Abstract
Current methodologies for global identification of microbial proteins that elicit host humoral immune responses have several limitations and are not ideally suited for use in the postgenomic era. Here we describe a novel application of proteomics, proteomics-based expression library screening, to rapidly define microbial immunoproteomes. Proteomics-based expression library screening is broadly applicable to any cultivable, sequenced pathogen eliciting host antibody responses and hence is ideal for rapidly mining microbial proteomes for targets with diagnostic, prophylactic, and therapeutic potential. In this report, we demonstrate “proof-of-principle” by identifying 207 proteins of the Escherichia coli O157:H7 immunome in bovine reservoirs in only 3 weeks.
Proteins constituting microbial immunomes (the subset of microbial antigens that elicit host immune responses) have excellent diagnostic, prophylactic, and therapeutic potential because a subset of such immunogenic proteins is part of the repertoire of microbial factors that function to help pathogens counter host defenses, facilitate niche adaptation, and survive and replicate in these hosts. Methodologies to rapidly identify such protein targets would facilitate exploitation of microbial genome sequence data and expedite the development of novel management strategies against infectious diseases.
Traditional methodologies for proteome wide identification of immunogenic microbial proteins (IMPs)1 involve screening microbial recombinant genomic expression libraries in plasmid/phage expression vectors and laboratory host strains with sera from colonized or infected hosts. However, colony immunoscreening and in vivo induced antigen technology (IVIAT) (1), a variation of colony immunoscreening that defines only partial immunoproteomes, and bacterial surface display coupled with magnetic cell sorting (2) are laborious and require several months or more for definitive IMP identification. Immunoproteomics of pathogens cultured in vitro under either standard laboratory conditions or those that attempt to mimic the host environment are also popular; however, pathogens cultured in vitro might not express the entire spectrum of virulence proteins. In view of the challenging task of accurately reproducing the host environment, such approaches might overlook those immunogenic virulence proteins that are expressed exclusively in response to host environmental cues and contribute significantly to pathogenicity (3). Although these limitations may be circumvented by immunoproteomics of pathogens isolated directly from either biological specimens or host anatomical sites of infection, consistent recovery of sufficient numbers of suitable organisms for analysis presents a significant challenge (4). Protein microarray/chip technology has tremendous potential for rapid, global definition of IMPs but is constrained by bottlenecks in proteome scale purification of microbial proteins and currently permits immunological characterization of only a partial proteome (5). Newer formats such as nucleic acid programmable protein arrays (6) are still experimental, and neither nucleic acid programmable protein arrays nor antibody arrays/protein chips utilizing SELDI-TOF mass spectrometry for antigen identification (7) have been demonstrated to rapidly define microbial immunoproteomes.
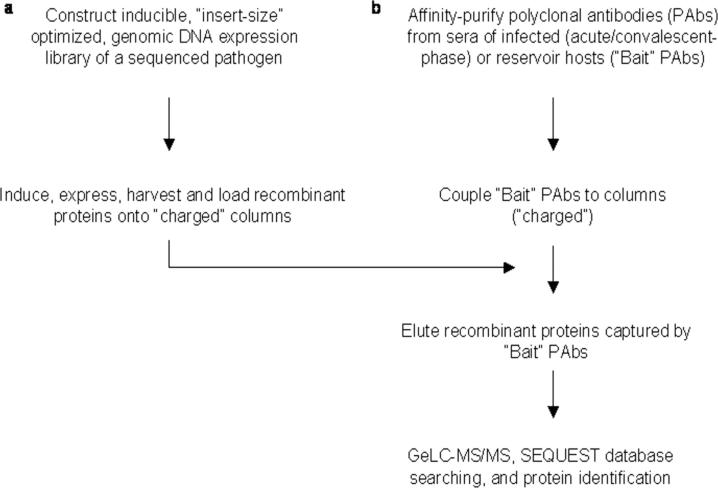
To rapidly identify proteins comprising microbial immunomes, we have developed a novel technique, proteomics-based expression library screening (PELS), that couples standard recombinant DNA and immunochemistry techniques with proteomics. The principle of PELS is outlined in Fig. 1 and involves capture of recombinant proteins expressed from an inducible, microbial genomic DNA expression library using polyclonal antibodies (pAbs) affinity-purified from acute/convalescent sera of infected hosts or sera from reservoirs colonized by the cognate pathogen (“bait” pAbs) coupled to a solid support. Proteins captured by the bait pAbs are subjected to one-dimensional (1D) SDS-PAGE ESI nano-LC-MS/MS (GeLC-MS/MS) and identified via SEQUEST database searching (8) (Fig. 1). The entire process, from recombinant genomic expression library construction to definitive protein identification, is accomplished in only 3 weeks without biases inherent to manual screening. To our knowledge, this is the first application of proteomics for rapid, global identification of IMPs from among proteins expressed from genes on inserts within recombinant clones comprising microbial genomic expression DNA libraries.
