Abstract
The continued success of genome sequencing projects has resulted in a wealth of information, but 40-50% of identified genes correspond to hypothetical proteins or proteins of unknown function. The Functional Annotation Screening Technology by NMR (FAST-NMR) screen was developed to assign a biological function for these unannotated proteins with a structure solved by the Protein Structure Initiative. FAST-NMR is based on the premise that a biological function can be described by a similarity in binding sites and ligand interactions with proteins of known function. The resulting co-structure and functional assignment may provide a starting point for a drug discovery effort.
Keywords: NMR High-throughput Screens, NMR, Structural Biology, Protein Structure Initiative, Functional Genomics, Hypothetical Proteins
The completion of the human genome project is spurring tremendous progress in cell biology, development, evolution and physiology [1]. The expanding number of protein structures emerging from the Protein Structure Initiative (PSI) is contributing to these advancements [2]. As of January 2007, the sequencing of 607 genomes has been completed with 1676 ongoing projects. Also, nearly 2,500 protein structures have been solved by PSI [3,4]. Drug discovery is benefiting from these successes through the identification of novel therapeutic targets and the development of new tools to optimize chemical leads [5-7]. As an example, the identification of novel anti-infectious targets may aid in avoiding common mechanisms of resistance and extend the lifetime of new antibiotics [8,9].
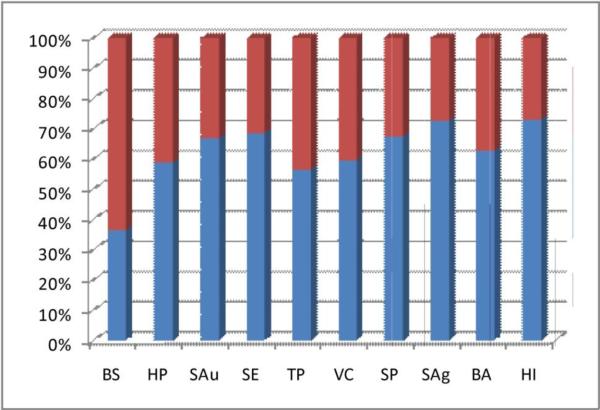
An underlying challenge to capitalizing on genome sequencing efforts is the abundance of hypothetical proteins, proteins that lack a functional annotation. Our recent analysis of various bacterial genomes from the August 2007 Gold release shows that, even with improved computational methods, approximately 40% of bacterial proteins have not been assigned to a functional category (Figure 1) [3]. There are more than 11,000 proteins from the ten bacterial organisms listed in Figure 1 that lack a functional annotation. Considering this list is only from a small segment of currently sequenced genomes, the prospect of obtaining experimental functional information for all hypothetical proteins identified from completed and ongoing sequencing efforts is a daunting proposition. Valuable information is hidden among this multitude of unannotated proteins that could be associated with cell viability, biofilm formation, infection, and pathogenesis. These proteins may provide key information for developing new antibiotics, where drug discovery efforts would benefit greatly from new functional annotations methodology.
Figure 1.
Functional analysis of bacterial genomes. The blue partitions are the percentage of proteins that have assigned functional categories; the red partitions are the percentage of unannotated proteins. Bacillus subtlus (BS); Heliobactor pylori (HP); Staphylococcus aureus (SAu); Staphylococcus epidermis (SE); Treponema pallidum (TP); Vibrio cholerae (VC); Streptococcus pneumoniae (SP); Streptococcus agalactiae (SAg); Bacillus anthracis (BA); Haemophilus influenzae (HI).
Most high-throughput experimental methods to assign function have focused primarily on generating knockout libraries to analyze cell phenotypes, monitoring changes in gene expression or determining protein interaction maps [10-12]. These methods generally do not provide functional information for a specific protein without additional detailed bioinformatics [13,14]. Global sequence similarity is routinely used to infer the function of hypothetical proteins, despite analysis that suggest error rates are as high as 30% [15,16]. Conversely, amino-acid residues associated with the active-sites and biological activities of proteins are stable evolutionarily relative to the remainder of the protein's sequence and provide an alternative approach for functional annotation [17,18]. A basic definition of biological function is derived from a protein's interaction with small molecules and other biomolecules. Thus, the identification of functional ligand(s), an active site and a corresponding protein-ligand co-structure is instrumental to defining a function for a hypothetical protein. The comparison and prediction of ligand binding sites from both structural and sequence information is a proven approach for functional assignments of proteins [19]; however, these predictions may lead to ambiguous or incorrect annotations [20,21]. A combination of experimental protein-ligand binding data with bioinformatic analysis will minimize the uncertainties commonly associated with pure computational approaches.
FAST-NMR (Functional Annotation Screening Technology by NMR) provides functional information for hypothetical proteins by experimentally characterizing and analyzing ligand binding sites [22]. Once the functional ligands [23] are identified and the binding site is located, a co-structure is obtained. A functional assignment is deduced by comparing the ligand-defined active-site from FAST-NMR to a database of protein-ligand binding sites for proteins of known function using CPASS (Comparison of Protein Active-Site Structures) [24]. The information obtained from FAST-NMR furthers our understanding of the basic biological role of hypothetical proteins and provides a potential starting point for drug discovery. A summary of some common applications for the functional annotation of proteins of unknown proteins and a comparison to our FAST-NMR method are listed in Table 1.
Table 1.
