Abstract
Active macromolecular transport between the nucleus and cytoplasm proceeds through nuclear pore complexes and is mostly mediated by transport receptors of the importin β-superfamily. Here we identify exportin 6 (Exp6) as a novel family member from higher eukaryotes and show that it mediates nuclear export of profilin·actin complexes. Exp6 appears to contact primarily actin, but the interaction is greatly enhanced by the presence of profilin. Profilin thus functions not only as the nucleotide exchange factor for actin, but can also be regarded as a cofactor of actin export and hence as a suppressor of actin polymerization in the nucleus. Even though human and Drosophila Exp6 share only ∼20% identical amino acid residues, their function in profilin·actin export is conserved. A knock-down of Drosophila Exp6 by RNA interference abolishes nuclear exclusion of actin and results in the appearance of nuclear actin paracrystals. In contrast to a previous report, we found no indications of a major and direct role for CRM1 in actin export from mammalian or insect nuclei.
Keywords: actin/exportin/nuclear pore complex/profilin/RanGTPase
Introduction
The nuclear envelope separates eukaryotic cells into nuclear and cytoplasmic compartments. The nucleus is specialized in DNA replication and transcription, RNA processing and assembly of ribonucleoprotein particles (RNPs), while the cytoplasm harbours the translation machinery, for example, and most cytoskeletal elements such as microtubules and actin filaments. This compartmentalization certainly provides many advantages, but also necessitates nucleocytoplasmic transport: the nucleus lacks protein synthesis and therefore has to import each and every protein from the cytoplasm. Conversely, the cytoplasm relies on a nuclear supply of tRNAs, mRNA and ribosomes.
All nucleocytoplasmic exchange proceeds through nuclear pore complexes (NPCs), which permit passage of material through two modes: passive diffusion and facilitated translocation (reviewed in Mattaj and Englmeier, 1998; Görlich and Kutay, 1999; Conti and Izaurralde, 2001; Weis, 2003). Passive diffusion is fast for small molecules, but becomes increasingly inefficient as the mass of the diffusing species approaches a limit of 20–40 kDa. In contrast, facilitated translocation can accommodate the transport of even very large objects. It is often coupled to an input of metabolic energy (active transport), which permits import or export cargoes to accumulate even against steep gradients of chemical activity (Görlich et al., 2003).
Most of these active nuclear transport pathways are mediated by nuclear transport receptors of the importin β-superfamily (reviewed in Wozniak et al., 1998; Görlich and Kutay, 1999; Imamoto, 2000; Macara, 2001; Strom and Weis, 2001). These receptors circulate between nucleus and cytoplasm, recognize cargo molecules and transfer them from one side of the nuclear envelope to the other. To achieve unidirectional transport, positional information is provided by a RanGTP gradient across the nuclear membrane (Görlich et al., 1996; Izaurralde et al., 1997; Kalab et al., 2002; Smith et al., 2002; Görlich et al., 2003). Nuclear transport receptors are RanGTP-binding proteins and respond to this gradient by loading and unloading their cargo in the appropriate compartment. Nuclear import mediators (importins) recruit cargo at low RanGTP levels in the cytoplasm and release it upon RanGTP binding in the nucleus (Rexach and Blobel, 1995; Görlich et al., 1996). They return as RanGTP complexes to the cytoplasm, where the Ran-bound GTP is hydrolyzed, the complex dissociated and the importin enabled to bind and import another substrate (Bischoff and Görlich, 1997; Floer et al., 1997). Exportins function in exactly the opposite manner. They recruit cargo at high RanGTP levels in the nucleus, forming ternary cargo·exportin·RanGTP complexes (Fornerod et al., 1997b; Kutay et al., 1997); these complexes are transferred through NPCs into the cytoplasm, where GTP is hydrolyzed and the export complex disassembled. The cargo-free and Ran-free exportin can then re-enter the nucleus and export another cargo molecule.
Human cells employ at least 11 different importins as well as 10 adaptor·importin β-heterodimers to distinguish the thousands of different nuclear import cargoes from proteins with a non-nuclear destination (reviewed by Strom and Weis, 2001). The export side appears no less demanding. Several highly abundant export substrates, such as ribosomes, mRNAs, tRNAs and import adaptors, as well as a much larger number of regulatory factors must be exported from the nuclei (Izaurralde, 2002;Johnson et al., 2002; Lei and Silver, 2002; Simos et al., 2002). The nuclear export machinery also antagonizes the constant leakage of cytoplasmic proteins into the nuclear compartment and expels translation factors, for example, into the cytoplasm (Mingot et al., 2001; Bohnsack et al., 2002; Calado et al., 2002).
The nuclear export mediators known so far comprise the mRNA export factors (reviewed in Izaurralde, 2002) and six members of the importin β-superfamily. CRM1/exportin 1 (Fornerod et al., 1997a; Stade et al., 1997) is the exportin with the broadest substrate specificity and exports a large number of proteins with leucine-rich nuclear export signals (NESs) and several RNPs such as ribosomes, SRP and U snRNAs. CAS/exportin 2 accounts for the recycling of importin α adaptors to the cytoplasm (Kutay et al., 1997). Exportin-t mediates export of tRNA (Arts et al., 1998; Kutay et al., 1998), exportin 5 (Exp5) exports tRNA, the translation elongation factor 1A and certain minihelix-containing RNAs (Bohnsack et al., 2002; Calado et al., 2002; Gwizdek et al., 2003), while exportin 4 (Exp4) has so far only a single substrate, eIF5A (Lipowsky et al., 2000). Finally, importin 13 is quite unique in that it moves different cargoes in opposite directions (Mingot et al., 2001). It imports several proteins, but also exports the translation initiation factor eIF1A from nuclei.
While characterizing a novel human exportin, we identified β-actin and profilin I as export cargoes. β-Actin is one of three actin types and the predominant form in non-muscle cells (Pollard and Earnshaw, 2002). It forms the cytoplasmic microfilaments, which in turn serve as tracks for myosin-type motor proteins, control cell shape, aid phagocytosis and form the contractile ring that separates daughter cells after nuclear division. Actin filaments (F-actin) can be highly dynamic, whereby the barbed (+) end of the filament grows by the addition of ATP-charged actin monomers (G-actin), and the pointed (–) end shrinks by releasing G-actin in its ADP form. A great number of regulatory factors control these dynamics by either sequestering G-actin, stabilizing or destabilizing the filaments, or seeding new filaments. Profilin is one of these regulators and a central and highly abundant constituent of the actin-system (Schluter et al., 1997; Pollard and Borisy, 2003). It binds selectively to G-actin and causes two types of effects: First, it suppresses the spontaneous (i.e. non-nucleated) polymerization into F-actin (Carlsson et al., 1977; Reichstein and Korn, 1979). Secondly, profilin functions as a nucleotide-exchange factor and catalyzes the efficient conversion of ADP– G-actin to the ATP form, which in turn is required for the controlled growth of actin filaments (Mockrin and Korn, 1980; Wolven et al., 2000; Lu and Pollard, 2001). Profilin·actin·ATP complexes represent the preferred substrate for filament elongation. Profilin itself interacts with numerous additional regulators, which typically bind profilin via proline-rich domains and thereby recruit profilin·actin complexes to sites of actin-filament growth (Holt and Koffer, 2001).
We identify here exportin 6 (Exp6) as a novel nuclear export receptor and profilin·actin complexes as its predominant or even sole cargo. It is well established that nuclei are void of phalloidin-stainable actin filaments. This could be achieved, at least in part, by confining nucleators of actin polymerization to the cytoplasm. In addition, however, free nuclear actin must be kept below the critical concentration for polymerization and steady nuclear export of profilin·actin complexes along the Exp6 pathway appears an elegant solution to this problem.
Results
Exportin 6—a novel member of the importin β-family
Importin β-type nuclear transport receptors recognize a wide range of substrates and are therefore rather diverse in sequence. Their N-terminal RanGTP-binding site is still the most conserved region and thus a diagnostic feature of the superfamily (Fornerod et al., 1997b; Görlich et al., 1997). To identify the complete set of human family members, we therefore searched the databases for ESTs coding for proteins with putative importin β-like RanGTP-binding sites. Thereby we identified partial sequences of a novel family member that, for reasons detailed below, will be referred to as Exp6. A cDNA containing the entire coding region was finally obtained by a combination of 5′ RACE and RT–PCR using HeLa mRNA as a template.