Fig. 1.
Principle of PELS.
EXPERIMENTAL PROCEDURES
Recombinant DNA Methods and Proteomic Analysis
Isolation of plasmid DNA, restriction digestions, and agarose gel electrophoresis were performed using standard procedures (9). Enzymes for restriction digestions, DNA modifications, and ligations were procured from New England Biolabs, Beverly, MA. Oligonucleotides for PCR were obtained from the DNA Synthesis Core Facility, Department of Molecular Biology, Massachusetts General Hospital. Plasmids were electroporated into Escherichia coli DH5α or BL21(DE3) using a Gene Pulser (Bio-Rad) as instructed by the manufacturer. Electroporation conditions were 2,500 V at 25-micro-faraday capacitance, producing time constants of 4.8 − 4.9 ms. Proteomic analysis, namely GeLC-MS/MS, was performed at the Harvard Partners Center for Genetics and Genomics, Cambridge, MA.
Construction of an Inducible E. coli O157:H7 (O157) Genomic DNA Expression Library Comprising Clones Containing DNA Inserts of Optimal Size (“Optimized” O157 Expression Library)
We generated an O157 genomic DNA expression library as described earlier (10) using genomic DNA isolated from the O157 strain 43894 (an isolate from a human patient with hemorrhagic colitis in the United States) in the pET-30abc series of expression vectors (Novagen, Madison, WI). Vector and insert DNA in the size range of 0.5−3.0 kbp were prepared as described previously (10). Because accuracy of protein identification using tandem MS/MS data and SEQUEST database searching increases with the number of peptides generated following trypsin digestion of the cognate protein (8), we sought to preferentially ligate insert DNA fragments that were larger than the average size of ORFs in this pathogen. The rationale was that larger DNA fragments were more likely to include genes encoding full-length or near full-length recombinant proteins containing potentially a larger number of recognition sites for cleavage by trypsin such that multiple peptide fragments resulting from trypsin cleavage of such proteins might facilitate robust protein identification. We accomplished this by performing multiple ligation reactions with each containing a different mole ratio of insert to vector. Each ligation reaction was then used to transform competent E. coli DH5α via electroporation according to standard protocols (9). To minimize overrepresentation of sister clones in the O157 expression library, transformants were directly plated onto LB plates supplemented with 50 μg/ml kanamycin (LB-Kan) without allowing for phenotypic expression and incubated overnight at 37 °C. To determine both the percentage of transformants containing inserts as well as the insert size, 100 colonies were randomly picked from each O157 expression library and analyzed by colony PCR using vector-specific primers. Greater than 90% of all transformants examined contained inserts, and >80% of clones of one O157 expression library included inserts that exceeded 1.7 kbp in size. Recombinant clones comprising this particular O157 expression library were scraped off LB-Kan plates, plasmid DNA was isolated using the QIAprep Spin miniprep kit (Qiagen Sciences) and used to transform electrocompetent E. coli BL21(DE3) (Novagen), the recommended expression host, and transformants were plated as described above. The resultant O157 expression library comprised >105 recombinant clones. Given the genome sizes of the sequenced O157 strains EDL933 (11) and Sakai (12) and that the average size of ORFs in these strains is 1 kbp (13), we concluded that this expression library, comprising recombinant clones containing inserts of optimal size (optimized O157 expression library), was adequate for defining components of the O157 immunoproteome (14).
