Abstract
The hairpin ribozyme cleaves phosphodiester bond within a cognate substrate. Structural and biochemical data indicate the conserved A9 and A10 bases reside close to the scissile bond but make distinct contributions to catalysis. To investigate these residues, we replaced the imino moiety of each base with N1-deazaadenosine. This single-atom change resulted in an 8-fold loss in kobs for A9 and displacement of the base from the active site; no effects were observed for A10. We propose the imino moiety of A9 promotes a key water-mediated contact that favors transition-state formation, which suggests an enhanced chemical repertoire for RNA.
RNA enzymes pervade biological systems and are integral to intron splicing, tRNA maturation, genetically encoded peptide-bond formation and gene regulation (1-4). As such, a fundamental understanding of the principles by which RNA molecules control chemical reactivity is of substantial interest. Probing functional groups within an RNA active site can be challenging due to the paucity of natural bases that are isosteric and methods for facile incorporation of such bases into RNA at specific locations.
The hairpin ribozyme (HPRZ) is a self-catalyzing RNA motif innate to tobacco ringspot virus satellite RNA (5). It is a member of the small ribozyme family whose members perform a reversible phosphoryl-transfer reaction (6). Cleavage occurs when the 2′-OH of residue A-1 is transiently deprotonated, thus facilitating SN2 attack of the scissile phosphorus of G+1 (Figure 1A) (7). Products include a 2′,3′-cyclic phosphodiester and a free 5′-OH. The reaction proceeds without need of metal-hydroxide, implicating the local nucleobases directly in catalysis (8). As such, the HPRZ is ideal to elucidate the general principles by which RNA bases govern active site formation and reactivity to promote biological function.
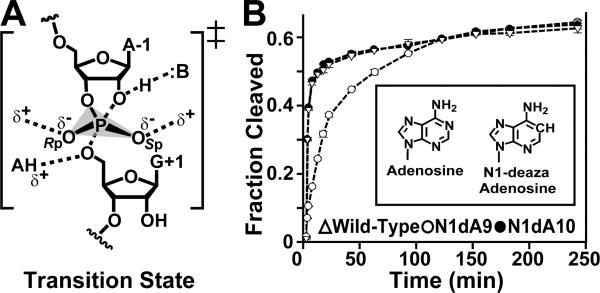
Figure 1.
Transition-state schematic and activity profiles for adenosine and N1-deazaadenosine at positions 9 and 10. (A) Transition-state stabilization of small ribozymes. (B) Single-turnover cleavage of adenosine and N1dA at position 9 and 10 of the HPRZ crystallization construct (Figure S1). The reaction was monitored for 4 h. Inset: bases at 9 and 10 of this investigation.
The HPRZ is one of the most well studied functional RNA motifs. Considerable evidence suggests that proton transfer is facilitated by G8 and A38 (:B and HA respectively, Figure 1A) (9-12). Crystal structures demonstrate that these bases are within hydrogen-bonding distance of the scissile bond (13), which supports a critical role for G8 and A38 in structural organization and catalysis by this small RNA enzyme. In addition to G8 and A38, residues A9 and A10 also reside in the HPRZ active site (13). Nucleotide analog interference mapping indicated that modifications that alter the N1 pKa values at A9 or A10 influence ribozyme activity (14, 15). Less conservative purine and pyrimidine substitutions at these sites affected tertiary folding and ligation with little impact on cleavage (16). Several high-resolution crystal structures revealed ordered waters in the HPRZ active site that coordinate the N1-imino groups of A9 and A10 in conformations representative of the reaction coordinate (17, 18). The observation of a water molecule trapped between the N1 group of A9 and the pro-Sp oxygen of a transition-state analogue (TSA) led to the hypothesis that this interaction favors phosphorane stabilization (18) in a manner comparable to the exocylic amines of G8 and A38 (13).
At present, no investigation has sought to directly probe the importance of the N1 moiety at A9 or A10 of the HPRZ. We therefore synthesized (19) and incorporated (20) N1-deazaadenosine (N1dA) – an inert, structural isostere of adenosine – at these respective positions. A prior investigation using this strategy resulted in no detectable cleavage activity for the N1dA38 variant even though the core RNA fold was preserved (10). We utilized a comparable approach herein to probe the contributions of the N1-imino atoms of A9 and A10 on HPRZ activity and structure. The results provide direct evidence that the imino groups of A9 and A10 function asymmetrically in cleavage and active site formation. These effects are considered in the context of prior structure-function studies that support a role for N1 of A9 in transition-state formation (18).