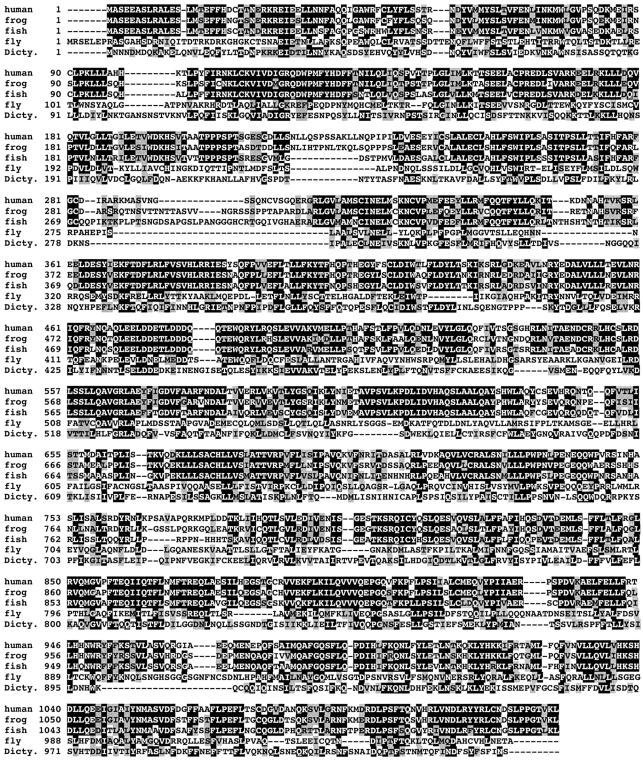
Detailed homology searches confirmed Exp6 as a genuine member of the importin β-like transport receptor family. Orthologous proteins are found in all vertebrates and insects examined (see Figure 1 and data not shown). There is no Exp6 in the nematode Caernorhabditis elegans, in plants or in fungi; however, the slime mould Dictyostelium discoideum contains a clear orthologue to human Exp6 (Figure 1). Within the receptor family, proteins of the exportin-t and the Exp5/MSN5 groups show the highest degree of similarity to Exp6 proteins (not shown).
Fig. 1. shows the multiple alignment of Exp6 from human, frog (X.laevis), fish (D.rerio), fruitfly (D.melanogaster) and slime mould (D.discoideum). Identical residues are shaded in black, similar ones in grey.
Identification of Exportin 6-specific nuclear transport substrates
It was initially unclear if Exp6 would function in nuclear import, export or indeed in nucleocytoplasmic transport at all. To clarify the issue, we recombinantly expressed human Exp6 and immobilized the protein in order to enrich potential cargoes from a HeLa extract (Figure 2). In the absence of RanGTP, i.e. under cytoplasmic conditions, only background binding occurred. When, however, RanGTP (Q69L-mutant) was added to mimic a nuclear environment, several potential export cargoes were recovered in the Exp6-bound fraction. These were β-actin, profilin I, Diaphanous 1, VASP and Mena. Strikingly, all five factors are functionally related and represent either constituents or regulators of the actin cytoskeleton. β-Actin and its adenine nucleotide exchange factor, profilin, constitute the major bands and both were recovered in a nearly stoicheiometric ratio to Ran. Their binding to Exp6 appeared indeed very specific as judged by the strict RanGTP dependence of the interaction (Figure 2) and because binding was not evident when Exp6 was replaced by any of the other 10 nuclear transport receptors tested (Figure 3).
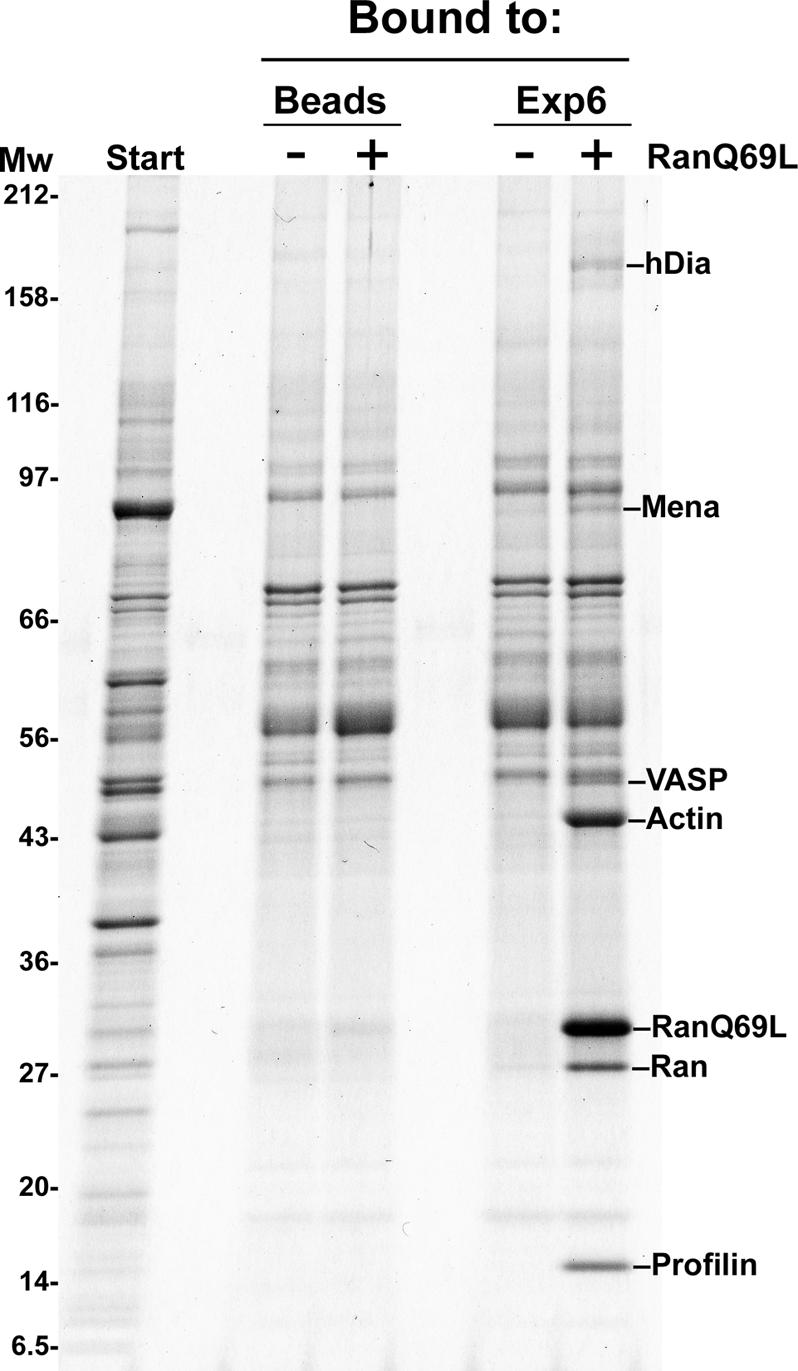
Fig. 2. Identification of the profilin I and β-actin as putative export substrates for Exp6. zz-tagged Exp6 was immobilized to IgG–Sepharose and used to enrich potential cargoes from a cytoplasmic HeLa cell extract. Binding was in the absence or presence of 5 µM RanGTP (Q69L-mutant). Starting material and bound fractions were analysed by SDS–gel electrophoresis, followed by Coomassie staining. Protein bands identified by mass spectrometry are indicated. The negative control was empty beads. The putative export cargoes profilin I, β-actin, and the actin-binding proteins Mena, VASP and Diaphanous 1 bound specifically to Exp6, provided RanGTP (Q69L-mutant) had been added to mimic a nuclear environment.
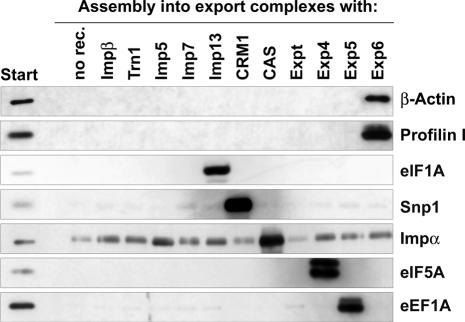
Fig. 3. A cytoplasmic HeLa cell extract was depleted of endogenous nuclear transport receptors and subsequently replenished with individual importins and exportins as indicated. To form export complexes between the added receptors and cargoes from the extract, zz-tagged RanGTP (Q69L) was added. The complexes were purified on IgG–Sepharose, and analysed by immunoblotting with anti β-actin and anti-profilin I antibodies. Note that both proteins formed export complexes with Exp6 and with no other transport receptor tested. As controls, we included immunoblots against characterized cargoes of other export pathways, namely eIF1A (exported by importin 13), snurportin 1 (exported by CRM1), importin α (exported by CAS), eIF5A (exported by Exp4) and eEF1A (exported by Exp5).