Expression of the Optimized O157 Expression Library and Preparation of Cell Lysate and Pellet Fractions Containing Recombinant Proteins for Immunoaffinity Capture
To expedite sample preparation and circumvent laborious experimentation to define the inducer concentration and postinduction growth conditions (such as incubation temperature and duration of growth after induction) that facilitated recombinant O157 protein expression in a form that permitted optimal capture by pAbs affinity-purified from pooled hyperimmune cattle sera (bait pAbs) and coupled to HiTrap NHS-activated HP columns (see below), several batches of the optimized O157 expression library were cultured simultaneously at 37 °C to an initial A600 of 0.6. Cultures were then induced with 0.25, 0.5, 1, or 2.0 mm isopropyl β-d-thiogalactopyranoside (IPTG) and incubated at 25, 30, or 37 °C for either 5 h or overnight. Cultures were pooled and centrifuged at 7,000 rpm for 10 min at 4 °C to harvest cell pellets, which were then washed three times with chilled, sterile PBS (pH 7.4). The “Complete” protease inhibitor mixture (Roche Diagnostics) was added to twice the concentration recommended by the manufacturer after which cells were lysed by three freeze-thaw cycles. Lysed cells were resuspended in 2 ml of chilled PBS and microcentrifuged at maximum speed for 10 min, and supernatant (lysate) fractions were decanted into fresh tubes. Pellet fractions were resuspended in 2 ml of PBS containing 2× Complete protease inhibitor mixture (Roche Diagnostics), and the nonionic detergent n-octyl β-d-glucopyranoside (NOG; 0.2% final concentration) was added to solubilize membrane proteins. Both the lysate and pellet fractions containing recombinant O157 proteins were stored at −70 °C until used.
Coupling of Bait pAbs to HiTrap NHS-activated HP Columns
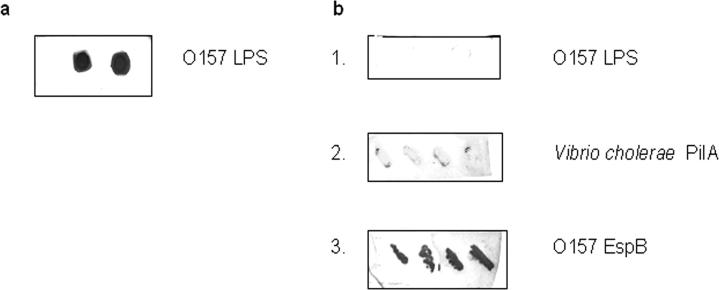
Prior to coupling, hyperimmune cattle sera against diverse O157 strains were generated and evaluated for reactivity against previously identified O157 antigens (supplemental material and Fig. 2, a and b). Polyclonal antibodies were affinity-purified from pooled hyperimmune cattle sera (bait pAbs) as detailed in the supplemental material. Affinity-purified bait pAb fractions were pooled together and dialyzed overnight against PBS (pH 7.4) at 4 °C and then for 4 h against coupling buffer consisting of 0.2 m NaHCO3, 0.5 m NaCl (pH 8.3) using a dialysis membrane with a molecular weight cutoff of 3500. Coupling of bait pAbs via amine groups was done as instructed by the manufacturer with minor modifications. Following removal of isopropanol, HiTrap NHS-activated columns were equilibrated with 10 column volumes of coupling buffer. Pooled bait IgG pAbs were slowly loaded on the column using a syringe and then recirculated through the column for 30 min at room temperature by attaching another syringe to the outlet. Active groups that did not couple to the ligand were quenched with 10 ml of 1 m tris(hydroxymethyl)aminomethane (pH 9.0), and nonspecifically bound pAbs were eluted with 10 ml of 1 m acetic acid. Columns with immobilized bait pAbs (charged columns) were then rinsed with 10 ml of deionized water to remove the quencher and the eluant.
Fig. 2. Reactivity of purified E. coli O157 LPS and recombinant E. coli BL21(DE3) (pET-30b) expressing various bacterial proteins with pooled hyperimmune sera from cattle experimentally infected with E. coli O157.
a, dot immunoblot of purified O157 lipopolysaccharide (LPS) spotted in duplicate and screened with pooled unadsorbed hyperimmune bovine sera. b, 1, dot immunoblot of O157 lipopolysaccharide spotted in duplicate (test); 2, colony immunoblot of four recombinant clones expressing PilA of Vibrio cholerae, an irrelevant protein (control); and 3, colony immunoblot of four recombinant clones expressing EspB of E. coli O157 (test) screened with pooled hyperimmune bovine sera adsorbed against purified E. coli O157 lipopolysaccharide.