Summary of Applications for Protein Functional Annotation
| Method | Advantages/Disadvantages | Website |
|---|---|---|
| FAST-NMR [22] |
Advantages
Disadvantages
|
http://bionmr-c1.unl.edu |
| eF-seek [69] |
Advantages
Disadvantages
|
http://ef-site.hgc.jp/eF-seek |
| JAFA [70] |
Advantages
Disadvantages
|
http://jafa.burnham.org |
| PDB-UF [27] |
Advantages
Disadvantages
|
http://pdbuf.bioinfo.pl |
| ProcFunc [71] |
Advantages
Disadvantages
|
http://www.ebi.ac.uk/thornton-srv/databases/profunc |
| SuMo [72] |
Advantages
Disadvantages
|
http://sumo-pbil.ibcp.fr |
The FAST-NMR Method
FAST-NMR was developed to assign biological functions to hypothetical protein structures solved by PSI. Recent statistical analysis indicates that ~20-50% of protein structures determined by PSI may be amenable to analysis by NMR [25,26]. The Protein Structure Database (PDB) currently contains ~2,200 proteins of unknown function [27]. These proteins tend to be “orphaned” from any further functional analysis because of a complete absence of information to guide a research project [28-31]. These orphaned proteins are ideal targets for analysis by FAST-NMR. An assigned 1H-15N Heteronuclear single quantum coherence (HSQC) NMR spectrum and a corresponding structure for a protein of unknown function are the primary requirements for the FAST-NMR methodology. In general, high-resolution NMR structures and assignments can be routinely obtained for proteins <25 kDa using standard 13C and 15N protein labeling techniques [32,33]. This molecular-weight upper limit may be extended by upwards of 900 kDa [34-40] by the application of deuterium labeling, specific methyl labeling and Transverse Relaxation-Optimized NMR Spectroscopy (TROSY)-based experiments [41-43].
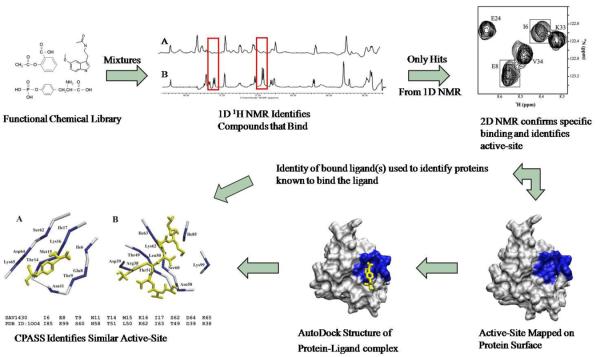
The FAST-NMR assay applies a tired approach to screening, where an overview of the methodology is illustrated in Figure 2. The first 1D 1H line-broadening (LB) experiments are amenable for screening a large number of compounds in a relatively short time (< 10 minutes/sample) and requiring a minimal amount of unlabeled protein material (< 0.1 mgs/sample). Only positive “hits” from the LB experiments are further screened in 2D 1H-15N HSQC spectra. In this manner, the tiered approach minimizes resources and increases throughput by funneling only the most promising candidates forward to the more resource intensive 2D 1H-15N HSQC experiments.
Figure 2.
Flow chart of FAST-NMR. The hypothetical proteins are screened against mixtures of ligands from the functional chemical library. Reference 1D 1H spectra of the mixtures are compared to those containing protein, where a hit is identified by changes in NMR line-width. Only the ligands identified as binding in the primary screen are further assayed in the secondary 2D 1H-15N experiment. Chemical shift changes confirm a specific interaction and identify the binding site from mapping of the CSPs on the protein's surface. The binding site and CSPs are utilized to determine a rapid co-structure using AutoDock. This co-structure is then used by CPASS to compare the ligand-defined binding site from the hypothetical protein to all other protein-ligand interactions present in the PDB. A general biological function can then be assigned based on an observed similarity to a ligand-defined binding site for a protein of known function.
In the first LB experiment, the formation of a protein-ligand complex can be determined by monitoring changes in the ligand's spectrum. Binding of the ligand to the protein causes line width broadening that may result in the complete disappearance of the ligand's NMR peak(s). This is caused by the large differences in molecular-weight and correlation time (τc, time it takes the molecule to rotate one radian) between the protein and small molecule. In the second NMR experiment, changes in the protein's NMR spectrum are followed to further confirm a specific interaction and identify a potential binding site. Ligand binding causes local environmental changes in the protein resulting in the observation of chemical shift perturbations (CSPs) in the 2D 1H-15N HSQC spectrum. Since each peak in the HSQC spectrum has been sequentially assigned to a specific amino acid in the protein's sequence, the CSPs can be mapped onto the surface of the protein. A consensus clustering of CSPs identifies the location of the ligand binding site. A lack of CSPs or a random distribution of CSPs over the protein's surface indicates non-specific binding of the ligand to the protein.
A rapid protein-ligand co-structure is determined by combining the experimental CSPs with molecular modeling. AutoDock [44] has been demonstrated to outperform two other well-known docking tools, FlexX and DOCK [45] in a virtual screen, and is currently the most cited of molecular modeling applications [46]. The experimental CSPs define a grid used by AutoDock to guide the ligand docking into the NMR defined binding site. Our AutoDockFilter (ADF) program is then used to filter the AutoDock conformers and select a pose that best fits the CSPs. In general, amino acid residues with the largest CSPs are expected to be closer to the bound ligand relative to residues with smaller CSPs. The best-structure is then used to determine a ligand-defined binding site for CPASS analysis. CPASS identifies ligand-defined binding sites for proteins of known function from a PDB derived database that best matches the sequence and structural details of the ligand-defined binding site determined by FAST-NMR. A similarity between these ligand-defined binding sites infers a function that can be assigned to the hypothetical protein.
