Summary
Identification of bona fide tumor suppressors is often challenging because of the large number of alterations present in most human cancers. To evaluate candidates present within regions recurrently deleted in human cancers we coupled high-resolution genomic analysis with a two-stage genetic study using RNA interference (RNAi). We found that Cyfip1, a subunit of the WAVE complex, which regulates cytoskeletal dynamics, is commonly deleted in human epithelial cancers. Reduced expression of Cyfip1 is commonly observed during invasion of epithelial tumors and it associated with poor prognosis in same tumor types. Silencing of Cyfip1 disturbed normal epithelial morphogenesis in vitro and cooperated with oncogenic Ras to produce invasive carcinomas in vivo. Mechanistically, we have linked alterations in WAVE-regulated actin dynamics with impaired cell-cell adhesion and cell-ECM interactions. Thus, we propose Cyfip1 as an invasion suppressor gene.
Introduction
Development of technologies that scan the genome, such as expression profiling (Fan et al., 2006; Trevino et al., 2007), analysis of copy number variations (Firestein et al., 2008; Hicks et al., 2006), or massively parallel sequencing (Campbell et al., 2008; Chiang et al., 2009) has provided us with the potential to molecularly profile the entire set of genetic alterations present in human cancers. However, the inability to discriminate which of these myriad genetic variations causally contribute to tumorigenesis remains a major barrier.
The use of RNAi to attenuate the expression of candidate genes represents a powerful strategy to link genotype to phenotype. To examine the impact of silencing candidates within regions of deletion in human tumors, we chose to use a three-dimensional (3D) culture model that recapitulates many aspects of epithelial morphogenesis in vitro (Debnath et al., 2003; Fischbach et al., 2007).
By coupling genomic and genetic analysis, we have identified Cyfip1 as a potential tumor suppressor that regulates invasive behavior. Cyfip1 is a Rac-1 interacting protein (Kobayashi et al., 1998), which transmits signals from Rac-1 to the ARP2/3 complex by modulating the activity of the Wasp family member, Wave, within the WAVE complex. Wave-mediated activation of ARP2/3 induces the nucleation of G-actin to form a membrane protrusion at the leading edge of cells growing in classical two-dimensional (2D) cultures, called a lamellipodium (Kunda et al., 2003; Stradal et al., 2004; Takenawa and Suetsugu, 2007). Although the function of the WAVE complex in site-directed actin polymerization and membrane protrusion formation is well characterized, it was unclear whether distortion of this process could influence tumorigenesis. Our data indicate that Cyfip1 can impact tumorigenesis through its effects on cytoskeletal dynamics and cell-cell and cell-substratum adhesion. These studies also provide a general path toward identifying underlying driver mutation in regions of genetic aberration in human cancers.
Results
Cyfip1 is a regulator of epithelial morphogenesis that is altered in tumors
We used high-resolution profiling of DNA copy number alterations (Hicks et al., 2006) to detect focal deletions (< 2Mb) in a panel of 293 primary human cancers and 71 cancer cell lines (Powers et al. in prep). More than 90% of these samples were from common epithelial tumor types, including 83 lung, 104 breast, and 71 colon cancers. Of 36 regions identified using our criteria, 10 harbored genes previously shown to be homozygously deleted in cancer, including RB1, PTEN, CDK2N, SMAD4/DPC4 (Ikediobi et al., 2006) LRP1B, FHIT, PARK2, WWOX (Smith et al., 2007), miR-16/miR-15 (Calin et al., 2002), RUNX1 (Silva et al., 2003). Two additional loci contained candidate tumor suppressor genes with observed bi-allelic point mutations, including the activin A type II receptor (ACVR2) (Hempen et al., 2003) and the phosphatase, PTPRT (Wang et al., 2004). We also detected 24 focally deleted loci that did not contain any known tumor suppressor (Table S1, Fig. 1A).
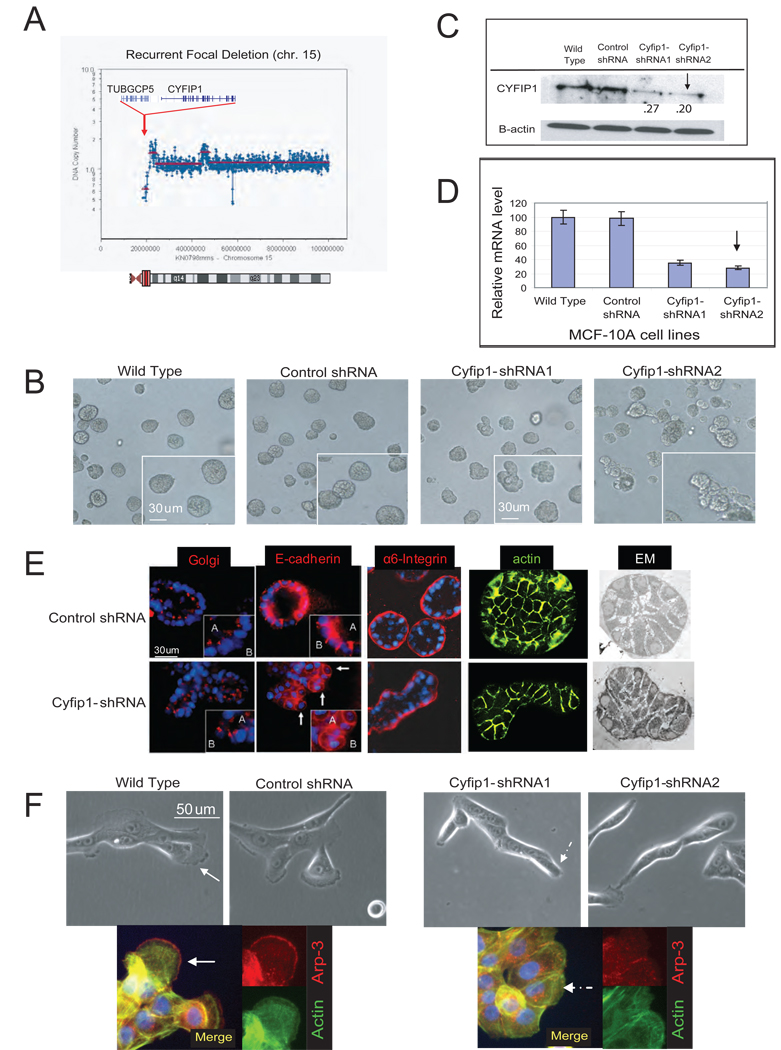
Figure 1. ROMA deletion profile of Cyfip1 and effect of knock-down on morphogenesis.
(A) Representative focal deletion found on chromosome 15 in a lung cancer sample. The genes shown correspond to the consensus epicenter of the deletion (red area in the chromosome ideogram). (B) Morphology of the acini formed by control cells (wild type and control shRNA) and Cyfip1 knock-down MCF-10A cells. (C) Level of Cyfip1 silencing in the MCF-10A cells analyzed by Western blotting and (D) quantitative RT-PCR (QRT-PCR). Error bars show standard deviation from the mean (n=3). The arrow indicates the shRNA selected for further studies. (E) Characterization of acinar architecture by Immunofluorescence and electromicroscopy (EM). (F) Morphology of the cells from panel B when grown in classical 2D cultures. The IF presents the same cells staining with Arp-3, a canonical marker of lamellipodia and actin (Boguslavsky et al., 2007). Lamellipodial structures are indicated by continuous arrows, lack of lamellipodia is indicated by discontinuous arrows..
MCF-10A is an immortalized but not transformed mammary epithelial cell line that forms 3D acinar structures when grown on extracellular matrix components (ECM, matrigel). This model recreates aspects of epithelial cellular organization that occur in tissues more than do classical 2D cultures. Alterations in several oncogenes or tumor suppressors involved in proliferation, polarization or apoptosis disturb normal acinar architecture, resulting in overtly abnormal morphologies that can be easily observed (Debnath et al., 2003; Muthuswamy et al., 2001)
Focusing on focal deletions without known tumor suppressors, we used stable shRNA expression to create cell lines in which twenty nine of the thirty five genes located in these regions were silenced to varying degrees (Table S1). These lines were individually tested in the 3D morphogenesis assay. Although the majority of shRNA-expressing MCF-10A derivatives produced acini with a normal appearance, cells expressing shRNAs against Cyfip1 generated abnormal structures (Fig. 1B). 20% of knock-down acini appeared as shapeless or oval structures instead of the normal symmetrical spheres (<5% in control acini;T-test, p<.01). This phenotype was reproduced using several distinct Cyfip1 shRNAs, and its severity correlated with the degree of Cyfip1 knock-down (Fig. 1C, D).
Although knock–down acini formed hollow lumens, they were smaller than those formed in controls due to the transition from spherical to oval shape (Fig. 1E). Examination of structural markers (Debnath et al., 2003) revealed that a majority of the cells in the knock–down acini maintained basal (a-6 Integrin) and lateral (E-cadherin) polarization and the apical/basal organization of the golgi and nuclei Interestingly, it was common to find individual cells that had broken out the acinar architecture. These cells displayed a less cubical, more spherical shape with abnormally intense basal E-cadherin staining and a random positioning of the golgi/nucleus. All of these characteristics were suggestive of polarization defects (Fig. 1E). Importantly, while wild-type acini presented an almost perfect symmetry, a disorganized and chaotic distribution was present in knock-down acini (Fig. 1E). Finally, we did not find significant differences in the levels of proliferation (Ki67) or apoptosis (activated caspase-3) in knock-down acini (data not shown). As expected, Cyfip1 knock-down cells grown in standard 2D culture conditions did not form lamellipodia (Fig. 1F).