Capture of Recombinant O157 Proteins
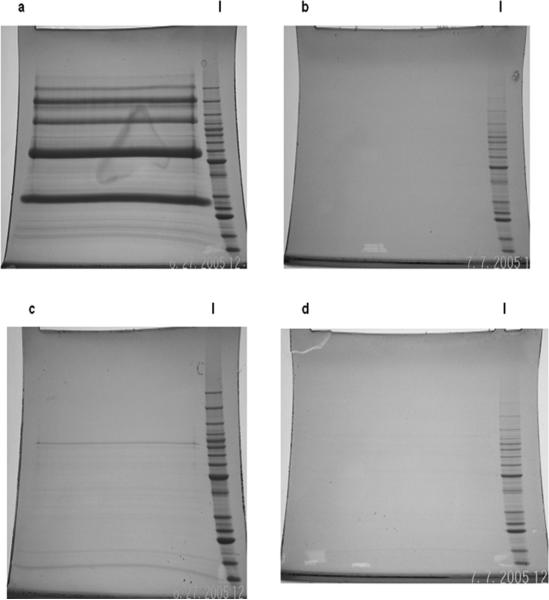
“Charged” columns were equilibrated with 10 column volumes of binding buffer consisting of PBS (pH 7.4), 0.2% NOG. Cell lysates and pellet fractions containing recombinant O157 proteins from the previous step were diluted with PBS, 0.2% NOG containing 2× concentration of Complete protease inhibitor mixture (Roche Diagnostics) in 20 ml and then loaded via a syringe separately onto the charged columns at a flow rate dictated by gravity (∼0.75−1 ml/min). Following a rinse with 20 volumes of loading buffer to remove loosely bound proteins, specifically captured recombinant proteins were eluted with 10 ml of 1 m acetic acid directly into 15-ml Falcon tubes containing 500 μl of ammonium hydroxide (elution with 1 m acetic acid facilitates multiple reuses of the charged column without a significant loss in specific binding capacity). This process was repeated three times to maximize yield of captured recombinant O157 proteins. Nonspecific adsorption of recombinant O157 proteins to the column matrix was assessed by passing optimized O157 expression library lysate and pellet fractions of cells of the O157 expression library through “uncharged” columns (HiTrap NHS-activated HP columns not coupled to bait pAbs) with quenched active groups. Specificity of capture and lack of nonspecific adsorption was confirmed by visualizing recombinant O157 proteins by fractionation of elutions from charged and uncharged columns on SDS-PAGE gels and subsequent staining (Fig. 3, a–d).
Fig. 3. SDS-PAGE profiles of recombinant O157 proteins in elutions from columns coupled to bait pAbs (charged) or uncharged columns.
Multiple O157 expression library cultures were induced with a range of IPTG concentrations at an initial A600 of 0.6. Following incubations at different temperatures for varying lengths of time, cell lysates and pellets prepared from pooled, pelleted cultures were loaded on charged or uncharged columns. Elutions were fractionated on SDS-PAGE Tris-Glycine 8−16% gradient gels and visualized by SimplyBlue Safe Stain. a, elutions from a charged column (test) loaded with lysate fractions. b, elutions from an uncharged column (control) loaded with lysate fractions. c, elutions from a charged column (test) loaded with pellet fractions. d, elutions from an uncharged column (control) loaded with pellet fractions. l, molecular weight ladder.
Proteomic Analysis of Elutions from Charged and Uncharged Columns Using GeLC-MS/MS
Elutions from charged and uncharged columns were subjected to GeLC-MS/MS (see below) at the Harvard Partners Center for Genetics and Genomics, Cambridge, MA. The advantages of GeLC-MS/MS over LC-MS/MS have been well documented (8). In preparation for 1D SDS-PAGE, elutions were concentrated using spin filters (molecular mass cutoff, 5000 daltons) (Viva-science Inc., Edgewood, NY) and reduced by incubation in 250 μl of 8 m urea, 100 mm ammonium bicarbonate, 1%SDS, 10 mm DTT at 37 °C for 1 h. Following a further incubation at room temperature for 20 min, each sample was alkylated by the addition of 15 μl of 500 mm iodoacetamide and incubation at room temperature in the dark for 60 min. Alkylation was then quenched with the addition of 3 μl of 2 m DTT to each sample. Following the addition of 150 μl of SDS-PAGE loading buffer/tube, each sample was centrifuged at 14,000 rpm for 10 min at room temperature, and 400 μl of each sample was fractionated on a 1D SDS-PAGE Tris-glycine 8 − 16% gradient gels (Invitrogen) for 2.5 h at 125 V, 20 mA, and 8 watts. Gels were alternatively shrunk for 12 h by the addition of 500 ml of 50% methanol, 5% acetic acid and then allowed to swell up for 1 h by the addition of 500 ml of deionized water for 60 min on a rotary shaker. Gels were then stained with SimplyBlue Safe Stain (Invitrogen) for 14 −16 h on a rotary shaker, imaged, sliced horizontally (using molecular weight standards as a guide) into multiple sections of equal size, and processed as described below.