Functional Chemical Library
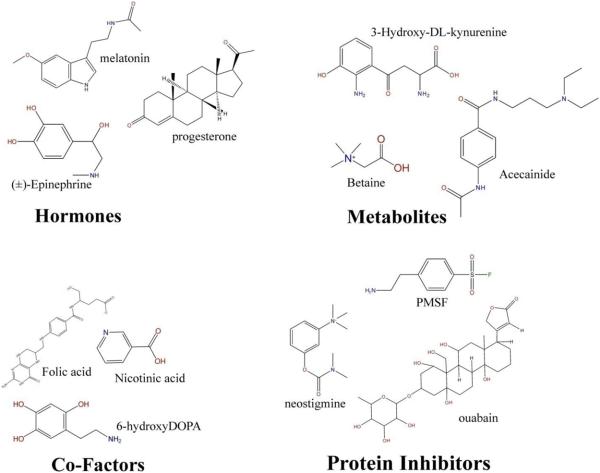
A critical component of the FAST-NMR methodology is the functional chemical library [47] (Figure 3). The library contains compounds with demonstrated protein affinity and, as completely as possible, covers the diversity of biological activity. The library is screened for binders by NMR to probe for protein function and includes known drugs, inhibitors and protein substrates that target a diverse set of protein functions covering a range of structural chemical classes. This list includes amino-acids, carbohydrates, co-factors, fatty-acids, hormones, metabolites, neurotransmitters, nucleic acids and vitamins. Importantly, the distribution of chemical and physical properties for the compounds in the functional library is similar to drug-like molecules (MW < 500 Da, number of heteroatoms < 10, cLogP < 5, and number of rings < 2). This indicates that compounds identified as binders in a FAST-NMR screen may also be useful starting points for structure-based drug design, if the hypothetical protein is identified as a valuable therapeutic target.
Figure 3.
Functional chemical library. A subset of compounds from four different functional categories from the functional chemical library is displayed. Proteins are screened against mixtures of compounds, and these mixtures were designed to have diverse structure and function to minimize spectral and functional overlap.
The design of the functional chemical library was optimized for screening by NMR. The compounds are soluble in water, do not aggregate, precipitate or react in mixtures and have unique NMR resonances for ready identification that avoids deconvolution. The selection process for the compounds in the functional library was extensive and based on a number of criteria: known biological activity, the existence of a co-structure in the Protein Database (PDB) [48], the likelihood of aqueous solubility (≥ 100μM), purity (≥90%), commercial availability (≥ 5mg) and cost (≤ $32). Each of the compounds are dissolved in D6-DMSO or D2O and stored in 96-well plates in a dessicator in a −80°C freezer. Compounds are screened in mixtures to further increase throughput while minimizing resources. The mixtures were designed to contain 3-4 compounds based on our statistical analysis of the optimal mixture size for NMR screens [49].
CPASS
CPASS incorporates a computer program with a structural database to compare ligand binding sites and provide a putative function for hypothetical proteins screened by FAST-NMR [24]. CPASS is a structure-based functional annotation program that differs from a variety of 3D template or sequence-based annotation programs (for a review see Watson et al. (2005) [50]) routinely used to predict ligand binding sites and protein function. These programs attempt to predict the location of ligand binding sites using various sequence and structure heuristics. Instead, CPASS aligns experimentally determined ligand-defined binding sites from FAST-NMR and the PDB using sequence and structural descriptors. Simply, CPASS identifies matches between functionally relevant ligand-binding sites to leverage an annotation.
CPASS may also aid the development of selective chemical leads. Drug toxicity is a common cause of clinical failures [51], where this toxicity is associated with non-specific in vivo protein activity [52]. In practice, it is not possible to screen against every potential protein target that may bind a chemical lead. Instead, a small panel of homologous proteins is used in secondary assays to infer compound specificity. The proteins are generally selected based on global sequence similarity to the protein target of interest. Unfortunately, there are also other proteins that share a high similarity in the ligand binding site that may lack global sequence similarity. Our previous CPASS analysis of ATP binding proteins indicates a significant cluster of proteins with sequence similarity < 20% that had high CPASS similarity of > 40% [24]. Similarly, we identified two alanine racemases that share only an 8% sequence similarity, but had essentially identical PLP binding sites. Clearly, proteins that share high ligand binding site similarity, but lack global sequence similarity pose serious risks of causing toxic side-effects in clinical trials unless identified using applications like CPASS.
Protein-Ligand Database
The CPASS database is continuously updated from the PDB and contains proteins in complex with small molecules, peptides, and oligonucleotides. Proteins may bind one ligand, multiple ligands, or the same ligand more than once. Each unique ligand-binding site (< 80% sequence similarity, distinct ligand) is incorporated into the CPASS database. There are ~55,000 protein-ligand binding sites currently present in the PDB, where ~21,000 are unique. These ligand-defined binding sites include all the amino acids in the protein sequences that have at least one atom within 6 Å of any atom of the ligand. Both the structure coordinates and the sequence identity are then used in a comparison with ligand-defined binding sites from other proteins. The ligand structure is not included in the ligand-defined binding site, but is used to classify the type of binding site (i.e. ATP binding site, FAD binding site, etc).
