,A.R. and Thorner,J. (1998) Repression of yeast Ste12 transcription factor by direct binding of unphosphorylated Kss1 MAPK and its regulation by the Ste7 MEK. Genes Dev., 12, 2887–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlmann R. (1996) Isolierung und Charakterisierung von filamentspezifisch exprimierten Genen aus Ustilago maydis. PhD thesis, Fakultät für Biologie, Ludwig-Maximilian-Universität München, Germany. [Google Scholar]
- Bölker M. (2001) Ustilago maydis—a valuable model system for the study of fungal dimorphism and virulence. Microbiology, 147, 1395–1401. [DOI] [PubMed] [Google Scholar]
- Bölker M., Urban,M. and Kahmann,R. (1992) The a mating type locus of U.maydis specifies cell signaling components. Cell, 68, 441–450. [DOI] [PubMed] [Google Scholar]
- Bölker M., Genin,S., Lehmler,C. and Kahmann,R. (1995) Genetic regulation of mating and dimorphism in Ustilago maydis. Can. J. Bot., 73, 320–325. [Google Scholar]
- Bottin A., Kämper,J. and Kahmann,R. (1996) Isolation of a carbon source-regulated gene from Ustilago maydis. Mol. Gen. Genet., 253, 342–352. [DOI] [PubMed] [Google Scholar]
- Brachmann A., Schirawski,J., Müller,P. and Kahmann,R. (2003) An unusual MAP kinase is required for efficient penetration of the plant surface by Ustilago maydis. EMBO J., 22, 2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasova V.A., McCully,R., Wang,Y., Hinnebusch,A. and Elion,E.A. (2003) A novel functional link between MAP kinase cascades and the Ras/cAMP pathway that regulates survival. Curr. Biol., 13, 1220–1226. [DOI] [PubMed] [Google Scholar]
- Clark K.L., Feldmann,P.J., Dignard,D., Larocque,R., Brown,A.J., Lee,M.G., Thomas,D.Y. and Whiteway,M. (1995) Constitutive activation of the Saccharomyces cerevisiae mating response pathway by a MAP kinase kinase from Candida albicans. Mol. Gen. Genet., 249, 609–621. [DOI] [PubMed] [Google Scholar]
- Davey J. (1998) Fusion of a fission yeast. Yeast, 14, 1529–1566. [DOI] [PubMed] [Google Scholar]
- Davidson R.C., Nichols,C.B., Cox,G.M., Perfect,J.R. and Heitman,J. (2003) A MAP kinase cascade composed of cell type specific and non-specific elements controls mating and differentiation of the fungal pathogen Cryptococcus neoformans. Mol. Microbiol., 49, 469–485. [DOI] [PubMed] [Google Scholar]
- Dhillon A.S., Meikle,S., Yazici,Z., Eulitz,M. and Kolch,W. (2002) Regulation of Raf-1 activation and signalling by dephosphorylation. EMBO J., 21, 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza C.A. and Heitman,J. (2001) Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol. Rev., 25, 349–364. [DOI] [PubMed] [Google Scholar]
- Dürrenberger F., Wong,K. and Kronstad,J.W. (1998) Identification of a cAMP-dependent protein kinase catalytic subunit required for virulence and morphogenesis in Ustilago maydis. Proc. Natl Acad. Sci. USA, 95, 5684–5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion E.A. (2000) Pheromone response, mating and cell biology. Curr. Opin. Microbiol., 3, 573–581. [DOI] [PubMed] [Google Scholar]
- Errede B., Gartner,A., Zhou,Z., Nasmyth,K. and Ammerer,G. (1993) MAP kinase-related FUS3 from S.cerevisiae is activated by STE7 in vitro. Nature, 362, 261–264. [DOI] [PubMed] [Google Scholar]
- Gillissen B., Bergemann,J., Sandmann,C., Schroeer,B., Bölker,M. and Kahmann,R. (1992) A two-component regulatory system for self/non-self recognition in Ustilago maydis. Cell, 68, 647–657. [DOI] [PubMed] [Google Scholar]
- Gold S., Duncan,G., Barrett,K. and Kronstad,J. (1994) cAMP regulates morphogenesis in the fungal pathogen Ustilago maydis. Genes Dev., 8, 2805–2816. [DOI] [PubMed] [Google Scholar]
- Gold S.E., Brogdon,S.M., Mayorga,M.E. and Kronstad,J.W. (1997) The Ustilago maydis regulatory subunit of a cAMP-dependent protein kinase is required for gall formation in maize. Plant Cell, 9, 1585–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyuris J., Golemis,E., Chertkov,H. and Brent,R. (1993) Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell, 75, 791–803. [DOI] [PubMed] [Google Scholar]
- Hamer J. and Talbot,N.J. (1998) Infection-related development in the rice blast fungus Magnaporthe grisea. Curr. Opin. Microbiol., 1, 693–697. [DOI] [PubMed] [Google Scholar]