Alteration of Cyfip1 in human cancers
Since Cyfip1 loss could alter acinar morphology, implicating it as a potential tumor suppressor, we examined a set of 841 additional human tumors where we catalogued both focal and larger deletions affecting the Cyfip1 locus. This revealed frequent alteration of Cyfip1 in epithelial tumors (Table S2). Most often, losses appeared to be heterozygous and affected most of 15q, although focal deletions affecting smaller regions were also observed (Fig. 1A). Importantly, a significant correlation existed between DNA copy number and Cyfip1 expression for all tumor types examined (Table S2).
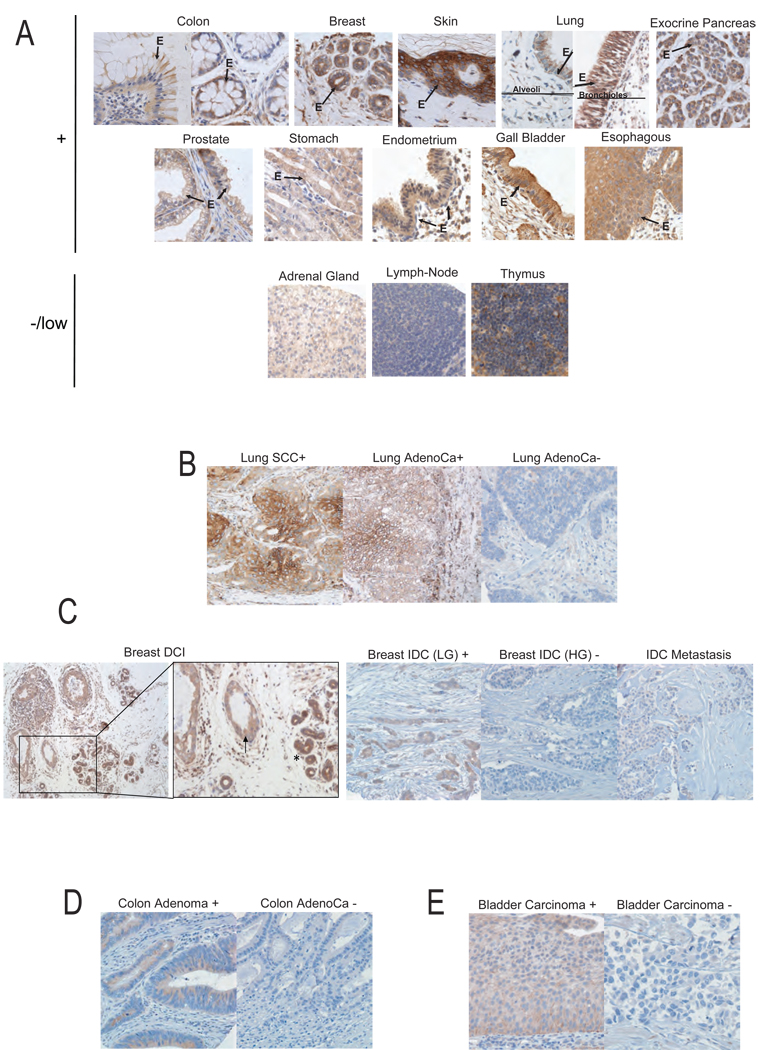
Frequently, the accumulation of genetic and/or epigenetic alterations in a tumor suppressor leads to loss of its expression. Immunohistochemistry showed robust Cyfip1 staining in the epithelium in human tissues (Fig. 2A). In contrast, an examination of 249 tumor samples representing four of the most common human epithelial tumor types (breast, colon, lung and bladder) indcated that loss of expression of Cyfip1 (defined as fewer than 10% positive tumor cells) was a common event. Up to 63% of lung (32/51) (Fig. 2B), 31% of breast (22/70) (Fig. 2C), 59% of colon (39/66) (Fig. 2D), and 24% of bladder (15/62) (Fig. 2E) cancers had negative staining. Importantly, 100% superficial bladder carcinomas (non-invasive) were Cyfip1-positive (30/30) and displayed strong staining (80% of samples had more than 40% positive cells), whereas 47% of invasive bladder tumors (15/32) were negative (p=1.65×10−5). It is also notable that, 75% of breast metastases analyzed (Fig. 2C) were Cyfip1 negative (3/4). Additionally, we analyzed two ductal carcinoma in situ (DCI) of the breast where normal ducts were surrounding the tumor lesion (Fig. 2C). This gave us the possibility of directly comparing the dysplastic cells with their normal counterparts. Although DCI lesions were positive for Cyfip1 staining, they did show a clear reduction in the staining as compared to normal ducts.
Figure 2. Cyfip1 expression in human normal and tumor tissues and its association with tumor progression.
Representative figures of Cyfip1 expression detected by immunohistochemistry (IH) in (A) normal tissues. The “E” and the arrows indicate the epithelium. (B) The majority of lung squamous cell carcinomas (SCC) and some adenocarcinomas (AdenoCa) showed a membranous-cytoplasmic expression of the protein (+), while most lung adenocarcinomas were found to have a negative phenotype (−). (C) Left panels show a ductal carcinoma in situ of the breast (DCI) (arrow) together with normal ducts (asterisk). Right panels show low grade (LG) invasive ductal breast carcinoma (IDC) exhibited intense and homogenous staining of Cyfip1, while most high grade (HG) IDC and bone metastatic adenocarcinoma lesions revealed a negative phenotype. (D) In colon tumor samples, high grade non-invasive adenomas maintain Cyfip1 while it is lost in a majority of invasive adenocarcinoma. (E) In bladder tumor samples, we observed that all low grade non-invasive superficial bladder carcinomas displayed a positive phenotype, while the majority of high grade invasive bladder carcinomas revealed undetectable Cyfip1 levels.
Globally, our results suggest that loss of Cyfip1 expression correlates with tumor progression in epithelial cancers and it raised the possibility that loss of Cyfip1 might correlate with clinical outcome. We therefore examined by quantitative RT-PCR (QRT-PCR) the expression of Cyfip1 in an additional one hundred and seventy human tumors (84 colons and 86 breasts) with available histological and pathological data (see Supplementary Methods). In order to select a metric to stratify tumor samples in a way comparable to our immunohistochemistry, we first selected 8 tumors with different expression levels of Cyfip1 (from very low to high) that also had available matched paraffin sections. We compared QRT-PCR and immunohistochemistry data to generate a standard curve (Fig. S1). Through this approach, we found that low expression of Cyfip1 (negative tumors) was significantly (p<0.05) associated with higher stage. This also (p<0.1) correlated with lymph node metastases in invasive ductal breast carcinomas (IDC). In colon cancers, low expression of Cyfip1 was significantly associated with vascular invasion and with higher stage (Table S3). It was therefore critical to probe mechanisms by which loss of Cyfip1 might contribute to tumorigenesis.
Cyfip1 regulates epithelial morphogenesis as part of the WAVE complex
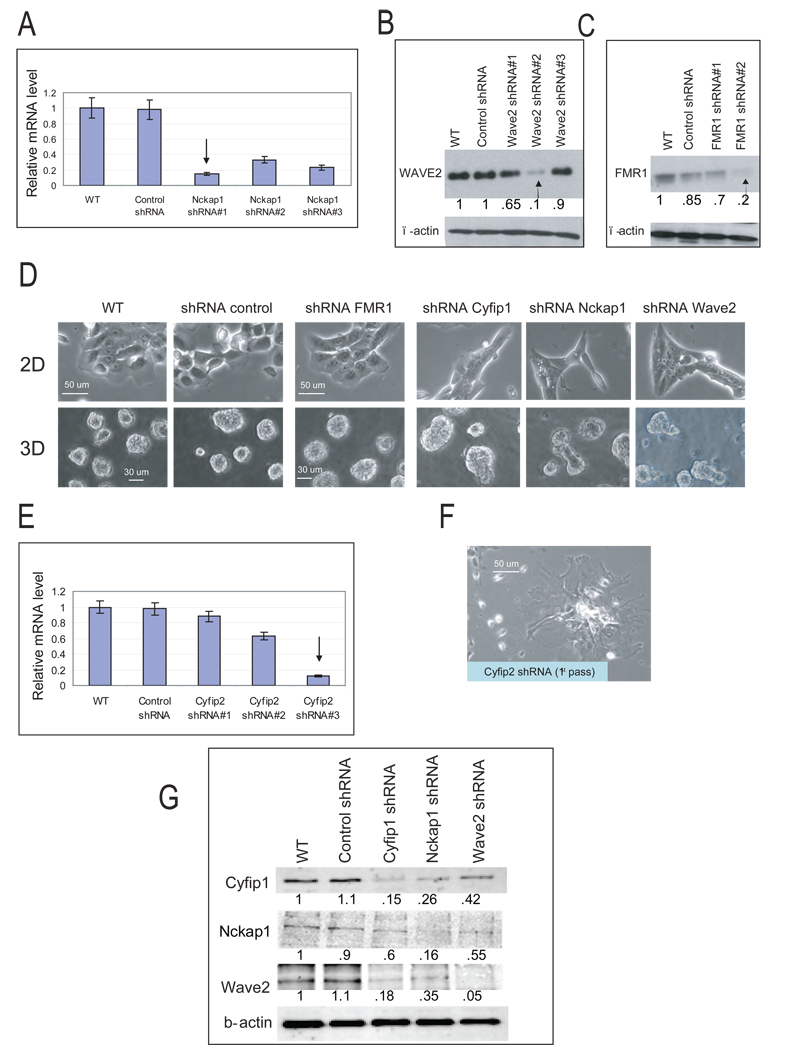
In addition to its role as a component of WAVE, Cyfip1 interacts with FMR1, an RNA binding protein (Billuart and Chelly, 2003). Although its function in that context is poorly understood, various observations suggest that it affects local protein translation and/or transport (Zalfa et al., 2006). We set out to determine whether our observed phenotypes were mediated by defects in actin remodeling (WAVE pathway), by alterations in mRNA metabolism (FMR1 pathway), or by both. ShRNAs targeting core components of the two pathways were selected (Fig. 3A–C) and stably expressed in MCF-10A cells to produce knock-down lines. Suppression of FMR1 did not produce any evident difference in 2D colonies or 3D acinar structures. In contrast, knock-down of WAVE pathway components, Nckap1 and Wave-2, generated phenotypes similar to those observed upon Cyfip1 silencing (Fig. 3D). The phenotype seen in Nckap1 knock-down cells was exceptionally penetrant, with almost 100% of colonies in 2D lacking lamellipodia (T-test, p<.01) and 50–70% (T-test, p<.01) of acini displaying abnormal morphologies (Fig. S2A). To verify the specificity of the observed phenotypes we transduced WAVE knock-down cells with a non-targeted form of the mouse Cyfip1 homolog. This rescued the formation of lamellipodia in Cyfip1 but not in Nckap-1 knock-down cells (Fig. S2B–D).