Exportin 6 has a strong preference for the actin·profilin complex
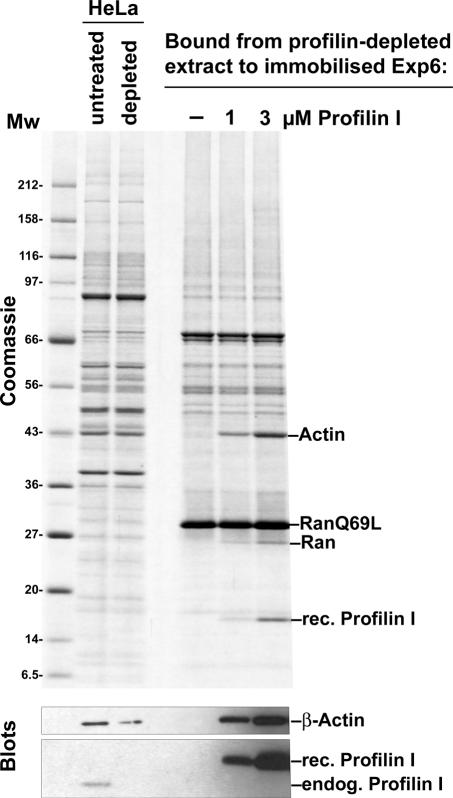
Profilin and actin were both recovered in the Exp6-bound fraction (Figures 2 and 3). This could be mere coincidence and mean that both proteins carry, independently of each other, the same type of Exp6-specific nuclear export signal. However, profilin is the adenine nucleotide exchange factor for actin (Mockrin and Korn, 1980); the two proteins can form a stable soluble complex (Carlsson et al., 1977) and this could indicate that Exp6 actually recognizes the profilin·actin complex. To clarify this issue, we exploited the tight binding of profilin to immobilized polyproline (Tanaka and Shibata, 1985) and depleted profilin from the HeLa extract. The remaining actin failed to form stable export complexes with Exp6 (Figure 4). Binding of β-actin to Exp6 was fully restored after addition of recombinant profilin (Figure 4). Thus, the interaction of β-actin with Exp6 is greatly enhanced by profilin.
Fig. 4. Profilin enhances actin binding to Exp6. Profilin was depleted from a cytoplasmic HeLa cell extract by means of polyproline–Sepharose. The resulting extract was used to assemble export complexes with RanGTP (Q69L-mutant) and immobilized Exp6. Binding of actin to Exp6 was only observed when profilin was re-added. Analysis was performed as described in Figure 2. Binding was performed in 50 mM Tris–HCl pH 7.5, 50 mM NaCl, 5 mM magnesium acetate.
We next prepared a HeLa cell extract from which actin had been depleted by a two-step procedure. The HeLa cells were lysed in the presence of phalloidin, which favours actin-polymerization and thereby removes G-actin from the solution (Dancker et al., 1975); the resulting actin polymers were removed by ultracentrifugation. This ‘low-actin extract’ was subsequently passed through a column of immobilized DNase I, which is known as a highly specific and avidly binding ligand for actin (Zechel, 1980). The resulting flow-through was essentially actin free, but still contained profilin (see blot in Figure 5). Strikingly, no profilin binding to Exp6 was observed in the absence of actin; however, re-addition of purified β-actin fully restored the profilin–Exp6 interaction. Thus, a stable profilin–Exp6 interaction requires actin and Exp6 therefore preferentially recognizes the profilin·actin complex.
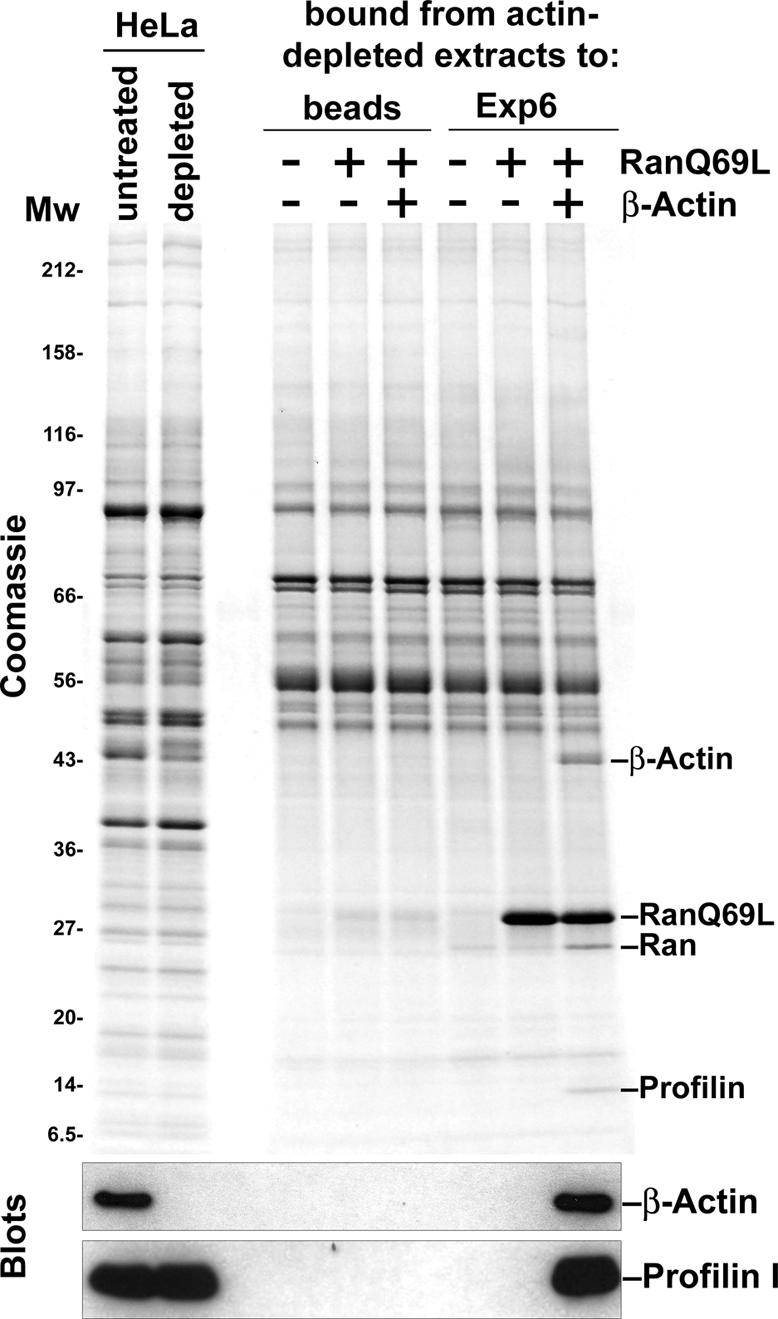
Fig. 5. Actin is required for a stable profilin–Exp6 interaction. Actin was depleted from a HeLa extract by phalloidin treatment, followed by passage through a DNase I column. The resulting actin-free extract was then used to assemble export complexes with immobilized Exp6. Binding of profilin to Exp6 was only observed when actin (1 µM) had been re-added.
Exportin 6 appears to contact the actin·profilin complex via actin
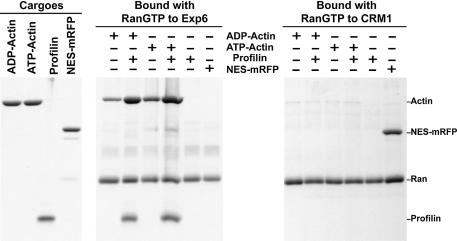
To test which component of the profilin·actin complex makes the greater contribution to Exp6 binding, we changed the conditions of the binding assay such that very weak interactions could also be detected (Figure 6). Full-length Ran was replaced by a C-terminal deletion mutant (RanQ69L 1–180) that interacts more tightly with nuclear transport receptors (Nilsson et al., 2002); we reduced the ionic strength in the binding assay and the washing time of the affinity beads. Also under these conditions, binding of profilin alone to Exp6 was undetectable. However, actin in either ADP or ATP form could now be recruited even in the absence of profilin, suggesting that Exp6 contacts the profilin·actin complex primarily via the actin component. As expected, this binding was enhanced by profilin addition and it should be emphasized that the actin–Exp6 interaction shows a strict profilin dependence at more physiological ionic strength (Figure 4). Profilin binding is known to cause significant changes in the actin conformation (Schutt et al., 1993) and we would assume that the profilin-bound conformation of actin is the high affinity form for Exp6 binding (for detailed discussion see below).
Fig. 6. Exp6 appears to contact the profilin·actin complex via the actin component. Export complexes were formed with purified cargoes (1 µM ADP–actin, ATP–actin or NES–mRFP), recombinant exportins (1 µM Exp6 or CRM1) and the RanQ69L 1–180 mutant (3 µM) that binds transport receptors more tightly than the full-length protein. Binding was performed in a low salt buffer (15 mM Tris–HCl pH 7.5, 1 mM magnesium acetate, 0.2% digitonin) to favour low-affinity interactions. Export complexes were retrieved with IgG–Sepharose via the zz-tag of the exportins, and exportin-bound components were eluted after very brief washing with 1.5 M magnesium chloride. Analysis of bound components was by SDS–PAGE/Coomassie staining. Note that actin assembled into export complexes with Exp6, although not as efficiently as in the presence of profilin. CRM1 bound the NES–mRFP but showed no significant interaction with any of the actin forms.