Similarity Scoring Function
Although the CPASS program will allow the user to search binding-sites based on the type of ligand that defines the binding site, it is not required. The comparison can be made against all ligand-defined binding sites present in the CPASS database or any ligand-type subset. The CPASS scoring function is based on the simultaneous structure and sequential alignments of two ligand-defined binding sites. A BLOSUM62 probability function weighted by root-mean square distance (rmsd) is used to compare the similarity of spatially aligned residues:
Active site a contains n residues and is compared to active site b of m residues from the CPASS database. pi,j is the BLOSUM62 probability for replacement of amino acid i from active site a with residue j from active site b, Δrmsdi,j is the corrected root-mean-square-difference in the Cα positions between the residues i and j, and dmin/di is the ratio of the shortest distance to an atom in the ligand from any atom in the residue i. This last term minimizes boundary effects. Small structural changes may result in residues entering or leaving the 6Å cut-off used to define a ligand-defined binding site. This may result in relatively large changes in the scoring function due to modest structural fluctuations.
The similarities between the active sites are then calculated by:
where S is the similarity score, Sab is the similarity score for the protein target against an active site from the CPASS database, and Saa is the similarity of the active site compared to itself used for normalization. In effect, a percent similarity is determined based on how well the sequence and structures of the two ligand-binding sites overlap. The scoring function is not symmetrical since it depends on the size of the binding site.
CPASS Functional Prediction of Hypothetical Proteins
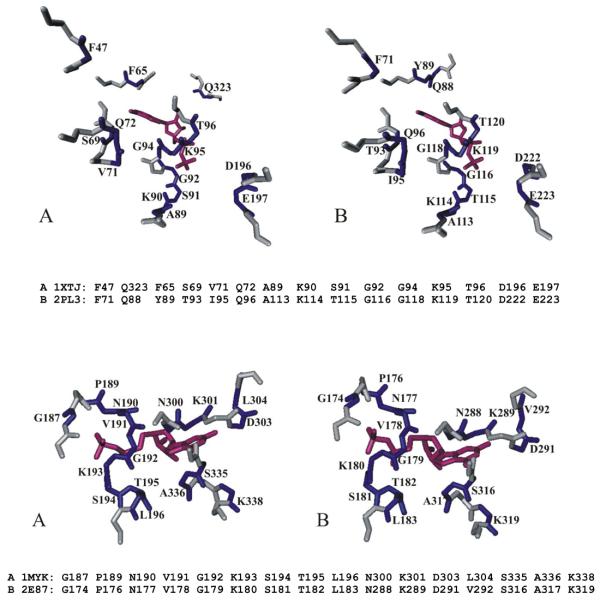
To illustrate further the utility of CPASS, a recent protein deposited in the PDB was chosen that only had a putative functional annotation. A human protein (PDB-ID 2PL3) was tentatively assigned as a probable ATP-dependent RNA helicase DDX10 and the structure contained a bound ADP molecule. CPASS analysis identified PDB-ID 2OXC as having the highest similarity (56.26%). Both proteins bind ADP and are hypothetical DEAD domains. The highest CPASS similarity score (50.30%) to a protein of known function was to PDB-ID 1XTJ, a DECD to DEAD mutation of human UAP56, which is also in complex with ADP [53]. Recently, the UAP56 protein has been shown experimentally to exhibit RNA-stimulated ATPase activity and ATP-dependent RNA helicase activity [54]. Thus, the CPASS analysis supports the prior putative assignments of hypothetical proteins 2PL3 and 2OXC as ATP-dependent RNA helicases. The top panel in Figure 4 shows the alignment of the ADP binding sites for the 2PL3 and 1XTJ structures. This figure clearly highlights the overall similarity in the structure and sequence alignments for the ADP binding sites.
Figure 4.
(top panel) Binding sites of 1XTJ and 2PL3. Binding-site residues for proteins (A) 1XTJ and (B) 2PL3. Residues within 6 Å of ADP are colored blue and the ligand ADP is colored pink. The amino acid alignment for the ADP binding sites is shown at the bottom of the figure. (bottom panel) Binding sites of 1MKY and 2E87. Binding-site residues for proteins (A) 1MKY and (B) 2E87. Residues within 6 Å of GDP are colored blue and the ligand GDP is colored pink. The amino acid alignment for the GDP binding sites is shown at the bottom of the figure.
A crystal structure of hypothetical protein PH1320 from Pyrococcus horikoshii OT3 was recently released by the PDB (PDB-ID 2E87). The protein is complexed to guanosine-5′-diphosphate (GDP), but completely lacks a functional assignment and a paper describing the structure has yet to be published. A CPASS analysis using only proteins complexed to GDP indicates hypothetical protein PH1320 has a very high similarity (70.47%) to an Escherichia coli elongation factor Der (PDB-ID 1MKY), an EngA homolog [55]. The bottom panel in figure 4 clearly demonstrates the high overall similarity in the structure and sequence alignments for the GDP binding sites between these two proteins. Hypothetical protein PH1320 shows CPASS similarity scores of 50-70% to EngB, EngC, EI-F2γ, EI-F5B, EF-Tu, EF-1α, EF-2 and EF-G, which are also members of the elongation factor super family. Hypothetical protein PH1320 also exhibits a slightly smaller similarity (50-60%) to Arf, Sar and Rab, members of the small GTPase super family that regulate a diverse range of cellular events [56]. Thus, the CPASS results suggest PH1320 is probably an elongation factor or potentially involved in GTP signal regulation similar to either Arf, Sar or Rab.