- Hartmann H.A., Kahmann,R. and Bölker,M. (1996) The pheromone response factor coordinates filamentous growth and pathogenicity in Ustilago maydis. EMBO J., 15, 1632–1641. [PMC free article] [PubMed] [Google Scholar]
- Hartmann H.A., Krüger,J., Lottspeich,F. and Kahmann,R. (1999) Environmental signals controlling sexual development of the corn smut fungus Ustilago maydis through the transcriptional regulator Prf1. Plant Cell, 11, 1293–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I. (1995) MAP kinase pathways in yeast: for mating and more. Cell, 80, 187–197. [DOI] [PubMed] [Google Scholar]
- Hull C.M. and Heitman,J. (2002) Genetics of Cryptococcus neoformans. Annu. Rev. Genet., 36, 557–615. [DOI] [PubMed] [Google Scholar]
- Hunter T. (2000) Signaling 2000 and beyond. Cell, 100, 113–127. [DOI] [PubMed] [Google Scholar]
- Jacobs D., Glossip,D., Xing,H., Muslin,A.J. and Kornfeld,K. (1999) Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev., 13, 163–175. [PMC free article] [PubMed] [Google Scholar]
- Kahmann R., Basse,C. and Feldbrügge,M. (1999) Fungal–plant signalling in the Ustilago maydis–maize pathosystem. Curr. Opin. Microbiol., 2, 647–650. [DOI] [PubMed] [Google Scholar]
- Kahmann R., Steinberg,G., Basse,C., Feldbrügge,M. and Kämper,J. (2000) Ustilago maydis, the causative agent of corn smut disease. In Kronstad,J.W. (ed.), Fungal Pathology. Kluwer Academic Publishers, Dordrecht, pp. 347–371. [Google Scholar]
- Kämper J., Reichmann,M., Romeis,T., Bölker,M. and Kahmann,R. (1995) Multiallelic recognition: nonself-dependent dimerization of the bE and bW homeodomain proteins in Ustilago maydis. Cell, 81, 73–83. [DOI] [PubMed] [Google Scholar]
- Keon J.P., White,G.A. and Hargreaves,J.A. (1991) Isolation, characterization and sequence of a gene conferring resistance to the systemic fungicide carboxin from the maize smut pathogen. Ustilago maydis. Curr. Genet., 19, 475–481. [DOI] [PubMed] [Google Scholar]
- Krüger J., Loubradou,G., Regenfelder,E., Hartmann,A. and Kahmann,R. (1998) Crosstalk between cAMP and pheromone signalling pathways in Ustilago maydis. Mol. Gen. Genet., 260, 193–198. [DOI] [PubMed] [Google Scholar]
- Kurjan J. (1992) Pheromone response in yeast. Annu. Rev. Biochem., 61, 1097–1129. [DOI] [PubMed] [Google Scholar]
- Lee N., D’Souza,C. and Kronstad,J.W. (2003) Of smuts, blasts, mildews and blights: cAMP signalling in phytopathogenic fungi. Annu. Rev. Phytopathol., 41, 399–427. [DOI] [PubMed] [Google Scholar]
- Lengeler K.B., Davidson,R.C., D’Souza,C., Harashima,T., Shen,W.C., Wang,P., Pan,X., Waugh,M. and Heitman,J. (2000) Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev., 64, 746–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo W.S. and Dranginis,A.M. (1998) The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell, 9, 161–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loubradou G., Brachmann,A., Feldbrügge,M. and Kahmann,R. (2001) A homolog of the transcriptional repressor Ssn6p antagonizes cAMP signalling in Ustilago maydis.Mol. Microbiol., 40, 719–730. [DOI] [PubMed] [Google Scholar]
- Madhani H.D. and Fink,G.R. (1998) The control of filamentous differentiation and virulence in fungi. Trends Cell Biol., 8, 348–353. [DOI] [PubMed] [Google Scholar]
- Madhani H.D., Styles,C.A. and Fink,G.R. (1997) MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell, 91, 673–684. [DOI] [PubMed] [Google Scholar]
- Mayorga M.E. and Gold,S.E. (1999) A MAP kinase encoded by the ubc3 gene of Ustilago maydis is required for filamentous growth and full virulence. Mol. Microbiol., 34, 485–497. [DOI] [PubMed] [Google Scholar]
- Mochizuki N. and Yamamoto,M. (1992) Reduction in the intracellular cAMP level triggers initiation of sexual development in fission yeast. Mol. Gen. Genet., 233, 17–24. [DOI] [PubMed] [Google Scholar]
- Müller P., Aichinger,C., Feldbrügge,M. and Kahmann,R. (1999) The MAP kinase Kpp2 regulates mating and pathogenic development in Ustilago maydis. Mol. Microbiol., 34, 1007–1017. [DOI] [PubMed] [Google Scholar]