In-gel Digestion/Peptide Extraction
Gel sections from the previous step were placed in 2.0-ml tubes (Axygen, Union City, CA) and moved into a dual isolation biosafety cabinet. Gel pieces were destained with two washes of 50% methanol, 5% acetic acid and rinsed with three alternating washes of 100 mm ammonium bicarbonate and 100% acetonitrile to remove the destain solution. After removal of the acetonitrile following the final wash, gel slices were dried for 10 min in a SpeedVac. Tubes containing the dried gel sections were placed on ice, and 200 μl of Promega sequencing grade trypsin at a concentration of 6.6 μg/ml in 50 mm ammonium bicarbonate was added to each sample. The gel pieces were allowed to swell for 60 min on ice after which the tubes were capped and incubated at 37 °C for 20 h. Peptides were extracted with two washes of 500 μl of 50 mm ammonium bicarbonate and two washes of 500 μl of 50% acetonitrile, 0.1% formic acid. All extracts were frozen at −80 °C, lyophilized to dryness, and redissolved in 60 μl of 5% acetonitrile, 0.1% formic acid. Samples were then loaded onto a 96-well plate for MS analysis (see below).
Mass Spectrometry
Samples for MS were run on either an LCQ DECA XP (LCQ) plus Proteome X work station as described in an earlier report from our group (10) or on a linear ion trap-Fourier transform mass spectrometer (LTQ-FT) from Thermo Finnigan. For each run using the LCQ, 10 μl of each reconstituted sample was injected with a Famos Autosampler, and the separation was done on a 75-μm (inner diameter) × 20-cm column packed with C18 media running at a flow rate of 250 nl/min provided from a Surveyor MS pump with a flow splitter with a gradient of 5−72% water, 0.1% formic acid, and 5% acetonitrile over the course of 240 min (4.0-h run). For each run using the LTQ-FT, 10 μl of each reconstituted sample was injected with a Famos Autosampler, and the separation was done on a 75-μm (inner diameter) × 20-cm column packed with C18 media running at a 225 nl/min flow rate provided from a Surveyor MS pump with a flow splitter with a gradient of 5−60% water, 0.1% formic acid, acetonitrile 0.1% formic acid over the course of 120 min (150-min total run). Between each set of samples, standards from a mixture of peptides, 5 Angiotensin (Michrom Bioresources), were run for 2.5 h to ascertain column performance and observe any potential carryover that might have occurred. The LCQ was run in a top five configuration with one MS scan and five MS/MS scans. Dynamic exclusion was set to 1 with a limit of 30 s. The LTQ-FT was run in a top nine configuration with one MS 200,000 resolution full scan and nine MS/MS scans. Dynamic exclusion was set to 1 with a limit of 180 s with early expiration set to two full scans.