Functional Annotation of Staphylococcus aureus Protein SAV1430
Staphylococcus aureus protein SAV1430, a hypothetical protein of unknown function, was selected to demonstrate the FAST-NMR methodology (Figure 2). SAV1430 is a typical target of the NorthEast Structural Genomic Consortium (NESG) [29], where a structure was previously determined [57,58]. A Dali analysis suggested that SAV1430 has a similar topology to a ferredoxin-like fold, but the Z-score of < 3 was insignificant [59]. The only proteins that had any significant sequence homology to SAV1430 were other hypothetical proteins, so a reliable function could not be assigned based on structure homology alone.
O-phospho-L-tyrosine (pTyr) was identified as one of 21 compounds that exhibited line-broadening and chemical shift perturbations in the FAST-NMR screen with SAV1430. The other compounds are chemically similar to pTyr and were all shown to interact in a consensus binding-site that comprises residues I6-P10, T14-K16 and I61-V63. This binding site contains a shallow cleft on the SAV1430 surface surrounded by relatively flat structural features strongly suggestive of a protein-protein interaction site. A rapid structure of the pTyr-SAV1430 complex was determined using CSPs and AutoDock for CPASS analysis.
CPASS identified PDB ID 1oo4 as a significant hit (37% similarity), a Src SH2 domain complexed with a pTyr containing peptide. SH2 domains are typically part of multi-domain proteins involved in cell signaling and form a protein-protein complex with a kinase after phosphorylation of a tyrosine [60]. Phosphorylation of Ser, Thr and Tyr are also common mechanisms for regulating protein activity in bacteria [61,62]. The similarity in the characteristics of the SAV1430 and Src SH2 ligand binding sites, and the fact that SAV1430 binds pTyr, further supports the general proposal that SAV1430 functions by forming a protein-protein complex.
Rosetta Stone [63] analysis suggests hypothetical protein SAV0936 may be a binding partner of SAV1430. SAV0936 exhibits 47% sequence identity with the N-terminal region of the C-terminal NifU domain. NifU is a multi-protein complex that is a critical component of the [Fe-S] cluster assembly pathway [64-66] and is essential for the viability of bacteria [67]. A more exhaustive sequence analysis of SAV1430, based on the results with SAV0936, indicates the protein shares ~30% sequence identity with the C-terminal region of the C-terminal domain of the NifU multi-domain structure. These results imply that SAV1430 may interact with SAV0936 to form a complex that exhibits similar activity as the full length NifU domain or may regulate NifU activity. Thus, inhibiting the SAV1430-SAV0936 complex formation may represent a novel target for developing next generation antibiotics.
Conclusion
FAST-NMR provides a high-throughput approach to obtain functional assignments for hypothetical proteins, based on experimentally-determined protein-ligand interactions. FAST-NMR also addresses the current lack in high-throughput experimental methods to obtain functional information [68] where current methods [14] primarily rely on sequence similarity to confer function despite error rates as high as 30% [15,16]. FAST-NMR is based on basic tenets of biochemistry where detailed structural information of a protein-ligand interaction is paramount for understanding the function of a protein. Active-site residues are evolutionarily stable relative to the remainder of the protein's sequence decreasing the likelihood of annotation errors [17,18]. FAST-NMR compliments the success of the Human Genome Project and the Protein Structure Initiative by providing a means to annotate functionally the rapidly expanding number (~2,200) of hypothetical proteins currently deposited in the PDB [31]. Understanding protein function is a paramount necessity for drug discovery programs ability to make successful contributions to human health issues.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Teaser: FAST-NMR is a high-throughput method to determine biological function of hypothetical proteins identified from the Protein Structure Initiative as a means to further biological knowledge and drug discovery efforts.