Figure 3. Silencing of WAVE complex components phenocopies the knock-down of Cyfip1.
(A) Representative morphologies of FMR1 and WAVE component knock-down cells growing in 2D and 3D culture. (B–D) Level of specific gene silencing in MCF-10A cells from panel A analyzed by Western blotting or QRT-PCR. (E) Level of Cyfip2 silencing in MCF-10A. Error bars show standard deviation from the mean (n=3). The arrow indicates the shRNA selected for further studies. (F) Representative picture of the phenotype generated in 2D cultures after silencing of Cyfip2. (G) Western blots show the steady state protein levels of Cyfip1, Nckap1 and Wave2 after individual WAVE components were silenced. The numbers inside the western-blot are the quantitative values of the bands normalized to the wild type.
Cyfip1 is highly conserved through evolution. In humans, two homologs, Cyfip1 and Cyfip2, share 88% amino acid sequence identity (Schenck et al., 2001). Interestingly, RNAi of Cyfip2 (Fig. 3E) induced abnormal spreading of MCF-10A cells (Fig. 3F) and a dramatic reduction in proliferation (Fig. S3A, B). Consequently, Cyfip2 was not studied further.
Silencing any WAVE component destabilizes the complex, leading to proteasome-mediated degradation of the other subunits (Innocenti et al., 2004; Kunda et al., 2003). We also observed this phenomenon in cultured MCF-10A cells (Fig. 3G) and in vivo in subcutaneous tumors formed in nude mice by injecting oncogene-transformed MCF-10A cells carrying different WAVE shRNAs (Fig. S4).
Three Wave family members (Wave1–3) are present in the mammalian genome (Takenawa and Suetsugu, 2007). Their Individual knock-down has been reported to increase the steady state levels of others family members, possibly as a compensatory mechanism (Zipfel et al., 2006). We noted a similar effect in our Wave-2 knock-down cells (Fig. S3C). Overall, these results suggest that interference with the WAVE complex and its function in regulating actin dynamics leads to the phenotypes similar to those generated by suppression of Cyfip1.
These data support the hypothesis that Cyfip1 might contribute to tumor suppression through its role in the WAVE complex. In accord with this notion, our ROMA analyses revealed that deletions of Wave2 were also present in human cancers, although the Nckap1 locus was highly stable (Table S4). We therefore set out to understand precisely how changes in WAVE activity might impact critical hallmarks of cancer cells.
Suppression of WAVE disrupts cell adhesion by altering E-cadherin distribution
Uncontrolled proliferation and increased resistance to stress stimuli are two hallmarks of cancer cells (Hanahan and Weinberg, 2000). WAVE knockdown cells, showed no significant difference in short-term growth in 2D or 3D cultures (Fig. S4A, B) in comparison to control cells. These results agree with our previous finding that the level of Ki67 was unchanged in matrigel-cultured Cyfip1 knock-down acini. Moreover, knock-down cells behaved similarly to controls during loss of anchorage (anoikis) or growth factor starvation (Fig. S4C, D).
MCF-10A cells are not tumorigenic, but they can be transformed by strong oncogenic stimuli (Datta et al., 2007). To test whether the growth of MCF-10A cells is affected in an in vivo setting by compromising WAVE activity, we knocked-down WAVE components in normal or oncogene-transformed MCF-10A cells (overexpressing a constitutively active form of ErbB2, MCF-10A/ErbB2) and injected these into nude mice. These experiments revealed that disruption of the WAVE pathway neither transformed wild-type MCF-10A cells nor altered the growth of transformed MCF-10A/ErbB2 as subcutaneous xenografts (Fig. S4E, F). Xenograft tumors were of high grade but showed no remarkable differences between controls and knock-downs. This result was not exclusive to MCF-10A cells, as silencing of Nckap1 did not alter the ability of three other tumorigenic breast cancer lines (MCF-7, T-47D and MDA-MB-231) to form subcutaneous tumors in immunocompromised mice (Fig. S4G, data not shown).
Along with a balance between proliferation and cell death, epithelial tissue homeostasis is enforced by its characteristic architecture (Hanahan and Weinberg, 2000; Perez-Moreno et al., 2003), and distortion of this organization, induced by defects in cell adhesion, is often associated with epithelial tumorigenesis (Shin et al., 2006). Therefore, we tested whether knocking down WAVE components had any effect on cell-cell and cell-substratum adhesion.
MCF-10A cells were plated on plastic and grown to confluence for 48 hours; in this scenario they form a typical epithelial sheet wherein the cells are tightly bound to each other by a variety of adhesion complexes and junctions (Underwood et al., 2006). Next, EDTA was added in order to chelate calcium, an essential modulator of cadherin-mediated cell-cell adhesion, and changes in the morphology of the culture were recorded by live imaging. After 1 hour, control cells started losing cell-cell adhesion and showed fewer surface contacts, though they still maintained significant interactions at this timepoint (Movie S1A, B, Fig. 4A). In contrast, EDTA treatment of MCF-10A cells engineered to have reduced cell adhesion capacity (overexpressing Snail) resulted in rapid rounding of cells and loss of almost all cell-cell contacts (Movie S1C, Fig. 4A). WAVE knock-down cells also lost cell-cell adhesion more rapidly than controls (Movies S1D–F, Fig. 4A). To obtain a more quantitative measurement of this defect we performed cell aggregation assays (Wilkemeyer et al., 2002). Here, the percentage of individual cells in suspension that form aggregates through time reflects their ability to form cell-cell contacts. Again, knock-down cells presented a significant reduction (p<.01) in the number of aggregates formed after 3 hours (Fig. 4B).
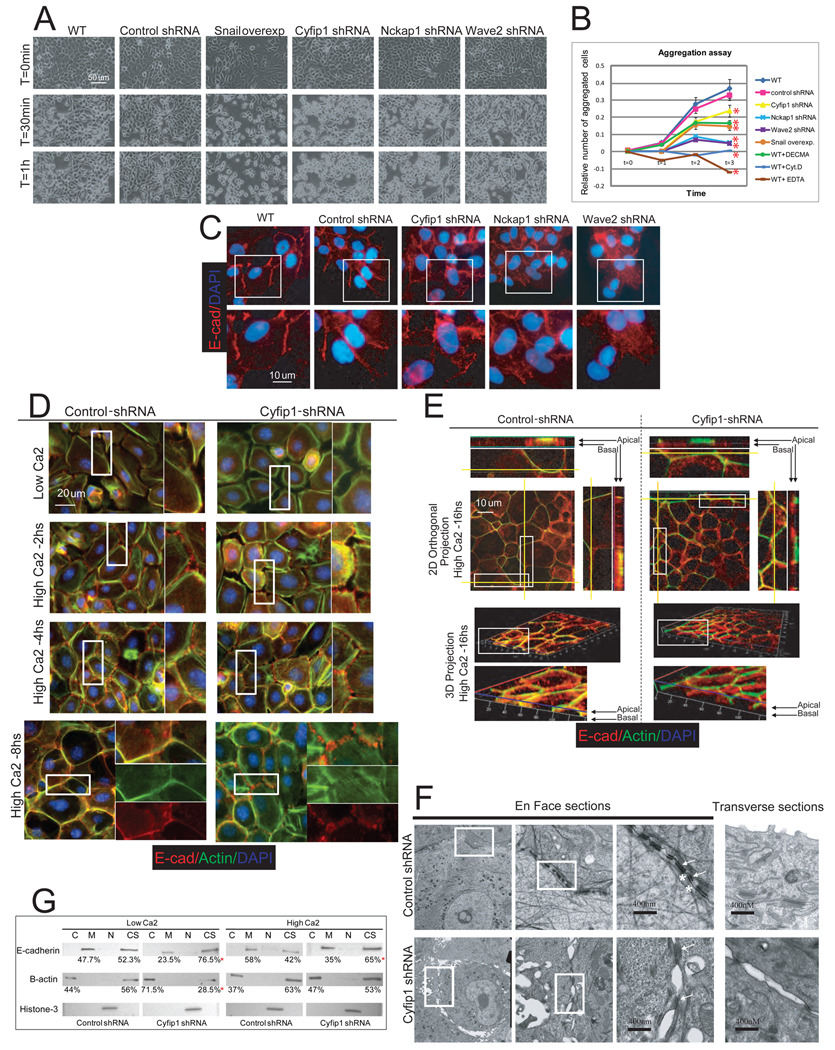
Figure 4. Silencing of WAVE complex components reduces cell-cell adhesion.