Peptide identifications were made using SEQUEST (Thermo Finnigan) through the Bioworks Browser 3.2. Sequential database searches were performed using the E. coli O157:H7 strain EDL933 FASTA database from the European Bioinformatics Institute (www.ebi.ac.uk/newt/display) using static carbamidomethyl-modified cysteines and differential oxidized methionines. A reverse E. coli O157:H7 strain EDL933 FASTA database was spiked in to provide noise and determine validity of peptide hits. In this fashion, the statistical relevance of all the data could be determined in a sample- and mass spectrometer-independent manner (15). LCQ data were searched with a 2-dalton window on the MS precursor with 0.8 dalton on the fragment ions, whereas the FT data were searched at 5 ppm for the precursor ion and 0.5 Da on the fragment ions. Peptide score cutoff values were chosen at cross-correlation values (Xcorr) of 1.8 for singly charged ions, 2.5 for doubly charged ions, and 3.0 for triply charged ions along with delta rank scoring preliminary cutoff (ΔCn) values of 0.1, peptide probability values of 1.00E—3 and cross-correlation-normalized values (RSp) of <10. The cross-correlation values chosen for each peptide assured a high confidence match for the different charge states, whereas the ΔCn ensured the uniqueness of the peptide hit. The RSp value of 1 was used on single peptide hits to ensure that the peptide matched the top hit in the preliminary scoring. At these peptide filter values, very few single peptide reverse database hits were observed, allowing us to place a higher confidence on the few single peptide protein identifications. Single hit proteins were also manually validated to ensure relevance.
Cellular localization and putative functions of hypothetical proteins, identified by querying the E. coli O157:H7 strain EDL933 FASTA database at European Bioinformatics Institute with tandem MS data, were determined using bioinformatics as follows. (i) Cellular localization of such proteins was determined using the PSORTb Version 2.0/PSLpred at psort.org/ and PSORTdb at db.psort.org. (ii) Extracytoplasmic location was confirmed by examining the N terminus of amino acid sequences of cognate proteins from the O157 strain EDL933 database at www.tigr.org for the presence of a signal sequence using the program SignalP 3.0 at www.cbs.dtu.dk/services/SignalP. (iii) Putative functions were designated using the Conserved Domain Database (CDD) at www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi.
RESULTS AND DISCUSSION
To validate PELS (Fig. 1), we sought to rapidly define a protein subset constituting the O157 immunome in bovine reservoirs for future evaluation as candidates for a vaccine for elimination of this pathogen from the gastrointestinal tracts of cattle, a principal source of human infection. For this, we constructed and induced recombinant protein expression from genes on inserts within clones comprising an optimized O157 expression library using a range of IPTG concentrations followed by growth at different incubation temperatures for varying time intervals. To capture recombinant proteins contained in cell lysate and pellet fractions of library clones, we affinity-purified bait pAbs from pooled hyperimmune cattle sera previously generated against diverse O157 strains following confirmation of reactivity of this hyperimmune cattle serum pool against previously identified O157 antigens (Fig. 2, a and b). We then charged HiTrap NHS-activated columns by coupling to bait pAbs after which pooled cell lysate and pellet fractions from above were applied separately to charged columns. Specifically captured O157 recombinant proteins (Fig. 3, a–d) were identified by subjecting pooled elution fractions to GeLC-MS/MS and SEQUEST database searching (Table I and Supplemental Table 1).
Table I.
Previously reported adhesins of O157 and other pathogens identified by PELS
| Protein | Locus ID in O157 EDL 933a/Sakaib strains | Mass | No. peptide hits | Protein coverage | Contributes to adherence in | Ref. |
|---|---|---|---|---|---|---|
| kDa | % | |||||
| OmpA; outer membrane protein 3a (II*;G;d); adhesin | Z1307/Ecs1041 | 38 | 42 | 43 | O157 | 16 |
| Iha; adhesin; exogenous ferric siderophore receptor R4 | Z1178/Ecs1360 | 77 | 4 | 8 | O157, uropathogenic E. coli | 17, 18 |