References
- 1.Venter C, et al. The Sequence of the Human Genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 2.Burley SK. An overview of structural genomics. Nat. Struct. Biol. 2000;7(Suppl):932–934. doi: 10.1038/80697. [DOI] [PubMed] [Google Scholar]
- 3.Bernal A, et al. Genomes online database (GOLD): a monitor of genome projects world-wide. Nucleic Acids Research. 2001;29(1):126–127. doi: 10.1093/nar/29.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Todd AE, et al. Progress of structural genomics initiatives: an analysis of solved target structures. Journal of Molecular Biology. 2005;348(5):1235–1260. doi: 10.1016/j.jmb.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 5.Sioud M. Main approaches to target discovery and validation. Methods in Molecular Biology (Totowa, NJ, United States) 2007;360:1–12. doi: 10.1385/1-59745-165-7:1. Target Discovery and Validation, Volume 1. [DOI] [PubMed] [Google Scholar]
- 6.Zheng CJ, et al. Therapeutic targets: progress of their exploration and investigation of their characteristics. Pharmacological Reviews. 2006;58(2):259–279. doi: 10.1124/pr.58.2.4. [DOI] [PubMed] [Google Scholar]
- 7.Manning AM. Impact of the human genome on the discovery of immune-modulatory therapeutics. Current Opinion in Investigational Drugs (Thomson Scientific) 2006;7(5):406–411. [PubMed] [Google Scholar]
- 8.Schrenzel J, et al. A randomized clinical trial to compare fleroxacin-rifampicin with flucloxacillin or vancomycin for the treatment of staphylococcal infection. Clinical Infectious Diseases. 2004;39(9):1285–1292. doi: 10.1086/424506. [DOI] [PubMed] [Google Scholar]
- 9.Natsch S, et al. Guidelines for the prevention of antimicrobial resistance in hospitals. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 1998;26(6):1482–1483. doi: 10.1086/517655. [DOI] [PubMed] [Google Scholar]
- 10.Lee Y-H, et al. Gene knockdown by large circular antisense for high-throughput functional genomics. Nature Biotechnology. 2005;23(5):591–599. doi: 10.1038/nbt1089. [DOI] [PubMed] [Google Scholar]
- 11.Michiels F, et al. One step further towards real high throughput functional genomics. Trends in Biotechnology. 2003;21(4):147–148. doi: 10.1016/S0167-7799(03)00029-5. [DOI] [PubMed] [Google Scholar]
- 12.Tucker CL. High-throughput cell-based assays in yeast. Drug Discovery Today. 2002;7(18 Suppl):S125–S130. doi: 10.1016/s1359-6446(02)02409-1. [DOI] [PubMed] [Google Scholar]
- 13.del Val C, et al. High-throughput protein analysis integrating bioinformatics and experimental assays. Nucleic Acids Research. 2004;32(2):742–748. doi: 10.1093/nar/gkh257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joshi T, et al. Genome-scale gene function prediction using multiple sources of high-throughput data in yeast Saccharomyces cerevisiae. Omics. 2004;8(4):322–333. doi: 10.1089/omi.2004.8.322. [DOI] [PubMed] [Google Scholar]
- 15.Rost B. Enzyme function less conserved than anticipated. Journal of Molecular Biology. 2002;318(2):595–608. doi: 10.1016/S0022-2836(02)00016-5. [DOI] [PubMed] [Google Scholar]
- 16.Devos D, Valencia A. Intrinsic errors in genome annotation. Trends in Genetics. 2001;17(8):429–431. doi: 10.1016/s0168-9525(01)02348-4. [DOI] [PubMed] [Google Scholar]
- 17.Zvelebil MJ, et al. Prediction of protein secondary structure and active sites using the alignment of homologous sequences. Journal of Molecular Biology. 1987;195(4):957–961. doi: 10.1016/0022-2836(87)90501-8. [DOI] [PubMed] [Google Scholar]
- 18.Livingstone CD, Barton GJ. Protein sequence alignments: a strategy for the hierarchical analysis of residue conservation. CABIOS, Computer Applications in the Biosciences. 1993;9(6):745–756. doi: 10.1093/bioinformatics/9.6.745. [DOI] [PubMed] [Google Scholar]
- 19.Campbell SJ, et al. Ligand binding: functional site location, similarity and docking. Current Opinion in Structural Biology. 2003;13(3):389–395. doi: 10.1016/s0959-440x(03)00075-7. [DOI] [PubMed] [Google Scholar]
- 20.Greaves R, Warwicker J. Active Site Identification through Geometry-based and Sequence Profile-based Calculations: Burial of Catalytic Clefts. Journal of Molecular Biology. 2005;349(3):547–557. doi: 10.1016/j.jmb.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 21.Bateman A, Birney E. Searching databases to find protein domain organization. Advances in Protein Chemistry. 2000;54:137–157. doi: 10.1016/s0065-3233(00)54005-4. Analysis of Amino Acid Sequences. [DOI] [PubMed] [Google Scholar]
- 22.Mercier KA, et al. FAST-NMR: Functional Annotation Screening Technology Using NMR Spectroscopy. Journal of the American Chemical Society. 2006;128(47):15292–15299. doi: 10.1021/ja0651759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mercier KA, et al. Design and characterization of a functional library for NMR screening against novel protein targets. Comb Chem High Throughput Screen FIELD Full Journal Title:Combinatorial chemistry & high throughput screening. 2006;9(7):515–534. doi: 10.2174/138620706777935342. [DOI] [PubMed] [Google Scholar]
- 24.Powers R, et al. Comparison of protein active site structures for functional annotation of proteins and drug design. Proteins: Structure, Function, and Bioinformatics. 2006;65(1):124–135. doi: 10.1002/prot.21092. [DOI] [PubMed] [Google Scholar]