(A) Representative snapshot from supplementary movies 1A-1F after EDTA (5mM final concentration) was added to equally confluent MCF-10A cultures. (B) Aggregation experiment showing the percentage of aggregates formed during 3 hours. MCF-10A cells where actin dynamics (cytochalasin-D, 1ug/ml) or E-cadherin mediated adhesion (5mM EDTA,10ug/ml of DECMA E-cadherin blocking antibodies, and Snail-overexpressing cells) was abolished represent controls. The red asterisks indicate p values<.05 (T-test of three replicas) when the aggregation values after 3 hours were compare with wild type cells. (C) Immunofluorescence showing E-cadherin and nuclear staining in controls and knock-down MCF-10A cells. Through the entire figure magnified panels represent images of the selected area (white rectangle). (D) Immunofluorescence showing the development of cell-cell adhesion in control and Cyfip1 knock-down keratinocytes after calcium exposure (2, 4 and 8 hours)..(E) Immunofluorescence of control and Cyfip1 knock-down keratinocytes 16 hours after calcium exposure. 4X magnified images of selected areas together with 2D orthogonal projections (yellow lines) are shown on the side of the upper half of the figure. A 3D reconstruction of the same keratinocyte layer is shown in the lower half. 4X magnified images of selected areas are displayed below. (F) The composition shows structural studies (EM) of control and Cyfip1 knock-down keratinocytes 16 hours after calcium switch. En Face sections present three 10X consecutive magnifications of the same area. Arrows indicate desmosomes, and asterisks indicate adherence junctions. The transverse section shows the apical part of two cells from the same cultures shown En Face. (G) Distribution of E-cadherin and B-actin in different fractions, cytoplasmic (C), membrane (M), nuclear (N) and cytoskeleton (CS) of control and Cyfip1 knocked-down cells growing in low and high calcium media. The numbers below the blots represent the average of the quantitative value of bands in the blots from 3 independent experiments. The red asterisks in (B) and (G) represent p values<.05 (T-test of three replicas) when the values from knock-down cells were compared with controls.
To investigate the mechanisms underlying reduced adhesion, we examined the localization of the key adherence junction component, E-cadherin. As expected, E-cadherin accumulated along cell-cell boundaries in control cells. In contrast, knock down of WAVE subunits generated very compacted colonies with a more diffuse pattern of staining in the cells located at the margins (Fig. 4C). In fact, 2D orthogonal projections showed that E-cadherin was more spread between overlapping areas of neighboring cells (Fig. S5A). This pattern changed near the center of the colonies. Here, it was common to see very intense E-cadherin signals (Fig. S5B). Remarkably, DAPI staining of the colonies showed that knock-down cell nuclei commonly overlap, revealing that the cells had in fact piled on top of each other (Fig. S5C). To unify these observations we monitored the growth of our engineered MCF-10A variants (Movies S2A–D). We found that colonies of wild-type cells formed by an orchestrated series of events in which the cells first divide and then spread apart from each other. This process is controlled by lamellipodia-mediated motility at the periphery of the colony. Knock-down cells divided normally but were unable to separate because of the absence of lamellipodia. This, in turn, led to the formation of a mass of highly compacted cells.
During immortalization and transformation of cultured cell lines, like MCF-10A, normal actin organization and formation of adhesion contacts become aberrant (Vasioukhin et al., 2000). Low-passage keratinocytes represent an elegant and well characterized model that more closely maintains the epithelial characteristics of normal epidermis (Vaezi et al., 2002; Vasioukhin et al., 2000). Briefly, when mouse keratinocytes are grown in media containing low calcium concentrations, they display low levels of cell-cell interaction and high motility. Increased calcium concentrations induce actin cytoskeleton remodeling that is essential to promote contacts between neighboring cells. These are eventually stabilized by cadherin-mediated intercellular adhesion. Thus, upon calcium switch filopodia-like projections emerge between neighboring cells and become decorated with E-cadherin to generate characteristic structures called adhesion zippers. With the passage of the time an increasing number of E-cadherin puncta accumulate at the zippers, which then form a continuous line at cell borders. Finally, keratinocytes polarize forming a honeycombed network of thick cortical actin belts and E-cadherin complexes in the apical plane and fluid membranes with less organized E-cadherin structures at the basal plane.
Using the keratinocyte model, we analyzed the formation of cell contacts and E-cadherin distribution after silencing the mouse homologs of Cyfip1 and Nckap1 (Fig. S5D, E). As expected, at low calcium concentrations, silencing of WAVE components compromised the formation of lamellipodia (Movies S3A, B, Fig. S5F). After calcium switch, control cells followed the normal pattern of adhesion (Movie S4A, Fig. 4D, E, Fig. S5G). Knock-down keratinocytes presented only finger-like protrusions that did not efficiently form stable interactions (Movie S4B), and numerous but transient contacts between neighboring cells predominated. This correlated with reduced E-cadherin immunostaining and fewer adhesion zippers (Fig. 4D, Fig. S5G). At 8 hours after calcium exposure, a wider distribution of E-cadherin complexes and a more diffuse actin staining was observed in knock-downs as compared to controls (Fig.4D, Fig.S5G). Finally, at later time points (16 hours), 2D orthogonal and 3D projections showed that in normal keratinocytes, E-cadherin coexists with a cortical actin ring in the apical part of cells. In contrast, knock-down keratinocytes presented a patched and diffuse distribution of actin and E-cadherin with a scattered overlap between them (Fig.4E). Ultrastructural studies at this latest time point provided more detailed information. We noted that the mature architecture, normally characterized by the alignment of alternating adherence junctions (AJ) and desmosomes along sealed membranes, was replaced in knock-down cells by immature desmosomes with fewer AJ. Importantly, this led to large gaps and spaces between closely opposed keratinocytes, (Fig. 4F, Fig. S5H).
We obtained cellular fractions from keratinocytes grown in low and high calcium to study the distribution of actin and E-cadherin. We found that, in control keratinocytes, cytosolic G-actin is mobilized to the cytoskeleton (F-actin) in the presence of calcium, and this correlates temporally with the enrichment of E-cadherin in the membrane fraction (Fig. 4G). These data are consistent with the immunofluorescence results where first, thick bundles filopodia and then, a cortical ring of actin are formed while E-cadherin accumulated at cell-cell contacts. In knock-down cells grown in low calcium media, a substantially larger amount of actin was found unpolymerized in the cytosol (Fig. 4G). After calcium switch, actin was still mobilized to the cytoskeletal fraction; however, this occurred to a lower degree than was observed in wild-type keratinocytes. Notably, E-cadherin accumulation at the membrane was aslo reduced (Fig. 4G).
Thus, as an integrated model to explain the defects observed in WAVE deficient cells, we propose that a lack of WAVE activity reduces the formation of AJ by reducing the formation and stabilization of filopodia-like structures and the accumulation of E-cadherin complexes at cell-cell boundaries. This physically compromises the drawing together of opposing cell surfaces, leaving the passive formation of desmosomes (Vasioukhin et al., 2000) as the only event that clamps two opposing membranes. As a result, the entire epithelial architecture fails to assemble properly. In accord with this model, silencing of WAVE components recapitulated the phenotype observed when AJ formation was compromised by α-catenin knock out or by perturbing VASP/Mena function (Vasioukhin et al., 2000). It is interesting to note that in our knock-down cells VASP localized normally at adhesion zippers (Supplementary Fig. 5I). However, as expected, its maturation towards a more continuous honeycomb-like belt failed.
To generate knock-down cells, we used viral transduction to produce pooled cell populations. Thus, different degrees of knock-down can be expected in individual cells, depending upon viral integration sites and copy number. IF staining of Cyfip1 in our pooled cells revealed that approximately 10% of cells still retained high expression levels, and these cells retained the ability to normally relocate E-cadherin upon calcium switch (Fig. S5J). Moreover, expression of exogenous Cyfip1 rescued the phenotype induced by silencing of the endogenous protein (Fig. S5K).