To assess the activity effects resulting from replacement of the respective A9 and A10 imino groups, we conducted single-turnover cleavage assays (Figure 1B). The wild-type (WT) and N1dA10 constructs exhibited equivalent activity described by a double-exponential equation (10). These molecules exhibited total reactive fractions of 63% and 60%, respectively, and similar rate constants in the fast cleavage phase of k1 = 0.84 ± 0.05 min−1 for WT, and k1 = 0.96 ± 0.04 min−1 for N1dA10; comparable rates were observed for the slow phase with k2 = 0.010 ± 0.002 min−1 for WT and k2 = 0.020 ± 0.003 min−1 for N1dA10. The N1dA9 variant exhibited a total reactive fraction of 62% indicating a level of folding comparable to WT. However, k1 was 0.12 ± 0.02 min−1 (with a k2 of 0.015 ± 0.003 min−1). This value represents an 8-fold loss in rate during the fast phase, suggesting the N1-imino of A9 is more important for catalysis than that of A10, even though neither group directly contacts the scissile bond (13, 17, 18).
Prior structures of the HPRZ in the pre-catalytic state showed the respective N1-imino nitrogens of A9 and A10 coordinate water molecules localized in an active site pocket (17). We hypothesized that the N1 atoms of A9 and A10 promote structural integrity via a hydrogen-bonding network to waters in the pre-cleavage state. The observation of water also supported a previously postulated mechanism involving specific-base catalysis (6, 11, 17). To provide insight into the prospect of local misfolding versus the absence of a key functional group in the active site, we determined structures of the N1dA9 and N1dA10 variants at 2.90 and 2.75 Å resolution, respectively, using pre-catalytic constructs in which the nucleophile was blocked by a 2′-O-methyl group (21, 22). Electron-density maps confirmed that the Watson-Crick edge of each base points into the active site similar to WT (Figures 2A and S2A).
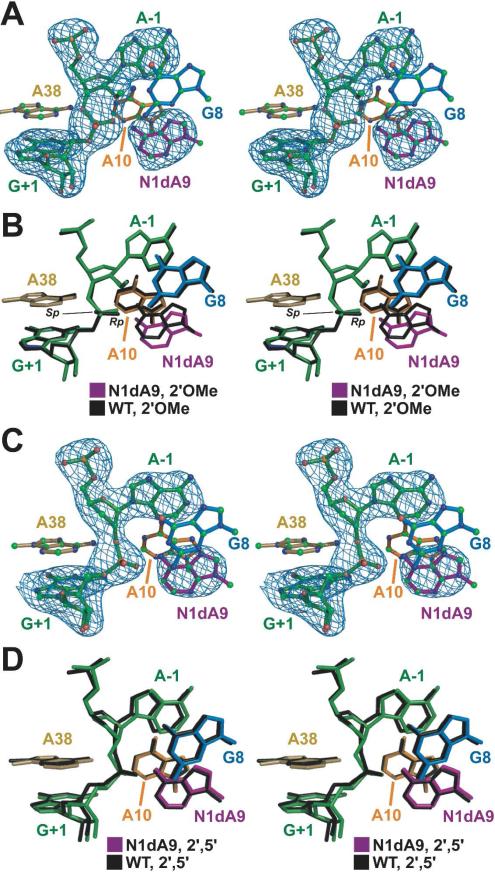
Figure 2.
Stereoviews of mFo– DFc omit electron-density maps and superpositions. (A) The N1dA9 variant in the context of the pre-catalytic construct. Maps are contoured at the 3σ level. (B) The pre-catalytic N1dA9 variant superimposed on the WT structure (PDB 2OUE). (C) The N1dA38 variant in the context of 2′,5′-linkage TSA. (D) The N1dA9 variant in the context of the 2′,5′-TSA superimposed on a matched WT structure (PDB 2P7F).
To reveal significant conformational changes, we superimposed the respective pre-catalytic N1dA9 and N1dA10 structures onto WT coordinates (Figures 2B and S2B). The N1dA10 structure fit well with WT. However, a significant difference was observed between WT and the N1dA9 base, whose position recoiled from the active site to the extent that the C1 atom shifted 1.7 Å relative to N1 of WT (Figure 2B). This shift exceeds the average coordinate error (Table S1), and reduced stacking between N1dA9 and the flanking A10 base by 51.6 Å2 (Figure S3). Absence of an N1 group at A9, and the accompanying conformational change of the base would preclude hydrogen bonding to water molecules that were observed previously between the A9 N1 atom and the exocyclic amine of G+1 (17). No other structural changes were noteworthy.
In the WT pre-catalytic structure, no active site RNA functional groups reside in the vicinity of the N1 atom of A9. This implies that the base shift of the N1-deazaA9 variant herein did not arise from a steric clash with the C1 atom. Possible sources of the structural change include: (i) loss of a key N1 imino hydrogen bond with the active site or (ii) perturbed stacking of the deaza base. However, because replacement of N1 with a CH moiety would make the N1dA9 base stack better due to greater hydrophobicity (23), it seems plausible that loss of a key hydrogen bond causes the observed base shift. It was shown previously that replacement of A9 with purine (P) reduced cleavage by 2-fold (16), whereas substitution with an abasic residue reduced activity 10-fold (11). These observations are not inconsistent with our assignment of an important role for the A9 N1 group in catalysis since both P9 and abasic variants would still be capable of coordinating or accommodating an ordered water molecule.