- Müller P., Weinzierl,G., Brachmann,A., Feldbrügge,M. and Kahmann,R. (2003) Both mating and pathogenic development of the smut fungus Ustilago maydis are regulated by one MAP kinase cascade. Eukaryot. Cell, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson K.A., Nelson,C., Tai,G., Hung,W., Yong,C., Astell,C. and Sadowski,I. (2000) Two regulators of Ste12p inhibit pheromone-responsive transcription by separate mechanisms. Mol. Cell. Biol., 20, 4199–4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X. and Heitman,J. (1999) Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 4874–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X. and Heitman,J. (2002) Protein kinase A operates a molecular switch that governs yeast pseudohyphal differentiation. Mol. Cell. Biol., 22, 3981–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi H., Chien,C.T. and Fields,S. (1997) Transcriptional activation upon pheromone stimulation mediated by a small domain of Saccharomyces cerevisiae Ste12p. Mol. Cell. Biol., 17, 6410–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regenfelder E., Spellig,T., Hartmann,A., Lauenstein,S., Bölker,M. and Kahmann,R. (1997) G proteins in Ustilago maydis: transmission of multiple signals? EMBO J., 16, 1934–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp S., Summers,E., Lo,H.J., Madhani,H. and Fink,G. (1999) MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J., 18, 1257–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Martínez C. and Pérez-Martín,J. (2001) Dimorphism in fungal pathogens: Candida albicans and Ustilago maydis—similar inputs, different outputs. Curr. Opin. Microbiol., 4, 214–221. [DOI] [PubMed] [Google Scholar]
- Saxena M., Williams,S., Taskén,K. and Mustelin,T. (1999) Crosstalk between cAMP-dependent kinase and MAP kinase through a protein tyrosine phosphatase. Nature Cell Biol., 1, 305–311. [DOI] [PubMed] [Google Scholar]
- Seger R. et al. (1991) Microtubule-associated protein 2 kinases, ERK1 and ERK2, undergo autophosphorylation on both tyrosine and threonine residues: implications for their mechanism of activation. Proc. Natl Acad. Sci. USA, 88, 6142–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snetselaar K.M., Bölker,M. and Kahmann,R. (1996) Ustilago maydis mating hyphae orient their growth toward pheromone sources. Fungal Genet. Biol., 20, 299–312. [DOI] [PubMed] [Google Scholar]
- Spellig T., Bölker,M., Lottspeich,F., Frank,R.W. and Kahmann,R. (1994) Pheromones trigger filamentous growth in Ustilago maydis. EMBO J., 13, 1620–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellig T., Bottin,A. and Kahmann,R. (1996) Green fluorescent protein (GFP) as a new vital marker in the phytopathogenic fungus Ustilago maydis. Mol. Gen. Genet., 252, 503–509. [DOI] [PubMed] [Google Scholar]
- Stork P.J.S. and Schmitt,J.M. (2002) Crosstalk between cAMP and MAP kinase signaling in the regulation of cell proliferation. Trends Cell Biol., 12, 258–266. [DOI] [PubMed] [Google Scholar]
- Sugimoto A., Iino,Y., Maeda,T., Watanabe,Y. and Yamamoto,M. (1991) Schizosaccharomyces pombe ste11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes Dev., 5, 1990–1999. [DOI] [PubMed] [Google Scholar]
- Szabo Z., Tönnis,M., Kessler,H. and Feldbrügge,M. (2002) Structure–function analysis of lipopeptide pheromones from the plant pathogen Ustilago maydis. Mol. Gen. Genet., 268, 362–370. [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Buechler,J.A. and Yonemoto,W. (1990) cAMP-dependent protein kinase: framework for a diverse family of regulatory enzymes. Annu. Rev. Biochem., 59, 971–1005. [DOI] [PubMed] [Google Scholar]
- Tedford K., Kim,S., Sa,D., Stevens,K. and Tyers,M. (1997) Regulation of the mating pheromone and invasive growth responses in yeast by two MAP kinase substrates. Curr. Biol., 7, 228–238. [DOI] [PubMed] [Google Scholar]
- Urban M., Kahmann,R. and Bölker,M. (1996) Identification of the pheromone response element in Ustilago maydis. Mol. Gen. Genet., 251, 31–37. [DOI] [PubMed] [Google Scholar]
- Whiteway M. (2000) Transcriptional control of cell type and morphogenesis in Candida albicans. Curr. Opin. Microbiol., 3, 582–588. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J., Simon,I., Harbison,C.T., Hannett,N.M., Volkert,T.L., Fink,G.R. and Young,R.A. (2003) Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell, 113, 395–404. [DOI] [PubMed] [Google Scholar]