Compromising WAVE reduces epithelial cell-ECM adhesion by disturbing focal adhesion complexes
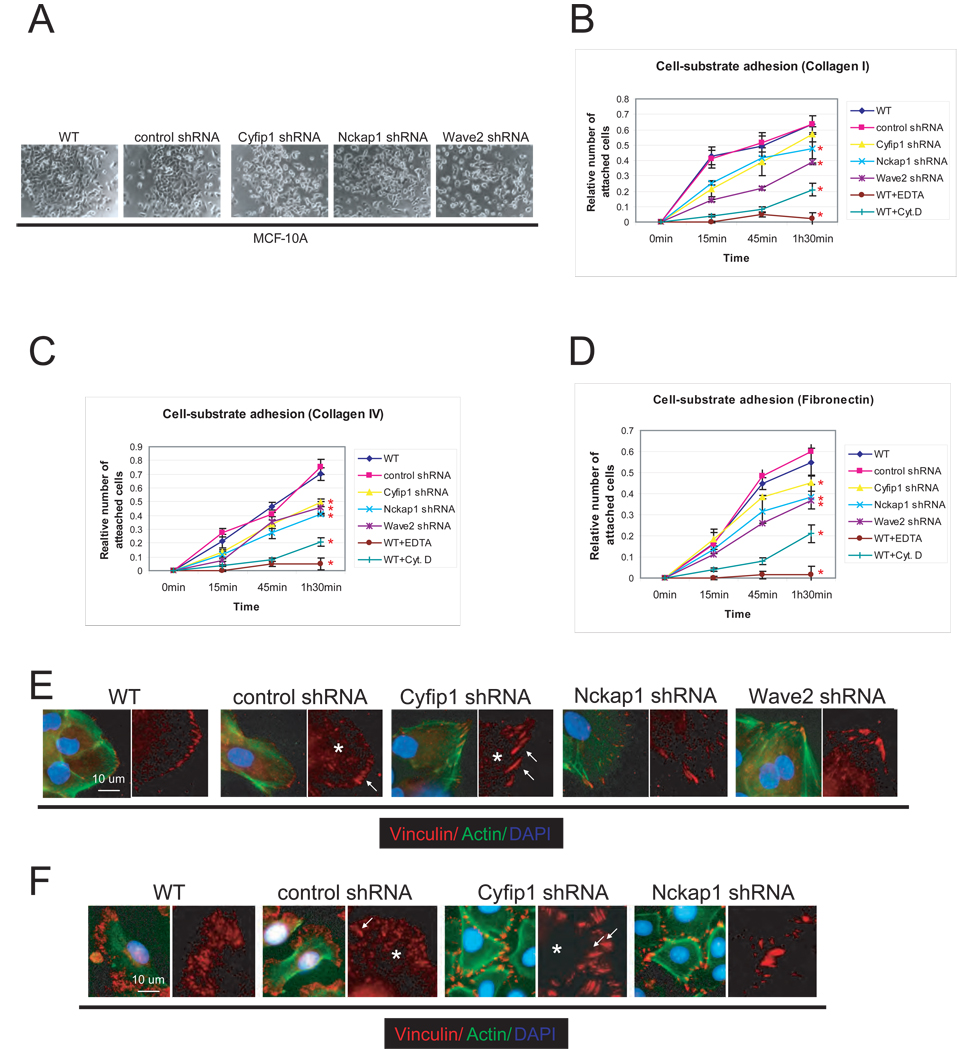
MCF-10A WAVE knock-down cells also displayed an obvious defect in their ability to attach to standard tissue culture plates. Even 6 hours after plating, the majority of cells were still rounded and showed very little spreading (Fig. 5A). Coating plates with Collagen-I, Collagen-IV, and Fibronectin accelerated the attachment of MCF-10A cells while no effect was observed with Laminin or Fibrinogen (Fig S6A). Cyfip1 knock-down cells showed a pleiotropic reduction in adhesion to all tested substrates (Fig. 5B–D).
Figure 5. Silencing of WAVE complex components reduces epithelial cell-ECM adhesion.
(A) Representative image of the attachment and spreading of MCF-10A WAVE knock-down cells 6 hours after plating. Kinetics of adhesion of cells from panel (A) in different ECM substrates, (B) Collagen-I, (C) Collagen-IV, (D) Fibronectin. As a negative control, wild-type cells were plated in the presence of 5mM of EDTA (WT+EDTA). The graphics also show the attachment kinetics of wild-type cells in the presence of an inhibitor of actin polymerization, Cytochalasin D, 2µgr/ml (WT+CytD). Error bars show standard deviation from the mean (n=3). Red asterisks indicate differences statistically significant (T-test, p<.05) compared with WT. Notice that although the p value for Cyfip1 knock-down in panel B is not significant, however it was <.1. Immunofluorescence studies showing the distribution and morphology of focal adhesions, labeled with anti-Vinculin antibody, in knock-down MCF-10A cells (E) and keratinocytes (F). The arrows indicate the focal contacts and the asterisks the focal complexes.
Interaction with ECM substrates is mediated by integrins, a superfamily of membrane receptors that in cultured cells are typically localized at cellular microdomains called focal adhesions (FA) (Hynes, 2002). Therefore, we assessed the status of FA formation in WAVE knock-down cells, both MCF-10A and keratinocytes grown in low calcium medium. Control cells were well spread and displayed two morphologically different types of focal adhesions: focal contacts (robust adhesion located at the periphery) and focal complexes (smaller adhesions situated centrally). Both are characterized by the presence of markers such as Vinculin (Fig. 5E, F) and Focal Adhesion Kinase/FAK (Fig. S6B, C) (Hynes, 2002; Raghavan et al., 2003). In contrast, WAVE knock-down cells showed very large focal contacts at the tips of unusually thick actin cables, and the number of the internal focal complexes was substantially reduced. This phenotype mirrors that observed in integrin β1-null (Raghavan et al., 2003) and FAK-null (Schober et al., 2007) keratinocytes, where these defects were characterized as a reflection of the inability to turn over focal adhesion complexes.
WAVE impairment cooperates with activated Ras to promote tumor progression
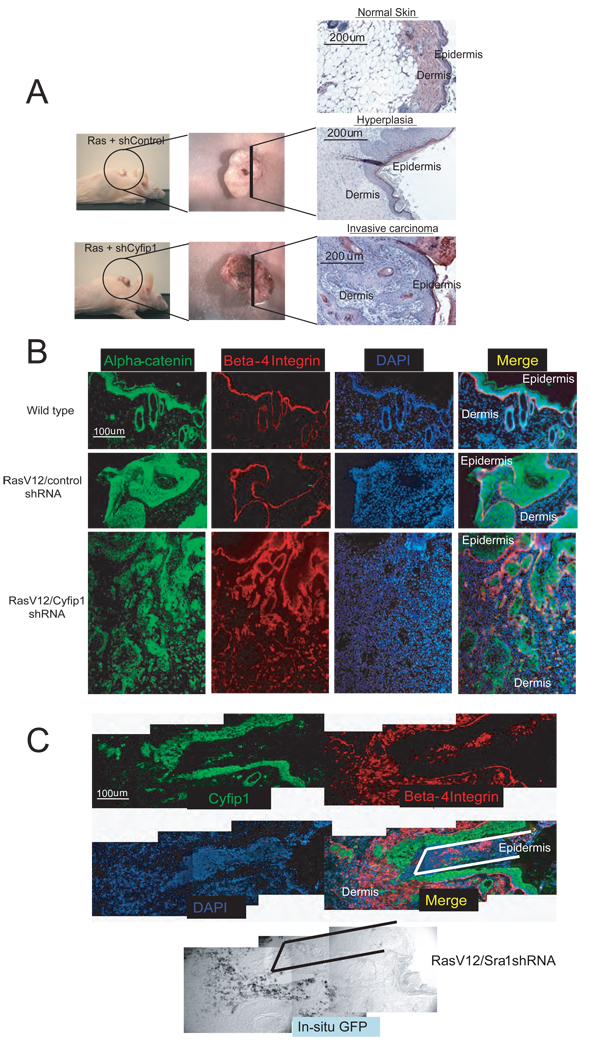
To probe the relevance of our findings in an in vivo setting, we took advantage of an orthotopic mouse model that has previously been used to study links between cell adhesion and tumor progression (Dajee et al., 2003; Weinberg et al., 1991). In this model, primary keratinocytes are co-transplanted with fibroblasts onto a wounded nude mouse to reconstitute skin (Fig. S7). If the keratinocytes are engineered ex vivo to express activated Ras (H-RasV12), the engrafted skin develops hyperplastic and dysplastic benign lesions (papillomas). Additional genetic alterations, such as loss of p53, permit progression to squamous cell carcinomas, which invade through the basal lamina into the dermis (Azzoli et al., 1998).
Accordingly, we found that keratinocytes that express activated Ras in combination with control shRNAs produced hyperplastic lesions in 5 out of 7 mice. In the remaining two cases, a focal area with few (1–3) individual keratinocytes invading into underlying dermis was observed. In contrast, shRNAs directed against Cyfip1 cooperated with Ras to induce rapid progression (3–4 weeks) to squamous cell carcinoma in 6 out of 7 animals (Fig. 6A). These lesions are characterized by disruption of the basal lamina as illustrated by staining with β4-integrin and α-catenin antibodies (Fig. 6B). Since our silencing constructs were engineered to co-express a fluorescent GFP marker we could verify that cells invading and colonizing the dermal compartment were indeed those with Cyfip1 knock-down (Fig. 6C).
Figure 6. Knock-down of Cyfip1 promotes invasion in vivo.