- 25.Snyder DA, et al. Comparisons of NMR Spectral Quality and Success in Crystallization Demonstrate that NMR and X-ray Crystallography Are Complementary Methods for Small Protein Structure Determination. Journal of the American Chemical Society. 2005;127(47):16505–16511. doi: 10.1021/ja053564h. [DOI] [PubMed] [Google Scholar]
- 26.Yee AA, et al. NMR and X-ray Crystallography, Complementary Tools in Structural Proteomics of Small Proteins. Journal of the American Chemical Society. 2005;127(47):16512–16517. doi: 10.1021/ja053565+. [DOI] [PubMed] [Google Scholar]
- 27.von Grotthuss M, et al. PDB-UF: database of predicted enzymatic functions for unannotated protein structures from structural genomics. BMC Bioinformatics. 2006;7 doi: 10.1186/1471-2105-7-53. No pp given. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lattman E. The state of the Protein Structure Initiative. Proteins: Structure, Function, and Bioinformatics. 2004;54(4):611–615. doi: 10.1002/prot.20000. [DOI] [PubMed] [Google Scholar]
- 29.Anon The protein target list of the Northeast Structural Genomics Consortium. Proteins: Structure, Function, and Bioinformatics. 2004;56(2):181–187. doi: 10.1002/prot.20091. [DOI] [PubMed] [Google Scholar]
- 30.Bonanno JB, et al. New York-structural genomiX research consortium (NYSGXRC): A large scale center for the protein structure initiative. Journal of Structural and Functional Genomics. 2005;6(23):225–232. doi: 10.1007/s10969-005-6827-0. [DOI] [PubMed] [Google Scholar]
- 31.von Grotthuss M, et al. PDB-UF: database of predicted enzymatic functions for unannotated protein structures from structural genomics. BMC Bioinformatics. 2006;7(1):53. doi: 10.1186/1471-2105-7-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lian L-Y, Middleton DA. Labeling approaches for protein structural studies by solution-state and solid-state NMR. Progress in Nuclear Magnetic Resonance Spectroscopy. 2001;39(3):171–190. [Google Scholar]
- 33.Ferentz AE, Wagner G. NMR spectroscopy: a multifaceted approach to macromolecular structure. Quarterly Reviews of Biophysics. 2000;33(1):29–65. doi: 10.1017/s0033583500003589. [DOI] [PubMed] [Google Scholar]
- 34.Sprangers R, et al. Quantitative NMR spectroscopy of supramolecular complexes: Dynamic side pores in ClpP are important for product release. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(46):16678–16683. doi: 10.1073/pnas.0507370102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jain NU, et al. Rapid Analysis of Large Protein-Protein Complexes Using NMR-derived Orientational Constraints: The 95kDa Complex of LpxA with Acyl Carrier Protein. Journal of Molecular Biology. 2004;343(5):1379–1389. doi: 10.1016/j.jmb.2004.08.103. [DOI] [PubMed] [Google Scholar]
- 36.Tugarinov V, et al. Four-Dimensional NMR Spectroscopy of a 723-Residue Protein: Chemical Shift Assignments and Secondary Structure of Malate Synthase G. Journal of the American Chemical Society. 2002;124(34):10025–10035. doi: 10.1021/ja0205636. [DOI] [PubMed] [Google Scholar]
- 37.Tugarinov V, Kay LE. Quantitative 13C and 2H NMR Relaxation Studies of the 723-Residue Enzyme Malate Synthase G Reveal a Dynamic Binding Interface. Biochemistry. 2005;44(49):15970–15977. doi: 10.1021/bi0519809. [DOI] [PubMed] [Google Scholar]
- 38.Peterson FC, Gettins PGW. Insight into the mechanism of serpin-proteinase inhibition from 2D [1H-15N] NMR studies of the 69 kDa a1-proteinase inhibitor Pittsburgh-trypsin covalent complex. Biochemistry. 2001;40(21):6284–6292. doi: 10.1021/bi010100x. [DOI] [PubMed] [Google Scholar]
- 39.Liu D, et al. Backbone resonance assignments of the 45.3 kDa catalytic domain of human BACE1. Journal of Biomolecular NMR. 2004;29(3):425–426. doi: 10.1023/B:JNMR.0000032509.81283.d3. [DOI] [PubMed] [Google Scholar]
- 40.Revington M, Zuiderweg ERP. Letter to the editor: TROSY-driven NMR backbone assignments of the 381-residue nucleotide-binding domain of the Thermus thermophilus DnaK molecular chaperone. Journal of Biomolecular NMR. 2004;30(1):113–114. doi: 10.1023/B:JNMR.0000042961.48233.f9. [DOI] [PubMed] [Google Scholar]
- 41.Riek R, et al. TROSY and CRINEPT: NMR with large molecular and supramolecular structures in solution. Trends Biochem. Sci. 2000;25(10):462–468. doi: 10.1016/s0968-0004(00)01665-0. [DOI] [PubMed] [Google Scholar]
- 42.Gardner KH, Kay LE. The use of 2H, 13C, 15N multidimensional NMR to study the structure and dynamics of proteins. Annu. Rev. Biophys. Biomol. Struct. 1998;27:357–406. doi: 10.1146/annurev.biophys.27.1.357. [DOI] [PubMed] [Google Scholar]
- 43.Kay LE. The development of NMR methods to study protein structure and dynamics. NATO ASI Ser., Ser. C. 1998;510:285–293. ew Methods for the Study of Biomolecular Complexes. [Google Scholar]
- 44.Morris GM, et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. Journal of Computational Chemistry. 1998;19(14):1639–1662. [Google Scholar]
- 45.Park H, et al. Critical assessment of the automated AutoDock as a new docking tool for virtual screening. Proteins: Structure, Function, and Bioinformatics. 2006;65(3):549–554. doi: 10.1002/prot.21183. [DOI] [PubMed] [Google Scholar]
- 46.Sousa SF, et al. Protein-ligand docking: current status and future challenges. Proteins: Structure, Function, and Bioinformatics. 2006;65(1):15–26. doi: 10.1002/prot.21082. [DOI] [PubMed] [Google Scholar]
- 47.Mercier KA, et al. Design and characterization of a functional library for NMR screening against novel protein targets. Combinatorial Chemistry & High Throughput Screening. 2006;9(7):515–534. doi: 10.2174/138620706777935342. [DOI] [PubMed] [Google Scholar]