PELS identified 207 proteins, comprising 3.8% of the proteome of the sequenced O157 strain EDL933 (11), as components of the immunome of this organism in bovine reservoirs (Table I and Supplemental Table 1). PELS was strongly validated by the fact that 35 of 207 (17%) proteins were also part of the O157 immunome in humans convalescing from extraintestinal O157 disease (10). Here we wish to emphasize that further experimentation is required to ascertain whether the rest of the PELS-identified proteins are unique to the O157 immunome in cattle. This is because the immunoproteome defined by IVIAT is a partial one in that it includes only proteins that are expressed either uniquely during human infection or at significantly higher levels in vivo than during in vitro growth because of a series of adsorptions of human convalescent sera against O157 grown in standard laboratory media (10). In contrast, the unadsorbed cattle hyperimmune sera used in PELS facilitated the definition of a more complete immunoproteome. Consequently, immunogenic O157 proteins expressed either equally well both in vitro and in vivo or at higher levels during laboratory growth than during human infection and are part of the PELS-identified protein repertoire remain excluded from the immunome identified by IVIAT. Further validation of PELS came from identification of previously identified adhesins of O157 (16, 17) and of other pathogens (18) (Table I) as well as several bovine colonization factors (19) (Supplemental Table 1); this is consistent with the fact that this human pathogen only colonizes the gastrointestinal tracts of cattle but does not cause disease in these reservoirs. Also validating PELS was the identification of two (EspB and EspP) of four secreted O157 proteins (EspA, EspB, Tir, and EspP) that are strongly immunogenic in cattle as constituents of an experimental vaccine (20). Parenteral administration of this vaccine formulated with an adjuvant reportedly results in a reduction in fecal shedding of O157 following oral challenge of bovine reservoirs (20). The reasons why EspA and Tir were not identified in the current study are unclear. Plausible explanations include the fact that such proteins might be (i) poorly expressed by wild-type O157 strains following oral inoculation and therefore engender suboptimal antibody responses in the bovine host and/or (ii) expressed at very low levels following induction by the E. coli host strain used in this study. As in the earlier report on the O157 immunome in humans identified by IVIAT (10), PELS identified outer membrane components of high affinity iron transport, bacteriophage proteins, biosynthetic and metabolic enzymes, and proteins involved in energy generation and anaerobic respiration in accordance with requirements for adaptation to the host environment, in vivo growth, and survival. PELS also identified several hypothetical and unknown proteins, including those unique to this study (Supplemental Table 1).
In conclusion, this report details a new application of proteomics and highlights the power of proteomics-based methodologies. A striking feature is the rapidity of proteome wide IMP identification, which renders PELS an ideal alternative/complement to emerging protein chip/array technologies. Other attractive features include broad applicability, robustness, and an elimination of subjective bias. Also because proteins are expressed from genes on inserts within clones of a genomic DNA expression library, a more comprehensive determination of the immunoproteome of the cognate pathogen is possible. Limitations relate to constraints on expression of heterologous proteins in E. coli hosts (1); however, improved prokaryotic cell-free in vitro translation systems coupled with the advent of technologies for facile generation of expression ORFeomes of microbes (21) should further enhance the power of this methodology.
Supplementary Material
Footnotes
The abbreviations used are: IMP, immunogenic microbial protein; PELS, proteomics-based expression library screening; IVIAT, in vivo induced antigen technology; pAb, polyclonal antibody; 1D, one-dimensional; GeLC-MS/MS, 1D SDS-PAGE ESI nano-LC-MS/MS; NHS, N-hydroxysuccinimide; IPTG, isopropyl β-d-thiogalactopyranoside; NOG, n-octyl β-d-glucopyranoside; HP, high performance.
This work was supported in part by National Institutes of Health Grants R21 AI055963 (to I. T. K.) and R01 DK52081 (to P. I. T.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Rollins SM, Peppercorn A, Hang L, Hillman JD, Calderwood SB, Handfield M, Ryan ET. In vivo induced antigen technology (IVIAT). Cell. Microbiol. 2005;7:1–9. doi: 10.1111/j.1462-5822.2004.00477.x. [DOI] [PubMed] [Google Scholar]