Panel (A) illustrates macroscopically and microscopically the appearance of skin lesions produced in nude mice after transplantation of primary keratinocytes engineered to express oncogenic Ras or Ras plus Cyfip1 shRNA as compared with wild-type skin. (B) Markers of the epithelial compartment of the skin (β4-integrin and α-catenin) were used to study tissue architecture in sections from panel (A). (C) Immunofluorescence showing that invasive keratinocytes (β4-integrin positive) express the construct that contains the hairpin targeting Cyfip1 (positive for GFP mRNA in the in situ panel) and show low Cyfip1 expression. The lines (black, in merged panel and white, in in situ panel) are used as a reference.
Reduced expression of Cyfip1 is common during tumor invasion
During invasion of the stroma by tumor cells, the expression of many proteins changes, including adhesion molecules (ex. E- and N- cadherin), cytoskeleton-associated proteins (ex. Vimentin) and transcriptional regulators (e.g., Snail) (Christofori, 2006; Grunert et al., 2003). Consequently, the expression levels of these genes can serve as marker of invasion. It was therefore critical to determine whether Cyfip1 expression is also generally affected as tumor cells gain invasive potential.
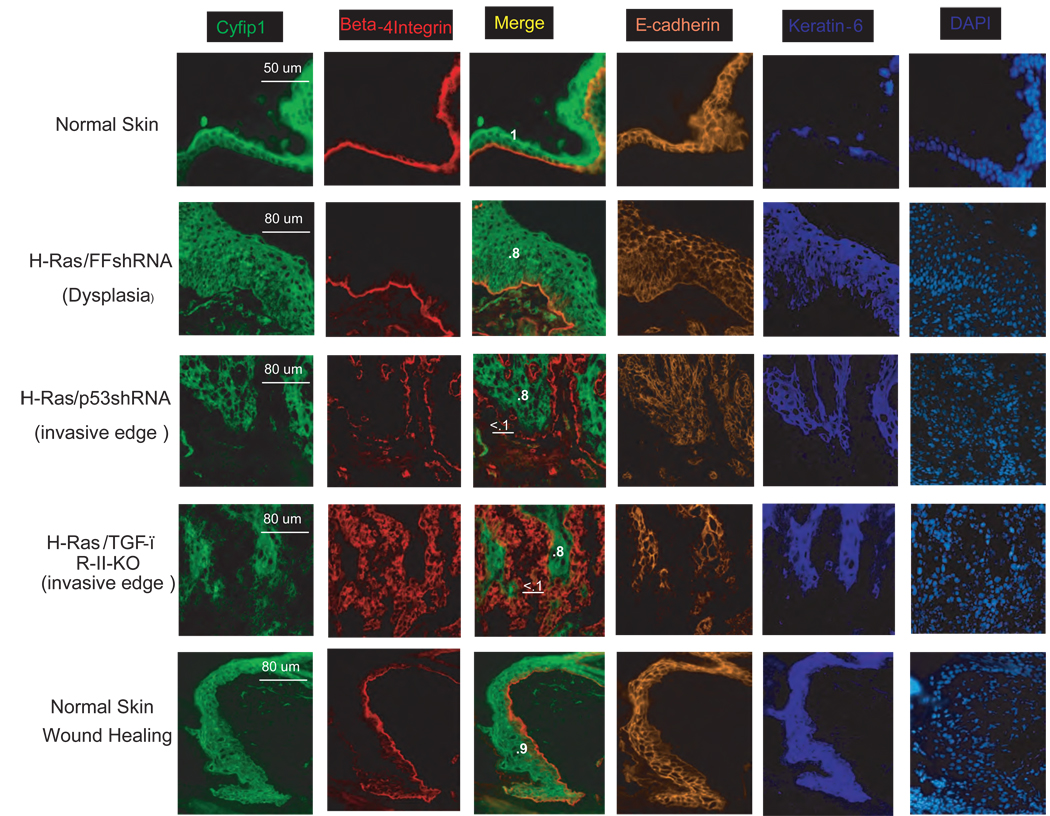
To probe this possibility, we generated several mosaic mouse models (using the orthotropic transplantation previously described) that reflect different stages of tumor progression and examined the expression of Cyfip1 (Fig. 7). Antibody staining revealed strong Cyfip1 expression in the epidermis that was more intense in terminally differentiated layers. In dysplastic lesions produced by the expression of oncogenic Ras, Cyfip1 was still obvious, but the lesions did not present the intense staining typical of normal, differentiated layers. When an invasive squamous cell carcinoma (SCC) was generated by silencing p53, the expression of Cyfip1 was greatly reduced at the invasive edge. Furthermore, in a more aggressive SCC generated by transplanting TGF-β-RII null keratinocytes carrying oncogenic Ras (Guasch et al., 2007), the expression of Cyfip1 was almost completely abolished in large invasive areas and coincided with a strong reduction of E-cadherin. Interesting we also observed reduction of Cyfip1 expression at the edge of human lung cancers (6 out of 6 comprising 3 SCC and 3 AdenoCa.) and breast tumors (1 out of 3 IDC) with well-defined limits between epithelial and stromal tissues. (Fig. S8)
Figure 7. Down-regulation of Cyfip1 occurs during invasion in vitro and in vivo.
Immunofluorescence staining comparing the expression and distribution of Cyfip1 and different characteristic markers of the epidermis, β-4 integrin and E-cadherin is shown. A continuous β-4 integrin staining is an indication of intact epidermal/dermal boundaries (Normal skin, dysplasia and wound healing). In contrast, patched and disorganized staining reflects invasion of tumor cells into the stroma (invasive edges). In this model keratin-6 is a marker that labels proliferating epidermis. Quantitative data comparing the expression of Cyfip1 at the leading edge of the invasion (underlined) and inside of the tumor (bold) are provided. The raw values were double normalized; first, internally to the staining of Beta-4 integrin and then, to the value of the normal skin, and these have been included in the figure.
We wondered whether loss of Cypfip1 expression was specific to the migratory and invasive responses that occur in tumors. We therefore tested whether changes in Cyfip1 were observed during epidermal wound healing, a non-tumorigenic tissue remodeling response. A small wound was created on the back of a mouse and wound closure was analyzed 2–4 days later. Although strong proliferation and mobilization of epidermal keratinocytes occurred at the area close to the wound, immunofluorescence staining revealed no changes in expression of Cyfip1 (Fig. 7). This result indicates that reduction of Cyfip1 expression coincides with invasiveness rather than with motility, per se.
Discussion
The ability to generate a polarized cytoskeletal network that is intricately associated with intercellular junctions allows epithelial cells to integrate tension-based movements and function coordinately within a tissue (Cau and Hall, 2005; Vaezi et al., 2002). The presence of calcium modulates the interaction of cadherin-mediated junctions with the cytoskeleton, (Gumbiner, 2005) and in some circumstances, WAVE-mediated actin reorganization has been implicated in the organization and maintenance of adhesion (Yamazaki et al., 2007). Based on this and on the striking abnormalities that we have characterized in cells with compromised WAVE function, we propose a model wherein depletion of WAVE components directly perturbs actin dynamics, which in turn reduces epithelial adhesion and leads to disorganization of tissue architecture.
Tissue architecture represents an important level of control, which transformed cells must disrupt, in order to become invasive. Correspondingly, a number of molecules that regulate tissue architecture are also commonly altered during tumor formation (e.g., E-cadherin (Cowin et al., 2005), α6 and β4 integrins (Gilcrease, 2007), Podoplanin (Wicki et al., 2006) or Scribble (Bilder, 2004)). Changes in genes encoding these regulators often show a strong correlation with poor prognosis and the capacity of tumor cells to invade stromal compartments (Cowin et al., 2005; Gumbiner, 1996). We now add Cyfip1 to those genes that modify the invasive phenotype, since its suppression can promote the development of invasive carcinomas in models that would normally yield benign, non-invasive lesions.
Our observations have reinforced connections between actin dynamics and tumorigenesis, since effects on the actin cytoskeleton impact tissue architecture and cell adhesion and consequently invasive potential. However, the catalog of downstream pathways potentially affected by disrupting WAVE may be far from complete. Remodeling of the actin-cytoskeleton influences multiple aspects of cellular behavior, including motility, adhesion, signal transduction, apoptosis, cytokinesis, endocytosis, and differentiation (Doherty and McMahon, 2008; Olson and Sahai, 2008; Perez-Moreno et al., 2003). Thus, further work will almost certainly be required to understand the complete mechanistic basis of the effects that we observe.
A remaining critical question is whether distortion of general actin dynamics can influence epithelial tumorigenesis or whether invasive potential is impacted specifically by alterations of the WAVE/lamellipodia pathway. There are several indications that support the former hypothesis. Although cytochalasin-D was toxic, low concentrations of this drug induced abnormalities in acinar development, and specific inhibition of Rho GTPases dramatically impacted acinar architecture without compromising viability (Fig. S9). Moreover, a number of genes controlling actin dynamics are altered in human cancers (Olson and Sahai, 2008), and the formation of actin–based membrane protrusions is essential during tumor progression and invasion (Yamaguchi and Condeelis, 2007). It seemly likely that there is no one unique route through which tumor cells gain the capacity to invade. Indeed, it is now clear that tumor cells are able to shift amongst a variety of pathways if one that fosters mobility becomes blocked (Friedl, 2004; Sanz-Moreno et al., 2008; Wolf et al., 2003). Nonetheless, our data demonstrate that alterations in either the Cyfip1 locus or its expression contribute to the loss of epithelial cell architecture and promote tumor progression, marking this locus a strong candidate for a bone fide human invasion suppressor.