To explore the compatibility of the N1-deaza base with transition-state geometry we examined A9 and A10 variants in the context of a transition-state analogue (TSA). We established previously that a 2′,5′-phosphodiester bond in lieu of the scissile bond restrains the pro-Rp and pro-Sp non-bridging oxygens to adopt a transition-state-like conformation (18). As such, we determined structures of N1dA9 and N1dA10 in the presence of the 2′,5′-linkage and refined each coordinate set to 2.8 Å resolution, (Figures 2C and S2C).
The results revealed that the Watson-Crick faces of N1dA10 and N1dA9 were pointed toward the active site, which is analogous to the TSA structure of WT. Also similar to WT was the anchoring of the pro-Rp and pro-Sp non-bridging oxygen equivalents of the scissile bond in the N1-deaza variants by the exocyclic amines of G8, A38 and the N6 group from position 9. A structural superposition of the N1dA10-TSA variant with its WT counterpart again revealed no substantial difference in the location of the A10 base (Figure S2D). Elimination of the A10 N6 amine by replacement with purine reduced cleavage by ∼2-fold whereas reversal of the amine location by 2-aminopurine (AP) substitution had little effect on activity (16). In contrast, the A10G variant enhanced cleavage by 5-fold, but decreased the folded population by 25%. Our kinetic data suggest N1dA10 retains folding as observed for other conservative base substitutions such as A10P or A10AP (16). These observations are supported by our N1dA10-TSA structure, which shows the deaza variant is compatible transition-state geometry. Prior nucleotide analog interference mapping demonstrated that N1-imino ionization at A10 favors ligation (15). Our substitution of the N1-imino for a methine (CH) mimics a protonated adenosine, but without localized charge. Although we did not analyze ligation, recent work suggested no benefit to ligation from an ionized imino at position 10 (16). This result appears consistent with the lack of a conformational change at A10 arising from N1-deaza substitution, although further analysis is necessary in the context of pre-ligation (i.e. post-cleavage) structures.
In contrast to the N1dA9 variant in the pre-cleavage state, the N1dA9 base in the context of the TSA did not change relative to WT (Figure 2D). A plausible explanation for this is that the interaction comprising the N6 group of N1dA9 and the pro-Rp oxygen of the scissile bond is sufficient to localize the base in the active site. Importantly, this interaction is highly favored by the 2′,5′-linkage constraint, which is not present in the pre-catalytic conformation in which the N1dA9 base shifts (Figure 2B). Thus, although the N1dA9 TSA structure appears compatible with transition-state geometry, the 8-fold loss of activity implies a deleterious effect. Even more interesting is the observation that crystal structures place the N1 moiety near an ordered water molecule rather than an RNA functional group (17, 18). Inspection of the substrate fraction cleaved by the N1dA9 variant indicates only half of the input ribozyme adopted a catalytically competent conformation in the fast (k1) reaction phase (i.e. the amplitudes for N1dA9 and WT were 0.26 versus 0.47, respectively) even though the total fraction cleaved was nearly identical for each. These observations, along with the pre-catalytic N1dA9 structure, imply that the imino group at A9 influences both folding and catalysis. By contrast, the A9P variant did not impact folding and had little effect on cleavage (16).
Our data support a more significant role for the N1-imino of A9 than its A10 counterpart in HPRZ folding and catalysis. Prior crystal structures revealed a water molecule (W5) within hydrogen bonding distance of the N1 imino of A9 in both the pre-catalytic (17) and oxo-vanadium TSA constructs (18). In the latter complex, W5 interacts with G+1 N2, A9 N1 and the pro-Rp oxygen of the scissile bond. This constellation of interactions was hypothesized to support phosphorane formation by donating a hydrogen bond to the developing oxyanion (Figure 1A). Replacement of the N1 group with CH presumably blocks water binding at this site, consistent with a key role for the imino of A9 in active site folding and function. Water may also restore interactions in instances where bases or their substituents are ablated, thus ameliorating the severity of RNA defects.
Overall, the single-atom analysis of the HPRZ here highlights distinct differences between flanking adenosine residues in the catalytic core. These striking differences originate from localized interactions that lead to exquisite functional group complementarity. The modest 8-fold loss in N1dA9 activity compared to the catastrophic loss of function for N1dA38 (10) suggests that the HPRZ relies mostly on A38 for rate acceleration. However our results suggest an important role for specific imino groups beyond Watson-Crick pairing, as well as a role for oriented water in promoting catalysis. These observations have broad implications for the evolved function and chemical repertoire of non-protein-coding RNAs.