- 48.Berman HM, et al. The Protein Data Bank. Nucleic acids research. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mercier KA, Powers R. Determining the optimal size of small molecule mixtures for high throughput NMR screening. Journal of Biomolecular NMR. 2005;31(3):243–258. doi: 10.1007/s10858-005-0948-4. [DOI] [PubMed] [Google Scholar]
- 50.Watson JD, et al. Prediction protein function from sequence and structural data. Curr. Opin. Struct. Biol. 2005;15:275–284. doi: 10.1016/j.sbi.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 51.Kubinyi H. Opinion: Drug research: myths, hype and reality. Nature Reviews Drug Discovery. 2003;2(8):665–668. doi: 10.1038/nrd1156. [DOI] [PubMed] [Google Scholar]
- 52.Ekins S. Predicting undesirable drug interactions with promiscuous proteins in silico. Drug Discovery Today. 2004;9(6):276–285. doi: 10.1016/S1359-6446(03)03008-3. [DOI] [PubMed] [Google Scholar]
- 53.Shi H, Cordin O, Minder CM, Linder P, Xu R-M. Crystal structure of the human ATP-dependent splicing and export factor UAP56. Proceeds of the National Academy of Sciences. 2004;101:17628–17633. doi: 10.1073/pnas.0408172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen J, et al. Biochemical Characterization of the ATPase and Helicase Activity of UAP56, an Essential Pre-mRNA Splicing and mRNA Export Factor. Journal of Biological Chemistry. 2007;282(31):22544–22550. doi: 10.1074/jbc.M702304200. [DOI] [PubMed] [Google Scholar]
- 55.Robinson VL, et al. Domain Arrangement of Der, a Switch Protein Containing Two GTPase Domains. Structure (Cambridge, MA, United States) 2002;10(12):1649–1658. doi: 10.1016/s0969-2126(02)00905-x. [DOI] [PubMed] [Google Scholar]
- 56.Wennerberg K, et al. The Ras superfamily at a glance. Journal of Cell Science. 2005;118(5):843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 57.Baran MC, et al. Solution Structure Determination of the Staphylococcus Aureus Hypothetical Protein SAV1430. Northeast Structure Consortium target ZR18. 5844. Department of Biochemistry, University of Wisconson-Madison; 2003. [Google Scholar]
- 58.Baran MC, et al. Solution Structure of the Hypothetical Staphylococcus Aureus protein SAV1430. Northest Strucutral Genomics Consortium target ZR18. 2003 [Google Scholar]
- 59.Dietmann S, et al. A fully automatic evolutionary classification of protein folds: Dali Domain Dictionary version 3. Nucleic Acids Research. 2001;29(1):55–57. doi: 10.1093/nar/29.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marengere LEM, Pawson T. Structure and function of SH2 domains. Journal of Cell Science, Supplement. 1994;18:97–104. doi: 10.1242/jcs.1994.supplement_18.14. Cell Biology of Cancer. [DOI] [PubMed] [Google Scholar]
- 61.Kennelly PJ, Potts M. Fancy meeting you here! A fresh look at “prokaryotic” protein phosphorylation. Journal of Bacteriology. 1996;178(16):4759–4764. doi: 10.1128/jb.178.16.4759-4764.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alzari PM. First Structural Glimpse at a Bacterial Ser/Thr Protein Phosphatase. Structure (Cambridge, MA, United States) 2004;12(11):1923–1924. doi: 10.1016/j.str.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 63.Suhre K, Claverie J-M. FusionDB: a database for in-depth analysis of prokaryotic gene fusion events. Nucleic Acids Research. 2004;32:D273–D276. doi: 10.1093/nar/gkh053. Database. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Frazzon J, Dean DR. Formation of iron-sulfur clusters in bacteria: an emerging field in bioinorganic chemistry. Current Opinion in Chemical Biology. 2003;7(2):166–173. doi: 10.1016/s1367-5931(03)00021-8. [DOI] [PubMed] [Google Scholar]
- 65.Dos Santos PC, et al. Iron-Sulfur Cluster Assembly: NifU-directed activation of the nitrogenase Fe protein. Journal of Biological Chemistry. 2004;279(19):19705–19711. doi: 10.1074/jbc.M400278200. [DOI] [PubMed] [Google Scholar]
- 66.Dos Santos PC, et al. Formation and Insertion of the Nitrogenase Iron-Molybdenum Cofactor. Chemical Reviews (Washington, DC, United States) 2004;104(2):1159–1173. doi: 10.1021/cr020608l. [DOI] [PubMed] [Google Scholar]
- 67.Olson JW, et al. Characterization of the NifU and NifS Fe-S Cluster Formation Proteins Essential for Viability in Helicobacter pylori. Biochemistry. 2000;39(51):16213–16219. doi: 10.1021/bi001744s. [DOI] [PubMed] [Google Scholar]
- 68.Flook PK, et al. Target validation through high throughput proteomics analysis. Targets. 2003;2(5):217–223. [Google Scholar]
- 69.Kinoshita K, et al. eF-seek: prediction of the functional sites of proteins by searching for similar electrostatic potential and molecular surface shape. Nucleic Acids Res. 2007;35:W398–402. doi: 10.1093/nar/gkm351. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Friedberg I, et al. JAFA: a protein function annotation meta-server. Nucleic Acids Res. 2006;34:W379–381. doi: 10.1093/nar/gkl045. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Laskowski RA, et al. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. 2005;33:W89–93. doi: 10.1093/nar/gki414. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jambon M, et al. The SuMo server: 3D search for protein functional sites. Bioinformatics. 2005;21(20):3929–3930. doi: 10.1093/bioinformatics/bti645. [DOI] [PubMed] [Google Scholar]