Experimental Procedures
ROMA
High-resolution genome profiling of DNA copy number alterations was performed as previously described (Hicks et al., 2006). Focal deletions were defined as regions of less than 2Mb.
Cell culture and cell line construction
ShRNAs cassettes from our published library (Silva et al., 2005) were cloned into a retroviral vector that links GFP to expression of the shRNA. All the available shRNAmirs targeting the same gene were pooled together to produce suppressed cell populations. The sequences for the shRNAs can be obtained at http://codex.cshl.edu/scripts/newmain.pl. or http://www.openbiosystems.com ) and in the supplementary methods. 3D cultures were performed as described previously (Debnath et al., 2003). For depletion of growth factors, all supplements were omitted from the negative media. For anoikis experiments, negative media was also used and the cells were plated on ultra low attachment plates (Packard). For cell-cell adhesion studies 5mM EDTA (final concentration) was added to equally confluent MCF-10A cell cultures (control and knockdown lines). For cell-ECM adhesion studies plates coated with different ECM substrates were purchased from Cell Biolabs, and protocols recommended by the vendor were followed. All assays were done at least two independent times in triplicate.
Western blotting, Immunofluoresce (IF) and QRT-PCR
IF on cells seeded on glass was performed using standard methods. IF on tissue sections was performed as previously described (Kobielak and Fuchs, 2006). QRT-PCR for Cyfip1 and Nckap1 was done after standard total RNA extraction and reverse transcription. The antibodies, dilutions and the oligo sequences used are described in detail in the supplementary methods.
Microscopy
Images of live cells were collected at 4 minute intervals for 24 hour using a Zeis observer Z1. Confocal image Z series (LSM format) were imported into the 3D module of Axiovision (version 4.6.3 Zeiss MicroImaging, Thornwood, NY) and maximum projection 3D images were generated. From the same Z series orthogonal perspectives were made using Zen LE (Zeiss). For standard immunofluorescence a Zeis Axioskop 2 plus was used. For Transmission Electron Microscopy cells were cultured on Thermanox Nunc coverslips and then processes following the procedures described in detail in the supplementary methods.
In vivo studies
Wild-type or knock-down cells were injected subcutaneously into nude mice, and tumor size was determined after 4–6 weeks. Engraftment experiments were performed as described previously (Azzoli et al., 1998; Weinberg et al., 1991) by infecting primary keratinocytes with retroviral constructs expressing Ha-RasV12, or different shRNAs. Mice were sacrificed and analyzed 4–5 weeks after grafting.
Human tumor studies
Pathological parameters are described in detail in the supplementary methods. Total RNA was extracted from 30 mg of tumor samples and was reverse transcribed using standard protocols. We calculated the expression level of Cyfip1 as the ratio: relative mRNA expression of target / relative mRNA expression of housekeeping control. The tumors were stratified based on their correlated immunohistochemistry value (explained in the main text and Fig. S7) The statistical correlations were analyzed using the T-test and the SPSS package.
Immunohistochemistry
Immunohistochemistry analysis was performed on formalin-fixed and paraffin-embedded tissue sections from Tissue MicroArrays (TMAs) following the standard avidin-biotin immunoperoxidase staining procedure (described in more detail in the supplementary methods).
Supplementary Material
The movies show the de-adhesion kinetics of confluent cultures of MCF-10A variants when 5mM EDTA was added to the media. (A) wild type, (B) shRNA control, (C) overexpressing Snail, (D) shRNA Cyfip1, (E) shRNA NcKap1 and (F) shRNA Wave2 were monitored over the course of 1 hour.
The movies show the proliferation and structure of the colonies generated by MCF10A variants. (A) shRNA control, (B) shRNA Cyfip1, (C) shRNA NcKap1 and (D) shRNA Wave2 were monitored over the course of 14 hours.
The movies show the proliferation of keratinocyte variants (A) wild type and (B) shRNA Cyfip1 when grown in media with low calcium for 24 hours.
The movies show the proliferation of keratinocyte variants (A) wild type and (B) shRNA Cyfip1 when grown in media with high calcium for 24 hours.
Acknowledgments
We are especially thankful to Victoria Aranda (CSHL) for help with the 3D model, to Alberto Muňoz (Instituto de Investigaciones Biomédicas “Alberto Sols”) for providing the Snail expression vector, and to Geraldine Guasch (Rockefeller Univ.) for providing tissue sections of HRas/TGF-β R-II-KO mouse tumors. We also thank June Racelis for help with in situ hybridizations, and Nicole Stokes for assistance with skin chamber grafting. We thank Ken Chang and Edie Davis for comments on the manuscript and for helpful discussion. This work was partially supported by the NCI with the K99/R00 project # 54010101-5411 (J.M.S.). G.J.H and E.F. are Investigators of the Howard Hughes Medical Institute and E.E. is a Life Sciences Foundation Postdoctoral Fellow supported by the New York Stem Cell Institute. This work was supported by grants from the N.I.H (G.J.H and E.F. R01-AR27883) and by a kind gift from Kathryn W. Davis (G.J.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Azzoli CG, Sagar M, Wu A, Lowry D, Hennings H, Morgan DL, Weinberg WC. Cooperation of p53 loss of function and v-Ha-ras in transformation of mouse keratinocyte cell lines. Mol Carcinog. 1998;21:50–61. doi: 10.1002/(sici)1098-2744(199801)21:1<50::aid-mc7>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Bilder D. Epithelial polarity and proliferation control: links from the Drosophila neoplastic tumor suppressors. Genes Dev. 2004;18:1909–1925. doi: 10.1101/gad.1211604. [DOI] [PubMed] [Google Scholar]
- Billuart P, Chelly J. From fragile X mental retardation protein to Rac1 GTPase: new insights from Fly CYFIP. Neuron. 2003;38:843–845. doi: 10.1016/s0896-6273(03)00362-3. [DOI] [PubMed] [Google Scholar]
- Boguslavsky S, Grosheva I, Landau E, Shtutman M, Cohen M, Arnold K, Feinstein E, Geiger B, Bershadsky A. p120 catenin regulates lamellipodial dynamics and cell adhesion in cooperation with cortactin. Proc Natl Acad Sci U S A. 2007;104:10882–10887. doi: 10.1073/pnas.0702731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ, Stephens PJ, Pleasance ED, O’Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cau J, Hall A. Cdc42 controls the polarity of the actin and microtubule cytoskeletons through two distinct signal transduction pathways. J Cell Sci. 2005;118:2579–2587. doi: 10.1242/jcs.02385. [DOI] [PubMed] [Google Scholar]
- Chiang DY, Getz G, Jaffe DB, O’Kelly MJ, Zhao X, Carter SL, Russ C, Nusbaum C, Meyerson M, Lander ES. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods. 2009;6:99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofori G. New signals from the invasive front. Nature. 2006;441:444–450. doi: 10.1038/nature04872. [DOI] [PubMed] [Google Scholar]
- Cowin P, Rowlands TM, Hatsell SJ. Cadherins and catenins in breast cancer. Curr Opin Cell Biol. 2005;17:499–508. doi: 10.1016/j.ceb.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Dajee M, Lazarov M, Zhang JY, Cai T, Green CL, Russell AJ, Marinkovich MP, Tao S, Lin Q, Kubo Y, et al. NF-kappaB blockade and oncogenic Ras trigger invasive human epidermal neoplasia. Nature. 2003;421:639–643. doi: 10.1038/nature01283. [DOI] [PubMed] [Google Scholar]
- Datta S, Hoenerhoff MJ, Bommi P, Sainger R, Guo WJ, Dimri M, Band H, Band V, Green JE, Dimri GP. Bmi-1 cooperates with H-Ras to transform human mammary epithelial cells via dysregulation of multiple growth-regulatory pathways. Cancer Res. 2007;67:10286–10295. doi: 10.1158/0008-5472.CAN-07-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Doherty GJ, McMahon HT. Mediation, modulation, and consequences of membrane-cytoskeleton interactions. Annu Rev Biophys. 2008;37:65–95. doi: 10.1146/annurev.biophys.37.032807.125912. [DOI] [PubMed] [Google Scholar]
- Fan C, Oh DS, Wessels L, Weigelt B, Nuyten DS, Nobel AB, van’t Veer LJ, Perou CM. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355:560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- Firestein R, Bass AJ, Kim SY, Dunn IF, Silver SJ, Guney I, Freed E, Ligon AH, Vena N, Ogino S, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach C, Chen R, Matsumoto T, Schmelzle T, Brugge JS, Polverini PJ, Mooney DJ. Engineering tumors with 3D scaffolds. Nat Methods. 2007;4:855–860. doi: 10.1038/nmeth1085. [DOI] [PubMed] [Google Scholar]
- Friedl P. Prespecification and plasticity: shifting mechanisms of cell migration. Curr Opin Cell Biol. 2004;16:14–23. doi: 10.1016/j.ceb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Gilcrease MZ. Integrin signaling in epithelial cells. Cancer Lett. 2007;247:1–25. doi: 10.1016/j.canlet.2006.03.031. [DOI] [PubMed] [Google Scholar]