Supplementary Material
ACKNOWLEDGEMENT
Portions of this research were conducted at the Stanford Synchrotron Radiation Lightsource (SSRL), a national user facility operated by Stanford on behalf of the U.S. DOE. The SSRL Structural Molecular Biology Program is supported by the DOE, Office of Biological and Environmental Research, and by the NIH/NCRR Biomedical Technology Program, and the NIGMS.
Footnotes
Support from the National Institutes of Health (NIH) grant R01 GM63162 and the donors of the Petroleum Research Fund (45534 AC4) to JEW is gratefully acknowledged. RCS was supported in part by an Elon Huntington Hooker graduate fellowship.
The atomic coordinates and structure factor amplitudes are deposited in the Protein Data Bank as entry codes 312Q, 312R, 312S and 312U.
SUPPORTING INFORMATION AVAILABLE
Supporting methods, tables and figures are available for kinetic analysis and crystallography. Materials are available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 2.Valadkhan S, Manley JL. Nature. 2001;413:701–707. doi: 10.1038/35099500. [DOI] [PubMed] [Google Scholar]
- 3.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 4.Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 5.Hampel A, Tritz R. Biochemistry. 1989;28:4929–4933. doi: 10.1021/bi00438a002. [DOI] [PubMed] [Google Scholar]
- 6.Fedor MJ. Annu Rev Biophys. 2009;38:271–299. doi: 10.1146/annurev.biophys.050708.133710. [DOI] [PubMed] [Google Scholar]
- 7.Fedor MJ. J Mol Biol. 2000;297:269–291. doi: 10.1006/jmbi.2000.3560. [DOI] [PubMed] [Google Scholar]
- 8.Nesbitt S, Hegg LA, Fedor MJ. Chem Biol. 1997;4:619–630. doi: 10.1016/s1074-5521(97)90247-7. [DOI] [PubMed] [Google Scholar]
- 9.Liu L, Cottrell JW, Scott LG, Fedor MJ. Nat Chem Biol. 2009;5:351–357. doi: 10.1038/nchembio.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spitale RC, Volpini R, Heller MG, Krucinska J, Cristalli G, Wedekind JE. J Am Chem Soc. 2009;131:6093–6095. doi: 10.1021/ja900450h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuzmin YI, Da Costa CP, Cottrell JW, Fedor MJ. J Mol Biol. 2005;349:989–1010. doi: 10.1016/j.jmb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 12.Kuzmin YI, Da Costa CP, Fedor MJ. J Mol Biol. 2004;340:233–251. doi: 10.1016/j.jmb.2004.04.067. [DOI] [PubMed] [Google Scholar]
- 13.Rupert PB, Massey AP, Sigurdsson ST, Ferre-D'Amare AR. Science. 2002;298:1421–1424. doi: 10.1126/science.1076093. [DOI] [PubMed] [Google Scholar]
- 14.Ryder SP, Strobel SA. J Mol Biol. 1999;291:295–311. doi: 10.1006/jmbi.1999.2959. [DOI] [PubMed] [Google Scholar]
- 15.Ryder SP, Oyelere AK, Padilla JL, Klostermeier D, Millar DP, Strobel SA. RNA. 2001;7:1454–1463. [PMC free article] [PubMed] [Google Scholar]
- 16.Gaur S, Heckman JE, Burke JM. RNA. 2008;14:55–65. doi: 10.1261/rna.716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salter J, Krucinska J, Alam S, Grum-Tokars V, Wedekind JE. Biochemistry. 2006;45:686–700. doi: 10.1021/bi051887k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torelli AT, Krucinska J, Wedekind JE. RNA. 2007;13:1052–1070. doi: 10.1261/rna.510807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cristalli G, Franchetti P, Grifantini M, Vittori S, Bordoni T, Geroni C. J Med Chem. 1987;30:1686–1688. doi: 10.1021/jm00392a029. [DOI] [PubMed] [Google Scholar]
- 20.Wedekind JE, McKay DB. Methods Enzymol. 2000;317:149–168. doi: 10.1016/s0076-6879(00)17013-2. [DOI] [PubMed] [Google Scholar]
- 21.Rupert PB, Ferre-D'Amare AR. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- 22.Alam S, Grum-Tokars V, Krucinska J, Kundracik ML, Wedekind JE. Biochemistry. 2005;44:14396–14408. doi: 10.1021/bi051550i. [DOI] [PubMed] [Google Scholar]
- 23.Guckian KM, Schweitzer BA, Ren RXF, Sheils CJ, Tahmassebi DC, Kool ET. J Am Chem Soc. 2000;122:2213–2222. doi: 10.1021/ja9934854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.