The binding assay shown in Figure 6 was performed with recombinant profilin, Exp6, Ran and highly purified actin and we can thus conclude that no additional component is required for export complex formation. The contact between actin and Exp6 is therefore direct. As a control, we replaced Exp6 with human CRM1. CRM1 efficiently bound a fusion between the NES of PKI (Wen et al., 1995) and mRFP (Campbell et al., 2002); however, it showed no significant interaction with either ADP–actin, ATP–actin or the profilin·actin complex (see below for discussion).
Exportin 6 mediates nuclear export of profilin
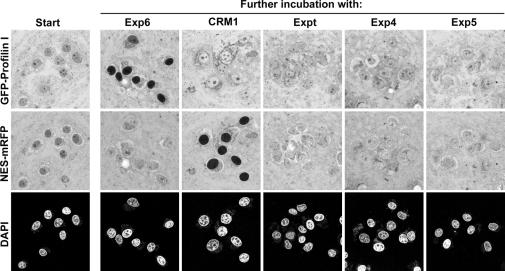
The aforementioned binding experiments are consistent with the assumption that Exp6 represents the nuclear export receptor for the profilin·actin complex. To test this directly, we performed export experiments from nuclei of permeabilized HeLa cells. The nuclei were incubated in a Xenopus egg extract, from which nuclear transport receptors had been depleted with the phenyl–Sepharose method (Ribbeck and Görlich, 2002). This extract not only stabilized nuclei, but also served as a source of (unlabelled) β-actin. Profilin was added as a GFP-fusion protein, which allowed detection by confocal laser scanning microscopy (Figure 7). Without further addition, the profilin equilibrated between nuclei and cytoplasm. This sample was then split and various nuclear transport receptors were added. Strikingly, upon Exp6-addition, rapid export occurred. This export was so efficient that nuclei appeared nearly black against the bright cytoplasmic signal. This effect was highly specific and was not observed when Exp6 was replaced by a different exportin, such as CRM1, Exp4, exportin-t or Exp5. Conversely, Exp6 had no effect on the NES–mRFP fusion, which is exported by CRM1 only (Figure 7).
Fig. 7. Exp6 mediates nuclear export of profilin. Nuclei were incubated in a Xenopus egg extract, which had been prior depleted of endogenous nuclear transport receptors and replenished with an energy-regenerating system and 0.3 µM NTF2. GFP–profilin fusion protein (3 µM) and an NES–mRFP fusion (4 µM) were added and allowed to equilibrate between nuclei and cytoplasm (‘Start’). The sample was then split, indicated nuclear transport receptors were added and the distributions of the export substrates were imaged 10 min later by confocal laser scanning microscopy. Exp6 promoted nuclear export of profilin, while the other exportins had no effect. Conversely, only CRM1 caused nuclear exclusion of the NES–mRFP fusion. Chromatin was detected by DAPI staining and excitation at 405 nm to indicate the positions of nuclei. GFP and mRFP were excited at 488 and 547 nm, respectively.
The interaction with actin is essential for Exportin 6-mediated export of profilin
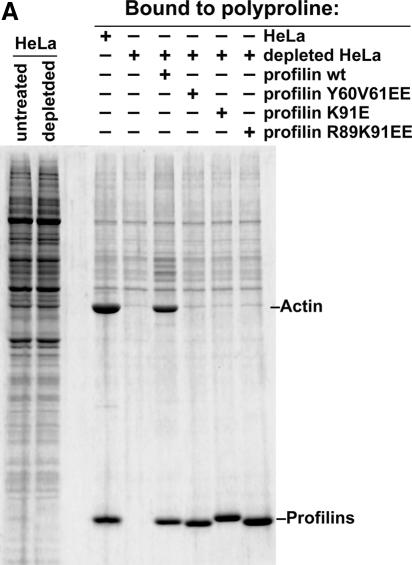
The binding experiments in Figures 4 and 5 demonstrate a strong preference of Exp6 for the profilin·actin complex. To test if the profilin–actin interaction is essential for profilin export, we used the known 3D structure of the profilin·actin complex (Schutt et al., 1993) to construct three different mutations in human profilin I (Y60V61→EE, K91→E and R89K91→EE), which all disrupt the charge complementarity of the binding interfaces and thereby reduce actin binding to background-levels (see Figure 8A). This loss of actin binding was specific by several criteria and not just a consequence of misfolding. First, these profilin mutants were perfectly soluble after expression in Escherichia coli. Secondly, they behaved as the wild-type protein during gel filtration (not shown). Thirdly, their polyproline-binding site remained functional (Figure 8A). Finally, their CD spectra are fully consistent with the assumption of proper folding (not shown).
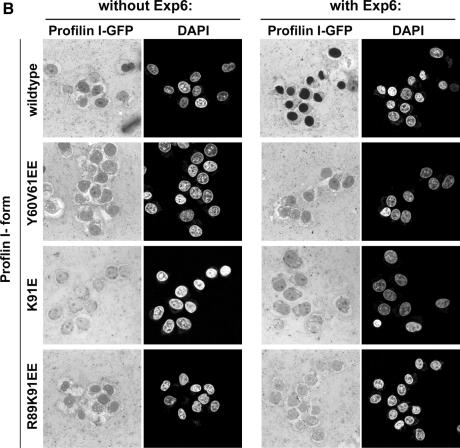
Fig. 8. Mutational analysis shows that Exp6 exports profilin only if profilin binds actin. (A) Binding to polyproline–Sepharose was performed from an unfractionated cytosolic HeLa extract, a profilin-depleted HeLa extract, or from depleted extracts supplemented with either wild-type profilin I or indicated profilin I mutants (untagged proteins). Panels show analysis of bound fractions by SDS–PAGE, followed by Coomassie staining. Note that all forms of profilin bound to polyproline–Sepharose. β-Actin co-purified on polyproline–Sepharose with endogenous and recombinant wild-type profilin, but not with the Y60V61→EE, K91→E and R89K91→EE profilin mutants, which had been designed to disrupt interaction with actin. (B) Nuclear export of GFP-tagged profilin derivatives was performed as in Figure 7. Addition of Exp6 resulted in nuclear export of wild-type profilin. In contrast, the Y60V61→EE, K91→E and R89K91→EE profilin mutants resisted Exp6-mediated export.
We then produced the mutants in a GFP-tagged form and incubated them with nuclei of permeabilized cells as described above, which allowed equilibration of the fusion proteins between nucleus and cytoplasm. Strikingly, Exp6 was unable to export any of the three mutants, while the wild-type protein was efficiently expelled from the nuclei (Figure 8B).
The mutated residues are located on the surface of uncomplexed profilin. Yet we can rule out the possibility that they directly contact Exp6, because they are deeply buried by the bound actin molecule and are thus inaccessible when Exp6 binds the profilin·actin complex. Thus, this failure of Exp6-dependent export cannot be explained by destruction of the Exp6-binding site on the profilin molecule, but instead has to be considered as a consequence of impaired actin binding. In summary, we can conclude that Exp6 exports profilin as a complex with actin and apparently only as such a complex.
Exportin 6 exports all human profilin isoforms
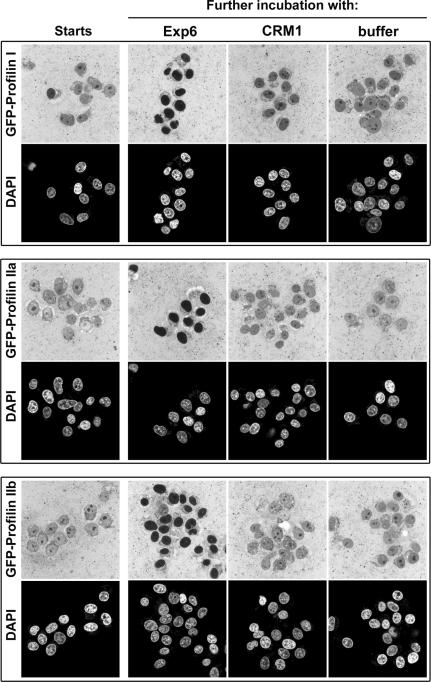
In human, three types of profilin can be found: profilin I, IIa and IIb. Isoform I is the prevalent form in HeLa cells and most other cell types, profilin IIa is ∼60% identical to profilin I and abundantly expressed in the brain, while profilin IIb appears to be a rare, alternatively spliced variant of profilin IIa (Lambrechts et al., 2000). To test whether all human isoforms represent export cargoes of Exp6, we produced them in a GFP-tagged form and subjected them to the export assay as described for profilin I. As seen in Figure 9, Exp6 addition caused nuclear exclusion of all three human profilins. This result supports our interpretation that Exp6 recognizes the actin component of the profilactin complex, which indeed predicts that sequence variation in profilin should not directly affect the interaction with the exportin.