- Grunert S, Jechlinger M, Beug H. Diverse cellular and molecular mechanisms contribute to epithelial plasticity and metastasis. Nat Rev Mol Cell Biol. 2003;4:657–665. doi: 10.1038/nrm1175. [DOI] [PubMed] [Google Scholar]
- Guasch G, Schober M, Pasolli HA, Conn EB, Polak L, Fuchs E. Loss of TGFbeta signaling destabilizes homeostasis and promotes squamous cell carcinomas in stratified epithelia. Cancer Cell. 2007;12:313–327. doi: 10.1016/j.ccr.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner BM. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol. 2005;6:622–634. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hempen PM, Zhang L, Bansal RK, Iacobuzio-Donahue CA, Murphy KM, Maitra A, Vogelstein B, Whitehead RH, Markowitz SD, Willson JK, et al. Evidence of selection for clones having genetic inactivation of the activin A type II receptor (ACVR2) gene in gastrointestinal cancers. Cancer Res. 2003;63:994–999. [PubMed] [Google Scholar]
- Hicks J, Krasnitz A, Lakshmi B, Navin NE, Riggs M, Leibu E, Esposito D, Alexander J, Troge J, Grubor V, et al. Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome Res. 2006;16:1465–1479. doi: 10.1101/gr.5460106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- Ikediobi ON, Davies H, Bignell G, Edkins S, Stevens C, O’Meara S, Santarius T, Avis T, Barthorpe S, Brackenbury L, et al. Mutation analysis of 24 known cancer genes in the NCI-60 cell line set. Mol Cancer Ther. 2006;5:2606–2612. doi: 10.1158/1535-7163.MCT-06-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti M, Zucconi A, Disanza A, Frittoli E, Areces LB, Steffen A, Stradal TE, Di Fiore PP, Carlier MF, Scita G. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat Cell Biol. 2004;6:319–327. doi: 10.1038/ncb1105. [DOI] [PubMed] [Google Scholar]
- Kobielak A, Fuchs E. Links between alpha-catenin, NF-kappaB, and squamous cell carcinoma in skin. Proc Natl Acad Sci U S A. 2006;103:2322–2327. doi: 10.1073/pnas.0510422103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunda P, Craig G, Dominguez V, Baum B. Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr Biol. 2003;13:1867–1875. doi: 10.1016/j.cub.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Muthuswamy SK, Li D, Lelievre S, Bissell MJ, Brugge JS. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat Cell Biol. 2001;3:785–792. doi: 10.1038/ncb0901-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson MF, Sahai E. The actin cytoskeleton in cancer cell motility. Clin Exp Metastasis. 2008 doi: 10.1007/s10585-008-9174-2. [DOI] [PubMed] [Google Scholar]
- Palmer HG, Larriba MJ, Garcia JM, Ordonez-Moran P, Pena C, Peiro S, Puig I, Rodriguez R, de la Fuente R, Bernad A, et al. The transcription factor SNAIL represses vitamin D receptor expression and responsiveness in human colon cancer. Nat Med. 2004;10:917–919. doi: 10.1038/nm1095. [DOI] [PubMed] [Google Scholar]
- Perez-Moreno M, Jamora C, Fuchs E. Sticky business: orchestrating cellular signals at adherens junctions. Cell. 2003;112:535–548. doi: 10.1016/s0092-8674(03)00108-9. [DOI] [PubMed] [Google Scholar]
- Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE, Tibshirani R, Botstein D, Borresen-Dale AL, Brown PO. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci U S A. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan S, Vaezi A, Fuchs E. A role for alphabeta1 integrins in focal adhesion function and polarized cytoskeletal dynamics. Dev Cell. 2003;5:415–427. doi: 10.1016/s1534-5807(03)00261-2. [DOI] [PubMed] [Google Scholar]
- Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S, Sahai E, Marshall CJ. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135:510–523. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- Schenck A, Bardoni B, Moro A, Bagni C, Mandel JL. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc Natl Acad Sci U S A. 2001;98:8844–8849. doi: 10.1073/pnas.151231598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober M, Raghavan S, Nikolova M, Polak L, Pasolli HA, Beggs HE, Reichardt LF, Fuchs E. Focal adhesion kinase modulates tension signaling to control actin and focal adhesion dynamics. J Cell Biol. 2007;176:667–680. doi: 10.1083/jcb.200608010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin K, Fogg VC, Margolis B. Tight junctions and cell polarity. Annu Rev Cell Dev Biol. 2006;22:207–235. doi: 10.1146/annurev.cellbio.22.010305.104219. [DOI] [PubMed] [Google Scholar]
- Silva FP, Morolli B, Storlazzi CT, Anelli L, Wessels H, Bezrookove V, Kluin-Nelemans HC, Giphart-Gassler M. Identification of RUNX1/AML1 as a classical tumor suppressor gene. Oncogene. 2003;22:538–547. doi: 10.1038/sj.onc.1206141. [DOI] [PubMed] [Google Scholar]
- Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- Smith DI, McAvoy S, Zhu Y, Perez DS. Large common fragile site genes and cancer. Semin Cancer Biol. 2007;17:31–41. doi: 10.1016/j.semcancer.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Stradal TE, Rottner K, Disanza A, Confalonieri S, Innocenti M, Scita G. Regulation of actin dynamics by WASP and WAVE family proteins. Trends Cell Biol. 2004;14:303–311. doi: 10.1016/j.tcb.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Takenawa T, Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol. 2007;8:37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- Trevino V, Falciani F, Barrera-Saldana HA. DNA microarrays: a powerful genomic tool for biomedical and clinical research. Mol Med. 2007;13:527–541. doi: 10.2119/2006-00107.Trevino. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood JM, Imbalzano KM, Weaver VM, Fischer AH, Imbalzano AN, Nickerson JA. The ultrastructure of MCF-10A acini. J Cell Physiol. 2006;208:141–148. doi: 10.1002/jcp.20639. [DOI] [PubMed] [Google Scholar]
- Vaezi A, Bauer C, Vasioukhin V, Fuchs E. Actin cable dynamics and Rho/Rock orchestrate a polarized cytoskeletal architecture in the early steps of assembling a stratified epithelium. Dev Cell. 2002;3:367–381. doi: 10.1016/s1534-5807(02)00259-9. [DOI] [PubMed] [Google Scholar]
- Vasioukhin V, Bauer C, Yin M, Fuchs E. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell. 2000;100:209–219. doi: 10.1016/s0092-8674(00)81559-7. [DOI] [PubMed] [Google Scholar]
- Wang Z, Shen D, Parsons DW, Bardelli A, Sager J, Szabo S, Ptak J, Silliman N, Peters BA, van der Heijden MS, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004;304:1164–1166. doi: 10.1126/science.1096096. [DOI] [PubMed] [Google Scholar]
- Weinberg WC, Morgan DL, George C, Yuspa SH. A comparison of interfollicular and hair follicle derived cells as targets for the v-rasHa oncogene in mouse skin carcinogenesis. Carcinogenesis. 1991;12:1119–1124. doi: 10.1093/carcin/12.6.1119. [DOI] [PubMed] [Google Scholar]
- Wicki A, Lehembre F, Wick N, Hantusch B, Kerjaschki D, Christofori G. Tumor invasion in the absence of epithelial-mesenchymal transition: podoplanin-mediated remodeling of the actin cytoskeleton. Cancer Cell. 2006;9:261–272. doi: 10.1016/j.ccr.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Wilkemeyer MF, Menkari CE, Spong CY, Charness ME. Peptide antagonists of ethanol inhibition of l1-mediated cell-cell adhesion. J Pharmacol Exp Ther. 2002;303:110–116. doi: 10.1124/jpet.102.036277. [DOI] [PubMed] [Google Scholar]
- Wolf K, Mazo I, Leung H, Engelke K, von Andrian UH, Deryugina EI, Strongin AY, Brocker EB, Friedl P. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J Cell Biol. 2003;160:267–277. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Condeelis J. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim Biophys Acta. 2007;1773:642–652. doi: 10.1016/j.bbamcr.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki D, Oikawa T, Takenawa T. Rac-WAVE-mediated actin reorganization is required for organization and maintenance of cell-cell adhesion. J Cell Sci. 2007;120:86–100. doi: 10.1242/jcs.03311. [DOI] [PubMed] [Google Scholar]
- Zalfa F, Achsel T, Bagni C. mRNPs, polysomes or granules: FMRP in neuronal protein synthesis. Curr Opin Neurobiol. 2006;16:265–269. doi: 10.1016/j.conb.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Zipfel PA, Bunnell SC, Witherow DS, Gu JJ, Chislock EM, Ring C, Pendergast AM. Role for the Abi/wave protein complex in T cell receptor-mediated proliferation and cytoskeletal remodeling. Curr Biol. 2006;16:35–46. doi: 10.1016/j.cub.2005.12.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The movies show the de-adhesion kinetics of confluent cultures of MCF-10A variants when 5mM EDTA was added to the media. (A) wild type, (B) shRNA control, (C) overexpressing Snail, (D) shRNA Cyfip1, (E) shRNA NcKap1 and (F) shRNA Wave2 were monitored over the course of 1 hour.
The movies show the proliferation and structure of the colonies generated by MCF10A variants. (A) shRNA control, (B) shRNA Cyfip1, (C) shRNA NcKap1 and (D) shRNA Wave2 were monitored over the course of 14 hours.
The movies show the proliferation of keratinocyte variants (A) wild type and (B) shRNA Cyfip1 when grown in media with low calcium for 24 hours.
The movies show the proliferation of keratinocyte variants (A) wild type and (B) shRNA Cyfip1 when grown in media with high calcium for 24 hours.