- 2.Etz H, Minh DB, Henics T, Dryla A, Winkler B, Triska C, Boyd AP, Sollner J, Schmidt W, von Ahsen U, Buschle M, Gill SR, Kolonay J, Khalak H, Fraser CM, von Gabain A, Nagy E, Meinke A. Identification of in vivo expressed vaccine candidate antigens from Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 2002;99:6573–6578. doi: 10.1073/pnas.092569199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahan MJ, Heithoff DM, Sinsheimer RL, Low DA. Assessment of bacterial pathogenesis by analysis of gene expression in the host. Annu. Rev. Genet. 2000;34:139–164. doi: 10.1146/annurev.genet.34.1.139. [DOI] [PubMed] [Google Scholar]
- 4.Xu Q, Dziejman M, Mekalanos JJ. Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc. Natl. Acad. Sci. U. S. A. 2003;100:1286–1291. doi: 10.1073/pnas.0337479100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li B, Jiang L, Song Q, Yang J, Chen Z, Guo Z, Zhou D, Du Z, Song Y, Wang J, Wang H, Yu S, Wang J, Yang R. Protein microarray for profiling antibody responses to Yersinia pestis live vaccine. Infect. Immun. 2005;73:3734–3739. doi: 10.1128/IAI.73.6.3734-3739.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramachandran N, Hainsworth E, Bhullar B, Eisenstein S, Rosen B, Lau AY, Walter JC, LaBaer J. Self-assembling protein microarrays. Science. 2004;305:86–90. doi: 10.1126/science.1097639. [DOI] [PubMed] [Google Scholar]
- 7.Hess JL, Blazer L, Romer T, Faber L, Buller RM, Boyle MD. Immunoproteomics. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2005;815:65–75. doi: 10.1016/j.jchromb.2004.07.047. [DOI] [PubMed] [Google Scholar]
- 8.Steen H, Mann M. The ABC's (and XYZ's) of peptide sequencing. Nat. Rev. Mol. Cell. Biol. 2004;5:699–711. doi: 10.1038/nrm1468. [DOI] [PubMed] [Google Scholar]
- 9.Sambrook J, Russel D. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- 10.John M, Kudva IT, Griffin RW, Dodson AW, McManus B, Krastins B, Sarracino D, Progulske-Fox A, Hillman JD, Handfield M, Tarr PI, Calderwood SB. Use of in vivo-Induced antigen technology for identification of Escherichia coli O157:H7 proteins expressed during human infection. Infect. Immun. 2005;73:2665–2679. doi: 10.1128/IAI.73.5.2665-2679.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perna NT, Plunkett G, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Posfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 12.Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T, Tanaka M, Tobe T, Iida T, Takami H, Honda T, Sasakawa C, Ogasawara N, Yasunaga T, Kuhara S, Shiba T, Hattori M, Shinagawa H. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 13.Ohnishi M, Tanaka C, Kuhara S, Ishii K, Hattori M, Kurokawa K, Yasunaga T, Makino K, Shinagawa H, Murata T, Nakayama K, Terawaki Y, Hayashi T. Chromosome of the enterohemorrhagic Escherichia coli O157:H7; comparative analysis with K-12 MG1655 revealed the acquisition of a large amount of foreign DNAs. DNA Res. 1999;6:361–368. doi: 10.1093/dnares/6.6.361. [DOI] [PubMed] [Google Scholar]
- 14.Stephenson HF. Calculations in Molecular Biology and Biotechnology—A Guide to Mathematics in the Laboratory. Academic Press; Boston, MA: 2005. pp. 208–212. [Google Scholar]
- 15.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 16.Torres AG, Kaper JB. Multiple elements controlling adherence of enterohemorrhagic Escherichia coli O157:H7 to HeLa cells. Infect. Immun. 2003;71:4985–4995. doi: 10.1128/IAI.71.9.4985-4995.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tarr PI, Bilge SS, Vary JC, Jr., Jelacic S, Habeeb RL, Ward TR, Baylor MR, Besser TE. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 2000;68:1400–1407. doi: 10.1128/iai.68.3.1400-1407.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson JR, Jelacic S, Schoening LM, Clabots C, Shaikh N, Mobley HLT, Tarr PI. The IrgA homologue adhesin Iha is an Escherichia coli virulence factor in murine urinary tract infection. Infect. Immun. 2005;73:965–971. doi: 10.1128/IAI.73.2.965-971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dziva F, van Diemen PM, Stevens MP, Smith AJ, Wallis TS. Identification of Escherichia coli O157:H7 genes influencing colonization of the bovine gastrointestinal tract using signature-tagged mutagenesis. Microbiology. 2004;150:3631–3645. doi: 10.1099/mic.0.27448-0. [DOI] [PubMed] [Google Scholar]
- 20.Potter AA, Klashinsky S, Li Y, Frey E, Townsend H, Rogan D, Erickson G, Hinkley S, Klopfenstein T, Moxley RA, Smith DR, Finlay BB. Decreased shedding of Escherichia coli O157:H7 by cattle following vaccination with type III secreted proteins. Vaccine. 2004;22:362–369. doi: 10.1016/j.vaccine.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 21.Brasch MA, Hartley JL, Vidal M. ORFeome cloning and systems biology: standardized mass production of the parts from the parts-list. Genome Res. 2004;14:2001–2009. doi: 10.1101/gr.2769804. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.