Fig. 9. Panels show nuclear export assays for human GFP–profilin I and the isoforms IIa and IIb, using human Exp6 or CRM1 as nuclear export receptors. Experimental set-up was as described in Figure 7.
Exportin 6-mediated export of profilin·actin complexes is conserved between vertebrates and insects
Exp6 orthologues can be found in most higher eukaryotes; however, the sequence conservation between species appears rather low compared with other nuclear transport receptors. Exp6 from the fish Danio rerio still shares ∼72% identical amino acid residues with the human protein, but the identity between Drosophila and human Exp6 is as low as 20% (see Figure 1; for comparison, human and Drosophila CRM1 are 70% identical). Nevertheless, our export assays clearly demonstrate that fish and Drosophila Exp6 represent fully functional export receptors for human profilins I, IIa and IIb (see Figure 10; data not shown). Also in these cases, profilin export required an intact actin-binding site (not shown). Exp6-mediated nuclear export of profilin·actin complexes appears therefore to be a general phenomenon in vertebrates and insects and it apparently applies to all profilin isoforms. It probably evolved already in protozoan animals, as the slime mould D.discoideum also contains an Exp6 orthologue that is, quite surprisingly, more closely related to the human sequence than is the Drosophila protein (Figure 1).
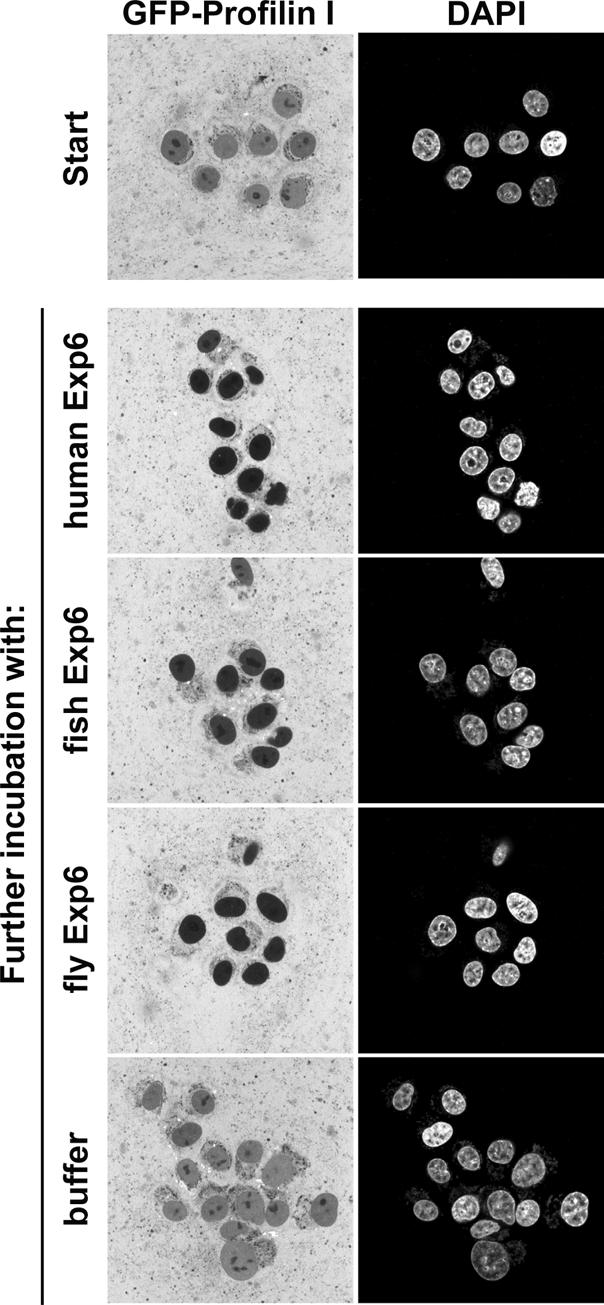
Fig. 10. Panels compare nuclear export of human GFP–profilin I mediated by Exp6 from human, zebrafish (D.rerio) or fruitfly (D.melanogaster). Experimental set-up was as described in Figure 7.
Exportin 6 is required for nuclear exclusion of actin in Drosophila cells
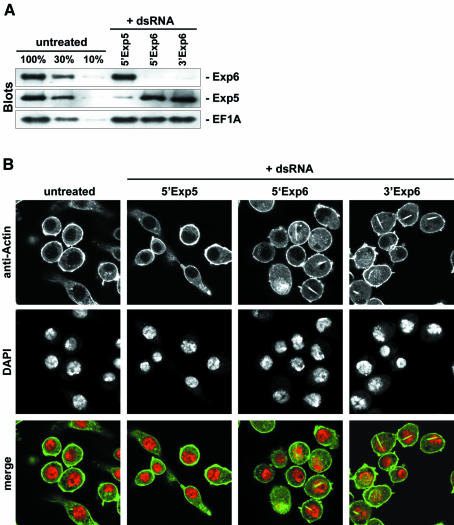
We so far used GFP–profilin as a tracer to follow Exp6-dependent export of the profilin·actin complex. These results together with the binding experiments strongly suggested, but did not prove, a role of Exp6 in nuclear exclusion of bulk actin. We therefore employed the RNA-interference technique (reviewed by Hannon, 2002) to deplete Exp6 from Drosophila Schneider cells and tested the effect on the cellular actin distribution. Exp6 turned out to be an exceptionally stable protein, but after 6 days of treatment the cellular Exp6 concentration was finally reduced to ≤10% of the original levels (Figure 11A). Untreated control cells or cells depleted of an unrelated exportin (Exp5) showed uniform nuclear exclusion of actin (Figure 11B). In Exp6-depleted cells, however, a dramatic effect was observed and ∼50% of the nuclei had accumulated large, elongated actin-containing aggregates that typically crossed the entire nucleus. These nuclear aggregates resemble actin paracrystals that are induced when cultured cells are treated with a high concentration of DMSO (Osborn and Weber, 1980; Sanger et al., 1980). They always occurred at areas of low chromatin density as if the surrounding chromatin was displaced by them. Taken together, these results clearly demonstrate that Exp6 is essential for nuclear exclusion of actin and mediates a non-redundant pathway of actin export.
Fig. 11. Exp6 is required for nuclear exclusion of actin in living cells. Drosophila Schneider cells were left either untreated or incubated for 6 days with double-stranded RNA to deplete Exp5 and Exp6 by RNA interference. (A) Immunoblot analysis with anti-Exp6 and anti-Exp5 antibodies of treated and untreated cells. The anti eEF1A immunoblot served as a loading control. (B) Actin distribution in untreated and RNA-interference cells as detected by anti-actin immuno-staining and confocal laser scanning microscopy. Positions of nuclei were detected by DAPI staining. In the merged images, actin is shown in green and DNA in red.
Discussion
A novel nuclear export pathway
Transport between nucleus and cytoplasm represents a major activity in eukaryotic cells, and because of the large variety of cargoes that need to be ferried between the two compartments, numerous pathways are employed to co-ordinate and mediate this vast amount of trafficking. With the exceptions of mRNA export (Izaurralde, 2002) and NTF2-mediated import of Ran (Ribbeck et al., 1998), all known pathways are driven by importin β-related nuclear transport receptors. Twenty such receptors exist in human cells and six of them (CRM1, CAS, exportin-t, Exp4, Exp5 and importin 13) have been known to function in nuclear export. We describe here Exp6 as a novel family member and show that it mediates nuclear export of the profilin·β-actin complex.
Why should the cytoplasmic localization of actin require active nuclear export?
It is well established that nuclei are void of (phalloidin-stainable) F-actin and contain typically only very low levels of G-actin. As actin operates in the cytoplasm and as nuclear actin filaments are likely to interfere with nuclear structure and functions, such cytoplasmic confinement of the actin system makes perfect sense. The existence of the Exp6 pathway for profilactin, however, implies that a significant influx of actin into nuclei must be counteracted, which in turn poses the question as to why actin enters nuclei in the first place. In higher eukaryotes, this will occur by at least two mechanisms. First, cytoplasm and nuclear contents mix during the open mitosis and actin could simply be enclosed by the nuclear envelope when nuclei reform in telophase. Secondly, nuclear pore complexes are not perfect barriers. They can restrict, but not completely prevent, passage of inert material. In the absence of nuclear export, it would just be matter of time until the nuclear and cytoplasmic concentrations have equilibrated and the nuclear actin pool exceeds the critical concentration for polymerization. This situation resembles translation factors, which are also excluded from the nuclear compartment (Bohnsack et al., 2002), probably to avoid an untimely translation of (pre-)mRNAs in the nucleus. Also in this case, leakage into nuclei can apparently not be fully prevented, instead the exclusive cytoplasmic distribution requires a second line of defence: steady nuclear export. One can generalize this concept in that a strict cytoplasmic localization of a given protein is unlikely to occur by default, i.e. because of a lack of a nuclear import signal. Instead, it requires explicit ‘signals’ that confer either nuclear export or binding to strictly cytoplasmic structures (cytoplasmic retention).
Nuclear actin has been reported for a few specialized cell types, such as amphibian oocytes (Clark and Merriam, 1977), and it has been assumed to play regulatory roles, e.g. in general transcription by RNA-Pol II (Scheer et al., 1984) and in the expression of specific genes. This could imply that the levels of residual nuclear actin might be regulated, possibly through a cell type-specific adjustment of the cellular Exp6 concentration. We are testing this possibility at present.
Actin needs to be exported in soluble form
Profilin greatly facilitates recruitment of actin to Exp6 (Figure 4) and it can therefore be regarded as an actin-specific co-factor for nuclear export. It indeed appears to be perfectly suitable for this function. First, one should assume that only the soluble G-actin and not the polymerized F-actin can be transferred out of nuclei. Profilin binds specifically to monomeric actin and keeps it in solution, and so this species appears to be a good target for the nuclear export machinery. Secondly, profilins are small proteins (14–18 kDa) that pass NPCs quite rapidly by passive diffusion (D.Mohr, T.Stüven and D.Görlich, unpublished). This should ensure a steady profilin supply of nuclei without the need for active import, and thereby make profilin very suitable for multi-round export. Finally, free profilin is not an Exp6 cargo and thus not a competitor of actin·profilin complexes for export capacity, and this feature certainly improves the efficiency of the system.
The great extent of co-operativity between profilin- and actin-binding to Exp6 (Figure 4) poses the question as to how the profilin·actin complex is recognized. One possibility is that Exp6 contacts both profilin and actin. In this scenario, the free energy released upon binding of the profilin·actin complex would equal the sum of the individual Exp6–profilin and Exp6–actin interactions, and as binding constants increase exponentially with binding energies (Gibbs’ law); such a model could easily explain why Exp6 binds the profilin·actin complex orders of magnitude more strongly than the individual components. However, actin clearly makes a greater contribution to Exp6 binding than profilin and the data are even consistent with the assumption that Exp6 contacts actin only. That model would also explain why Exp6 recognizes actin complexes with profilin I, profilin IIa and IIb equally well. The function of profilin in export complex formation would be the induction of a conformational change that brings actin into the high affinity form for Exp6 binding.
The rate of actin polymerization is not solely determined by the concentration of actin, but is regulated locally by nucleators, such as the Arp2/3 complex (Welch et al., 1997). Profilin apparently enhances the specificity of nucleators; it suppresses background polymerization, but also supplies the nucleators with a form of actin that is optimal for incorporation into growing microfilaments. A cytoplasmic confinement of the nucleators should therefore also be required to avoid filament formation in the nucleus. We do not yet know which exportin might exclude Arp2/3 from the nucleus. However, in the Exp6-bound fraction, we could detect several other factors that promote F-actin nucleation, namely Diaphanous 1, VASP and Mena. The three possess proline-rich domains that confer profilin binding (Holt and Koffer, 2001). We would therefore assume that this class of actin-binding proteins becomes co-exported with profilin·actin complexes via the Exp6-dependent pathway and, indeed, we could reconstitute a VASP·profilin·actin·Exp6·RanGTP export complex from the purified components (not shown). This in turn points to an additional role of Exp6 in nuclear exclusion of components that nucleate actin filaments.
Conservation of the Exportin 6 pathway
The facts that (i) a profilin·actin-specific nuclear export pathway evolved at all and (ii) this pathway remained functional from insects to vertebrates (and probably even in Dictyostelium) point to a considerable selective pressure to keep nuclear actin levels low. Interestingly, Drosophila mutants with a defective Exp6 gene (at the CG3923 locus of the X-chromosome) were identified already, 15 years ago (http://flybase.org/; Perrimon et al., 1989). The mutation is recessive lethal and homozygous mutant flies die before the pupal stage, probably when the stockpiles of Exp6 protein and mRNA supplied by the heterozygous mother become exhausted. How exactly a defective export of profilin·actin complexes kills the flies is unclear at present, but it appears likely that the huge nuclear actin aggregates that form in the absence of Exp6 cause severe problems.
What is the role of CRM1 in actin export?
Wada et al. (1998) suggested previously that nuclear export of actin is mediated by CRM1. We, however, found no supporting evidence for this scenario. Human CRM1 does not recognize actin as an export cargo (Figures 3 and 6). It is not required for profilin·actin export (Figure 7) and the fact that Exp6-deficient cells accumulate actin in the nucleus (Figure 11) demonstrates that CRM1 is not sufficient to prevent nuclear actin accumulation. We would therefore conclude that CRM1 is not directly involved in actin export from vertebrate or Drosophila nuclei.
The nematode C.elegans, plants and fungi such as the yeasts Saccharomyces cerevisiae and Schizosaccharo myces pombe, lack Exp6 orthologues. This could imply that these organisms also lack active nuclear exclusion of actin. On the other hand, it is possible that another exportin, such as CRM1, fulfils the function. In this case, it would be very interesting to know if actin alone, the profilin·actin heterodimer or yet another actin complex is recognized.
Materials and methods
Molecular cloning of Exportin 6
Sequence information of several ESTs from human, rat and mouse, deposited in the NCBI database, was used to reconstruct large parts of the human Exp6 coding region. The missing 5′ end was elucidated by several rounds of 5′ RACE, using HeLa cell cDNA as a template. The N-terminal sequence then also allowed to identify corresponding ESTs in other organisms. The coding regions of full-length human and mouse Exp6 were obtained by RT–PCR using mRNA isolated from HeLa and 3T3 cells, as template. cDNA clones covering the entire coding region of Exp6 orthologues from D.rerio (zebrafish) and Drosophila melanogaster were obtained from the Deutsches Ressourcenzentrum für Genomforschung GmbH (RZPD).
Recombinant protein expression in E.coli and purification
Expression and purification of the following proteins have been described before: his-tagged RanQ69L, zzRanQ69L, Impβ, transportin, Imp5, 7, 13, CAS, CRM1, Exp4, exportin-t, Exp5 and NTF2 (Mingot et al., 2001; Bohnsack et al., 2002). The RanQ69L 1–180 mutant was expressed with an N-terminal his-tag and purified by Ni–NTA chromatography followed by gel filtration. Exp6 was expressed as an N-terminally hexa-histidine-tagged protein from pET28a. Expression from zzTev80N yielded Exp6 with N-terminal zz-tag and C-terminal hexa-histidine-tag. Purification was by Ni–NTA chromatography followed by an anion-exchange step and gel filtration on Superdex200. Exp6 from Drosophila or zebrafish were expressed with N-terminal his-tags and purified as the human protein.
The open reading frame coding for human profilin I was amplified from HeLa cDNA and cloned into pQE60 derivatives that permit expression of profilin, either untagged, with a C-terminal his-tag, or with GFP- and his-tags. Purification of his-tagged profilin–fusion proteins was by chromatography on Ni–NTA–Sepharose, followed by gel filtration. Tagging of profilin did not interfere with actin binding or export complex formation with Exp6. However, tags on either N- or C-termini severely compromized interaction with polyproline. Therefore, untagged profilin was used for the experiment in Figure 7A. Purification was on polyproline–Sepharose.
NES–mRFP was constructed as a fusion between the NES of PKI (Wen et al., 1995), the monomeric version of the red fluorescent protein (Campbell et al., 2002) and a C-terminal his-tag. Purification was on Ni–NTA–Sepharose.
HeLa extracts and depleted HeLa extracts
Standard cytoplasmic HeLa cell extracts were prepared by hypotonic lysis, cleared by ultracentrifugation, and supplemented with 5 µg/ml cytochalasin B in order to prevent precipitation of filamentous actin. To deplete profilin, 12 ml extract was passed through a 7 ml polyproline column (MW 5000, coupled at 50 mg/ml to cyanogen bromide-activated Sepharose). To obtain actin-free extract, cells where lysed in the absence of cytochalasin and in the presence of 20 µg/ml phalloidin and 10 ml of the resulting extract was passed through a 1 ml DNase I column (30 mg DNase I coupled per ml of NHS-activated Sepharose).
Purification of β-actin
50 ml cytoplasmic extract (corresponding to 5 × 109 HeLa cells) was supplemented with 10 mM EDTA and 10 µM untagged recombinant profilin to promote formation of the stable nucleotide-free β-actin·profilin complex, which in turn was recovered on a 7 ml polyproline–Sepharose column, equilibrated in 20 mM Tris–HCl pH 7.5, 3 mM β-mercaptoethanol, 250 mM sucrose. After extensive washing, actin was dissociated from profilin and thus eluted in the ATP form from the column by applying the following buffer: 50 mM Bicine–HCl pH 8.0, 300 mM NaCl, 5 mM CaCl2 1 mM ATP, 250 mM sucrose, 5 µg/ml cytochalasin B, 0.2% BigChap. For preparation of ADP-actin, elution was performed with 2 mM ADP in the presence of 5 mM MgCl2 and in the absence of CaCl2.
Binding assays were essentially performed as described (Mingot et al., 2001; Bohnsack et al., 2002). Bait proteins were immobilized via zz-tags (IgG-binding domains from Staphylococcus aureus protein A) to IgG–Sepharose (Pharmacia) at a concentration of ∼2 mg/ml matrix. Each 20 µl matrix was rotated overnight with 500 µl extract. Elution was with 50 mM Tris–HCl pH 7.5, 1.5 M magnesium chloride, which disrupts interactions between cargoes, nuclear transport receptors and Ran, but does not elute the zz-tagged baits from the matrix. Eluted proteins were precipitated with 95% isopropanol (final concentration), and dissolved in SDS sample buffer. Further details and analysis are given in the figure legends.
In vitro nuclear export
A (low speed spin) interphase extract from Xenopus laevis eggs was prepared as described (Leno and Laskey, 1991). Nuclear transport receptors were depleted with phenyl–Sepharose (low substitution; Pharmacia), using 500 µl beads per ml of extract. Permeabilized HeLa B cells were prepared in suspension as described previously (Ribbeck et al., 1998). For export experiments, 0.5 µl nuclei were mixed with 20 µl of depleted extract, an energy-regenerating system, 0.3 µM NTF2, 3 µM profilin-GFP, 2 µg/ml DAPI and where indicated also with 4 µM NES–mRFP. Export reactions were performed at 18°C. For confocal laser scanning microscopy, 2 µl of each of the export sample was placed into a well of 10-well multi-test slide and covered with a coverslip. Other details are given in the figure legends.
Antibodies
The anti-actin antibodies were purchased from Sigma (A-5441, Figures 2–4; A2066, Figure 11); the monoclonal anti profilin-antibody (2H11) was a kind gift from B.M.Jockusch (Mayboroda et al., 1997). Antibodies against snurportin 1, importin α, eIF1A, eIF5A and eEF1A have been described previously (Bohnsack et al., 2002). Antibodies against Drosophila Exp5 were raised in rabbits against a C-terminal peptide, antibodies against Drosophila Exp6 were raised against the recombinant protein. The secondary antibody used for immunofluorescence was goat anti rabbit antibody conjugated to Alexa 568 (Molecular Probes).
Protein identification
Protein bands were identified in the ZMBH mass spectrometry facility after tryptic digests by MALDI–TOF peptide fingerprints (β-actin, profilin I, Diaphanous 1 and VASP) or by sequencing of internal peptides using an electrospray-Q-TOF mass spectrometer (MENA).
RNA-interference in Schneider cells
Drosophila melanogaster Schneider cells (ATCC CLR-1963) were cultured at 26°C in Drosophila SFM medium with 20 mM glutamine (Gibco) and 100 U/ml penicillin/streptomycin. For RNA-interference, 0.5 × 106 cells/ml were seeded with fresh medium into a 12-well-plate and dsRNA was immediately added to a final concentration of 8 µg/ml. The medium was changed 48 and 96 h later, whereby 8 µg/ml dsRNA was each time included. After 6 days, cells were resuspended and seeded onto coverslips. Cells were fixed 3 h later for 7 min in 3% PFA + 0.1% glutaraldehyde at 26°C and processed for immunofluorescence. Each 105 cells were analysed by western blotting for Exp5 and Exp6 levels.
Double-stranded RNA was obtained by in vitro transcription of PCR products that contained T7 promotors on both sides. The PCR products comprised nucleotides 378–936 or 2345–3031 of the Drosophila Exp6 ORF or nucleotides 38–1651 of the Drosophila Exp5 ORF. In vitro transcription was performed according to the manufacturer′s protocol (Promega). Ethanol-precipitated transcription products were dissolved in water to a final concentration of 3 mg/ml, annealed at 65°C and stored at –80°C until use.
DDBJ/EMBL/GenBank accession Nos
The nucleotide sequences of Exp6 (RanBP20) are listed under accession numbers AAK01471 (human), AAK40296 (mouse), AAL78259 (zebrafish) and AAL39935 (Drosophila).
Acknowledgments
Acknowledgements
We wish to thank Thomas Ruppert and Armin Bosserhof for mass spectrometry, Froso Paraskeva for initiating the project, Petra Schwarzmaier, Petra Rübmann and Ursula Jäkle for excellent technical help, B.M.Jockusch for the anti profilin antibody, R.Tsien for the mRFP construct, Jörg Großhans and B.M.Jockusch for helpful discussions, the members of our laboratory for critical reading of the manuscript and stimulating discussions. This work received financial support from the Alfried-Krupp Foundation, the DFG (SFB 352) and the Fonds der Chemischen Industrie.
References
- Arts G.J., Fornerod,M. and Mattaj,I.W. (1998) Identification of a nuclear export receptor for tRNA. Curr. Biol., 8, 305–314. [DOI] [PubMed] [Google Scholar]
- Bischoff F.R. and Görlich,D. (1997) RanBP1 is crucial for the release of RanGTP from importin β-related nuclear transport factors. FEBS Lett., 419, 249–254. [DOI] [PubMed] [Google Scholar]
- Bohnsack M.T., Regener,K., Schwappach,B., Saffrich,R., Paraskeva,E., Hartmann,E. and Görlich,D. (2002) Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J., 21, 6205–6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado A., Treichel,N., Muller,E.C., Otto,A. and Kutay,U. (2002) Exportin-5-mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J., 21, 6216–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell R.E., Tour,O., Palmer,A.E., Steinbach,P.A., Baird,G.S., Zacharias,D.A. and Tsien,R.Y. (2002) A monomeric red fluorescent protein. Proc. Natl Acad. Sci. USA, 99, 7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson L., Nystrom,L.E., Sundkvist,I., Markey,F. and Lindberg,U. (1977) Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J. Mol. Biol., 115, 465–483. [DOI] [PubMed] [Google Scholar]
- Clark T.G. and Merriam,R.W. (1977) Diffusible and bound actin nuclei of Xenopus laevis oocytes. Cell, 12, 883–891. [DOI] [PubMed] [Google Scholar]
- Conti E. and Izaurralde,E. (2001) Nucleocytoplasmic transport enters the atomic age. Curr. Opin. Cell Biol., 13, 310–319. [DOI] [PubMed] [Google Scholar]
- Dancker P., Low,I., Hasselbach,W. and Wieland,T. (1975) Interaction of actin with phalloidin: polymerization and stabilization of F-actin. Biochim. Biophys. Acta, 400, 407–414. [DOI] [PubMed] [Google Scholar]
- Floer M., Blobel,G. and Rexach,M. (1997) Disassembly of RanGTP-karyopherin β ?complex, an intermediate in nuclear protein import. J. Biol. Chem., 272, 19538–19546. [DOI] [PubMed] [Google Scholar]
- Fornerod M., Ohno,M., Yoshida,M. and Mattaj,I.W. (1997a) Crm1 is an export receptor for leucine rich nuclear export signals. Cell, 90, 1051–1060. [DOI] [PubMed] [Google Scholar]
- Fornerod M., van Deursen,J., van Baal,S., Reynolds,A., Davis,D., Murti,K.G., Fransen,J. and Grosveld,G. (1997b) The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J., 16, 807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Dabrowski,M., Bischoff,F.R., Kutay,U., Bork,P., Hartmann,E., Prehn,S. and Izaurralde,E. (1997) A novel class of RanGTP binding proteins. J. Cell. Biol., 138, 65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D. and Kutay,U. (1999) Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell. Dev. Biol., 15, 607–660. [DOI] [PubMed] [Google Scholar]
- Görlich D., Pante,N., Kutay,U., Aebi,U. and Bischoff,F.R. (1996) Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J., 15, 5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Seewald,M.J. and Ribbeck,K. (2003) Characterization of Ran-driven cargo transport and the RanGTPase system by kinetic measurements and computer simulation. EMBO J., 22, 1088–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwizdek C., Ossareh-Nazari,B., Brownawell,A.M., Doglio,A., Bertrand,E., Macara,I.G. and Dargemont,C. (2003) Exportin-5 mediates nuclear export of minihelix-containing RNAs. J. Biol. Chem., 278, 5505–5508. [DOI] [PubMed] [Google Scholar]
- Hannon G.J. (2002) RNA interference. Nature, 418, 244–251. [DOI] [PubMed] [Google Scholar]
- Holt M.R. and Koffer,A. (2001) Cell motility: proline-rich proteins promote protrusions. Trends Cell. Biol., 11, 38–46. [DOI] [PubMed] [Google Scholar]
- Imamoto N. (2000) Diversity in nucleocytoplasmic transport pathways. Cell Struct. Funct., 25, 207–216. [DOI] [PubMed] [Google Scholar]
- Izaurralde E. (2002) Nuclear export of messenger RNA. Results Probl. Cell Differ., 35, 133–150. [DOI] [PubMed] [Google Scholar]
- Izaurralde E., Kutay,U., von Kobbe,C., Mattaj,I.W. and Görlich,D. (1997) The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J., 16, 6535–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A.W., Lund,E. and Dahlberg,J. (2002) Nuclear export of ribosomal subunits. Trends Biochem. Sci., 27, 580–585. [DOI] [PubMed] [Google Scholar]
- Kalab P., Weis,K. and Heald,R. (2002) Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science, 295, 2452–2456. [DOI] [PubMed] [Google Scholar]
- Kutay U., Bischoff,F.R., Kostka,S., Kraft,R. and Görlich,D. (1997) Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell, 90, 1061–1071. [DOI] [PubMed] [Google Scholar]
- Kutay U., Lipowsky,G., Izaurralde,E., Bischoff,F.R., Schwarzmaier,P., Hartmann,E. and Görlich,D. (1998) Identification of a tRNA-specific nuclear export receptor. Mol. Cell, 1, 359–369. [DOI] [PubMed] [Google Scholar]
- Lambrechts A. et al. (2000) Profilin II is alternatively spliced, resulting in profilin isoforms that are differentially expressed and have distinct biochemical properties. Mol. Cell. Biol., 20, 8209–8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei E.P. and Silver,P.A. (2002) Protein and RNA export from the nucleus. Dev. Cell, 2, 261–272. [DOI] [PubMed] [Google Scholar]
- Leno G.H. and Laskey,R.A. (1991) DNA replication in cell-free extracts from Xenopus laevis. Methods Cell Biol., 36, 561–579. [DOI] [PubMed] [Google Scholar]
- Lipowsky G., Bischoff,F.R., Schwarzmaier,P., Kraft,R., Kostka,S., Hartmann,E., Kutay,U. and Görlich,D. (2000) Exportin 4: a mediator of a novel nuclear export pathway in higher eukaryotes. EMBO J., 19, 4362–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J. and Pollard,T.D. (2001) Profilin binding to poly-l-proline and actin monomers along with ability to catalyze actin nucleotide exchange is required for viability of fission yeast. Mol. Biol. Cell, 12, 1161–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macara I.G. (2001) Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev., 65, 570–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I.W. and Englmeier,L. (1998) Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem., 67, 265–306. [DOI] [PubMed] [Google Scholar]
- Mayboroda O., Schluter,K. and Jockusch,B.M. (1997) Differential colocalization of profilin with microfilaments in PtK2 cells. Cell Motil. Cytoskeleton, 37, 166–177. [DOI] [PubMed] [Google Scholar]
- Mingot J.M., Kostka,S., Kraft,R., Hartmann,E. and Görlich,D. (2001) Importin 13: a novel mediator of nuclear import and export. EMBO J., 20, 3685–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockrin S.C. and Korn,E.D. (1980) Acanthamoeba profilin interacts with G-actin to increase the rate of exchange of actin-bound adenosine 5′-triphosphate. Biochemistry, 19, 5359–5362. [DOI] [PubMed] [Google Scholar]
- Nilsson J., Weis,K. and Kjems,J. (2002) The C-terminal extension of the small GTPase Ran is essential for defining the GDP-bound form. J. Mol. Biol., 318, 583–593. [DOI] [PubMed] [Google Scholar]
- Osborn M. and Weber,K. (1980) Dimethylsulfoxide and the ionophore A23187 affect the arrangement of actin and induce nuclear actin paracrystals in PtK2 cells. Exp. Cell Res., 129, 103–114. [DOI] [PubMed] [Google Scholar]
- Perrimon N., Engstrom,L. and Mahowald,A.P. (1989) Zygotic lethals with specific maternal effect phenotypes in Drosophila melanogaster. I. Loci on the X chromosome. Genetics, 121, 333–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D. and Borisy,G.G. (2003) Cellular motility driven by assembly and disassembly of actin filaments. Cell, 112, 453–465. [DOI] [PubMed] [Google Scholar]
- Pollard T.D. and Earnshaw,W.C. (2002) Cell Biology. Elsevier Science, Philadelphia, PA. [Google Scholar]
- Reichstein E. and Korn,E.D. (1979) Acanthamoeba profilin. A protein of low molecular weight from Acanpthamoeba castellanii that inhibits actin nucleation. J. Biol. Chem., 254, 6174–6179. [PubMed] [Google Scholar]
- Rexach M. and Blobel,G. (1995) Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors and nucleoporins. Cell, 83, 683–692. [DOI] [PubMed] [Google Scholar]
- Ribbeck K. and Görlich,D. (2002) The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J., 21, 2664–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K., Lipowsky,G., Kent,H.M., Stewart,M. and Görlich,D. (1998) NTF2 mediates nuclear import of Ran. EMBO J., 17, 6587–6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger J.W., Sanger,J.M., Kreis,T.E. and Jockusch,B.M. (1980) Reversible translocation of cytoplasmic actin into the nucleus caused by dimethyl sulfoxide. Proc. Natl Acad. Sci. USA, 77, 5268–5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U., Hinssen,H., Franke,W.W. and Jockusch,B.M. (1984) Microinjection of actin-binding proteins and actin antibodies demonstrates involvement of nuclear actin in transcription of lampbrush chromosomes. Cell, 39, 111–122. [DOI] [PubMed] [Google Scholar]
- Schluter K., Jockusch,B.M. and Rothkegel,M. (1997) Profilins as regulators of actin dynamics. Biochim. Biophys. Acta, 1359, 97–109. [DOI] [PubMed] [Google Scholar]
- Schutt C.E., Myslik,J.C., Rozycki,M.D., Goonesekere,N.C. and Lindberg,U. (1993) The structure of crystalline profilin-β-actin. Nature, 365, 810–816. [DOI] [PubMed] [Google Scholar]
- Simos G., Grosshans,H. and Hurt,E. (2002) Nuclear export of tRNA. Results Probl. Cell Differ., 35, 115–131. [DOI] [PubMed] [Google Scholar]
- Smith A.E., Slepchenko,B.M., Schaff,J.C., Loew,L.M. and Macara,I.G. (2002) Systems analysis of Ran transport. Science, 295, 488–491. [DOI] [PubMed] [Google Scholar]
- Stade K., Ford,C.S., Guthrie,C. and Weis,K. (1997) Exportin 1 (Crm1p) is an essential nuclear export factor. Cell, 90, 1041–1050. [DOI] [PubMed] [Google Scholar]
- Strom A.C. and Weis,K. (2001) Importin-β-like nuclear transport receptors. Genome Biol., 2, REVIEWS3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M. and Shibata,H. (1985) Poly(l-proline)-binding proteins from chick embryos are a profilin and a profilactin. Eur. J. Biochem., 151, 291–297. [DOI] [PubMed] [Google Scholar]
- Wada A., Fukuda,M., Mishima,M. and Nishida,E. (1998) Nuclear export of actin: a novel mechanism regulating the subcellular localization of a major cytoskeletal protein. EMBO J., 17, 1635–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis K. (2003) Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell, 112, 441–451. [DOI] [PubMed] [Google Scholar]
- Welch M.D., Iwamatsu,A. and Mitchison,T.J. (1997) Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature, 385, 265–269. [DOI] [PubMed] [Google Scholar]
- Wen W., Meinkoth,J.L., Tsien,R.Y. and Taylor,S.S. (1995) Identification of a signal for rapid export of proteins from the nucleus. Cell, 82, 463–473. [DOI] [PubMed] [Google Scholar]
- Wolven A.K., Belmont,L.D., Mahoney,N.M., Almo,S.C. and Drubin,D.G. (2000) In vivo importance of actin nucleotide exchange catalyzed by profilin. J. Cell. Biol., 150, 895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak R.W., Rout,M.P. and Aitchison,J.D. (1998) Karyopherins and kissing cousins. Trends Cell Biol., 8, 184–188. [DOI] [PubMed] [Google Scholar]
- Zechel K. (1980) Isolation of polymerization-competent cytoplasmic actin by affinity chromatography on immobilized DNAse I using formamide as eluant. Eur. J. Biochem., 110, 343–348. [DOI] [PubMed] [Google Scholar]